In yeast, CDKs and the NDR kinase Cbk1 are regulators of polarized growth. It is found that the CDK Cdc28 regulates the function of Cbk1 in response to hypha-inducing conditions by direct phosphorylation of Mob2, a conserved regulatory subunit of Cbk1.
Abstract
Nuclear Dbf2-related (NDR) protein kinases are essential components of regulatory pathways involved in cell morphogenesis, cell cycle control, and viability in eukaryotic cells. For their activity and function, these kinases require interaction with Mob proteins. However, little is known about how the Mob proteins are regulated. In Candida albicans, the cyclin-dependent kinase (CDK) Cdc28 and the NDR kinase Cbk1 are required for hyphal growth. Here we demonstrate that Mob2, the Cbk1 activator, undergoes a Cdc28-dependent differential phosphorylation on hyphal induction. Mutations in the four CDK consensus sites in Mob2 to Ala significantly impaired hyphal development. The mutant cells produced short hyphae with enlarged tips that displayed an illicit activation of cell separation. We also show that Cdc28 phosphorylation of Mob2 is essential for the maintenance of polarisome components at hyphal tips but not at bud tips during yeast growth. Thus we have found a novel signaling pathway by which Cdc28 controls Cbk1 through the regulatory phosphorylation of Mob2, which is crucial for normal hyphal development.
INTRODUCTION
The ability to modify cell morphology in response to external cues is a fundamental property of cell development. The dynamic modification of cell architecture is the result of signal transduction mechanisms that reorganize the cell polarity machinery in response to such cues. Members of the evolutionarily conserved nuclear Dbf2-related (NDR) kinase family are essential components of signaling pathways involved in cell morphogenesis from yeast to humans (Hergovich et al., 2006). These kinases require interaction with regulatory Mob proteins for their activity and function (Komarnitsky et al., 1998; Hou et al., 2000, 2003; Colman-Lerner et al., 2001; Weiss et al., 2002; Hergovich et al., 2005). A well-conserved trait of these signaling pathways is the existence of a group of proteins that interact physically and functionally and together constitute the kinase core of the pathway (Sudol and Harvey, 2010). This group of proteins is formed by a scaffolding protein, a Ste20-like kinase, an NDR kinase, and its Mob coactivator. In metazoans, the Wts/LATS NDR kinase and its coactivator Mats/MOB are members of the Hippo pathway, a tumor suppressor pathway that restricts cell growth and proliferation and promotes apoptosis (Tapon et al., 2002; Harvey and Tapon, 2007). In Drosophila melanogaster, the tricornered NDR kinase is essential for neuronal morphogenesis and wing-hair development (Geng et al., 2000; Emoto et al., 2004; He et al., 2005). The tricornered homologue in Caenorhabditis elegans, designated SAX-1, is also important in dendritic outgrowth (Gallegos and Bargmann, 2004). In fungi, the most closely related NDR kinases to the Drosophila Tricornered kinase involved in the control of polarized growth are Cbk1 from Saccharomyces cerevisiae, COT1 from Neurospora crassa, and Orb6 from Schizosaccharomyces pombe (Yarden et al., 1992; Verde et al., 1998; Racki et al., 2000).
In S. cerevisiae, the regulation of Ace2 and morphogenesis (RAM) network is a signaling pathway that controls cell separation and polarized growth. The kinase core of the RAM pathway is constituted by the scaffolding protein Tao3, the Ste20-like kinase Kic1, and the complex formed by the NDR kinase Cbk1 and its coactivator Mob2 (Nelson et al., 2003). All of these proteins localize to the cortical sites of growth during budding and mating (Racki et al., 2000; Bidlingmaier et al., 2001; Colman-Lerner et al., 2001; Weiss et al., 2002; Nelson et al., 2003). In addition to cell cortex localization, the Cbk1/Mob2 complex also localizes to the daughter cell nucleus at the M/G1 transition (Colman-Lerner et al., 2001; Weiss et al., 2002). Cells lacking any of these proteins exhibit two independent phenotypes: a cell separation defect and poor maintenance of polarized growth. The failure in cell separation is due to the loss of Ace2-dependent transcription (Bidlingmaier et al., 2001; Colman-Lerner et al., 2001). The Ace2 transcription factor specifically localizes to the daughter nucleus in late M and G1, where it directs the expression of daughter-specific genes, such as ENG1, CTS1, and SCW11, all of them involved in the separation of daughter and mother cells after cytokinesis (Colman-Lerner et al., 2001; Baladrón et al., 2002; Weiss et al., 2002). Recently, it has been shown that Cbk1 directs the daughter-specific nuclear accumulation of Ace2 by blocking its export from the daughter cell nucleus (Mazanka et al., 2008). The role of the RAM pathway in the regulation of polarized growth is poorly understood. Recent evidence suggests that Cbk1 regulates polarized growth by controlling Golgi- and Sec2/Sec4-dependent processes (Kurischko et al., 2008). In fission yeast, Orb6 controls polarized cell growth by spatial regulation of the small GTPase Cdc42 (Das et al., 2009).
The nuclear localization and activity of the Cbk1/Mob2 complex are dependent on all known RAM proteins, suggesting that it functions downstream from the other network components (Nelson et al., 2003). Like other NDR kinases, Cbk1 kinase activity is regulated by phosphorylation at multiple conserved residues. The phosphorylation of a highly conserved C-terminal hydrophobic motif (known as HM site or CT motif) is essential for Cbk1 function and requires all of the RAM proteins (Jansen et al., 2006). Recent work has identified Cbk1 as a basophilic kinase with a strong preference for histidine at position −5 and the putative recognition site H-X-[K/R]-[K/R]-X-[S/T] (Mazanka et al., 2008). This substrate specificity is very well conserved in other NDR/LATS kinases. Drosophila Warts/Lats and human LATS1 kinases phosphorylate the transcriptional coactivator Yorkie/YAP at sequences that match the Cbk1 consensus motif (Dong et al., 2007; Zhao et al., 2007; Hao et al., 2008).
Candida albicans is able to switch from unicellular yeast to multicellular filamentous growth forms in response to environmental signals (Biswas et al., 2007), providing a framework for understanding how cell morphology is modified in response to external stimuli. Hyphal growth is characterized by robust polarized growth at cell tips and by inhibition of cell separation after cytokinesis. In C. albicans, cell separation is probably regulated by a mechanism similar to that present in S. cerevisiae. Deletion of ACE2 or any of the RAM genes results in the impairment of cell separation (Kelly et al., 2004; Clemente-Blanco et al., 2006; Song et al., 2008). In addition, in both yeasts Ace2 localization to the daughter cell nucleus depends on the protein phosphatase Cdc14 (Weiss et al., 2002; Clemente-Blanco et al., 2006).
Because polarized growth and cell separation, the two major outputs of the RAM pathway, are differentially regulated in yeast and hyphae, we wanted to investigate whether the developmental signals that induce hyphal growth modulate the function of Cbk1/Mob2, the major downstream component of the RAM network.
RESULTS
Cbk1 and Mob2 localize to sites of polarized growth
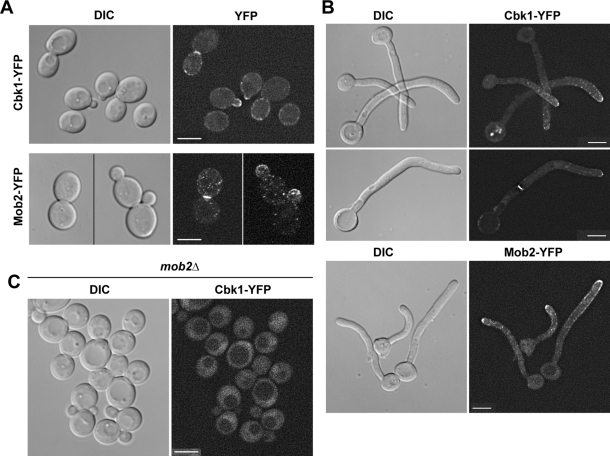
To investigate the localization of Cbk1 and Mob2 during C. albicans yeast and hyphal growth, strains expressing functional Cbk1–yellow fluorescent protein (YFP) and Mob2-YFP alleles under the control of their native promoters were constructed. In C. albicans yeast cells Cbk1-YFP and Mob2-YFP localized to the cell cortex of small- and medium-budded cells and to the bud neck in large-budded cells (Figure 1A), as observed previously in S. cerevisiae (Bidlingmaier et al., 2001; Weiss et al., 2002). During hyphal growth, Cbk1-YFP and Mob2-YFP were present at the tip of hyphae and in the region of the septum (Figure 1B). Therefore Cbk1 and Mob2 localize to sites of polarized growth in both yeast and hyphal cells. As expected, Mob2 was essential for Cbk1 localization to regions of polarized growth (Figure 1C).
FIGURE 1:
Cbk1 and Mob2 localize to sites of polarized growth. The JC369 (CBK1-YFP/CBK1) and JC871 (MOB2-YFP/MOB2) strains were grown as yeast (A) or hyphae (B) for 2 h and visualized using a Deltavision microscope. (C) mob2Δ cells expressing Cbk1-YFP (JC524) were grown to midlogarithmic phase and analyzed on a Nikon microscope. Differential interference contrast (DIC) and YFP images are shown. Throughout the figures the scale bars represent 5 μm, unless indicated otherwise.
Mob2 undergoes growth mode–dependent phosphorylation
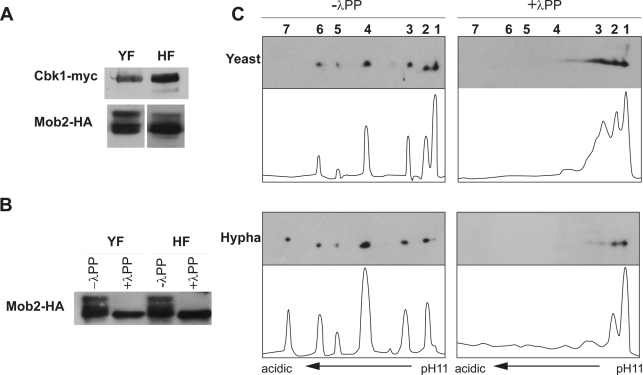
To determine whether Cbk1 and Mob2 were regulated in a growth-dependent manner, a strain expressing functional Cbk1- and Mob2-tagged alleles (CBK1-myc MOB2-HA) was constructed to analyze the proteins by Western blotting. Whereas Cbk1 migrated as a single band with no evident modifications in either the yeast or hyphal extracts, a fraction of Mob2 exhibited reduced electrophoretic mobility (Figure 2A). The treatment of the extracts with λ-phosphatase eliminated the slow-migrating forms (Figure 2B), indicating that they were different phosphorylated isoforms. To analyze these phosphorylation patterns in greater detail, we used two-dimensional (2D) gel electrophoresis followed by Western blotting (2D-WB). On the basis of this approach, different Mob2 isoforms were clearly distinguished. In yeast cells, six Mob2 spots were detected (Figure 2C, 1–6, from the basic to the acidic end). In hyphal extracts, the most basic spot disappeared and a new spot was observed (spot 7) at the acidic end. In addition, the relative abundance of the different isoforms also varied from yeast to hyphae, isoform 4 being the most abundant in hyphae. Spots 4–7 correspond to different phospho-isoforms, since treatment with λ-phosphatase abolished them (Figure 2C). Therefore Mob2 undergoes a growth mode–dependent phosphorylation.
FIGURE 2:
Mob2 undergoes growth mode–dependent phosphorylation. (A) Asynchronous cultures of a CBK1/CBK1-myc MOB2/MOB2-HA strain (JC413) were grown under yeast-inducing (YF) or hypha-inducing conditions (HF) for 2 h. Protein extracts were analyzed by Western blot using anti-myc or anti-HA antibodies. (B) A fraction from the same lysates was pretreated with λ-phosphatase (λPP). (C) Cell lysates from strain JC413 obtained in yeast- and hypha-inducing conditions were subjected to 2D-WB and probed with anti-HA. Half of each lysate was pretreated with λ-phosphatase. Histograms obtained using ImageJ show the quantification of the relative intensity of each spot of the blot.
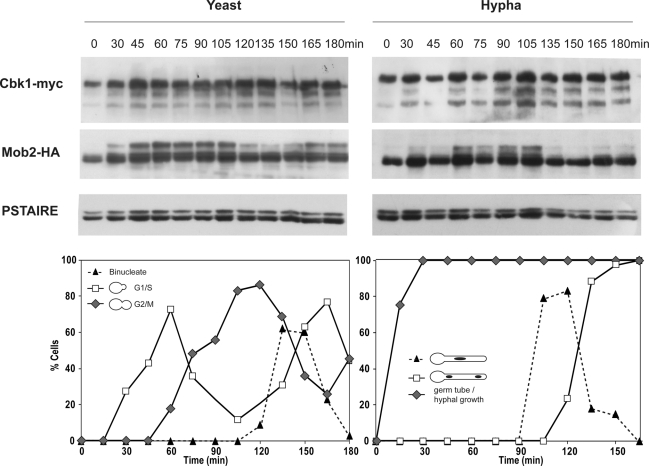
To investigate whether Mob2 phosphorylation was cell cycle regulated, yeast cells carrying Cbk1-myc and Mob2-HA were synchronized in G1 by elutriation (Figure 3). In unbudded G1 cells (time 0), Mob2 was seen as a single band in SDS–PAGE. The phosphorylated forms started to appear during bud emergence, and this pattern was maintained until the G2/M transition. A decrease in the slower-migrating forms was correlated with the appearance of binucleate cells (120 min), suggesting that Mob2 is dephosphorylated at the end of mitosis. In hyphae, the accumulation of Mob2 phosphorylated forms was delayed in comparison to yeast cells, but once the phosphorylated forms were present they persisted until the first mitosis took place within the germ tube (135 min), suggesting that dephosphorylation also occurs at the end of mitosis, as in yeast cells. In contrast to these results, no evident modifications were observed for Cbk1 during the cell cycle of either yeast or hyphal cells. Taken together, these results show that Mob2 is phosphorylated in a cell cycle–dependent manner in both types of growth.
FIGURE 3:
Mob2 phosphorylation is cell cycle regulated. Small G1 cells carrying Cbk1-myc and Mob2-HA (JC413) were isolated by elutriation and released into YPD at 30°C (yeast growth) or YPD plus 10% serum at 37°C (hyphal growth). Samples were collected at 15-min intervals after release and assayed by Western blot with anti-myc or anti-HA antibodies. Anti-PSTAIRE antibodies were used as loading control. DAPI staining was used as a marker of cell cycle progression.
Mob2 phosphorylation is dependent on cyclin-dependent kinase activity
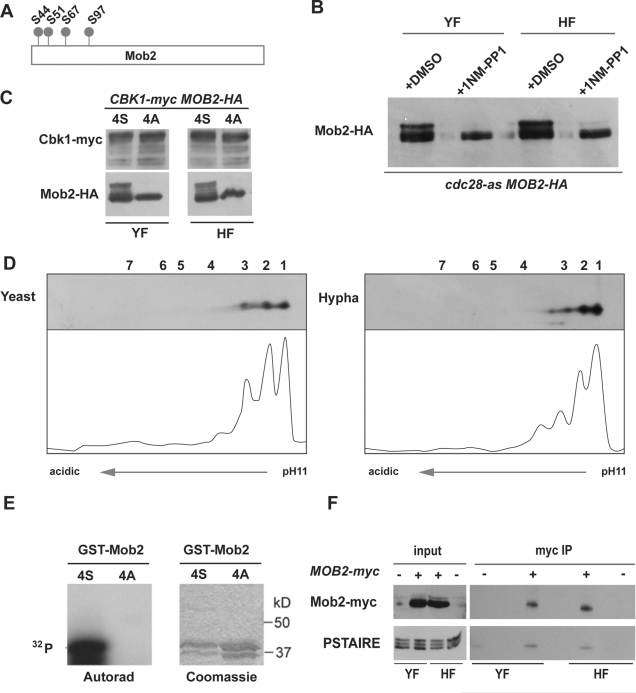
The Mob2 protein contains four putative cyclin-dependent kinase (CDK) consensus phosphorylation sites (Figure 4A; S/T-P-X-K/R), suggesting that Mob2 could be a CDK1 substrate. To explore this possibility, Mob2 was tagged with hemagglutinin (HA) in a strain containing the cdc28-as allele, a mutated form of Cdc28 that is sensitive to the ATP analogue 1NM-PP1 (Bishop et al., 2000; Sinha et al., 2007). In contrast to untreated cells, the addition of 1NM-PP1 resulted in a dramatic decrease of the slower-migrating forms in both yeast and hyphae (Figure 4B), suggesting that Mob2 phosphorylation is Cdc28 dependent, although we cannot rule out the possibility that this could have been an indirect effect due to a cell cycle arrest imposed by the inactivation of Cdc28.
FIGURE 4:
Mob2 phosphorylation depends on CDK activity. (A) Schematic representation of CDK consensus phosphorylation sites in Mob2. (B) JC877 (MOB2/MOB2-HA cdc28-as/cdc28Δ) cells were grown as yeast or hyphae in the presence of 25 μM 1NM-PP1 or DMSO for 30 min. Protein extracts were probed with anti-HA antibodies. (C) Cells from the MOB2-HA/mob2Δ (wt, JC413) and mob2-4A-HA/mob2Δ (JC964) strains expressing Cbk1-myc were grown under yeast- and hypha-inducing conditions. Protein extracts were probed with anti-myc or anti-HA antibodies. (D) Lysates of JC785 (mob2-4A-HA/mob2Δ) were subjected to 2D-WB and probed with anti-HA antibodies. (E) In vitro kinase assays of purified Cdc28 complex using GST-Mob21-115-4S or GST-Mob21-115-4A as substrate. (F) Protein extracts from yeast and hyphal cultures of a MOB2/MOB2-myc strain (JC482) were immunoprecipitated using anti-myc antibodies. Samples separated by SDS–PAGE were analyzed with anti-HA or anti-PSTAIRE antibodies. An untagged BWP17 strain was used as a negative control of immunoprecipitation.
To further investigate the role of Cdc28 in Mob2 phosphorylation, the four putative phosphoacceptor Ser residues were changed to Ala, creating the quadruple mutant mob2S44A/S51A/S67A/S97A allele (hereafter referred to as mob2-4A) tagged with HA. The mutant allele was used to replace the wild-type (WT) copy of a heterozygous MOB2/mob2Δ strain, resulting in a strain containing mob2-4A-HA under the control of its own promoter as the sole source of Mob2 in the cell. Mob2 protein levels were similar in the MOB2-4S (WT) and mob2-4A strains (Figure 4C). When cell extracts from the mob2-4A strain were analyzed by 2D-WB, only spots 1–3 were present in the yeast and hyphal extracts (Figure 4D), indicating that isoforms 4–7 depend on the CDK phosphorylation sites. Together the data suggest that the Mob2 CDK sites are phosphorylated in vivo in yeast and hyphal cells.
To determine whether Cdc28 was able to phosphorylate Mob2 directly, we performed an in vitro kinase assay using as substrate a Mob2 N-terminal fragment expressed as a GST fusion in Escherichia coli with or without the 4 CDK consensus sites (GST-Mob21-115-4S and GST-Mob21-115-4A, respectively). Figure 4E shows that immunoprecipitated Cdc28 was able to phosphorylate GST-Mob21-115-4S, whereas no detectable phosphorylation was observed for GST-Mob21-115-4A. Thus Mob2 is an in vitro substrate for Cdc28.
To obtain further evidence that Mob2 was a Cdc28 substrate in vivo, we performed coimmunoprecipitation experiments to test whether a physical interaction between Mob2 and Cdc28 occurred. Cell extracts from yeast and hyphal cultures of Mob2-myc or control untagged Mob2 strains were immunoprecipitated using anti-myc antibody, and then anti-PSTAIRE antibody was used to detect the presence of Cdc28 in the precipitates. Cdc28 was indeed enriched in the Mob2-myc immunoprecipitates (Figure 4F). Thus the foregoing results strongly suggest that Cdc28 regulates Mob2 in vivo.
The Mob2 CDK phosphorylation sites are required for apical hyphal growth
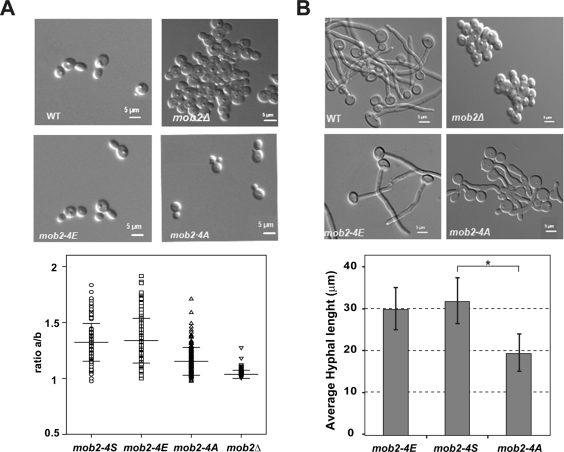
To determine the biological significance of these CDK phosphorylation sites, the phenotype of the mob2-4A mutants was analyzed during yeast and hyphal growth. We also constructed a phosphomimetic version of Mob2 by replacing the four Ser residues with Glu (the mob2S44E/S51E/S67E/S97E allele, designated mob2-4E). Similar to the mob2-4A strain, the mob2-4E allele was integrated in a MOB2/mob2Δ background, resulting in a strain in which it was the sole source of Mob2. It has been reported that mob2Δ S. cerevisiae cells have two independent phenotypes (Bidlingmaier et al., 2001; Colman-Lerner et al., 2001; Weiss et al., 2002). The cells are round as a consequence of the loss of polarity, and they also have cell separation defects, producing clumps of cells. In contrast to the Camob2Δ strain, yeast cells expressing Mob2-4A or Mob2-4E were able to separate after cytokinesis, indicating that the four CDK phosphorylation sites were not essential for cell separation (Figure 5A). However, cells from the mob2-4A strain were slightly rounder than those of the wild-type and mob2-4E strains. To quantify this phenotype, the length/width ratio (axial ratio) was determined by measuring 150 cells from each strain in three independent experiments. This ratio is a measure of the roundness of a cell. Wild-type and mob2-4E cells were oval shaped, with an average mean of 1.32 ± 0.17 and 1.33 ± 0.2, respectively, whereas cells from the mob2Δ strain were round, and hence their axial ratio was close to 1 (1.03 ± 0.03). In mob2-4A cells, the average axial ratio was 1.15 ± 0.12, that is, an intermediate phenotype between the null mutant and the wild-type strain. Thus phosphorylation of Mob2 CDK sites produces mild effects on polarized growth in yeast cells.
FIGURE 5:
Mob2 CDK consensus phosphorylation sites are required for hyphal growth. DIC images of wild-type (MOB2-4S-HA/mob2Δ, JC613), mob2-4E-HA/mob2Δ (JC620), mob2Δ (JC502), and mob2-4A-HA/mob2Δ (JC785) strains grown under yeast-inducing (A) or hypha-inducing conditions (B). (A) The graph represents the axial ratio (length/width) of yeast cells of the indicated strains. Error bars are SD (n = 150 cells). (B) The graph represents the average hyphal length of the indicated strains, and the error bars indicate SD (n = 30). The pairwise t test is shown above the bracket (*p < 0.0001).
We next examined the morphology of the mob2 mutants during hyphal growth. As expected, mob2Δ cells were unable to polarize their growth in response to hypha-inducing conditions (Song et al., 2008), whereas the mob2-4A and mob2-4E strains produced germ tubes at the same rate as the wild-type cells. Of interest, with prolonged incubation, the germ tubes of the Mob2-4A cells were significantly shorter and wider than those of the wild-type and mob2-4E cells (Figure 5B). The replacement of the mob2-4A allele by the wild-type MOB2 (mob2-4A/S-HA) rescued the phenotype, indicating that the loss of polarity was specifically due to a modification of the Mob2 CDK sites (Supplemental Figure S1).
Phosphorylation of Mob2 CDK sites is required for the maintenance of polarisome components in hyphae
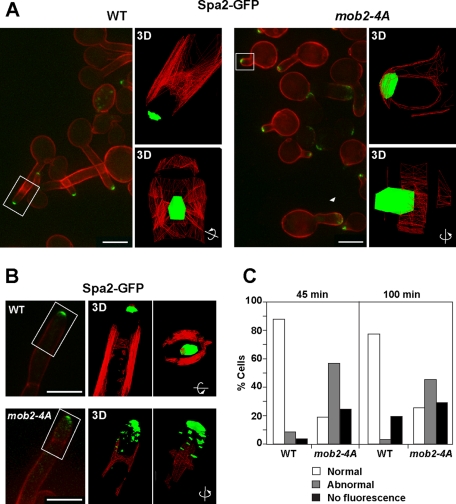
In C. albicans, Spa2 is a component of the polarisome complex that localizes to the hyphal tip in a cell cycle–independent manner (Zheng et al., 2003; Crampin et al., 2005). Deletion of SPA2 results in wider and shorter germ tubes than those of the wild-type strain (Zheng et al., 2003), a phenotype similar to that of mob2-4A. Thus we analyzed whether Mob2 played any role in regulating Spa2 localization in hyphae. In wild-type germ tubes, Spa2 concentrated as a bright spot at the center of the growing tip 45 min after hyphal induction (Figure 6A). In contrast, in mob2-4A cells Spa2 had an abnormal localization in more than 50% of the hyphae (Figure 6C). This abnormal distribution included hyphae in which the Spa2 spot was off center and localized to the sides of the tip (Figure 6A, rectangle) or was not concentrated at the tip (Figure 6A, arrowhead). In addition, there were some hyphae in which Spa2 fluorescence could not be detected. three-dimensional (3D) models of Spa2 localization in wild-type and mutant hyphae are shown in Figure 6A. It can be seen that in wild-type hyphae, Spa2 was organized as a single spot at the middle of the apex, whereas in the mob2-4A hyphae the main spot was wider and was localized laterally. By 100 min, the apical and centered localization of the Spa2–green fluorescent protein (GFP) signal was perfectly maintained in the wild-type filaments (Figure 6B), whereas in the mob2-4A hyphae the same defects were observed: hyphae with Spa2 fluorescence off center or diffuse over the tip and hyphae without the Spa2 signal (Figure 6C). Again, 3D model reconstructions showed that in wild-type hyphae Spa2 remained perfectly organized as a single spot at the middle of the apex, whereas it was spread laterally in many small dots in Mob2-4A hyphae. Because no differences in Spa2 localization were observed in yeast cells (Supplemental Figure S2A), these results indicate that CDK1 phosphorylation sites in Mob2 are required for the proper maintenance of the spatial organization of Spa2 in hyphae.
FIGURE 6:
Localization of the polarisome component Spa2 is defective in the mob2-4A mutant. Localization of Spa2-GFP in wild-type (JC961) and mob2-4A (JC921) cells 45 min (A) and 100 min (B) after hypha induction. Spa2-GFP is shown in green, whereas calcofluor is shown in red. To the right of the images, 3D models generated using softWoRx from the regions indicated by rectangles are shown. The fluorescent signal is shown in green, and calcofluor is represented as a red wire frame. The second frame of each 3D model was generated by rotating the first image 90º on the indicated axis. (C) Quantification of the localization of Spa2-GFP at the cell tip from the experiment shown in A and B (n ≥ 80).
To investigate whether Cbk1/Mob2 phosphorylated Spa2 directly, we performed in vitro kinase assays. The C. albicans Spa2 is 1416 amino acids in length and contains a cluster of four potential Cbk1 phosphorylation sites (S121, S143, S153, and S163) in the proximity of the Spa2 direct-repeat elements (Roemer et al., 1998; Zheng et al., 2003) (Supplemental Figure S3A). As a substrate for Cbk1 kinase assays, we constructed a tandem of three copies of the 25–amino acid peptide corresponding to amino acids 138–163 of Spa2, which was expressed as a GST fusion in E. coli (GST-3xSpa2138–163). We performed the in vitro kinase reactions from hyphal cells because CDK phosphorylation of Mob2 is essential for Spa2 localization in hyphal forms but not in yeast cells. Anti-HA immunoprecipitates from MOB2-4S and mob2-4A strains, but not mock immunoprecipitates, were able to phosphorylate GST-3xSpa2138-163 (Figure S3B). Substitution of putative Cbk1 phosphoacceptor residues to Ala abolished the 32P labeling of GST-3xSpa2. Thus, Spa2 is an in vitro target of Mob2-associated kinase activity.
To study whether this phenotype was specific for Spa2, the localization of another putative cell end marker was analyzed in mob2-4A hyphae. In Aspergillus nidulans, TeaA is a Kelch-domain protein that concentrates as a single dot at the center of the hyphal tip and whose deletion results in a zigzag growing phenotype (Takeshita et al., 2008). TeaA is an orthologue of S. cerevisiae Kel1 (Philips and Herskowitz, 1998), and in C. albicans, the Candida Genome Database (http://www.candidagenome.org/) identifies orf19.6092 as the putative CaKEL1 gene. To analyze Kel1 localization, a strain expressing a functional Kel1-YFP was constructed. Similar to what has been described in S. cerevisiae (Philips and Herskowitz, 1998), Kel1 localized to the cell cortex of small- and medium-budded cells and at the bud neck in large-budded cells (Supplemental Figure S2B). In hyphae, Kel1 localized as a single dot at the tip of the hypha in a pattern similar to that of Spa2 (Supplemental Figure S2C). Therefore the Kelch-domain protein Kel1 accumulates at sites of polarized growth in C. albicans yeast and hyphal cells. Examination of cells expressing Kel1-YFP and Spa2-CFP revealed that both proteins colocalized in yeast cells and filaments, suggesting that Kel1 might be a new component of the polarisome. As described earlier for Spa2, the maintenance of Kel1 at the hyphal tip was dependent on Mob2 CDK phosphorylation sites, since Kel1 was disorganized in the filaments of cells expressing Mob2–4A after 120 min of incubation in hypha-inducing conditions (Supplemental Figure S2D). Thus these results indicate that phosphorylation of Mob2 CDK sites is required for the maintenance of the distinctive organization of some components of the polarisome during hyphal growth.
Characterization of the mob2-4A mutant
The actin cytoskeleton is important for polarized growth (Pruyne et al., 2004). The fact that mob2-4A cells produced shorter hyphae could be an indication that they were not able to properly organize cortical actin patches at the hyphal tip. To explore this possibility, wild-type and mob2-4A hyphae were stained with Alexa-phalloidin to visualize the actin cytoskeleton. Both strains showed a similar pattern, in which cortical actin patches were highly polarized at the hyphal tips (Supplemental Figure S4B). These results therefore suggest that CDK phosphorylation sites are not essential for actin cytoskeleton polarization during hyphal growth. Next we wondered whether Mob2-4A could localize to sites of polarized growth. Fluorescence microscopy of yeast and hyphae showed that the Ser-to-Ala substitutions did not alter Mob2 localization (Supplemental Figure S4A). Accordingly, we studied whether the Cbk1/Mob2 interaction was affected by the presence of the Mob2-4A allele by coimmunoprecipitation experiments, but no defects in the interaction were found (Supplemental Figure S4C). Thus elimination of the Mob2 CDK sites does not affect actin cytoskeleton organization, the localization of Mob2, or the interaction between Cbk1 and Mob2.
Phosphorylation of Mob2 CDK sites is required for inhibition of cell separation in hyphae
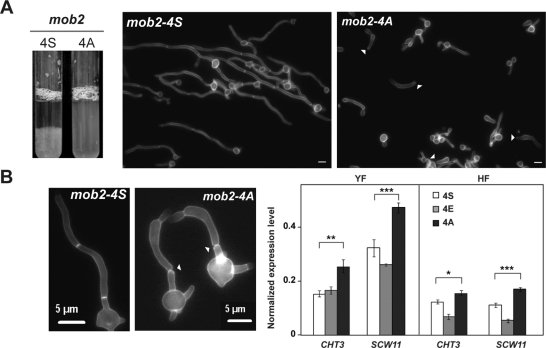
An essential feature of hyphal growth is the inhibition of cell separation, which allows the formation of chains of elongated cells. After 5 h of hyphal growth, wild-type cells flocculated very easily, whereas the mob2-4A cells remained in suspension (Figure 7A), with a mixture of short hyphae and elongated individual cells. The aspect of such cells suggested that cell separation had been activated. To examine this possibility, we determined the percentage of hyphae in which the separation of cell compartments at the first septum was observed. In contrast to the wild-type hyphae, where no cell separation was observed, 20% of the mob2-4A hyphae showed cell separation at the first septum (n = 200) at 3 h after induction (Figure 7B). These results thus suggest that the CDK phosphorylation sites in Mob2 are important for inhibiting cell separation during hyphal growth.
FIGURE 7:
mob24A hyphae activate cell separation. (A) Flocculation of JC613 and JC785 hyphal cultures after 5 h of serum induction. Cells from these cultures were recovered 3 h after induction and stained with calcofluor white (A, B). Arrowheads, separated hyphal compartments. (B) The graph on the right shows the expression of Ace2-target genes CHT3 and SCW11 measured by quantitative RT-PCR in the wild-type, mob24E, and mob24A strains normalized using ADE2. The data are means of two independent experiments, and the error bars indicate standard deviations. The results of pairwise t tests are shown above the brackets: ***p < 0.0001, **p = 0.0005, *p = 0.0054.
Cell separation depends on the Ace2 transcription factor (Kelly et al., 2004; Clemente-Blanco et al., 2006). During hyphal growth, cell chain formation is achieved by the down-regulation of Ace2-target genes through CDK phosphorylation of the developmental regulator Efg1 (Wang et al., 2009). To assess the effect of Mob2 CDK sites on the expression of Ace2 target genes, the levels of CHT3 and SCW11 transcripts were measured by RT-qPCR in the wild-type, mob2-4E, and mob2-4A strains. Whereas high levels of CHT3 and SCW11 transcripts were observed in the mob2-4A mutant in both yeast and hyphae in comparison to the control MOB2-4S (Figure 7B), the levels in the mob2-4E strain, especially those of SCW11 transcripts, were lower. Because repression of CHT3 and SCW11 transcripts was detected in mob2-4A and mob2-4E hyphae, CDK phosphorylation of Mob2 might play a role in the Cbk1/Mob2-dependent activation of Ace2-target genes but not in the hypha-specific repression. Therefore mob2-4A hyphae activated an illicit cell separation but did not display a severe increase in the expression level of CHT3 and SCW11.
DISCUSSION
Our results demonstrate that CDK-dependent phosphorylation of Mob2 is required to maintain two essential traits of hyphal development in C. albicans: continuous polarized apical growth and cell chain formation.
Mob2 is phosphorylated by the CDK Cdc28
In S. cerevisiae, the Cbk1/Mob2 complex is regulated by the phosphorylation of two highly conserved residues present in Cbk1 (Jansen et al., 2006). In C. albicans, we did not observe evidence of Cbk1 phosphorylation, although the presence of these highly conserved residues suggests that it is probably regulated by a similar mechanism(s). However, we did find that CaMob2 is a phosphoprotein whose phosphorylation responds to external and internal inputs. Our data provide strong evidence that the CDK Cdc28 regulates Mob2 function by direct phosphorylation of a cluster of four CDK consensus sites at its N-terminus (S44, S51, S67, and S97). Of interest, S44, S51 and S67 were identified as phosphoserines in a large-scale study of the C. albicans phosphoproteome (Beltrao et al., 2009). In S. cerevisiae, CDK phosphorylation of Mob1 regulates the activity of the Dbf2/Mob1 complex in mitosis (Konig et al., 2010). In metazoans, the phosphorylation of MOB/Mats proteins increases their binding to the NDR/LATS1 kinases (Wei et al., 2007; Praskova et al., 2008). In C. albicans, we observed that the Mob2 phosphorylation status did not affect its binding capacity to Cbk1, its localization to sites of polarized growth, or its in vitro kinase activity. One interesting possibility is that the differential phosphorylation of Mob2 could modify the interaction of the complex with its substrates. In S. cerevisiae, the transcription factor Ace2 is a target of Cbk1, and phosphorylation of Cbk1 at its CT motif is necessary for the formation of a productive complex between the kinase and its substrate (Jansen et al., 2006; Mazanka et al., 2008).
CDK phosphorylation of Mob2 is required for maintenance of apical hyphal growth
During the yeast-to-hyphal transition, different components of the polarization machinery undergo a dramatic spatial reorganization in order to sustain the extreme polarized growth of the hyphae (Jones and Sudbery, 2010). In this study, we have shown that CDK phosphorylation of Mob2 is essential for the maintenance of the polarisome architecture at the hyphal tip but not in yeast cells.
Our results suggest that the onset and maintenance of hyphal growth require different Cbk1/Mob2 complexes that could be regulated by differential phosphorylation of the Mob2 subunit. This is consistent with the observation that Mob2 undergoes a growth mode–dependent phosphorylation. It is therefore possible that temporally controlled Mob2 phosphorylation might regulate the interaction of Cbk1/Mob2 with different targets.
In agreement with previous reports (McNemar and Fonzi, 2002; Song et al., 2008), our results suggest that Cbk1/Mob2 could be involved in cell surface remodeling during polarized growth in C. albicans. In good correlation with this function, Cbk1 and Mob2 localized to sites of active growth. However, the relevant targets in the cell cortex are unknown. In this report, we demonstrate that Spa2 and Kel1 were disorganized in mob2-4A hyphae, suggesting that unphosphorylated Mob2 could impair the interaction between Cbk1/Mob2 with Spa2 and/or Kel1. The search for putative Cbk1 phosphorylation sites in these proteins revealed that Kel1 (1018 amino acids) has no sites, whereas Spa2 (1416 amino acids) contains a cluster of four sites (S121, S143, S153, and S163) that can be phosphorylated in vitro by Cbk1/Mob2. These sites are located in a disordered region, absent in S. cerevisiae, immediately after the Spa2 direct repeat elements, which in S. cerevisiae interact with components of the MAPK pathways that are important for polarized growth (Roemer et al., 1998; Sheu et al., 1998; Zheng et al., 2003). In addition, S163 is phosphorylated in C. albicans (Beltrao et al., 2009). Together these data suggest that Spa2 could be a direct target of the Cbk1/Mob2 complex. The abnormal Kel1 localization in mob2-4A hyphae could be an indirect effect of the impaired Spa2 function. Although we have not shown that Kel1 localization depends on Spa2, both proteins colocalized to sites of polarized growth. Because the gain or loss of phosphorylation sites in rapidly evolving regions could facilitate the evolution of kinase signaling circuits (Holt et al., 2009), it is possible that the appearance of these Cbk1 phosphorylation sites in the scaffolding protein CaSpa2 has been an adaptation to modify polarisome function in response to the developmental signals that promote hyperpolarized growth.
CDK phosphorylation of Mob2 prevents cell separation during hyphal growth
We show that Mob2-4A increases the expression of Ace2-target genes in yeast cells, suggesting that CDK phosphorylation of Mob2 inhibits the Cbk1-dependent activation of Ace2. If the inhibitory effect of the CDK on the Cbk1/Mob2 complex were solely dependent on the phosphorylation of Mob2, then the mob2-4E allele should show a defect in cell separation. However, the transcript levels of CHT3 and SCW11 were slightly reduced and cell separation was not impaired in mob2-4E cells, suggesting an additional level of regulation. CaCbk1 also contains six potential CDK sites. Therefore the absence of phenotype of mob2-4E could be explained if the negative regulation by CDK were executed by the phosphorylation of both Cbk1 and Mob2. This is consistent with recent work carried out in S. cerevisiae indicating that CDK phosphorylation of Cbk1 inhibits cell separation (Brace et al., 2011). However, those authors did not analyze the physiological relevance of the CDK sites present in Mob2. Taken together, we believe that the CDK-driven control of both components of the Cbk1/Mob2 complex could represent a general mechanism to regulate the NDR kinase in these two diverged yeasts.
During hyphal growth, cell chain formation is achieved by the down-regulation of Ace2-target genes through CDK phosphorylation of the developmental regulator Efg1 (Wang et al., 2009). We show that mob2-4A cells, like the wild type, are able to down-regulate Ace2-target genes in response to hypha-inducing conditions, which indicates that the Efg1 regulation of Ace2-target genes might not be affected in mob2-4A cells. However, although cell separation was inhibited in wild-type hyphae, it was activated in mob2-4A cells. For the mother–daughter separation to occur, polarized exocytosis of cell wall hydrolases to the septum region is required. Septins constitute a group of GTP-binding proteins involved in cytokinesis and other essential cellular functions (Weirich et al., 2008; González-Novo et al., 2009; Gladfelter, 2010). As in S. cerevisiae, septins play a role in guiding exocytosis in C. albicans (Gladfelter et al., 2005; Li et al., 2007). It has been shown that hyphal development has a Sec3-dependent mechanism that restricts exocytosis to the tip after the first septin ring has formed within the germ tube, and it has been suggested that whereas septin rings located at the bud neck of yeast cells are able to target secretion to the bud base, those located at the division plate in hyphae fail to attract exocytosis through an unknown mechanism (Li et al., 2007; Sudbery, 2007). Furthermore, septins are required to prevent the activation of cell separation in hyphae (González-Novo et al., 2008). Therefore two possible scenarios could explain the cell separation phenotype of mob2-4A hyphae. First, the aberrant polarisome architecture of mob2-4A hyphae could impair the Sec3-dependent mechanism that eliminates the intrinsic ability of septin rings to attract exocytosis, allowing the delivery of the hydrolases to the septum. However, deletion of CaSPA2 does not activate cell separation in hyphae (Zheng et al., 2003). Alternatively, the pool of Cbk1/Mob2 located at the septum could be required to modify the septin ring, changing its ability to attract polarized secretion. Indeed, Cdc12 contains a putative Cbk1 site (S64) close to the threonine (T68) that is essential for GTP binding and hydrolysis in mammalian Sept2 (Sirajuddin et al., 2007, 2009). In S. cerevisiae, this residue is conserved in septins Cdc10 (T74) and Cdc12 (T75), and its change to alanine causes a temperature-sensitive growth defect (Sirajuddin et al., 2009), demonstrating its importance in septin function. Because the GTP binding region plays a structural role in septin function, future experiments will address whether Cbk1/Mob2 regulates Cdc12 function through the phosphorylation of S64.
MATERIALS AND METHODS
Strains and culture conditions
The strains used in this study are listed in Supplemental Table 1. C. albicans was grown at 30ºC in YPD medium (2% Bacto Peptone, 2% dextrose, 1% yeast extract) or in MM (2% dextrose, 0.67% YNB) supplemented with amino acids if required. For hyphal growth, 10% serum was added to liquid medium, and cells were incubated at 37ºC. G1 cells were synchronized by elutriation as described (Clemente-Blanco et al., 2006).
Strain construction
Disruption and epitope-tagged strains were made according to the PCR-mediated system using pFA plasmids (Gola et al., 2003; Schaub et al., 2006). Strains were confirmed by PCR. Oligonucleotides were obtained from biomers.net (Ulm, Germany) and are listed in Supplemental Table 2. The functionality of the tagged alleles was tested in a heterozygous (+/D) background. Mob2 point mutants were generated by PCR and cloned into pFA-URA3 with the 3′UTR-MOB2 region (+441 to +973 from the stop codon) to direct the integration of the plasmid in the MOB2 locus. The construction was transformed into a MOB2 heterozygous strain and analyzed by PCR and sequencing.
Microscopy
Actin staining was performed using Alexa Fluor 488 phalloidin (Invitrogen, Carlsbad, CA) using standard procedures (Marks and Hyams, 1985). Fluorescence microscopy was performed with a Nikon Eclipse i90 microscope controlled with MetaMorph (Molecular Devices, Downingtown, PA) or with a Personal Deltavision microscope running softWoRx (Applied Precision Instruments, Seattle, WA). Z-stack images were collected with step sizes of 0.3 μm and deconvoluted using softWoRx. Except where stated otherwise, images are projections of the deconvoluted Z-stacks. Three-dimensional model building was done using the softWoRx modeling module. To facilitate GFP visualization, calcofluor fluorescence is represented as a wire frame.
Protein extracts, immunoprecipitation, and Western blot
Protein extracts were prepared from 1.6 × 108 cells resuspended in 50 μl of lysis buffer (150 mM NaCl, 50 mM, Tris pH 7.4, 15 mM ethylene glycol tetraacetic acid, 1% Triton 100-X, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 60 mM β-glycerophosphate) containing EDTA-free protease inhibitor mix and Phosphatase Inhibitor Cocktail (Roche, Indianapolis, IN), and 300 μl of glass beads (0.4 mm; Sigma-Aldrich, St. Louis, MO) was added. Cells were broken for 40 s in a RiboLyser (Hybaid, Teddington, United Kingdom), and the extract was recovered by washing twice with 400 μl of lysis buffer. Soluble proteins were obtained by centrifugation of the extracts at 13,200 × g for 10 min at 4°C. For immunoprecipitation, protein extracts were immunoprecipitated using anti-HA or anti-myc μMACS Epitope Tag Protein Isolation kits (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's instructions. For Western blot, 30-μg protein extracts were separated by 8% SDS–PAGE, transferred to Hybond-P (Amersham Bioscience, Buckinghamshire, United Kingdom) membranes, and probed with anti-myc (9E10, 1/5000), anti-HA (3F10 Roche, 1:20,000), or anti-PSTAIRE (Sigma-Aldrich, 1:5000) antibodies. Secondary antibodies conjugated to horseradish peroxidase (immunoglobulin G–HRP; Santa Cruz Biotechnology, Santa Cruz, CA) were diluted 1:20,000. Immunoblots were developed using the Supersignal West Femto Kit (Pierce Biotechnology, Thermo Fisher Scientific, Rockford, IL). Phosphatase treatment of cell extracts was performed using λ-PPTase (New England Biolabs, Ipswich, MA). To inhibit the cdc28-as allele, cdc28-as yeast cells were grown as yeast or hyphae in MM supplemented with histidine in the presence of 25 μM 1NM-PP1 (Calbiochem, San Diego, CA) or dimethyl sulfoxide (DMSO).
2D Western blot
Cells extracts prepared as described were precipitated using methanol-chloroform and resuspended in hydration buffer (8 mM urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, 1 mM dithiothreitol, 0.5% IPG buffer, pH 6–11, and 1.2% DeStreak Reagent [GE Healthcare Bio-Sciences, Piscataway, NJ] at a 2 μg/μl final concentration. Approximately 75 μg of protein was resolved on 2D gels using 7-cm Immobiline Drystrip pH 6–11 and the Ettan IPGphor 3 Isoelectric Focusing System (GE Healthcare).
In vitro kinase assays
CDK in vitro kinase assays were performed as previously described (Sinha et al., 2007). Briefly, to affinity purify cyclin/Cdc28 kinases, 1 l of log-phase cells expressing Myc-tagged Cdc28 (Cdc28–-6Myc) were lysed and extracted in 5 ml of high-salt lysis buffer containing 50 mM Tris-HCl, pH 7.4, 1% NP-40, 0.5 M KCl, and EDTA-free protease inhibitor mix (Roche). The lysate was mixed and incubated with 200 μl of appropriate antibody-coupled beads at 4°C for 2 h. Then, the beads were washed four times with the same buffer to remove nonspecific bound proteins. Kinase reactions were performed by mixing the beads with 5 μg of GST-4S or GST-4A in a final volume of 100 μl of standard kinase reaction buffer containing γ-32P-ATP and incubating them at 30°C for 30 min. The reaction was stopped by washing twice with 300 μl of kinase buffer, adding 5× gel loading buffer, and boiling for 5 min. Labeled proteins were visualized by SDS–PAGE and autoradiography. Mob2-associated kinase activity was determined as described (Jansen et al., 2006). Briefly, 500 OD600 equivalents of Mob2-HA–expressing cells were resuspended in 1 ml of ice-cold lysis buffer and broken as described earlier. Lysates were cleared by centrifugation, and the supernatant was normalized to 8 mg/ml. To immunoprecipitate Mob2, 1 ml of normalized lysates was incubated with 1 μg of anti-HA antibodies (3F10; Roche) and processed as described (Jansen et al., 2006). For kinase reactions, the immunoprecipitates were incubated with 3 μg of purified GST-3xSpa2138-163 as substrate in 30 μl of kinase reaction buffer (20 mM Tris, pH 8.0, 150 mM NaCl. and 5 mM MnCl2) containing 20 mM cold ATP and 0.33 μCi/μl γ-32P-ATP. Kinase reactions were incubated for 60 min at room temperature and stopped by the addition of 7 μl of 5× SDS–PAGE. Proteins were separated by SDS–PAGE, and kinase activity was quantified using a Molecular Imager FX (Bio-Rad, Hercules, CA).
Protein purification
Wild-type and mutant Mob2-4A N-terminal fragment (1–115) were cloned into the pGEX-5x-3 (GE Healthcare) and expressed as GST fusions in BL21(DE) cells. To express the fragment of Spa2, a DNA sequence encoding a tandem of three copies of the 25–amino acid peptide corresponding to the amino acids 138–163 of Spa2 was synthesized (Eurofins MWG Operon, Ebersberg, Germany). As a control for the kinase assays, the same fragment with substitutions of phosphoacceptor residues S143, S153, and S163 to Ala was synthesized. These DNA fragments were cloned into the pGEX-5x-3 (GE Healthcare) and expressed as GST fusions in BL21(DE) cells. All of the GST-fusion proteins were induced in 2xYT medium with 0.5 mM IPTG for 4 h at 37ºC and purified with glutathione-Sepharose beads (GE Healthcare), following the manufacturer's instructions.
RT quantitative PCR
Mid-log cultures grown in MM were harvested, frozen in liquid nitrogen, and stored at –80ºC. For RNA extraction, 50 mg of cells was lysed in a FastPrep cell disruptor in the presence of 50 μl of TRIzol reagent (Invitrogen). Total RNA was isolated according to the manufacturer's instructions. The quantity, quality, and integrity of the RNA were analyzed in an Agilent 2100 Bioanalyzer system (Agilent Technologies, Santa Clara, CA). cDNA synthesis was performed with the SuperScript II First-Strand Synthesis System (Invitrogen) using oligo(dT), from 3 μg of total RNA previously treated with DNase I (Invitrogen). qPCR assays were done using SYBR Premix Ex Taq (Takara Bio, Shiga, Japan). For each reaction, 1 μl of cDNA was used. Controls without template or without reverse transcriptase were included. The assays were carried out in duplicate under generic cycle conditions (95°C for 45 s, 40 cycles of 95°C for 5 s, and 60°C for 31 s, followed by a dissociation step at 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s) in an Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA). Relative gene expression was quantified using the standard curve method.
Supplementary Material
Acknowledgments
We thank Pilar Pérez for critical comments and Nick Skinner for supervising the English version of the manuscript. We also thank Eric Weiss and Stephan Seiler for reagents and helpful suggestions about Mob2-associated kinase assays. This work was supported by grants from the Spanish Ministry of Science and Innovation (BFU2006-10318 and BFU2009-11251 to J.C.-B and BFU2007-60390 and BFU2010-15884 to C.R.V.) and from the Regional Government of Extremadura (PRI08A017 and GRU09001) to J.C.-B. All Spanish funding was cosponsored by the European Union Fonds Européen de Développement Régional program. P.G-E. was supported by a predoctoral fellowship (Formación de Profesorado Universitario program) from the government of Spain. Y.W. and C.-R.L. were funded by the Agency of Science, Technology and Research of Singapore.
Abbreviations used:
- 2D
two-dimensional
- CDK
cyclin-dependent kinase
- NDR
nuclear Dbf2-related
- RAM
regulation of Ace2 and morphogenesis
- WB
Western blot
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-03-0205) on May 18, 2011.
REFERENCES
- Baladrón V, Ufano S, Dueñas E, Martín-Cuadrado AB, del Rey F, Vázquez de Aldana CR. Eng1p, an endo-1,3-β-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae. Eukaryot Cell. 2002;1:774–786. doi: 10.1128/EC.1.5.774-786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrao P, Trinidad JC, Fiedler D, Roguev A, Lim WA, Shokat KM, Burlingame AL, Krogan NJ. Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol. 2009;7:e1000134. doi: 10.1371/journal.pbio.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidlingmaier S, Weiss EL, Seidel C, Drubin DG, Snyder M. The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:2449–2462. doi: 10.1128/MCB.21.7.2449-2462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- Biswas S, Van Dijck P, Datta A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol Mol Biol Rev. 2007;71:348–376. doi: 10.1128/MMBR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brace J, Hsu J, Weiss EL. Mitotic exit control of the Saccharomyces cerevisiae Ndr/LATS kinase Cbk1 regulates daughter cell separation after cytokinesis. Mol Cell Biol. 2011;31:721–735. doi: 10.1128/MCB.00403-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente-Blanco A, González-Novo A, Machín F, Caballero-Lima D, Aragón L, Sánchez M, Vázquez de Aldana CR, Jiménez J, Correa-Bordes J. The Cdc14p phosphatase affects late cell-cycle events and morphogenesis in Candida albicans. J Cell Sci. 2006;119:1130–1143. doi: 10.1242/jcs.02820. [DOI] [PubMed] [Google Scholar]
- Colman-Lerner A, Chin TE, Brent R. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell. 2001;107:739–750. doi: 10.1016/s0092-8674(01)00596-7. [DOI] [PubMed] [Google Scholar]
- Crampin H, Finley K, Gerami-Nejad M, Court H, Gale C, Berman J, Sudbery P. Candida albicans hyphae have a Spitzenkorper that is distinct from the polarisome found in yeast and pseudohyphae. J Cell Sci. 2005;118:2935–2947. doi: 10.1242/jcs.02414. [DOI] [PubMed] [Google Scholar]
- Das M, Wiley DJ, Chen X, Shah K, Verde F. The conserved NDR kinase Orb6 controls polarized cell growth by spatial regulation of the small GTPase Cdc42. Curr Biol. 2009;19:1314–1319. doi: 10.1016/j.cub.2009.06.057. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto K, He Y, Ye B, Grueber WB, Adler PN, Jan LY, Jan YN. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell. 2004;119:245–256. doi: 10.1016/j.cell.2004.09.036. [DOI] [PubMed] [Google Scholar]
- Gallegos ME, Bargmann CI. Mechanosensory neurite termination and tiling depend on SAX-2 and the SAX-1 kinase. Neuron. 2004;44:239–249. doi: 10.1016/j.neuron.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Geng W, He B, Wang M, Adler PN. The tricornered gene, which is required for the integrity of epidermal cell extensions, encodes the Drosophila nuclear DBF2-related kinase. Genetics. 2000;156:1817–1828. doi: 10.1093/genetics/156.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter AS. Guides to the final frontier of the cytoskeleton: septins in filamentous fungi. Curr Opin Microbiol. 2010;13:720–726. doi: 10.1016/j.mib.2010.09.012. [DOI] [PubMed] [Google Scholar]
- Gladfelter AS, Kozubowski L, Zyla TR, Lew DJ. Interplay between septin organization, cell cycle and cell shape in yeast. J Cell Sci. 2005;118:1617–1628. doi: 10.1242/jcs.02286. [DOI] [PubMed] [Google Scholar]
- Gola S, Martin R, Walther A, Dunkler A, Wendland J. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast. 2003;20:1339–1347. doi: 10.1002/yea.1044. [DOI] [PubMed] [Google Scholar]
- González-Novo A, Correa-Bordes J, Labrador L, Sánchez M, Vázquez de Aldana CR, Jiménez J. Sep7 is essential to modify septin ring dynamics and inhibit cell separation during Candida albicans hyphal growth. Mol Biol Cell. 2008;19:1509–1518. doi: 10.1091/mbc.E07-09-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Novo A, Vázquez de Aldana CR, Jiménez J. Fungal septins: one ring to rule it all? Cent Eur J Biol. 2009;4:274–289. [Google Scholar]
- Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- Harvey K, Tapon N. The Salvador-Warts-Hippo pathway—an emerging tumour-suppressor network. Nat Rev Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- He Y, Fang X, Emoto K, Jan YN, Adler PN. The tricornered Ser/Thr protein kinase is regulated by phosphorylation and interacts with furry during Drosophila wing hair development. Mol Biol Cell. 2005;16:689–700. doi: 10.1091/mbc.E04-09-0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A, Bichsel SJ, Hemmings BA. Human NDR kinases are rapidly activated by MOB proteins through recruitment to the plasma membrane and phosphorylation. Mol Cell Biol. 2005;25:8259–8272. doi: 10.1128/MCB.25.18.8259-8272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A, Stegert MR, Schmitz D, Hemmings BA. NDR kinases regulate essential cell processes from yeast to humans. Nat Rev Mol Cell Biol. 2006;7:253–264. doi: 10.1038/nrm1891. [DOI] [PubMed] [Google Scholar]
- Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. Global analysis of CDK1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou MC, Salek J, McCollum D. Mob1p interacts with the Sid2p kinase and is required for cytokinesis in fission yeast. Curr Biol. 2000;10:619–622. doi: 10.1016/s0960-9822(00)00492-9. [DOI] [PubMed] [Google Scholar]
- Hou MC, Wiley DJ, Verde F, McCollum D. Mob2p interacts with the protein kinase Orb6p to promote coordination of cell polarity with cell cycle progression. J Cell Sci. 2003;116:125–135. doi: 10.1242/jcs.00206. [DOI] [PubMed] [Google Scholar]
- Jansen JM, Barry MF, Yoo CK, Weiss EL. Phosphoregulation of Cbk1 is critical for RAM network control of transcription and morphogenesis. J Cell Biol. 2006;175:755–766. doi: 10.1083/jcb.200604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LA, Sudbery PE. Spitzenkorper, exocyst, and polarisome components in Candida albicans hyphae show different patterns of localization and have distinct dynamic properties. Eukaryot Cell. 2010;9:1455–1465. doi: 10.1128/EC.00109-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly MT, MacCallum DM, Clancy SD, Odds FC, Brown AJ, Butler G. The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol Microbiol. 2004;53:969–983. doi: 10.1111/j.1365-2958.2004.04185.x. [DOI] [PubMed] [Google Scholar]
- Komarnitsky SI, Chiang YC, Luca FC, Chen J, Toyn JH, Winey M, Johnston LH, Denis CL. DBF2 protein kinase binds to and acts through the cell cycle-regulated MOB1 protein. Mol Cell Biol. 1998;18:2100–2107. doi: 10.1128/mcb.18.4.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig C, Maekawa H, Schiebel E. Mutual regulation of cyclin-dependent kinase and the mitotic exit network. J Cell Biol. 2010;188:351–368. doi: 10.1083/jcb.200911128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurischko C, Kuravi VK, Wannissorn N, Nazarov PA, Husain M, Zhang C, Shokat KM, McCaffery JM, Luca FC. The yeast LATS/Ndr kinase Cbk1 regulates growth via Golgi-dependent glycosylation and secretion. Mol Biol Cell. 2008;19:5559–5578. doi: 10.1091/mbc.E08-05-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CR, Lee RT, Wang YM, Zheng XD, Wang Y. Candida albicans hyphal morphogenesis occurs in Sec3p-independent and Sec3p-dependent phases separated by septin ring formation. J Cell Sci. 2007;120:1898–1907. doi: 10.1242/jcs.002931. [DOI] [PubMed] [Google Scholar]
- Marks J, Hyams J. Localization of F-actin through the cell-division cycle of Schizosaccharomyces pombe. Eur J Cell Biol. 1985;39:27–32. [Google Scholar]
- Mazanka E, Alexander J, Yeh BJ, Charoenpong P, Lowery DM, Yaffe M, Weiss EL. The NDR/LATS family kinase Cbk1 directly controls transcriptional asymmetry. PLoS Biol. 2008;6:e203. doi: 10.1371/journal.pbio.0060203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNemar MD, Fonzi WA. Conserved serine/threonine kinase encoded by CBK1 regulates expression of several hypha-associated transcripts and genes encoding cell wall proteins in Candida albicans. J Bacteriol. 2002;184:2058–2061. doi: 10.1128/JB.184.7.2058-2061.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B, et al. RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol Biol Cell. 2003;14:3782–3803. doi: 10.1091/mbc.E03-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips J, Herskowitz I. Identification of Kel1p, a kelch domain-containing protein involved in cell fusion and morphology in Saccharomyces cerevisiae. J Cell Biol. 1998;143:375–389. doi: 10.1083/jcb.143.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praskova M, Xia F, Avruch J. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr Biol. 2008;18:311–321. doi: 10.1016/j.cub.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D, Legesse-Miller A, Gao L, Dong Y, Bretscher A. Mechanisms of polarized growth and organelle segregation in yeast. Annu Rev Cell Dev Biol. 2004;20:559–591. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- Racki WJ, Becam AM, Nasr F, Herbert CJ. Cbk1p, a protein similar to the human myotonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisiae. EMBO J. 2000;19:4524–4532. doi: 10.1093/emboj/19.17.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer T, Vallier L, Sheu YJ, Snyder M. The Spa2-related protein, Sph1p, is important for polarized growth in yeast. J Cell Sci. 1998;111(4):479–494. doi: 10.1242/jcs.111.4.479. [DOI] [PubMed] [Google Scholar]
- Schaub Y, Dunkler A, Walther A, Wendland J. New pFA-cassettes for PCR-based gene manipulation in Candida albicans. J Basic Microbiol. 2006;46:416–429. doi: 10.1002/jobm.200510133. [DOI] [PubMed] [Google Scholar]
- Sheu YJ, Santos B, Fortin N, Costigan C, Snyder M. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol Cell Biol. 1998;18:4053–4069. doi: 10.1128/mcb.18.7.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha I, Wang YM, Philp R, Li CR, Yap WH, Wang Y. Cyclin-dependent kinases control septin phosphorylation in Candida albicans hyphal development. Dev Cell. 2007;13:421–432. doi: 10.1016/j.devcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Sirajuddin M, Farkasovsky M, Hauer F, Kuhlmann D, Macara IG, Weyand M, Stark H, Wittinghofer A. Structural insight into filament formation by mammalian septins. Nature. 2007;449:311–315. doi: 10.1038/nature06052. [DOI] [PubMed] [Google Scholar]
- Sirajuddin M, Farkasovsky M, Zent E, Wittinghofer A. GTP-induced conformational changes in septins and implications for function. Proc Natl Acad Sci USA. 2009;106:16592–16597. doi: 10.1073/pnas.0902858106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Cheon SA, Lee KE, Lee SY, Lee BK, Oh DB, Kang HA, Kim JY. Role of the RAM network in cell polarity and hyphal morphogenesis in Candida albicans. Mol Biol Cell. 2008;19:5456–5477. doi: 10.1091/mbc.E08-03-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery P. Morphogenesis of a human fungal pathogen requires septin phosphorylation. Dev Cell. 2007;13:315–316. doi: 10.1016/j.devcel.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Sudol M, Harvey KF. Modularity in the Hippo signaling pathway. Trends Biochem Sci. 2010;35:627–633. doi: 10.1016/j.tibs.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Takeshita N, Higashitsuji Y, Konzack S, Fischer R. Apical sterol-rich membranes are essential for localizing cell end markers that determine growth directionality in the filamentous fungus Aspergillus nidulans. Mol Biol Cell. 2008;19:339–351. doi: 10.1091/mbc.E07-06-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DA, Hariharan IK. salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Verde F, Wiley DJ, Nurse P. Fission yeast orb6, a ser/thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase, is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc Natl Acad Sci USA. 1998;95:7526–7531. doi: 10.1073/pnas.95.13.7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Raniga PP, Lane S, Lu Y, Liu H. Hyphal chain formation in Candida albicans: Cdc28-Hgc1 phosphorylation of Efg1 represses cell separation genes. Mol Cell Biol. 2009;29:4406–4416. doi: 10.1128/MCB.01502-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Shimizu T, Lai ZC. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. EMBO J. 2007;26:1772–1781. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirich CS, Erzberger JP, Barral Y. The septin family of GTPases: architecture and dynamics. Nat Rev Mol Cell Biol. 2008;9:478–489. doi: 10.1038/nrm2407. [DOI] [PubMed] [Google Scholar]
- Weiss EL, Kurischko C, Zhang C, Shokat K, Drubin DG, Luca FC. The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J Cell Biol. 2002;158:885–900. doi: 10.1083/jcb.200203094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden O, Plamann M, Ebbole DJ, Yanofsky C. cot-1, a gene required for hyphal elongation in Neurospora crassa, encodes a protein kinase. EMBO J. 1992;11:2159–2166. doi: 10.1002/j.1460-2075.1992.tb05275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XD, Wang YM, Wang Y. CaSPA2 is important for polarity establishment and maintenance in Candida albicans. Mol Microbiol. 2003;49:1391–1405. doi: 10.1046/j.1365-2958.2003.03646.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.