Abstract
Environmental and genetic factors define the susceptibility of an individual to autoimmune disease. Although common genetic pathways affect general immunological tolerance mechanisms in autoimmunity, the effects of such genes could vary under distinct immune challenges within different tissues. Here we demonstrate this by observing that autoimmune type 1 diabetes (T1D) protective haplotypes at the susceptibility region 10 (Idd10) introgressed from chromosome 3 of B6 and A/J mice onto the NOD background increase the severity of autoimmune primary biliary cirrhosis (PBC) induced by infection with Novosphingobium aromaticivorans (N. aro), an ubiquitous alphaproteobacterium, when compared to mice having the NOD and NOD.CAST Idd10 T1D susceptible haplotypes. Substantially increased liver pathology in mice having the B6 and A/J Idd10 haplotypes correlates with reduced expression of CD101 on dendritic cells (DCs), macrophages and granulocytes following infection, delayed clearance of N. aro and the promotion of overzealous, IFN-γ- and IL-17-dominated T cell responses essential for the adoptive transfer of liver lesions. CD101-knockout mice generated on the B6 background also exhibit substantially more severe N.aro-induced liver disease correlating with increased IFN-γ and IL-17 responses compared to wild type mice. These data strongly support the hypothesis that allelic variation of the Cd101 gene, located in the Idd10 region, alters the severity of liver autoimmunity induced by N. aro.
Keywords: rodent, cytokines, dendritic cells, T cells, autoimmunity, costimulation, inflammation
Introduction
Complex interactions between environmental factors and genetic traits (1–4) underpin the etiology of autoimmune diseases and the functioning of the immune system. Although certain genetic regions are associated with multiple autoimmune diseases, in some cases the protective allele for one autoimmune disease is the susceptibility allele for another (5–9). The nature of environmental triggers, effector mechanisms mediating various autoimmune pathologies, or tissue-specific events that enable an allele to be protective for one or more autoimmune diseases on the one hand but increase susceptibility to different autoimmune targets on the other requires definition. The IL2RA locus, for example, is associated with both type 1 diabetes (T1D) and multiple sclerosis (MS); however, an allele that is protective in one disease is a susceptibility allele for the other (10). NOD congenic mice mimic this situation in humans: when the diabetogenic H2g7 of the NOD mouse is replaced with the H2h4 haplotype, the congenic mice are protected from T1D but now develop autoimmunity directed against the thyroid gland, which is exacerbated by exposure to dietary iodine (11–13). It is believed that the underlying single nucleotide polymorphisms (SNPs) that redirect the organ specific autoimmune manifestation likely evolved due to microbial pressure and reveal a consequence of natural selection for altered susceptibility to certain pathogens (14–16); selective pressure to promote functional variation in immune-related genes to resist infectious challenge could add to the pool of variants that alter autoimmune disease susceptibility. Here, we have investigated the effect of alleles that protect from T1D in our infection-induced mouse model of primary biliary cirrhosis (PBC).
The diagnostic hallmark of PBC is the development of autoantibodies to mitochondrial antigens that bind the E2 subunit of the pyruvate dehydrogenase complex (PDC-E2). Histopathological lesions in PBC patients are characterized by mixed lymphoid/mononuclear portal infiltrates and immune-mediated destruction of intrahepatic small bile ducts (17–18). While inflammatory cytokines including IFN-γ and IL-17 are enhanced, immunomodulatory ones such as IL-10 are reduced (19–26).
Although the stimuli that trigger autoreactivity are often uncharacterized, recent studies indicate that undetected bacterial or viral infections may underlie various forms of autoimmunity (27–36). We have previously shown that patients with PBC exhibit chronic immune reactions against lipoylated proteins of Novosphingobium aromaticivorans (N. aro) (37–41), and we have established a model in which infection of mice with N. aro induces anti-PDC-E2 IgG responses and liver lesions resembling PBC in humans (42). Studies in this model suggest that the specific development of lesions in the liver during the more acute phase result from: (a) a greater persistence of N. aro in the liver than in other organs; and (b) the activation of natural killer T (NKT) cells, which are particularly abundant in the liver of mice, by N. aro-glycosphingolipids (GSLs) presented by dendritic cells (DCs). Once established, liver disease can be adoptively transferred by CD4+ and CD8+ T cells from wild type, but not NKT-deficient mice in the chronic phase. This illustrates the importance of early microbial activation of NKT cells in initiating autonomous, organ-specific autoimmunity and in propagating autoreactive T cells, at least on the B6 genetic background. Although we have begun to identify the environmental agent and cellular mechanisms that induce liver specific immunopathology, the factors underlying genetic susceptibility to this model of PBC remain unknown. Our previous studies suggested that NOD.B6 Idd10/18 mice, which are partially protected from T1D as compared to the NOD parental strain (43–44), develop more severe liver lesions following N. aro infection than NOD mice (42). Therefore, we reasoned that the Idd10/18 region might harbor susceptibility alleles that determine the severity of bile duct disease upon infection with N. aro.
Here, we show that the B6-derived Idd10 region present in NOD.B6 Idd10 mice primarily accounts for the increased PBC risk of NOD.B6 Idd10/18 congenic mice upon infection and correlates with genotype-dependent protein expression of Cd101, the primary T1D candidate gene of the Idd10 region (44–47). Analyses of additional Idd10 NOD congenic strains, NOD.A/J Idd10 and NOD.CAST Idd10, confirm the correlation of differential control of T1D (44, 47) and PBC with Cd101 gene expression. A newly developed CD101-deficient B6 knockout strain develops PBC with similar severity to that observed in NOD.B6 Idd10 mice and significantly more severe liver disease than parental NOD and B6 mice. Enhanced suppression of CD101 protein expression on DCs, macrophages and granulocytes from mice carrying the B6 Idd10 allele correlates with prolonged N. aro persistence, hyperreactive T cell responses and extensive liver autoimmunity, indicating that the allelic variations within the NOD and B6 alleles of Cd101 are likely the causal genetic variants for susceptibility and resistance to PBC, and defining Cd101 as the first susceptibility gene for infection-induced PBC. The differential regulation of CD101 in various cell types within tissues undergoing distinct immune challenges could explain how Idd10 alleles that protect from T1D increase PBC susceptibility.
Materials and Methods
Mice
B6 CD45.1, B6 CD101−/−, B6 CD1d−/−, NOD, NOD.B6 Ptprc CD45.2 (line 6908) (48), NOD.B6 Idd10/18 (lines 1101 and 7754, both having the same Idd10/18 segment (49–50), NOD.B6 Idd10 (line 3538), and NOD.B6 Idd18 (line 3539), NOD.A/J Idd10, and NOD.CAST Idd10 mice were maintained in our laboratories. Additional information about the NOD congenic strains can be obtained at http://www.t1dbase.org/page/DrawStrains. NOD congenic strains having line numbers were obtained from the Emerging Models program at Taconic (Germantown, NY). B6, B6 MHCII−/−, B6β2M−/−, and B6 IFN-γR−/− were purchased from the Jackson Laboratory (Bar Harbor, ME); B6 IL-17R−/− mice were obtained from Amgen (Thousand Oaks, CA). The generation of the CD101-knockout B6, NOD.B6 Idd10 (line 3538), NOD.A/J Idd10 and NOD.CAST Idd10 strains is detailed in the co-submitted manuscript (47). NOD.B6 Idd18 (line 3539) N16 mice were developed by selectively breeding a mouse having a recombination event in the distal region of the Idd10/18 congenic segment present in the NOD.B6 Idd3 Idd10 Idd18 (line 1538) strain (49) that was discovered when line 1538 was being backcrossed to the NOD strain. The T1D phenotype of line 3539 will be detailed in a separate publication.
All mice were raised in a specific pathogen-free environment according to the Institutional Animal Care and Use Committee guidelines.
Bacterial strains and live infection experiments
N. aro (ATCC 700278) was grown overnight in LB broth, inoculated in fresh medium, grown for 8 hours at 37°C to an OD of 0.5 at 600nm, washed and resuspended in sterile PBS. 100µl of bacterial suspension containing 5×107 N. aro or PBS only (control) was injected intravenously into 4–7 week old mice on day 0 and on day 14. At indicated time points after infection, bacterial counts were performed after tissue homogenization in 0.5% Triton X-100 and cultured for colony formation.
Cell preparation and co-culture assays
Bone marrow-derived mouse macrophages, and DCs were collected after 7 days of culture in RPMI medium supplemented with glutamine, antibiotics, and 10% FCS with 2 ng/ml of recombinant mouse macrophage colony-stimulating factor (macrophages) and 2 ng/mL GM-CSF (DCs) (all from R&D systems, Minneapolis, MN). Splenocytes and liver lymphocytes were prepared as described (42, 51) and 250,000 of the specified APCs were co-cultured with 250,000 lymphocytes. Cell culture supernatant were assayed after exposure to the indicated stimuli for cytokine release by ELISA (R&D Systems, Minneapolis, MN).
Gentamicin protection assay
Bone marrow-derived DCs from wild-type B6 and CD101−/− mice were exposed to N. aro at a ratio of 1:50 for 12 hours. After rinsing the cultures three times in phosphate-buffered saline (PBS; Gibco-BRL) to remove non-adherent bacteria, cells were incubated for 1 h in fresh prewarmed medium containing 100 µg/ml of gentamicin (Gibco-BRL) to kill extracellular bacteria. Gentamicin was removed, and the cells were gently rinsed three times in PBS. DCs were lifted and disrupted by incubating them for three minutes in PBS containing 2 mM EDTA and 0.5% saponin (Sigma), followed by transfer to Eppendorf tubes and high-speed vortexing for 30 s. Viable intracellular bacteria were quantitated by gentle lysis of the DCs and subsequent plating on LB agar.
Real time quantitative PCR (RT-qPCR) analysis for N.aro copy number
Total DNA was extracted from different organs using the DNeasy® tissue extraction kit (Qiagen, Valencia, CA). The following primers were used for detection of N. aro: forward: 5'-TCCGAGTGTAGAGGTGAAAT-3', reverse: 5'-CGTCAATACTTGTCCAGTCA-3'. RT-qPCR was performed with RT2 qPCR SYBR Green Master Mix (SABiosciences, Frederick, MD) on a Biorad iCycler. A 25 µl reaction volume was used for each sample analyzed, using 12.5 µl RT2 SYBR green mix, 1 µl of each 10 µM primer (400 nM final concentration), 4 µl DNA template (100 ng/ul) and 6.5 µl of nuclease free water. N. aro copy number per 400 ng of total DNA isolated from each tissue was calculated by reference to a standard of pure N. aro genomic DNA (700278D-5, ATCC). PCR products were analyzed by melting curve analysis and agarose gel electrophoresis to confirm that the amplicons were N. aro specific.
Tissue collection and RNA preparation
Spleen and liver samples from uninfected and infected mice were harvested at the indicated time points and immediately immersed in RNAlater® solution (Ambion/Applied Biosystems, Austin, TX) to prevent RNA degradation. Total RNA was isolated using RNAqueous 4-PCR® kit (Ambion/Applied Biosystems, Austin, TX) following manufacturer’s instructions. Isolated RNA was treated with DNase I (Ambion/Applied Biosystems, Austin, TX) to eliminate DNA contamination during the subsequent PCR steps. The concentration and purity of the isolated RNA was determined using a Nanodrop-1000 spectrophotometer (Thermoscientific, Waltham, MA).
cDNA synthesis and RT-qPCR for mRNA expression
cDNA was synthesized using a first strand cDNA synthesis kit for RT-PCR (Roche, Indianapolis, IN) following manufacturer’s instructions, with slight modifications where necessary. Briefly, 1 µg of total RNA and 2 µl random hexamers were incubated at 65°C for 5 minutes to remove secondary structures in the RNA template and enhance primer annealing, then added to the Reverse-transcription reaction mix. The reaction was carried out at 25°C for 10 min (annealing) followed by a 1hr reverse transcription step at 42°C. Finally, the Reverse transcriptase enzyme was heat-inactivated at 99°C for 10 minutes. Quantitative real-time PCR was performed as mentioned before. The primers used for the analysis were as follows: CCL-21c (forward: 5’-ACCCAAGGCAGTGATGGA-3’; Reverse: 5’-TCCGGGGTAAGAACAGGATT-3’); IL-17A (forward: 5’-CTCCAGAAGGCCCTCAGACTAC −3’; Reverse: 5’- AGCTTTCCCTCCGCATTGACACAG-3’), IFN-γ (forward: 5’- TCAAGTGGCATAGATGTGGAAGAA −3’; Reverse: 5’-TGGCTCTGCAGGATTTTCATG −3’). The PCR-products were analyzed by melting curve analysis and also by Agarose gel electrophoresis for a single target-specific amplicon (results not shown). mRNA of Hypoxanthine Guanine Phosphoribosyl Transferase (HGPRT) were used for normalization (Forward: 5’-ACCTCTCGAAGTGTTGGATA-3’, Reverse: 5’-CAACAACAAACTTGTCTGGA-3’).
Flow cytometry and intracellular cytokine staining
CD1d-lipid tetramers were prepared as described (52). Anti-Nkp46, -CD4, -CD8, -TCRβ, -CD25, -γ/δ TCR, -CD69, -CD44, -CD45.1, -CD45.2, - CD11b, - CD11c, -F4/80 and -MHCII antibodies were purchased from eBioscience (San Diego, CA). Tregs and intracellular cytokines were analyzed by using a staining kit from eBioscience (San Diego, CA) following manufacture’s instructions. Anti-CD101 was obtained from R&D Systems (Minneapolis, MN). Cells were analyzed on a LSR II (BD Biosciences, San Diego, CA) with FlowJo software.
Bone marrow radiation chimeras
For the preparation of mixed bone marrow radiation chimeras a mixture of 5×106 CD45.2+ NOD.B6 Ptprc and 5×106 CD45.1+ NOD.B6 Idd10 bone marrow cells was injected intravenously into 7–12 week old CD45.2+ NOD.B6 Ptprc which had been irradiated twice with a cesium source (Gammacell 40, Nordion International Inc. Ontario, Canada) one day before within a 4 hour interval (1st dose: 700 Rad and 2nd dose: 435 Rad). For the preparation of reciprocal bone marrow radiation chimeras either 5×106 CD45.2+ NOD.B6 Ptprc or 5×106 CD45.1+ NOD.B6 Idd10 bone marrow cells were injected intravenously into 7–12 week old (NOD.B6 Ptprc x NOD.B6 Idd10) F1 mice which had been irradiated twice with a cesium source (Gammacell 40, Nordion International Inc. Ontario, Canada) one day before within a 4 hour interval (1st dose: 700 Rad and 2nd dose: 435 Rad). Similarly B6 CD45.1+ / B6 CD101−/− mixed and reciprocal bone marrow chimeras have been prepared. The reconstitution of the mixed bone marrow chimeras with the different cell populations was determined 6–8 weeks after bone marrow injection by FACS analysis of blood samples using CD45.1 and CD45.2 antibodies and the respective surface markers. The chimeras were intravenously injected with 5×107 N. aro at days 0 and 14 and their livers and spleens analyzed at the indicated time points by flow cytometry.
Cell transfer experiments
1×107 TCRβ+, 1×107 NKp46+ or 1×107 γ/δ TCR+ cells mixed from spleen and liver of diseased (week 10 post infection) B6 or NOD.B6 Idd10 mice were injected iv into syngeneic adult irradiated NOD.B6 Idd10, B6, B6 CD1d−/−, B6 MHCII−/−, B6β2M−/−, and B6 IFN-γR−/− and B6 IL-17R−/− recipients, respectively. Livers were harvested 6 weeks after transfer for the histopathological analysis of inflammatory bile duct lesions.
Diabetes frequency studies
Diabetes frequency studies were conducted using female mice. The presence of T1D was tested every 10 to 14 days beginning at approximately 80 days of age by the detection of urinary glucose >500 mg/dl using Diastix (Miles, Elkhart, IN). Kaplan-Meier survival curves were plotted for each mouse strain and these were compared using the logrank test (Prism® 4 software).
Histology
Liver tissue was fixed in 10% buffered formalin, embedded in paraffin, and cut into 5-µm sections. Liver sections were deparaffinized, stained with hematoxylin and eosin by the Cincinnati Children’s Hospital Medical Center’s Histology Research laboratories, and evaluated microscopically in double blind studies for leukocytic and lymphocytic infiltration as described before (42).
Statistical analysis
Statistical significance of the data was analyzed by either one-way Analysis Of Variance (one-way ANOVA), student’s t-test or Mann-Whitney test as indicated in the respective experiments. A sample size of at least 3 (n=3) was used for each sample group in a given experiment, and a p-value of less than 5% (p<0.05; *) or less than 1% (p<0.01, **) was considered as significant to accept the alternate hypothesis.
Results
The B6 Cd101 allele is associated with severe autoimmune liver disease and prolonged bacterial persistence on the NOD background upon infection with N. aro
Our previous study characterized NOD.B6 Idd10/18 congenic mice as particularly susceptible to autoimmune liver disease upon N. aro-infection (42). To identify the PBC-susceptibility genes within the Idd10/18 congenic genetic region, we study here NOD.B6 congenic mice having only the Idd10 or the Idd18 T1D-protective regions (Fig. S1).
Infection with 5×107 N. aro cfus triggered similar massive liver pathology in NOD.B6 Idd10 as in NOD.B6 Idd10/18 mice (Fig. 1A). Although considerably more prominent than in parental NOD mice liver lesions remained significantly less severe in NOD.B6 Idd18 compared to NOD.B6 Idd10 and NOD.B6 Idd10/18 mice (Fig. 1A and Fig. S2A) suggesting a more critical role for Idd10 in liver disease progression. Infection of (NOD x NOD.B6 Idd10) F1 mice also induced significantly more severe liver disease compared to parental NOD mice suggesting that allelic variations within the Idd10 region drive susceptibility to PBC in a gene dose dependent manner (not shown). As introgression of the B6 Idd10 allele protects from spontaneous type 1 diabetes (T1D) (43, 44, 47), we tested the effect of N. aro infection on T1D in NOD.B6 Idd10 mice (Fig. 1B). N. aro-infection did not alter the B6 Idd10 allele-mediated protection from T1D. As Cd101 is a prime candidate T1D gene in the Idd10 region (44–46, 53) and haplotype analysis comparing NOD, NOD.B6 Idd10, NOD.A/J Idd10, and NOD.CAST Idd10 mice demonstrated a correlation between protection from spontaneous T1D and variation in CD101 expression (47), we tested the severity of liver lesions in NOD.A/J Idd10 and NOD.CAST Idd10 mice, which have T1D-protective and T1D-susceptible Idd10 haplotypes, respectively. NOD.A/J Idd10 mice developed severe liver disease indistinguishable from that of NOD.B6 Idd10 mice, while liver disease remained comparably mild in NOD and NOD.CAST Idd10 mice (Fig. 1C). As the Cd101 allele in NOD.A/J Idd10 mice is identical to the Cd101 allele in NOD.B6 Idd10 mice whereas other Idd10 genes have sequence differences between the A/J and B6 Idd10 haplotypes (47), we concluded that genetic variations within Cd101 were likely the causal genetic event in triggering severe PBC during infection. If Cd101 is the Idd10 gene responsible for altering susceptibility to PBC, we would predict that a mouse having a disrupted CD101 gene would have altered disease susceptibility. Therefore, we compared the severity of liver lesions after infection in heterozygous and homozygous B6 CD101-knockout and wild type mice. Wild-type, heterozygous and homozygous CD101−/− offspring were born in the expected Mendelian ratios and uninfected, homozygous CD101−/− mice appear healthy for more than 18 months without developing any spontaneous infiltration at any organ site. Upon infection, however, B6 CD101−/− mice developed more severe liver disease compared to wild type B6 mice (Fig. 1 D). B6 CD101+/− littermates exhibited also more severe PBC than B6 wild type mice indicating that a 50% reduction of CD101 expression already impacts disease (Fig. 1D and Fig. S2B). Severity of liver disease in infected CD101+/− and CD101−/− mice in a F2 cross was comparable to F1 and F0 mice (not shown) excluding any influence of unknown, randomly segregating genes in the B6 background of the CD101−/− strain on PBC pathogenesis. Collectively, these results strongly support allelic variations within Cd101 as the major causal genetic events for infection-induced PBC within the Idd10 region, which contains six other protein coding genes (44, 47).
Figure 1. Cd101 is the major PBC-susceptibility gene within the Idd10/18 region.
(A) The B6 allele at the Idd10 region is sufficient for severe liver disease.
5 week-old female B6, NOD, NOD.B6 Idd10/18, NOD.B6 Idd10 and NOD.B6 Idd18 mice were infected intravenously with 5×107 N. aro cfus. H&E stained liver sections of the indicated mouse strains were analyzed at week 19 after infection for the severity of portal inflammation in blinded studies. Statistical significance was calculated using a Mann-Whitney test (a rank test) for the median score of five liver sections from 10 individual mice per strain and indicated as * for p<0.05. Data of two independent experiments were combined and respective pictures for the determination of inflammation in NOD and NOD.B6 Idd10 mice are displayed in Fig. S2.
(B) Infection does not reverse the protection from T1D in NOD.B6 Idd10 mice.
Blood glucose levels in infected and age- and sex-matched uninfected female NOD.B6 Idd10 mice were determined every other week and analyzed until 200 days after infection. Mice with blood glucose levels > 300 mg/mL were considered diabetic and the pancreas was harvested for the analysis of insulitis after reconfirmation of blood glucose levels > 500 mg/mL. Statistical significance was calculated using a survival curve analysis for 19 mice in the uninfected control group and 30 infected mice (p=0.1225).
(C) NOD.A/J Idd10 mice containing the B6 Cd101 haplotype develop similar severe liver disease as NOD.B6 Idd10 mice.
Liver sections of the indicated mouse strains were analyzed in blinded studies for the severity of portal inflammation. Statistical significance was calculated using a Mann-Whitney test for 8 individual mice 4 months after infection and indicated as * for p<0.05.
(D) Genetic deletion of CD101 enhances the severity of liver lesions in B6 mice upon infection with N. aro.
Histopathological scores for portal inflammation two months after infection were determined in blinded studies. Statistical significance was calculated using a Mann-Whitney test for eight individual mice in each group and indicated as * for p<0.05 and ** for p<0.01. The experiments were repeated twice with similar results.
(E) Increased numbers of N. aro in the livers of NOD.B6 Idd10 mice.
Bacterial burden in different organs from individual NOD and NOD.B6 Idd10 mice on day two post-infection (each bar represents mean ± standard deviation of three individual mice per strain) was assessed by cfu plating assays and limiting dilution. One experiment representative of three is shown. Statistical significance was calculated using a student’s t-test and indicated as * for p<0.05 comparing bacterial load in the livers of NOD.B6 Idd10 and NOD mice.
(F) Prolonged bacterial persistence in mice containing the B6 CD101 allele.
Copy numbers of N. aro in individual livers of the indicated mouse strains were analyzed by a N. aro-specific 16S rRNA qPCR. N. aro copies in the livers of NOD.B6 Idd10 and NOD.B6 Idd10/Idd18 mice were compared to parental NOD mice and analyzed at weeks 7 (left panel) and 9 (right panel) after infection. Statistical significance for two combined independent experiments was calculated with organs from 3 individual mice for each strain using a student’s t-test and indicated as * for p<0.05.
As persistence of N. aro is absolutely required for the induction of liver lesions and the long-term evolution to autoimmunity (42), we tested if introgression of the B6 Cd101 allele influences the course of N. aro-infection, and subsequently impacts the outcome of disease. As observed previously (42), we detected the preferential accumulation of N. aro in the livers of infected mice compared to other organs. N. aro was observed at much lower levels in the spleen and kidneys, but was completely absent from the pancreas and the heart (Fig. 1E). Although bacterial persistence was prolonged in the livers of NOD.B6 Idd10 and NOD.B6 Idd10/18 congenic compared to parental NOD mice (Fig. 1F, left panel), bacterial infection was cleared 2 weeks later (Fig. 1F, right panel). Similarly, bacterial infection was more slowly cleared from the livers of B6 CD101−/− compared to B6 wild type mice (Fig. S2C). As clearance of bacteria was delayed in NOD.B6 Idd10 mice compared to B6 mice correlating with substantially enhanced liver pathology, we concluded that the increased susceptibility to autoimmune liver disease in NOD.B6 Idd10 mice requires both non-Idd10 NOD alleles and the B6 Idd10 haplotype.
Overall, these results point to the B6 allele at Idd10 as being the primary liver disease-promoting subregion within the larger Idd10/18 region upon infection; increased liver pathology may be associated with the prolonged bacterial persistence in the livers of NOD mice with the B6 and A/J Idd10 haplotypes compared to NOD mice with the NOD and CAST haplotypes.
N. aro infection does not alter CD101 expression on T cells, but enhances their activation status in the liver
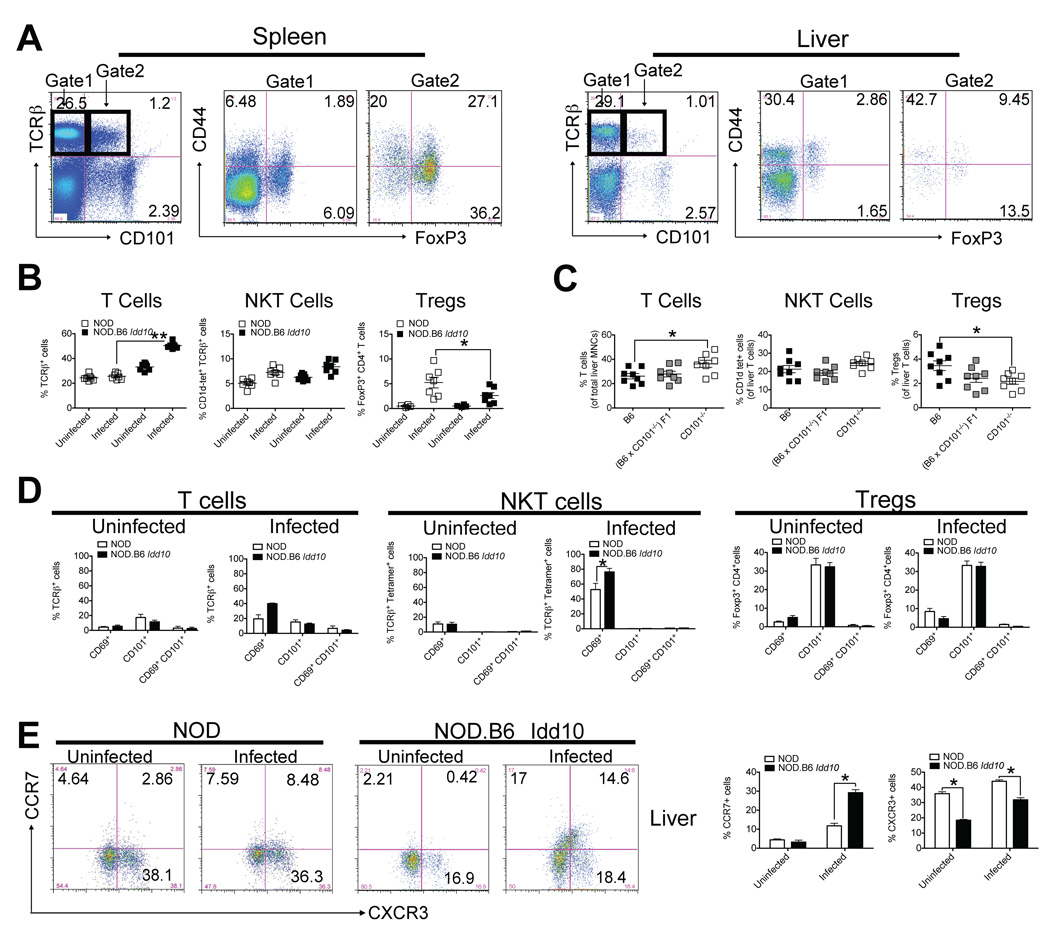
Expression of CD101 has been described on 25–30% of CD4+FoxP3+ regulatory T cells, approximately 10–20% of conventional CD4+ and CD8+ memory T cells and few naïve CD4+ and CD8+ T cells from splenocytes of B6 mice (54, 58, 59, 60). In contrast to the spleen, where most of the CD101+ T cells express FoxP3, most of the CD101+ T cells in the liver do not express FoxP3 (Fig. 2A). As the presence of the B6 CD101 allele as part of the B6 haplotype spanning the Idd10 region is associated not only with protection from T1D, but also with more severe PBC upon exposure of mice to N. aro, we evaluated the distribution of the different T cell populations and the expression of CD101 expression on these different T cell populations in the liver by flow cytometry (Fig. 2B–D) upon infection, the organ site where N. aro preferentially persists (42) (Fig. 1E and F). T cell numbers, especially in NOD.B6 Idd10 mice significantly increased at day 18 after infection (Fig. 2B, left panel). FoxP3+ Tregs were only found in the livers of infected mice, preferentially NOD mice, whereas in uninfected mice they were rarely detected (Fig. 2B, right panel). B6 CD101−/− mice recruited significantly fewer hepatic Tregs than B6 mice upon infection, but significantly more T cells (Fig. 2C); thus, changes on the B6 CD101−/− background closely resemble the phenotype observed in NOD.B6 Idd10 mice. NKT cells from neither naïve nor infected mice expressed CD101 at any time in the liver (Fig. 2D). Infection did not change the percentage of CD101 expressing cells in any of these T cell populations (Fig. 2D) or the mean of CD101 expression on these T cells (Fig. S3A). Most of the T cells that expressed CD69, IFN-γ or IL-17 were CD101-negative (Fig. 2D and S3C). Few CD44+ T cells in the livers of NOD.B6 Idd10 mice expressed CD101 (Fig. S3D); however, these T cells expressed CCR7 (Fig. 2E). CCR7 binds to CCL21 (see also Fig. 4E) - a chemokine expressed during lymphoneogenesis and in inflamed portal tracts (55) that has been implicated in the pathogenesis of PBC in humans (56). CXCR3, which recruits T cells with regulatory functions upon NKT cell activation to the liver (57) was preferentially expressed on T cells from NOD compared to NOD.B6 Idd10 mice (Fig. 2E). Livers of B6 CD101−/− mice revealed higher numbers of CCR7+ and lower percentages of CXCR3+ T cells compared to wild type mice (not shown) suggesting that the genetic deletion of CD101 or the introgression of the B6 Cd101 allele on the NOD background affects the upregulation of different chemokines within the liver environment and subsequently the recruitment of different T cell populations. Albeit these data indicate that expression of the B6 Cd101 allele or genetic deletion of CD101 affects T cell activation, this in an indirect process regulated by DCs as outlined later (Fig. 4 and 6) in this study.
Figure 2. N. aro induces an overzealous T cell response in the liver without modulating the expression of CD101 on T cells.
(A) CD101-expressing T cells in the livers are mainly CD44 positive, while CD101-expressing T cells in the spleen preferentially express FoxP3.
Representative FACS dot plots for the expression of CD101 in spleen and liver of uninfected 6 week-old NOD.B6 Idd10 mice are shown. Cells were distinguished within CD101+ and CD101− T cells and the percentage of CD44+ and FoxP3+ populations in spleen and liver are displayed. Note that the majority of CD101 expressing immune cells in the liver are not T cells.
(B+C) N. aro-infection increases total T cell percentages in the livers of NOD.B6 Idd10 (B) and B6 CD101−/− (C) mice, while Tregs preferentially accumulate in the livers of infected parental NOD (B) and B6 (C) mice
T, NKT and Treg percentages in uninfected and infected 5 week-old NOD and NOD.B6 Idd10 mice (B) as well as in infected B6 CD101+/+, B6 CD101+/− and B6 CD101−/− (B) mice representative of 7 (B) and 8 (C) individual mice each were determined at day 18 after infection. Statistical significance was calculated using a student’s t-test and indicated as * for p<0.05 and as ** for p<0.01. Note that there are very few Tregs in the livers of uninfected mice.
(D) Infection of mice does not alter CD101 expression levels and distribution on liver T lymphocytes
A representative summary of CD69 and/or CD101 expression by T cells (gated on TCRβ+) (left two panels), NKT cells (gated on TCRβ+ CD1d-tetramer+) (middle two panels) and Tregs (gated on TCRβ+ CD4+ FoxP3+) (right two panels) for 8 individual mice at day 1 after infection is shown. Statistical significance was calculated using a student’s t-test and indicated as * for p<0.05.
(E) NOD.B6 Idd10 mice recruit enhanced numbers of CCR7-expressing T cells specifically to the liver upon N. aro-infection.
Representative FACS dot plots for CCR7- and CXCR3- expression by T cells (gated on TCRβ+) for 8 individual mice are displayed at day 24 after infection. Data for 8 individual mice are summarized in the bar graphs on the right. Statistical significance was determined using a student’s t-test and indicated as * for p<0.05.
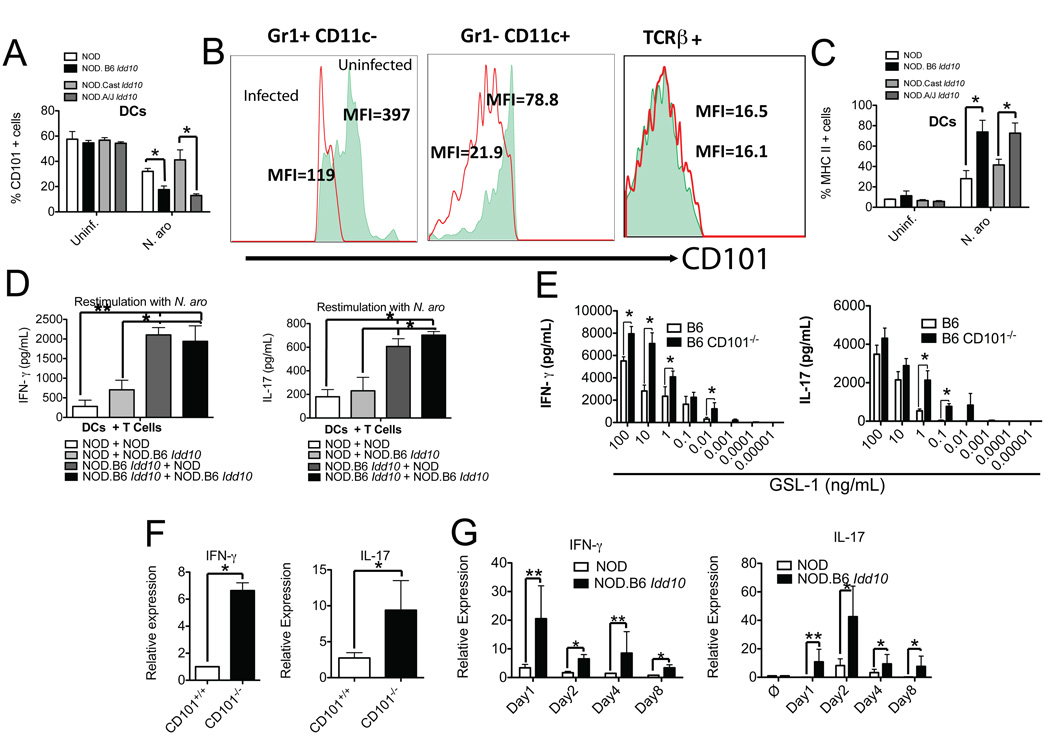
Figure 4. Infection of mice with N. aro selectively suppresses the expression of CD101 on APCs and phagocytes.
(A–C) Enhanced downregulation of CD101 expression on liver cells from NOD.B6 Idd10 mice upon infection with N. aro.
Representative FACS plots for the percentages and mean fluorescence intensities (MFIs) of Gr1+ (A), CD11c+ (B) and F4/80+ cells (C) obtained from the livers of naïve and infected (five days after infection) NOD.B6 Idd10 mice are displayed. Data describing the number of CD101+ Gr1+ (A), CD101+ CD11c+ (B) and CD101+ F4/80+ cells (C) in NOD, NOD.B6 Idd10 and B6 mice from two independent experiments were summarized in the right panels. Statistical significance was calculated using a student’s t-test and indicated as * for p<0.05. Note the increased accumulation of all three cell populations upon infection.
(D) Enhanced recruitment of DCs, but reduced accumulation of granulocytes in the livers of infected NOD.B6 Idd10 and CD101-deficient B6 compared to parental NOD and B6 mice.
Mice (n=6) were infected with N. aro five days prior to analysis by flow cytometry and the granulocyte and DC percentages in their livers were compared to age- and sex matched control mice. Each dot represents the percentage of granulocytes (Gr1+) and DCs (CD11c+) in the liver of an individual mouse. Statistical significance was calculated using a student’s t-test and indicated as * for p<0.05.
(E+F) Persistent N. aro-infection correlates with increased CCL21 expression in the liver.
CCL21 mRNA copies were analyzed in 5×106 liver lymphocytes (E) and splenocytes (F) from infected mice at week 4 after infection by qPCR. HGPRT mRNA copies were used to normalize expression levels and the quotient of the respective cytokine over the HGPRT copies was calculated. The quotient cytokine/HGPRT was set as 1 for the liver of naïve, uninfected NOD mice. Spleen cells from uninfected NOD mice have a quotient of 0.53, while liver and spleen cells from uninfected NOD.B6 Idd10 mice have a quotient of 1.18 and 0.41, respectively. The relative increase of CCL21 copy numbers in infected NOD and NOD.B6 Idd10 is displayed. Mean and standard deviation of four individual mice were compared. Statistical significance was calculated using a student’s t-test and indicated as * for p<0.05.
Figure 6. Expression of the B6 Cd101 allele enhances IFN-γ and IL-17 responses and defines the severity of liver disease.
(A–E) Allele dependent suppression of CD101 on DCs enhances IFN-γ- and IL-17-release by T cells.
(A–C) Suppression of CD101 expression and enhancement of MHC II expression after in vitro exposure to N. aro.
Bone marrow-derived DCs (after seven days of in vitro culture) from the indicated mouse strains (A+C) or liver mononuclear cells from 3 individual NOD.B6 Idd10 mice depleted of B and NK cells using MACS (B) were exposed to N. aro at a ratio of 1:50 (A+C) or 1:10 (B) or remained uninfected. 48 hours later, infected and uninfected DCs (A+C) as well as Gr1+, CD11c+ and TCRβ+ cells (B) were analyzed for CD101 (A+B) and MHC II (C) expression. The summary of four independent experiments is displayed (A+C). Statistical significance was calculated using a student’s t-test and indicated as * for p<0.05. Note that the suppression of CD101 protein expression on DCs containing the B6 Cd101 allele is enhanced (A) and the CD101 expression on both Gr1+ and CD11c+ cells is reduced upon infection while CD101 expression on T cells remains unaffected (B). The MFIs for CD101 expression in uninfected cells are represented by the filled areas in the histograms, while infected cells are represented by the red line (B).
(D) N. aro stimulated IFN-γ and IL-17 release by T cells is augmented by the B6.Idd10 allele in a DC intrinsic manner
Bone marrow-derived DCs from NOD and NOD.B6 Idd10 mice were pulsed with N. aro. at a ratio N. aro to APCs of 50:1 and co-cultured with liver lymphocytes from infected NOD and NOD.B6 Idd10 mice. Cell culture supernatants were assayed 72 hours later for the release of IFN-γ (left panel) and IL-17 (right panel) by ELISA. Results of two independent experiments with cells from 6 mice in total were combined and statistical significance calculated using a student’s t-test and indicated as * for p<0.05 and as ** for p<0.01.
(E) CD101-deficiency enhances IFN-γ and IL-17 release from T cells
Bone marrow-derived DCs from CD101-deficient and wild type B6 mice were pulsed with the indicated N. aro GSL-1 concentrations and incubated with liver T cells from uninfected mice. Cell culture supernatants were assayed 72 hours later for the release of IFN-γ (left panel) and IL-17 (right panel) by ELISA. Results of two independent experiments with cells from 6 mice in total were combined and statistical significance calculated using a student’s t-test and indicated as * for p<0.05.
(F+G) The induction of IFN-γ and IL-17 responses upon N. aro-infection is a consequence of CD101 suppression.
Expression of mRNA copies for IFN-γ and IL-17 was analyzed in 5×106 liver lymphocytes each from infected B6 wild type and CD101-deficient mice 19 days after infection (F) and in a kinetic experiment comparing NOD and NOD.B6 Idd10 mice (G) by qPCR. The relative increase in cytokine copy numbers of CD101-deficient and NOD.B6 Idd10 over parental B6 and NOD mice is displayed. Mean and standard deviation for cytokine copies of four individual mice were compared and statistical significance was determined using a student’s t-test and indicated as * for p<0.05 and as ** for p<0.01.
MHC I and MHC II restricted T cells that produce IFN-γ and IL-17 are required for the adoptive transfer of liver lesions
As we previously published, chronic liver inflammation in the absence of long-term microbial persistence, and the adoptive transfer of disease by TCRα/β+ CD4+ and CD8+ T cells suggest an autoimmune etiology of the liver disease (42). To identify critical antigen presenting molecules and cytokines necessary for the transfer of liver lesions by T cells, we transferred 1×107 TCRβ+ liver and spleen cells from diseased B6 mice into irradiated recipients that cannot present antigens via CD1d, MHC I or MHC II or do not respond to cytokines such as IFN-γ or IL-17 (Fig. 3A) 10 weeks after infection when all persisting bacteria have been cleared (Fig. 1F, right panel and Fig.S2C). Notably, IFN-γ and IL-17 were critical for the adoptive transfer of liver disease (Fig. 3A). The requirement for antigen presentation by MHC class I and II, but not CD1d confirmed the need for CD4+ as well as CD8+ T cells for the transfer of liver disease (Fig. 3A) (42). Although γ/δ+ T cells and NK cells accumulated in the livers of diseased mice (not shown), these cells did not adoptively transfer liver lesions (Fig. 3B).
Figure 3. MHC class I and II-restricted TCRα/β+ T cells secreting IFN-γ and IL-17, but not γ/δ T cells or NK cells, are required for the adoptive transfer of liver lesions.
Transfer of 1×107 purified TCRβ+ T cells (A), TCRγ/δ+ or NKp46+ cells (B) from spleens and livers of 12 individual N. aro-infected B6 (A) or NOD.B6 Idd10 (B) mice into the indicated, irradiated 12 week-old recipient mice (cells from two individual mice are combined for the adoptive transfer into one recipient each). Livers from recipient mice were harvested 6 weeks after the adoptive transfer and the severity of liver lesions was scored in blinded studies. Recipients and donors were N. aro-free as determined by a N. aro-specific 16S rRNA qPCR. Results of two independent experiments (A+B) have been combined and statistical significance of the differences in histopathological scores was calculated using a Mann-Whitney test for 6 individual recipients per strain and indicated as * for p<0.05 and as ** for p<0.01.
Collectively, our data suggest that the dominant IFN-γ and IL-17 cytokine profile correlates with the induction of severe liver disease and promotes autoreactive CD4+ and CD8+ T cells that are responsible for the adoptive transfer of liver lesions.
N. aro-infection suppresses CD101 expression on granulocytes, DCs and macrophages in the liver
CD101 is not only expressed on subsets of T cells, but also on macrophages, DCs and granulocytes (54, 58), and has been implicated in the maintenance of peripheral tolerance in humans (58, 59). As N. aro localizes to the liver in our infection-induced mouse model (Fig. 1 E and F) (42) and as CD101 expression is not changed on hepatic T cells after infection (Fig. 2D and Fig. S3A), we focused on these other immune cell subsets in the hepatic compartment that constitute the majority of CD101 expressing cells in the liver (Fig. 2A). To evaluate if N. aro infection affects CD101 expression on these cell populations, we analyzed mononuclear cells from the liver of NOD and NOD.B6 Idd10 mice. Around 40–70% of Gr1+, CD11c+ and F4/80+ cells expressed CD101 in naïve, uninfected NOD and NOD.B6 Idd10 mice to similar levels and frequencies (Fig. 4A–C). Absolute cell numbers and the relative distribution of cell populations were comparable between the analyzed mouse strains (not shown). All DC subpopulations, including CD8+, B220+ and CD11b+ increased upon infection without a significant change in the proportion of the subpopulations (Fig. S4A). CD101 was preferentially expressed on CD11b+ CD11c+ myeloid DCs in the spleen, while CD101 positive DCs also co-expressed B220 in the liver (Fig. S4B); very few CD8+ lymphoid DCs (CD8+ CD11c+) expressed CD101 in either tissue (Fig. S4B).
Surface expression of CD101 (Fig. S3B) and the number of CD101 positive cells were reduced on Gr1+ (Fig. 4A), CD11c+ (Fig. 4B), and F4/80+ cells upon infection (Fig. 4C). Although the mean fluorescence intensity on Gr1high CD101high in the livers of uninfected NOD.B6 Idd10 was higher than in uninfected NOD mice as observed in the spleen (47), Gr1+ cells from NOD.B6 Idd10 mice had reduced CD101 levels compared to cells from parental NOD mice after infection (Fig. 4A); a similar enhanced CD101 suppression was also observed on DCs and macrophages from NOD.B6 Idd10 mice (Fig. 4B and C). CD101 levels on DCs did not return to baseline levels up to 8 weeks after infection in the liver (Fig. S5A). However, infection did not influence the distribution of CD101 expression (Fig. S5B) on DCs in pancreatic lymph nodes, while CD101 expression was only transiently reduced in the spleen returning to baseline levels about 3–4 weeks after infection (Fig. S5C and not shown). These data suggest that suppression of CD101 expression correlates with the presence/persistence of N. aro at these different organ sites (Fig. 1E and F). Accordingly, increased copy numbers of N. aro were preferentially detected by a 16S rRNA specific qPCR in cell-sorted, purified CD101-negative DCs, whereas CD101-positive DCs contained in general less N. aro 16S rRNA (Fig. S5D). As CD101 deficiency did not alter the rate of infection (Fig. S5E), these data suggest that either infection suppresses CD101 expression on APCs or APCs downregulate the negative costimulatory molecule CD101 upon bacterial contact. Whereas greater numbers of granulocytes accumulated in the livers of NOD compared to NOD.B6 Idd10 mice, a reduced influx of DCs into the livers of NOD mice was observed (Fig. 4D). Similarly, reduced numbers of granulocytes accumulated in the livers of infected B6 CD101−/− mice as compared to B6 mice, while DCs were increased (Fig. 4D). CD101 may not only have a negative costimulatory role (54), but may also be involved in the recruitment or containment of certain cell populations within a particular tissue. DCs and neutrophils represent resident cell populations in the liver (Fig. 4D); DC and granulocyte numbers increase upon exposure to N. aro suggesting that both are recruited to the liver upon infection (Fig. 4D). Infection of neutrophils occurs during the systemic response to N. aro and subsequent downregulation of CD101 may impair their recruitment to the liver, which is exacerbated in NOD.B6 Idd10 and B6 CD101−/− mice. Alternatively, CD101 negative granulocytes may be recruited preferentially to the livers of NOD.B6 Idd10 and B6 CD101−/− mice responding to the increased expression of chemotactic factors and/or inflammatory cytokines in these mice. Since the survival time of DCs is longer than that of granulocytes, the magnitude of CD101 expression on DCs is lower than on granulocytes and DC numbers are not reduced in the livers of CD101-deficient mice without infection, subsequent upregulation of chemotactic factors by infection may be contributing to the accumulation of DCs at the site of inflammation despite CD101 deficiency. We found that CCL21 mRNA expression was most prominently enhanced in the livers, but not in the spleens, of NOD.B6 Idd10 mice (Fig. 4E and F) compared to parental NOD mice. The increased expression of CCL21, the ligand for CCR7, may be responsible for the preferential recruitment of CCR7-expressing T cells to the infected and inflamed livers of NOD.B6 Idd10 and B6 CD101−/− mice (Fig. 2E). In summary, the suppression of CD101 on APCs from NOD.B6 Idd10 mice upon infection is correlated with an inflammatory profile in the liver environment that resembles the phenotype in B6 CD101−/− mice.
Cell intrinsic differences in CD101 specific for the genetic background determine the recruitment of granulocytes and DCs to the infected liver
To determine if Idd10/Cd101 gene haplotype-dependent expression differences are mediated in a cell-intrinsic manner or rather secondary events, we measured variations in the accumulation of different cell populations in N. aro-infected NOD.B6 Ptprc CD45.2+ / NOD.B6 Idd10 as well as B6 CD45.1+ / B6 CD101-deficient mixed bone marrow chimeras. These mixed bone marrow chimeras expressed similar numbers of CD45.1 and CD45.2 positive cells as determined by FACS analysis (55.3±4.9 vs 44.7±6.1 and 49.7±8.8 vs 50.3±9.1, respectively) in the blood. We found that the altered accumulation of granulocytes by genotype (Fig. 4A) is cell-intrinsic as CD45.2 positive granulocytes originating from NOD.B6 Ptprc mice accumulated to greater numbers in the livers of infected mixed bone marrow chimeras than granulocytes from NOD.B6 Idd10 mice (Fig. 5A). Similar results were obtained with reciprocal bone marrow chimeras (not shown). The increased influx of Gr1+ cells expressing the NOD Idd10 allele correlated with increased CD101 expression on these Gr1+ cells as compared to CD101 levels on Gr1+ cells from NOD.B6 Idd10 mice (Fig. S6A). Accumulation of granulocytes derived from CD101-deficient B6 bone marrow was also reduced as compared to those from the wild type B6 bone marrow (Fig. 5B); these data strongly suggest that the intrinsic reduction of CD101 on NOD.B6 Idd10 granulocytes upon infection leads to the reduced accumulation of granulocytes in the liver. The differential accumulation of DCs in the liver following infection (Fig. 5C and D) was also cell-intrinsic: introgression of the B6 Idd10 region on the NOD background and CD101-deficiency on the B6 background both enhanced the accumulation of DCs in the respective mixed bone marrow chimeras (Fig. 5C and D). As an intrinsic consequence of reduced CD101 expression, DCs of NOD.B6 Idd10 and CD101-deficient B6 origin expressed higher levels of MHC class II than parental NOD and B6 mice (Fig. 5E and F).
Figure 5. Background specific regulation of DC and granulocyte recruitment and activation upon infection.
(A–D) The influx of granulocytes and DCs in the livers of infected, mixed bone marrow chimeras is background-intrinsic.
Irradiated NOD.B6 Ptprc CD45.2+ mice were reconstituted with a 1:1 mixture of NOD.B6 Idd10 (CD45.1+) and CD45.2+ NOD.B6 Ptprc bone marrow cells (A+C). Similarly, irradiated B6 CD45.1+ mice were reconstituted with a 1:1 mixture of B6 CD101−/− (CD45.2+) and CD45.1+ B6 bone marrow cells (B+D). Bone marrow chimeras were infected 6 weeks after bone marrow reconstitution with N. aro. At day 1 (not shown) and day 8, percentages of granulocytes (A+B) and DCs (C+D) into infected and inflamed livers was assessed using CD45.1 allotype-specific antibodies to assign their origin to parental NOD and B6 or congenic NOD.B6 Idd10 and CD101-deficient B6 mice. Statistical significance was calculated using a Linear-Mixed-Effect Model for eight (A+C) and six (B+D) individual naïve and infected chimeric mice and indicated as * for p < 0.05. The summary of data as well as the representative FACS dot plots for infected and uninfected control mice are displayed. Similar results were obtained using reciprocal bone marrow chimeras (data not shown).
(E+F) DCs from NOD.B6 Idd10 and CD101-deficient B6 mice express higher levels of MHC II compared to parental NOD and B6 mice.
CD11c+ DCs were analyzed for their CD101 and MHC II expression. The origin of CD11c+ cells was distinguished by gating on CD45.1+ and CD45.2+ cells in the respective bone marrow chimeras as described above. Representative FACS dot plots are displayed for day 2 after infection for cells of NOD and NOD.B6 Idd10 (E) origin. Data for six individual NOD.B6 Ptprc CD45.2/NOD.B6 Idd10 (E) and six individual B6 CD45.1/ CD101−/− (F) bone marrow chimeras are summarized in the respective bar graphs. Statistical significance was calculated using a student’s t-test and indicated as * for p<0.05.
Overall, CD101 expression influences in a cell-intrinsic manner the accumulation of granulocytes and DCs in the infected liver and the upregulation of MHC class II on the accumulated DCs.
Suppression of CD101 on DCs triggers a IFN-γ and IL-17 dominated cytokine profile
Interactions with antigen presenting cells (APCs) are required for T cell activation. Even though conventional T cells express small amounts of CD1d in the periphery, they cannot present glycosphingolipid (GSL) antigen directly to NKT cells (61–65) in contrast to APCs. Therefore, conventional T cell activation by NKT cells responding to GSL antigens is an indirect process that requires NKT cell stimulation via APCs. The expression of co-stimulatory molecules by APCs that are induced by productive interactions with NKT and T cells and the local cytokine milieu are critical for the subsequent induction/suppression of T cell responses (61, 63). Since CD101 has been described as a negative co-stimulatory molecule (59, 65, 66), it is possible that CD101 contributes to the ability of APCs to activate the T cell response. To evaluate if infection induced suppression of CD101 on APCs contributes to the overzealous T cell response in infected mice, bone marrow derived DCs from NOD and NOD.B6 Idd10 mice were left untreated or exposed to N. aro for 48 hours (Fig. 6A). Untreated cells showed no major differences in the distribution of CD101 expression. However, as observed in vivo for liver CD11c+ cells (Fig. 4B), CD101 expression was suppressed on bone marrow-derived DCs (Fig. 6A), but not T cells (not shown) when exposed to N. aro. This suppression of CD101 expression was more prominent in NOD.B6 Idd10 and NOD.A/J Idd10 mice than in NOD and NOD.CAST Idd10 mice. Similarly, CD101 expression was suppressed on CD11c+ and Gr1+ cells in liver cell preparations one day after exposure to N. aro in vitro, while CD101 expression on T cells in the same cell preparations remained unaffected (Fig. 6B and Fig. S6B). As observed in vivo (Fig. 5E, F), MHC II expression was increased preferentially on DCs of NOD.B6 Idd10 or NOD.A/J Idd10 origin compared to DCs from NOD or NOD.CAST Idd10 mice (Fig. 6C). A comparable increase in MHC II expression was detected on DCs from CD101-deficient mice compared to DCs from B6 mice (81.1± 7.8 vs. 44.3±6.6). Cocultures of N. aro-pulsed DCs with liver lymphocytes (containing NKT and T cells) from infected mice demonstrated that the secretion of IFN-γ and IL-17 was enhanced in a NOD.B6 Idd10 DC-dependent manner (Fig. 6D). Similarly, cocultures of CD101-deficient DCs pulsed with GSL-1, a NKT cell ligand expressed in the cell wall of N. aro, and T/NKT cells from uninfected mice induced enhanced IFN-γ and IL-17 responses compared to cocultures with wild type DCs (Fig. 6E). These results suggest that infection-mediated reduced CD101 expression is not required for NKT cell-dependent DC activation; however, the lack of CD101 or the suppression of CD101 expression on APCs is sufficient to drive the overzealous Th1 and Th17 dominated conventional T cell response. mRNA copy numbers of IFN-γ and IL-17 in livers from infected CD101-deficient B6 (Fig. 6F) and NOD.B6 Idd10 mice (Fig. 6G) were increased compared to parental B6 and NOD mice respectively, suggesting a role of the B6 Cd101 allele in the induction of these Th1/Th17 dominated immune responses on the NOD background.
Collectively, our data suggest that enhanced IFN-γ and IL-17 responses in NOD.B6 Idd10 and NOD A/J Idd10 mice promote severe liver disease. Reduced CD101 expression on DCs following N. aro infection is therefore critical for the induction of these overzealous T cell responses.
Discussion
Autoimmune diseases often cluster in the same family; this indicates that there are common genetic pathways affecting immunological tolerance mechanisms, although the autoimmune assault may occur in diverse tissue types. Genome-wide association (GWAS) studies have now revealed very significant sharing of mapped susceptibility loci (67–69); in this study we have shown that a gene region previously shown to influence susceptibility to T1D in the NOD mouse model (44–46, 53), Idd10, also influences an autoimmune disease involving a different tissue, the liver, which follows an infection with N. aro (42). Using a CD101-deficient B6 mouse combined with haplotype analysis studies in NOD.CAST Idd10 and NOD.A/J Idd10 mice, our data suggest that allelic variations within Cd101 drive susceptibility for infection-induced PBC. Intravenous infection of mice with N. aro leads to the allele dependent downregulation of CD101 protein expression on DCs, macrophages and granulocytes in the liver, the preferential site of bacterial persistence. Since downregulation of CD101 is more pronounced in NOD congenic strains that express the B6 Cd101 allele than in parental NOD mice, and the clinical and histopathological phenotypes in these mice resemble those of CD101-deficient B6, we conclude that the allele-specific downregulation of CD101 protein underlies the induction of severe liver autoimmunity in NOD.B6 Idd10 mice (Fig. 7). The substantially enhanced liver disease in these mice is associated with overzealous Th1 and Th17 responses propagated by NKT cells responding with an enhanced release of inflammatory cytokines to GSL antigens of N. aro in the absence of CD101 expression on APCs. Whereas genetic deletion of CD101 in the B6 background mimics the phenotype in the NOD.B6 Idd10 congenic strain, the B6 haplotype of CD101 on the NOD background alone is not sufficient for triggering severe liver disease. As we observed greater downregulation of CD101 expression in NOD.B6 Idd10 compared to B6 mice after N. aro-infection (Fig. 4A–C), it is likely that the combination of multiple loci on the NOD background together with the B6 Idd10 haplotype confer increased susceptibility to infection induced PBC, which likely accounts for the more profound downregulation of CD101 in NOD.B6 Idd10 mice. That downregulation of CD101 is particular striking in Gr1+ cells: Gr1+ cells in NOD.B6 Idd10 mice start with higher CD101 expression than Gr1+ cells from naïve NOD mice, but downregulate CD101 more significantly upon infection (Fig. S6A). In contrast, B6 mice exhibit compensatory mechanisms that better control bacterial infection and T cell activation, two major mechanisms that are lost once the CD101 allele is deleted. Since B6 CD101−/− and NOD.B6 Idd10 congenic mice develop overzealous, Th1 and Th17 dominated T cell responses and clear bacterial infection more slowly than parental NOD and B6 mice, and GWAS studies in PBC patients identified genes affecting the Th1 response and T cell signaling as PBC risk factors including IL-12 (70), Spi-B and IRF5 (71, 72), it is tempting to speculate that bacterial infection triggers inappropriate T cell responses in genetically predisposed individuals. The murine orthologs of IL-12, Spi-B and IRF5 may be also the non-Idd10 NOD alleles in NOD.B6 Idd10 mice that interfere with the B6 Cd101 haplotype.
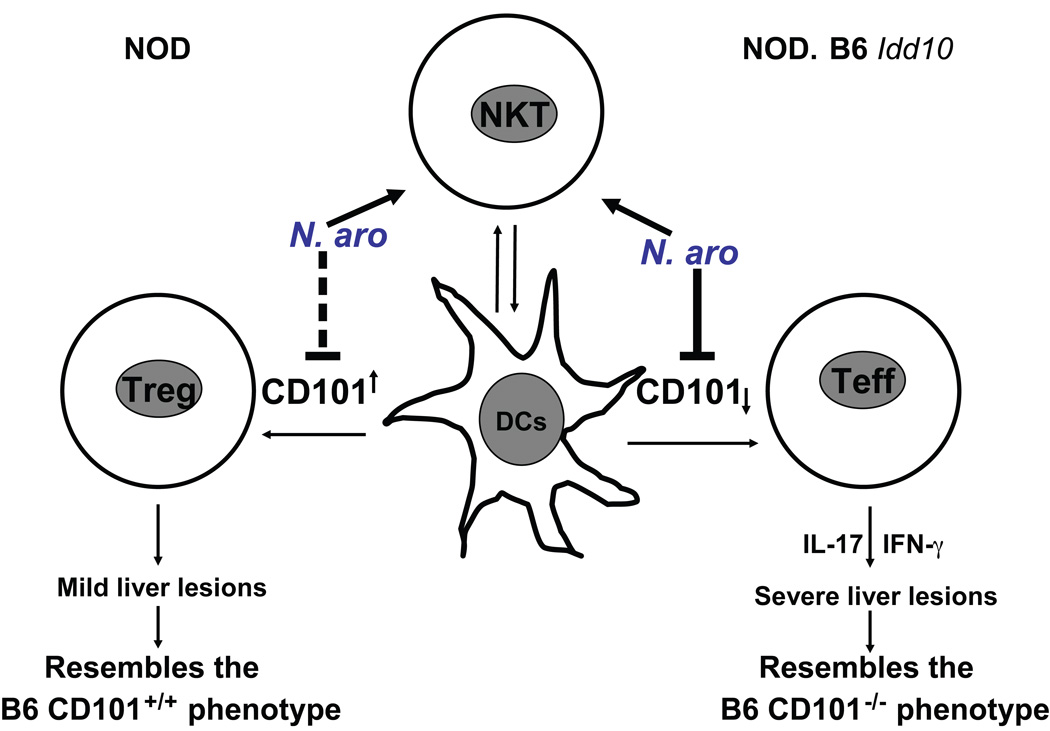
Figure 7. Cd101 alleles regulate the resistance and susceptibility to liver disease induced by N. aro-infection.
Expression of the NOD Cd101 allele induces a more tolerogenic milieu in the liver by promoting Treg responses whereas expression of the B6 Cd101 allele triggers an overzealous T cell response upon infection with N. aro. The loss of CD101 expression on DCs drives the enhanced IFN-γ and IL-17 production by T cells and subsequently the induction of liver disease upon N. aro-infection.
Downregulation of CD101 on granulocytes correlates with their reduced accumulation in the liver, an intrinsic property to this subset, likely contributing to the prolonged persistence of N. aro; downregulation of CD101 on DCs is closely associated with the evolution of overzealous, IFN-γ-/IL-17- dominated T cell responses (Fig. 7). These latter data confirm a critical role of CD101 as a negative costimulatory molecule in T cell activation (73–74) and define its action on T cells and T cell homeostasis as APC-intrinsic. The reduced accumulation of granulocytes upon infection in the liver and the intrinsic reduction of granulocytes in the bone marrow of CD101-deficient mice (47) suggest an additional role of CD101 in the containment and/or trafficking of granulocytes in/to defined organ sites.
DCs, however, still accumulate in the livers of infected mice despite CD101 downregulation or deficiency. As DCs express lower levels of CD101, survive longer upon infection and are preferentially infected in the liver compared to granulocytes that respond to systemic infection, secondary upregulation of inflammatory molecules during prolonged infection may overcome the lack of CD101 expression as suggested by the liver-specific upregulation of the chemokine CCL21. Alternatively, DCs may not require CD101 expression for trafficking in contrast to granulocytes and preferentially CD101-negative DCs may be recruited to the site of inflammation in order to induce an effector instead of a regulatory T cell response. As CCL21 is known to recruit DCs to the site of infection (75–76) and to be upregulated in portal tracts of chronically inflamed livers (55), induction of CCL21 may be also responsible for the recruitment of CCR7-expressing T cells to the infected and inflamed liver, an effect that may be augmented by CD101 suppression. Downregulation of CD101 on DCs and macrophages occurred upon bacterial contact: intracellular bacteria either suppress CD101 expression or APCs downregulate CD101 upon bacterial encounter. However, since CD101-deficient DCs and macrophages are infected similarly to wild type cells, we exclude a role of CD101 as a receptor involved in bacterial uptake. Alternatively, infection may induce the expression of a less functional CD101 splice variant in NOD.B6 Idd10 mice that is not recognized by our antibodies. This may be an allele intrinsic effect or reflect the alternative amino acid sequence of the B6 allotype. As one or more of the variations in the Cd101 allele are causal genetic events in the mouse model and may translate into human disease, future studies need to define if SNPs in the Cd101 gene, or in genes encoding proteins associated with CD101, are associated with PBC and whether there is a correlation with reduced CD101 expression and an inflammatory cytokine profile.
The correlation of severe pathological phenotypes in NOD.B6 Idd10, NOD.A/J Idd10 and B6 CD101 KO mice, along with the downregulation of CD101 in NOD.B6 Idd10 and NOD.A/J Idd10 mice mimicking the deletion of CD101 in the B6 background, strongly support the hypothesis that Cd101 variation within the Idd10 region is primarily responsible for altering the severity of liver autoimmunity induced by N. aro. However, on the NOD background, other genes within the B6-derived Idd10 region may contribute to severe liver disease such as B7h4 (encoded by Vtcn1), known for inhibiting T cell functions and granulocyte responses (77–79). While a loss of B7h4 function contributed by the B6 Idd10 region may contribute to the overzealous T cell response, it does not explain the reduced recruitment of granulocytes to the livers of NOD.B6 Idd10 mice that is observed in B6 CD101−/− mice. In addition, the development of comparably severe liver disease in B6 CD101−/− and NOD.B6 Idd10 as well as NOD.A/J Idd10 mice that expresses similar, but different B7h4 alleles strongly support the candidacy of Cd101 within Idd10. Since B7h4 was only very transiently expressed on some macrophages in the spleen on day 1 after infection (not shown), the blockade of B7h4 by neutralizing antibodies (80) had no impact on the amount of cytokines produced in the co-culture system of T cells with DCs independent of the Idd10 alleles expressed (not shown) and no genotype-dependent differences between NOD and NOD.B6 Idd10 mice in the expression of B7h4 protein were detected, we excluded a major contribution of the B7h4 alleles for influencing the severity of liver disease.
Even though several Idd-encoded molecules might be involved in different autoimmune diseases, the functional consequences of introgression of these alleles into the NOD background may differ from disease to disease and may change also the susceptibility of the host to infectious challenges. Therefore, the mechanisms through which allelic variations cause disease need to be unraveled individually for each disease. This is strikingly reflected by the fact that the B6 Cd101 allele protects from spontaneous T1D (43, 44, 46, 47) but exacerbates the PBC-like autoimmune process induced by N. aro infection. However, as introgression of the B6 Idd10 haplotype on the NOD background delays N. aro clearance similar that observed in B6 CD101−/− mice, CD101 should not only be considered as an autoimmune gene, but also an infectious disease gene regulating the persistence of bacterial infection. Not only the distribution of CD101 expression, but also its magnitude may contribute to these tissue-specific effects. Additionally, as cellular composition and total cell numbers differ between organs, even though the percentages of CD101 expressing cells may remain similar, altered cellular interactions may occur due to the absolute increase/decrease in CD101 available for the respective cells. In addition, different cell populations may express CD101 in various tissues; for example, most CD101+ T cells in the livers are CD44+, while they express predominantly FoxP3 in the spleen. Based on compelling evidence from various laboratories that development of diabetes in NOD mice is tightly controlled by Tregs (81–85), the interactions of FoxP3+ T cells with DCs likely influence the incidence of spontaneous T1D in our congenic mice. There are also only few Gr1+ myeloid cells or NKT cells found in pancreatic lymph nodes and there is no clear association of NKT cells with spontaneous T1D (61). In addition, no endogenous GSL antigen influencing the progression of T1D has been described to date. In contrast, we hypothesize that the interplay of DCs with NKT cells that are most abundant in the liver and respond to GSL antigens of N. aro (42, 51) propagate an overzealous T cell response. This inflammatory environment is also sustained due to the prolonged persistence of N. aro in the liver following the reduced recruitment of Gr1+ cells. These observations and the fact that environmental factors like infection may influence CD101 expression may all contribute to the tissue and/or cell specific effects of CD101 and determine if a Cd101 allele acts as a protective or susceptibility gene for a particular autoimmune disease. Although the effects of CD101 in promoting severe disease are ultimately T cell-mediated in our PBC model, the role of B cells and the anti-PDC-E2 response remains unclear and will be addressed in a separate study. It also needs to be explored if CD101 has a receptor or if it is involved in homotypic interactions.
Collectively, our data identify CD101 as the likely candidate molecule for N. aro-induced PBC and dissect the mechanisms by which N. aro-infection triggers liver inflammation. The defined diabetes susceptibility locus Idd10 that encodes CD101 as major candidate uncovers a key tool for understanding the circuits of cell-cell interactions in PBC. We confirm in this study not only the negative costimulatory role of CD101 in T cell activation, but also uncover a novel function of CD101 in the containment and/or trafficking of granulocytes to defined organ sites. Although not within the scope of the current study, dissecting the pathways in each autoimmune entity will allow us to understand the regulation of molecules in autoimmunity in general and their complex interplay as diabetes susceptibility loci have been associated with other autoimmune diseases as well. Consequently, our work will help to identify therapeutic targets and approaches that can be used to guide the development of effective therapies for PBC as well as to identify shared targets in autoimmune disease for clinical intervention in the future.
Supplementary Material
Acknowledgements
We thank the DHC core facility for the sectioning of histological slides, and Jorge Bezzera for critical reading of the manuscript. Kankana Chava for her help with the flow cytometry and qPCR analysis, Stacey Burgess and Christina S. Sexton for data collection and their help with in vivo experiments; Patrick S. Leung and Eric M. Gershwin for their support with the anti-PDC-E2 ELISA studies.
We would like to thank the NIH Tetramer Facility for providing α-GalCer CD1d-tetramers.
LSW and JAT are supported by a joint grant from the Juvenile Diabetes Research Foundation (JDRF), the Wellcome Trust and the National Institute for Health Research (NIHR) Biomedical Research Centre. Cambridge Institute for Medical Research (CIMR) is in receipt of a Wellcome Trust Strategic Award (079895). HIF was funded by a Wellcome Trust 4-year studentship. This work was also supported by a National Institutes of Health (USA) grant P01 AI39671 awarded to LSW and JT and R01DK074768 awarded to WMR and LSW. JM is supported by the Lupus Research Institute, in parts by PHS Grant P30 DK078392, a grant from the UC microbial pathogenesis core center, by Award Number R01DK084054 from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), the German Research Foundation DFG (MA 2621/2-1) and by the Interdisciplinary Center for Clinical Research of the Universitätsklinikum Erlangen (IZKF_JB10_A48). The availability of NOD congenic mice through the Taconic Emerging Models Program has been supported by grants from the Merck Genome Research Institute, NIAID, and the JDRF.
Abbreviations
- Idd
insulin-dependent diabetes
- SNP
single nucleotide polymorphism
- T1D
type 1 diabetes
- NOD
non-obese diabetic
- B6
C57BL/6
- MFI
mean fluorescence intensity
- DC
dendritic cell
- APC
antigen presenting cell
- GSL
GlycoSphingoLipid
- NKT
natural killer T cell
- PBC
primary biliary cirrhosis
References
- 1.Rioux JD, Abbas AK. Paths to understanding the genetic basis of autoimmune disease. Nature. 2005;435:584–589. doi: 10.1038/nature03723. [DOI] [PubMed] [Google Scholar]
- 2.Bjorses P, Aaltonen J, Horelli-Kuitunen N, Yaspo ML, Peltonen L. Gene defect behind APECED: a new clue to autoimmunity. Hum. Mol. Genet. 1998;7:1547–1553. doi: 10.1093/hmg/7.10.1547. [DOI] [PubMed] [Google Scholar]
- 3.Walker LS, Abbas AK. The enemy within: keeping self-reactive T cells at bay in the periphery. Nat. Rev Immunol. 2002;2:11–19. doi: 10.1038/nri701. [DOI] [PubMed] [Google Scholar]
- 4.He XS, Ansari AA, Ridgway WM, Coppel RL, Gershwin ME. New insights to the immunopathology and autoimmune responses in primary biliary cirrhosis. Cell Immunol. 2006;239:1–13. doi: 10.1016/j.cellimm.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 6.Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, Ardlie KG, Huang Q, Smith AM, Spoerke JM, Conn MT, Chang M, Chang SY, Saiki RK, Catanese JJ, Leong DU, Garcia VE, McAllister LB, Jeffery DA, Lee AT, Batliwalla F, Remmers E, Criswell LA, Seldin MF, Kastner DL, Amos CI, Sninsky JJ, Gregersen PK. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am. J. Hum. Genet. 2004;75:330–337. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kyogoku C, Langefeld CD, Ortmann WA, Lee A, Selby S, Carlton VE, Chang M, Ramos P, Baechler EC, Batliwalla FM, Novitzke J, Williams AH, Gillett C, Rodine P, Graham RR, Ardlie KG, Gaffney PM, Moser KL, Petri M, Begovich AB, Gregersen PK, Behrens TW. Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am. J. Hum. Genet. 2004;75:504–507. doi: 10.1086/423790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orozco G, Sanchez E, Gonzalez-Gay MA, Lopez-Nevot MA, Torres B, Caliz R, Ortego-Centeno N, Jimenez-Alonso J, Pascual-Salcedo D, Balsa A, de Pablo R, Nunez-Roldan A, Gonzalez-Escribano MF, Martin J. Association of a functional single-nucleotide polymorphism of PTPN22, encoding lymphoid protein phosphatase, with rheumatoid arthritis and systemic lupus erythematosus. Arthritis Rheum. 2005;52:219–224. doi: 10.1002/art.20771. [DOI] [PubMed] [Google Scholar]
- 9.Rose NR, Saboori AM, Rasooly L, Burek CL. The role of iodine in autoimmune thyroiditis. Crit. Rev. Immunol. 1997;17:511–517. [PubMed] [Google Scholar]
- 10.Maier LM, Lowe CE, Cooper J, Downes K, Anderson DE, Severson C, Clark PM, Healy B, Walker N, Aubin C, Oksenberg JR, Hauser SL, Compston A, Sawcer S, De Jager PL, Wicker LS, Todd JA, Hafler DA. IL2RA genetic heterogeneity in multiple sclerosis and type 1 diabetes susceptibility and soluble interleukin-2 receptor production. PLoS Genet. 2009;5:e1000322. doi: 10.1371/journal.pgen.1000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braley-Mullen H, Sharp GC, Medling B, Tang H. Spontaneous autoimmune thyroiditis in NOD.H-2h4 mice. J. Autoimmun. 1999;12:157–165. doi: 10.1006/jaut.1999.0272. [DOI] [PubMed] [Google Scholar]
- 12.Rasooly L, Burek CL, Rose NR. Iodine-induced autoimmune thyroiditis in NOD-H-2h4 mice. Clin. Immunol. Immunopathol. 1996;81:287–292. doi: 10.1006/clin.1996.0191. [DOI] [PubMed] [Google Scholar]
- 13.Vladutiu AO, Rose NR. Autoimmune murine thyroiditis relation to histocompatibility (H-2) type. Science. 1971;174:1137–1139. doi: 10.1126/science.174.4014.1137. [DOI] [PubMed] [Google Scholar]
- 14.Cooke A. Infection and autoimmunity. Blood Cells Mol. Dis. 2009;42:105–107. doi: 10.1016/j.bcmd.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 15.Willcocks LC, Smith KG, Clatworthy MR. Low-affinity Fcgamma receptors, autoimmunity and infection. Expert Rev. Mol. Med. 2009;11:e24. doi: 10.1017/S1462399409001161. [DOI] [PubMed] [Google Scholar]
- 16.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N. Engl. J. Med. 2002;347:911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 17.Kaplan MM, Gershwin ME. Primary biliary cirrhosis. N. Engl. J. Med. 2005;353:1261–1273. doi: 10.1056/NEJMra043898. [DOI] [PubMed] [Google Scholar]
- 18.Gershwin ME, Ansari AA, Mackay IR, Nakanuma Y, Nishio A, Rowley MJ, Coppel RL. Primary biliary cirrhosis: an orchestrated immune response against epithelial cells. Immunol. Rev. 2000;174:210–225. doi: 10.1034/j.1600-0528.2002.017402.x. [DOI] [PubMed] [Google Scholar]
- 19.Lan RY, Salunga TL, Tsuneyama K, Lian ZX, Yang GX, Hsu W, Moritoki Y, Ansari AA, Kemper C, Price J, Atkinson JP, Coppel RL, Gershwin ME. Hepatic IL-17 responses in human and murine primary biliary cirrhosis. J. Autoimmun. 2009;32:43–51. doi: 10.1016/j.jaut.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honda M, Kawai H, Shirota Y, Yamashita T, Kaneko S. Differential gene expression profiles in stage I primary biliary cirrhosis. Am. J. Gastroenterol. 2005;100:2019–2030. doi: 10.1111/j.1572-0241.2005.41662.x. [DOI] [PubMed] [Google Scholar]
- 21.Baba N, Kobashi H, Yamamoto K, Terada R, Suzuki T, Hakoda T, Okano N, Shimada N, Fujioka S, Iwasaki Y, Shiratori Y. Gene expression profiling in biliary epithelial cells of primary biliary cirrhosis using laser capture microdissection and cDNA microarray. Transl. Res. 2006;148:103–113. doi: 10.1016/j.trsl.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 22.Shackel NA, McGuinness PH, Abbott CA, Gorrell MD, McCaughan GW. Identification of novel molecules and pathogenic pathways in primary biliary cirrhosis: cDNA array analysis of intrahepatic differential gene expression. Gut. 2001;49:565–576. doi: 10.1136/gut.49.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harada K, Nakanuma Y. Biliary innate immunity and cholangiopathy. Hepatol. Res. 2007;37 Suppl 3:S430–S437. doi: 10.1111/j.1872-034X.2007.00247.x. [DOI] [PubMed] [Google Scholar]
- 24.Harada K, Isse K, Kamihira T, Shimoda S, Nakanuma Y. Th1 cytokine-induced downregulation of PPARgamma in human biliary cells relates to cholangitis in primary biliary cirrhosis. Hepatology. 2005;41:1329–1338. doi: 10.1002/hep.20705. [DOI] [PubMed] [Google Scholar]
- 25.Morishita A, Murota M, Fujita J, Ohtsuki Y, Nishioka M. Helper T lymphocytes specific for cytokeratin 19 in patients with primary biliary cirrhosis. Hepatol. Res. 2001;21:108–116. doi: 10.1016/s1386-6346(01)00086-9. [DOI] [PubMed] [Google Scholar]
- 26.Nagano T, Yamamoto K, Matsumoto S, Okamoto R, Tagashira M, Ibuki N, Matsumura S, Yabushita K, Okano N, Tsuji T. Cytokine profile in the liver of primary biliary cirrhosis. J. Clin. Immunol. 1999;19:422–427. doi: 10.1023/a:1020511002025. [DOI] [PubMed] [Google Scholar]
- 27.Pisetsky DS. The role of innate immunity in the induction of autoimmunity. Autoimmun. Rev. 2008;8:69–72. doi: 10.1016/j.autrev.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 28.Bergman M, Del Prete G, van Kooyk Y, Appelmelk B. Helicobacter pylori phase variation, immune modulation and gastric autoimmunity. Nat. Rev. Microbiol. 2006;4:151–159. doi: 10.1038/nrmicro1344. [DOI] [PubMed] [Google Scholar]
- 29.Benoist C, Mathis D. Autoimmunity provoked by infection: how good is the case for T cell epitope mimicry? Nat. Immunol. 2001;2:797–801. doi: 10.1038/ni0901-797. [DOI] [PubMed] [Google Scholar]
- 30.Marshak-Rothstein A, Rifkin IR. Immunologically active autoantigens: the role of toll-like receptors in the development of chronic inflammatory disease. Annu. Rev. Immunol. 2007;25:419–441. doi: 10.1146/annurev.immunol.22.012703.104514. [DOI] [PubMed] [Google Scholar]
- 31.Lunardi C, Tinazzi E, Bason C, Dolcino M, Corrocher R, Puccetti A. Human parvovirus B19 infection and autoimmunity. Autoimmun. Rev. 2008;8:116–120. doi: 10.1016/j.autrev.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Holmoy T, Hestvik AL. Multiple sclerosis: immunopathogenesis and controversies in defining the cause. Curr. Opin. Infect. Dis. 2008;21:271–278. doi: 10.1097/QCO.0b013e3282f88b48. [DOI] [PubMed] [Google Scholar]
- 33.Niller HH, Wolf H, Minarovits J. Regulation and dysregulation of Epstein-Barr virus latency: implications for the development of autoimmune diseases. Autoimmunity. 2008;41:298–328. doi: 10.1080/08916930802024772. [DOI] [PubMed] [Google Scholar]
- 34.Bach JF. Infections and autoimmune diseases. J. Autoimmun. 2005;25 Suppl:74–80. doi: 10.1016/j.jaut.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 35.Neisser A, Schwerer B, Bernheimer H, Moran AP. Ganglioside-induced antiganglioside antibodies from a neuropathy patient cross-react with lipopolysaccharides of Campylobacter jejuni associated with Guillain-Barre syndrome. J. Neuroimmunol. 2000;102:85–88. doi: 10.1016/s0165-5728(99)00159-9. [DOI] [PubMed] [Google Scholar]
- 36.Yuki N, Susuki K, Koga M, Nishimoto Y, Odaka M, Hirata K, Taguchi K, Miyatake T, Furukawa K, Kobata T, Yamada M. Carbohydrate mimicry between human ganglioside GM1 and Campylobacter jejuni lipooligosaccharide causes Guillain-Barre syndrome. Proc. Natl. Acad. Sci. U S A. 2004;101:11404–11409. doi: 10.1073/pnas.0402391101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selmi C, Balkwill DL, Invernizzi P, Ansari AA, Coppel RL, Podda M, Leung PS, Kenny TP, Van De Water J, Nantz MH, Kurth MJ, Gershwin ME. Patients with primary biliary cirrhosis react against a ubiquitous xenobiotic-metabolizing bacterium. Hepatology. 2003;38:1250–1257. doi: 10.1053/jhep.2003.50446. [DOI] [PubMed] [Google Scholar]
- 38.Kaplan MM. Novosphingobium aromaticivorans: a potential initiator of primary biliary cirrhosis. Am. J. Gastroenterol. 2004;99:2147–2149. doi: 10.1111/j.1572-0241.2004.41121.x. [DOI] [PubMed] [Google Scholar]
- 39.Padgett KA, Selmi C, Kenny TP, Leung PS, Balkwill DL, Ansari AA, Coppel RL, Gershwin ME. Phylogenetic and immunological definition of four lipoylated proteins from Novosphingobium aromaticivorans, implications for primary biliary cirrhosis. J. Autoimmun. 2005;24:209–219. doi: 10.1016/j.jaut.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 40.Bogdanos DP, Vergani D. Bacteria and Primary Biliary Cirrhosis. Clin. Rev. Allergy. Immunol. 2008;36:30–39. doi: 10.1007/s12016-008-8087-9. [DOI] [PubMed] [Google Scholar]
- 41.Olafsson S, Gudjonsson H, Selmi C, Amano K, Invernizzi P, Podda M, Gershwin ME. Antimitochondrial antibodies and reactivity to N. aromaticivorans proteins in Icelandic patients with primary biliary cirrhosis and their relatives. Am .J. Gastroenterol. 2004;99:2143–2146. doi: 10.1111/j.1572-0241.2004.40397.x. [DOI] [PubMed] [Google Scholar]
- 42.Mattner J, Savage PB, Leung P, Oertelt SS, Wang V, Trivedi O, Scanlon ST, Pendem K, Teyton L, Hart J, Ridgway WM, Wicker LS, Gershwin ME, Bendelac A. Liver autoimmunity triggered by microbial activation of natural killer T cells. Cell Host Microbe. 2008;3:304–315. doi: 10.1016/j.chom.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maier LM, Smyth DJ, Vella A, Payne F, Cooper JD, Pask R, Lowe C, Hulme J, Smink LJ, Fraser H, Moule C, Hunter KM, Chamberlain G, Walker N, Nutland S, Undlien DE, Ronningen KS, Guja C, Ionescu-Tirgoviste C, Savage DA, Strachan DP, Peterson LB, Todd JA, Wicker LS, Twells RC. Construction and analysis of tag single nucleotide polymorphism maps for six human-mouse orthologous candidate genes in type 1 diabetes. BMC. Genet. 2005;6:9. doi: 10.1186/1471-2156-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Penha-Goncalves C, Moule C, Smink LJ, Howson J, Gregory S, Rogers J, Lyons PA, Suttie JJ, Lord CJ, Peterson LB, Todd JA, Wicker LS. Identification of a structurally distinct CD101 molecule encoded in the 950-kb Idd10 region of NOD mice. Diabetes. 2003;52:1551–1556. doi: 10.2337/diabetes.52.6.1551. [DOI] [PubMed] [Google Scholar]
- 45.Podolin PL, Denny P, Lord CJ, Hill NJ, Todd JA, Peterson LB, Wicker LS, Lyons PA. Congenic mapping of the insulin-dependent diabetes (Idd) gene, Idd10, localizes two genes mediating the Idd10 effect and eliminates the candidate Fcgr1. J. Immunol. 1997;159:1835–1843. [PubMed] [Google Scholar]
- 46.Yamaji K, Ikegami H, Fujisawa T, Noso S, Nojima K, Babaya N, Itoi-Babaya M, Makino S, Sakamoto T, Ogihara T. Evidence for Cd101 but not Fcgr1 as candidate for type 1 diabetes locus, Idd10. Biochem. Biophys. Res. Commun. 2005;331:536–542. doi: 10.1016/j.bbrc.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 47.Rainbow DB, Moule C, Fraser HI, Clark J, Howlett SK, Burren O, Christensen M, Moody V, Steward CA, Mohammed JP, Fusakio ME, Masteller EL, Finger EB, Houchins JP, Naf D, Koentgen F, Ridgway WM, Todd JA, Bluestone JA, Peterson LB, Mattner J, Wicker LS. Evidence that Cd101 is a susceptibility gene in non obese diabetic mice. J. Immunol. 2011;187:325–336. doi: 10.4049/jimmunol.1003523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamilton-Williams EE, Martinez X, Clark J, Howlett S, Hunter KM, Rainbow DB, Wen L, Shlomchik MJ, Katz JD, Beilhack GF, Wicker LS, Sherman LA. Expression of diabetes-associated genes by dendritic cells and CD4 T cells drives the loss of tolerance in nonobese diabetic mice. J. Immunol. 2009;183:1533–1541. doi: 10.4049/jimmunol.0900428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fraser HI, Dendrou CA, Healy B, Rainbow DB, Howlett S, Smink LJ, Gregory S, Steward CA, Todd JA, Peterson LB, Wicker LS. Nonobese diabetic congenic strain analysis of autoimmune diabetes reveals genetic complexity of the Idd18 locus and identifies Vav3 as a candidate gene. J. Immunol. 2010;184:5075–5084. doi: 10.4049/jimmunol.0903734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Podolin PL, Denny P, Armitage N, Lord CJ, Hill NJ, Levy ER, Peterson LB, Todd JA, Wicker LS, Lyons PA. Localization of two insulin-dependent diabetes (Idd) genes to the Idd10 region on mouse chromosome 3. Mamm. Genome. 1998;9:283–286. doi: 10.1007/s003359900749. [DOI] [PubMed] [Google Scholar]
- 51.Mattner J, Debord KL, Ismail N, Goff RD, Cantu C, 3rd, Zhou D, Saint-Mezard P, Wang V, Gao Y, Yin N, Hoebe K, Schneewind O, Walker D, Beutler B, Teyton L, Savage PB, Bendelac A. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 52.Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J. Exp. Med. 2000;191:1895–1903. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maier LM, Wicker LS. Genetic susceptibility to type 1 diabetes. Curr. Opin. Immunol. 2005;17:601–608. doi: 10.1016/j.coi.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 54.Fernandez I, Zeiser R, Karsunky H, Kambham N, Beilhack A, Soderstrom K, Negrin RS, Engleman E. CD101 surface expression discriminates potency among murine FoxP3+ regulatory T cells. J. Immunol. 2007;179:2808–2814. doi: 10.4049/jimmunol.179.5.2808. [DOI] [PubMed] [Google Scholar]
- 55.Bonacchi A, Petrai I, Defranco RM, Lazzeri E, Annunziato F, Efsen E, Cosmi L, Romagnani P, Milani S, Failli P, Batignani G, Liotta F, Laffi G, Pinzani M, Gentilini P, Marra F. The chemokine CCL21 modulates lymphocyte recruitment and fibrosis in chronic hepatitis C. Gastroenterology. 2003;125:1060–1076. doi: 10.1016/s0016-5085(03)01194-6. [DOI] [PubMed] [Google Scholar]
- 56.Borchers AT, Shimoda S, Bowlus C, Keen CL, Gershwin ME. Lymphocyte recruitment and homing to the liver in primary biliary cirrhosis and primary sclerosing cholangitis. Semin Immunopathol. 2009;31:309–322. doi: 10.1007/s00281-009-0167-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santodomingo-Garzon T, Han J, Le T, Yang Y, Swain MG. Natural killer T cells regulate the homing of chemokine CXC receptor 3-positive regulatory T cells to the liver in mice. Hepatology. 2009;49:1267–1276. doi: 10.1002/hep.22761. [DOI] [PubMed] [Google Scholar]
- 58.Ruegg CL, Rivas A, Madani ND, Zeitung J, Laus R, Engleman EG. V7, a novel leukocyte surface protein that participates in T cell activation. II. Molecular cloning and characterization of the V7 gene. J. Immunol. 1995;154:4434–4443. [PubMed] [Google Scholar]
- 59.Bouloc A, Bagot M, Delaire S, Bensussan A, Boumsell L. Triggering CD101 molecule on human cutaneous dendritic cells inhibits T cell proliferation via IL-10 production. Eur. J.Immunol. 2000;30:3132–3139. doi: 10.1002/1521-4141(200011)30:11<3132::AID-IMMU3132>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 60.Bouloc A, Boulland ML, Geissmann F, Fraitag S, Andry P, Teillac D, Bensussan A, Revuz J, Boumsell L, Wechsler J, Bagot M. CD101 expression by Langerhans cell histiocytosis cells. Histopathology. 2000;36:229–232. doi: 10.1046/j.1365-2559.2000.00827.x. [DOI] [PubMed] [Google Scholar]
- 61.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu. Rev. Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 62.Brigl M, Brenner MB. CD1: antigen presentation and T cell function. Annu. Rev. Immunol. 2004;22:817–890. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- 63.Kronenberg M. Toward an understanding of NKT cell biology: progress and paradoxes. Annu. Rev. Immunol. 2005;23:877–900. doi: 10.1146/annurev.immunol.23.021704.115742. [DOI] [PubMed] [Google Scholar]
- 64.Van Kaer L. NKT cells: T lymphocytes with innate effector functions. Curr. Opin. Immunol. 2007;19:354–364. doi: 10.1016/j.coi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 65.Soares LR, Tsavaler L, Rivas A, Engleman EG. V7 (CD101) ligation inhibits TCR/CD3-induced IL-2 production by blocking Ca2+ flux and nuclear factor of activated T cell nuclear translocation. J. Immunol. 1998;161:209–217. [PubMed] [Google Scholar]
- 66.Shao L, Jacobs AR, Johnson VV, Mayer L. Activation of CD8+ regulatory T cells by human placental trophoblasts. J. Immunol. 2005;174:7539–7547. doi: 10.4049/jimmunol.174.12.7539. [DOI] [PubMed] [Google Scholar]
- 67.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J. Clin. Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hirschhorn JN. Genomewide association studies--illuminating biologic pathways. N. Engl. J. Med. 2009;360:1699–1701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- 69.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 70.Hirschfield GM, Liu X, Xu C, Lu Y, Xie G, Lu Y, Gu X, Walker EJ, Jing K, Juran BD, Mason AL, Myers RP, Peltekian KM, Ghent CN, Coltescu C, Atkinson EJ, Heathcote EJ, Lazaridis KN, Amos CI, Siminovitch KA. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. N. Engl. J. Med. 2009;360:2544–2555. doi: 10.1056/NEJMoa0810440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu X, Invernizzi P, Lu Y, Kosoy R, Bianchi I, Podda M, Xu C, Xie G, Macciardi F, Selmi C, Lupoli S, Shigeta R, Ransom M, Lleo A, Lee AT, Mason AL, Myers RP, Peltekian KM, Ghent CN, Bernuzzi F, Zuin M, Rosina F, Borghesio E, Floreani A, Lazzari R, Niro G, Andriulli A, Muratori L, Muratori P, Almasio PL, Andreone P, Margotti M, Brunetto M, Coco B, Alvaro D, Bragazzi MC, Marra F, Pisano A, Rigamonti C, Colombo M, Marzioni M, Benedetti A, Fabris L, Strazzabosco M, Portincasa P, Palmieri VO, Tiribelli C, Croce L, Bruno S, Rossi S, Vinci M, Prisco C, Mattalia A, Toniutto P, Picciotto A, Galli A, Ferrari C, Colombo S, Casella G, Morini L, Caporaso N, Colli A, Spinzi G, Montanari R, Gregersen PK, Heathcote EJ, Hirschfield GM, Siminovitch KA, Amos CI, Gershwin ME, Seldin MF. Genome-wide meta-analyses identify three loci associated with primary biliary cirrhosis. Nat. Genet. 2010;42:658–660. doi: 10.1038/ng.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hirschfield GM, Liu X, Han Y, Gorlov IP, Lu Y, Xu C, Chen W, Juran BD, Coltescu C, Mason AL, Milkiewicz P, Myers RP, Odin JA, Luketic VA, Speiciene D, Vincent C, Levy C, Gregersen PK, Zhang J, Heathcote EJ, Lazaridis KN, Amos CI, Siminovitch KA. Variants at IRF5-TNPO3, 17q12-21 and MMEL1 are associated with primary biliary cirrhosis. Nat. Genet. 2010;42:655–657. doi: 10.1038/ng.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rivas A, Ruegg CL, Zeitung J, Laus R, Warnke R, Benike C, Engleman EG. V7, a novel leukocyte surface protein that participates in T cell activation. I. Tissue distribution and functional studies. J. Immunol. 1995;154:4423–4433. [PubMed] [Google Scholar]
- 74.Soares LR, Rivas A, Tsavaler L, Engleman EG. Ligation of the V7 molecule on T cells blocks anergy induction through a CD28-independent mechanism. J. Immunol. 1997;159:1115–1124. [PubMed] [Google Scholar]
- 75.Grant AJ, Goddard S, Ahmed-Choudhury J, Reynolds G, Jackson DG, Briskin M, Wu L, Hubscher SG, Adams DH. Hepatic expression of secondary lymphoid chemokine (CCL21) promotes the development of portal-associated lymphoid tissue in chronic inflammatory liver disease. Am J Pathol. 2002;160:1445–1455. doi: 10.1016/S0002-9440(10)62570-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yoneyama H, Ichida T. Recruitment of dendritic cells to pathological niches in inflamed liver. Med. Mol. Morphol. 2005;38:136–141. doi: 10.1007/s00795-005-0289-0. [DOI] [PubMed] [Google Scholar]
- 77.Sica GL, Choi IH, Zhu G, Tamada K, Wang SD, Tamura H, Chapoval AI, Flies DB, Bajorath J, Chen L. B7-H4, a molecule of the B7 family, negatively regulates T cell immunity. Immunity. 2003;18:849–861. doi: 10.1016/s1074-7613(03)00152-3. [DOI] [PubMed] [Google Scholar]
- 78.Suh WK, Wang S, Duncan GS, Miyazaki Y, Cates E, Walker T, Gajewska BU, Deenick E, Dawicki W, Okada H, Wakeham A, Itie A, Watts TH, Ohashi PS, Jordana M, Yoshida H, Mak TW. Generation and characterization of B7-H4/B7S1/B7x-deficient mice. Mol. Cell. Biol. 2006;26:6403–6411. doi: 10.1128/MCB.00755-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu G, Augustine MM, Azuma T, Luo L, Yao S, Anand S, Rietz AC, Huang J, Xu H, Flies AS, Flies SJ, Tamada K, Colonna M, van Deursen JM, Chen L. B7-H4-deficient mice display augmented neutrophil-mediated innate immunity. Blood. 2009;113:1759–1767. doi: 10.1182/blood-2008-01-133223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen Y, Yang C, Xie Z, Zou L, Ruan Z, Zhang X, Tang Y, Fei L, Jia Z, Wu Y. Expression of the novel co-stimulatory molecule B7-H4 by renal tubular epithelial cells. Kidney Int. 2006;70:2092–2099. doi: 10.1038/sj.ki.5001867. [DOI] [PubMed] [Google Scholar]
- 81.You S, Leforban B, Garcia C, Bach JF, Bluestone JA, Chatenoud L. Adaptive TGF-beta-dependent regulatory T cells control autoimmune diabetes and are a privileged target of anti-CD3 antibody treatment. Proc. Natl. Acad. Sci. USA. 2007;104:6335–6340. doi: 10.1073/pnas.0701171104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.You S, Belghith M, Cobbold S, Alyanakian MA, Gouarin C, Barriot S, Garcia C, Waldmann H, Bach JF, Chatenoud L. Autoimmune diabetes onset results from qualitative rather than quantitative age-dependent changes in pathogenic T-cells. Diabetes. 2005;54:1415–1422. doi: 10.2337/diabetes.54.5.1415. [DOI] [PubMed] [Google Scholar]
- 83.Herbelin A, Gombert JM, Lepault F, Bach JF, Chatenoud L. Mature mainstream TCR alpha beta+CD4+ thymocytes expressing L-selectin mediate “active tolerance” in the nonobese diabetic mouse. J. Immunol. 1998;161:2620–2628. [PubMed] [Google Scholar]
- 84.Billiard F, Litvinova E, Saadoun D, Djelti F, Klatzmann D, Cohen JL, Marodon G, Salomon B. Regulatory and effector T cell activation levels are prime determinants of in vivo immune regulation. J. Immunol. 2006;177:2167–2174. doi: 10.4049/jimmunol.177.4.2167. [DOI] [PubMed] [Google Scholar]
- 85.Salomon B, Lenschow DJ, Rhee L, Ashourian N, Singh B, Sharpe A, Bluestone JA. B7/CD28 Costimulation is essential for the homeostasis of the CD4+CD25+ immunoregulatory T cells that control autoimmune diabetes. Immunity. 2000;12:431–440. doi: 10.1016/s1074-7613(00)80195-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.