Abstract
The p53 family members, which consist of 3 transcription factors—p53, p63, and p73—are conserved during evolution. The p53 family proteins are involved in many important cellular functions, including tumor suppression (p53 and p73), the development of epithelial cell layers (p63), and the development of central nervous system and immune system (p73). Studies on p53-like proteins in low organisms have demonstrated that their primordial functions are to maintain the genomic integrity of germ cells and ensure faithful development and reproduction. In vertebrates, the p53 family proteins retain these functions in reproduction and at the same time have developed additional important functions in reproduction, such as the regulation of embryonic implantation (p53). p53 regulates embryonic implantation through transcriptional regulation of leukemia inhibitory factor (LIF). p63, in particular TAp63, is a main regulator to protect the fidelity of female germ cells during meiotic arrest. p73, in particular TAp73, regulates the ovary function and the quality of oocytes. Loss of p53, p63, or p73 genes in female mice leads to a significant decrease in fertility. These functions of the p53 family proteins in reproduction provide a plausible explanation for positive evolutionary selection observed in a group of single nucleotide polymorphisms and haplotypes in the p53 family genes. A better understanding of the functions of the p53 family proteins in reproduction may lead to new strategies for fertility treatment.
Keywords: p53, p63, p73, implantation, female germ cells
Introduction
The p53 family proteins consist of 3 transcription factors: p53, p63, and p73. These 3 proteins have a very similar domain organization. They all contain an amino-terminal transactivation domain (TAD), a prolin-enriched region (PR), a central DNA binding domain (DBD), and a carboxy-terminal oligomerization domain (OD). p63 and p73 proteins have additional domains, including a carboxy-terminal sterile-α motif (SAM), which is thought to mediate protein–protein interactions, and a transcription inhibition domain (TID).1 The p53 family proteins have limited overall homology but share strong similarity in the DBD (approximately 55%-58% between p53 and p63/p73 and approximately 87% between p63 and p73).2 The p53 family proteins bind to similar or identical DNA sequences to regulate the transcription of a subset of genes in common and a subset of genes unique to each protein, which in turn are involved in similar as well as unique cellular functions. p53, p63, and p73 are all involved in tumor suppression. In addition, p63 is essential for epidermal morphogenesis and limb development, and p73 is involved in the development of central nervous system and immune system. Studies on p53-like proteins in low organisms, including Caenorhabditis elegans and Drosophila, have revealed that the primordial functions of p53 family proteins are to maintain germ cell integrity and faithful reproduction. Recent studies in mice and humans have demonstrated that the p53 family proteins play important roles in regulation of reproduction, including maintaining primordial and primary follicular pool size, germ cell genomic integrity, ovulatory ability, pre-implantation development, and embryonic implantation. The regulation of reproduction appears to be an evolutionarily conserved and important function of the p53 family proteins.
The Isoforms and Functions of the p53 Family Proteins
The p53 family proteins exist as multiple functionally diverse proteins that originate from a combination of alternative promoter usage and alternative mRNA splicing.3-7 Each gene has 2 promoters: the first is located in a noncoding region upstream of exon 1, and the alternative promoter is located in intron 4 of the p53 gene and intron 3 of the p63 and p73 genes, respectively. The promoter upstream of exon 1 generates the transactivating (TA) isoforms, which are transcriptionally proficient, and the alternative promoter leads to the expression of amino-terminal truncated (ΔN) isoforms that lack the transactivation domain and are transcriptionally inactive. For both p53 and p73, additional N-terminal variants are generated by alternative splicing or internal initiation of translation.8 ΔN isoforms can actively repress the transactivation of TA isoforms by competing for binding to the DNA binding elements in regulated genes, and thus they function as dominant negative inhibitors of TAp53, TAp63, and TAp73. To add further complexity, splice variants of the C-terminal region generate 3 additional isoforms of p53 and p63 (α, β, γ) and at least 8 splice variants of p73 (α, β, γ, δ, ϵ, ζ, η, φ). Splicing at the C-terminal end of the p53, p63, and p73 genes gives rise to isoforms that regulate stability of the protein and DNA binding. Thus, p53, p63, and p73 each have many different isoforms with a number of diverse properties. The full-length and truncated isoforms of the p53 family generally exhibit reciprocal biological functions: ΔN isoforms support proliferation, whereas TA isoforms promote cell cycle arrest, cellular senescence, and apoptosis.8 The existence of p53 family proteins with opposing activities implies that the balance of various isoforms may determine cell fate.
p53 protein plays an important role in tumor suppression. In response to a variety of stress signals, p53 is activated and transcriptionally induces its target genes that lead to various cellular responses, including cell cycle arrest, apoptosis, and senescence, to inhibit the proliferation of nascent cancer cells.9 The p53 gene is the most frequently mutated gene in human cancer; ~50% of human cancers have mutations in the p53 gene, and ~80% of human cancers have dysfunction of the p53 pathway.9 p63 and p73 are also involved in tumor suppression. Both TAp63 and TAp73 isoforms can bind to canonical p53 DNA binding elements and transcriptionally activate a subset of p53-target genes, which induce p53-like functions including cell cycle arrest, apoptosis, and senescence.3,6,10,11 A study examining the tumorigenic effects of p63 and p73 in heterozygous mouse models showed that p63+/− and p73+/− mice are predisposed to develop spontaneous tumors. p63 and p73 also cooperate with p53 in tumor suppression. p53+/− p63+/− and p53+/− p73+/− mice have higher tumor burden and increased metastasis compared with p53+/− mice.12 Loss of TAp73 in vivo leads to increased incidence of spontaneous and carcinogen-induced tumors, suggesting that TAp73 is a tumor suppressor. In human tumors, although mutations of p63 and p73 are rare, overexpression of ΔN isoforms of p63 and p73 has been often observed.13-15 It has been suggested that this imbalance between TA and ΔN isoforms could inhibit the function of TA isoforms in tumor suppression, which promotes tumorigenesis.
Although the p53 family proteins are all involved in the regulation of tumor suppression, the most important role of p63 and p73 appears to be the regulation of differentiation and development. p63 has been shown to be essential for epidermal morphogenesis and limb development. Mice deficient for all p63 isoforms die within hours of birth, presumably due to dehydration. The developmental abnormalities of these mice include craniofacial malformations, limb truncation and lack of epidermis, and squamous epithelia (prostate, urothelium) and epithelial appendages, such as hair follicles, teeth, and mammary, salivary, and lachrymal glands.16,17 In humans, germ line mutations in the p63 gene cause several rare autosomal dominant developmental diseases, including ectrodactyly ectodermal dysplasia-clefting syndrome (EEC), acro-dermato-ungual-lacrimal-tooth malformations (ADULT), limb-mammary syndrome (LMS), Hay-Wells syndrome (AEC), split-hand/foot malformations (SHFM), and Rapp-Hodgkin syndromes.18-20 These diseases are characterized by various degrees of limb abnormalities, ectodermal dysplasia, and facial clefts, indicating the relevance of p63 to normal epidermal development in humans.21 p73 is involved in the regulation of neuronal development and differentiation and of immune function. Mice deficient for all p73 isoforms are viable but are runt and have high rates of mortality. A majority of the p73 null mice die before 4 weeks of age, and only 25% of them survive to adulthood. These mice exhibit profound developmental defects, including hippocampal dysgenesis, hydrocephalus, chronic infections and inflammation, and abnormalities in pheromone sensory pathways.22 It has been shown that ΔNp73 is expressed in developing brain, sympathetic ganglia, and adult neurons and plays an important role in neuronal survival during development and in adult neurons by blocking p53/TAp63/TAp73-mediated apoptosis.22-24 In addition, TAp73 is important for the maintenance of neural stem cell (NSC) pool and may support neuronal differentiation.25 Increased expression of TAp73 has been observed in NSCs following differentiation and in neuroblastoma cells that have been induced to differentiate by retinoic acid.26 Ectopic TAp73 overexpression can induce neuronal differentiation, probably by antagonizing Notch signaling.27
The Evolution of the p53 Family Genes
The p53 family is evolutionarily conserved. The homologues of the p53 family genes have been described in many different organisms, including mollusce, sea anemone, clams, C. elegans, Drosophila, frogs, and zebrafish.28-35 The DBD is the most conserved domain among different organisms. All p53-like family proteins that have been identified so far contain a DBD. The residues in DBD that contact DNA and the DNA binding sequences are highly conserved. All members of the p53-like family proteins that have been investigated so far demonstrate very similar DNA binding specificities.36,37 Based on sequence similarity, p53-like family proteins in primitive organisms and invertebrates, including sea anemone, C. elegans, and Drosophila, are more closely related to p63/p73 than to p53.38,39 Furthermore, many p53-like family proteins identified in invertebrates contain a SAM domain, which is a protein interaction domain existing in p63/p73 protein but not in p53 protein. Therefore, p63/p73 is likely to be evolutionarily ancient, and p53 is possibly evolved from a p63/p73-like gene.29 The high conservation of residues that contact DNA and of the DNA binding specificity suggests that the original function of this protein family reaches far back in evolution.
The existence of ancient p53 homologue proteins in organisms that have no threat from cancer within their short life spans suggests that the primordial functions of the p53 family proteins may not be tumor suppression, and the tumor suppression function may be derived from their primordial functions. Evidence suggests that maintaining the genomic integrity of germ cells to ensure the faithful reproduction could be an important primordial function of the p53 family proteins. In sea anemone (Nematostella vectensis), the p53-like family protein nvp63 is highly expressed in germ cells but not in somatic cells. nvp63 is activated by genotoxic stress signals to induce apoptosis in germ cells, which in turn eliminates damaged germ cells. In this way, nvp63 maintains the integrity of the genome transmitted to the next generation and avoids defective progeny.30 C. elegans contains a single p53-like family protein, CEP-1 (C. elegans p53-like-1). CEP-1 is commonly expressed in germ cells. In response to genotoxic stress, CEP-1 induces 2 target genes that encode BH-3 (Bcl-2 homology 3) domain–only proteins, EGL-1 (egg laying abnormal-1) and CED-13 (cell death abnormality protein 13), to activate apoptosis in germ cells and eliminate defective offspring from the population.32,40-42 In response to UV, CEP-1 can induce phg-1 (pharynx associated Gas1), which is a cell cycle inhibitor that results in the arrest of germ cell proliferation to allow time for DNA repair. In addition, CEP-1 is required for normal chromosome segregation during meiosis in germ cells.32 In Drosophila, the only p53-like family protein, Dmp53 (Drosophila p53), also plays a similar role in female germ cells. Dmp53 is highly expressed in germ cells.40 In response to DNA damage and other genotoxic stress, Dmp53 protein is activated and induces apoptosis in primordial germ cells. Dmp53 null mutants exhibit genomic instability, especially after γ-irradiation.43 In Drosophila, caspases that cause apoptosis are normally maintained in an inactive state by inhibitors of apoptosis (IAPs). In response to apoptotic stimuli, IAP antagonists, including reaper, hid, grim, sickle, and jafrac2, are transcriptionally activated to degrade IAPs and induce apoptosis. Among them, reaper, sickle, and hid are Dmp53-regulated genes.36,43,44 Another important pathway in Dmp53-induced apoptosis is the Hippo (Hpo) pathway, which includes the kinases Hpo and Warts/Lats. It has been shown that in response to irradiation, Hpo is activated by phosphorylation, which in turn promotes apoptosis, in a Dmp53-dependent manner.45 In addition to apoptosis, Dmp53 may be involved in DNA repair. It has been suggested that a group of DNA double-strand break (DSB) repair genes, including Ku70, Ku80, Mre11, and Rad50, are potential target genes of Dmp53.44,46 Dmp53 is also required for proper meiotic recombination. Dmp53 null mutants have reduced meiotic recombination frequencies.47 It has been shown that Dmp53 proteins are activated by the DSBs in oocyte precursors during the initiating steps of meiotic recombination. In cells that are defective for meiotic DNA repair, Dmp53 is persistently activated. Furthermore, Dmp53 is involved in the development of the ovary. Dmp53 null mutants have degenerative ovarioles and decreased fertility compared with their parental strains.48 It is clear from these observations that the maintenance of genomic integrity of germ cells to ensure faithful development and fecundity is an important primordial function of the p53 family proteins.
The Role of the p53 Protein in Reproduction in Mice
The primordial function of the p53 family proteins is evolutionarily conserved. Similar function of the p53 family proteins in reproduction is observed in mice. In mice, p53 protein is highly expressed in testes with increased activity during spermatogenesis, and it regulates both spermatogenesis and radiation response. Meiotic recombination appears to be an important signal to activate p53 protein during spermatogenesis. p53 is transiently activated in early spermatocytes; the expression of p53 peaks between the leptotene and zygotene stages and disappears after the early pachytene stage.47 p53 null mice or mice with reduced levels of p53 have a high frequency of multinucleated giant cells within the testicular seminiferous tubules, which is a degenerative syndrome most likely due to the impaired meiotic divisions during the meiotic prophase.49 p53 also mediates DNA damage-induced spermatogonial apoptosis.50 Irradiation can induce p53 activation in spermatocytes, which results in apoptosis to remove damaged spermatocytes.51,52 In p53 null mice, irradiation induces very limited apoptosis, which in turn results in the production of an increased number of giant-size spermatogonial stem cells. p53 protein is also expressed in mouse embryos during development to monitor and respond to DNA damage. p53 is highly expressed in mouse embryos until the midgestation stage.53 In response to irradiation, p53 protein is activated to induce apoptosis to efficiently eliminate the damaged offspring, whereas p53 null embryos show a very small percentage of death, which results in a high percentage of developmental abnormalities.54,55 These observations demonstrate that the p53-dependent DNA damage response (apoptosis) is highly active in germ cells and during development to ensure faithful development and fecundity in mice.
Mammals have a much longer and more complex gestation period compared with other vertebrates. The p53 family proteins gained new functions in regulation of reproduction in mammals during evolution. Our recent work has revealed an additional important function of p53 in reproduction in mice; p53 regulates embryonic implantation through its target gene, leukemia inhibitory factor (LIF) (Figure 1).56,57 Although p53 null mice seem to be fertile and are able to give birth of intact offspring, detailed analysis of fertility has shown that p53 loss in female but not in male mice significantly decreases fertility. p53 null female mice have much lower pregnancy rates and smaller litter size. This function of p53 in regulation of maternal reproduction is mediated by LIF. LIF is a multifunctional cytokine that plays a crucial role in embryonic implantation. LIF functions through its binding to the LIF receptor complex, composed of LIF receptor (LIF-R) and glycoprotein gp-130, which in turn activates selective pathways, including JAK/STAT3, MAPK, and PI3K/ATP pathways.58,59 At the onset stage of implantation, the LIF expression levels increase significantly in the endometrial glands to induce decidualization of uterine tissue.60-62 Decidualization is a process that changes endometrium into decidua by modifying endometrial stromal cells, uterine glands, and vessels as well as population of uterine immune cells, thus preparing the uterus for embryonic implantation. LIF-deficient female mice have a defect in reproduction due to the complete lack of uterine decidualization at the implantation stage that leads to the failure of blastocyst implantation. This reproduction defect in LIF-deficient female mice can be rescued by injection of exogenous LIF at the implantation stage.61 The increased uterine LIF expression at the implantation stage requires the coordinated regulation of p53 and estrogen.63 p53 can bind to the p53-binding element in the first intron of mouse LIF gene and regulate the expression levels of LIF under both nonstressed and stressed conditions. At the implantation stage, p53 protein levels and activity are increased selectively in the endometrial tissues, including endometrial glands, where the highest levels of LIF expression are observed. It is unclear what signals activate p53 in endometrial tissues at the implantation stage. Implantation occurs under the hypoxic condition,34 and hypoxic signals are known to activate p53.35 It is possible that a hypoxic condition is the stimulus for p53 activation in the uterus at the implantation stage. Another regulator of LIF expression is estrogen. The estrogen levels and the activity of estrogen receptor, which mediates the estrogen signaling, increase at the implantation stage in parallel with the increase of LIF expression.64 In primary cultured mouse endometrial epithelial cells, the highest expression levels of LIF were observed in p53 wild-type cells in the presence of estrogen, suggesting the coordinated regulation of LIF by p53 and by estrogen signaling. In p53 null mice, uterine LIF levels at the implantation stage are much lower than those in wild-type mice, which results in impaired implantation. The implantation can be restored in p53 null female mice by injection of exogenous LIF at the implantation stage, which greatly improves fertility in these mice.
Figure 1.
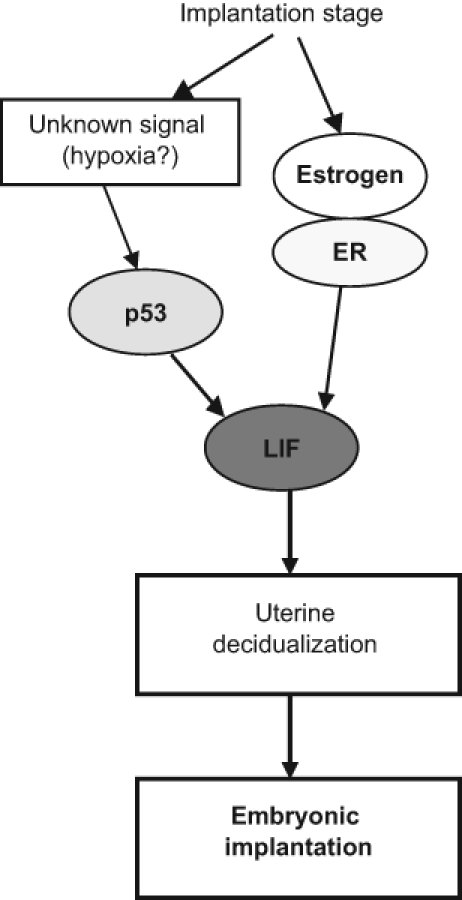
p53 regulates embryonic implantation through LIF in mice. p53 is selectively activated in endometrial tissues at the implantation stage. Estrogen levels also increase at the implantation stage, which in turn activates the estrogen signaling pathway in endometrial tissues through ERα. Selective activation of p53 and ERα coordinately regulates LIF expression at the implantation stage to induce uterine decidualization and ensure embryonic implantation.
The Role of p63 Protein in Protecting Female Germ Line Fidelity in Mice
As described above, p63 is a more ancestral member of the p53 family during evolution. Phylogenetic analysis consistently show that the single p53-like family gene in genetically tractable organisms such as C. elegans or Drosophila is related to the p63 gene.38 In mice, the central role of p63 in epidermal development, which is contributed mainly by ΔN isoforms of p63, was established by the observation made in mice null for both TA and ΔN isoforms. Using mice that were deficient only for TAp63 isoforms, studies have shown that TAp63 is responsible for maintaining genomic integrity of oocytes through eliminating damaged oocytes.65 This function of TAp63 in mammalian female germ cells is similar to the function of the p53-like family proteins in C. elegans and Drosophila, which is consistent with the notion that p63 is the ancestral member of the p53 family.
In mice, TAp63α plays an important role in maintaining the genome integrity of female germ cells during meiotic arrest. TAp63 is mainly expressed in the nuclei of oocytes but not in male germ cells. The expression of TAp63α in oocytes is initiated in the late prophase I from late pachytene stage at embryonic day 18.5 and peaks in the diplotene stage shortly after birth.65,66 TAp63α is activated in oocytes in response to DNA damage induced by γ-irradiation and chemotherapeutic agents, such as cisplatin. TAp63α is activated through phosphorylation by c-Abl, a nonreceptor tyrosine kinase, which in turn induces a group of proapoptotic genes, including Puma and Noxa, and leads to p53-independent apoptosis to efficiently remove damaged cells (Figure 2).67 Loss of p63 in oocytes prevents the cleavage of caspases-9 and caspases-3. The oocytes without TAp63 or p63 are completely resistant to DNA damage–induced apoptosis.65,68 These results clearly demonstrate that TAp63 retains the primordial function of the p53 family proteins in mice, which protects the female germ line fidelity by eliminating damaged cells and, therefore, controls maternal reproduction.
Figure 2.
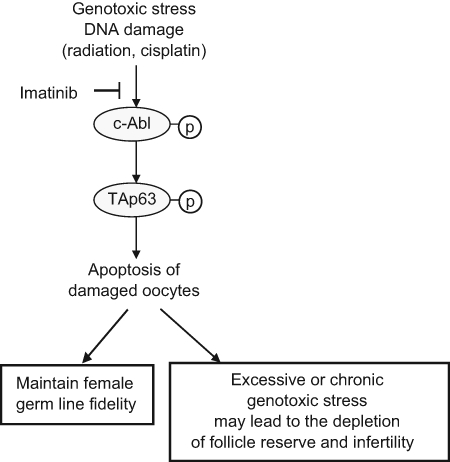
TAp63 protects female germ cells in response to genotoxic stress. In response to genotoxic stress signals, TAp63 is activated through phosphorylation by c-Abl. Activated TAp63 induces p53-independent apoptosis to remove damaged cells and protect the fidelity of female germ cells. Under the conditions of excessive or chronic genotoxic stress, TAp63-dependent apoptosis may lead to the depletion of follicle reserve and infertility. The activation of TAp63 and TAp63-dependent apoptosis can be blocked by imatinib, the c-Abl kinase inhibitor.
The Role of p73 in Ovary Function and Quality of Female Germ Line Cells
Studies on mice deficient for p73 have shown that p73 regulates reproduction at several different steps.22,69 Mice deficient for both TA and ΔN isoforms of p73 are infertile. As described above, p73 is involved in the regulation of neuronal development and differentiation. Among many neuronal defects, p73 null mice have a dysfunction of the vomeronasal organ, an accessory olfactory structure involved in pheromone detection, which normally expresses high levels of p73.22 Mice deficient for p73 are not interested in mating with sexually mature females and are infertile. Mice null for TAp73 only are infertile in female mice but through different mechanisms. TAp73-deficient mice have defects in several steps in maternal reproduction, including reduced follicular pool size, decreased ovulatory ability, and poor oocyte quality (Figure 3).69 First, the number of primordial (quiescent) and primary (early growing) follicles is significantly decreased in TAp73-deficient ovaries. Because the duration of female fertility is determined by the initial size of the primordial follicular pool and by the rate of its activation and subsequent depletion, the lower number of primordial follicles in TAp73-deficient mice results in a faster reproductive failure. Second, TAp73-deficient female mice have a reduced ovulation rate. These female mice ovulate much fewer oocytes compared with wild-type mice. Furthermore, these few ovulated oocytes are trapped under the bursa and cannot progress toward the fallopian tubes as they should. Third, TAp73- deficient oocytes are of poorer quality than wild-type oocytes. It has been reported that when germinal vesicle oocytes as well as ovulated oocytes from 3- to 4-week-old mice were induced to mature in vitro, TAp73-deficient oocytes exhibited a striking increase in aneuploidy and spindle abnormalities, including multipolar spindles, spindle relaxation, and spindle scattering accompanied by varying degrees of misalignment. During meiosis and mitosis, accurate chromosome segregation is critical to prevent aneuploidy. The spindle assembly checkpoint (SAC) complex plays an important role in ensuring accurate chromosome segregation by sensing the improper attachment of sister chromatids to the mitotic or meiotic spindle and delaying anaphase until all chromosomes are correctly oriented for segregation. SAC complex contains over 20 protein partners, including Bub1, Bub3, BubR1, MAD2, cyclin B, Rae1, and Aurora B. Defect of proteins in the SAC complex, such as Bub1, BubR1, or Aurora B, has been reported to cause an increase of aneuploidy, which contributes to poor oocyte quality and infertility.70-72 It has been shown that TAp73 is able to directly interact with several partners of the SAC complex (Bub1, Bub3, and BubR1) and is involved in SAC complex localization and activities. In ovulated oocytes, TAp73 is localized in the cytoplasm and associated specifically with the meiotic spindle at metaphase II. In TAp73-deficient oocytes, the localization of Bub1 and BubR1 to the meiotic spindle and its associated chromosomes is diminished. BubR1, an important component of the SAC complex, is a protein kinase that binds and inhibits p55cdc20 (p55), a major anaphase-promoting complex regulatory protein, to control the activation of the anaphase-promoting complex.73 TAp73 is found to be able to potentiate BubR1 activity; loss of TAp73 compromises BubR1 function. The weakened SAC response in TAp73-deficient oocytes could be an important mechanism for aneuploidy and genomic instability that accounts for infertility in females. The poor quality of TAp73-deficient oocytes has been demonstrated also by their poor developmental competence. Comparing the ability of TAp73-deficient and wild-type oocytes to undergo in vitro fertilization (IVF) and preimplantation development showed that TAp73-deficient oocytes often fail in preimplantation development. Although the fertilization rate is similar in both genotypes, embryos obtained from TAp73-deficient oocytes often arrest during early cleavage, resulting in embryos with multinucleated blastomeres and blastocysts of inferior quality with abnormal cell number.69 Together, TAp73 regulates female reproduction through maintaining the fidelity of female germ cells and proper ovary functions (Figure 3).
Figure 3.
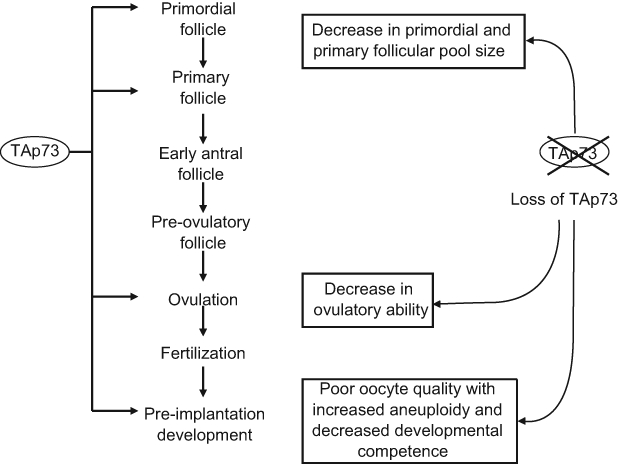
TAp73 regulates ovary function and quality of female germ cells. TAp73 is involved in several steps to regulate female reproduction, including maintaining the sizes of primordial and primary follicular pool, the proper ovulatory function, and genomic integrity of female germ cells. Loss of TAp73 results in reduced follicular pool size, decreased ovulatory ability, and poor oocyte quality.
Increased maternal age has a significant negative impact upon fertility, and a major underlying mechanism for decreased fertility in aged females is the poor quality of oocytes.74 Interestingly, the expression of TAp73 in oocytes declines with natural aging. Furthermore, oocytes from aged wild-type mice and young TAp73 knockout mice exhibit similar abnormalities, including increased aneuploidy and spindle abnormalities,69 suggesting that TAp73 is involved in maintaining genomic stability in oocytes, and decreased TAp73 function may contribute to the increased genomic instability and aneuploidy observed in aged normal oocytes that have compromised developmental capacity.
The Role of p53 and Its Pathway in Implantation and Fertility in Humans
In humans, p53 appears to retain its function in embryonic implantation and is involved in the regulation of reproduction. The regulation of LIF by p53 is conserved from mice to humans.56 As in mice, in humans the LIF gene is a p53 target gene, which contains a p53 DNA-binding element in the first intron. p53 transcriptionally regulates LIF expression levels under both nonstressed and stressed conditions. LIF is also an important player in implantation in humans. The levels of LIF protein increase at the time of expected implantation in fertile women. It has been shown that women with higher endometrial LIF levels during midsecretory phase are more likely to get pregnant than are women with lower LIF levels.75 Deceased uterine LIF levels are often associated with decreased fertility in humans; women with unexplained infertility have lower LIF levels compared with fertile women.76,77 Furthermore, high endometrium LIF levels are associated with a successful implantation after an IVF cycle in IVF patients.78 The regulation of human LIF gene by p53 suggests that p53 may regulate the efficiency of implantation and reproduction in humans.
In humans, naturally occurring single nucleotide polymorphisms (SNPs) with functional consequences exist in genes at critical nodes in the p53 pathway. p53 codon 72 SNP is a common coding SNP in the p53 gene that results in either an arginine (R72) or a proline (P72) residue at codon 72. The p53 P72 allele is weaker in inducing apoptosis and suppressing cellular transformation compared with the p53 R72 allele.79 Individuals with the p53 P72 allele have increased cancer risk compared with the p53 R72 allele.79 The allele frequency of this SNP varies among populations with different ethnic backgrounds and also changes as a function of the latitude where people live. The p53 P72 allele is an ancestral allele with ~60% frequency in African populations. However, in Caucasian and Asian populations, which arose more recently, the p53 P72 allele becomes the minor allele, with ~30% frequency. The change of the allele frequency of this SNP in the p53 gene suggests that certain alleles are under evolutionary selection pressure depending upon the ethnic background. Cancer arises late in life, so it is therefore unlikely that the tumor suppression function of p53 is the major cause for the observed evolutionary selection. Instead, the role of p53 in implantation and reproduction is more likely to be the cause for the observed evolutionary selection. The p53 P72 allele has lower transcriptional activity toward a subset of p53 target genes compared with the p53 R72 allele. We found that LIF is one of the p53 target genes that are differentially induced by the p53 codon 72 SNP. The p53 P72 allele has ~2-fold lower transcriptional activity toward the LIF gene compared with the p53 R72 allele in cultured human cells.80 Recently, mice carrying SNPs at p53 codon 72 have been generated by knocking in wild-type human p53 gene carrying either the p53 P72 allele or R72 allele in place of the corresponding mouse p53 gene.81 Interestingly, the p53 P72 allele produces much less uterine LIF protein in mice, especially at the implantation stage, compared with the p53 R72 allele.63 This impact of the p53 codon 72 SNP upon the uterine LIF production suggests that this SNP may affect implantation and fertility in humans. Indeed, the association of the p53 P72 allele with decreased reproduction efficiency has been observed in IVF patients. Coulam and her colleagues reported that the p53 P72 allele is enriched in women with recurrent implantation failure.82 Consistently, we found that the p53 P72 allele is enriched in IVF patients (Table 1) and is a risk factor for implantation failure after an IVF procedure.80 Interestingly, it has been suggested that environmental stresses, such as cold winter temperature, select the p53 R72 allele, which could increase the levels of LIF to promote a more efficient embryo implantation under adverse conditions.83
Table 1.
Significant Association of Selected Alleles of Genes in the p53 Family Pathways with Decreased Fertility in Humans
| IVF patients | |||||||
|---|---|---|---|---|---|---|---|
| <35 years | ≥35 years | ||||||
| Gene (SNP) | Genotype | Control, % | % | Pa | % | Pa | |
| The p53 pathway | p53 (rs1042522) | G | 77.3 | 64.8 | 1.8E-06b | 70.3 | 0.03b |
| C | 22.7 | 35.2 | 29.7 | ||||
| MDM2 (rs2279744) | T | 62.7 | 55.7 | 0.03b | 58 | 0.35 | |
| G | 37.3 | 44.3 | 42 | ||||
| MDM4 (rs1563828) | C | 68.4 | 61.7 | 0.05b | 59 | 0.02b | |
| T | 31.6 | 38.3 | 41 | ||||
| HAUSP (rs1529916) | G | 68.5 | 56 | 0.004b | 65.1 | 0.46 | |
| A | 31.5 | 44 | 34.9 | ||||
| LIF (rs929271) | T | 71.4 | 63.3 | 0.008b | 68.9 | 0.43 | |
| G | 28.6 | 36.7 | 31.1 | ||||
| p63 | p63 (rs17506395) | T | 70.9 | 78.3 | 0.02b | 79.2 | 0.01b |
| G | 29.1 | 21.7 | 20.8 | ||||
| p73 | p73 (rs4648551) | G | 56.9 | 58.4 | 0.70 | 66.5 | 0.004b |
| A | 43.1 | 41.6 | 33.5 | ||||
| p73 (rs6695978) | G | 95.8 | 94.7 | 0.31 | 92.1 | 0.03b | |
| A | 4.2 | 5.3 | 7.9 | ||||
Note: IVF patients were recruited at Weill Cornell Medical College. DNA from 200 healthy Caucasian individuals was obtained from the Coriell Cell Repositories, and women recruited as controls for the Women’s Insights and Shared Experiences (WISE) study were used as controls.
χ2 test.
Significant difference between IVF patients and controls.
In addition to p53 codon 72 SNP, the p53 pathway contains other functional SNPs in genes that regulate the levels and activity of p53. Recently, we identified a SNP (SNP309) in the MDM2 gene, which encodes a key negative regulator of p53. SNP309 (a T to G change) in the regulatory region in the first intron of MDM2 gene creates a stronger Sp1 binding site, which results in 2- to 4-fold increased transcriptional levels of MDM2 and, therefore, the attenuation of p53. In humans, SNP309 is associated with increased risk for cancer.84,85 MDM2 SNP309 also appears to be under evolutionary selection pressure, with its allele frequency differing greatly among populations with different ethnic backgrounds. The T allele is the ancestral allele with more than 90% frequency in African populations. However, in Caucasian and Asian populations, the frequency of the G allele, which arose more recently, is increased to ~40%. Similar evolutionary selection of certain alleles has been observed in MDM4, HAUSP (herpesvirus-associated ubiquitin-specific protease), and LIF genes.86-88 MDM4 is a structural homolog of MDM2, which is also an important negative regulator of p53 through binding to the amino terminus of p53 protein. HAUSP is a deubiquitinating enzyme that plays a crucial role in regulating the levels of p53, MDM2, and MDM4. HAUSP binds and stabilizes p53 by deubiquitination. Meanwhile, HAUSP can also deubiquitinate Mdm2 and Mdm4 and down-regulate p53 levels.89 Interestingly, selected alleles of MDM2, MDM4, HAUSP, and LIF genes have been found to be strongly associated with decreased fertility in IVF patients (Table 1).
Advanced maternal age is usually associated with decreased ovarian reserve and quality of oocytes and has a significant negative impact upon fertility. Impaired implantation is a more prominent cause for infertility in young patients with unexplained infertility compared with patients of advanced maternal age. Interestingly, the association of these at-risk alleles in the p53 pathway with decreased fertility mainly occurs in young patients, and the association disappears or is largely reduced in patients with advanced maternal age (Table 1).88 These observations strongly suggest a role of the p53 pathway in regulation of human reproduction through LIF and indicate the presence of candidate functional SNPs in these alleles that influence p53 function in reproduction. These observations also suggest that the regulation of reproduction is an important cause for the observed positive selection on certain alleles in the p53 pathway.
The Roles of p63 and p73 in Reproduction in Humans
Although TAp63 has an important function in maintaining the fidelity of female germ cells in mice, TAp63 also plays a similar role in humans. Infertility in patients with p63 mutation has been reported. Four females from the same family with RHS syndrome due to a germline p63 mutation all exhibited premature menopause and infertility.90 These patients have a heterozygous single nucleotide deletion at codon 595, within exon 14 of the p63 gene, which results in a modification of the reading frame and leads to a stop codon 65 bp downstream to the canonic TGA stop codon. These patients all have premature menopause at age of around 30.
The involvement of p63 and p73 in human reproduction has been further demonstrated by our recent study showing that the selected alleles of the p63 and p73 genes are associated with decreased fertility in IVF patients.63 In human p63 and p73 genes, there are SNPs that appear to be under evolutionary selection pressure. These SNPs include a SNP in the p63 gene (rs17506395) and 2 SNPs in the p73 gene (rs4648551 and rs6695978). A clear enrichment of selected alleles of these SNPs has been observed in IVF patients ( Table 1 ). Interestingly, the at-risk alleles in the p63 and p73 genes for human fertility are enriched in patients of advanced maternal age, who are more likely to have decreased oocyte quality and ovary function. These data are consistent with the idea that p63 and p73 may play important roles in maintaining the quality of oocytes and ovary function in humans.
Conclusion and Future Perspectives
The p53 family proteins are evolutionarily conserved, and an ancestral gene of p53/p63/p73 existed at early times during evolution. The primordial functions of this gene family are the surveillance of the genomic integrity of germ cells to ensure faithful development and reproduction. These functions appear to be evolutionarily conserved and can be found in mice and humans. In vertebrates, the p53 family proteins have developed additional important functions in reproduction, such as the regulation of embryonic implantation by p53. These functions of p53 family proteins in reproduction result in strong evolutionary selection pressure on this family of genes. Later in evolution, when the average lifetime of the individual exceeded the average lifetime of a cell, thus making tissue renewal mandatory, the new functions of the p53 family proteins were evolved, such as the tumor suppressive function of p53 and p73.
In humans, causes of infertility can be related to both male and female conditions. Approximately 60% of infertility is caused by female conditions. Among the causes of female infertility, ovulatory dysfunction is the single most frequent cause. The quality and quantity of the primordial follicle pool are a key of female fertility conservation. Oocytes exist as a limited population largely arrested in a potentially vulnerable, tetraploid state for a prolonged period. As women age, their ovary function declines and aneuploidy rate in oocytes increases. The effects of reproductive aging on women have become more important as women increasingly become pregnant at older ages. Studies in mice have shown that TAp73 is involved in maintaining genomic stability in oocytes. TAp73 levels decline with aging, which may contribute to the increased aneuploidy in aged oocytes. It is currently unclear what causes the decrease of TAp73 expression with natural aging. It will be interesting to study whether increasing TAp73 levels in aged oocytes will improve ovary function and the quality of oocytes.
Preserving female fertility is also an increasingly urgent issue for female cancer patients. Ovarian failure and infertility are major adverse effects of anticancer therapy in children and young women. Chemotherapeutic agents can activate TAp63 in oocytes and induce apoptosis to remove damaged cells. This function of TAp63 in protecting the female germ line fidelity is crucial to ensure the integrity of the genome transmitted to the next generation; however, under conditions of excessive or chronic genotoxic stress, the same function of TAp63 may be responsible for the depletion of the oocyte population, which has a negative consequence of fertility. Germ cells are very sensitive to DNA damage insults. It has been shown that oocytes from mouse primordial follicles can be almost completely removed even at low doses of radiation.91 A recent study demonstrated that the c-Abl kinase inhibitor imatinib, which blocks activation of TAp63, can prevent cisplatin-induced depletion of the follicle reserve and premature ovarian failure in mice (Figure 2).67 It is possible that imatinib or other c-Abl inhibitors could be used to preserve female fertility during anticancer therapy. The challenging question is how to ensure that such treatment will not affect the antitumor activity of chemotherapeutic agents and at the same time not save damaged germ cells that may potentially increase mutation frequency. Further investigation is needed to gain a better understanding of the molecular mechanisms used by damaged germ cells to repair lesions and survive, which will help to develop effective approaches that preserve female fertility and at the same time maintain the genomic integrity of germ cells during anticancer therapy.
Inefficient implantation is another important cause of infertility, especially in a portion of patients with unexplained infertility. Inefficient implantation is often a major cause of pregnancy failure after an IVF procedure. Administering recombinant human LIF may improve implantation in patients with inefficient implantation. Indeed, in a small group of patients with pure female factor infertility with unexplained recurrent implantation failure, the use of recombinant LIF in an IVF procedure improved implantation and increased pregnancy rates.92 However, in an extended study, LIF treatment failed to show improved implantation in patients with implantation failure.93 These inconsistent results suggest that multiple gene products and signaling pathways could be involved in the regulation of implantation. Therefore, it is important to carefully select patient groups who are more likely to have LIF deficiency for further study. Our finding that the p53 pathway regulates uterine LIF levels suggests that analyzing SNPs of the p53 pathway in IVF patients may help to identify patients who could potentially benefit from therapy with recombinant human LIF.
In summary, the discoveries of the functions of the p53 family proteins in maintaining female germ cell genomic integrity and regulating embryonic implantation have demonstrated that the p53 family proteins are important regulators of female reproduction. Further understanding of the functions of the p53 family proteins in reproduction could lead to the development of new strategies for fertility preservation and fertility treatment and could reduce the incidence of infertility.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
W.H. is supported by grants from the U.S. National Institutes of Health (1P30CA147892-01) and U.S. Department of Defense (W81XWH-10-1-0435).
References
- 1. Dotsch V, Bernassola F, Coutandin D, Candi E, Melino G. p63 and p73, the ancestors of p53. Cold Spring Harb Perspect Biol. 2010;2:a004887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yang A, Kaghad M, Caput D, McKeon F. On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet. 2002;18:90-5 [DOI] [PubMed] [Google Scholar]
- 3. De Laurenzi V, Costanzo A, Barcaroli D, et al. Two new p73 splice variants, gamma and delta, with different transcriptional activity. J Exp Med. 1998;188:1763-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. De Laurenzi VD, Catani MV, Terrinoni A, et al. Additional complexity in p73: induction by mitogens in lymphoid cells and identification of two new splicing variants epsilon and zeta. Cell Death Differ. 1999;6:389-90 [DOI] [PubMed] [Google Scholar]
- 5. Bourdon JC, Fernandes K, Murray-Zmijewski F, et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19:2122-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang A, Kaghad M, Wang Y, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305-16 [DOI] [PubMed] [Google Scholar]
- 7. Moll UM, Slade N. p63 and p73: roles in development and tumor formation. Mol Cancer Res. 2004;2:371-86 [PubMed] [Google Scholar]
- 8. Murray-Zmijewski F, Lane DP, Bourdon JC. p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006;13:962-72 [DOI] [PubMed] [Google Scholar]
- 9. Levine AJ, Hu W, Feng Z. The P53 pathway: what questions remain to be explored? Cell Death Differ. 2006;13:1027-36 [DOI] [PubMed] [Google Scholar]
- 10. Jost CA, Marin MC, Kaelin WG., Jr p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature. 1997;389:191-4 [DOI] [PubMed] [Google Scholar]
- 11. Keyes WM, Wu Y, Vogel H, Guo X, Lowe SW, Mills AA. p63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes Dev. 2005;19:1986-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Flores ER, Sengupta S, Miller JB, et al. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7:363-73 [DOI] [PubMed] [Google Scholar]
- 13. Casciano I, Mazzocco K, Boni L, et al. Expression of DeltaNp73 is a molecular marker for adverse outcome in neuroblastoma patients. Cell Death Differ. 2002;9:246-51 [DOI] [PubMed] [Google Scholar]
- 14. Crook T, Nicholls JM, Brooks L, O’Nions J, Allday MJ. High level expression of deltaN-p63: a mechanism for the inactivation of p53 in undifferentiated nasopharyngeal carcinoma (NPC)? Oncogene. 2000;19:3439-44 [DOI] [PubMed] [Google Scholar]
- 15. Puig P, Capodieci P, Drobnjak M, et al. p73 Expression in human normal and tumor tissues: loss of p73alpha expression is associated with tumor progression in bladder cancer. Clin Cancer Res. 2003;9:5642-51 [PubMed] [Google Scholar]
- 16. Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708-13 [DOI] [PubMed] [Google Scholar]
- 17. Yang A, Schweitzer R, Sun D, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714-8 [DOI] [PubMed] [Google Scholar]
- 18. Celli J, Duijf P, Hamel BC, et al. Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell. 1999;99:143-53 [DOI] [PubMed] [Google Scholar]
- 19. van Bokhoven H, Hamel BC, Bamshad M, et al. p63 Gene mutations in eec syndrome, limb-mammary syndrome, and isolated split hand-split foot malformation suggest a genotype-phenotype correlation. Am J Hum Genet. 2001;69:481-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brunner HG, Hamel BC, Van Bokhoven H. The p63 gene in EEC and other syndromes. J Med Genet. 2002;39:377-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van Bokhoven H, Brunner HG. Splitting p63. Am J Hum Genet. 2002;71:1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang A, Walker N, Bronson R, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99-103 [DOI] [PubMed] [Google Scholar]
- 23. Pozniak CD, Barnabe-Heider F, Rymar VV, Lee AF, Sadikot AF, Miller FD. p73 is required for survival and maintenance of CNS neurons. J Neurosci. 2002;22:9800-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Walsh GS, Orike N, Kaplan DR, Miller FD. The invulnerability of adult neurons: a critical role for p73. J Neurosci. 2004;24:9638-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Agostini M, Tucci P, Chen H, et al. p73 regulates maintenance of neural stem cell. Biochem Biophys Res Commun. 2010;403:13-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. De Laurenzi V, Raschella G, Barcaroli D, et al. Induction of neuronal differentiation by p73 in a neuroblastoma cell line. J Biol Chem. 2000;275:15226-31 [DOI] [PubMed] [Google Scholar]
- 27. Hooper C, Tavassoli M, Chapple JP, et al. TAp73 isoforms antagonize Notch signalling in SH-SY5Y neuroblastomas and in primary neurones. J Neurochem. 2006;99:989-99 [DOI] [PubMed] [Google Scholar]
- 28. Fernandes AD, Atchley WR. Biochemical and functional evidence of p53 homology is inconsistent with molecular phylogenetics for distant sequences. J Mol Evol. 2008;67:51-67 [DOI] [PubMed] [Google Scholar]
- 29. Nedelcu AM, Tan C. Early diversification and complex evolutionary history of the p53 tumor suppressor gene family. Dev Genes Evol. 2007;217:801-6 [DOI] [PubMed] [Google Scholar]
- 30. Pankow S, Bamberger C. The p53 tumor suppressor-like protein nvp63 mediates selective germ cell death in the sea anemone Nematostella vectensis. PLoS ONE. 2007;2:e782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kelley ML, Winge P, Heaney JD, et al. Expression of homologues for p53 and p73 in the softshell clam (Mya arenaria), a naturally-occurring model for human cancer. Oncogene. 2001;20:748-58 [DOI] [PubMed] [Google Scholar]
- 32. Derry WB, Putzke AP, Rothman JH. Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science. 2001;294:591-5 [DOI] [PubMed] [Google Scholar]
- 33. Jin S, Martinek S, Joo WS, et al. Identification and characterization of a p53 homologue in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2000;97:7301-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cheng R, Ford BL, O’Neal PE, et al. Zebrafish (Danio rerio) p53 tumor suppressor gene: cDNA sequence and expression during embryogenesis. Mol Mar Biol Biotechnol. 1997;6:88-97 [PubMed] [Google Scholar]
- 35. Soussi T, Caron de Fromentel C, Mechali M, May P, Kress M. Cloning and characterization of a cDNA from Xenopus laevis coding for a protein homologous to human and murine p53. Oncogene. 1987;1:71-8 [PubMed] [Google Scholar]
- 36. Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM. Drosophila p53 binds a damage response element at the reaper locus. Cell. 2000;101:103-13 [DOI] [PubMed] [Google Scholar]
- 37. Huyen Y, Jeffrey PD, Derry WB, et al. Structural differences in the DNA binding domains of human p53 and its C. elegans ortholog Cep-1. Structure. 2004;12:1237-43 [DOI] [PubMed] [Google Scholar]
- 38. Rutkowski R, Hofmann K, Gartner A. Phylogeny and function of the invertebrate p53 superfamily. Cold Spring Harb Perspect Biol. 2010;2:a001131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Belyi VA, Ak P, Markert E, et al. The origins and evolution of the p53 family of genes. Cold Spring Harb Perspect Biol 2010;2:a001198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ollmann M, Young LM, Di Como CJ, et al. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell. 2000;101:91-101 [DOI] [PubMed] [Google Scholar]
- 41. Derry WB, Bierings R, van Iersel M, Satkunendran T, Reinke V, Rothman JH. Regulation of developmental rate and germ cell proliferation in Caenorhabditis elegans by the p53 gene network. Cell Death Differ. 2007;14:662-70 [DOI] [PubMed] [Google Scholar]
- 42. Stergiou L, Doukoumetzidis K, Sendoel A, Hengartner MO. The nucleotide excision repair pathway is required for UV-C-induced apoptosis in Caenorhabditis elegans. Cell Death Differ. 2007;14:1129-38 [DOI] [PubMed] [Google Scholar]
- 43. Sogame N, Kim M, Abrams JM. Drosophila p53 preserves genomic stability by regulating cell death. Proc Natl Acad Sci U S A. 2003;100:4696-701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Akdemir F, Christich A, Sogame N, Chapo J, Abrams JM. p53 directs focused genomic responses in Drosophila. Oncogene. 2007;26:5184-93 [DOI] [PubMed] [Google Scholar]
- 45. Colombani J, Polesello C, Josue F, Tapon N. Dmp53 activates the Hippo pathway to promote cell death in response to DNA damage. Curr Biol. 2006;16:1453-8 [DOI] [PubMed] [Google Scholar]
- 46. Brodsky MH, Weinert BT, Tsang G, et al. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol Cell Biol. 2004;24:1219-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lu WJ, Chapo J, Roig I, Abrams JM. Meiotic recombination provokes functional activation of the p53 regulatory network. Science. 2010;328:1278-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee JH, Lee E, Park J, Kim E, Kim J, Chung J. In vivo p53 function is indispensable for DNA damage-induced apoptotic signaling in Drosophila. FEBS Lett. 2003;550:5-10 [DOI] [PubMed] [Google Scholar]
- 49. Rotter V, Schwartz D, Almon E, et al. Mice with reduced levels of p53 protein exhibit the testicular giant-cell degenerative syndrome. Proc Natl Acad Sci U S A. 1993;90:9075-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hasegawa M, Zhang Y, Niibe H, Terry NH, Meistrich ML. Resistance of differentiating spermatogonia to radiation-induced apoptosis and loss in p53-deficient mice. Radiat Res. 1998;149:263-70 [PubMed] [Google Scholar]
- 51. Sjoblom T, Lahdetie J. Expression of p53 in normal and gamma-irradiated rat testis suggests a role for p53 in meiotic recombination and repair. Oncogene. 1996;12:2499-505 [PubMed] [Google Scholar]
- 52. Beumer TL, Roepers-Gajadien HL, Gademan IS, et al. The role of the tumor suppressor p53 in spermatogenesis. Cell Death Differ. 1998;5:669-77 [DOI] [PubMed] [Google Scholar]
- 53. Schmid P, Lorenz A, Hameister H, Montenarh M. Expression of p53 during mouse embryogenesis. Development. 1991;113:857-65 [DOI] [PubMed] [Google Scholar]
- 54. Nicol CJ, Harrison ML, Laposa RR, Gimelshtein IL, Wells PG. A teratologic suppressor role for p53 in benzo[a]pyrene-treated transgenic p53-deficient mice. Nat Genet. 1995;10:181-7 [DOI] [PubMed] [Google Scholar]
- 55. Norimura T, Nomoto S, Katsuki M, Gondo Y, Kondo S. p53-dependent apoptosis suppresses radiation-induced teratogenesis. Nat Med. 1996;2:577-80 [DOI] [PubMed] [Google Scholar]
- 56. Hu W, Feng Z, Teresky AK, Levine AJ. p53 regulates maternal reproduction through LIF. Nature. 2007;450:721-4 [DOI] [PubMed] [Google Scholar]
- 57. Hu W, Feng Z, Atwal GS, Levine AJ. p53: a new player in reproduction. Cell Cycle. 2008;7:848-52 [DOI] [PubMed] [Google Scholar]
- 58. Gonzalez RR, Rueda BR, Ramos MP, Littell RD, Glasser S, Leavis PC. Leptin-induced increase in leukemia inhibitory factor and its receptor by human endometrium is partially mediated by interleukin 1 receptor signaling. Endocrinology. 2004;145:3850-7 [DOI] [PubMed] [Google Scholar]
- 59. Terakawa J, Hondo E, Sugiyama M, Wakitani S, Stewart CL, Kiso Y. Agrin pathway is controlled by leukemia inhibitory factor (LIF) in murine implantation. J Reprod Dev. 2009;55:293-8 [DOI] [PubMed] [Google Scholar]
- 60. Stewart CL, Kaspar P, Brunet LJ, et al. Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature. 1992;359:76-9 [DOI] [PubMed] [Google Scholar]
- 61. Chen JR, Cheng JG, Shatzer T, Sewell L, Hernandez L, Stewart CL. Leukemia inhibitory factor can substitute for nidatory estrogen and is essential to inducing a receptive uterus for implantation but is not essential for subsequent embryogenesis. Endocrinology. 2000;141:4365-72 [DOI] [PubMed] [Google Scholar]
- 62. Vogiagis D, Salamonsen LA. Review: the role of leukaemia inhibitory factor in the establishment of pregnancy. J Endocrinol. 1999;160:181-90 [DOI] [PubMed] [Google Scholar]
- 63. Feng Z, Zhang C, Kang HJ, et al. et al. The regulation of female reproduction by p53 and its family members. FASEB J. 2011. Forthcoming [DOI] [PMC free article] [PubMed]
- 64. Yamashita S, Newbold RR, McLachlan JA, Korach KS. The role of the estrogen receptor in uterine epithelial proliferation and cytodifferentiation in neonatal mice. Endocrinology. 1990;127:2456-63 [DOI] [PubMed] [Google Scholar]
- 65. Suh EK, Yang A, Kettenbach A, et al. p63 protects the female germ line during meiotic arrest. Nature. 2006;444:624-8 [DOI] [PubMed] [Google Scholar]
- 66. Kurita T, Cunha GR, Robboy SJ, Mills AA, Medina RT. Differential expression of p63 isoforms in female reproductive organs. Mech Dev. 2005;122:1043-55 [DOI] [PubMed] [Google Scholar]
- 67. Gonfloni S, Di Tella L, Caldarola S, et al. Inhibition of the c-Abl-TAp63 pathway protects mouse oocytes from chemotherapy-induced death. Nat Med. 2009;15:1179-85 [DOI] [PubMed] [Google Scholar]
- 68. Livera G, Petre-Lazar B, Guerquin MJ, Trautmann E, Coffigny H, Habert R. p63 null mutation protects mouse oocytes from radio-induced apoptosis. Reproduction. 2008;135:3-12 [DOI] [PubMed] [Google Scholar]
- 69. Tomasini R, Tsuchihara K, Wilhelm M, et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 2008;22:2677-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Baker DJ, Jeganathan KB, Cameron JD, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36:744-9 [DOI] [PubMed] [Google Scholar]
- 71. Leland S, Nagarajan P, Polyzos A, et al. Heterozygosity for a Bub1 mutation causes female-specific germ cell aneuploidy in mice. Proc Natl Acad Sci U S A. 2009;106:12776-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shuda K, Schindler K, Ma J, Schultz RM, Donovan PJ. Aurora kinase B modulates chromosome alignment in mouse oocytes. Mol Reprod Dev. 2009;76:1094-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhang Y, Lees E. Identification of an overlapping binding domain on Cdc20 for Mad2 and anaphase-promoting complex: model for spindle checkpoint regulation. Mol Cell Biol. 2001;21:5190-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Spandorfer SD, Davis OK, Barmat LI, Chung PH, Rosenwaks Z. Relationship between maternal age and aneuploidy in in vitro fertilization pregnancy loss. Fertil Steril. 2004;81:1265-9 [DOI] [PubMed] [Google Scholar]
- 75. Serafini P, Rocha AM, Osorio CT, da Silva I, Motta EL, Baracat EC. Endometrial leukemia inhibitory factor as a predictor of pregnancy after in vitro fertilization. Int J Gynaecol Obstet. 2008;102:23-7 [DOI] [PubMed] [Google Scholar]
- 76. Mikolajczyk M, Wirstlein P, Skrzypczak J. The impact of leukemia inhibitory factor in uterine flushing on the reproductive potential of infertile women—a prospective study. Am J Reprod Immunol. 2007;58:65-74 [DOI] [PubMed] [Google Scholar]
- 77. Tsai HD, Chang CC, Hsieh YY, Lo HY. Leukemia inhibitory factor expression in different endometrial locations between fertile and infertile women throughout different menstrual phases. J Assist Reprod Genet. 2000;17:415-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Serafini PC, Silva ID, Smith GD, Motta EL, Rocha AM, Baracat EC. Endometrial claudin-4 and leukemia inhibitory factor are associated with assisted reproduction outcome. Reprod Biol Endocrinol. 2009;7:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Murphy ME. Polymorphic variants in the p53 pathway. Cell Death Differ. 2006;13:916-20 [DOI] [PubMed] [Google Scholar]
- 80. Kang HJ, Feng Z, Sun Y, et al. Single-nucleotide polymorphisms in the p53 pathway regulate fertility in humans. Proc Natl Acad Sci U S A. 2009;106:9761-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Reinbold M, Luo JL, Nedelko T, et al. Common tumour p53 mutations in immortalized cells from Hupki mice heterozygous at codon 72. Oncogene. 2008;27:2788-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kay C, Jeyendran RS, Coulam CB. p53 tumour suppressor gene polymorphism is associated with recurrent implantation failure. Reprod Biomed Online. 2006;13:492-6 [DOI] [PubMed] [Google Scholar]
- 83. Shi H, Tan SJ, Zhong H, et al. Winter temperature and UV are tightly linked to genetic changes in the p53 tumor suppressor pathway in Eastern Asia. Am J Hum Genet. 2009;84:534-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bond GL, Hu W, Bond EE, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591-602 [DOI] [PubMed] [Google Scholar]
- 85. Hu W, Feng Z, Ma L, et al. A single nucleotide polymorphism in the MDM2 gene disrupts the oscillation of p53 and MDM2 levels in cells. Cancer Res. 2007;67:2757-65 [DOI] [PubMed] [Google Scholar]
- 86. Atwal GS, Kirchhoff T, Bond EE, et al. Evolutionary selection and altered tumor formation of genetic variants in the human Mdm4 oncogene. Proc Natl Acad Sci U S A. 2009. Forthcoming [DOI] [PMC free article] [PubMed]
- 87. Atwal GS, Bond GL, Metsuyanim S, et al. Haplotype structure and selection of the MDM2 oncogene in humans. Proc Natl Acad Sci U S A. 2007;104:4524-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kang HJ, Feng Z, Sun Y, et al. Single-nucleotide polymorphisms in the p53 pathway regulate fertility in humans. Proc Natl Acad Sci U S A. 2009;106:9761-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Brooks CL, Li M, Hu M, Shi Y, Gu W. The p53–Mdm2–HAUSP complex is involved in p53 stabilization by HAUSP. Oncogene. 2007;26:7262-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Holder-Espinasse M, Martin-Coignard D, Escande F, Manouvrier-Hanu S. A new mutation in TP63 is associated with age-related pathology. Eur J Hum Genet. 2007;15:1115-20 [DOI] [PubMed] [Google Scholar]
- 91. Morita Y, Perez GI, Paris F, et al. Oocyte apoptosis is suppressed by disruption of the acid sphingomyelinase gene or by sphingosine-1-phosphate therapy. Nat Med. 2000;6:1109-14 [DOI] [PubMed] [Google Scholar]
- 92. Brinsden PR, Ndukwe G, Engrand P, et al. Does recombinant human leukaemia inhibitory factor improve implantation in women with recurrent failure of assisted reproduction treatment? Hum Reprod. 2003;18:18 [Google Scholar]
- 93. Brinsden PR, Alam V, de Moustier B, Engrand P. Recombinant human leukemia inhibitory factor does not improve implantation and pregnancy outcomes after assisted reproductive techniques in women with recurrent unexplained implantation failure. Fertil Steril. 2009;91:1445-7 [DOI] [PubMed] [Google Scholar]


