Abstract
Circadian clocks sustain daily oscillations in gene expression, physiology, and behavior, relying on transcription–translation feedback loops of clock genes for rhythm generation. Cultured astrocytes display daily oscillations of extracellular ATP, suggesting that ATP release is a circadian output. We hypothesized that the circadian clock modulates ATP release via mechanisms that regulate acute ATP release from glia. To test the molecular basis for circadian ATP release, we developed methods to measure in real-time ATP release and Bmal1::dLuc circadian reporter expression in cortical astrocyte cultures from mice of different genotypes. Daily rhythms of gene expression required functional Clock and Bmal1, both Per1 and Per2, and both Cry1 and Cry2 genes. Similarly, high-level, circadian ATP release also required a functional clock mechanism. Whereas blocking IP3 signaling significantly disrupted ATP rhythms with no effect on Bmal1::dLuc cycling, blocking vesicular release did not alter circadian ATP release or gene expression. We conclude that astrocytes depend on circadian clock genes and IP3 signaling to express daily rhythms in ATP release.
Introduction
Circadian pacemakers coordinate the daily timing of physiology and behavior with environmental cycles. These oscillators rely on transcription–translation feedback loops of canonical clock genes (e.g., Bmal1, Clock, Period1, Period2, Cryptochrome1, and Cryptochrome2) to generate overt circadian rhythms (Siepka et al., 2007). Clock genes are ubiquitous, and most mammalian cells and tissues express circadian rhythms both in vitro and in vivo (Yoo et al., 2004; Kowalska and Brown, 2007). Neuronal cultures from the hypothalamic suprachiasmatic nuclei (SCN) exhibit sustained oscillations in PERIOD2-driven luciferase activity (PER2::LUC) (Liu et al., 2007). Although single SCN neurons are intrinsically capable of generating sustained circadian oscillations, intercellular signals synchronize them to each other, sustaining and enhancing the precision of the rhythms at the population level (Liu et al., 2007; Webb et al., 2009). Individual fibroblasts also display sustained circadian oscillations in PER2::LUC (Welsh et al., 2004), but they do not synchronize to each other, leading to damping of the population rhythms (Welsh et al., 2004). Cultured astrocytes exhibit damped circadian oscillations that are sustained by SCN explants (Prolo et al., 2005) and can be entrained by the neuropeptide vasoactive intestinal polypeptide (VIP) (Marpegan et al., 2009). However, the biological significance of circadian oscillations in astrocytes is unknown.
Mammalian astrocytes regulate neuronal networks through the reuptake and release of various transmitters including glutamate (Perea et al., 2009; Halassa and Haydon, 2010). Glutamate reuptake is modulated by clock genes but is not circadian in cultured cortical astrocytes (Beaulé et al., 2009). In Drosophila, the glia-specific gene ebony is involved in the synthesis of biogenic amines and is required for circadian rhythms in locomotor behavior (Suh and Jackson, 2007), suggesting that glial cells can modulate neuronal networks responsible for complex behaviors through the circadian control of secreted neuroactive molecules.
A recent report showed that cultured mammalian astrocytes display circadian rhythms in extracellular ATP accumulation (Womac et al., 2009). It is possible that ATP is a circadian signal released by astrocytes. ATP and its dephosphorylated products, ADP, AMP, and adenosine, regulate synaptic communication (Burnstock, 2006) through a variety of purinergic receptors on astrocytes and neurons (Fields and Burnstock, 2006). Astrocytes release ATP through hemichannels (Cotrina et al., 1998), vesicle-dependent mechanisms (Coco et al., 2003; Pascual et al., 2005), the large pore of the P2X7 purinergic receptor (Suadicani et al., 2006), and through ATP-binding cassette proteins (multidrug resistance protein and p-glycoprotein) (Declèves et al., 2000; Darby et al., 2003). There is also evidence that release of transmitters, including ATP, from glia is calcium dependent (for a recent review, see Parpura and Zorec, 2010). Little is known about circadian modulation of any of these mechanisms for ATP release.
We tested the hypotheses that key clock genes in astrocytes drive circadian oscillations in ATP release through either calcium- or vesicle-dependent mechanisms. We developed a novel, real-time assay and found that circadian ATP release depends on Clock, Per1, and Per2 expression and IP3-dependent calcium signaling in astrocytes.
Materials and Methods
Animals.
Mice were bred and maintained in the Danforth Campus Animal Facility under a 12 h light/dark cycle. All procedures were approved by the Animal Care and Use Committee at Washington University in St. Louis and conformed to National Institutes of Health guidelines. Unless noted, mice were maintained on a C57BL/6 genetic background by backcrossing annually with mice from Charles River.
Astrocytes were derived from mice of different genotypes to assess whether the molecular basis of daily timing in astrocytes is similar to other cells and to determine whether circadian rhythms in ATP release are controlled by the expression of circadian genes. Founders for the circadian mutants used in this work were generously donated by Drs. J. Takahashi, University of Texas-Southwestern Medical Center (Dallas, TX) (Clock/Clock), S. McKnight, University of Texas-Southwestern Medical Center (Neuronal PAS domain 2 knock-out, Npas2−/−), S. M. Reppert, University of Massachusetts Medical School (Boston, MA) (Period1ldcPeriod2ldc double mutant mice referred as Per1mPer2m), R. Van Gelder, University of Washington Medical Center (Seattle, WA) (Cryptochome1 and Cryprochrome2 homozygote double knock-out mice, Cry1−/−Cry2−/−), and C. Bradfield, University of Wisconsin (Madison, WI) (Bmal1 knock-out, Bmal1−/−). Cry1−/−Cry2−/− mice were maintained on a mixed background of 129 × C57BL/6J, and BMAL1−/− mice were obtained from heterozygote breeders.
To test possible mechanisms of circadian regulation of ATP release, we used two strains of mice with astrocyte-specific gliotransmission deficiencies. Both strains were obtained by crosses between a hemizygote hGFAP.tTA mouse (“tet-Off” tetracycline transactivator protein controlled by the human glial fibrillary acidic protein promoter) and either a hemizygote tetO.SNARE or tetO.VIPP. In the absence of tetracycline (or analogs), the hGFAP.tTA::tetO.SNARE mice [dominant-negative (dn) SNARE] express a fragment of the cytosolic portion of the SNARE domain of synaptobrevin 2, thus blocking vesicular release from astrocytes. dnSNARE mice were characterized previously and show a phenotype following sleep deprivation (Pascual et al., 2005; Halassa et al., 2009). In addition, tetO.SNARE mice contain a tetO.EGFP (enhanced green fluorescent protein) reporter used to validate transgene expression (Pascual et al., 2005; Halassa et al., 2009). The hGFAP.tTA::tetO.VIPP mice (VIPP) express high levels of type I Ins(1,4,5)P3 5-phosphatase (IPP), the enzyme that terminates IP3 signaling (De Smedt et al., 1997), thus impairing IP3-dependent calcium signaling pathways. VIPP mice contain a tetO. Venus yellow fluorescent protein (YFP) reporter fused to the IPP component to validate transgene expression. Complete characterization of the VIPP mouse is the subject of another study (S.-Y. Lee, M. M. Halassa, D. Hines, L. Gainey, Q. Deng, O. Pascual, C. Erneux, P. J. Atkinson, N. J. Brandon, P. G. Haydon, unpublished observations). For both genotypes, we confirmed transgene expression by measuring the expression of the fluorescent reporters EGFP for dnSNARE, and Venus-YFP for VIPP in our astrocyte cultures. To prevent transgene expression in the pups, female mice were fed the tetracycline analog doxycycline (40 mg/kg of rodent food, BioServ) from 2 weeks before mating until the astrocyte cultures were prepared.
Cell cultures.
Cortical astrocytes were obtained from postnatal day 2–6 mice according to a previously published protocol (Beaulé et al., 2009). Each culture was made from 6–15 neonatal mice pooled from 1–3 litters of the same age and genotype. Replicates were performed with cultures made from the same and different litters. Astrocyte cultures were made from wild-type (WT), Clock/Clock, Clock/+, Npas2−/−, Per1mPer2m, Cry1−/−Cry2−/−, Cry1−/−Cry2+/−, Bmal1−/−, Bmal1+/−, dnSNARE, and VIPP mice. NPAS2 is highly expressed in the cortical areas we harvested for astrocyte cultures (Garcia et al., 2000). Absence of NPAS2 in these areas results in behavioral deficits that are not compensated by the presence of its analog (CLOCK) that forms functional complexes with BMAL1 (Reick et al., 2001). For these reasons, we included astrocytes derived from Npas2−/− mice in our evaluation of genes required for circadian rhythmicity. For comparison with dnSNARE and VIPP astrocytes, we used astrocytes from wild-type mice or littermates expressing only the hGFAP.tTA transgene. All astrocytes were incubated at 37°C in DMEM supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin. Medium changes were performed twice a week. Cells grew to confluence before passaging into new flasks. Astrocyte cultures between passage 1 and 3 were used for all experiments.
Gene reporter constructs.
We used Bmal1::dLuc and Per2::dLuc plasmids for transfection of pure astrocyte cultures (generous gifts from Dr. Hiroki R. Ueda, RIKEN Center for Developmental Biology, Kobe, Japan). Plasmids were each inserted into the 4818 bp pGL3 vector (Promega). The Bmal1::dLuc plasmid contained 530 bp of the Bmal1 promoter driving luciferase. The Per2::dLuc plasmid contained 252 bp of the Per2 promoter driving luciferase (Sato et al., 2004). Transfection was performed with TransIT-LT1 reagent according to the manufacturer's instructions (Mirus). Briefly, 12 μl of the transfection reagent and 2 μg of plasmid DNA were diluted in DMEM up to a final volume of 100 μl. Transfection medium was added to 100 μl of a suspension containing between 9 × 104 and 1.8 × 105 astrocytes in DMEM with 10% FBS and penicillin/streptomycin. Transfection efficiency was 0.5–5% based on the number of cells expressing EGFP divided by the number of DAPI-stained nuclei in cultures cotransfected with pCMS-EGFP (Clontech). Astrocytes were immediately plated in 35 mm BD Primaria tissue culture dishes (BD Biosciences). The medium was replaced 48 h after transfection. Bioluminescence recordings were started ∼3 weeks later.
We performed lentiviral infections of pure astrocyte cultures using a lentiviral construct expressing a Bmal1::dLuc reporter (Liu et al., 2008; Zhang et al., 2009). Astrocytes were incubated with the viral particles for 12 h, washed, and passaged twice over a 2 week period before plating and imaging. Viral transduction resulted in 30–50% of the cells expressing Bmal1::dLuc.
ATP reporter system.
We used the BioThema ATP Reagent HS, which includes a more thermostable recombinant luciferase to measure real-time, extracellular ATP levels for up to 72 h. The ability of the ATP reagent to detect ATP over a 72 h period was greatly decreased as temperatures approached 37°C. We were thus required to lower the incubator temperature to 32°C to allow detection of ATP release for up to 72 h. This temperature also yielded high-amplitude oscillation in clock-gene driven bioluminescence.
Astrocytes were first plated into a 96-well plate (104 cells per well) and allowed to grow in a 5% CO2 incubator for 48 h at 37°C. The culture medium was then changed to HEPES-buffered DMEM with B27, and plates were sealed with silicone grease, moved to the imaging chamber, and cultured for an additional 48 h at 32°C. After 48 h, 20 μl of ATP reagent (10% v/v final) and 5 μl of VIP (500 nm) were added to each well to induce high-amplitude oscillations as previously reported (see Marpegan et al., 2009 for the description of the effects of VIP on circadian reporter-driven bioluminescence rhythms).
We generated a standard curve of bioluminescence as a function of ATP concentration at 24 h to account for temperature-dependent degradation of ATP reagent (Fig. 1D). We confirmed that the bioluminescence measured was driven by extracellular ATP by supplementing cultures with the ATPase apyrase (Sigma, catalog #A6535–200UN). Apyrase was diluted in MilliQ water (Millipore, 1 mg/ml), sterile filtered, and stored at −20°C until use. Apyrase (5 μl/well) was first administered and allowed to act for 15 min before the addition of ATP reagent containing a second dose of apyrase (5 μl/well). Control cultures received 5 μl of sterile-filtered MilliQ water (Millipore) for 15 min, followed by ATP reagent with 5 μl of sterile-filtered MilliQ water (Millipore). ATP bioluminescence measurements were started immediately after the addition of the ATP reagent and lasted 72 h.
Figure 1.
ATP release from astrocytes is circadian. A, B, Representative circadian and arrhythmic ATP-induced bioluminescence from WT astrocytes. Numbers above the x-axis are the proportion of rhythmic cultures. C, Representative 6 min frame of bioluminescence induced by extracellular ATP in 32 wells of a 96-well plate. Each well was seeded with 104 wild-type astrocytes. Light flashes (arrow) and luminescence within each well are proportional to ATP concentration. D, Standard curve of bioluminescence as a function of ATP concentration performed immediately after the addition of ATP to a 96-well plate containing ATP reagent incubated at 32°C for 24 h. E, Addition of apyrase decreased ATP-induced bioluminescence to background levels. F, Only cultures with RelAmp values <0.2 were considered rhythmic. Graph shows the percentage of WT cultures scored as rhythmic as a function of RelAmp values determined by FFT-NLLS.
Bioluminescence recordings.
Light from clock-gene reporters (Bmal1::dLuc or Per2::dLuc) and ATP release were detected in customized light-tight incubators equipped with either photomultiplier tubes (HC135--11; Hamamatsu Corp.) (Prolo et al., 2005; Beaulé et al., 2009; Marpegan et al., 2009) or with an intensified CCD camera (XR/Mega-10Z; Stanford Photonics Inc.) controlled by Micro-Manager imaging software (http://www.micro-manager.org). Single-cell bioluminescence imaging was performed with the same camera attached to an inverted microscope using a 10× objective (TE-2000, Nikon). Recordings of gene expression were conducted at 34°C and ATP release was recorded at 32°C. Bioluminescence was integrated every 6 min for whole cultures or every hour for single cells over 4–7 d after culture medium was exchanged with HEPES-buffered DMEM supplemented with B27 and 0.1 μm d-luciferin (BioThema).
Immunocytochemistry.
Following bioluminescence data collection, astrocytes were stained to assess cell density, astrocyte purity, and transfection efficiency. Cells were rinsed once with 0.01 m PBS and fixed with 4% paraformaldehyde in 0.01 m phosphate buffer at room temperature for 15 min. Cells were then rinsed three times with PBS and stored at 4°C until staining. Astrocytes were blocked by incubating in 0.25% Triton X-100 in PBS (Triton-PBS) supplemented with 10% bovine serum albumin (BSA) for 30 min at room temperature. After blocking, cells were incubated with a rabbit polyclonal antibody raised against glial fibrillary acidic protein (GFAP) (Dako North America) diluted 1:1000 in Triton-PBS with 3% BSA. Cells were incubated for 24 h at 4°C in the primary solution. On the following day, cells were rinsed three times for 5 min each with PBS and incubated for 90 min at room temperature with a donkey anti-rabbit antibody conjugated to Cy3 (1:200 dilution in Triton-PBS and 3% BSA, Jackson ImmunoResearch). Cells were washed three times for 5 min with PBS, stained with 300 nm DAPI (Invitrogen) for 10 min, and rinsed in PBS. Fluorescence images were obtained with a Retiga-EX camera at 10× magnification using the Northern Eclipse software.
Cell density analysis.
Six nonoverlapping photomicrographs were obtained for each culture at 10× magnification, and DAPI-stained nuclei were counted with the Nucleus Counter plugin in the NIH ImageJ image processing software (http://rsbweb.nih.gov/ij). The average cell density (cells/mm2) was calculated and the total number of cells estimated by multiplying the cell density by the total area covered by cells
Data analysis.
Bioluminescence data were analyzed as previously published with modifications (Abe et al., 2002). Raw data were detrended by subtracting a 24 h running average and then smoothed using a 3 h running average. Each smoothed trace was baseline adjusted by adding the raw bioluminescence value at 24 h. The presence of circadian rhythms was determined by fast Fourier transform, followed by nonlinear least-squares fitting (FFT-NLLS). Because FFT-NLLS will fit data with different goodness of fit levels, we selected a criterion relative amplitude (RelAmp) value ≤0.2 to score cultures as rhythmic. Data not fitted by FFT-NLLS or with RelAmp values >0.2 were scored as arrhythmic. The range of dominant periods allowed for FFT-NLLS was set between 16 and 32 h. Serial dilutions of ATP were used to generate ATP standard curve after fitting the data (grayscale intensity vs ATP concentration) with a linear regression model. This curve was used to approximate the concentration of ATP released by astrocytes. Using a standard curve generated after 24 h at 32°C, we compared ATP release between genotypes by converting bioluminescence values to ATP concentration, and normalizing this value by the number of cells in each well at the end of the experiments. These ATP values are reported as femtomolar (fm) per cell. One-way ANOVA with Scheffé post hoc test was used to compare ATP levels between genotypes. Chi-square (χ2) was used to compare the proportion of cultures showing ATP release rhythms. Alpha was set at 0.05 for all analyses.
Results
ATP release from cultured astrocytes is circadian
We developed a real-time assay to continuously monitor ATP release using stabilized recombinant firefly luciferase and luciferin that allowed us to detect extracellular ATP for >72 h. We found 72% (23 of 32 cultures) of WT astrocyte cultures were circadian for at least 3 d based on strict period and amplitude criteria (Fig. 1A,B). Thus, ATP release from murine cortical astrocytes is circadian, consistent with findings in rat astrocytes, immortalized SCN 2.2 cells, and the in vivo rat SCN (Womac et al., 2009).
Cultured astrocytes showed diffused long-lasting bioluminescence as well as brief flashes (∼0.75–2.5 mm in diameter) lasting up to 40 min (Fig. 1C). Light emission exceeded background levels by 1000-fold at 0.015 nm ATP and increased linearly to 2.5 nm at 32°C when measured 24 h after addition of the ATP reagent (Fig. 1D). The addition of apyrase, which catalyzes ATP hydrolysis, decreased bioluminescence levels to background values (Fig. 1E; n = 16 wells for vehicle- and apyrase-treated astrocytes, n = 8 for empty wells), demonstrating the specificity of the assay. Astrocytes released sufficient ATP so that bioluminescence exceeded background levels (wells without cells or without ATP reagent) for >72 h. The FFT-NLLS criterion we selected to score cultures as rhythmic is shown in Figure 1F.
Astrocyte circadian rhythms depend on clock genes
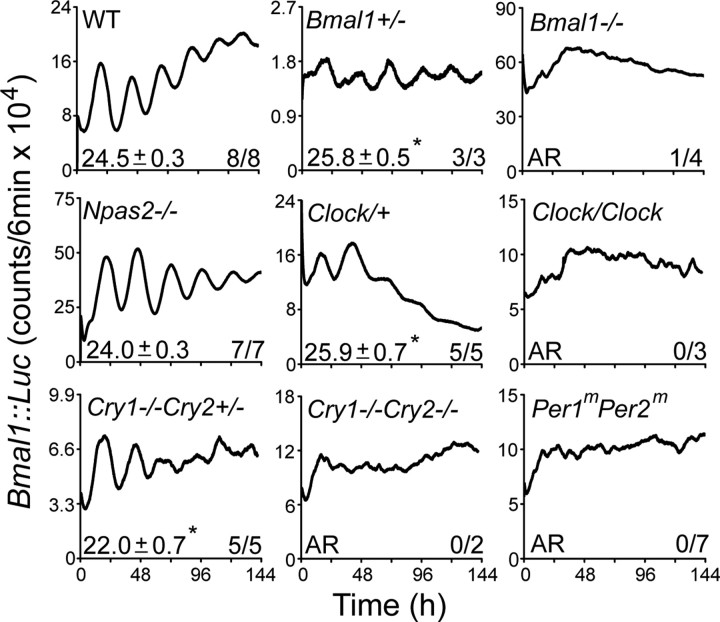
To assess the molecular basis of circadian timing in astrocytes, we tested the roles of canonical clock genes (Bmal1, Clock, Npas2, Per1 and Per2, Cry1 and Cry2). All WT astroglial cultures transfected with Bmal1::dLuc reporters displayed circadian oscillations in bioluminescence (Fig. 2). High-amplitude rhythms were also present in Npas2−/− astrocytes. In contrast, Bmal1−/−, Clock/Clock, Cry1−/−Cry2−/−, and Per1mPer2m astrocyte cultures were arrhythmic. Bmal1+/− and Clock/+ cultures had longer circadian periods (25.8 ± 0.5 h and 25.9 ± 0.7 h, respectively), and Cry1−/−Cry2+/− astrocytes had shorter periods (22.0 ± 0.7 h) than WT (24.5 ± 0.3 h; p < 0.05, Dunnett's test). We found similar clock phenotypes with a transfected Per2::dLuc reporter (data not shown).
Figure 2.
Clock genes regulate circadian rhythms in astrocytes. Representative raw bioluminescence traces for pure astrocyte cultures transfected with Bmal1::dLuc. Numbers above the x-axis provide the period (mean ± SEM) and the proportion of rhythmic cultures for each genotype. AR, Arrhythmic. *p < 0.05, significantly different from WT.
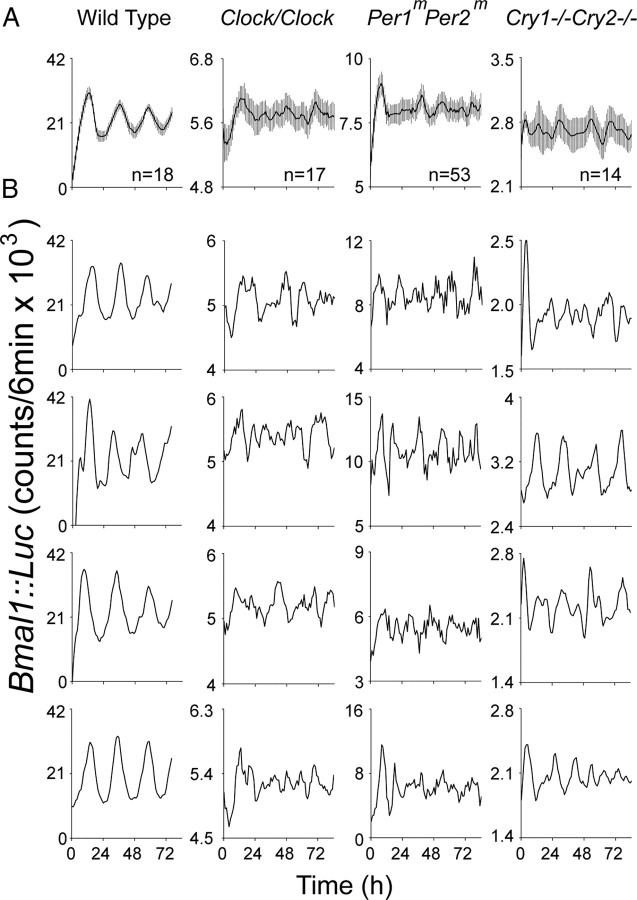
Loss of rhythmicity in knock-out cells could reflect desynchrony or arrhythmicity of individual cells. To differentiate the two possibilities, we measured bioluminescence in individual astrocytes expressing a lentiviral Bmal1::dLuc reporter, and found that the majority of Clock/Clock (n = 25 of 26, 2 cultures), Per1mPer2m (53 of 53, 1 culture), and Cry1−/−Cry2−/− (24 of 26, 2 cultures) cells were not circadian, in contrast to the proportion of WT astrocytes that displayed rhythmicity (8 of 18, 1 culture; Fig. 3). We conclude that cultured individual astrocytes, like many other types of mammalian cells, rely on functional Bmal1, Clock, both Per1 and Per2, and both Cry1 and Cry2 for circadian oscillations.
Figure 3.
Individual astrocytes are circadian oscillators. Representative bioluminescence traces for astrocytes infected with lenti-Bmal1::dLuc. A, Mean (±SEM) bioluminescence for all single astrocytes recorded in one culture for each genotype. Numbers above x-axis represent the number of cells analyzed per culture. Note that the ensemble of WT astrocytes showed circadian cycling. B, Representative bioluminescence traces reveal reliable circadian rhythms in individual WT astrocytes and weak or arrhythmic expression in astrocytes of the other three genotypes.
Circadian rhythms in ATP release depend on clock genes
To test whether the circadian clock mechanism underlies circadian ATP release, we assayed extracellular ATP in astrocytes of different clock genotypes. We observed that daily cycling of ATP release was severely blunted in astrocytes from two representative clock mutants, Clock/Clock (28% circadian; 9 of 32 cultures) and Per1mPer2m mice (38% circadian; 9 of 24) compared with WT astrocytes (72% circadian; 23 of 32) (Fig. 4A,B). The proportions of Clock/Clock and Per1mPer2m cultures that showed rhythmic ATP release were significantly different from WT (χ2 = 13.403, p < 0.01), but the mutant genotypes did not differ from each other (χ2 = 0.533, N.S.). Furthermore, the Clock/Clock and Per1mPer2m mutations more than halved the amount of ATP released compared with WT cultures (101.4 ± 3.6 fm per cell for Clock/Clock, 94.2 ± 1.7 fm for Per1mPer2m, 207.6 ± 5.3 fm for WT; when measured 24 h after the start of the recording; p < 0.05, Scheffé test; Fig. 4B). Thus, the circadian pattern and overall magnitude of ATP release in astrocytes depend on functional Clock and both Per1 and Per2 genes.
Figure 4.

Rhythmic ATP release depends on a functional clock. A, Representative ATP bioluminescence traces for circadian (top) and arrhythmic (bottom) cultures of WT (left), Clock/Clock (middle), and Per1mPer2m (right) astrocytes. Numbers above x-axis represent proportion of rhythmic cultures for each genotype. B, The proportion of circadian cultures was calculated based on the RelAmp values determined by FFT-NLLS (rhythmic: RelAmp ≤0.2). C, ATP release per cell (mean ± SEM) in Clock/Clock and Per1mPer2m cultures was significantly less than that in WT astrocytes measured 24 h after the start of the recording (*p < 0.05, Scheffé test).
ATP release, but not Bmal1 expression, depends on gliotransmission
Gliotransmitters can be released through mechanisms including vesicular release, hemichannels, and transporters. Using astrocytes cultured from two transgenic mouse models designed to interfere with astrocytic transmitter release, we studied possible targets for circadian regulation of ATP release. The dnSNARE mouse overexpresses a dominant-negative form of synaptobrevin 2 only in astrocytes, blocking vesicular gliotransmission (Pascual et al., 2005; Halassa et al., 2009). The VIPP mouse overexpresses type I Ins(1,4,5)P3 5-phosphatase and therefore lacks IP3-dependent intracellular calcium signaling, a mechanism proposed for gliotransmission. Expression of dnSNARE was confirmed by the identification of EGFP-positive cells, and VIPP was confirmed by Venus-positive cells (Fig. 5A). Both dnSNARE (n = 5) and VIPP (n = 8) astrocyte cultures exhibited high-amplitude circadian rhythms in Bmal1-driven bioluminescence (Fig. 5A). In contrast, ATP release rhythms were found in only 6% of the VIPP cultures (2 of 32), compared with 82% of WT (n = 23 of 28) and 84% of dnSNARE (n = 27 of 32) cultures (Fig. 5B,C). This reduction in the proportion of rhythmic cultures for VIPP astrocytes differed significantly from the WT and dnSNARE cultures (χ2 = 52.204, p < 0.0001) and, importantly, WT and dnSNARE did not differ from each other (χ2 = 0.007, N.S.). We conclude that while vesicular release is not critical for circadian rhythms in ATP release in astrocytes, IP3 signaling contributes to the amplitude of ATP release.
Figure 5.

Gliotransmission affects ATP release but not Bmal1::Luc rhythms. A, Representative raw bioluminescence traces for dnSNARE and VIPP cortical astrocyte cultures infected with the lenti-Bmal1::dLuc reporter. Numbers above the x-axis are the proportion of rhythmic cultures for each genotype. EGFP (dnSNARE cells) and Venus (VIPP cells) fluorescence emitted from astrocyte cultures at the end of the recordings confirmed the phenotype of the cells. Insets, EGFP and Venus signals in grayscale. Scale bar, 100 μm. B, Representative ATP-induced bioluminescence traces for circadian (top) and arrhythmic (bottom) cultures of WT, dnSNARE, and VIPP astrocytes. C, The proportion of circadian cultures was calculated based on the RelAmp values determined by FFT-NLLS (rhythmic: RelAmp ≤0.2).
Discussion
We found that cultured mouse cortical astrocytes display circadian rhythms in extracellular ATP, in agreement with recent results from rat astrocyte cultures, SCN and SCN2.2 cells (Womac et al., 2009). We used a stabilized form of luciferase that allowed long-term recordings of extracellular ATP from the same cells without perturbations that can affect circadian clock-gene expression in astrocytes (Prolo et al., 2005). We found that Clock/Clock, Per1m Per2m, Cry1−/−Cry2−/− and Bmal1−/− astrocytes are arrhythmic, similar to the locomotor behavior deficits of these mice (Vitaterna et al., 1994; van der Horst et al., 1999; Bunger et al., 2000; Zheng et al., 2001). We found that Bmal1, Clock−/+ and Cry1−/−Cry2−/+ glia have abnormal periods, much like the heterozygous mouse behavior. The correlations between rhythmicity in clock genes, extracellular ATP in glia and locomotor behavior suggest they may be tightly related.
Most of the astrocyte cultures deficient for functional CLOCK or PERIOD proteins lacked circadian rhythms in Bmal1::dLuc and Per2::dLuc reporter expression and in extracellular ATP. In addition, a low percentage of Clock/Clock or Per1mPer2m glial cultures displayed disrupted circadian rhythms in ATP release. It is unclear whether the mutant cultures scored as circadian were able to generate weak circadian cycling. Importantly, their ATP release was constitutively low, suggesting that the circadian clock functions to upregulate ATP release at specific times of day. Although circadian rhythms in extracellular ATP metabolism and intracellular ATP production may contribute to the rhythms we observed, the ATP-dependent flashes recorded here, the reduction of extracellular ATP levels, and the loss of (high-amplitude) rhythmicity following genetic disruption of IP3 signaling provide evidence for a circadian control of ATP release. In contrast, genetic disruption of vesicular release had no effect on circadian ATP release suggesting that vesicular release of ATP is not under circadian control or that its deficiency can be compensated by other releasing mechanisms.
It is unknown how the circadian clock modulates pathways involved in purinergic signaling. We observed tonic and discrete changes in bioluminescence, suggesting that there are at least two mechanisms that contribute to glial release of ATP. Future studies examining the frequency of flashes as a function of genotype and circadian time may be able to resolve the role of the circadian clock in the discrete release events. Extracellular ATP concentration depends on diverse mechanisms including cell–cell interactions, activity of ectonucleotidases, cell-volume regulation and ionic homeostasis (Reetz et al., 1997; Arcuino et al., 2002; Corriden and Insel, 2010). There are also over 20 purinergic receptors sensitive to ATP or its metabolites, some of which can feedback on ATP release. Although none of these were studied for circadian variation, the activity of enzymes involved in the synthesis of adenosine display daily rhythms in areas of the brain related to sleep (Mackiewicz et al., 2003). ATP release could also be affected by the intracellular concentration of ATP. Although circadian and metabolic pathways can be interrelated (Yang et al., 2006; Lin et al., 2008; Nakahata et al., 2009; Ramsey et al., 2009), no circadian rhythms in intracellular ATP have been reported. Since purines are common signals between cells in the CNS and peripheral organs, future studies should address the relationship between circadian systems and ATP release.
The role of circadian rhythms in ATP release is unclear. Daily increases in extracellular ATP must not be critical for circadian rhythms in cortical astrocytes because VIPP astrocytes show normal circadian rhythms in Bmal1::dLuc expression despite having arrhythmic extracellular ATP. ATP or its metabolites may be a circadian output from cortical astrocytes that will affect other cells. Whether this is a universal property of astrocytes from all brain regions is unknown. Rhythms in ATP content and release from the SCN and SCN derived cells (Yamazaki et al., 1994; Womac et al., 2009), as well as retinal adenosine (Ribelayga and Mangel, 2005), also have no known function. Extracellular ATP, 5′-AMP and adenosine are associated with many brain functions which are also circadian, including thermoregulation, hormonal secretion, and sleep (Hill et al., 1976; Scaccianoce et al., 1989; Gourine et al., 2002; Jones, 2009; Stojilkovic, 2009). ATP-induced activation of P2X receptors in the hypothalamus regulates both hormone secretion and body temperature while adenosine is associated with sleep need (Porkka-Heiskanen et al., 1997; Blanco-Centurion et al., 2006). Adenosine content in basal ganglia shows daily variations that were proposed to be related to changes in sleep–wake cycles during aging while A1 receptors and 5′-ectonucleotidase activity are rhythmic in cortex presumably playing a role in sleep (Virus et al., 1984; Florio et al., 1991; Rosati et al., 1993; Mackiewicz et al., 2003; Murillo-Rodriguez et al., 2004). Sleep deprivation can shift circadian rhythms (Antle and Mistlberger, 2000; Antle et al., 2001), possibly through adenosine that accumulates with sleep need, a mechanism supported by the findings that light-induced phase shifts can be blocked by both, A1 agonists and IP3 blockers (Watanabe et al., 1996; Marpegán et al., 2005; Leone et al., 2006; Krueger, 2008). Although ATP receptors are expressed throughout the brain in both neurons and glial cells (North, 2002), differential expression of the numerous subtypes of receptors can lead to different responses and roles for ATP in different areas. This is illustrated by the expression of P2X5 in the hypothalamus, where it is coexpressed with arginine vasopressin and neuronal nitric oxide synthase in several nuclei but not in the SCN (Xiang et al., 2006) indicating that circadian rhythms in ATP release could lead to multiple responses depending on the combinations of receptors, dynamics of release, and cell types involved. Corriden and Insel (2010) proposed that ATP release and the consequent activation of purinergic receptors could help establish the basal level of activation for signal transduction pathways, regulating a wide array of responses including blood flow, ion transport, cell volume regulation, neuronal signaling, and host-pathogen interactions. All these responses certainly require a temporal organization that could be provided by the circadian control of basal ATP release in astrocytes and other cell types.
Interestingly previous in vivo studies using dnSNARE mice have shown a critical role for the glial SNARE-sensitive purinergic pathway in the modulation of sleep homeostasis (Halassa et al., 2009). In these studies, sleep deprivation induced changes in slow-wave activity as well as compensatory behavioral changes including total sleep time, and these changes were shown to require an astrocytic dnSNARE-sensitive pathway of ATP/adenosine. Given that the current study shows that circadian changes in ATP are insensitive to astrocytic dnSNARE expression, we conclude that sleep homeostasis and circadian oscillations may be regulated by different signaling mechanisms.
In summary, continuous measurements of extracellular ATP levels reveal circadian release from mammalian astroglia which depends on key clock genes and IP3 signaling. These results indicate that extracellular ATP levels are augmented at specific times of day by CLOCK, PER and BMAL1 activity and suggest a clock-induced increase in energy metabolism and glial activity which may participate in sleep–wake changes in the brain.
Footnotes
This work was supported by NIMH Grant 63104, NIH Grant P30 NS057105, and a grant from the Children's Discovery Institute. We thank Sungwon An for support with the Bmal1::dLuc and Per2::dLuc plasmids. We thank Drs. Joseph Takahashi, Steven McKnight, Steven Reppert, Russell Van Gelder, Christopher Bradfield, Christopher Colwell, and Anthony Harmar for founder mice and Hiroki Ueda for plasmids.
The authors declare no conflict of interest.
References
- Abe M, Herzog ED, Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, Block GD. Circadian rhythms in isolated brain regions. J Neurosci. 2002;22:350–356. doi: 10.1523/JNEUROSCI.22-01-00350.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antle MC, Mistlberger RE. Circadian clock resetting by sleep deprivation without exercise in the Syrian hamster. J Neurosci. 2000;20:9326–9332. doi: 10.1523/JNEUROSCI.20-24-09326.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antle MC, Steen NM, Mistlberger RE. Adenosine and caffeine modulate circadian rhythms in the Syrian hamster. Neuroreport. 2001;12:2901–2905. doi: 10.1097/00001756-200109170-00029. [DOI] [PubMed] [Google Scholar]
- Arcuino G, Lin JH, Takano T, Liu C, Jiang L, Gao Q, Kang J, Nedergaard M. Intercellular calcium signaling mediated by point-source burst release of ATP. Proc Natl Acad Sci U S A. 2002;99:9840–9845. doi: 10.1073/pnas.152588599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulé C, Swanstrom A, Leone MJ, Herzog ED. Circadian modulation of gene expression, but not glutamate uptake, in mouse and rat cortical astrocytes. PLoS One. 2009;4:e7476. doi: 10.1371/journal.pone.0007476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Centurion C, Xu M, Murillo-Rodriguez E, Gerashchenko D, Shiromani AM, Salin-Pascual RJ, Hof PR, Shiromani PJ. Adenosine and sleep homeostasis in the Basal forebrain. J Neurosci. 2006;26:8092–8100. doi: 10.1523/JNEUROSCI.2181-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnstock G. Historical review: ATP as a neurotransmitter. Trends Pharmacol Sci. 2006;27:166–176. doi: 10.1016/j.tips.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Coco S, Calegari F, Pravettoni E, Pozzi D, Taverna E, Rosa P, Matteoli M, Verderio C. Storage and release of ATP from astrocytes in culture. J Biol Chem. 2003;278:1354–1362. doi: 10.1074/jbc.M209454200. [DOI] [PubMed] [Google Scholar]
- Corriden R, Insel PA. Basal release of ATP: an autocrine-paracrine mechanism for cell regulation. Sci Signal. 2010;3:re1. doi: 10.1126/scisignal.3104re1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotrina ML, Lin JH, Alves-Rodrigues A, Liu S, Li J, Azmi-Ghadimi H, Kang J, Naus CC, Nedergaard M. Connexins regulate calcium signaling by controlling ATP release. Proc Natl Acad Sci U S A. 1998;95:15735–15740. doi: 10.1073/pnas.95.26.15735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby M, Kuzmiski JB, Panenka W, Feighan D, MacVicar BA. ATP released from astrocytes during swelling activates chloride channels. J Neurophysiol. 2003;89:1870–1877. doi: 10.1152/jn.00510.2002. [DOI] [PubMed] [Google Scholar]
- Declèves X, Regina A, Laplanche JL, Roux F, Boval B, Launay JM, Scherrmann JM. Functional expression of P-glycoprotein and multidrug resistance-associated protein (Mrp1) in primary cultures of rat astrocytes. J Neurosci Res. 2000;60:594–601. doi: 10.1002/(SICI)1097-4547(20000601)60:5<594::AID-JNR4>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- De Smedt F, Missiaen L, Parys JB, Vanweyenberg V, De Smedt H, Erneux C. Isoprenylated human brain type I inositol 1,4,5-trisphosphate 5-phosphatase controls Ca2+oscillations induced by ATP in Chinese hamster ovary cells. J Biol Chem. 1997;272:17367–17375. doi: 10.1074/jbc.272.28.17367. [DOI] [PubMed] [Google Scholar]
- Fields RD, Burnstock G. Purinergic signalling in neuron-glia interactions. Nat Rev Neurosci. 2006;7:423–436. doi: 10.1038/nrn1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florio C, Rosati AM, Traversa U, Vertua R. Circadian rhythm in adenosine A1 receptor of mouse cerebral cortex. Life Sci. 1991;48:PL25–PL29. doi: 10.1016/0024-3205(91)90476-r. [DOI] [PubMed] [Google Scholar]
- Garcia JA, Zhang D, Estill SJ, Michnoff C, Rutter J, Reick M, Scott K, Diaz-Arrastia R, McKnight SL. Impaired cued and contextual memory in NPAS2-deficient mice. Science. 2000;288:2226–2230. doi: 10.1126/science.288.5474.2226. [DOI] [PubMed] [Google Scholar]
- Gourine AV, Melenchuk EV, Poputnikov DM, Gourine VN, Spyer KM. Involvement of purinergic signalling in central mechanisms of body temperature regulation in rats. Br J Pharmacol. 2002;135:2047–2055. doi: 10.1038/sj.bjp.0704679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halassa MM, Haydon PG. Integrated brain circuits: astrocytic networks modulate neuronal activity and behavior. Annu Rev Physiol. 2010;72:335–355. doi: 10.1146/annurev-physiol-021909-135843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halassa MM, Florian C, Fellin T, Munoz JR, Lee SY, Abel T, Haydon PG, Frank MG. Astrocytic modulation of sleep homeostasis and cognitive consequences of sleep loss. Neuron. 2009;61:213–219. doi: 10.1016/j.neuron.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill MK, Macleod RM, Orcutt P. Dibutyryl cyclic AMP, adenosine and guanosine blockade of the dopamine, ergocryptine and apomorphine inhibition of prolactin release in vitro. Endocrinology. 1976;99:1612–1617. doi: 10.1210/endo-99-6-1612. [DOI] [PubMed] [Google Scholar]
- Jones BE. Glia, adenosine, and sleep. Neuron. 2009;61:156–157. doi: 10.1016/j.neuron.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Kowalska E, Brown SA. Peripheral clocks: keeping up with the master clock. Cold Spring Harbor Symp Quant Biol. 2007;72:301–305. doi: 10.1101/sqb.2007.72.014. [DOI] [PubMed] [Google Scholar]
- Krueger JM. The role of cytokines in sleep regulation. Curr Pharm Des. 2008;14:3408–3416. doi: 10.2174/138161208786549281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone MJ, Marpegan L, Bekinschtein TA, Costas MA, Golombek DA. Suprachiasmatic astrocytes as an interface for immune-circadian signalling. J Neurosci Res. 2006;84:1521–1527. doi: 10.1002/jnr.21042. [DOI] [PubMed] [Google Scholar]
- Lin JD, Liu C, Li S. Integration of energy metabolism and the mammalian clock. Cell Cycle. 2008;7:453–457. doi: 10.4161/cc.7.4.5442. [DOI] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, Doyle FJ, 3rd, Takahashi JS, Kay SA. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackiewicz M, Nikonova EV, Zimmerman JE, Galante RJ, Zhang L, Cater JR, Geiger JD, Pack AI. Enzymes of adenosine metabolism in the brain: diurnal rhythm and the effect of sleep deprivation. J Neurochem. 2003;85:348–357. doi: 10.1046/j.1471-4159.2003.01687.x. [DOI] [PubMed] [Google Scholar]
- Marpegán L, Bekinschtein TA, Costas MA, Golombek DA. Circadian responses to endotoxin treatment in mice. J Neuroimmunol. 2005;160:102–109. doi: 10.1016/j.jneuroim.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Marpegan L, Krall TJ, Herzog ED. Vasoactive intestinal polypeptide entrains circadian rhythms in astrocytes. J Biol Rhythms. 2009;24:135–143. doi: 10.1177/0748730409332042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo-Rodriguez E, Blanco-Centurion C, Gerashchenko D, Salin-Pascual RJ, Shiromani PJ. The diurnal rhythm of adenosine levels in the basal forebrain of young and old rats. Neuroscience. 2004;123:361–370. doi: 10.1016/j.neuroscience.2003.09.015. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North RA. Molecular physiology of P2X receptors. Physiol Rev. 2002;82:1013–1067. doi: 10.1152/physrev.00015.2002. [DOI] [PubMed] [Google Scholar]
- Parpura V, Zorec R. Gliotransmission: Exocytotic release from astrocytes. Brain Res Rev. 2010;63:83–92. doi: 10.1016/j.brainresrev.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual O, Casper KB, Kubera C, Zhang J, Revilla-Sanchez R, Sul JY, Takano H, Moss SJ, McCarthy K, Haydon PG. Astrocytic purinergic signaling coordinates synaptic networks. Science. 2005;310:113–116. doi: 10.1126/science.1116916. [DOI] [PubMed] [Google Scholar]
- Perea G, Navarrete M, Araque A. Tripartite synapses: astrocytes process and control synaptic information. Trends Neurosci. 2009;32:421–431. doi: 10.1016/j.tins.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Porkka-Heiskanen T, Strecker RE, Thakkar M, Bjorkum AA, Greene RW, McCarley RW. Adenosine: a mediator of the sleep-inducing effects of prolonged wakefulness. Science. 1997;276:1265–1268. doi: 10.1126/science.276.5316.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prolo LM, Takahashi JS, Herzog ED. Circadian rhythm generation and entrainment in astrocytes. J Neurosci. 2005;25:404–408. doi: 10.1523/JNEUROSCI.4133-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, Bass J. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reetz G, Wiesinger H, Reiser G. ATP-induced oscillations of cytosolic Ca2+ activity in cultured astrocytes from rat brain are modulated by medium osmolarity indicating a control of [Ca2+]i oscillations by cell volume. Neurochem Res. 1997;22:621–628. doi: 10.1023/a:1022430305491. [DOI] [PubMed] [Google Scholar]
- Reick M, Garcia JA, Dudley C, McKnight SL. NPAS2: an analog of clock operative in the mammalian forebrain. Science. 2001;293:506–509. doi: 10.1126/science.1060699. [DOI] [PubMed] [Google Scholar]
- Ribelayga C, Mangel SC. A circadian clock and light/dark adaptation differentially regulate adenosine in the mammalian retina. J Neurosci. 2005;25:215–222. doi: 10.1523/JNEUROSCI.3138-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati AM, Traversa U, Florio C, Vertua R. Circadian rhythm of cortical and striatal adenosine receptors. Life Sci. 1993;52:1677–1684. doi: 10.1016/0024-3205(93)90475-i. [DOI] [PubMed] [Google Scholar]
- Sato F, Kawamoto T, Fujimoto K, Noshiro M, Honda KK, Honma S, Honma K, Kato Y. Functional analysis of the basic helix-loop-helix transcription factor DEC1 in circadian regulation. Eur J Biochem. 2004;271:4409–4419. doi: 10.1111/j.1432-1033.2004.04379.x. [DOI] [PubMed] [Google Scholar]
- Scaccianoce S, Navarra D, Di Sciullo A, Angelucci L, Endröczi E. Adenosine and pituitary-adrenocortical axis activity in the rat. Neuroendocrinology. 1989;50:464–468. doi: 10.1159/000125264. [DOI] [PubMed] [Google Scholar]
- Siepka SM, Yoo SH, Park J, Lee C, Takahashi JS. Genetics and neurobiology of circadian clocks in mammals. Cold Spring Harbor Symp Quant Biol. 2007;72:251–259. doi: 10.1101/sqb.2007.72.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojilkovic SS. Purinergic regulation of hypothalamopituitary functions. Trends Endocrinol Metab. 2009;20:460–468. doi: 10.1016/j.tem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suadicani SO, Brosnan CF, Scemes E. P2X7 receptors mediate ATP release and amplification of astrocytic intercellular Ca2+ signaling. J Neurosci. 2006;26:1378–1385. doi: 10.1523/JNEUROSCI.3902-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh J, Jackson FR. Drosophila ebony activity is required in glia for the circadian regulation of locomotor activity. Neuron. 2007;55:435–447. doi: 10.1016/j.neuron.2007.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, Buijs R, Bootsma D, Hoeijmakers JH, Yasui A. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- Virus RM, Baglajewski T, Radulovacki M. Circadian variation of [3H]N6-(l-phenylisopropyl)adenosine binding in rat brain. Neurosci Lett. 1984;46:219–222. doi: 10.1016/0304-3940(84)90445-2. [DOI] [PubMed] [Google Scholar]
- Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A, Moriya T, Nisikawa Y, Araki T, Hamada T, Shibata S, Watanabe S. Adenosine A1-receptor agonist attenuates the light-induced phase shifts and fos expression in vivo and optic nerve stimulation-evoked field potentials in the suprachiasmatic nucleus in vitro. Brain Res. 1996;740:329–336. doi: 10.1016/s0006-8993(96)00881-5. [DOI] [PubMed] [Google Scholar]
- Webb AB, Angelo N, Huettner JE, Herzog ED. Intrinsic, nondeterministic circadian rhythm generation in identified mammalian neurons. Proc Natl Acad Sci U S A. 2009;106:16493–16498. doi: 10.1073/pnas.0902768106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womac AD, Burkeen JF, Neuendorff N, Earnest DJ, Zoran MJ. Circadian rhythms of extracellular ATP accumulation in suprachiasmatic nucleus cells and cultured astrocytes. Eur J Neurosci. 2009;30:869–876. doi: 10.1111/j.1460-9568.2009.06874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z, He C, Burnstock G. P2X5 receptors are expressed on neurons containing arginine vasopressin and nitric oxide synthase in the rat hypothalamus. Brain Res. 2006;1099:56–63. doi: 10.1016/j.brainres.2006.04.126. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Ishida Y, Inouye S. Circadian rhythms of adenosine triphosphate contents in the suprachiasmatic nucleus, anterior hypothalamic area and caudate putamen of the rat—negative correlation with electrical activity. Brain Res. 1994;664:237–240. doi: 10.1016/0006-8993(94)91978-x. [DOI] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, Menaker M, Takahashi JS. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, Liu X, Atwood A, Huss JW, 3rd, Janes J, Su AI, Hogenesch JB, Kay SA. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139:199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, Lee CC. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]