Abstract
Human papillomavirus (HPV) causes cervical cancer and other hyperproliferative diseases. There currently are no approved antiviral drugs for HPV that directly decrease viral DNA load and that have low toxicity. We report the potent anti-HPV activity of two N-methylpyrrole-imidazole polyamides of the hairpin type, polyamide 1 (PA1) and polyamide 25 (PA25). Both polyamides have potent anti-HPV activity against 3 different genotypes when tested on cells maintaining HPV episomes. The compounds were tested against HPV16 (in W12 cells), HPV18 (in Ker4-18 cells), and HPV31 (tested in HPV31 maintaining cells). From a library of polyamides designed to recognize AT-rich DNA sequences such as those in or near E1 or E2 binding sites of the HPV16 origin of replication (ori), 4 polyamides were identified that possessed apparent IC50s ≤ 150 nM with no evidence of cytotoxicity and we report two highly-active compounds here. Treatment of epithelia engineered in organotypic cultures with these compounds also causes a dose-dependent loss of HPV episomal DNA that correlates with accumulation of compounds in the nucleus. Bromodeoxyuridine (BrdU) incorporation demonstrates that DNA synthesis in organotypic cultures is suppressed upon compound treatment, correlating with a loss of HPV16 and HPV18 episomes. PA1 and PA25 are currently in preclinical development as an antiviral compound for treatment of HPV-related disease, including cervical dysplasia. PA1 and related polyamides offer promise as antiviral agents and the ability to regulate HPV episomal levels in cells for the study of HPV biology. We also report that anti-HPV16 activity for Distamycin A, a natural product related to PA1, is accompanied by significant cellular toxicity.
Keywords: papillomavirus, antiviral, polyamide, episome, cervical cancer
Introduction
Human papillomavirus (HPV) causes the vast majority of cervical cancers, with HPV16 and HPV18 being responsible for greater than 65% of these cases (Munoz et al., 2003; zur Hausen, 2002). HPV is highly trophic for epithelia. Initial infection, which is believed to occur within the basal or proliferative cell layers by way of micro-lesions within the epithelium, establishes viral episomes within the proliferative, basal cells. During productive infections (i.e. warts) where progeny virions are made, episomes are amplified within the differentiating keratinocytes via an unscheduled round of DNA synthesis. This unscheduled round of synthesis produces viral DNA for packaging in virions and eventual shedding within the most apical, differentiated cells of the epithelium (Cheng et al., 1995; Garner-Hamrick et al., 2004; Howley, 1996).
Most HPV infections, whether caused by high- or low-risk viruses, clear on their own. Progression to high-grade disease of the cervix and invasive cervical cancer is thought to be dependent upon several factors including persistence of infection, HPV viral DNA load, and HPV DNA integration (Stanley et al., 2007; Woodman et al., 2007). Antiviral compounds that decrease or eliminate human papillomavirus (HPV) levels and reverse associated disease pathology are not available (Fradet-Turcotte and Archambault, 2007). An antiviral agent effective at reducing HPV load or persistence has the potential to impact public health, especially given that vaccines protect against a small number of viral subtypes and widespread use of HPV DNA testing in cervical cancer screening programs is anticipated (Sankaranarayanan et al., 2009; Schiffman and Wacholder, 2009). In addition, such compounds have the potential to be used as tools to manipulate and study HPV episomes in cells.
Episome-maintaining keratinocytes have been isolated from low-grade HPV lesions (Doorbar et al., 1990; Meyers et al., 1992) and from keratinocytes transfected with cloned HPV genomes (Flores et al., 2000; Frattini et al., 1996; Garner-Hamrick and Fisher, 2002). In general, high-risk HPV episomes appear to be required for robust establishment of episome-maintaining cell lines in primary keratinocytes. This is likely due to the ability of high-risk HPVs to impart effects upon keratinocytes such as extension of cell lifespan, immortalization, circumvention of innate immunity, and promotion of DNA synthesis (Hebner and Laimins, 2006; Howley, 1996; Longworth and Laimins, 2004; Thomas et al., 2001), which are required for episome maintenance and increased survival of the host cells.
Virally encoded DNA polymerases such as HIV reverse transcriptase have been successful medicinal chemistry targets, but HPV encodes few proteins that resemble traditional targets for medicinal chemistry. Thus, the small HPV genome does not encode a DNA polymerase but depends on the viral proteins E1 and E2 to initiate viral DNA replication and recruit host cell DNA replication machinery to the viral origin of replication (ori). However, some possible drug strategies include targeting the helicase and ATPase activities of the HPV E1 protein and disrupting the E1/E2 protein-protein interactions that are of particularly high importance for a small virus encoding so few proteins (Fradet-Turcotte and Archambault, 2007), or targeting activities of the viral oncogenes such as E7 (He and Fisher, 2003). To our knowledge, these strategies have been effective only on low-risk HPV. To attempt a fresh approach against high-risk HPV, we based our initial chemical synthesis of potential antiviral agents on the proposition that targeting long AT-rich regions of the viral genome with DNA-binding polyamides would interfere with viral maintenance, in part because such regions occur near viral DNA binding sites for important viral E1 and/or E2 proteins, including sites at the ori essential for HPV replication. Here we describe the activity and use of two synthetic N-methyl-pyrrole/N-methyl-imidazole containing polyamides (Dervan, 2001; Dervan and Edelson, 2003; White, 1998), polyamide 1 (PA1) and polyamide 25 (PA25), which is active against HPV16 in W12E cells. We show that PA1 decreased the HPV16 episome count in W12E cells with a pseudo IC50 of 100 nM. In addition, PA1 decreased the HPV16 episome levels of organotypic cultures made from W12E cells.
Materials and Methods
Synthesis of Polyamide 1 and Polyamide 25
Polyamide 1 (PA1) has the sequence dIm-Py-Py-β-Py-Py-Py-γ-Py-Py-β-Py-Py-Py-Py-β-Ta, where the amino acid building blocks and terminating amine are denoted as follows: dIm is desamino-N-methylimidazole, Py is N-methylpyrrole, β is β–alanine, γ is γ–aminobutyric acid, and Ta is CH3N(CH2CH2CH2NH2)2 (Dervan, 2001; Dervan and Edelson, 2003; White, 1998). PA1 was prepared by a variety of methods, including manual and automated solid-phase methods and by combined solid-phase and solution methods (Baird and Dervan, 1996; Krutzik and Chamberlin, 2002; Wang et al., 2001; Wurtz et al., 2001; Xiao et al., 2000). Automated solid phase synthesis was performed on an ABI 433A peptide synthesizer (Applied Biosystems, Foster City, California) by t-Boc methods (Baird and Dervan, 1996) while manual solid phase synthesis used Fmoc methods (Wurtz et al., 2001). Purification of the crude products was accomplished as previously reported (Baird and Dervan, 1996; Wurtz et al., 2001). Desired product and pure HPLC fractions were identified initially by ultraviolet and visible light detection (UV/Vis) and Matrix-Assisted Laser Desorption Ionization Mass Spectrometry (MALDI, Global Peptides, Fort Collins, Colorado), and later by HPLC/mass spectrometry with diode array UV/Vis detection and electrospray ionization (ESI HPLC/MS, Agilent Technologies, Santa Clara, California). Purity ≥ 95% by reverse phase (RP) HPLC/mass spec); high resolution mass spectrometry (HRMS, Danforth Plant Science Center, St. Louis, MO) of the purified product was carried out to confirm composition: HRMS found 1894.89331; calc. 1894.89038 for [M+H]+ (C91H112N31O16).
Polyamide 25 (PA25) has the sequence dIm-Py-Py-β-Py-Py-Im-β-Py-Py-γ-Py-Py-β-Py-Py-Py-β-Py-Py-Py-β-Ta and was prepared and purified by analogy with PA1 (purity 95% by RP HPLC/mass spec); high resolution mass spectrometry (HRMS, Danforth Plant Science Center, St. Louis, MO) of the purified product was carried out to confirm its composition: HRMS found 2526.15570; calc. 2526.15186 for [M+H]+ (C120H145N42O22).
Reagents were prepared according to the literature (Xiao et al., 2000) or purchased from Wako Chemicals (Richmond, Virginia), Oakwood (West Columbia, South Carolina), Chem-Impex (Wood Dale, Illinois), Peptides International (Louisville, Kentucky), Aldrich (Milwaukee, Wisconsin) and EMD-NovaBiochem (Gibbstown, New Jersey).
Cells and Cell Culture
The extensively analyzed W12E cervical keratinocyte cell line that maintains HPV16 episomes was derived from a low grade cervical dysplasia (15, 23, 32). Ker4-18 cells maintaining HPV18 episomes were derived from cloned HPV18 DNA as previously described for HPV31 cells (23). Cells maintaining HPV31 were the gift of Dr. Lou Laimins (Northwestern University). All three cells were cultured on mitomycin C-treated J2 3T3 cells in media containing three parts Dulbecco’s modified Eagle medium (DMEM) and one part F12 media as previously described (23). Culture media (E media) was supplemented with 0.4 µg/mL hydrocortisone, 10 ng/mL cholera toxin, 5 µg/mL insulin, 24 µg/mL adenine, 5 µg/mL transferrin, 20 pM 3,3’, 5-triiodothyronine (T3), 5 ng/mL epidermal growth factor (EGF), 100 U/mL penicillin,100 µg/mL streptomycin, and 5% fetal bovine serum (FBS). All cells were passaged at 70% confluence using a split ratio of 1:10 (W12E) and 1:20 (Ker4-18).
Compound Efficacy Testing on Episome Harboring Keratinocyte Monolayer Cultures
Lyophilized single-use aliquots of polyamide were dissolved at 10 mM in 100% DMSO and diluted with dH2O to 1 mM. Polyamides were added to cells at concentrations spanning 0.001–10 µM with final DMSO concentration of 0.1% in E media. Cells were also incubated with normal E media or media with 0.1% DMSO as controls. After 48 h incubation, cells were harvested from the plates for isolation of total DNA. Cells were lysed with DNAzol reagent (Invitrogen), precipitated with ethanol, and DNA resuspended in 8 mM NaOH. After resuspension, the DNA solution was neutralized by the addition of 1M Hepes buffer.
Taqman® (Real-Time) PCR and Determination of IC50 Values
Quantitative PCR (Q-PCR) was performed using real-time technology on the ABI PRISM 7700 Sequence Detector (Applied Biosystems, Foster City, CA) as previously described (Garner-Hamrick and Fisher, 2002). All HPV probes and primers were designed within the relevant HPV L1 gene using Primer Express 1.0 software (Applied Biosystems, Foster City, CA). The probes were labeled with the 5’ reporter dye FAM (6-carboxyfluorescein) and the 3’ quencher dye TAMRA (6-carboxytetramethylrhodamine) (Integrated DNA Technologies, IDT). PCR reactions contained a final concentration of 1× JumpStart Taq ReadyMix (Sigma), 200 nM of each primer (IDT), and 250 nM probe (IDT) in a reaction volume of 25 µL. A standard curve was generated using cloned HPV16, 18, or 31 DNA. Total copies of episomal DNA in samples were calculated using the following formula:
(1.82 × 1015 µg/µL stock DNA) / (length in base pairs) × (2) = (copies of DNA) / (µL stock DNA)
For drug efficacy screening, 20 ng of total DNA was added to the PCR reaction and the effect on HPV copy number was plotted as a percent decrease compared to vehicle-treated cells. The log dose-response was plotted for each compound that showed any activity. Each experiment was conducted in triplicate, with an independent experimental at least n = 3 for both PA1 and PA25. The data from each independent experiment were combined, IC50s calculated, and variance determined to reveal the fit of the data to the model and its reliability using a best-fit curve generated by the Microsoft Excel add-in, XLfit (version 2.0; ID Business Solutions, Guildford, UK).
For polyamide efficacy analysis against HPV in the context of organotypic cultures, total DNA was extracted from the rafts with DNAzol reagent and episome copies determined via Q-PCR for each drug-treated and vehicle-treated sample. The reduction in HPV16 or 18 DNA was plotted as a function of drug concentration and expressed as % decrease in HPV episomes relative to the vehicle-treated controls.
Several quality control tests, including use of alternative qPCR amplicons, Southern blotting, and isolation of low molecular weight (“Hirt”) DNA, were routinely conducted through the course of these studies to ensure that the measurements of HPV episomal DNA were accurate.
Cell Viability Assay
Cells were plated in triplicate at 4.5 × 103 cells per well in 96-well tissue culture plates in 100 µL of E-media. Cells were treated 24 h. later with E media, 0.1% DMSO (vehicle control), or PA1 or 25 at doses ranging from 10 nM to 200 µM, and the cells cultured for an additional 48 h. Cell viability was assessed using an MTT assay kit (ATCC, cat# 30-1010K) as per manufacturer’s recommendations. Briefly, 10 µL tetrazolium MTT reagent (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) was added to each well and incubated 4 h at 37° C in a tissue culture incubator. The intracellular purple formazan precipitate was solubilized in detergent according to kit directions, and absorbance was read at 595 nm on a Bio-Rad plate reader. The data was plotted as the percentage of viable cells following 48 h of drug treatment compared with control-treated cells.
Organotypic Cultures
Organotypic cultures were constructed using a dermal equivalent seeded with W12E cells or Ker4-18 cells, and grown at the air liquid interface according to modifications of previously published procedures (Flores et al., 1999; Meyers and Laimins, 1994). Rat tail type I collagen (5.2 mg/mL; Invitrogen cat# A10483-01) was gently mixed on ice with 10× reconstitution buffer (2.2 g NaHCO3 and 4.77 g Hepes in 100 mL 0.05 N NaOH) and 10× DMEM (Invitrogen) at a ratio of 8:1:1. To this mixture, we added 1.5 × 105 per mL J2 3T3 cell fibroblasts. Corning Transwell Inserts (Cat. # 3450; Corning, NY) with a permeable base were transferred to Deep Well Trays (BD Biosciences) and 2 mL of the collagen-fibroblast mixture were transferred into each insert (2 mg/mL final collagen concentration). The trays were placed in a 37°C incubator (5% CO2) where they were allowed to gel for 30 min. DMEM (18 mL) was added to each deep well tray and the gels allowed to contract for 4 days. DMEM was removed and 5 × 105 W12E cells or Ker4-18 cells in 1 mL of E media were added to the surface of each dermal equivalent. After 2 hours, an additional 18 mL of E media was added to the deep well reservoir. After reaching confluence (4 days post-plating of keratinocytes), the cells were raised to the air-liquid interface by placing the transwell inserts upon 2 × 2 inch sterile gauze pads placed in the bottom of the deep well trays. Enough E media (without EGF) was pipetted into the deep well trays to permeate the dermal equivalent but leave the keratinocytes exposed to the atmosphere within the tray. Raft cultures were fed with fresh media every 2 days. All organotypic cultures were routinely treated with 50µM bromodeoxyuridine (BrdU; Sigma) added directly to the culture media 24 h. prior to harvest. The rafts were harvested by carefully bisecting each raft with a scalpel and fixing one half in 3% neutral buffered formalin for histology. The stratified epithelium from the remaining half was carefully stripped from the collagen gel and total DNA extracted with DNAzol.
Treatment of W12E rafts with PA1 occurred at the time of cell plating onto the collagen gel; cells were plated in E media in the presence of a final concentration of 0.1% DMSO containing 10, 50, or 100 µM polyamide. Any compound remaining in the E media from W12E cell plating was removed two days later by washing with media and 1 mL fresh E media was applied to the top of the raft cultures. Rafts were raised to the air-liquid interface upon confluence 2 days later. The rafts were harvested fifteen days post-raise.
Treatment of Ker4-18 rafts was topical in a mixture of 50% DMSO and 50% PEG 400. Polyamide 25 (1 mM) was topically applied to the surface of the raft cultures following stratification in a formulation of 50%PEG-400/50%DMSO either once (1X-at day 6 after raising to the air-liquid interface) or twice (2X-at days two and six after raising).
All organotypic cultures were treated with 50 µM 5-bromodeoxyuridine (BrdU; Sigma) for hours prior to harvesting.
Tissue Processing, Immunohistochemistry, and Cell Counts
The formalin-fixed tissues were embedded in paraffin following dehydration through a graded ethanol series and xylene, and 5 µM sections cut with a Leica rotary microtome. Sections were mounted on poly-L-lysine coated slides. Immunohistochemistry was conducted after de-waxing in xylene and re-hydrating through a graded ethanol series. Sections were incubated for 30 min in 2N HCl at 37°C, rinsed in tap water, and transferred to Tris-buffered saline with Tween (TBST: 50 mM Tris-HCl, 150 mM NaCl, and 0.025% Tween-20) and blocked with 3% normal rabbit serum (MP Biomedicals) for 1 h. Slides were drained and incubated 1 h with antibromodeoxyuridine (BrdU) antibody (clone BU-33; Sigma) at 2.5 µg/mL in diluent buffer (DB: 50 mM Tris-HCl, 150 mM NaCl, 1% bovine serum albumin (BSA), 0.1% Tween-20, pH 7.2). After 3 rinses (5 min each) with TBST, a biotinylated rabbit anti-mouse IgG (Vector Labs, cat. # BA-9200) was applied to the sections at 3 µg/mL for 30 min at r.t. Sections were rinsed 3 times with TBS (TBST without Tween-20) for 5 min. each and were incubated with an avidin-biotinhorseradish peroxidase complex (Vectastain ABC Kit, Vector Labs), prepared according to manufacturer’s recommendations, for 30 min. Sections were developed, and peroxidase was detected with diaminobenzidene (Sigma; cat. # D-0426).
The % BrdU-labeled cells in organotypic cultures was determined by direct counting of cells from matched brightfield and phase contrast photomicrographs of BrdU-stained sections. Multiple sections separated by at least 20 µM were examined, and a minimum of 12 fields were photographed in brightfield and in phase contrast for each culture. These images were printed and nuclei within each field were counted in the phase contrast micrographs (minimum of 5700 nuclei were counted per culture) and total BrdU-stained nuclei were counted in the matching brightfield images. Total labeled nuclei were counted for W12E rafts. In the case of Ker4-18 rafts, which stratified and differentiated much better, nuclei in the basal and suprabasal compartments were analyzed separately. P-values were determined by Student’s t-Test: two-sample unequal variance, two-tailed distribution.
Results
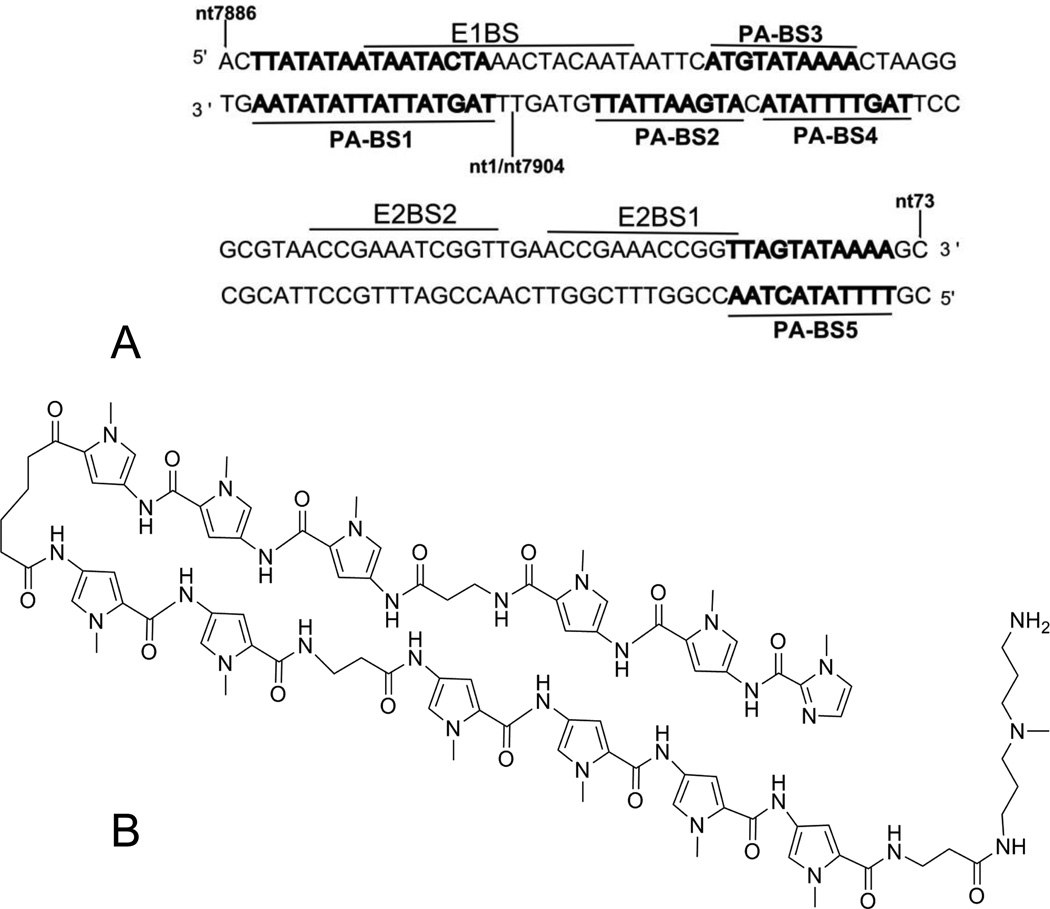
To identify candidate compounds, we designed and screened a library of polyamides that target AT-rich regions of the ori containing or adjacent to the HPV16 E1 and E2 binding sites (Figure 1). Ten negative control compounds were constructed by adding imidazole building blocks, ensuring that increased GC content would be required for strong DNA binding, and a total of twenty-four compounds were initially prepared and tested. Five binding sites for the polyamide library are present within the minimal HPV16 ori (Fig. 1A). Two such sites (PA-BS1, PA-BS2) are within the consensus E1 binding site (E1BS), two overlapping binding sites (PA-BS3, PABS4) fall between the E1BS and an E2 binding site (E2BS2), and one binding site (PA-BS5) overlaps with E2BS1. Furthermore, one site (PA-BS1) can accommodate longer polyamides than the other four sites. Similar sites are conserved in HPV18 and HPV31. Polyamides were initially tested for their effects on episomal DNA levels in W12E cells. A significant decrease in HPV16 episome levels was seen with PA1, targeted to AT-rich regions containing only one GC base pair, while the negative control compounds designed for sequences richer in GC showed no effects on HPV16 episome levels. Any compounds showing a significant initial decrease in HPV episomes at the highest dose (10 µM) were re-tested for a dose response so that an apparent IC50 value could be calculated. A drug concentration of less than 250 nM gave a 50% decrease in viral DNA (apparent IC50) for seven library compounds and a full report of a series of libraries will be published separately. Reproducibility of the results was high with three or more independent experiments performed in triplicate for each active compound (n ≥ 3). Four library compounds had apparent IC50 values equal to or below 150 nM, and one compound, PA1 (Fig. 1B) and PA25, were chosen for further studies. Table 1 summarizes the activity of PA1 and PA25 against 3 different HPV genotypes.
Figure 1. Polyamide binding sites within the HPV16 origin of replication (ori) and the structure of PA1.
(A) The minimal origin of replication (ori) of HPV16 genome is shown with the polyamide binding sites (PA-BS1-5, in bold) that were used in part to design a targeted polyamide library. The nucleotide 1/7904 boundary of the circular HPV16 genome is indicated. Numerous other binding sites for polyamides 1–24 exist within the HPV16 genome. (B) Structure of polyamide 1, selected for further study based upon its potent ability to decrease levels of HPV16 DNA episomes.
Table 1.
Apparent IC50’s of polyamides 1 and 25 against 3 different high risk HPV episomes maintained in cells. The structures of the polyamides are represented by the following symbols: Im: Imidazole, P: Pyrrole, and β: beta-alanine.
| Polyamide 1 | Polyamide 25 | |
|---|---|---|
| ImPPβPPPγPPβPPPPβTa | ImPPβPPImβPPγPPβPPPβPPPβTa | |
| HPV16 | 0.100 ± 0.02 (n=4) | 0.036 ± 0.0004 (n=3) |
| HPV18 | 0.657 ± 0.16 (n=3) | 0.056 ± 0.01 (n=6) |
| HPV31 | 0.108 ± 0.02 (n=4) | 0.030 ± 0.001 (n=3) |
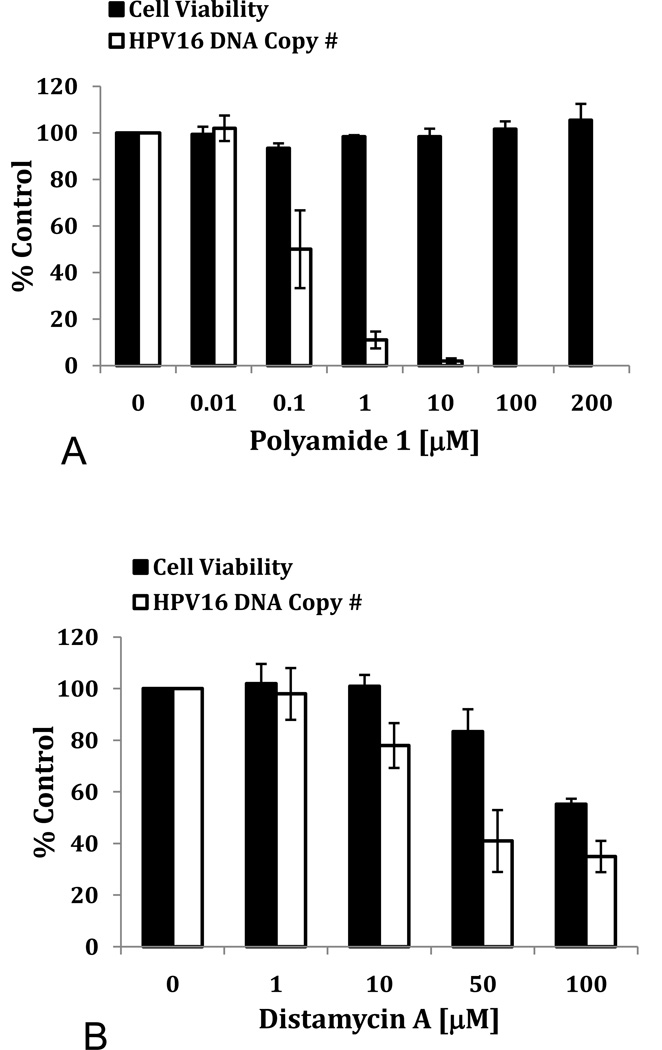
To determine if decreases in HPV16 DNA were associated with cytotoxicity, W12E cells were treated with increasing concentrations of PA1 and cell viability was measured at 48 hours. No toxicity was observed for PA1 up to the highest concentration studied: 200 µM (Fig. 2A). This observation was consistent with additional observations: no changes in cell morphology or apparent growth rate of cells, or other evidence of toxicity have been noted with hairpin polyamides throughout the course of our work or in the literature (Harki et al., 2008; Hsu and Dervan, 2008; Matsuda et al., 2006; Ueno et al., 2009). No cytotoxicity has been observed for PA1 or PA25 against any HPV episome-maintaining cell line.
Figure 2. Effects of PA1 and distamycin A on HPV16 DNA levels and viability of W12E cells.
(A) The effects of PA1 on HPV16 DNA copy number and cellular viability are shown. (B) Distamycin A effects upon HPV16 DNA levels and cellular viability are shown.
Numerous Southern blots have been run to confirm the reduction in HPV episomes by polyamides, and to see if integration of the viral DNA is occurring during the loss of viral DNA over the short culture period. One example is provided is supplemental data (SFig.1a). A dose-dependent loss of episomal DNA in noted. In addition, while loss of the 8 kb episome occurs, there is no subsequent appearance of higher molecular weight DNA hybridizing with the probe, indicating that integration of the viral DNA is not occurring concurrent with loss of episomes. Numerous experiments using Q-PCR indicate that polyamides are reducing cell episomal DNA content in the absence of integration (SFig1b). These separate experiments all show that the final cellular concentration of HPV DNA copies is well below one copy per cell.
PA1 is a synthetic, higher homolog of naturally-occurring oligoamides, such as Distamycin A, which has previously been reported to have antiviral activity against DNA viruses (Becker and Weinber, 1972; Broyles et al., 2004). For this reason, we examined the effects of Distamycin A on monolayer cultures of W12E cells. Distamycin A showed evidence of weak anti-HPV activity with an apparent IC50 of 33 µM. To the best of our knowledge, this is the first report of anti-HPV activity for Distamycin A. However, Distamycin A activity largely tracked with its cellular toxicity (Fig. 2B). Thus, PA1 has a selectivity index (SI) more than 1000-fold superior to a natural product from which its basic structure is derived (SI = IC50/TD50, where TD50 is the dose at which 50% of cells are killed).
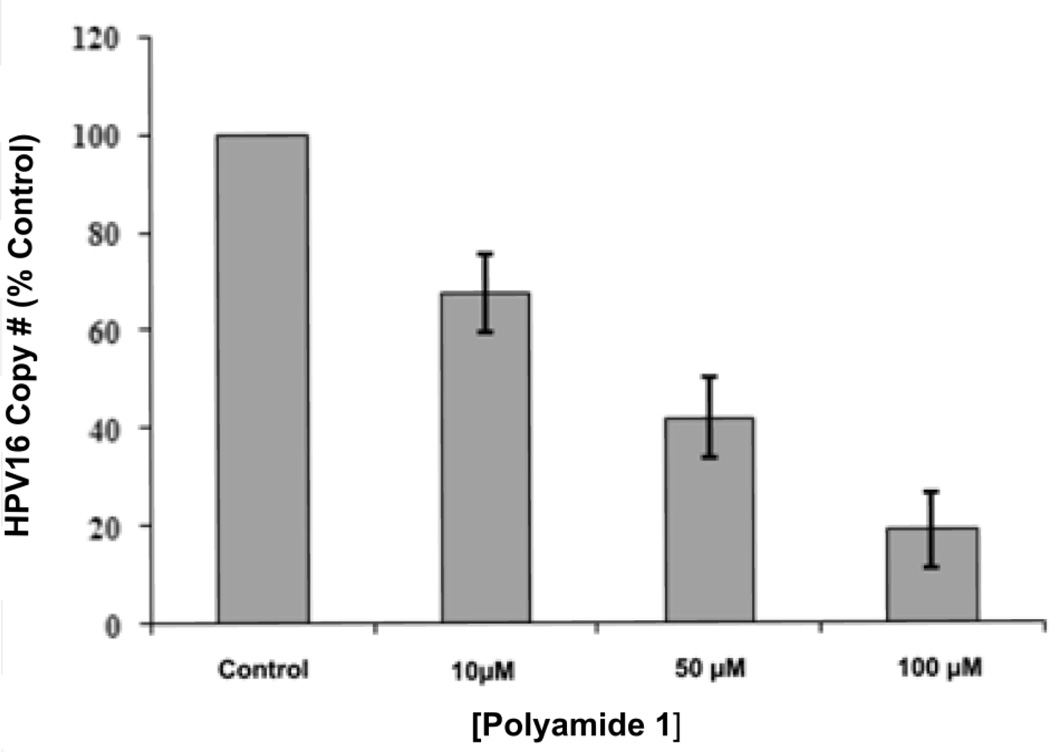
We next examined the effects of PA1 on HPV16 episome levels in an organotypic culture of W12E cells. W12E cells were plated on an engineered “dermal equivalent” in the presence of a single treatment of 10, 50, or 100 µM of PA1. Two days post-plating of W12E cells, the tissue culture media containing PA1 was changed (washing out any unabsorbed drug), the cultures were incubated another two days until confluent, and then raised to the air-liquid interface. The W12E cells were allowed to differentiate for an additional fifteen days. Q-PCR examination of HPV16 levels revealed that the single treatment with PA1 caused a dose-dependent decrease in HPV16 episome levels within the W12E cell-derived epithelium (Fig. 3). The highest dose of 100 µM resulted in an 80% loss of viral DNA at the time of harvest compared to vehicle-treated control raft cultures. While 100 µM is well above the apparent IC50 determined in monolayer cultures, the doses are not inconsistent: we have not yet determined the intracellular concentrations in rafts or monolayers and they may be considerably lower than the nominal concentrations used for apparent IC50 calculations. The 10 µM and 50 µM doses resulted in losses of 35% and 65% of HPV16 DNA from the rafts, respectively (Fig. 3).
Figure 3. Effect of Polyamide 1 on HPV16 DNA levels in an organotypic epithelium.
HPV16 copy number is represented as a percent of vehicle control. Average copy number and standard deviation were calculated from duplicate DNA input amounts using increasing amounts of total DNA (5, 10, 20, 40, 80, 160 ng) in the PCR reaction.
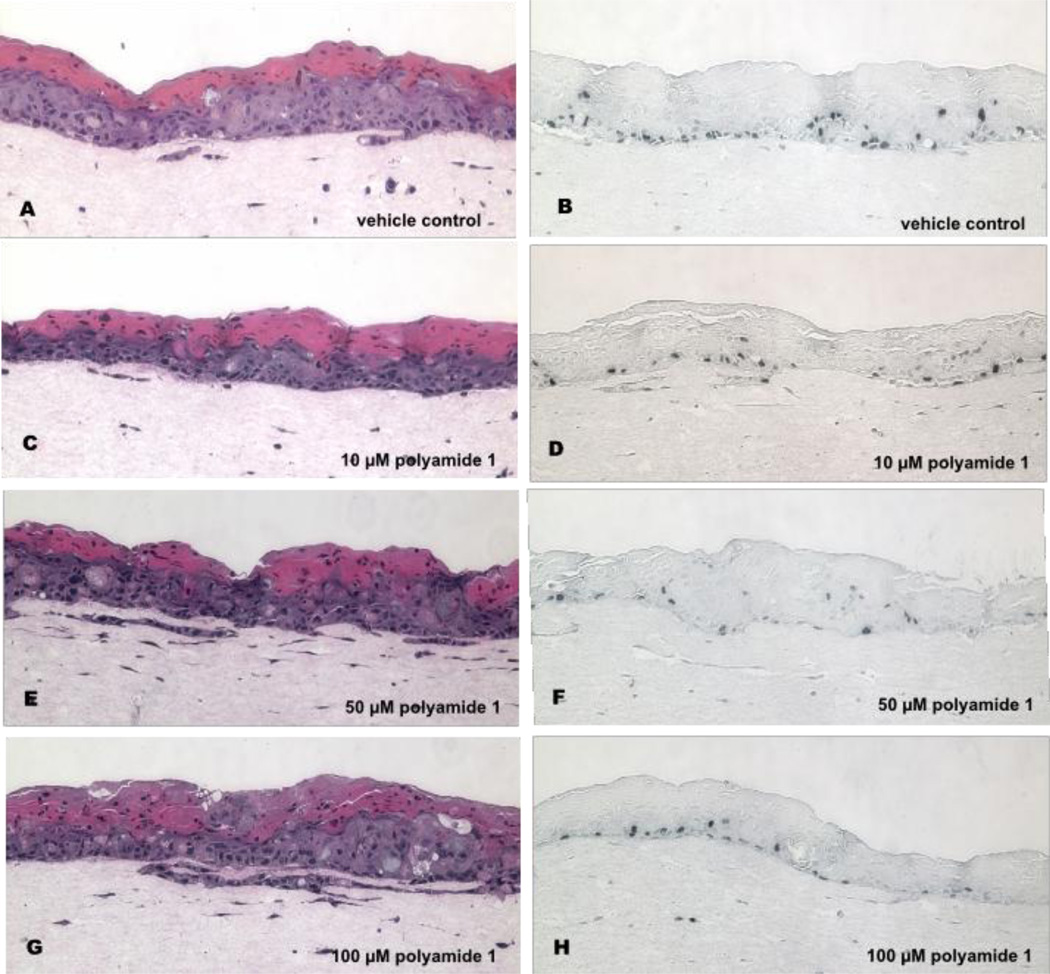
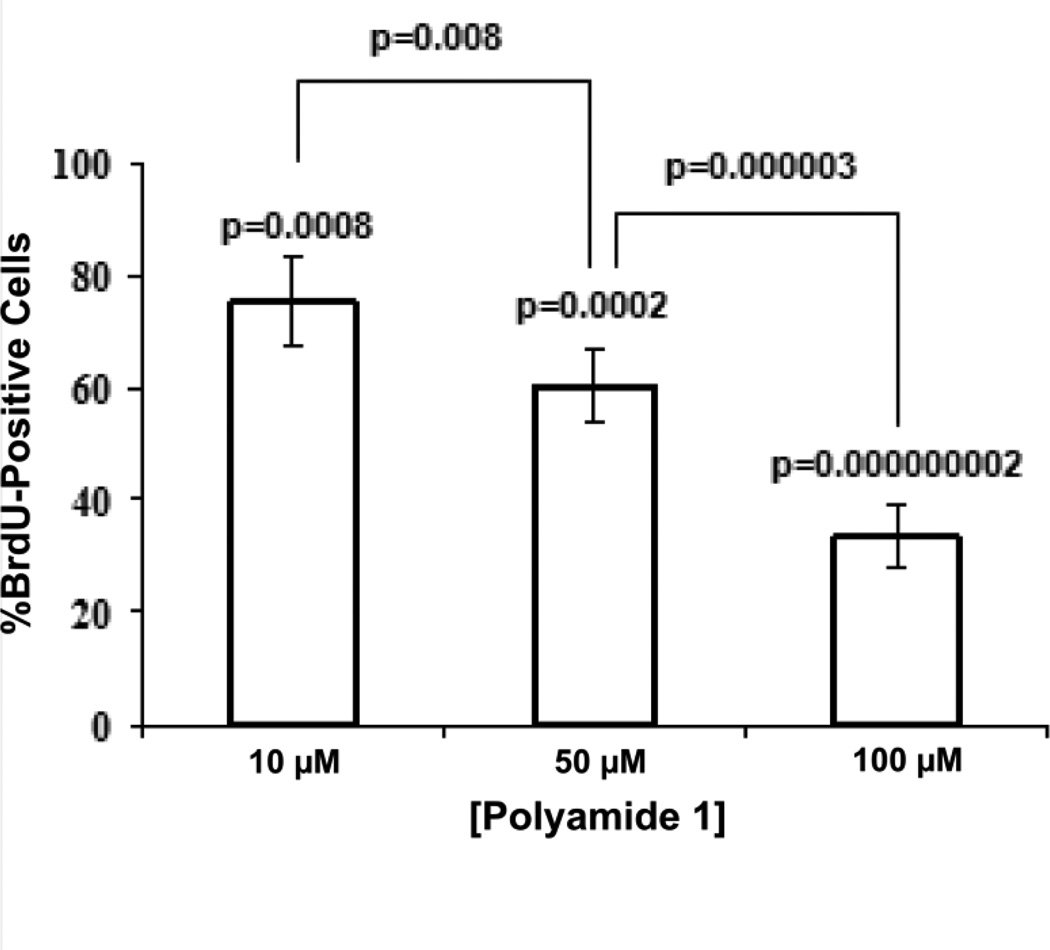
Histological examination of the W12E organotypic cultures demonstrated that both the vehicle-treated controls (treated with the delivery buffer--0.1% DMSO in E-media minus polyamide 1) and the polyamide 1-treated raft cultures differentiated over the course of the experiment, with all cultures showing evidence of stratification and cornification (Fig. 4). The effects of decreases in W12E episome levels on DNA synthesis were next tested by probing adjacent, multiple histological sections of the treated epithelia with a BrdU antibody. BrdU incorporating cells were in both the basal cell layer and in more differentiated cell layers of the epithelium, although the limited thickness and poor organization of the W12E epithelium made it difficult to conclusively assign labeled cells to a particular compartment. Nevertheless, fewer cells incorporated BrdU within the polyamide 1-treated epithelia than within the vehicle-treated controls (Fig. 4), and qualitatively this seems to hold for both suprabasal and basal layers. We therefore quantified the total number of nuclei versus the number of BrdU-labeled nuclei in order to compare BrdU uptake among samples. The percentage of epithelial cells whose nuclei stained positive for BrdU in the control samples was 9.8 ± 1.15%, while the BrdU labeling of all three treatment groups was lower than the control (p < 0.05; Fig 5). The difference in BrdU staining was also statistically significant between dosage groups (Fig. 5). These results demonstrate that a single treatment of W12E cells with polyamide 1 results in a significant, dose-dependent decrease in HPV16 DNA load, and in a loss of cells in S-phase within the engineered epithelia.
Figure 4. Hemotoxylin and eosin (H&E) staining and BrdU incorporation in organotypic cultures treated with vehicle and increasing doses of polyamide 1.
The panel shows vehicle-treated control cultures (A, B), and cultures treated initially with 10 µM (C, D), 50 µM (E, F), and 100 µM (G, H) of polyamide 1. Micrographs of H&E stained cultures are on the left (A, C, E, G) and BrdU labeled sections (B, D, F, H) on the right. Magnification = 100×.
Figure 5.
Cell counts of total and BrdU-stained nuclei from the samples exhibited in Figure 4.
Examination of LDH levels in the organotypic culture media following treatment showed no elevation over vehicle-treated controls (data not shown). Fluorescence microscopy of the PA1-treated rafts at the end of the culture period revealed prominent nuclear staining attributable to PA1 (SFig. 4). This result is consistent with the long-term ability of PA1 to diminish HPV DNA levels.
W12E cell derived epithelia are not ideal for histological studies for several reasons, notably poor differentiation and a lack of organization relative to other HPV episome-positive cell lines, particularly those lines that have been derived by transfection and been in culture for shorter periods of time. For this reason, we next set out to examine the effects of topical-treatment of highly organized, well-differentiated raft cultures derived from an HPV18 episome-positive cell line (Ker4-18). Since PA1 had lower activity against HPV18 (IC50 ~650 nM; Table 1), we employed a different polyamide (PA25) that potently reduced HPV18 episomes in Ker4-18 cell monolayers with an IC50 of 56 ± 5 nM. As observed in the W12E study with active anti-HPV polyamides, PA25 had no detectable effects on cell viability in MTT assays (data not shown).
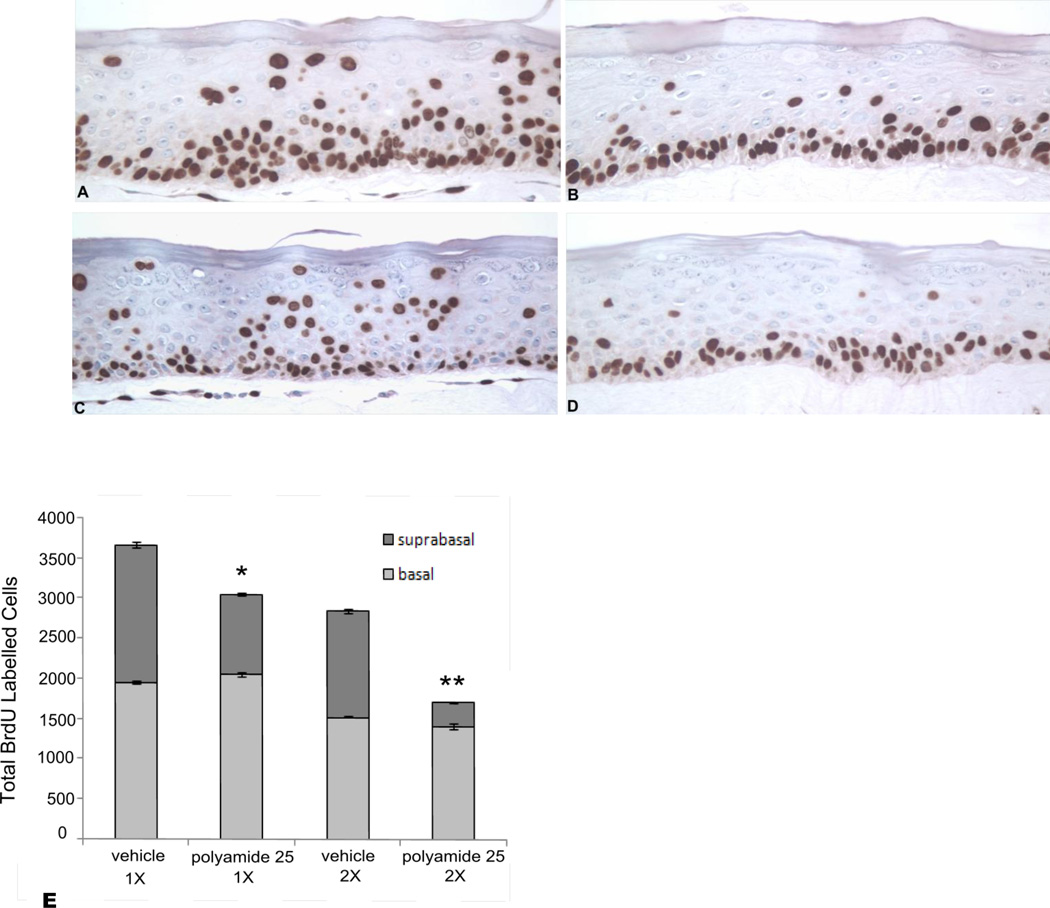
Polyamide 25 (1 mM) was topically applied to the surface of HPV18 raft cultures as described. Q-PCR confirmed that both the 1X and 2X treatments resulted in significant reductions of HPV18 episome levels of approximately 70% and 80%, respectively, in the cultured epithelia relative to the vehicle-treated controls (S Fig. 3). Due to the excellent stratification, differentiation, and histological organization of the Ker4-18 cells in raft cultures, cells in both the basal and suprabasal compartments of the epithelia were easily identified and counted. BrdU labeling decreased within the suprabasal compartments of the Ker4-18 epithelia following both the 1X and 2X treatments with PA25 relative to vehicle-treated controls (Fig. 6). Cell counts confirmed that PA25 caused a statistically significant reduction in suprabasal labeling in both treatment groups relative to controls (Fig. 6E). In contrast, no significant effect on basal cell labeling was observed between treatment groups. The difference in labeling index between basal and suprabasal layers was not attributable to polyamide bioavailability because polyamide nuclear labeling was found throughout all Ker4-18 epithelial cell layers at the end of the culture period (S Fig. 4). LDH levels in the organotypic culture media of Ker4-18 rafts following treatment showed no elevation over vehicle-treated controls (data not shown).
Figure 6.
An average of 9,000 cells was counted for each raft. BrdU-positive cells were counted in the basal and suprabasal layers and the labeling index calculated as a percent of the total cell population. No significant change between controls (A,C) and rafts (B,D) treated with polyamide 25 was noted for basal cell labeling. For suprabasal labeling, a significant decrease was noted relative to controls for rafts treated with polyamide 25: * p=0.002; ** p=0.0000001. Basal cell labeling was not significantly affected (E).
Discussion
We report a novel polyamide compound that controls and decreases the HPV episome content of keratinocytes for the major high-risk virus HPV16. Minor groove binding agents such as Distamycin A have long been considered as possible DNA antiviral agents (Becker and Weinber, 1972; Verini, 1964), although problems such as toxicity and lack of specificity have hindered their clinical usefulness. We describe here the synthesis and testing of PA1, a synthetic minor groove DNA binding agent initially designed to target E1 and E2 binding sites on the HPV genome, including those at the HPV16 ori. PA1 is a higher homolog of naturally occurring oligoamides such as Distamycin A. PA1 shows no evidence of cytotoxicity at doses up to 200 µM. We attribute this improvement over Distamycin A to the much greater sequence-selectivity of PA1 for DNA binding and possibility to a decreased accessibility to host DNA of the relatively large PA1 vs. Distamycin. PA1 is currently under development as an HPV16 antiviral compound. In addition, it and related reagents may also prove to be useful for controlling HPV episomal levels in cells and thereby promote experimentation on various aspects of the HPV biology.
The difference in cellular uptake of compounds is currently under investigation as part of the SAR. We previously reported that uptake and nuclear localization of polyamides are influenced by multidrug resistance efflux pumps for certain cell lines, including human rheumatoid synovial fibroblasts (RSFs). Our report showed that verapamil, an inhibitor of p-glycoprotein-dependent ABC transporter systems, blocked the accumulation of certain polyamides in vesicles and led to nuclear accumulation of the polyamides in RSFs (Crowley et al., 2003). We further showed that uptake can change dramatically with small changes in chemical structure, for example in HCT116 colon cancer cells, where altering molecular properties by changing one dye (BODIPY) for another (fluorescein) led to efficient uptake and nuclear localization (Crowley et al., 2003). Subsequent studies on polyamide uptake have reported similar effects, including successful uptake and biological activity with appropriate molecular designs, in a wide range of cell types, both in vitro and in vivo (Belitsky et al., 2002; Best et al., 2003; Harki et al., 2008; Nickols et al., 2007; Takahashi et al., 2008; Ueno et al., 2009; Xiao et al., 2007).
For the keratinocytes under study here, we have routinely observed successful polyamide uptake. For example, we show uptake into cells in engineered, differentiated epithelial tissues by the inherent fluorescence of polyamides localized in the nuclei of target cells (S Fig. 4). It should be noted that polyamide fluorescence is greatly enhanced upon binding DNA, so the absence of the characteristic polyamide emission from the cytoplasm does not necessarily mean that the cytoplasm is polyamide-free (Rucker et al., 2004, Hsu and Dervan, 2008). Recently, it was reported that polyamides up to a molecular weight (MW) of 3115 are taken up well by two cell lines as long as the imidazole content is not high (Nishijima et al., 2010). The value 3115 is significantly above the MW of compounds we report here. This same uptake study (Nishijima et al., 2010) noted that uptake into cells did not correlate at all with MW: large polyamides were taken up by cells just as readily as small polyamides, and the “standard” ideas about MW and cell permeability do not apply for polyamides. Note that our ultimate use for PA1 or its analogs is as a topical agent, not as a systemic drug. We also note that a previous report demonstrated blocking of E2 binding to HPV DNA by a tandem hairpin polyamide in a cell free system (Schaal et al., 2003), an encouraging observation.
Organotypic cultures (or “raft” cultures) have been useful for investigations of the HPV lifecycle because they allow formation and differentiation of a stratified squamous epithelium very similar to that found in vivo and they support the entire life cycle of the virus (Flores et al., 1999; Frattini et al., 1997; Garner-Hamrick et al., 2004; Meyers and Laimins, 1994). We therefore set out to determine if we could also reduce HPV16 episome content with PA1 in this more complex culture system, using engineered epithelia derived from W12E cells. Surprisingly, we found that a single treatment of PA1 at the time of initial raft construction resulted in a dose-dependent decrease in episome content of the rafts at the time of harvest 19 days later. W12 organotypic cultures are highly dysplastic, which makes it difficult to precisely assign BrdU uptake to basal versus suprabasal cell populations. However, Ker4-18 organotypic cultures are highly stratified and differentiated. Topical treatment of these cultures with PA25 causes a dramatic loss of suprabasal BrdU uptake while basal cell BrdU labeling is apparently unaffected. These changes are consistent with the treated epithelium reverting to a phenotype that is more consistent with a normal, rather than an HPV-infected, epithelium We are currently examining the effects of episome loss following PA1 and PA25 treatment on DNA synthesis and cell cycle progression.
The observed, polyamide-driven decrease in HPV DNA is not attributable to cellular toxicity. We have searched extensively for signs of toxic effects from our compounds using multiple assay systems, including the MTT assays reported here, and have yet to see evidence of polyamide toxicity in cells up to 200 µM concentrations (ca. 2000-fold higher than the apparent IC50). In addition, we routinely monitor the media of all raft cultures at the end of our long-term polyamide treatment periods for the presence of lactate dehydrogenase (LDH) activity, a widely used assay for assessment of cell or tissue damage in vivo and in vitro (Babson and Babson, 1973). These studies are part of an ongoing pre-clinical development program for identifying the best formulation for topical delivery of our lead compound. We have never observed short- or long-term toxicity of polyamides using the LDH assay with our organotypic culture system or the MTT assay with monolayer cultures. Finally, many others have reported on the use and nontoxic nature of synthetic polyamides in a variety of in vitro and animal models (Fukuda, 2006; Harki et al., 2008; Matsuda et al., 2006; Shinohara et al., 2005). Together, these results strongly indicate that the reduction of HPV DNA episomes by our active polyamides is specific.
Maintenance of HPV episomes in cultured human keratinocytes is a poorly understood phenomenon that primarily occurs in “high-risk” or cancer-causing forms of the virus. Furthermore, establishing episome-maintaining cells, whether from cultured clinical tissue or following transfection of normal keratinocytes, generally requires a “high risk” genotype (Doorbar et al., 1990; Frattini et al., 1997; Garner-Hamrick and Fisher, 2002; Meyers et al., 1992; Stanley et al., 2007). However, exceptions to this generalization do exist (Fang et al., 2006; Pittayakhajonwut and Angeletti, 2008; Thomas et al., 2001). Numerous viral proteins and interaction domains have been implicated in the maintenance of HPV episomes (Cote-Martin et al., 2008; Hebner and Laimins, 2006; Ilves et al., 1999; Pittayakhajonwut and Angeletti, 2008; Thomas et al., 1999). For this reason, it is understood that episome maintenance is complicated, involving episome replication, host cell lifespan extension, evasion of innate immunity, chromosomal tethering and nuclear sequestration, and equitable segregation of viral DNA during cell replication. Since PA1 binds with high affinity adjacent to E1 and E2 binding sites within the HPV16 ori, a current working model for the potent action of these compounds is interruption of E1 and/or E2 binding activities, which are known to substantially contribute to the ori structure and function. For example, E2 is a multi-functional protein implicated in tethering to mitotic chromosomes as well as regulating viral DNA transcription and replication (Botchan, 2004; McBride et al., 2006; Van Tine et al., 2004). E1 possesses well described ATPase and helicase activities that are of interest as antiviral targets because of their requisite role in unwinding the papillomavirus ori during initiation of viral DNA synthesis (Fradet-Turcotte and Archambault, 2007). Additional E1 activities have also been identified that are required for viral episome maintenance or for promoting HPV amplification (Cote-Martin et al., 2008; Moody et al., 2007). Finally, both the E1 and E2 proteins are implicated in the higher order structure of the viral ori. While the role of the ori DNA structure in replication and maintenance is unknown, it is likely to be important for both replication and maintenance of the HPV genome (Abbate et al., 2004; Sim et al., 2008; Wilson et al., 2002). Importantly, the binding of polyamides is likely to alter the viral DNA conformation profoundly, including possibly at the ori, due to the well-known biophysical effects of polyamides on DNA such as widening the minor groove, shrinking the major groove, and stiffening the double helix (Dervan and Edelson, 2003).
Loss of HPV episomes following treatment of cells with polyamide is not associated with integration. Southern blots have not revealed HPV-containing bands other than the 8 kb episomal DNA, so we observed none of the bands expected if integration of viral DNA were occurring. Furthermore, we observed, following treatment of cells with polyamide, that the total remaining HPV DNA is far below 1 copy per cell. Others have previously shown, using W12 cells, that integration in vitro occurs only over many passages (Jeon et al., 1995; Grey et. al., 2010). These reported results on integration in W12 cells are consistent with populations of W12 cells that carry HPV integrants out-competing episome-maintaining cells: and these circumstances require considerable time in culture to be achieved. It is therefore highly improbable that integration of viral DNA would occur during the short treatment times employed in the experiments reported here and that resulted in a robust decline in episomal DNA even though the cells are dividing rapidly.
In summary, HPV16 causes greater than 50% of cervical cancers worldwide and contributes to other cancers and diseases as well. Antiviral compounds that specifically reduce or eliminate human papillomavirus (HPV) and reverse the disease pathology are currently not available. An antiviral compound effective against HPV has potential for an impact on public health, especially in light of the anticipated widespread use of HPV DNA testing in cervical cancer screening programs. This paper describes a synthetic pyrrole-imidazole containing polyamide designed to bind the HPV16 ori DNA sequence and related AT-rich viral sequences. The compounds potently decrease levels of episomal HPV DNA while showing no evidence of toxicity to human cells or tissue cultures. Thus, we believe that the compounds reported here have potential utility as research tools for controlling HPV DNA levels in vitro and as therapeutics for HPV-related diseases. PA1 and PA25 are currently under preclinical development as topical agents for the prevention of HPV-related disease.
Supplementary Material
Acknowledgements
This work was supported by NIH grants R41AI062182 and R42AI062182 and by funds from the State of Michigan awarded by the Biosciences Research and Commercialization Center of Western Michigan University. The University of Missouri-St. Louis provided support for equipment in the form of a Small Institutional Research Grant. C.F. and J.K.B. are co-founders of NanoVir and hold significant ownership positions in NanoVir. We thank L. Hicks and Z. Liu of the Donald Danforth Plant Science Center for HRMS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbate EA, Berger JM, Botchan MR. The X-ray structure of the papillomavirus helicase in complex with its molecular matchmaker E2. Genes Dev. 2004;18:1981–1996. doi: 10.1101/gad.1220104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babson AL, Babson SR. Kinetic colorimetric measurement of serum lactate dehydrogenase activity. Clin. Chem. 1973;19:766–769. [PubMed] [Google Scholar]
- Baird EE, Dervan PB. Solid Phase Synthesis of Polyamides Containing Imidazole and Pyrrole Amino Acids. J. Am. Chem. Soc. 1996;118:6141–6146. [Google Scholar]
- Becker Y, Weinber A. Distamycin A inhibition of Epstein-Barr virus replication in arginine-deprived Burkitt lymphoblasts. Isr. J. Med. Sci. 1972;8:75–78. [PubMed] [Google Scholar]
- Belitsky JM, Leslie SJ, Arora PS, Beerman TA, Dervan PB. Cellular uptake of N-methylpyrrole/N-methylimidazole polyamide-dye conjugates. Bioorg. Med. Chem. 2002;10:3313–3318. doi: 10.1016/s0968-0896(02)00204-3. [DOI] [PubMed] [Google Scholar]
- Best TP, Edelson BS, Nickols NG, Dervan PB. Nuclear localization of pyrrole-imidazole polyamide-fluorescein conjugates in cell culture. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12063–12068. doi: 10.1073/pnas.2035074100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchan M. Hitchhiking without covalent integration. Cell. 2004;117:280–281. doi: 10.1016/s0092-8674(04)00410-6. [DOI] [PubMed] [Google Scholar]
- Broyles SS, Kremer M, Knutson BA. Antiviral Activity of Distamycin A against Vaccinia Virus Is the Result of Inhibition of Postreplicative mRNA Synthesis. J. Virol. 2004;78:2137–2141. doi: 10.1128/JVI.78.4.2137-2141.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Schmidt-Grimminger DC, Murant T, Broker TR, Chow LT. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 1995;9:2335–2349. doi: 10.1101/gad.9.19.2335. [DOI] [PubMed] [Google Scholar]
- Cote-Martin A, Moody C, Fradet-Turcotte A, D'Abramo CM, Lehoux M, Joubert S, Poirier GG, Coulombe B, Laimins LA, Archambault J. Human papillomavirus E1 helicase interacts with the WD repeat protein p80 to promote maintenance of the viral genome in keratinocytes. J. Virol. 2008;82:1271–1283. doi: 10.1128/JVI.01405-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley KS, Phillion DP, Woodard SS, Schweitzer BA, Singh M, Shabany H, Burnette B, Hippenmeyer P, Heitmeier M, Bashkin JK. Controlling the intracellular localization of fluorescent polyamide analogues in cultured cells. Bioorg. Med. Chem. Lett. 2003;13:1565–1570. doi: 10.1016/s0960-894x(03)00152-5. [DOI] [PubMed] [Google Scholar]
- Dervan PB. Molecular recognition of DNA by small molecules. Bioorg. Med. Chem. 2001;9:2215–2235. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- Dervan PB, Edelson BS. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr. Opin. Struct. Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- Doorbar J, Parton A, Hartley K, Banks L, Crook T, Stanley M, Crawford L. Detection of novel splicing patterns in a HPV16-containing keratinocyte cell line. Virology. 1990;178:254–262. doi: 10.1016/0042-6822(90)90401-c. [DOI] [PubMed] [Google Scholar]
- Fang L, Meyers C, Budgeon LR, Howett MK. Induction of productive human papillomavirus type 11 life cycle in epithelial cells grown in organotypic raft cultures. Virology. 2006;347:28–35. doi: 10.1016/j.virol.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Flores ER, Allen-Hoffmann BL, Lee D, Lambert PF. The human papillomavirus type 16 E7 oncogene is required for the productive stage of the viral life cycle. J. Virol. 2000;74:6622–6631. doi: 10.1128/jvi.74.14.6622-6631.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores ER, Allen-Hoffmann BL, Lee D, Sattler CA, Lambert PF. Establishment of the human papillomavirus type 16 (HPV-16) life cycle in an immortalized human foreskin keratinocyte cell line. Virology. 1999;262:344–354. doi: 10.1006/viro.1999.9868. [DOI] [PubMed] [Google Scholar]
- Fradet-Turcotte A, Archambault J. Recent advances in the search for antiviral agents against human papillomaviruses. Antivir. Ther. 2007;12:431–451. [PMC free article] [PubMed] [Google Scholar]
- Frattini MG, Lim HB, Doorbar J, Laimins LA. Induction of human papillomavirus type 18 late gene expression and genomic amplification in organotypic cultures from transfected DNA templates. J. Virol. 1997;71:7068–7072. doi: 10.1128/jvi.71.9.7068-7072.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frattini MG, Lim HB, Laimins LA. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc. Natl. Acad. Sci. U.S.A. 1996;93:3062–3067. doi: 10.1073/pnas.93.7.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda N. Development of gene therapies for cardiovascular and renal diseases by nucleic acid medicines. Med. Chem. 2006;2:13–19. doi: 10.2174/157340606775197723. [DOI] [PubMed] [Google Scholar]
- Garner-Hamrick PA, Fisher C. HPV episomal copy number closely correlates with cell size in keratinocyte monolayer cultures. Virology. 2002;301:334–341. doi: 10.1006/viro.2002.1558. [DOI] [PubMed] [Google Scholar]
- Gray EMR, Pett D, Ward DM, Winder MA, Stanley I, Roberts C, Scarpini G, Coleman N. In Vitro Progression of HPV16 Episome-Associated Cervical Neoplasia Displays Fundamental Similarities to Integrant-Associated Carcinogenesis. Cancer Res. 2010;70(10):4081–4091. doi: 10.1158/0008-5472.CAN-09-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner-Hamrick PA, Fostel JM, Chien WM, Banerjee NS, Chow LT, Broker TR, Fisher C. Global effects of human papillomavirus type 18 E6/E7 in an organotypic keratinocyte culture system. J. Virol. 2004;78:9041–9050. doi: 10.1128/JVI.78.17.9041-9050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harki DA, Satyamurthy N, Stout DB, Phelps ME, Dervan PB. In vivo imaging of pyrroleimidazole polyamides with positron emission tomography. Proc. Natl. Acad. Sci. U. S. A. 2008;105:13039–13044. doi: 10.1073/pnas.0806308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Fisher C. Direct activation of cyclin-dependent kinase 2 (CDK2) by human papillomavirus (HPV) E7. J. Virol. 2003;77:10566–11577. doi: 10.1128/JVI.77.19.10566-10574.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebner CM, Laimins LA. Human papillomaviruses: basic mechanisms of pathogenesis and oncogenicity. Rev. Med. Virol. 2006;16:83–97. doi: 10.1002/rmv.488. [DOI] [PubMed] [Google Scholar]
- Howley PM. Papillomaviridae: the viruses and their replication. In: Fields BN, Knipe DM, Howley PM, editors. Fundamental Virology. 3rd ed. Philadelphia, PA: Lippencott-Raven; 1996. [Google Scholar]
- Hsu CF, Dervan PB. Quantitating the concentration of Py-Im polyamide-fluorescein conjugates in live cells. Bioorg. Med. Chem. Lett. 2008;18:5851–5855. doi: 10.1016/j.bmcl.2008.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilves I, Kivi S, Ustav M. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J. Virol. 1999;73:4404–4412. doi: 10.1128/jvi.73.5.4404-4412.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon S, Allen-Hoffmann BL, Lambert PF. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 1995;69:2989–2997. doi: 10.1128/jvi.69.5.2989-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzik PO, Chamberlin AR. Rapid solid-Phase synthesis of DNA-Binding pyrrole-Imidazole polyamides. Bioorg. Med. Chem. Lett. 2002;12:2129–2132. doi: 10.1016/s0960-894x(02)00359-1. [DOI] [PubMed] [Google Scholar]
- Longworth MS, Laimins LA. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol. Mol. Biol. Rev. 2004;68:362–372. doi: 10.1128/MMBR.68.2.362-372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda H, Fukuda N, Ueno T, Tahira Y, Ayame H, Zhang W, Bando T, Sugiyama H, Saito S, Matsumoto K, Mugishima H, Serie K. Development of gene silencing pyrrole-imidazole polyamide targeting the TGF-beta1 promoter for treatment of progressive renal diseases. J. Amer. Soc. Nephrol. 2006;17:422–432. doi: 10.1681/ASN.2005060650. [DOI] [PubMed] [Google Scholar]
- McBride AA, Oliveira JG, McPhillips MG. Partitioning viral genomes in mitosis: same idea, different targets. Cell Cycle. 2006;5:1499–1502. doi: 10.4161/cc.5.14.3094. [DOI] [PubMed] [Google Scholar]
- Meyers C, Frattini MG, Hudson JB, Laimins LA. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science. 1992;257:971–973. doi: 10.1126/science.1323879. [DOI] [PubMed] [Google Scholar]
- Meyers C, Laimins LA. In vitro systems for the study and propagation of human papillomaviruses. Curr Top Microbiol. Immunol. 1994;186:199–215. doi: 10.1007/978-3-642-78487-3_11. [DOI] [PubMed] [Google Scholar]
- Moody CA, Fradet-Turcotte A, Archambault J, Laimins LA. Human papillomaviruses activate caspases upon epithelial differentiation to induce viral genome amplification. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19541–19546. doi: 10.1073/pnas.0707947104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, Snijders PJ, Meijer CJ. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- Nickols NG, Jacobs CS, Farkas ME, Dervan PB. Improved nuclear localization of DNA-binding polyamides. Nucleic Acids Res. 2007;35:363–370. doi: 10.1093/nar/gkl1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishijima S, Shinohara K-i, Bando T, Minoshima M, Kashiwazaki G, Sugiyama H. Cell permeability of Py-Im-polyamide-fluorescein conjugates: Influence of molecular size and Py/Im content. Bioorg. Med. Chem. 2010;18:978–983. doi: 10.1016/j.bmc.2009.07.018. [DOI] [PubMed] [Google Scholar]
- Pittayakhajonwut D, Angeletti PC. Analysis of cis-elements that facilitate extrachromosomal persistence of human papillomavirus genomes. Virology. 2008;374:304–314. doi: 10.1016/j.virol.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rucker VC, Dunn AR, Sharma S, Dervan PB, Gray HB. Mechanism of sequence-specific fluorescent detection of DNA by N-Methyl-imidazole, N-Methyl-pyrrole, and β-alanine linked polyamides. J. Phys. Chem. B. 2004;108:7490–7494. [Google Scholar]
- Sankaranarayanan R, Nene BM, Shastri SS, Jayant K, Muwonge R, Budukh AM, Hingmire S, Malvi SG, Thorat R, Kothari A, Chinoy R, Kelkar R, Kane S, Desai S, Keskar VR, Rajeshwarkar R, Panse N, Dinshaw KA. HPV screening for cervical cancer in rural India. N. Engl. J. Med. 2009;360:1385–1394. doi: 10.1056/NEJMoa0808516. [DOI] [PubMed] [Google Scholar]
- Schaal TD, Mallet WG, McMinn DL, Nguyen NV, Sopko MM, John S, Parekh BS. Inhibition of human papilloma virus E2 DNA binding protein by covalently linked polyamides. Nucleic Acids Res. 2003;31:1282–1291. doi: 10.1093/nar/gkg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffman M, Wacholder S. From India to the World - A Better Way to Prevent Cervical Cancer. N. Engl. J. Med. 2009;360:1453–1455. doi: 10.1056/NEJMe0901167. [DOI] [PubMed] [Google Scholar]
- Shinohara K-I, Sasaki S, Bando T, Sugiyama H. Sequence-specific gene silencing by alkylating Py-Im polyamide. Nucl. Acids Symp. Ser. 2005:75–76. doi: 10.1093/nass/49.1.75. [DOI] [PubMed] [Google Scholar]
- Sim J, Ozgur S, Lin BY, Yu JH, Broker TR, Chow LT, Griffith J. Remodeling of the human papillomavirus type 11 replication origin into discrete nucleoprotein particles and looped structures by the E2 protein. J Mol Biol. 2008;375:1165–1177. doi: 10.1016/j.jmb.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley MA, Pett MR, Coleman N. HPV: from infection to cancer. Biochem Soc Trans. 2007;35:1456–1460. doi: 10.1042/BST0351456. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Asami Y, Kitamura E, Suzuki T, Wang X, Igarashi J, Morohashi A, Shinojima Y, Kanou H, Saito K, Takasu T, Nagase H, Harada Y, Kuroda K, Watanabe T, Kumamoto S, Aoyama T, Matsumoto Y, Bando T, Sugiyama H, Yoshida-Noro C, Fukuda N, Hayashi N. Development of Pyrrole-Imidazole Polyamide for Specific Regulation of Human Aurora Kinase-A and -B Gene Expression. Chem. Biol. 2008;15:829–841. doi: 10.1016/j.chembiol.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Thomas JT, Hubert WG, Ruesch MN, Laimins LA. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc Natl Acad Sci U S A. 1999;96:8449–8454. doi: 10.1073/pnas.96.15.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JT, Oh ST, Terhune SS, Laimins LA. Cellular changes induced by low-risk human papillomavirus type 11 in keratinocytes that stably maintain viral episomes. J. Virol. 2001;75:7564–7571. doi: 10.1128/JVI.75.16.7564-7571.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T, Fukuda N, Tsunemi A, Yao E-H, Matsuda H, Tahira K, Matsumoto T, Matsumoto K, Matsumoto Y, Nagase H, Sugiyama H, Sawamura T. A novel gene silencer, pyrrole-imidazole polyamide targeting human lectin-like oxidized low-density lipoprotein receptor-1 gene improves endothelial cell function. Journal of Hypertension. 2009;27:508–516. doi: 10.1097/hjh.0b013e3283207fe1. [DOI] [PubMed] [Google Scholar]
- Van Tine BA, Dao LD, Wu SY, Sonbuchner TM, Lin BY, Zou N, Chiang CM, Broker TR, Chow LT. Human papillomavirus (HPV) origin-binding protein associates with mitotic spindles to enable viral DNA partitioning. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4030–4035. doi: 10.1073/pnas.0306848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verini MA, Ghione M. Activity of distamycin A on vaccinia virus infection in culture. Chemotherapi. 1964;9:144–157. doi: 10.1159/000220355. [DOI] [PubMed] [Google Scholar]
- Wang CCC, Ellervik U, Dervan PB. Expanding the recognition of the minor groove of DNA by incorporation of β-alanine in hairpin polyamides. Bioorg. Med. Chem. 2001;9:653–657. doi: 10.1016/s0968-0896(00)00282-0. [DOI] [PubMed] [Google Scholar]
- White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB. Recognition of the four Watson-Crick base pairs in the DNA minor groove by synthetic ligands. Nature. 1998;391:468–470. doi: 10.1038/35106. [DOI] [PubMed] [Google Scholar]
- Wilson VG, West M, Woytek K, Rangasamy D. Papillomavirus E1 proteins: form, function, and features. Virus Genes. 2002;24:275–290. doi: 10.1023/a:1015336817836. [DOI] [PubMed] [Google Scholar]
- Woodman CB, Collins SI, Young LS. The natural history of cervical HPV infection: unresolved issues. Nat. Rev. Cancer. 2007;7:11–22. doi: 10.1038/nrc2050. [DOI] [PubMed] [Google Scholar]
- Wurtz NR, Turner JM, Baird EE, Dervan PB. Fmoc Solid Phase Synthesis of Polyamides Containing Pyrrole and Imidazole Amino Acids. Org. Lett. 2001;3:1201–1203. doi: 10.1021/ol0156796. [DOI] [PubMed] [Google Scholar]
- Xiao J, Yuan G, Huang W, Chan ASC, Lee KLD. A Convenient Method for the Synthesis of DNA-Recognizing Polyamides in Solution. J. Org. Chem. 2000;65:5506–5513. doi: 10.1021/jo000135n. [DOI] [PubMed] [Google Scholar]
- Xiao X, Yu P, Lim H-S, Sikder D, Kodadek T. Design and synthesis of a cell-permeable synthetic transcription factor mimic. J. Comb. Chem. 2007;9:592–600. doi: 10.1021/cc070023a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.