Abstract
How distal transcriptional enhancer sequences interact with proximal promoters is poorly understood within the context of chromatin. In this report, we have used the immunoglobulin heavy chain locus to address the role of the PTIP protein in transcription regulation and class switch recombination in B cells, a process that depends on regulated transcription and DNA recombination via Pax5 and distal 3′ enhancer sequences. We first show that PTIP is recruited to a Pax5 binding site to promote histone H3 lysine 4 (H3K4) methylation. Using a CD19-Cre driver strain, we deleted PTIP in mature B cells. Loss of PTIP inhibited class switch recombination by suppressing transcription and histone H3K4 methylation at the germ line transcript promoters. In the absence of PTIP, Pax5 binding to the promoter regions is reduced and long-range chromatin interactions between the distal enhancer at the 3′ regulatory region and the germ line transcript promoters are not detected. We propose a model whereby PTIP stabilizes the Pax5 DNA interactions that promote chromatin looping and regulate transcriptional responses needed for class switch recombination.
INTRODUCTION
Long-range interactions between distal enhancers and promoters are known to regulate many multigene loci through DNA looping mechanisms that bring enhancer sequences and their respective binding proteins close to a transcription start site (14, 18, 27). The immunoglobulin heavy chain locus (IgH) is an example of a large locus with multiple transcripts, splice variants, regulatory elements, and genomic rearrangements that can produce different proteins in response to a variety of signaling inputs. In B lymphocytes, class switch recombination (CSR) rearranges the immunoglobulin heavy chain (IgH) locus such that new constant regions replace the IgM isotype (35). In response to specific stimuli, CSR requires the initiation of germ line transcripts (GLTs) upstream of the new constant region, before double-strand break formation and recombination. B cell differentiation and IgH transcription depend on the 3′ enhancer sequences (12), which are known to interact with the switch regions through a chromatin looping mechanism (17).
The Pax5 gene, a member of the paired-box (Pax) gene family first identified as homologues of the Drosophila melanogaster paired and gooseberry segmentation genes, encodes a protein that is essential for B cell specification and immunoglobulin gene expression (8). Genetic loss-of-function experiments in mice provide strong evidence that Pax5 is necessary not only for the transition from pre-B cell to IgM-positive B cells (36) but also for the maintenance of B cell phenotypic stability (24) and for isotype switching and generation of Ig-secreting B cells (15). Within the IgH locus, Pax5 binds to the 3′ enhancer (23), although how Pax5 contributes to chromatin looping and GLT regulation is unclear. Pax5 is a member of the Pax2/5/8 subfamily, whose amino-terminal DNA binding domains are virtually identical and whose carboxy-terminal sequences share large regions of identity (2). In humans and mice, there are 9 Pax genes, all of which are expressed in embryonic development, where they are known to regulate cell lineage pathways and morphogenesis (1, 31). Pax genes are also associated with human diseases because of either haploinsufficiency or aberrant expression (5). Despite their critical roles in development and their association with clinical disease, how Pax proteins function to specify cell lineages is poorly understood.
A close relative of Pax5, the Pax2 protein is known to interact with a histone H3 lysine 4 (H3K4) methyltransferase complex through the ubiquitously expressed nuclear protein PTIP (28). The PTIP protein is encoded by the Paxip1 gene and contains 3 pairs of BRCT domains, at least one of which is a strong P-serine binding domain (22). The PTIP protein is part of a KMT2C/D (MLL3/4) complex that contains the mammalian homologues of Drosophila Trithorax group proteins and can methylate H3K4 (H3K4me) (7, 16, 28). Loss of PTIP in mice (6) and flies (13) is embryonic lethal and results in a global reduction of trimethylated H3K4 (H3K4me3), an epigenetic mark that correlates with actively expressed genes. Methylation at H3K4 depends on the mammalian homologues of the yeast (Saccharomyces cerevisiae) COMPASS complex, which contain the SET domain encoding methyltransferases (KMT2s) and cofactors such as Rbbp5, WDR5, and Ash2L (33). High levels of H3K4me3 correlate strongly with transcription start sites and are thought to recruit PHD domain-containing proteins important for assembly of the initiation complex (34) and/or nucleosome remodeling (41).
Given the high degree of sequence identity between Pax2 and Pax5 and that the genes are functionally interchangeable (2), we asked whether PTIP is also important for Pax5-mediated functions in developing and mature B cells. Using an integrated reporter system, Pax5 recruits PTIP and the KMT2D complex to DNA, resulting in high levels of H3K4me3. Furthermore, we used a conditional allele of Paxip1 and a CD19-Cre driver to delete PTIP in B cells and assessed the ability of B cells to undergo isotype switching and chromatin remodeling. The loss of PTIP significantly reduced isotype switching, the levels of H3K4me3 around the promoter of the GLTs, the initiation of transcription, and the ability of the IgH 3′ enhancer to interact with the GLT promoters. These data point to a critical role for PTIP and its interaction with Pax5 in B-cell-specific chromatin-remodeling events.
MATERIALS AND METHODS
Mice.
The Paxip1 conditional floxed allele contains exon 1 and the 5′ regulatory sequences flanked with loxP sites and has been previously described (19). The Cd19-Cre mice were used to delete PTIP in the B cell lineage (30). Experiments were performed with Paxip1fl/+ mice or Cd19-Cre/+; Paxip1fl/fl mice.
B cell cultures.
Mature B cells were isolated from the spleens of 6- to 16-week-old mice by using a B lymphocyte enrichment kit (BD Biosciences). Enriched B cells were cultured in RPMI medium supplemented with 10% (vol/vol) fetal bovine serum (FBS), 1% (vol/vol) antibiotic/antimycotic, 1% (vol/vol) glutamine, 1% (vol/vol) sodium pyruvate, and 50 μM β-mercaptoethanol. Mature B cells were stimulated to undergo class switch recombination with 25 μg/ml lipopolysaccharide (LPS) (Sigma) or 1 μg/ml anti-CD40 (BD Bioscience) with 25 ng/ml interleukin-4 (IL-4) (R&D Systems).
Flow cytometry.
Cells were washed with phosphate-buffered saline (PBS) supplemented with 10% (vol/vol) FBS and incubated on ice in the dark for 30 to 60 min with anti-B220 (1:200; eBioscience), anti-IgG1 (1:100; BD Pharmingen), and anti-IgG2b (1:100; Southern Biotech) antibodies. Cells were then washed with PBS, resuspended, and counted with a Beckman Coulter FC500 flow cytometer using Cytomic RXP software. Data were analyzed with FlowJo software.
Western blotting.
Whole-cell lysates were prepared by resuspending cell pellets in 2× SDS-PAGE loading dye and boiling for 5 min. Proteins were separated by 8 or 10% Tris-glycine SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. Antibodies for Western blotting were used at the following dilutions: rabbit anti-PTIP, 1:1,000 (28); goat anti-Pax5, 1:1,000 (C-20; Santa Cruz); antiactin, 1:5,000 (C-11; Sigma); and antitubulin, 1:5,000 (Sigma). Extracts were normalized to actin as a loading control.
Preparation of cDNA and qPCR.
Total RNA was isolated from unstimulated and stimulated cells by using Trizol (Invitrogen) according to the manufacturer's protocol. The cDNAs were prepared with oligo(dT) primers according to the SuperScript II kit (Invitrogen). Diluted cDNAs were amplified with the iTaq Sybr green master mix (Bio-Rad) in a Prism 7500 (Applied Biosystems). Hypoxanthine phosphoribosyltransferase (HPRT) was used an endogenous control to normalize targets. All primer pairs used for quantitative PCR (qPCR) for RNA, chromatin immunoprecipitation (ChIP), and chromosomal conformation capture (3C) analysis are listed in Table 1.
Table 1.
List of primer pairs used in this study
| Primer type and/or name | Sequence (5′→3′)a |
|
|---|---|---|
| Forward | Reverse | |
| PRS4 flank, HEK293 cells | GCTACCGGACTCAGATCTCG | TGCGAAGTGGACCTCGGACC |
| B cell ChIP | ||
| D (IgG1-GLT promoter 12) | TGCTCCAGAAGGTTCCCTAA | CTGCTGTGTGGGATCTGCTA |
| E (IgG1-GLT promoter 11) | TTTCCTGACCACCTCATTGG | ACTCTGTCACAGCCCTCAGC |
| F (IgG1-GLT promoter 10) | CATTCTGGGGGTTTCTGTGT | CAGGAGTACAGCCAGGCTTC |
| G (IgG1-GLT promoter 9) | GGCTGGTCTGTCAACTCCTT | CTGCTTTCACAGCTTCCACA |
| H [g1-GLT 5′ (JAD)] | TCTCTTTCCCTGCAGGCTTGA | CTGAGGAATGTGTTTGGCATGG |
| J (LR1-LPS response element) | CTTTCCCTACTCCCCTGGTC | GGGGATAGCAGGGCTAAGAA |
| K (3RR-HS4-A) | GAGGAGGTTGACCTGATGGA | CAGGAACCACAGAGCAGAGG |
| L (IgG2b-pk2-B) | AATGTTTTTCCCAGCACCAA | CAAGCGTGTGTCCTGTTTGT |
| B cell 3C assay | ||
| A′ (IgG1-GLT R + H3b-4 F) | CGACACTGGGCAGTTCATTTTG | ACCCAACCTGTGTCCCTAGAG |
| B′ (3C-Ig2b-2b-L + H3b-4-F) | TGGTAACAAACAGGACACACG | ACCCAACCTGTGTCCCTAGAG |
| C′ (IgG1-GLT R + Eu-F) | CGACACTGGGCAGTTCATTTTG | AGACTCTGGACCTCTCCGAAAC |
| D′ (H3b-4 F + Eu R) | ACCCAACCTGTGTCCCTAGAG | CCCTAAAGCAATGACTGAAGACTC |
| E′ (Gapdh 3C control) | CAGTAGACTCCACGACATAC | AGTAGTGCGTTCTGTAGATTCC |
| F′ (flanking Eu HindIII site) | AGACTCTGGACCTCTCCGAAAC | CCCTAAAGCAATGACTGAAGACTC |
| PCR products for EMSA | ||
| (IgG1-GLT promoter 9-G | GGCTGGTCTGTCAACTCCTT | CTGCTTTCACAGCTTCCACA |
| IgG1-GLT promoter 10-F | CATTCTGGGGGTTTCTGTGT | CAGGAGTACAGCCAGGCTTC |
| IgG1-GLT promoter 11-E | TTTCCTGACCACCTCATTGG | ACTCTGTCACAGCCCTCAGC |
| IgG1-GLT promoter 12-D | TGCTCCAGAAGGTTCCCTAA | CTGCTGTGTGGGATCTGCTA |
| IgG2b-pk1-A | TGTGGCTCTCTCCAGCTTCT | GGACTGTGAATCCTGGAAGG |
| IgG2b-pk1-B | TTTCAGTGTCTGGGGTAGGG | CCAGGAACACTGGGTTGAGA |
| IgG2b-pk2-A | AAACCCTAACCCTGCTGCTT | CCGTCCTACAGAGGTTCCAG |
| IgG2b-pk2-B | AATGTTTTTCCCAGCACCAA | CAAGCGTGTGTCCTGTTTGT |
The sequences (top strand) of the oligonucleotides for EMSA are as follows: H2A, CAGGGTTGTGACGCAGCGGTGGGTGACGACTGTCGG; CS1, AGGCATTTCCTGACAGGGTTCCCCTTCATTCTGGGGGTTTCTGTGTCAGGG; and CS2, GGGCCTTTGGGGGTCCCTGGGCTGGCTGAGGCTGAGTGATTATGCCCACTCT.
ChIP.
HEK293 cells with an integrated PRS4EGFP reporter were transfected with 1 μg cytomegalovirus (CMV)-Pax5 and harvested 48 h posttransfection. A fraction of cells were lysed by 2× SDS-PAGE to determine the expression of Pax5, tubulin, and green fluorescent protein (GFP). ChIP was performed as previously described (28). Resting unstimulated B cells or B cells stimulated for 2 or 3 days were harvested for chromatin preparation and immunoprecipitation. ChIP was performed as previously described, except for a few modifications (28). Resting or stimulated B cells were fixed in culture media by the addition of formaldehyde to 1% for 8 min at room temperature. Chromatin was sheared by five 15-s pulses with a microtip probe sonicator (Branson Sonifier 250). Ten micrograms of chromatin was immunoprecipitated with 2 to 6 μg of antibodies. Precipitated DNA was reconstituted in sterile water, and real-time PCR quantitation of DNA relative to inputs was performed with iTaq Sybr green master mix (Bio-Rad) in a Prism 7500 (Applied Biosystems) with the primers listed in Table 1.
Antibodies.
Rabbit IgG and goat IgG (Bethyl Laboratories) were used as control antibodies for ChIP. Anti-Pax5 (C-20) and anti-polymerase II (Pol II) RNA were obtained from Santa Cruz. Rabbit anti-PTIP has been previously described (28). Anti-Ash2l and anti-Rbbp5 were obtained from Bethyl Laboratories. Anti-KMT2D, anti-H3K4me1, and anti-H3K4me2 were purchased from Abcam. Anti-H3K4me3 was obtained from Diagenode.
3C analysis.
The use of the 3C technique for monitoring the association of the 3′ enhancer site hs4 with sites in the IgH locus has been described previously (17, 40). Briefly, unstimulated resting wild-type (WT) B cells and LPS/IL-4-stimulated WT and PTIP− B cells were fixed with 2% formaldehyde in culture medium for 8 min, quenched with glycine, and lysed with 1× cell membrane lysis buffer. A total of 1 × 107 unstimulated nuclei or 0.5 × 107 stimulated nuclei were solubilized and then digested with HindIII overnight. The cross-linked DNA was ligated under dilute conditions overnight, followed by cross-link reversal and DNA extraction. Two hundred nanograms of DNA was used for each PCR under semiquantitative conditions, ensuring amplification was in the linear phase. Experiments were performed with at least three different biological replicates. The primers used in this study are listed in Table 1. Primers previously published were used to measure the association of the hs4 of the 3′ RR with either the Eμ region or upstream of the γ1 GLT (17, 40). Controls include a primer set measuring the ligation efficiency of a 3.5-kb fragment containing Gapdh and a primer set measuring the uncut DNA flanking the HindIII site of the Eμ. Semiquantitative PCR was performed for 32 and 35 cycles to ensure the reaction was in the linear phase.
For quantitation, primer pairs were labeled with [γ-32P]ATP and polynucleotide kinase and added directly to the PCR at a ratio of 1:10 labeled to unlabeled primers. PCRs were run on 6% polyacrylamide gels and dried, and specific bands were quantitated with a phosphorimager.
EMSAs.
For electrophoretic mobility shift assays (EMSAs), the paired domain from Pax2 (amino acids 1 to 170), containing a homologous paired domain to Pax5, was expressed in Escherichia coli and purified as previously described (3). The oligonucleotides and PCR fragments used are identified in Table 1. Oligonucleotides were end labeled with [γ-32P]ATP and polynucleotide kinase. Unlabeled fragments or double-stranded oligonucleotides were used as cold competitors. Binding reactions were performed in Z buffer (25 mM HEPES, pH 7.8, 20% glycerol, 12.5 mM MgCl2, 0.1 M KCl, 1 mM dithiothreitol [DTT]). Free DNA and DNA-protein complexes were resolved at room temperature on 6% neutral polyacrylamide gels in 0.5× Tris-borate-EDTA (TBE) at 150 V.
RESULTS
Pax5 recruits PTIP and an H3K4 methyltransferase complex to chromatin.
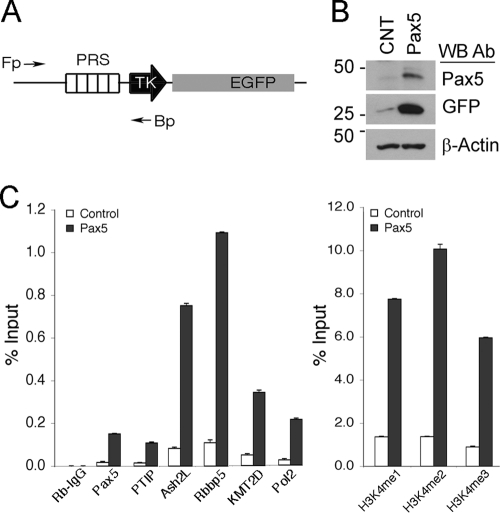
In order to assess the effects of Pax5 on a DNA target site, we utilized an HEK293 derivative cell line (PRS89), which contained an integrated reporter gene (Fig. 1 A). The reporter consists of 5 copies of a strong Pax2/5/8 response sequence (PRS) upstream of the minimal thymidine kinase (TK) promoter driving enhanced green fluorescent protein (EGFP). Transient transfection of Pax5 into PRS89 cells resulted in upregulation of EGFP (Fig. 1B). More importantly, alterations in chromatin at the PRS were consistent with the recruitment of a histone H3K4 methyltransferase complex (Fig. 1C). Chromatin immunoprecipitation (ChIP) experiments revealed Pax5 binding to the PRS. Endogenous PTIP protein was also bound to the PRS in a Pax5-dependent manner. Similarly, components of the KMT2D complex, including Rbbp5, Ash2L, and KMT2D, also localized to the PRS in a Pax5-dependent manner. Antibodies against H3K4 mono-, di-, and trimethyl modifications showed significant 4- to 6-fold increases in H3K4 methylation in response to Pax5 expression. These data demonstrate Pax5-dependent PTIP recruitment and H3K4 methylation at a strong Pax5 DNA binding sequence.
Fig. 1.
Pax5 recruits the PTIP histone methylation complex to a DNA target sequence. (A) Schematic of the Pax-dependent reporter gene integrated into HEK293 cells. PRS, Pax-responsive sequence; TK, minimal thymidine kinase promoter; EGFP, enhanced green fluorescent protein. The forward (Fp) and backward (Bp) primers for ChIP are indicated. (B) Western blots (WB) of control and Pax5-transfected cells performed with the antibodies (Ab) indicated. Note that Pax5 expression activates EGFP relative to the control. (C) ChIP assays for PRS binding sites performed with antibodies against Pax5, PTIP, Ash2l, Rbbp5, KMT2D, and RNA Pol II (left panel). Note that Pax5 recruits PTIP, Ash2l, Rbbp5, KMT2D, and Pol II to the PRS binding site. ChIP with antibodies against H3K4 methylation states (right panel) shows significant increases at the PRS in Pax5-expressing cells.
PTIP deletion in B cells affects gene expression and CSR.
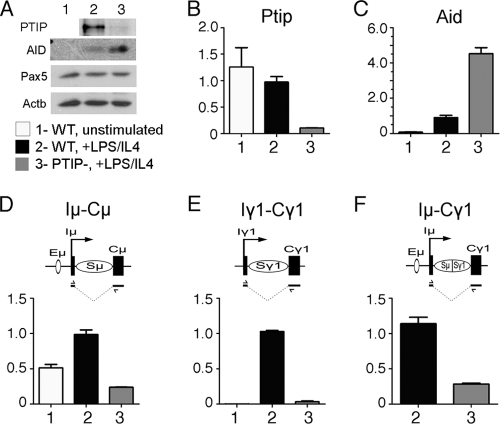
In order to assess the necessity of PTIP for Pax5-dependent functions in mature B cells, we generated a B-cell-specific deletion of PTIP by crossing the floxed allele (Paxip1fl/fl) to a well-characterized CD19-Cre driver strain (30). Splenic B cells from mice carrying the Paxip1fl/fl or Paxip1fl/− alleles with and without the CD19-Cre were purified and stimulated with LPS and IL-4 to assess proliferation and CSR. For simplicity, we designated the B cells either WT (Paxip1fl/fl or Paxip1fl/−) or PTIP− (Paxip1fl/fl; CD19-Cre or Paxip1fl/−; CD19-Cre). The PTIP− B cells did not contain detectable amounts of PTIP protein, although Pax5 protein was unaffected (Fig. 2 A). Upon stimulation with LPS/IL-4, B cells undergo CSR to IgG1 and IgG2b. CSR requires the activation of activation-induced deaminase (AID) and the initiation of GLTs near the switch region, upstream of the new constant region. In PTIP− B cells, AID was activated to even higher levels than in WT cells. However, the GLTs corresponding to the new constant regions were significantly reduced. These data suggest a defect in the transcriptional response of the GLTs after stimulation with LPS/IL-4.
Fig. 2.
Expression analysis in wild-type and PTIP mutant B cells. (A) Western blot analysis for AID, Pax5, and Actb proteins in whole-cell lysates from unstimulated splenic B cells and from WT and PTIP− B cells 72 h after stimulation with LPS/IL-4. (B to F) Quantitative reverse transcription (RT)-PCR of cDNAs from unstimulated WT B cells, WT B cells stimulated with LPS/IL-4, and PTIP− B cells stimulated with LPS/IL-4 is shown as relative expression units. The transcripts are AID mRNA, Iμ-Cμ (μ GLT), Iγ1-Cγ1 (γ1 GLT), and Iμ-Cγ1 (postrecombination transcript for γ1).
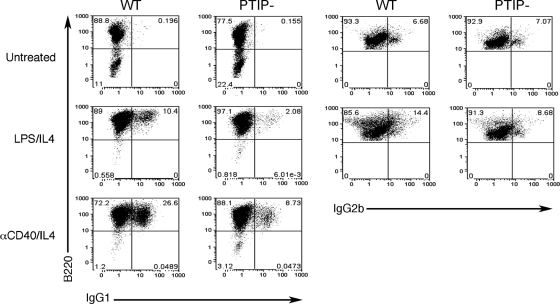
Since activation of the GLTs is a prerequisite for CSR, we assessed the ability of PTIP− B cells to undergo CSR by using either LPS/IL-4 or anti-CD40/IL-4 (Fig. 3). With either stimulus, CSR was significantly inhibited, but not completely abolished, in the PTIP− B cells. After LPS/IL-4 stimulation, the percentage of WT double-positive B220/IgG1 cells was 10.4%, compared to 2% in PTIP− B cells. The percentages of B220/IgG2b double positives were 14.4 and 8.7% in WT and PTIP− B cells, respectively, although untreated cells had a high background (7%) for the IgG2b marker. After anti-CD40/IL-4 treatment, the percentages of B220/IgG1 double positives were 26.6 and 8.7% in the WT and PTIP− B cell populations, respectively. Thus, loss of PTIP function significantly suppresses the ability of B cells to undergo CSR in response to LPS/IL-4 or anti-CD40/IL-4.
Fig. 3.
Flow cytometry of untreated or stimulated B cells. WT and PTIP− resting B cells were cultured for 4 days either untreated or stimulated with LPS/IL-4 or anti-CD40/IL-4 (αCD40/IL4) as indicated. Cells were stained for the B cell marker B220 and for surface expression of IgG1 (left) or IgG2b (right). WT cells efficiently undergo CSR when stimulated with LPS/IL-4 or anti-CD40/IL-4 compared to cells cultured without B cell receptor cross-linking agents. Note that the loss of PTIP results in a significant decrease in CSR to IgG1 compared to that in the WT when stimulated with LPS/IL-4 (from 10.4% to 2.08%) or anti-CD40/IL-4 (from 26.6% to 8.73%). CSR to IgG2b was also decreased in the PTIP− B cells compared to the WT (from 14.4% to 8.68%).
PTIP mediates histone methylation within the IgH locus.
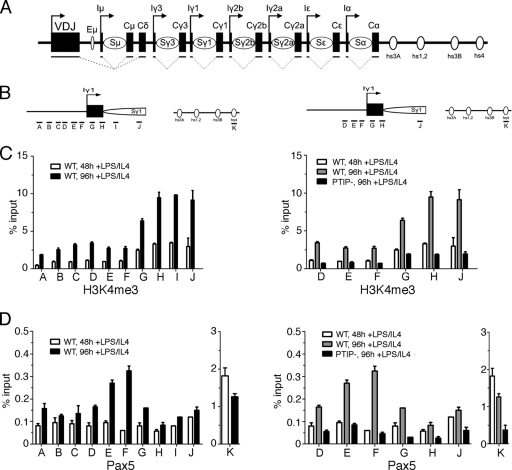
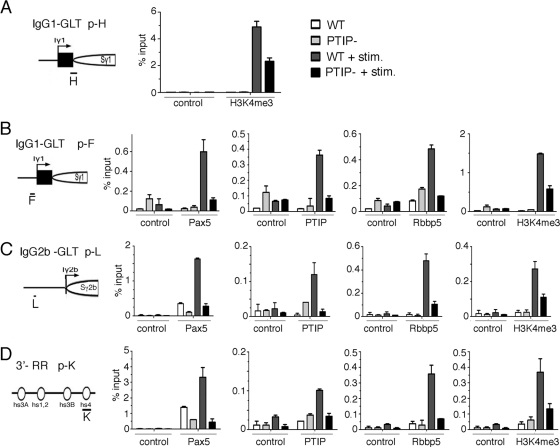
Given the association of PTIP with a KMT2C/D histone methylation complex, we examined the patterns of H3K4me3 by chromatin immunoprecipitation (ChIP) near the γ1 promoter for the GLT, upstream of the IgG1 constant region after stimulation with LPS/IL-4 (Fig. 4). Primer pairs spanning the region around the transcription start site showed the highest levels of H3K4me3 near the start site and immediately downstream (Fig. 4C, primer pairs G to J). Levels of H3K4me3 increased from 48 to 96 h after stimulation with LPS/IL-4. However, PTIP− B cells did not show an increase in H3K4me3 at the same sites, even 96 h after stimulation (Fig. 4B). We also examined the localization of Pax5 by ChIP in WT and PTIP− B cells. As described previously, Pax5 bound strongly to the hs4 site of the 3′ enhancer region of the IgH locus (Fig. 4D, primer pair K). Pax5 was also found close to the γ1 GLT promoter after stimulation (Fig. 4D, primer pairs E and F). However, in PTIP− B cells, Pax5 binding to the γ1 promoter was reduced to background levels. These data suggest that PTIP cooperates with Pax5 to impart H3K4me3 marks near the promoters of the GLTs to regulate CSR. We next examined PTIP, Pax5, Rbbp5, and H3K4me3 levels around 2 different GLT promoter regions and the 3′ enhancer before and after stimulation by LPS/IL-4 (Fig. 5). At both γ1 and the γ2b GLT promoters, stimulation of WT B cells resulted in increased Pax5, PTIP, and Rbbp5 localization and increased H3K4me3 near the GLT promoters (Fig. 5A to C). However, PTIP− B cells showed significantly reduced levels of PTIP, Rbbp5, and H3K4me3 at the same regions. Surprisingly, in the PTIP− cells the γ1 and γ2b promoter regions also showed a reduction of Pax5 localization, as did the 3′ enhancer (Fig. 5D). These data are inconsistent with a simple model whereby Pax5 recruits PTIP and suggest a more complex scenario in which PTIP may stabilize Pax5 interactions with the switch region promoters.
Fig. 4.
H3K4 methylation near the switch region of the Ig locus. (A) Diagram of IgH locus (not to scale) in resting splenic B cells. Each heavy chain isotype (except D) possesses an intervening exon, I; a switch region containing repeats, S; and a constant region, C. The μ constant region has an upstream enhancer (Eμ). The spliced GLTs are below the I and C regions of each isotype. Arrows indicate the direction of VDJ and GLT transcription. The open circles represent the 4 hypersensitive sites present at the 3′ regulatory region. (B) Location of the primer pairs used for ChIP assays around the Iγ1 transcription start site. (C) ChIP assay for H3K4me3 with untreated B cells, WT B cells stimulated by LPS/IL-4 for 48 or 96 h, or PTIP− B cells stimulated for 96 h. Primer pairs surrounding the Iγ1 transcription start site show H3K4me3 enrichment at the transcriptional start site was increased in WT cells treated for 96 h compared to cells treated for 48 h. The highest peaks of enrichment were found in primer sets G to H, which cover the Iγ1 and Sγ1 promoters. The PTIP− B cells showed decreased H3K4me3 even after 96 h of stimulation with levels at or below those of the WT cells after 48 h of stimulation. Untreated cells had levels below 0.2% of input. (D) ChIP assays of Pax5 as in panel C with additional primer pair K, which shows Pax5 binding to hs4 of the 3′ regulatory region. Pax5 enrichment at Iγ1 is maximal with primer set F and correlates with LPS/IL-4 stimulation and CSR. Loss of PTIP in resting B cells results in a decrease of H3K4me3 of the Iγ1 and Sγ1 and decreased Pax5 binding.
Fig. 5.
PTIP and Pax5 colocalize to the switch regions. ChIP assay demonstrating LPS/IL-4-dependent H3K4me3 and Pax5-PTIP complex association at Iγ1, Iγ2b, and hs4 in PTIP− and WT B cells, either untreated or stimulated for 72 h. (A) H3K4me3 enrichment is decreased by more than 50% in PTIP− compared to WT controls at the Iγ1/5′ Sγ1 region (primer pair H). Note that H3K4me3 is LPS/IL-4 dependent. (B) Pax5, PTIP, Rbbp5, and H3K4me3 are enriched after stimulation using primer pair F, which is upstream of the Iγ1 region. Loss of PTIP results in a decrease of H3K4me3 enrichment correlating with the decrease of Pax5 and Rbbp5 at primer F. (C) Changes in enrichment of Pax5, PTIP, Rbbp5, and H3K4me3 at primer pair L upstream of Iγ2b in PTIP− B cells. Pax5, PTIP, Rbbp5, and H3K4me3 association at L is stimulation dependent. PTIP− cells show loss of enrichment of Pax5, Rbbp5, and H3K4me3 upstream of Iγ2b, as seen with Iγ1. (D) ChIP assays for Pax5 binding site hs4 at the 3′ regulatory region using primer pair K. Pax5 enrichment at K is increased upon stimulation, but is significantly decreased in PTIP− cells. LPS/IL-4 stimulation of cultured WT cells results in PTIP and Rbbp5 being recruited to the hs4 site, but this complex is decreased in PTIP− cells along with H3K4me3.
PTIP mediates long-range chromatin interactions.
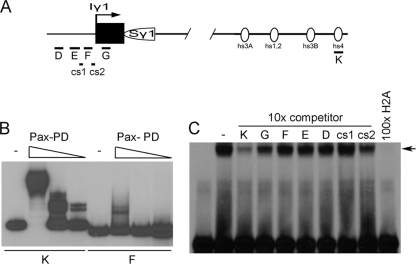
To more directly examine the relationship between Pax5 and PTIP and its impact on the GLT promoter regions, we tested for Pax5 binding to the sequences where Pax5 was detected by ChIP analyses (Fig. 6 A to C). Using recombinant paired domain proteins and fragments corresponding to the γ1 promoter and the 3′ enhancer, strong in vitro Pax5 binding was observed to the hs4 3′ enhancer fragment, as described previously (Fig. 6B). However, there was at least 50-fold less Pax5 binding to the γ1 fragment F, which showed Pax5 localization by ChIP. We also examined specificity by binding the paired domain to an optimal Pax5 site and used DNA sequences from the switch region GLT promoters or the hs4 site from the 3′ enhancer as competitors (Fig. 6C). Only the 3′ enhancer sequence at hs4 was able to significantly compete for Pax5 binding. Thus, very poor or no binding of Pax5 was observed to the DNA sequences in and around the GLT promoters. Yet in WT B cells, our ChIP assays show that Pax5 localizes to the GLT promoter regions upon stimulation.
Fig. 6.
Pax5 in vitro binding assays. (A) Probes used for binding assays are shown schematically and correspond essentially to the PCR primer pairs used for the ChIP assays. In addition, two evolutionarily conserved sequences, cs1 and cs2, were tested. (B) Electrophoretic mobility shift assay using probes K and F and decreasing amounts of the recombinant Pax2/5/8 paired domain (Pax-PD). Note strong binding to the 3′ enhancer sequence hs4, with poor binding to the Iγ1 transcription start region. (C) Electrophoretic mobility shift assay using an optimal Pax5 binding sequence H2A, recombinant Pax-PD, and competitor DNAs. At a 10-fold molar excess, only the 3′ enhancer sequence K is able to effectively compete for binding to the Pax-PD.
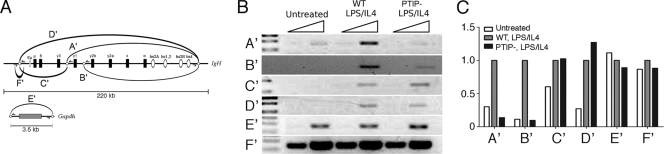
The localization of Pax5 to the GLT start sites could be the result of long-range chromatin interactions involving Pax5 from the 3′ enhancer and the GLT promoters. Such chromatin looping has been described for the IgH locus (17). Thus, we examined whether PTIP deletion affects the ability of the 3′ enhancer hs4 site to interact with the GLT promoters using the 3C chromatin capture assay (Fig. 7). Chromatin from WT and PTIP− B cells was cross-linked, digested with HindIII, religated under dilute conditions, and subject to PCR analysis with primers specifically designed to detect interactions between distant elements (Fig. 7A). Upon stimulation with LPS/IL-4, the interactions between the 3′ enhancer and both the γ1 and γ2b promoter regions are readily detected in WT B cells but are significantly reduced in PTIP− B cells (Fig. 7B and C). Interactions involving the Eμ enhancer are unaffected in the PTIP− cells, consistent with the levels of transcription observed from the Cμ region in PTIP− B cells. These data suggest that PTIP helps promote H3K4 methylation in part by stabilizing higher-order chromatin conformations between the 3′ enhancer and the switch region GLT promoters in response to LPS/IL-4 stimulation.
Fig. 7.
Chromatin conformation capture (3C) assays. (A) Schematic of the 3C assay with primer pairs designed to measure the chromatin configurations A′ to E′ after HindIII digestion and religation. The IgH locus (top) shows locations of relevant HindIII sites (open circles) near Eμ, Iγ1, and hs44. A′ to D′ are primer pairs that span the cut and religated HindIII sites, demonstrating potential close interactions. Control F′ spans the Eμ HindIII site amplifying the uncut DNA. The Gapdh locus (bottom) is used as a positive control, where the E′ primer pairs measure the ligation efficiency of the 3.5-kb fragment. (B) Agarose gels of primer pairs used in panel A. Note, the A′ and B′ PCR products are reduced in PTIP− B cells after stimulation, indicating that the 3′ enhancer is not interacting with the Iγ1 and Iγ2b start site efficiently. Primer pairs C′ to E′ are unaffected in PTIP− B cells, suggesting that the Eμ/hs4, and Eμ/Iγ1 interactions are not affected in PTIP− B cells. The ligation efficiencies of a 3.5-kb Gapdh fragment are similar in all samples (E′), and equal amounts of DNA were used in all reactions (F′). E′ allows for equal loading by measuring uncut DNA in all samples. (C) Quantitative PCR analysis of primer pairs A′ to F′ using 32P-labeled primers and phosphorimaging of the appropriately sized PCR products. The PCR products A′ and B′ are reduced 8- to 10-fold. Experiments were performed independently in duplicate, with representative data shown.
DISCUSSION
The BRCT domain containing protein PTIP has been linked to both gene transcription and DNA repair (26). In this report, we utilize a B-cell-specific deletion to address the role of PTIP in CSR, a process that requires both transcription regulation and DNA recombination (35). The loss of PTIP significantly attenuates CSR in B cells after treatment with LPS/IL-4, a stimulus that promotes switching to the IgG1 and IgG2b isotypes. This defect in switching is most likely due to a transcriptional block, as the GLTs corresponding to the new constant regions are suppressed. Consistent with the role for PTIP in assembling a histone methyltransferase complex, localized H3K4me3 at the GLT promoters is also reduced in PTIP− B cells. These data are consistent with a recent report from Daniels et al. (10), which also demonstrated an effect on CSR in PTIP mutant B cells. However, we have taken this observation further and now demonstrate a specific link to Pax5 and to higher-order chromatin-remodeling events that are likely to determine transcription from the switch region promoter.
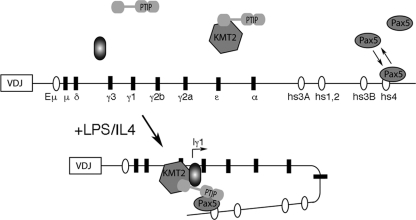
In response to cytokine stimulation and receptor cross-linking, the GLTs that correspond to specific isotype constant regions are transcribed and are required for the initiation of CSR. In our PTIP− B cells, the failure to switch to IgG1 is most likely due to a transcriptional defect of the γ1 GLT. Activation of the γ1 promoter appears to be mediated by NF-κB (20, 38) and STAT6 (21) binding near the GLT start site. We have shown significant increases in H3K4me3 at the GLT promoter regions in response to LPS/IL-4 stimulation. For IgG1, this increase is dependent upon PTIP and is coincident with PTIP and Pax5 localization to the γ1 promoter region in vivo. Given that this promoter does not contain functional Pax5 binding sites, based on our in vitro assays, we propose that Pax5 bound at the 3′ enhancer site hs4 is brought to the γ1 promoter by chromatin looping in a PTIP-dependent manner. Upon stimulation of B cells, Pax5 chromatin immunoprecipitates to the γ1 promoter, while the ChIP signal at the 3′ enhancer is actually decreased over time. This is likely to reflect limited cross-linking such that upon DNA looping, Pax5 is linked to either the 3′ enhancer or the γ1 promoter complex but not both. Thus, our model would predict that upon stimulation, PTIP helps stabilize Pax5 binding at the 3′ regulatory region and brings this region to the promoter of the GLTs, perhaps through interactions with proteins bound at the promoter, such as STAT6 or NF-κB (Fig. 8). This is consistent with the inability to see higher-order DNA associations between hs4 and the γ1 or γ2b promoters by using the chromatin conformation capture assay in PTIP-deficient B cells. Given the phospho-serine binding domains of PTIP, it is possible that stimulation phosphorylates Pax5 and the promoter-bound proteins to facilitate PTIP recruitment to chromatin. The need for stimulation in HEK293 cells is not evident for PTIP-Pax interactions, because Pax protein overexpression already results in a strong basal level of phosphorylation (4).
Fig. 8.
Model of PTIP-dependent chromatin interactions. In untreated B cells, Pax5 binds to the hs4 site with lower affinity. Upon LPS/IL-4 stimulation, PTIP stabilizes Pax5 binding and promotes chromatin looping to either the Iγ1 or Iγ2b transcription start site, where the KMT2D histone methyltransferase complex is recruited. Interactions at the specific start sites are likely to depend on other factors that bind directly to sequences upstream of the start sites and may also interact with PTIP.
Our model would predict that CSR and the GLTs are also dependent on the 3′ enhancer, which contains 4 DNase I-hypersensitive sites. Indeed, there is significant experimental evidence that the 3′ enhancer is essential for CSR and for regulating the GLTs. An initial deletion spanning 3.5 kb of the 3′ enhancer inhibited switching to all isotypes except IgG1 (9). Subsequent deletion of the DNase-hypersensitive sites hs3A and hs1,2 did not show significant attenuation of CSR (32). However, deletion of hs3b and hs4 significantly affected IgG3 and IgA isotypes and corresponded to a decrease in transcription from the GLT promoters (29). Further definition of the 3′ enhancer revealed that deletion of the entire 28-kb sequence containing all four hypersensitive sites affected GLTs from most, but not all, switch region promoters, with the α promoter being the least affected (11, 12). Interestingly, in a mouse line carrying an IgH bacterial artificial chromosome (BAC) transgene inserted near the endogenous IgH locus, the endogenous 3′ enhancer could rescue some of the CSR defects by rearranging the transgenic VDJ with the endogenous Cγ. These data, argue for an essential role of the 3′ enhancer in regulating GLTs. This point is underscored by the observation that the 3′ enhancer is able to directly interact with upstream promoter elements through potential chromatin looping mechanisms (17, 40).
The data presented here argue for a model in which Pax5, bound to the 3′ enhancer, complexes with the different upstream GLT promoter elements, depending upon which stimulus and which isotype are selected for switching. This model may be analogous to that of the locus control region of the globin genes, which has the ability to regulate multiple genes in cis, but only at one gene at a time through direct DNA looping mechanisms (14, 39). Alternatively, the H3K4me3 marks may be prerequisites for interactions between Pax5 and GLT promoters, independent of PTIP localization to the actual DNA sequences. In other words, the failure to maintain H3K4me3 levels could prevent higher-order chromatin conformations.
Mammals have multiple H3K4 methylation protein complexes, including the KMT2A-D (MLL1 to -4) and the KMT2F/G (hSet1A/B) complexes that are homologous to the COMPASS complex first described in yeast (25). To date, the best studied of these is KMT2A (MLL1), which regulates Hox gene expression and is responsible for H3K4me at about 5% of all promoters in mouse embryo fibroblasts (37). Thus, the remaining promoters are likely to interact with different KMT2s. However, PTIP has only been associated with the KMT2C/D complexes (7, 16). In cell culture assays, PTIP links the H3K4 methyltransferase complex to sequence specific DNA binding proteins (28), such as Pax2 and Pax5. The PTIP germ line mouse mutant phenotype is more severe and earlier in embryonic development than any single Pax gene mutation or KMT2 gene mutation (6), suggesting that PTIP may interact with multiple DNA binding proteins and methyltransferase complexes. This is consistent with the effects of PTIP deletion on global levels of H3K4 methylation in mouse embryos (6), stem cells (19), and Drosophila embryos (13). In our B-cell-specific PTIP knockout, the KMT2A/B complexes that do not contain PTIP appear unable to rescue the H3K4 methylation defect at the GLT promoters, in all likelihood because they lack the locus-specific DNA binding component to target their activity to the IgH locus promoters. Precisely how this locus specificity is achieved remains to be determined, but the data in this report clearly indicate a role for PTIP in recruiting H3K4me3 activity to the GLTs within the IgH locus.
Since the discovery of distal enhancers, how such enhancer elements communicate with more proximal promoters is poorly understood. In this report, we have used the IgH locus as a model to investigate the role of PTIP in transcription regulation and CSR, events that depend on both proximal and distal chromosomal elements. PTIP is part of a KMT2C/D H3K4 methyltransferase complex and is essential for development and lineage specification in mice and flies. The data presented provide the first evidence that PTIP is important for recruiting a KMT2 complex to an endogenous transcription start site and for the communication between a promoter and a distal enhancer. This communication is likely to require Pax5 and results in epigenetic modifications such as H3K4 methylation at the transcription unit. Still, many questions remain unanswered, as additional proteins are likely to be involved for regulating and stabilizing such chromatin interactions.
ACKNOWLEDGMENTS
We thank D. Ferguson, M. Dinkelmann, and L. Spehalski for help with the B cell cultures and flow cytometry; W. Dunnick for discussions; and M. Cascalho for critical reading of the manuscript.
This work was supported in part by NIH grants DK073722 to G.R.D. and DK079473 to K.R.S.
Footnotes
Published ahead of print on 31 January 2011.
REFERENCES
- 1. Blake J. A., Thomas M., Thompson J. A., White R., Ziman M. 2008. Perplexing Pax: from puzzle to paradigm. Dev. Dyn. 237:2791–2803 [DOI] [PubMed] [Google Scholar]
- 2. Bouchard M., Pfeffer P., Busslinger M. 2000. Functional equivalence of the transcription factors Pax2 and Pax5 in mouse development. Development 127:3703–3713 [DOI] [PubMed] [Google Scholar]
- 3. Brophy P. D., Ostrom L., Lang K. M., Dressler G. R. 2001. Regulation of ureteric bud outgrowth by Pax2-dependent activation of the glial derived neurotrophic factor gene. Development 128:4747–4756 [DOI] [PubMed] [Google Scholar]
- 4. Cai Y., et al. 2002. Phosphorylation of Pax2 by the c-Jun N-terminal kinase and enhanced Pax2-dependent transcription activation. J. Biol. Chem. 277:1217–1222 [DOI] [PubMed] [Google Scholar]
- 5. Chi N., Epstein J. A. 2002. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 18:41–47 [DOI] [PubMed] [Google Scholar]
- 6. Cho E. A., Prindle M. J., Dressler G. R. 2003. BRCT domain-containing protein PTIP is essential for progression through mitosis. Mol. Cell. Biol. 23:1666–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cho Y. W., et al. 2007. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 282:20395–20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cobaleda C., Schebesta A., Delogu A., Busslinger M. 2007. Pax5: the guardian of B cell identity and function. Nat. Immunol. 8:463–470 [DOI] [PubMed] [Google Scholar]
- 9. Cogne M., et al. 1994. A class switch control region at the 3′ end of the immunoglobulin heavy chain locus. Cell 77:737–747 [DOI] [PubMed] [Google Scholar]
- 10. Daniel J. A., et al. 2010. PTIP promotes chromatin changes critical for immunoglobulin class switch recombination. Science 329:917–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dunnick W. A., et al. 2009. Switch recombination and somatic hypermutation are controlled by the heavy chain 3′ enhancer region. J. Exp. Med. 206:2613–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dunnick W. A., Shi J., Graves K. A., Collins J. T. 2005. The 3′ end of the heavy chain constant region locus enhances germline transcription and switch recombination of the four gamma genes. J. Exp. Med. 201:1459–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fang M., et al. 2009. Drosophila ptip is essential for anterior/posterior patterning in development and interacts with the PcG and trxG pathways. Development 136:1929–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fraser P. 2006. Transcriptional control thrown for a loop. Curr. Opin. Genet. Dev. 16:490–495 [DOI] [PubMed] [Google Scholar]
- 15. Horcher M., Souabni A., Busslinger M. 2001. Pax5/BSAP maintains the identity of B cells in late B lymphopoiesis. Immunity 14:779–790 [DOI] [PubMed] [Google Scholar]
- 16. Issaeva I., et al. 2007. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol. Cell. Biol. 27:1889–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ju Z., et al. 2007. Evidence for physical interaction between the immunoglobulin heavy chain variable region and the 3′ regulatory region. J. Biol. Chem. 282:35169–35178 [DOI] [PubMed] [Google Scholar]
- 18. Kadauke S., Blobel G. A. 2009. Chromatin loops in gene regulation. Biochim. Biophys. Acta 1789:17–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim D., Patel S. R., Xiao H., Dressler G. R. 2009. The role of PTIP in maintaining embryonic stem cell pluripotency. Stem Cells 27:1516–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lin S. C., Stavnezer J. 1996. Activation of NF-κB/Rel by CD40 engagement induces the mouse germ line immunoglobulin Cγ1 promoter. Mol. Cell. Biol. 16:4591–4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Linehan L. A., Warren W. D., Thompson P. A., Grusby M. J., Berton M. T. 1998. STAT6 is required for IL-4-induced germline Ig gene transcription and switch recombination. J. Immunol. 161:302–310 [PubMed] [Google Scholar]
- 22. Manke I. A., Lowery D. M., Nguyen A., Yaffe M. B. 2003. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 302:636–639 [DOI] [PubMed] [Google Scholar]
- 23. Michaelson J. S., Singh M., Birshtein B. K. 1996. B cell lineage-specific activator protein (BSAP). A player at multiple stages of B cell development. J. Immunol. 156:2349–2351 [PubMed] [Google Scholar]
- 24. Mikkola I., Heavey B., Horcher M., Busslinger M. 2002. Reversion of B cell commitment upon loss of Pax5 expression. Science 297:110–113 [DOI] [PubMed] [Google Scholar]
- 25. Miller T., et al. 2001. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. U. S. A. 98:12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Munoz I. M., Rouse J. 2009. Control of histone methylation and genome stability by PTIP. EMBO Rep. 10:239–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nolis I. K., et al. 2009. Transcription factors mediate long-range enhancer-promoter interactions. Proc. Natl. Acad. Sci. U. S. A. 106:20222–20227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Patel S. R., Kim D., Levitan I., Dressler G. R. 2007. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev. Cell 13:580–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pinaud E., et al. 2001. Localization of the 3′ IgH locus elements that effect long-distance regulation of class switch recombination. Immunity 15:187–199 [DOI] [PubMed] [Google Scholar]
- 30. Rickert R. C., Roes J., Rajewsky K. 1997. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 25:1317–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Robson E. J., He S. J., Eccles M. R. 2006. A PANorama of PAX genes in cancer and development. Nat. Rev. Cancer 6:52–62 [DOI] [PubMed] [Google Scholar]
- 32. Saleque S., Singh M., Birshtein B. K. 1999. Ig heavy chain expression and class switching in vitro from an allele lacking the 3′ enhancers DNase I-hypersensitive hs3A and hs1,2. J. Immunol. 162:2791–2803 [PubMed] [Google Scholar]
- 33. Shilatifard A. 2008. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 20:341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sims R. J., III, et al. 2007. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell 28:665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stavnezer J., Guikema J. E., Schrader C. E. 2008. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 26:261–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Urbanek P., Wang Z.-Q., Fetka I., Wagner E. F., Busslinger M. 1994. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell 79:901–912 [DOI] [PubMed] [Google Scholar]
- 37. Wang P., et al. 2009. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol. Cell. Biol. 29:6074–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Warren W. D., Roberts K. L., Linehan L. A., Berton M. T. 1999. Regulation of the germline immunoglobulin Cgamma1 promoter by CD40 ligand and IL-4: dual role for tandem NF-kappaB binding sites. Mol. Immunol. 36:31–44 [DOI] [PubMed] [Google Scholar]
- 39. Wijgerde M., Grosveld F., Fraser P. 1995. Transcription complex stability and chromatin dynamics in vivo. Nature 377:209–213 [DOI] [PubMed] [Google Scholar]
- 40. Wuerffel R., et al. 2007. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity 27:711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wysocka J., et al. 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442:86–90 [DOI] [PubMed] [Google Scholar]