Ipl1/Aurora-mediated phosphorylation of the CPC component Sli15/INCENP promotes microtubule dynamics by restricting CPC–spindle association.
Abstract
Dynamic microtubules facilitate chromosome arrangement before anaphase, whereas during anaphase microtubule stability assists chromosome separation. Changes in microtubule dynamics at the metaphase–anaphase transition are regulated by Cdk1. Cdk1-mediated phosphorylation of Sli15/INCENP promotes preanaphase microtubule dynamics by preventing chromosomal passenger complex (CPC; Sli15/INCENP, Bir1/Survivin, Nbl1/Borealin, Ipl1/Aurora) association with spindles. However, whether Cdk1 has sole control over microtubule dynamics, and how CPC–microtubule association influences microtubule behavior, are unclear. Here, we show that Ipl1/Aurora-dependent phosphorylation of Sli15/INCENP modulates microtubule dynamics by preventing CPC binding to the preanaphase spindle and to the central spindle until late anaphase, facilitating spatiotemporal control of microtubule dynamics required for proper metaphase centromere positioning and anaphase spindle elongation. Decreased Ipl1-dependent Sli15 phosphorylation drives direct CPC binding to microtubules, revealing how the CPC influences microtubule dynamics. We propose that Cdk1 and Ipl1/Aurora cooperatively modulate microtubule dynamics and that Ipl1/Aurora-dependent phosphorylation of Sli15 controls spindle function by excluding the CPC from spindle regions engaged in microtubule polymerization.
Introduction
Cell division entails a series of complex events in order to ensure the equal segregation of replicated chromosomes from a mother cell into two daughter cells. In eukaryotes, accurate chromosome segregation is facilitated by microtubules (MTs). MTs are nucleated from spindle poles and also from kinetochores, proteinaceous complexes that form on centromeres. Spindle MTs either connect spindle poles to kinetochores (kinetochore MTs [kMTs]) or connect the two spindle poles (interpolar MTs [iMTs]). Before chromosomes separate (preanaphase/metaphase), MTs undergo rapid cycles of polymerization and depolymerization to facilitate attachment of each sister chromatid pair to opposite poles (chromosome biorientation; Holy and Leibler, 1994; Huang and Huffaker, 2006). Once chromosomes separate (anaphase), iMTs become less prone to depolymerization. This promotes iMT elongation, resulting in spindle pole separation, and also maintains spindle stability under the forces generated by pulling chromosomes to the opposite poles.
This significant alteration in MT dynamics at the metaphase–anaphase transition is triggered by decreased Cdk1-dependent substrate phosphorylation (Wheatley et al., 1997; Higuchi and Uhlmann, 2005). Cdk1 activity peaks in metaphase and declines after anaphase onset (Sullivan and Morgan, 2007). One Cdk1 substrate that supports anaphase spindle stabilization is the chromosomal passenger complex (CPC; Murata-Hori et al., 2002; Pereira and Schiebel, 2003). These observations raise some important questions that are addressed here, including whether Cdk1 solely governs cell cycle–dependent MT dynamics and how CPC association with MTs influences MT behavior.
The CPC is conserved among eukaryotes and consists of Ipl1/Aurora B kinase and its regulatory subunits: Sli15/INCENP, Bir1/Survivin, and Nbl1/Borealin/Dasra, each required for proper localization, and thus function, of the CPC (Carmena et al., 2009). The CPC regulates many functions including chromosome biorientation and condensation, spindle assembly checkpoint activation, anaphase spindle stability, and cytokinesis (Ruchaud et al., 2007). The CPC localizes to centromeres in metaphase and relocalizes to the spindle in anaphase (Earnshaw and Cooke, 1991), and this localization pattern is controlled by Cdk1 in both vertebrates and yeast (Pereira and Schiebel, 2003; Gruneberg et al., 2004; Hümmer and Mayer, 2009; Tsukahara et al., 2010).
Not only is the CPC modified by Cdk1, but three of the CPC subunits (Sli15/INCENP, Bir1/Survivin, and Ipl1/Aurora B) are also phosphorylated by Ipl1/Aurora (Kang et al., 2001; Bishop and Schumacher, 2002; Wheatley et al., 2004). Some of these phosphorylation sites are essential for kinase activation (Murata-Hori et al., 2002; Xu et al., 2009). However, little is known about the role of Ipl1/Aurora-dependent CPC phosphorylation at sites not directly involved in Aurora B activation. To uncover the role of this phosphorylation, we used chemical genetics and mutagenesis of phosphorylated residues, combined with cell biological, biochemical, and biophysical approaches. Here we describe how Ipl1/Aurora-dependent Sli15 phosphorylation modulates MT dynamics during cell division by regulating direct binding of the CPC to bundled MTs.
Results
Phosphorylation of Sli15/INCENP by Ipl1/Aurora is essential for CPC exclusion from preanaphase spindles
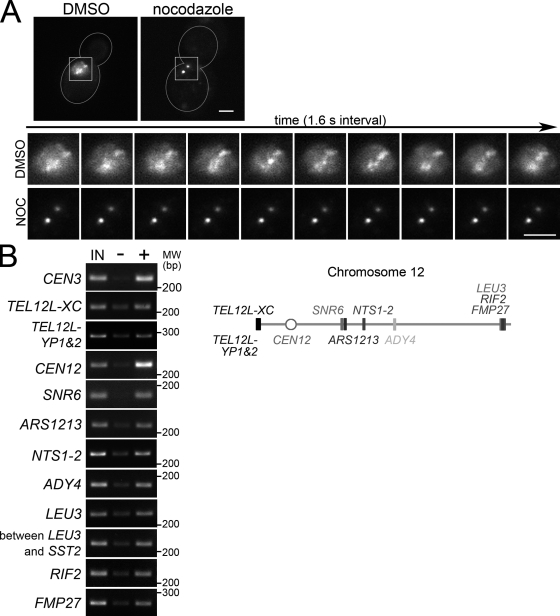
The yeast CPC, like vertebrate CPCs, associates with centromeres in preanaphase (Tanaka et al., 2002; Buvelot et al., 2003). In addition, the yeast CPC has a diffuse, nonuniform nuclear localization in preanaphase (Fig. 1 A; Tanaka et al., 2002; Shimogawa et al., 2009). During this period, CPC interacts with chromosome arms in addition to centromeres (Fig. 1 B). Interestingly, CPC localization during this period is dynamic and requires MTs (Fig. 1 A and Video 1).
Figure 1.
The CPC has a diffuse nuclear localization in preanaphase that requires MTs. (A, top) MTs regulate preanaphase CPC localization. Representative cells containing BIR1-3GFP treated with DMSO or nocodazole (NOC). Bar, 2 µm. (bottom) Time-lapse Bir1-3GFP images from Video 1 for the cells above. (B) CPC localizes to chromatin arms as well as centromeres in preanaphase. PCR products amplified from DNA obtained by chromatin immunoprecipitation from cells containing BIR1-13myc (left) and primer pair locations (right). IN, input DNA; −, no antibody; +, anti-myc antibody.
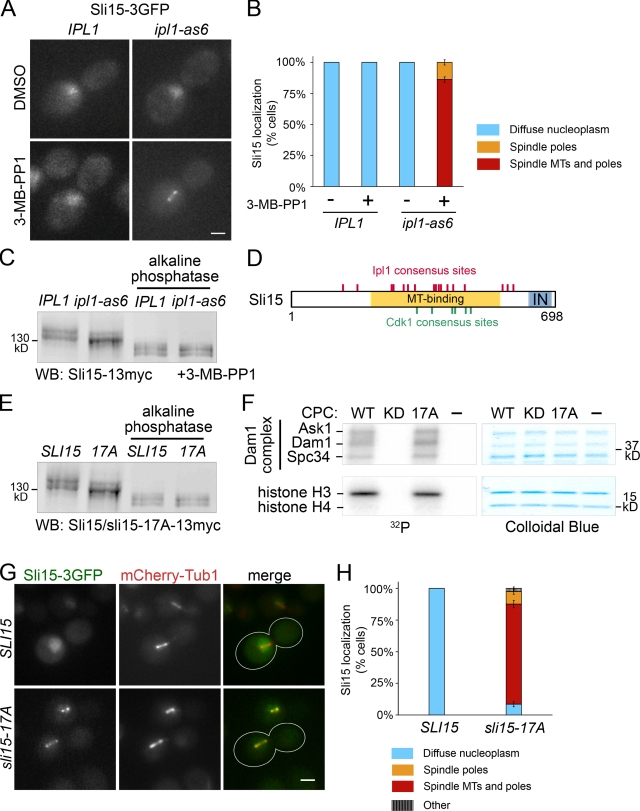
Because MTs regulate Aurora B (Rosasco-Nitcher et al., 2008; Tseng et al., 2010), and Ipl1/Aurora B phosphorylates CPC subunits (Kang et al., 2001; Bishop and Schumacher, 2002; Wheatley et al., 2004), we hypothesized that Ipl1/Aurora-dependent CPC subunit phosphorylation might regulate CPC function via changes in its localization. To investigate whether phosphorylation by Ipl1 affects CPC localization, we used an analogue-sensitive IPL1 allele (ipl1-as6; Pinsky et al., 2006b), and assessed preanaphase CPC localization when Ipl1 is inhibited. In wild-type cells, Sli15-3GFP was diffusely localized in the nucleus upon DMSO (carrier) or 3-MB-PP1 (inhibitor) treatment (Fig. 2, A [left] and B). In contrast, in all inhibitor-treated ipl1-as6 cells, Sli15-3GFP became concentrated on the spindle and around the spindle poles, whereas DMSO had no effect (Fig. 2, A [right] and B). The two other CPC components examined, Bir1 and Nbl1, also relocalized to the spindle when Ipl1 is inhibited (Fig. S1, A and B). As seen for preanaphase arrest, Sli15-3GFP became focused to the spindle and around spindle poles in inhibitor-treated ipl1-as6 cells arrested in metaphase by Cdc20 depletion (Uhlmann et al., 2000; Fig. S1 C). Similar results were also seen for the CPC in a temperature-sensitive (ts) ipl1 background (Fig. S1 E), validating the results obtained using a small-molecule inhibitor. To test whether a change in Sli15 phosphorylation coincides with CPC relocalization, we examined the electrophoretic mobility of Sli15 from wild-type and ipl1-as6 cells treated with inhibitor. In wild-type cells, Sli15 migrates as two major bands. At 15 min after adding inhibitor, the intensity of the top Sli15 band was significantly reduced in ipl1-as6 compared with wild-type cells (Fig. 2 C).
Figure 2.
Sli15/INCENP phosphorylation by Ipl1/Aurora is required for CPC exclusion from preanaphase spindles. (A) Ipl1 inhibition targets Sli15 to the preanaphase spindle. Representative cells containing IPL1 or ipl1-as6 and SLI15-3GFP, treated with DMSO or 40 µM 3-MB-PP1 for 15 min. Bar, 2 µm. (B) Quantification of observations represented in A. More than 200 cells from each treatment were analyzed using mCherry-Tub1 as a reference. Error bars show standard errors. (C) A subset of preanaphase Sli15 phosphorylation depends on Ipl1 activity in vivo. Sli15-13myc mobility shift upon Ipl1 inhibition was detected by Western blot of lysates from ipl1-as6 or IPL1 cells with or without alkaline phosphatase treatment. Cells were treated as described in A. (D) Diagram of Sli15 showing consensus sites for Ipl1/Aurora B ([RK]x[ST][ILVST]; Hsu et al., 2000; Bishop and Schumacher, 2002; Cheeseman et al., 2002) and Cdk1 ([ST]Px[RK]; Holmes and Solomon, 1996). MT-binding region (aa 227–559; Kang et al., 2001). T, three-helix bundle domain (aa 3–46; Jeyaprakash et al., 2007; Nakajima et al., 2009); IN, IN-box (aa 630–698; Adams et al., 2000). (E) Ipl1 phosphorylation site mutations in SLI15 diminish its phosphorylation in vivo. Western blot of whole-cell lysates from SLI15-13myc or sli15-17A-13myc cells. Cells were treated as described in A. (F) CPC containing sli15-17A retains normal kinase activity. In vitro kinase assay of the CPC containing only wild-type subunits (WT), ipl1-K133R (KD, kinase dead; Murata-Hori and Wang, 2002), or sli15-17A (17A) using the Dam1 complex and a histone H3/H4 tetramer as substrates. (G) sli15-17A shows a preanaphase spindle localization similar to that of Sli15 upon Ipl1 inhibition. Cells containing mCherry-TUB1, and SLI15-3GFP or sli15-17A-3GFP, were treated as described in A. Bar, 2 µm. (H) Quantification (as described in B) of the observations represented in G. “Other” denotes cells with foci that were not along spindle MTs or near the spindle poles. More than 200 cells from each treatment were analyzed. Error bars show standard errors.
Intriguingly, the observed premature spindle localization of the CPC resembles CPC localization in cells containing SLI15 mutations that reduce Cdk1 phosphorylation (Pereira and Schiebel, 2003). Our results demonstrate that Cdk1 is not the sole regulator of preanaphase CPC localization and that proper CPC localization also requires Ipl1/Aurora activity. The Cdk1 phosphorylation sites are within the MT-binding region of Sli15 (Mackay et al., 1993; Kang et al., 2001). Notably, Ipl1 consensus sites in Sli15 are concentrated within the MT-binding region (Fig. 2 D). Therefore, we investigated whether Ipl1-dependent phosphorylation of these sites also regulates CPC-spindle association. We mutated 17 serines or threonines that occur within the Ipl1 consensus sequence in Sli15 to alanines, generating sli15-17A. None of these residues are expected to be phosphorylated by Cdk1 (Pereira and Schiebel, 2003). The sli15-17A protein showed faster electrophoretic mobility compared with wild-type Sli15 (Fig. 2 E), as was seen for Sli15 upon Ipl1 inhibition (Fig. 2 C). To eliminate the possibility that the mutations in sli15-17A affect Ipl1 activity, we generated recombinant CPC containing either wild-type Sli15 (WT CPC) or sli15-17A (17A CPC) and examined their ability to phosphorylate two known in vivo Ipl1 substrates, the Dam1 complex and histone H3 in vitro (Hsu et al., 2000; Cheeseman et al., 2002). The 17A CPC phosphorylated Dam1 complex components (Ask1, Dam1, and Spc34) and histone H3 to a level comparable to the WT CPC (Fig. 2 F and Fig. S2 A). To test whether CPC in sli15-17A cells mimics the CPC localization in ipl1-as6 cells, we examined sli15-17A-3GFP localization during preanaphase. In 88% of cells, sli15-17A-3GFP localized to the spindle and/or around spindle poles compared with 0% for Sli15-3GFP (Fig. 2, G and H). This observation, together with the in vitro kinase activity of the 17A CPC, indicates that phosphorylation of these serines and threonines prevents CPC association with preanaphase spindles.
Ectopic localization of active Ipl1/Aurora to the metaphase spindle decreases MT dynamics
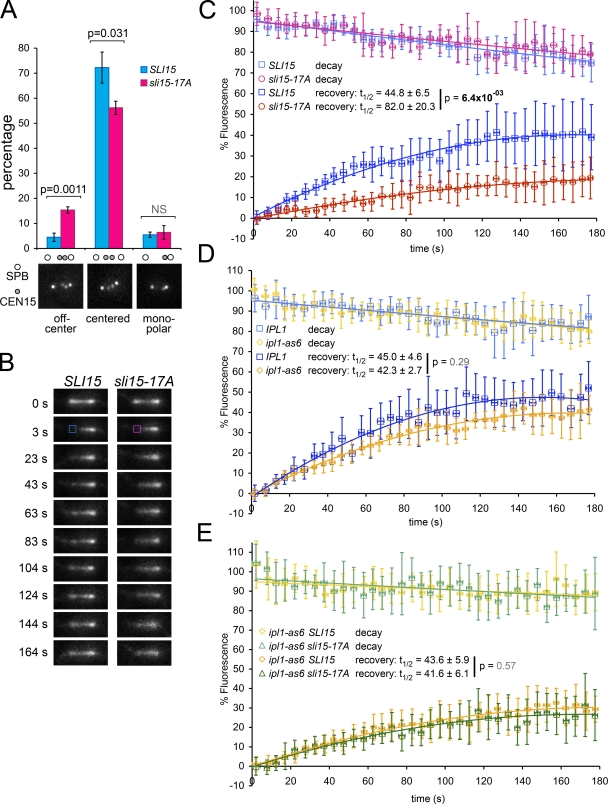
CPC localization in sli15-17A cells was similar to CPC localization upon Ipl1 inhibition, even though the 17A CPC retained normal kinase activity in vitro. These observations prompted us to investigate two ways in which the sli15-17A mutations might affect CPC function: (1) sli15-17A could cause an IPL1 loss of function phenotype by removing Ipl1 from where it normally acts, or (2) sli15-17A could give rise to a new phenotype by forcing active kinase to localize on the metaphase spindle, which may disrupt mechanisms operating under the spatiotemporal control of the CPC. To explore these possibilities, we first tested whether sli15-17A caused chromosome biorientation defects similar to those seen for loss of Ipl1 function by examining the position of CEN15 relative to the spindle pole protein Spc42 in wild-type and sli15-17A cells. In wild-type metaphase cells, the two CEN15 dots are 0.5–0.8 µm apart in the center of the spindle (Goshima and Yanagida, 2000). In contrast, ∼70% of cells in ts ipl1 and sli15 mutants have unseparated CEN dots near one spindle pole, an indication of monooriented chromosomes (Chan and Botstein, 1993; Biggins et al., 1999; Tanaka et al., 2002). The sli15-17A mutation caused no significant monopolar attachment compared with wild-type (6.4% and 5.5%, respectively; Fig. 3 A), indicating that the sli15-17A mutations are different from IPL1 or SLI15 loss-of-function alleles.
Figure 3.
Ectopic localization of active Ipl1/Aurora to metaphase spindles decreases MT dynamics. (A) sli15-17A increases centromere mispositioning in metaphase. Percentage of cells showing different classes of CEN15 (marked with GFP) positioning relative to Spc42-GFP in SLI15 and sli15-17A cells. (B) sli15-17A reduces MT polymerization in metaphase. Representative FRAP images of GFP-Tub1–marked spindles in SLI15 and sli15-17A cells. Photobleached areas are indicated with squares (at 3 s). (C) FRAP quantification of B reveals decreased MT dynamics in sli15-17A cells. (D) Ipl1 inhibition has no statistically significant effect on MT dynamics. FRAP quantification for metaphase spindles from IPL1 and ipl1-as6 cells treated with 40 µM 3-MB-PP1 for 15 min. (E) Ipl1 inhibition alleviates the decreased MT polymerization caused by sli15-17A. FRAP quantification for metaphase spindles from SLI15 ipl1-as6 and sli15-17A ipl1-as6 cells treated with 40 µM 3-MB-PP1.
Interestingly, sli15-17A cells did show a statistically significant increase in off-center pairs of CEN15 signals compared with wild type (15.4% and 4.5%, respectively; Fig. 3 A). Kinetochore mispositioning in metaphase is seen in cells containing hyper-stable tubulin mutations (Huang and Huffaker, 2006). Because localization of the CPC to MTs stabilizes the anaphase spindle (Pereira and Schiebel, 2003), we hypothesized that premature spindle localization of the CPC may perturb MT dynamics necessary for proper metaphase chromosome positioning.
To test this hypothesis, we assessed MT dynamics by performing FRAP analysis on metaphase spindles marked with GFP-Tub1 in wild-type and sli15-17A cells. We also analyzed fluorescence decay within the corresponding unbleached region of these spindles to assess MT depolymerization rates. There were no significant differences in decay kinetics between wild-type and sli15-17A cells. In contrast, signal recovery is significantly slower in sli15-17A than in wild-type cells (Fig. 3, B and C). The half recovery time for sli15-17A spindles is almost twice as long as for wild-type spindles (82.0 ± 20.3 s and 44.8 ± 6.5 s, respectively), indicating that the sli15-17A mutations decrease tubulin turnover. Furthermore, because there are no obvious differences in decay rate, decreased turnover seems to be primarily due to decreased MT polymerization. This defect coincides with a small delay in anaphase entry in sli15-17A cells (Fig. S2 B). A deficiency in MT polymerization would result in a gradual decrease in the amount of polymerized tubulin, which could explain why sli15-17A recovery did not reach the wild-type level. In contrast, Ipl1 inhibition caused no statistically significant effect on either signal decay or half recovery time (Fig. 3 D). However, the magnitude of the recovery was slightly lower for ipl1-as6 spindles, indicating that more MTs are failing to turn over. This difference could reflect the requirement for Aurora B in kMT turnover (Lampson et al., 2004; DeLuca et al., 2006). To test whether the decrease in MT dynamics caused by sli15-17A depends on Ipl1 activity, we analyzed wild-type and sli15-17A spindles in cells in which Ipl1 was inhibited. Ipl1 inhibition mostly restored the MT polymerization rate in sli15-17A cells (Fig. 3 E). Based on the above observations, we conclude that premature Ipl1 localization to the metaphase spindle reduces MT growth in sli15-17A cells. This is the first demonstration that premature targeting of Ipl1/Aurora activity to the metaphase spindle reduces MT dynamics.
Reduced Ipl1/Aurora-mediated phosphorylation of Sli15/INCENP promotes CPC accumulation on the central spindle in anaphase
The yeast CPC forms foci distributed along the spindle in early anaphase B and becomes focused at the spindle midzone toward the end of anaphase when spindle elongation slows as the spindle becomes fully elongated (Cooke et al., 1987; Buvelot et al., 2003; Nakajima et al., 2009). Despite the CPC’s known anaphase spindle localization, the reduced MT dynamics caused by ectopic targeting of Ipl1 to the preanaphase spindle prompted us to look for effects of reduced Sli15 phosphorylation by Ipl1 on the anaphase spindle. In vertebrates, Aurora B activity is required for CPC targeting to or retention at the central spindle (Murata-Hori et al., 2002; Xu et al., 2009). Therefore, we first examined Sli15’s anaphase localization upon Ipl1 inhibition. Unexpectedly, Ipl1 inhibition led to Sli15-3GFP accumulation around the central spindle when the spindle was not fully elongated (<8 µm long; Fig. 4, A and B). To eliminate the possibility of incomplete kinase inhibition in ipl1-as6 cells, we also examined the anaphase CPC localization in cells bearing a well-characterized ts allele ipl1-2 (Chan and Botstein, 1993), and an Ipl1 activation defective allele sli15-3 (Kim et al., 1999). We found that the CPC localizes to central spindles in both mutants at the restrictive temperature (Fig. S2 B), confirming that Aurora kinase activity is not essential for CPC targeting to the central spindle in yeast.
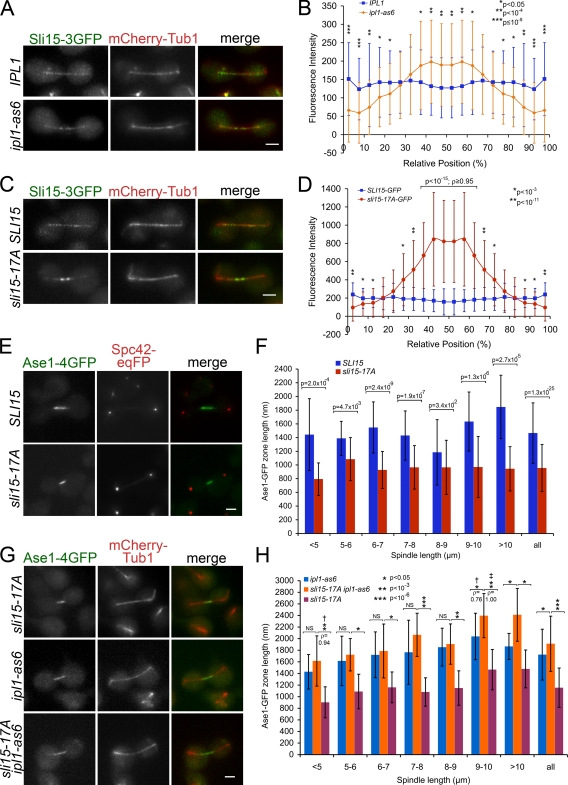
Figure 4.
Premature accumulation of active Ipl1/Aurora along the central spindle reduces midzone length. (A) CPC is prematurely targeted to the central spindle upon Ipl1 inhibition. Sli15-3GFP localization relative to mCherry-Tub1 during early/mid-anaphase B (spindle <8 µm long) in wild-type and ipl1-as6 cells treated with 40 µM 3-MB-PP1. Bar, 2 µm. (B) Quantification of A; P values determined using the Mann-Whitney U test. (C) sli15-17A prematurely targets CPC to the central spindle. Localization of Sli15-3GFP and sli15-17A-3GFP relative to mCherry-Tub1 during early/mid-anaphase B (spindle <8 µm long). Bar, 2 µm. (D) Quantification of C as described in B. (E) sli15-17A reduces spindle midzone length. Representative images of Ase1-4GFP and Spc42-eqFP in SLI15 and sli15-17A cells during anaphase B. Bar, 2 µm. (F) Quantification of Ase1-4GFP zone lengths from cells represented in E. (G) Ipl1 inhibition alleviates the reduced midzone length caused by sli15-17A. Representative images of Ase1-4GFP relative to mCherry-Tub1 in SLI15 ipl1-as6, sli15-17A ipl1-as6, and sli15-17A IPL1 cells treated with 40 µM 3-MB-PP1. Bar, 2 µm. (H) Quantification of Ase1-4GFP zone lengths from cells represented in G. † and ‡, P values determined using the Mann-Whitney U test. ‡, test was of the null hypothesis that ρ = 0.5.
To test whether premature CPC targeting to the central spindle is caused by reduced Sli15 phosphorylation, we examined sli15-17A localization in anaphase. Significantly more sli15-17A-3GFP concentrated around the central spindle compared with Sli15-3GFP in cells for which the spindle was not fully elongated (<8 µm long; Fig. 4, C and D). It is unclear why signal intensity for sli15-17A-3GFP is consistently stronger than for Sli15-3GFP. Nonetheless, the above observations support the idea that early focusing of the CPC toward the midzone is an effect of reduced Sli15 phosphorylation by Ipl1.
Premature accumulation of active Ipl1/Aurora along the central spindle reduces apparent midzone length
Intriguingly, upon Ipl1 inhibition, Sli15-3GFP was more widely distributed along the central spindle than was sli15-17A-3GFP. IPL1 loss of function widens the distribution of midzone proteins along the anaphase spindle and increases the frequency of off-centered spindle midzone proteins (Khmelinskii et al., 2007). The sharp focusing of sli15-17A-3GFP around the central spindle could therefore result from changes in central spindle organization caused by premature targeting of active Ipl1 to the vicinity of the spindle midzone. To assess whether central spindle accumulation of CPC changes midzone structure, we examined the localization of the midzone component Ase1 in sli15-17A cells. Ase1/PRC1, a conserved regulator of cytokinesis, selectively binds the midzone and functions in midzone stabilization and spindle elongation (Juang et al., 1997). Ase1-4GFP zone length on the anaphase spindle in sli15-17A was significantly diminished compared with wild type for all spindle lengths (Fig. 4, E and F). To test whether ectopic targeting of active Ipl1 to the central spindle is responsible for the reduced Ase1 zone size in sli15-17A cells, we analyzed Ase1 zone length in sli15-17A and SLI15 cells upon Ipl1 inhibition. There was no significant difference in Ase1 zone length for spindles that have not finished elongating (<8 µm long; Fig. 4, G and H), suggesting that Ipl1 kinase activity is responsible for the reduced Ase1 zone in sli15-17A cells. We therefore conclude that premature targeting of active Ipl1 to the spindle midzone reduces apparent midzone size.
Premature targeting of active Ipl1/Aurora to the spindle midzone slows spindle elongation
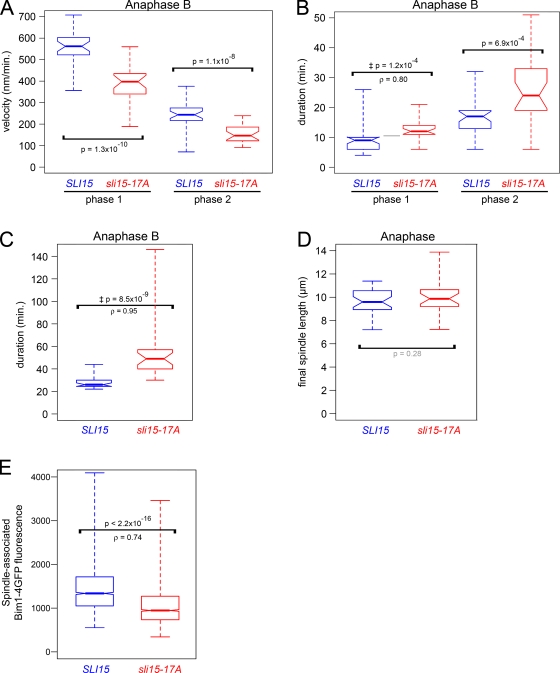
The spindle midzone is the site of iMT polymerization, bundling, and sliding, which are essential for anaphase spindle elongation and stability. The reduced midzone size in sli15-17A cells motivated us to ask whether premature CPC targeting to the central spindle affects midzone function. Because sli15-17A cells showed neither collapsed anaphase spindles nor mispositioned or elongated spindle midzones, we concluded that sli15-17A spindles were capable of iMT bundling and sliding, but could have defects in iMT polymerization. Therefore, we attempted FRAP to test whether premature CPC targeting to the midzone decreases the iMT polymerization rate in anaphase, as seen when the CPC localized to MTs in metaphase. However, we were unable to quantify the iMT polymerization rate at the iMTs plus ends due to deleterious effects of the laser intensity necessary for high-resolution imaging. Instead, we analyzed spindle elongation rate as a measure of iMT polymerization in sli15-17A cells during anaphase B. Anaphase B consists of two phases characterized by the speed of spindle elongation: the fast growing phase 1 and the slow growing phase 2 (Straight et al., 1998). The sli15-17A mutations significantly decreased both phase 1 and phase 2 elongation rates compared with wild-type cells (Fig. 5 A). There was also a corresponding shift toward longer phases 1 and 2 for sli15-17A compared with wild-type cells (Fig. 5 B). As a result of these delays, sli15-17A cells had longer anaphase B durations than wild-type cells (Fig. 5 C). However, wild-type and sli15-17A cells showed no significant difference in final spindle length (Fig. 5 D). When we attempted to test whether the decreased iMT elongation rate in sli15-17A cells requires Ipl1 kinase activity, the high degree of phenotypic variability present in these strains, likely caused by genetic interactions among the multiple mutations present (sli15-17A, ipl1-as6, and deletions of the multi-drug resistance genes PDR5 and SNQ2), made the results ambiguous. Nonetheless, when combined with our results for metaphase MT dynamics, the abovementioned observations suggest that decreased Sli15 phosphorylation by Ipl1 in anaphase B lowers iMT elongation rates by targeting the CPC to the spindle midzone.
Figure 5.
Premature targeting of Ipl1/Aurora activity to the spindle midzone slows anaphase spindle elongation. (A) sli15-17A reduces the anaphase B spindle elongation rate. Box plot showing rates of fast and slow spindle elongation (phases 1 and 2) in SLI15 and sli15-17A cells. (B) sli15-17A increases anaphase B phase 1 and 2 durations. Box plot showing durations of phases 1 and 2 in SLI15 and sli15-17A cells. (C) sli15-17A increases total anaphase B duration. Box plot showing anaphase B durations in SLI15 and sli15-17A cells. (D) sli15-17A does not significantly affect final anaphase spindle length. Box plot showing anaphase spindle lengths just before spindle breakdown in SLI15 and sli15-17A cells. (E) sli15-17A reduces Bim1/EB1 association with the anaphase spindle. Box plot showing mean Bim1-4GFP fluorescence intensities along anaphase spindles in cells containing sli15-17A or SLI15, BIM1-4GFP, and mCherry-TUB1. The distributions’ medians differed significantly (Mann-Whitney U test = 27613141, nWT = 1858, n17A = 2013, P < 2.2 × 10−16, two-tailed test; ρ = 0.74). See Materials and methods. ‡, P values determined using the Mann-Whitney U test for the null hypothesis that ρ = 0.5. Bim1-4GFP sizes and expression levels were comparable (see Fig. S2 D).
How does CPC targeting to the spindle midzone slow spindle elongation? Ipl1 partly regulates spindle midzone organization by controlling the localization of multiple midzone proteins (Pereira and Schiebel, 2003; Khmelinskii et al., 2007). Therefore, the reduced spindle elongation rate in sli15-17A cells might be caused by changes in localization and/or activity of proteins that are regulated by Ipl1. Therefore, we sought to identify downstream factors involved in Ipl1-dependent iMT dynamics and spindle elongation. Such factors might be expected to reduce anaphase spindle elongation and/or midzone length when improperly regulated. Reduced midzone length has been observed in cells in which the gene encoding the EB1-related plus end–binding protein Bim1 was deleted (Gardner et al., 2008). Furthermore, EB1 proteins modulate MT dynamics and are essential for anaphase spindle elongation (Rogers et al., 2002). Because Ipl1 phosphorylation diminishes Bim1 interaction with MTs (Zimniak et al., 2009; Woodruff et al., 2010), targeting active Ipl1 to the central spindle could decrease Bim1 binding to spindle MTs. To test this hypothesis, we examined anaphase spindle–associated Bim1-4GFP fluorescence intensity in sli15-17A cells. Bim1-4GFP signals decreased in sli15-17A compared with wild-type cells (Fig. 5 E). This result, combined with the aforementioned data, seems to imply that the CPC controls anaphase iMT dynamics in part by affecting Bim1 association with the spindle. It is unclear, however, whether the decreased Bim1 in sli15-17A cells was a direct consequence of increased Bim1 phosphorylation by Ipl1 because we could not detect changes in Bim1 phosphorylation using electrophoretic mobility shift assays (Fig. S2 E). It is possible that the difference in phosphorylation is too subtle to detect with this assay, or that additional factors controlled by the CPC affect Bim1 association with spindle MTs.
Decreased Sli15/INCENP phosphorylation by Ipl1/Aurora promotes direct binding of the CPC to MTs
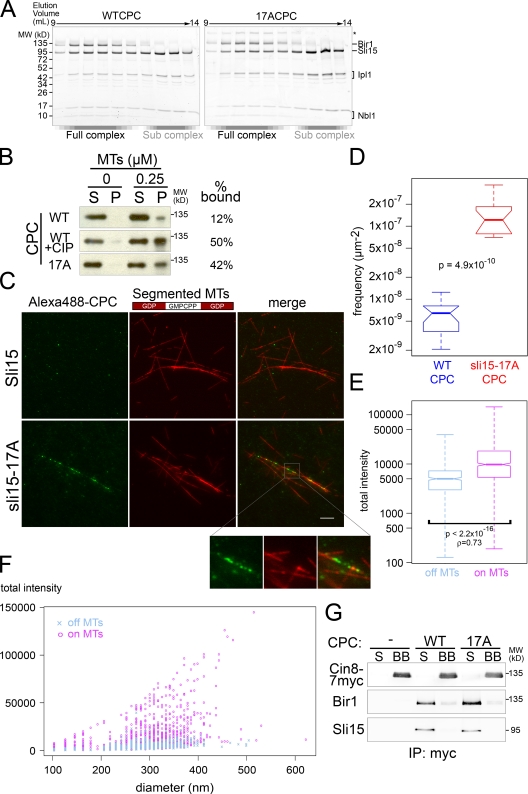
To understand the mechanism underlying the regulation of CPC targeting to the central spindle, we used an in vitro system to determine whether Ipl1/Aurora phosphorylation of Sli15 directly affects CPC affinity for spindle midzone MTs. To this end, recombinant WT and 17A CPCs that contain all four subunits were purified. The gel filtration profiles and subunit stoichiometries of the WT and 17A CPCs were comparable (Fig. 6 A).
Figure 6.
Decreased Sli15/INCENP phosphorylation by Ipl1/Aurora promotes direct CPC binding to bundled MTs. (A) Gel filtration profile of recombinant WT CPC or sli15-17A (17A) CPC. Void volume was ∼8.0 ml. (B) Reduced Sli15 phosphorylation by Ipl1 increases CPC affinity for MTs. MT cosedimentation assay using WT, 17A, and alkaline phosphatase–treated WT CPCs. The supernatant (S) and pellet (P) were analyzed by Western blot using anti-Bir1 antibody. (C) Reduced Sli15 phosphorylation by Ipl1 increases CPC affinity for bundled MTs. Segmented MTs (see Materials and methods) were bundled with Cin8 before incubation with Alexa 488–labeled WT or 17A CPC. Bar, 5 µm. (D) Quantification of CPC association with bundled MTs represented in C. See Materials and methods for details. Significance was determined using the two-sample Kolmogorov–Smirnov test (D = 1; P = 4.9 × 10−10). (E) MTs facilitate CPC multimerization. Box plot showing fluorescence intensities for 17A CPC spots represented in C that were not associated (off MTs) or associated (on MTs) with MTs. The distributions’ medians differed significantly (Mann-Whitney U test = 322910, noff_MTs = 445, non_MTs = 993, P < 2.2 × 10−16, two-tailed test; ρ = 0.73). (F) Plot of total fluorescence intensity versus spot diameter for 17A CPC that was found off MTs or on MTs. (G) 17A CPC accumulation on bundled MTs is not due to enhanced interaction with Cin8. After incubation with WT CPC or 17A CPC, Dynabeads-Cin8-7myc conjugate (BB) was separated from the supernatant (S). Cin8, Sli15, and Bir1 were detected by Western blot.
To assess whether Ipl1-dependent phosphorylation of Sli15 affects CPC binding to MTs, we performed MT cosedimentation assays using the recombinant WT and 17A CPCs. When 0.25 µM tubulin-derived MTs were used, 42% of 17A CPC cosedimented with MTs while 12% of WT CPC bound to MTs (Fig. 6 B). Moreover, when dephosphorylated by alkaline phosphatase, WT CPC cosedimented with MTs to a level similar to 17A CPC (50%: Fig. 6 B). These results demonstrate that removal of Ipl1/Aurora-dependent phosphorylation increases CPC affinity for MTs.
To study how Ipl1/Aurora-dependent phosphorylation affects CPC midzone recognition, we visualized Alexa 488–labeled WT and 17A CPCs mixed with rhodamine-labeled MTs. To validate this approach, we asked whether the difference between WT and 17A CPCs seen in the MT cosedimentation assay is also seen using the fluorescence-based assay. Consistent with the MT cosedimentation result, 17A CPC was ∼26 times more concentrated on MTs than the WT CPC when 0.2 µM tubulin-derived MTs were bundled with the kinesin-5 motor Cin8 (Fig. 6, C and D). Cin8 binds and bundles MTs, and is part of the spindle midzone (Hoyt et al., 1992; Roof et al., 1992; Sawin et al., 1992; Khmelinskii et al., 2009). Note that under these conditions, neither WT nor 17A CPCs showed apparent association with MTs when they were not bundled by Cin8 (Fig. S3 B).
Because sli15-17A prematurely targeted the CPC to the spindle midzone in vivo (Fig. 4, C and D), the efficient CPC interaction with bundled MTs in the fluorescence-based assay motivated us to investigate the mode of CPC association with these MTs. Interestingly, Alexa 488–labeled CPCs displayed a tendency to form bright distinctive foci when bound to MTs, rather than distributing homogeneously along MTs (Fig. 6 C). The median fluorescence intensity was significantly higher for MT-associated CPC spots than for CPC not bound to MTs (Fig. 6 E). In addition, a positive correlation between fluorescence intensity and spot diameter was seen for MT-associated CPC, but not for unassociated CPC (Fig. 6 F), suggesting that MT binding facilitates CPC multimerization.
Because the C. elegans kinesin-5 motor BMK-1 associates with Air-1/Aurora (Bishop et al., 2005), we tested whether CPC enrichment at bundled MTs was caused by direct interaction between Cin8 and the WT or 17A CPC. Under the same conditions used for our fluorescent analysis, we did not detect significant interaction between Cin8 and either WT or 17A CPC (Fig. 6 G). Based on these observations, we concluded that the CPC prefers the surface of bundled MTs and that when phosphorylation of Sli15 by Ipl1 is diminished, the affinity of the CPC for bundled MTs increases.
The CPC localizes to different subdomains of the spindle in response to Sli15/INCENP phosphorylation by Ipl1/Aurora and Cdk1
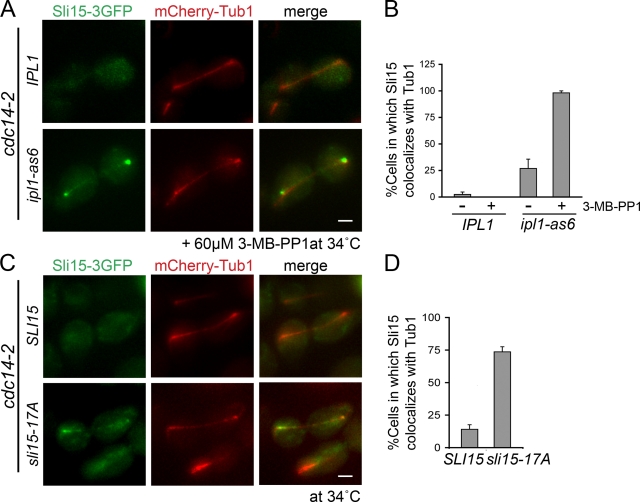
Cdc14 is the major counteracting phosphatase for Cdk1 in yeast, whereas the corresponding activity remains unclear in vertebrates (Queralt and Uhlmann, 2008; Mocciaro and Schiebel, 2010). Loss of Cdc14 prevents CPC binding to the anaphase spindle; this result led to the conclusion that Cdk1-dependent phosphorylation of Sli15 prevents CPC association with the preanaphase spindle (Pereira and Schiebel, 2003). Our observation that Ipl1 also prevents CPC binding to preanaphase spindles prompted us to test whether decreased Ipl1 activity promotes CPC association with anaphase spindles when the CPC remains phosphorylated by Cdk1. We examined Sli15-3GFP localization in cells containing the ts cdc14-2 allele (Pereira and Schiebel, 2003) and either ipl1-as6 or IPL1. Upon inhibitor treatment, Sli15-3GFP became associated with anaphase spindles in ipl1-as6 cells, whereas IPL1 cells showed no spindle localization (Fig. 7, A and B). To determine if this was a direct effect of Sli15 phosphorylation, we examined sli15-17A association with anaphase spindles in cdc14-2 cells. sli15-17A partially localizes to anaphase spindles in cdc14-2 cells (Fig. 7, C and D). These observations confirm that in addition to Cdk1, Ipl1/Aurora also regulates CPC association with spindle MTs. It is noteworthy that both Sli15-3GFP in ipl1-as6 cells and sli15-17A-3GFP accumulate most strongly near the spindle poles in cdc14-2 cells rather than being distributed along the spindle. This localization pattern, which we also saw for ipl1-as6 and sli15-17A cells in metaphase, is distinct from the reported localization of Sli156A, which lacks Cdk1 phosphorylation sites and localizes along the entire length of the spindle (Pereira and Schiebel, 2003). Our observations imply that, in the presence of phosphorylation at Cdk1 sites, Ipl1 phosphorylation functions to prevent the CPC from localizing to spindle poles. These observations also suggest that there may be combinatorial regulation of CPC localization based on its phosphorylation pattern.
Figure 7.
The CPC localizes to different spindle subdomains in response to Sli15/INCENP phosphorylation by Ipl1/Aurora and Cdk1. (A) Ipl1 inhibition partially restores CPC localization to the anaphase spindle in cdc14-2 cells. Localization of Sli15-3GFP and Tub1-mCherry in IPL1 cdc14-2 or ipl1-as6 cdc14-2 cells exposed to the restrictive temperature (34°C) for 2 h and then treated with 60 µM 3-MB-PP1 for 15 min. Bar, 2 µm. (B) Quantification of observations represented in A; error bars show standard errors. (C) sli15-17A partially restored its localization to the anaphase spindle in cdc14-2 cells. Localization of Sli15-3GFP or sli15-17A-3GFP and Tub1-mCherry in cdc14-2 strains exposed to 34°C for 2 h. Bar, 2 µm. (D) Quantification of observations represented in C; error bars show standard errors.
Discussion
We have demonstrated that Ipl1/Aurora-dependent phosphorylation of Sli15/INCENP prevents CPC association with specific mitotic spindle regions and that this self-regulation facilitates localized MT growth. Aurora B increases kMT turnover and destabilizes incorrect kinetochore–MT attachments to promote chromosome biorientation (Cheeseman et al., 2002; Lampson et al., 2004; Lan et al., 2004; DeLuca et al., 2006; Pinsky et al., 2006b). Premature CPC accumulation in sli15-17A cells on the spindle does not interfere with chromosome biorientation, but delays spindle MT growth. CPC autophosphorylation may thus contribute to the known Ipl1/Aurora-dependent mechanisms for chromosome misattachment correction by maintaining the MT dynamics essential for kinetochore capture.
Mechanisms for targeting CPC to the central spindle may vary in different organisms. Aurora B activity is required for CPC targeting to the spindle midzone in chicken DT40 cells (Xu et al., 2009), and for the retention of the CPC at the spindle midzone in rat NRK cells (Murata-Hori et al., 2002). In contrast, yeast CPC prematurely localized to spindle midzones upon Ipl1 inhibition. These differences are not surprising because yeast has only one Aurora kinase whereas vertebrates have Aurora A and B, which have distinct localization patterns and functions (Carmena et al., 2009). Presumably the behavior in yeast is ancestral whereas the vertebrate behavior reflects an elaboration subsequent to an ancient gene duplication. Although the best-characterized yeast Ipl1/Aurora functions parallel known Aurora B functions (Biggins et al., 1999; Kim et al., 1999; Tanaka et al., 2002; Pinsky et al., 2006b), Ipl1 also plays a role in bipolar spindle assembly similar to Aurora A (Kotwaliwale et al., 2007). It is thus plausible that the yeast CPC carries out the functions of both Aurora A and B, and consequently requires different patterns of regulation. In support of this idea, the spindle pole accumulation of the CPC in sli15-17A cells lacking Cdc14 phosphatase activity resembles Aurora A localization (Fig. 8 A [right] and B [bottom right]; Stenoien et al., 2003). This observation is consistent with our hypothesis that Cdk1 and Ipl1/Aurora combinatorially regulate CPC localization. Differential phosphorylation patterns could specifically and independently target the CPC to distinct places (centromeres, spindle poles, chromatin, and spindle MTs) at precise times (metaphase, anaphase, and cytokinesis) in order to perform the combined functions of Aurora A and B.
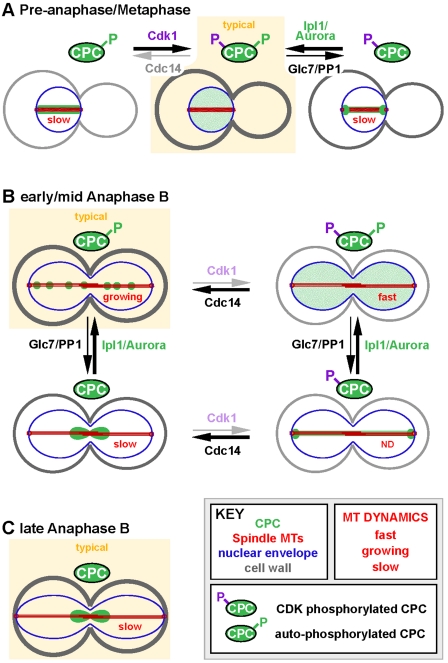
Figure 8.
A model illustrating the regulation of CPC localization in preanaphase and anaphase by Cdk1 and Ipl1/Aurora in yeast. A key to cellular components and states of spindle MT dynamics (red text) is shown. Predominant CPC localization patterns in wild-type cells are indicated as “typical” (yellow background). Other localization patterns are atypical or represent a minor part of the wild-type localization. Arrows indicate balance between Sli15 phosphorylation states. Centromere-localized CPC is not shown. (A) Regulation of CPC localization in preanaphase/metaphase. (B) Regulation of CPC localization in early/mid anaphase B. (C) Regulation of CPC localization in late anaphase B. See Discussion for details.
Ipl1/Aurora-mediated phosphorylation of Sli15/INCENP regulates CPC localization
It was suggested that Ipl1 phosphorylation of Sli15 has little effect on CPC localization based on abolishing eight Ipl1 consensus sites in Sli15 (Pereira and Schiebel, 2003). This result differs from our observations using sli15-17A, a protein lacking 17 Ipl1 consensus sites, which targets the CPC to the metaphase spindle ectopically. For the previous study the [RK]x[TS][ILV] Ipl1 consensus site was used (Cheeseman et al., 2002), whereas in this study we used [RK]x[ST][ILVST] based on the in vivo phosphorylation sites of known Ipl1/Aurora substrates such as histone H3 and Sli15/INCENP (Hsu et al., 2000; Bishop and Schumacher, 2002; Cheeseman et al., 2002). Consequently, sli15-17A contains nine more mutations, seven of which fall within the previously defined MT-binding domain of Sli15 (Kang et al., 2001).
The following observations provide additional support for our conclusions about CPC auto-regulation. First, the yeast CPC sometimes localizes to preanaphase spindles or spindle poles under mildly perturbed conditions (Kim et al., 1999; Buvelot et al., 2003; Pinsky et al., 2006a). Second, we observed that the electrophoretic mobility of sli15-17A resembles Sli15 upon Ipl1 inhibition. Third, the CPC was targeted to the metaphase spindle not only in response to Ipl1 chemical inhibition, but also using a ts allele of IPL1. Lastly, as seen for sli15-17A cdc14-2 cells, the anaphase spindle localization of the CPC was partially restored in cells in which both Ipl1 kinase and Cdc14 phosphatase activities were compromised. We conclude that the CPC auto-regulates its localization to the spindle through phosphorylation of Sli15.
Autophosphorylation of the CPC prevents its direct binding to bundled MTs
We demonstrated that the full CPC is sufficient to bind directly to MTs and that this binding is greatly weakened when Sli15/INCENP is phosphorylated by Ipl1/Aurora. Because 17A CPC showed increased binding to bundled MTs, we speculate that MT bundling promotes multiple interactions with the CPC when Sli15 is not phosphorylated by Ipl1. This model provides a mechanistic explanation for how CPC autophosphorylation controls CPC distribution along the anaphase spindle. We can imagine two possible mechanisms for self-regulation of CPC affinity for bundled MTs. First, phosphorylation makes the surface charge of MT-binding sites on Sli15 more negative and thus may inhibit interaction with the negatively charged MT surface. Second, phosphorylation might also induce conformational changes within the complex that preclude binding to midzone-like MT arrays.
The yeast CPC forms bright foci along the spindle during anaphase. This native CPC distribution resembles the in vitro distribution of CPC foci within MT bundles observed in this study, implying that MT-dependent cooperative multimerization of the CPC in vitro may also occur in vivo. We speculate that the CPC binds MTs most stably in a cooperative manner and that when Sli15 phosphorylation by Ipl1 is reduced the increased CPC affinity for bundled MTs recruits a critical amount of CPC. The positive feedback created by this cooperative binding could help explain how the CPC concentrates onto the spindle midzone in late anaphase in vivo. Future structural analyses and in vitro reconstitution of the CPC and its interactions with MTs will be informative.
Self-regulation of the CPC is necessary for the proper control of MT dynamics
The highly dynamic metaphase/preanaphase spindle MTs promote formation of bipolar chromosome attachments. This MT dynamicity is promoted by high Cdk1 and low Cdc14 activities (Higuchi and Uhlmann, 2005). A component of this mechanism is the Cdk1-dependent phosphorylation of Sli15 that helps prevent CPC binding to the spindle (Fig. 8 A; Pereira and Schiebel, 2003). Based on our observations, Ipl1-dependent phosphorylation of Sli15 is also crucial for maintenance of normal metaphase MT dynamics by preventing CPC binding to the spindle and spindle poles. Glc7/PP1, the counteracting phosphatase for Ipl1, possibly contributes to the CPC’s dynamic localization during this period by allowing rapid turnover of Sli15 phosphorylation.
At anaphase onset, Cdk1 activity becomes low, whereas Cdc14 phosphatase activity becomes high (Sullivan and Morgan, 2007). Loss of Cdk1-dependent phosphorylation of Sli15 targets the CPC to the anaphase spindle, where the CPC forms foci along the spindle and contributes to spindle midzone integrity by regulating the localization of midzone proteins (Pereira and Schiebel, 2003; Khmelinskii et al., 2007). Ipl1 is active throughout anaphase (Buvelot et al., 2003) and, based on our observations, the resultant CPC autophosphorylation prevents premature CPC focusing to the spindle midzone (Fig. 8 B), facilitating iMT polymerization by allowing recruitment of factors promoting MT polymerization. We hypothesize that, when the spindle becomes fully elongated in late anaphase, Glc7/PP1-dependent dephosphorylation of Sli15 promotes CPC movement toward the spindle midzone (Fig. 8 C). This accumulation of active Ipl1/Aurora at the spindle midzone could limit MT polymerization and thereby prevent spindle overgrowth by phosphorylating and removing MT-binding proteins that are involved in MT polymerization.
Why do cells need more than one kinase to regulate a single substrate in the context of a single biological process? It may be that multiple regulatory pathways allow cells to achieve fine spatiotemporal control over the CPC. Consistent with this idea, Cdk1 and Ipl1/Aurora are regulated differently. Cdk1 activity is primarily subject to temporal control governed by the level of cyclins (Sullivan and Morgan, 2007). In contrast, substrate phosphorylation by Ipl1/Aurora is mostly influenced by the spatial arrangement of the substrates relative to the kinase and Glc7/PP1 phosphatase (Fuller et al., 2008; Liu et al., 2009). It is therefore possible that Glc7/PP1 or one of its regulatory factors might localize along the anaphase spindle and reduce Ipl1/Aurora-dependent Sli15/INCENP phosphorylation in order to drive the CPC toward the midzone. Recently, an intermediate filament protein, Fin1 (van Hemert et al., 2003), was shown to localize along anaphase spindles and to activate Glc7/PP1 (Woodbury and Morgan, 2007; Akiyoshi et al., 2009). This localization would facilitate increased central spindle accumulation of the CPC without decreasing Ipl1-dependent substrate phosphorylation in the central spindle. In future work, it will be important to test whether Fin1-Glc7 activity is necessary for CPC focusing to the central spindle and to identify other factors involved in this process.
Materials and methods
Strains and plasmids
All yeast strains were derived from the S288C background, except for a protease-compromised strain (DDY1810), and were grown in yeast extract/peptone or synthetic medium supplemented with appropriate nutrients and 2% glucose at 25°C unless otherwise noted. All epitope- and fluorescently tagged genes were expressed at their endogenous loci under their native promoters as described previously (Longtine et al., 1998). The mutations in sli15-17A (S137A, S177A, S276A, T277A, S298A, S305A, S326A, S382A, S383A, S391A, T395A, S413A, S432A, S460A, T564A, S578A, and T589A) were generated by gene synthesis (GenScript) in combination with site-directed mutagenesis (QuikChange Mutagenesis kit; Agilent Technologies). Wild-type SLI15 and sli15-17A were cloned into pBSKS+ and then a nourseothricin resistance gene cassette (Goldstein and McCusker, 1999) was inserted 100 bp 3′ of each ORF. These inserts were transformed into a diploid strain heterozygous for a SLI15 deletion to target each construct to the endogenous locus. The 10-kb LacO array (Straight et al., 1997) was inserted into an intergenic region 1.8 kb away from CEN15 as described previously (Goshima and Yanagida, 2000). The Cin8 expression plasmid was constructed by cloning the CIN8 ORF (BamHI–NotI) into the pRS426 plasmid containing the GAL1-10 promoter, a TEV protease recognition site, and encoding seven repeats of the myc epitope (Rodal et al., 2003). All yeast strains and plasmids used in this study are listed in Table S1.
Yeast cell cycle synchronization
To create preanaphase spindle conditions, cells were treated with 0.1 M hydroxyurea (HU) for 2.5 h, which maintains spindle MT dynamicity and biorientation (Kosco et al., 2001). To synchronize cells in metaphase by depletion of Cdc20, an activator of the anaphase-promoting complex (Uhlmann et al., 2000), cells containing CDC20 driven by MET3 promoter were first arrested in G1 with 10 µg/ml α-factor for 3 h and then released into media containing 10 mM methionine for 3 h. To induce metaphase arrest by depolymerizing MTs, cells were treated with 15 µg/ml nocodazole for 2 h.
Statistics
Error bars show standard deviations except where otherwise indicated. Whiskers on box plots show the extreme values of the data. Top and bottom of box indicate the 75th and 25th percentiles. Dark bar at the waist of the box indicates the median (50th percentile). If the notches (indentations in the sides of the box) of two datasets do not overlap then this is considered strong evidence that the two medians are different (Chambers et al., 1983).
Statistical analyses and box plots were performed using R (http://www.r-project.org; R Development Core Team, 2010). Unless otherwise noted, significances (P values) were determined using the two-tailed Student’s t test (two-sample, unequal variance). Normality of data was evaluated using the Shapiro-Wilk test. For tests using the Mann-Whitney U test (two-tailed test), similarity of the data distributions was evaluated using quantile–quantile plots. Also calculated were ρ values (= U/(n1*n2), where nis are the sample sizes) that give an estimate of the probability that a randomly chosen value from the first population will be larger than a randomly chosen value from the second population (Hernstein et al., 1976). Unless otherwise noted, P values obtained from the Mann-Whitney U test were for tests of the null hypothesis (H0) that the medians were identical.
In vitro kinase assay
In vitro kinase assays were conducted as described previously (Cheeseman et al., 2002) with the following modifications. BSA was added to the reaction to a final concentration of 67 µg/ml. 1 pmol of CPC (containing only wild-type subunits, ipl1-K133R, or sli15-17A) and 100 pmol of substrate (Dam1 complex or histone H3-H4 tetramer) were used for each reaction. The reaction was conducted at ambient temperature and samples were collected every 10 min for 30 min. The Dam1 complex was expressed and purified as described previously (Westermann et al., 2005). In brief, BL21Rosetta cells (EMD) containing the plasmid pC43HSK3H (Miranda et al., 2005) was grown at 37°C to OD0.4, and then protein expression was induced in the presence of 1 mM IPTG for 4 h. After the cells were harvested and washed with phosphate-buffered saline (PBS) the cells were lysed in 20 mM sodium phosphate (pH 6.8), 500 mM NaCl, 1 mM EDTA, 20 mM imidazole, and 0.5% (vol/vol) Triton X-100 by sonication, and the lysate was cleared by centrifugation. Ni-NTA agarose beads (QIAGEN) were incubated with the cleared lysate for 2 h at 4°C and then were collected and washed with the lysis buffer. The proteins were eluted in 6 ml 20 mM sodium phosphate, 500 mM NaCl, 1 mM EDTA, and 200 mM imidazole. After overnight dialysis against 20 mM sodium phosphate, 150 mM NaCl, and 1 mM EDTA, the dialysate was loaded onto a 1-ml HiTrap SP Sepharose column (GE Healthcare), and proteins were eluted with a 10-ml linear gradient of 0.15 to 1 M NaCl in 20 mM sodium phosphate (pH 6.8) 1 mM EDTA buffer. The Xenopus histone H3/H4 tetramer was a kind gift from P. Kaufmann (University of Massachusetts, Worcester, MA).
Chromatin immunoprecipitation
Chromatin immunoprecipitation from preanaphase cells was conducted based on previously described methods (Braunstein et al., 1993; Kang et al., 2001). The yeast cells were grown to early log phase (OD600 0.2–0.4) and then treated with 0.1 M hydroxyurea for 2.5 h at 30°C. The protein–DNA complexes were then cross-linked with 1% formaldehyde (37% stock; Thermo Fisher Scientific) for 2 h at 25°C. The reaction was terminated by addition of glycine (Sigma-Aldrich) to a final concentration of 125 mM and incubating for 5 min. After washing twice with 50 ml of cold TBS, the cells were resuspended in 900 ml of the lysis buffer (all buffer compositions were as described in Braunstein et al., 1993) containing proteinase inhibitor cocktail IV (EMD). The cells were lysed using a Mini-Beadbeater (Biospec Products) with an equal volume of glass beads at 4°C at the maximum speed. Cycles of 2 min of beating followed by 1 min on ice were performed until >80% cells were lysed. After collecting the lysate by centrifugation, the lysate was sonicated six times for 10 s on ice (at 20% duty cycle at 2.5 output setting; Sonicator W-385 [Misonix]). The lysates were then cleared by centrifugation and split into three tubes: input, mock treatment, and target sample. The target sample was incubated with anti-myc antibody (9E10) for 1 h before the addition of preblocked Protein G–Sepharose (GE Healthcare) and 4 µg/µL lambda DNA (sonicated to 200–1000 bp). The mock-treated sample was treated as the target sample except no antibody was added. The protein G–Sepharose was preblocked in a solution consisting of 1x TE 1.54 mM NaN3 1 mg/ml BSA. After a 3-h incubation at 4°C, the beads were washed and resuspended in ChIP elution buffer containing proteinase K (Fermentas) to a final concentration of 250 µg/ml and incubated at 37°C for 12 h. The input DNA (the sonicated lysate) was also treated with proteinase K at this time. All samples were then incubated at 65°C for 6 h. DNA was recovered by phenol-chloroform extraction followed by ethanol precipitation. DNA was resuspended in TE and subjected to PCR analysis.
Western blotting
Yeast whole-cell extracts were prepared as described previously (Keogh et al., 2006). Target proteins were detected using anti-myc antibody (9E10), anti-GFP antibody (Torrey Pines Biolabs), anti-Pgk1 antibody (Invitrogen), or as otherwise noted.
FRAP and live-cell microscopy
Cells suspended in minimal growth medium were mounted on a coverslip coated with concanavalin A (Sigma-Aldrich). FRAP was conducted using a confocal microscope (LSM710; Carl Zeiss) equipped with a 63x 1.4 NA Plan Apochromat oil objective. Pinhole size was set to two Airy units. After photobleaching, images were acquired using ZEN 2009 software (Carl Zeiss) at ambient temperature every 2.6 s as a stack of four images with a 700-nm axial separation. These images were converted to a series of 2D images by maximum intensity projection using ZEN 2009 software. Image processing and the intensity measurements were conducted using iVision (BD) and Imaris (Bitplane) software. Initial corrections to fluorescence values were conducted by established methods (http://www.embl.de/eamnet/frap/FRAP6.html; Phair et al., 2004). Half-time for fluorescence recovery (t1/2) values were determined by plotting ln(ymax − y) vs. time, where y is the corrected fluorescence and ymax was determined from the binomial regression curve fit to the corrected fluorescence values. The slopes of these lines (k) were then converted to t1/2 values: t1/2 = ln(0.5)/k.
To display the averaged curves, fluorescence decay values were converted to a percentage of the prebleach fluorescence. Recovery values were converted to a percentage of the difference between the pre- and post-bleach fluorescence. The means and standard deviations for values falling in 5-s intervals were then determined and plotted; where necessary, curves were shifted vertically to align y-intercepts.
Other live-cell microscopy experiments were performed using a microscope (IX81-OMAC; Olympus) equipped with 100x 1.4 NA Plan Apochromat oil objective. Cells suspended in minimal growth medium were mounted on a coverslip coated with concanavalin A (Sigma-Aldrich). Images were acquired using MetaMorph software (Molecular Devices) at ambient temperature unless otherwise noted as a stack of seven images with 400-nm axial separation using an Orca II camera (Hamamatsu Photonics) with 2 × 2 binning (1 pixel, ∼129 nm). Alexa 488–labeled CPC and TRITC rhodamine-labeled MTs were visualized using the same equipment described above with 1 × 1 binning (1 pixel, ∼64.5 nm). Images were processed using Imaris (Bitplane) and ImageJ software (http://rsb.info.nih.gov/ij).
CPC purification
CPC purification was conducted as described previously (Nakajima et al., 2009), with the following modifications. Nbl1 was purified from bacteria and denatured in buffer containing 6 M guanidine HCl for 30 min on ice and then added to the cleared lysate from insect cells (ES-Sf9) expressing Ipl1, Sli15, and Bir1. For fluorescently labeled CPC, the complex was incubated with Alexa 488 C5 maleimide (Invitrogen) on ice in the dark for 2 h before gel filtration.
Size exclusion chromatography of the CPC
For analysis of the native yeast-CPC, a protease-compromised strain (DDY1810) expressing SLI15-13myc at the endogenous locus under the native promoter was grown to early log phase (OD600, 0.3–0.35) before addition of either 15 µg/ml nocodazole or 0.1 M hydroxyurea. After being spheroplasted, the cells were lysed through homogenization into buffer supplemented with metabolic, protease, and phosphatase inhibitors (50 mM Bis-Tris propane, pH 7.4, 0.1 M KCl, 5 mM EGTA, 5 mM EDTA, 10 mM NEM, 20 mM β-glycerophosphate, 40 µM cantharidin, 20 mM Na pyrophosphate, 10 mM NaN3, 20 mM NaF, 0.8 mM Na orthovanadate, 1 mM PMSF [all from Sigma-Aldrich], and 1x protease inhibitor cocktail IV [EMD]) to preserve the stability and post-translational modifications. The cleared lysates were subject to gel filtration using a Superose 6 10/300 column (GE Healthcare) with a fraction size of 0.5 ml. The same gel filtration condition was used for molecular weight markers (HMW; GE Healthcare, Bio-Rad Laboratories). Detection of Sli15-13myc in gel filtration fractions was done by Western blot using anti-myc (9E10) antibody. Gel filtration of recombinant CPC and molecular weight markers (Bio-Rad Laboratories) was conducted using a Superose 6 10/300 column (GE Healthcare) with a fraction size of 0.5 ml.
Fluorescence-based MT association assay
To generate segmented MTs, unlabeled guanylyl-(a,b)-methylene-diphosphonate (GMPCPP)-MT seeds were polymerized by incubating 2 µM tubulin (purified from bovine brain) with 1 mM GMPCPP in BRB80 buffer (80 mM Pipes-KOH, pH 6.8, 1 mM MgCl2, and 1 mM EGTA) for 30 min at 37°C. These seeds were then used to elongate TRITC rhodamine-labeled GDP-MTs by diluting 10-fold and incubating with a 10-µM labeled/unlabeled tubulin mix (5% TRITC rhodamine-labeled porcine tubulin [Cytoskeleton] and 95% unlabeled tubulin) and 100 µM GTP in BRB80 buffer for 20 min at 37°C. To stabilize the MTs, taxol (Paclitaxel; Sigma-Aldrich) was then added to a final concentration of 20 µM, and the MTs were incubated for another 10 min at 37°C.
Alexa 488–labeled CPCs were diluted into CPC buffer (50 mM Bis-Tris propane, pH 6.8, 100 mM arginine, 100 mM glutamic acid, 300 mM NaCl, 2 µM Zn(CH3CO2)2, 2 µM CaCl2, 100 µM MgCl2, 100 µM ATP, 1 mM DTT, and 20% glycerol) supplemented with 1 mg/ml BSA and dialyzed into CPC buffer containing 50 mM NaCl at 4°C for 2 h. After dialysis, taxol was added to the CPCs to a final concentration of 10 µM. Segmented MTs (0.2 µM tubulin) in CPC buffer were incubated with 2 nM Cin8 for 10 min at ambient temperature, then 2 nM final concentration of CPC was added to the reaction for 3 min before imaging in the presence of oxygen scavengers (0.2 mg/ml glucose oxidase, 35 µg/ml catalase, 0.25% β-mercaptoethanol, and 4.5 mg/ml glucose; Desai et al., 1999).
Fluorescent spot detection and analysis were done with Imaris software (Bitplane) using default settings except for setting the minimum spot diameter to 100 nm. CPC spots on unlabeled seeds (in gaps aligned with TRITC MTs) were manually classified as MT bound. Frequency (µm−2) of Alexa 488 CPCs on MTs was determined for each MT bundle examined by dividing the number of Alexa 488–labeled CPC dots associated with the bundle by the total MT area in the field containing the MT bundle. A total of 14 microscope fields containing MT bundles were examined for both the WT and 17A CPCs (containing 17 and 16 separate bundles, respectively) and no fields containing bundles were ignored. The total MT areas that were analyzed for the WT and 17A CPCs were 69.7 × 109 µm2 and 54.109 µm2, respectively. Total Alexa 488 fluorescence intensity over the area of the spot was determined for each spot.
MT cosedimentation assay
The recombinant CPCs were supplemented with 1 mg/ml BSA and dialyzed into CPC buffer containing 50 mM NaCl for 2 h at 4°C. After dialysis, taxol was added to a final concentration of 20 µM. MTs in CPC buffer containing 100 mM NaCl and 20 µM taxol were added to a final concentration of 0.25 µM tubulin. CPCs and MTs were incubated at ambient temperature for 20 min before ultracentrifugation at 50 krpm at 25°C for 15 min. The supernatant and pellet were separated and analyzed by Western blot using anti-Bir1 antibody. Signal intensity was quantified using ImageJ software.
Cin8 purification and immunoprecipitation of CPC
Cin8-7myc was overexpressed and purified as described previously (Rodal et al., 2003) with the following modifications. Cin8-7myc purification buffer consisted of 20 mM Hepes, pH 7.5, 1 mM EGTA, 5 mM MgCl2, 1 mM ATP, and 100 mM KCl. Cin8-7myc–bound IgG Sepharose (GE Healthcare) was washed with buffer containing 500 mM KCl to remove contaminants before myc-tag removal by TEV protease. TEV protease (pRK793) was expressed and purified as described previously (Kapust et al., 2001).
For immunoprecipitation of CPC with Cin8, myc antibody (9E10) was conjugated to Dynabeads (Invitrogen) following the company’s instructions. WT and 17A CPCs were prepared as described for the fluorescence-based MT association assay. The Dynabeads-bound Cin8 was incubated with CPC in CPC buffer (50 mM NaCl) containing 1 mg/ml BSA for 30 min at 4°C. The beads were washed three times with CPC buffer and the bound and unbound proteins were analyzed by Western blot using anti-Sli15, anti-Bir1 (generous gifts from A. Desai, University of California, San Diego, La Jolla, CA), and anti-myc (9E10) antibodies.
Online supplemental material
Fig. S1 shows that Ipl1 kinase activity is required for CPC exclusion from preanaphase spindles. Fig. S2 shows the time course of CPC kinase reactions with the Dam1 complex and histone H3 as substrates, the requirement for Ipl1-dependent phosphorylation of Sli15 for normal cell cycle progression, CPC targeting to the central spindle in ipl1 and sli15 temperature-sensitive mutants, Bim1 protein level and electrophoretic band shift in wild-type and sli15-17A cells, and electrophoretic mobility of Sli15- and sli15-17A-13myc throughout the cell cycle. Fig. S3 shows the low affinity of the Alexa 488–labeled CPC for rhodamine-labeled MTs. Video 1 shows MT-dependent nuclear localization of the CPC. Table S1 lists the yeast strains and plasmids used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201009137/DC1.
Acknowledgments
We thank A.C. Martin, V. Okreglak, and I.M. Cheeseman for critical reading of this manuscript; K. Shokat, C. Zhang, C. Kung, A. Desai, E. Schiebel, P. Kaufman, I. Le Blanc, J.B. Woodruff, R. Schekman, and M. Yanagida for sharing reagents; S. Ruzin and the UCB CNR imaging facility, and A. Chan and the SFSU molecular imaging center for imaging advice.
This research was supported by National Institutes of Health RO1 grant GM47842 to G. Barnes.
Footnotes
Abbreviations used in this paper:
- 17A CPC
- recombinant CPC containing sli15-17A
- CPC
- chromosomal passenger complex
- iMT
- interpolar microtubule
- kMT
- kinetochore microtubule
- MT
- microtubule
- ts
- temperature sensitive
- WT CPC
- recombinant CPC containing all wild-type subunits
References
- Adams R.R., Wheatley S.P., Gouldsworthy A.M., Kandels-Lewis S.E., Carmena M., Smythe C., Gerloff D.L., Earnshaw W.C. 2000. INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol. 10:1075–1078 10.1016/S0960-9822(00)00673-4 [DOI] [PubMed] [Google Scholar]
- Akiyoshi B., Nelson C.R., Ranish J.A., Biggins S. 2009. Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev. 23:2887–2899 10.1101/gad.1865909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S., Severin F.F., Bhalla N., Sassoon I., Hyman A.A., Murray A.W. 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13:532–544 10.1101/gad.13.5.532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop J.D., Schumacher J.M. 2002. Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B Kinase stimulates Aurora B kinase activity. J. Biol. Chem. 277:27577–27580 10.1074/jbc.C200307200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop J.D., Han Z., Schumacher J.M. 2005. The Caenorhabditis elegans Aurora B kinase AIR-2 phosphorylates and is required for the localization of a BimC kinesin to meiotic and mitotic spindles. Mol. Biol. Cell. 16:742–756 10.1091/mbc.E04-08-0682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein M., Rose A.B., Holmes S.G., Allis C.D., Broach J.R. 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 7:592–604 10.1101/gad.7.4.592 [DOI] [PubMed] [Google Scholar]
- Buvelot S., Tatsutani S.Y., Vermaak D., Biggins S. 2003. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell Biol. 160:329–339 10.1083/jcb.200209018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M., Ruchaud S., Earnshaw W.C. 2009. Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr. Opin. Cell Biol. 21:796–805 10.1016/j.ceb.2009.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers J., Cleveland W., Kleiner B., Tukey P. 1983. Graphical Methods for Data Analysis. Wadsworth International Group, Belmont, CA [Google Scholar]
- Chan C.S., Botstein D. 1993. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics. 135:677–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Anderson S., Jwa M., Green E.M., Kang J., Yates J.R., III, Chan C.S., Drubin D.G., Barnes G. 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 111:163–172 10.1016/S0092-8674(02)00973-X [DOI] [PubMed] [Google Scholar]
- Cooke C.A., Heck M.M., Earnshaw W.C. 1987. The inner centromere protein (INCENP) antigens: movement from inner centromere to midbody during mitosis. J. Cell Biol. 105:2053–2067 10.1083/jcb.105.5.2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., Salmon E.D. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- Desai A., Verma S., Mitchison T.J., Walczak C.E. 1999. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 96:69–78 10.1016/S0092-8674(00)80960-5 [DOI] [PubMed] [Google Scholar]
- Earnshaw W.C., Cooke C.A. 1991. Analysis of the distribution of the INCENPs throughout mitosis reveals the existence of a pathway of structural changes in the chromosomes during metaphase and early events in cleavage furrow formation. J. Cell Sci. 98:443–461 [DOI] [PubMed] [Google Scholar]
- Fuller B.G., Lampson M.A., Foley E.A., Rosasco-Nitcher S., Le K.V., Tobelmann P., Brautigan D.L., Stukenberg P.T., Kapoor T.M. 2008. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 453:1132–1136 10.1038/nature06923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M.K., Haase J., Mythreye K., Molk J.N., Anderson M., Joglekar A.P., O’Toole E.T., Winey M., Salmon E.D., Odde D.J., Bloom K. 2008. The microtubule-based motor Kar3 and plus end-binding protein Bim1 provide structural support for the anaphase spindle. J. Cell Biol. 180:91–100 10.1083/jcb.200710164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A.L., McCusker J.H. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 15:1541–1553 [DOI] [PubMed] [Google Scholar]
- Goshima G., Yanagida M. 2000. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 100:619–633 10.1016/S0092-8674(00)80699-6 [DOI] [PubMed] [Google Scholar]
- Gruneberg U., Neef R., Honda R., Nigg E.A., Barr F.A. 2004. Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. J. Cell Biol. 166:167–172 10.1083/jcb.200403084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernstein R.J., Loveland D.H., Cable C. 1976. Natural concepts in pigeons. J. Exp. Psychol. Anim. Behav. Process. 2:285–302 10.1037/0097-7403.2.4.285 [DOI] [PubMed] [Google Scholar]
- Higuchi T., Uhlmann F. 2005. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature. 433:171–176 10.1038/nature03240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes J.K., Solomon M.J. 1996. A predictive scale for evaluating cyclin-dependent kinase substrates. A comparison of p34cdc2 and p33cdk2. J. Biol. Chem. 271:25240–25246 10.1074/jbc.271.41.25240 [DOI] [PubMed] [Google Scholar]
- Holy T.E., Leibler S. 1994. Dynamic instability of microtubules as an efficient way to search in space. Proc. Natl. Acad. Sci. USA. 91:5682–5685 10.1073/pnas.91.12.5682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M.A., He L., Loo K.K., Saunders W.S. 1992. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J. Cell Biol. 118:109–120 10.1083/jcb.118.1.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J.Y., Sun Z.W., Li X., Reuben M., Tatchell K., Bishop D.K., Grushcow J.M., Brame C.J., Caldwell J.A., Hunt D.F., et al. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 102:279–291 10.1016/S0092-8674(00)00034-9 [DOI] [PubMed] [Google Scholar]
- Huang B., Huffaker T.C. 2006. Dynamic microtubules are essential for efficient chromosome capture and biorientation in S. cerevisiae. J. Cell Biol. 175:17–23 10.1083/jcb.200606021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hümmer S., Mayer T.U. 2009. Cdk1 negatively regulates midzone localization of the mitotic kinesin Mklp2 and the chromosomal passenger complex. Curr. Biol. 19:607–612 10.1016/j.cub.2009.02.046 [DOI] [PubMed] [Google Scholar]
- Jeyaprakash A.A., Klein U.R., Lindner D., Ebert J., Nigg E.A., Conti E. 2007. Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell. 131:271–285 10.1016/j.cell.2007.07.045 [DOI] [PubMed] [Google Scholar]
- Juang Y.L., Huang J., Peters J.M., McLaughlin M.E., Tai C.Y., Pellman D. 1997. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 275:1311–1314 10.1126/science.275.5304.1311 [DOI] [PubMed] [Google Scholar]
- Kang J., Cheeseman I.M., Kallstrom G., Velmurugan S., Barnes G., Chan C.S. 2001. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J. Cell Biol. 155:763–774 10.1083/jcb.200105029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapust R.B., Tözsér J., Fox J.D., Anderson D.E., Cherry S., Copeland T.D., Waugh D.S. 2001. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 14:993–1000 10.1093/protein/14.12.993 [DOI] [PubMed] [Google Scholar]
- Keogh M.C., Mennella T.A., Sawa C., Berthelet S., Krogan N.J., Wolek A., Podolny V., Carpenter L.R., Greenblatt J.F., Baetz K., Buratowski S. 2006. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 20:660–665 10.1101/gad.1388106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A., Lawrence C., Roostalu J., Schiebel E. 2007. Cdc14-regulated midzone assembly controls anaphase B. J. Cell Biol. 177:981–993 10.1083/jcb.200702145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A., Roostalu J., Roque H., Antony C., Schiebel E. 2009. Phosphorylation-dependent protein interactions at the spindle midzone mediate cell cycle regulation of spindle elongation. Dev. Cell. 17:244–256 10.1016/j.devcel.2009.06.011 [DOI] [PubMed] [Google Scholar]
- Kim J.H., Kang J.S., Chan C.S. 1999. Sli15 associates with the ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 145:1381–1394 10.1083/jcb.145.7.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosco K.A., Pearson C.G., Maddox P.S., Wang P.J., Adams I.R., Salmon E.D., Bloom K., Huffaker T.C. 2001. Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol. Biol. Cell. 12:2870–2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwaliwale C.V., Frei S.B., Stern B.M., Biggins S. 2007. A pathway containing the Ipl1/aurora protein kinase and the spindle midzone protein Ase1 regulates yeast spindle assembly. Dev. Cell. 13:433–445 10.1016/j.devcel.2007.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson M.A., Renduchitala K., Khodjakov A., Kapoor T.M. 2004. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 6:232–237 10.1038/ncb1102 [DOI] [PubMed] [Google Scholar]
- Lan W., Zhang X., Kline-Smith S.L., Rosasco S.E., Barrett-Wilt G.A., Shabanowitz J., Hunt D.F., Walczak C.E., Stukenberg P.T. 2004. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 14:273–286 [DOI] [PubMed] [Google Scholar]
- Liu D., Vader G., Vromans M.J., Lampson M.A., Lens S.M. 2009. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 323:1350–1353 10.1126/science.1167000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Mackay A.M., Eckley D.M., Chue C., Earnshaw W.C. 1993. Molecular analysis of the INCENPs (inner centromere proteins): separate domains are required for association with microtubules during interphase and with the central spindle during anaphase. J. Cell Biol. 123:373–385 10.1083/jcb.123.2.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda J.J., De Wulf P., Sorger P.K., Harrison S.C. 2005. The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 12:138–143 10.1038/nsmb896 [DOI] [PubMed] [Google Scholar]
- Mocciaro A., Schiebel E. 2010. Cdc14: a highly conserved family of phosphatases with non-conserved functions? J. Cell Sci. 123:2867–2876 10.1242/jcs.074815 [DOI] [PubMed] [Google Scholar]
- Murata-Hori M., Wang Y.L. 2002. The kinase activity of aurora B is required for kinetochore-microtubule interactions during mitosis. Curr. Biol. 12:894–899 10.1016/S0960-9822(02)00848-5 [DOI] [PubMed] [Google Scholar]
- Murata-Hori M., Tatsuka M., Wang Y.L. 2002. Probing the dynamics and functions of aurora B kinase in living cells during mitosis and cytokinesis. Mol. Biol. Cell. 13:1099–1108 10.1091/mbc.01-09-0467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima Y., Tyers R.G., Wong C.C., Yates J.R., III, Drubin D.G., Barnes G. 2009. Nbl1p: a Borealin/Dasra/CSC-1-like protein essential for Aurora/Ipl1 complex function and integrity in Saccharomyces cerevisiae. Mol. Biol. Cell. 20:1772–1784 10.1091/mbc.E08-10-1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Schiebel E. 2003. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 302:2120–2124 10.1126/science.1091936 [DOI] [PubMed] [Google Scholar]
- Phair R.D., Gorski S.A., Misteli T. 2004. Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol. 375:393–414 10.1016/S0076-6879(03)75025-3 [DOI] [PubMed] [Google Scholar]
- Pinsky B.A., Kotwaliwale C.V., Tatsutani S.Y., Breed C.A., Biggins S. 2006a. Glc7/protein phosphatase 1 regulatory subunits can oppose the Ipl1/aurora protein kinase by redistributing Glc7. Mol. Cell. Biol. 26:2648–2660 10.1128/MCB.26.7.2648-2660.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky B.A., Kung C., Shokat K.M., Biggins S. 2006b. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat. Cell Biol. 8:78–83 10.1038/ncb1341 [DOI] [PubMed] [Google Scholar]
- Queralt E., Uhlmann F. 2008. Cdk-counteracting phosphatases unlock mitotic exit. Curr. Opin. Cell Biol. 20:661–668 10.1016/j.ceb.2008.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2010. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Rodal A.A., Manning A.L., Goode B.L., Drubin D.G. 2003. Negative regulation of yeast WASp by two SH3 domain-containing proteins. Curr. Biol. 13:1000–1008 10.1016/S0960-9822(03)00383-X [DOI] [PubMed] [Google Scholar]
- Rogers S.L., Rogers G.C., Sharp D.J., Vale R.D. 2002. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J. Cell Biol. 158:873–884 10.1083/jcb.200202032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof D.M., Meluh P.B., Rose M.D. 1992. Kinesin-related proteins required for assembly of the mitotic spindle. J. Cell Biol. 118:95–108 10.1083/jcb.118.1.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosasco-Nitcher S.E., Lan W., Khorasanizadeh S., Stukenberg P.T. 2008. Centromeric Aurora-B activation requires TD-60, microtubules, and substrate priming phosphorylation. Science. 319:469–472 10.1126/science.1148980 [DOI] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W.C. 2007. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 8:798–812 10.1038/nrm2257 [DOI] [PubMed] [Google Scholar]
- Sawin K.E., LeGuellec K., Philippe M., Mitchison T.J. 1992. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 359:540–543 10.1038/359540a0 [DOI] [PubMed] [Google Scholar]
- Shimogawa M.M., Widlund P.O., Riffle M., Ess M., Davis T.N. 2009. Bir1 is required for the tension checkpoint. Mol. Biol. Cell. 20:915–923 10.1091/mbc.E08-07-0723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenoien D.L., Sen S., Mancini M.A., Brinkley B.R. 2003. Dynamic association of a tumor amplified kinase, Aurora-A, with the centrosome and mitotic spindle. Cell Motil. Cytoskeleton. 55:134–146 10.1002/cm.10120 [DOI] [PubMed] [Google Scholar]
- Straight A.F., Marshall W.F., Sedat J.W., Murray A.W. 1997. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 277:574–578 10.1126/science.277.5325.574 [DOI] [PubMed] [Google Scholar]
- Straight A.F., Sedat J.W., Murray A.W. 1998. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 143:687–694 10.1083/jcb.143.3.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M., Morgan D.O. 2007. Finishing mitosis, one step at a time. Nat. Rev. Mol. Cell Biol. 8:894–903 10.1038/nrm2276 [DOI] [PubMed] [Google Scholar]
- Tanaka T.U., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M.J., Nasmyth K. 2002. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 108:317–329 10.1016/S0092-8674(02)00633-5 [DOI] [PubMed] [Google Scholar]
- Tseng B.S., Tan L., Kapoor T.M., Funabiki H. 2010. Dual detection of chromosomes and microtubules by the chromosomal passenger complex drives spindle assembly. Dev. Cell. 18:903–912 10.1016/j.devcel.2010.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara T., Tanno Y., Watanabe Y. 2010. Phosphorylation of the CPC by Cdk1 promotes chromosome bi-orientation. Nature. 467:719–723 10.1038/nature09390 [DOI] [PubMed] [Google Scholar]
- Uhlmann F., Wernic D., Poupart M.A., Koonin E.V., Nasmyth K. 2000. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 103:375–386 10.1016/S0092-8674(00)00130-6 [DOI] [PubMed] [Google Scholar]
- van Hemert M.J., Deelder A.M., Molenaar C., Steensma H.Y., van Heusden G.P. 2003. Self-association of the spindle pole body-related intermediate filament protein Fin1p and its phosphorylation-dependent interaction with 14-3-3 proteins in yeast. J. Biol. Chem. 278:15049–15055 10.1074/jbc.M212495200 [DOI] [PubMed] [Google Scholar]
- Westermann S., Avila-Sakar A., Wang H.W., Niederstrasser H., Wong J., Drubin D.G., Nogales E., Barnes G. 2005. Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Mol. Cell. 17:277–290 10.1016/j.molcel.2004.12.019 [DOI] [PubMed] [Google Scholar]
- Wheatley S.P., Hinchcliffe E.H., Glotzer M., Hyman A.A., Sluder G., Wang Y. 1997. CDK1 inactivation regulates anaphase spindle dynamics and cytokinesis in vivo. J. Cell Biol. 138:385–393 10.1083/jcb.138.2.385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley S.P., Henzing A.J., Dodson H., Khaled W., Earnshaw W.C. 2004. Aurora-B phosphorylation in vitro identifies a residue of survivin that is essential for its localization and binding to inner centromere protein (INCENP) in vivo. J. Biol. Chem. 279:5655–5660 10.1074/jbc.M311299200 [DOI] [PubMed] [Google Scholar]
- Woodbury E.L., Morgan D.O. 2007. Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat. Cell Biol. 9:106–112 10.1038/ncb1523 [DOI] [PubMed] [Google Scholar]
- Woodruff J.B., Drubin D.G., Barnes G. 2010. Mitotic spindle disassembly occurs via distinct subprocesses driven by the anaphase-promoting complex, Aurora B kinase, and kinesin-8. J. Cell Biol. 191:795–808 10.1083/jcb.201006028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Ogawa H., Vagnarelli P., Bergmann J.H., Hudson D.F., Ruchaud S., Fukagawa T., Earnshaw W.C., Samejima K. 2009. INCENP-aurora B interactions modulate kinase activity and chromosome passenger complex localization. J. Cell Biol. 187:637–653 10.1083/jcb.200906053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimniak T., Stengl K., Mechtler K., Westermann S. 2009. Phosphoregulation of the budding yeast EB1 homologue Bim1p by Aurora/Ipl1p. J. Cell Biol. 186:379–391 10.1083/jcb.200901036 [DOI] [PMC free article] [PubMed] [Google Scholar]