Abstract
The early transcriptional defense responses and reactive oxygen species (ROS) production in Arabidopsis (Arabidopsis thaliana) cell suspension culture (ACSC), containing functional chloroplasts, were examined at high light (HL). The transcriptional analysis revealed that most of the ROS markers identified among the 449 transcripts with significant differential expression were transcripts specifically up-regulated by singlet oxygen (1O2). On the contrary, minimal correlation was established with transcripts specifically up-regulated by superoxide radical or hydrogen peroxide. The transcriptional analysis was supported by fluorescence microscopy experiments. The incubation of ACSC with the 1O2 sensor green reagent and 2′,7′-dichlorofluorescein diacetate showed that the 30-min-HL-treated cultures emitted fluorescence that corresponded with the production of 1O2 but not of hydrogen peroxide. Furthermore, the in vivo photodamage of the D1 protein of photosystem II indicated that the photogeneration of 1O2 took place within the photosystem II reaction center. Functional enrichment analyses identified transcripts that are key components of the ROS signaling transduction pathway in plants as well as others encoding transcription factors that regulate both ROS scavenging and water deficit stress. A meta-analysis examining the transcriptional profiles of mutants and hormone treatments in Arabidopsis showed a high correlation between ACSC at HL and the fluorescent mutant family of Arabidopsis, a producer of 1O2 in plastids. Intriguingly, a high correlation was also observed with ABA deficient1 and more axillary growth4, two mutants with defects in the biosynthesis pathways of two key (apo)carotenoid-derived plant hormones (i.e. abscisic acid and strigolactones, respectively). ACSC has proven to be a valuable system for studying early transcriptional responses to HL stress.
Oxygenic photosynthesis is the biological process that sustains life on Earth. In this light-driven reaction, water is split and molecular oxygen is released as a by-product. The molecular oxygen that accumulates in the atmosphere is vital for aerobic organisms, but it can also become a precursor of (undesirable) reactive oxygen species (ROS) that can induce oxidative damage in cells and therefore place the life of aerobic organisms in jeopardy (Halliwell, 2006). In plants, ROS can be generated during photochemical energy conversion. High light (HL) is a stress factor responsible for direct inhibition of the photosynthetic electron transport chain in chloroplasts, leading to the generation of ROS in several locations: singlet oxygen (1O2) in PSII, superoxide radical (O2·−) in PSI, and hydrogen peroxide (H2O2) in the chloroplast stroma and also in peroxisomes through the photorespiratory cycle (Niyogi, 1999; Asada, 2006). Consequently, plants are obliged to cope with ROS generation in order to maintain plastid redox homeostasis. Together with the ROS detoxification pathways in chloroplasts, there are other active ROS fronts in organelles such as mitochondria and peroxisomes as well as in other plant cell compartments such as the cytosol, the apoplast, and the cell wall that also require strict control (Apel and Hirt, 2004; Gechev et al., 2006). Although all these detoxification pathways seem to indicate that ROS play a detrimental role in plant cells, ROS generation can become an advantage rather than a drawback in the regulation of multiple biological processes in plants (Mittler et al., 2004; van Breusegem et al., 2008). ROS are known to be key signaling molecules in growth, developmental processes, stress adaptation, and programmed cell death in plants (Foyer and Noctor, 2005a; Bell et al., 2009).
Over the last decade, much progress has been made in plant ROS signaling under stress conditions that provoke changes in the redox homeostasis of plant cells (Foyer and Noctor, 2005b). Since several ROS are generated under HL conditions, a direct correspondence between the accumulation of a specific ROS and the observed changes in transcript expression is not straightforward. The analysis of various transcriptional profiles of transgenic Arabidopsis (Arabidopsis thaliana) plants with compromised levels of specific antioxidant enzymes and the identification of the conditional fluorescent (flu) mutant have shed much light on the specific effects of 1O2, O2·−and H2O2 (Gadjev et al., 2006). In that study, various transcripts were specifically regulated by 1O2, O2·−or H2O2, but others were identified as general ROS response markers. At the same time, a significant number of transcripts encoding WRKY, zinc finger type, MYB, AP2, and ERF transcription factors were overrepresented. Some of these transcription factors are known to play multiple roles in regulating redox homeostasis, cell development, and cell defense, all biological processes tied to ROS signaling events (Mittler et al., 2004, Ülker and Somssich, 2004; Davletova et al., 2005; Khandelwal et al., 2008).
Recent studies in photooxidative stress in plants have shown that 1O2 is the main ROS involved in the damage of leaf tissues (Triantaphylidès and Havaux, 2009). The main sites of 1O2 production are chlorophyll-containing photosynthetic complexes and the reaction center of PSII (Rinalducci et al., 2004; Krieger-Liszkay et al., 2008). By contrast, 1O2 does not seem to be produced at any significant level in PSI (Hideg and Vass, 1995). The 1O2 production in the flu mutant occurs because protochlorophyllide accumulates in the thylakoids (Meskauskiene et al., 2001), but unlike in wild-type plants, the production is not associated with excess energy excitation in PSII (Mullineaux and Baker, 2010). In this study, we present Arabidopsis cell suspension cultures (ACSCs), containing functional chloroplast, as a valuable cellular system in which to investigate ROS production at HL. These conditions are expected to overreduce the photosynthetic electron transport chain in ACSC and to launch the production of 1O2, O2·−and H2O2 in chloroplasts. Some of the ideal characteristics of ACSC for the analysis of early transcriptional responses include uniformity, homogeneity, repeatability, the absence of developmental processes, and slow systemic effects between cells (Menges et al., 2003).
Our transcriptional analysis has led us to conclude that 1O2 is the major ROS in ACSC under HL stress and that the 1O2 production is responsible for a transcriptional response that notably resembles the transcriptional profile of the flu mutant family and, intriguingly, the ABA deficient1 (aba1) and more axillary growth4 (max4) mutants, characterized with the blockade of the biosynthesis pathways of two (apo)carotenoid-derived phytohormones: abscisic acid (ABA) and strigolactones, respectively. Similarities and differences between ACSC under HL stress and the above Arabidopsis mutants are discussed.
RESULTS
Functioning of Chloroplasts in ACSC
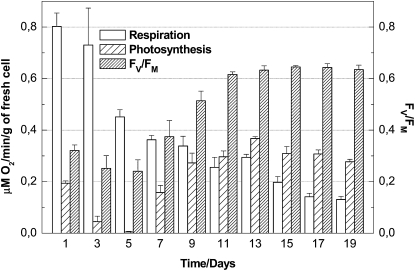
The functioning of the photosynthetic electron transport chain in ACSC was evaluated by following changes in oxygen evolution rate and PSII quantum efficiency during cellular growth. In brief, the “green” ACSC grew with a doubling time of about 5 d and had a cell density of 150 to 200 mg mL–1 at the beginning of the stationary phase. In the dark, the mitochondrial respiration rate of ACSC was very active during the lag phase but gradually diminished during cellular growth (Fig. 1). In contrast to the respiration rate, the oxygen evolution rate was slow during the lag phase, achieving a minimum level around day 5 after subculturing. From that point onward, the oxygen evolution rate increased gradually during the log phase, reaching a maximum level when the cellular growth moved into the stationary phase, where the oxygen evolution rate exceeded the respiration rate (Fig. 1). The chlorophyll a/b ratio was 3.8 ± 0.2 after 9 d of growth. The quantum efficiency of PSII (Fv/Fm) in ACSC followed a pattern similar to the one described for the oxygen evolution rate, and its maximum level reached a value of 0.65 (Fig. 1). The above results provide evidence to support that the photosynthetic electron transport chain in chloroplasts of ACSC is functional, although a large percentage of nonreducing QB (for secondary electron-accepting plastoquinone of PSII) complexes are expected to be present in their thylakoid membranes (see “Discussion”).
Figure 1.
Respiration rate, oxygen evolution, and PSII quantum efficiency of ACSC grown at 24ºC under continuous illumination (50 μE m–2 s–1).
Additionally, the oxygen evolution of ACSC after the HL treatment (1,800 μE m–2 s–1) was monitored to determine the extent of the overreduction of the photosynthetic electron chain. The results depicted in Supplemental Figure S1 show that the rate of oxygen evolution of ACSC did not change when it was measured at a light irradiance of 300 μE m–2 s–1, but it decreased to about 75% of the initial value if the light irradiance of 1,800 μE m–2 s–1 was maintained in the oxygen electrode chamber. This means that the photosynthetic electron chain partly became overreduced during the HL treatment, but ACSC could recover the original rate of oxygen evolution when the HL treatment ceased.
Reverse Transcription-PCR Analysis of Selected Transcripts Responding to Early ROS Generation
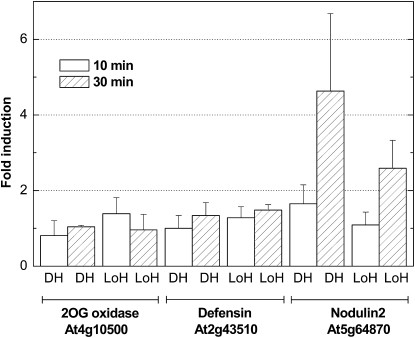
Changes in the expression profile for the selected ROS markers are shown in Figure 2. Short times of 10 and 30 min were selected to avoid the accumulation of slowly induced ROS that could blur the evolution of early changes in transcript expression. Additionally, the changes for each marker were examined when cultures shifted from low light (LL; 50 μE m–2 s–1) to HL (1,800 μE m–2 s–1) and from 1 h of dark to HL conditions. Two main conclusions were drawn from the results depicted in Figure 2. First, the fold change in expression for the 1O2 marker At5g64870 (a nodulin-like protein transcript, NOD) was higher than that for the other two markers after 30 min under HL stress; second, the fold change for NOD was even more pronounced when shifting from 1 h of dark to HL. The H2O2 marker At4g10500 [a 2-oxoglutarate-Fe(II) oxygenase family transcript, 2-OXO] as well as the general abiotic stress marker At2g43510 (a defensin-like family transcript, DEF) reached 2-fold changes in transcript expression after 1 h of HL treatment. Therefore, the incubation time of 30 min was found to be optimum for further transcriptional analysis of the 1O2-mediated stress responses in ACSC using DNA microarrays.
Figure 2.
Fold changes in the expression of the selected transcripts responding to ROS production in ACSC under HL stress. HL stress (1,800 μE m–2 s–1) started shifting from 1-h dark to HL (DH) or directly from control (or LL) conditions (50 μE m–2 s–1) to HL (LoH).
Transcriptional Profiling of ACSC under HL Stress
To further characterize the ROS-mediated responses of ACSC to HL, a whole-genome transcriptional profile analysis was performed using Affymetrix GeneChip Arabidopsis genome ATH1 arrays. Basically, nine microarray experiments were designed and classified as follows: control (1–3) with a light irradiance of 50 μE m–2 s–1, 1-h dark (1–3), and HL (1–3) with a light irradiance of 1,800 μE m–2 s–1, where the numbers 1 to 3 indicate the number of biological replicates (Gene Expression Omnibus database under accession no. GSE22671). Principal components analysis (PCA) was used in exploratory data analysis and showed that much of the variability of the nine microarray experiments could be depicted in a PCA plot of two components, where the three HL conditions were grouped together in the top right quadrant (Supplemental Fig. S2). After data normalization, the package Limma identified a total of 449 transcripts differentially expressed with an adjusted value of P < 0.05 when ACSC shifted from 1-h dark to HL for 30 min (Supplemental Table S1). There were 418 up-regulated transcripts (approximately 93%) and only 31 down-regulated transcripts (approximately 7%) among the total number of transcripts exhibiting differential expression. Intriguingly, it is worth noting that there were no significant changes in the transcriptional profile (adjusted P < 0.05) when ACSC shifted from control conditions (50 μE m–2 s–1) to 1-h dark (data not presented).
Functional Classification of Transcripts
The independent studies by Mittler et al. (2004) and Gadjev et al. (2006) compile comprehensive lists of ROS transcripts. Some of these transcripts are specifically up-regulated by one type of ROS, whereas other transcripts can be up-regulated by several types of ROS. The list of the 418 up-regulated transcripts in ACSC at HL was compared with the above lists, and the common transcripts are displayed in Table I. It is notable that 41 out of the 418 up-regulated transcripts in ACSC under HL stress were specifically activated by 1O2. Seven out of 418 corresponded with transcripts up-regulated by general ROS production, but only four out of 418 were transcripts specifically activated by O2·− or H2O2. Additionally, transcripts encoding enzymes with an active role in O2·− or H2O2 scavenging (or production as NADPH oxidases) were poorly represented (six out of 418). A direct inspection of the up-regulated transcripts in our study with the 30-min early-induced, 2-fold up-regulated transcripts in the flu mutant of Arabidopsis (op den Camp et al., 2003) showed that more than 50% of the up-regulated transcripts in the flu mutant also formed part of the 418 up-regulated transcripts in ACSC at HL. This result was further confirmed in a hierarchical clustering analysis (see below), where the coexpression relationship between transcripts in a subset of different conditions was scrutinized.
Table I. Transcripts up-regulated in ACSC under HL stress defined as specific ROS markers.
Adjusted P < 0.05.
| ROS Type or Enzyme | Gene Identifier | Log2 Ratio | Description |
| Up-regulated with 1O2 | At2g39200 | 2.49 | Mildew resistance locus O 12; calmodulin binding |
| At1g58420 | 3.45 | Unknown protein | |
| At4g27654 | 4.30 | Unknown protein | |
| At1g73540 | 3.43 | Nudix hydrolase homolog 21; hydrolase | |
| At5g64660 | 3.18 | U-box domain-containing protein | |
| At3g44260 | 3.89 | CCR4-associated factor 1-like protein | |
| At4g36500 | 4.17 | Unknown protein | |
| At4g27657 | 2.28 | Unknown protein | |
| At5g47230 | 1.65 | Ethylene-responsive element-binding factor 5 | |
| At4g34410 | 4.38 | Redox-responsive transcription factor 1 | |
| At3g57760 | 1.76 | Protein kinase family protein | |
| At3g18690 | 1.93 | Mitogen-activated protein kinase substrate 1; protein binding | |
| At3g46620 | 2.03 | Zinc finger (C3HC4-type RING finger) family protein | |
| At5g51190 | 3.01 | AP2 domain transcription factor, putative | |
| At3g56880 | 1.57 | VQ motif-containing protein | |
| At1g19770 | 1.03 | Purine transmembrane transporter | |
| At5g58430 | 1.42 | Exocyst subunit EXO70 family protein B1; protein binding | |
| At1g30370 | 3.83 | Lipase class 3 family protein | |
| At3g57530 | 1.14 | Calcium-dependent protein kinase 32 | |
| At5g05140 | 0.90 | Transcription elongation factor-related | |
| At2g44370 | 1.65 | DC1 domain-containing protein | |
| At3g46930 | 1.65 | Protein kinase 6-like protein | |
| At5g44070 | 1.31 | CAD1, glutathione γ-glutamylcysteinyltransferase | |
| At3g26980 | 0.95 | Membrane-anchored ubiquitin-fold protein 4 precursor | |
| At2g44840 | 3.21 | Ethylene-responsive element-binding factor 13 | |
| At4g33985 | 1.28 | Unknown protein | |
| At3g25600 | 1.40 | Calcium ion binding | |
| At2g35710 | 2.34 | Glycogenin glucosyltransferase | |
| At1g74450 | 1.71 | Unknown protein | |
| At1g67970 | 0.64 | Heat shock transcription factor A8 | |
| At3g16720 | 1.68 | ATL2; protein binding, zinc ion binding | |
| At4g28350 | 0.93 | Lectin protein kinase family protein | |
| At1g33590 | 1.68 | Disease resistance protein-related, LRR protein-related | |
| At5g04760 | 0.69 | MYB transcription factor | |
| At1g44830 | 1.35 | AP2 domain-containing transcription factor TINY, putative | |
| At4g01250 | 2.34 | WRKY22; transcription factor | |
| At5g28630 | 1.95 | Gly-rich protein | |
| At3g45640 | 1.27 | Mitogen-activated protein kinase 3 | |
| At1g11050 | 1.21 | Ser/Thr protein kinase | |
| At2g47060 | 0.90 | Ser/Thr protein kinase, putative | |
| At5g56980 | 1.87 | Unknown protein | |
| Up-regulated with O2·− | At2g17840 | 0.87 | Early responsive to dehydration 7 |
| Up-regulated with H2O2 | At3g05650 | 0.56 | Receptor-like protein 32; kinase, protein binding |
| At2g47190 | 2.55 | MYB2 | |
| At2g30500 | 1.06 | Kinase-interacting family protein | |
| Up-regulated with ROS | At4g37370 | 1.88 | CYP81D8; electron carrier, heme binding, monooxygenase |
| At1g19020 | 2.80 | Unknown protein | |
| At4g39670 | 1.80 | Glycolipid binding/glycolipid transporter | |
| At5g13080 | 1.21 | WRKY75 | |
| At3g28210 | 1.72 | Putative zinc finger protein | |
| At3g54150 | 2.19 | Embryonic abundant protein-like | |
| At1g13340 | 0.87 | Unknown protein | |
| ROS scavenger | At1g28480 | 1.95 | Glutaredoxin family, cytosol/mitochondria |
| At5g20230 | 1.17 | Blue copper protein | |
| At3g08710 | 0.79 | Thioredoxin family, cytosol | |
| At1g32350 | 0.58 | Alternative oxidase, mitochondria | |
| At3g09940 | 1.18 | Monodehydroascorbate reductase 2, cytosol | |
| ROS producer | At5g47910 | 0.86 | NADPH oxidase, membrane |
Only 31 transcripts were down-regulated in ACSC under HL stress; however, none of these were found in the list of transcripts specifically down-regulated in the flu mutant. Instead, three out of 31 down-regulated transcripts in ACSC under HL stress were present in the list of transcripts specifically up-regulated by O2·−H2O2 (At4g03510, At1g14890, and At1g19200; Gadjev et al., 2006).
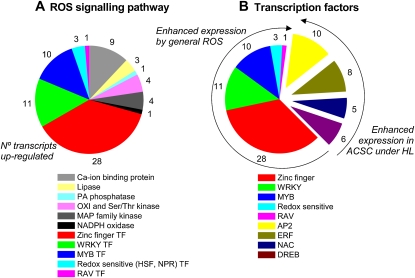
For a better understanding of the effect of the HL treatment on ACSC, we extracted further information through several Web-based applications. FatiGO was used to find Gene Ontology (GO) terms that were overrepresented and underrepresented in the list of the 418 up-regulated transcripts with regard to the rest of the Arabidopsis genome. A summary of the GO biological processes that were overrepresented and underrepresented is given in Supplemental Table S2. The most represented terms in the functional enrichment were associated with responses to several types of abiotic and biotic stresses, water stress deficit and hormone stimuli, and the hypersensitive response. A search for key components of the plant ROS signaling cascade (Mittler et al., 2004) in the list of the 418 up-regulated transcripts revealed several transcripts encoding calcium-binding proteins, lipases, kinases, and transcription factors (Fig. 3). The number of transcripts encoding transcription factors was remarkably large (approximately 80), representing several transcription factor families (i.e. HSF, NPR, zinc finger, WRKY, MYB, RAV, AP2, ERF, NAC, DRE/CRT, and DREB). The motif names for the transcription factors are given in Supplemental Table S3. Whereas the transcript expression of HSF, NPR, zinc finger, WRKY, MYB, and RAV transcription factor families is known to be enhanced by several types of ROS (Mittler et al., 2004; Wu et al., 2009), the transcript expression of the rest of the transcription factor families was mediated by 1O2 (AP2, ERF, DREB, and NAC) or by water stress (DREB, DRE/CRT; and NAC; see “Discussion”). Additionally, we also made use of FatiScan in an attempt to find a more general functional interpretation. This method detects significantly up- or down-regulated blocks of functionally related transcripts in the complete list of Arabidopsis genes (approximately 22,000) ordered by differential expression. The results summarized in Supplemental Figure S3 show that the most overrepresented biological processes in ACSC under HL stress were those associated with signaling mediated by hormones such as ethylene (ET), jasmonate (JA), and salicylic acid (SA), water stress, and cell death.
Figure 3.
Key components of the ROS signaling cascade in ACSC under HL stress (A) with special attention to the up-regulated transcription factors (B). The number of identified transcripts is indicated beside each wedge. Displaced wedges in B represent transcription factors not included in the generalized model of ROS signaling cascade proposed by Mittler et al. (2004), but their transcripts are induced under our experimental conditions. TF, Transcription factor.
Microarray Data Validation by Reverse Transcription-PCR
As depicted in Supplemental Figure S4, all the transcripts selected for the microarray validation showed similar fold changes in expression after the dark-HL transition when compared with their levels of expression on the microarrays. Except for the transcripts 2-OXO and DEF, all the selected transcripts showed statistically significant changes in their expression under HL stress (adjusted P < 0.05).
ROS Detection in ACSC under HL Stress
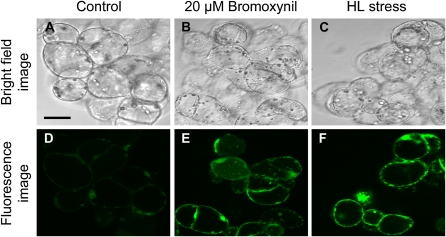
The production of 1O2 and H2O2 was evaluated in situ by fluorescence microscopy using the singlet oxygen sensor green (SOSG) reagent and 2′,7′-dichlorofluorescein diacetate (DCFH-DA) as fluorescence probes. As shown in Figure 4, the SOSG reacted to the presence of 1O2 after 30 min of HL treatment. In order to establish whether the observed green fluorescence was indeed associated with the photogeneration of 1O2, the phenolic herbicide bromoxynil (Fufezan et al., 2002) at a concentration of 20 μm was used as a positive control at LL. The result with bromoxynil was very similar to the HL treatment, and green fluorescence emission among cells was newly observed. Control cells showed very faint green fluorescence in comparison with the HL and bromoxynil treatments.
Figure 4.
Representative confocal micrographs illustrating the production of 1O2 in ACSC after 30 min under LL (50 μE m–2 s–1; A and D), LL and 20 μm bromoxynil (B and E), and HL (1,800 μE m–2 s–1; C and F) conditions. The production 1O2 was detected using the SOSG reagent. The top panels are bright-field images and the bottom panels are fluorescence images following excitation at 488 nm with an argon laser and an RSP500 excitation beam splitter. Bar = 25 μm.
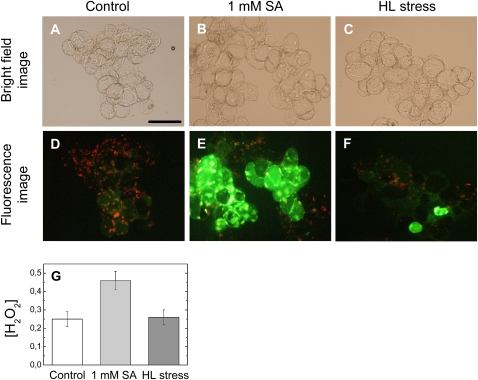
In contrast to SOSG, no fluorescence emission from DCFH-DA was detected in Arabidopsis cells subjected to a 30-min HL treatment. When ACSC was exposed to longer HL treatments (i.e. 45 min), a few Arabidopsis cells had the distinctive green fluorescence emission of DCFH-DA (Fig. 5). The effect of the HL treatment was contrasted with the effect caused by 1 mm SA at LL (positive control). The comparison showed that Arabidopsis cells treated with 1 mm SA exhibited a more intense green fluorescence emission. The intracellular concentration of H2O2 was also measured spectrophotometrically in ACSC under HL stress. The results indicated that the intracellular concentration of H2O2 was 0.25 ± 0.04 nmol g–1 (wet weight) ACSC at LL; however, no significant variation in the H2O2 concentration was observed after the HL treatment: 0.26 ± 0.04 nmol g–1 (wet weight) ACSC. In contrast, a 2-fold increase in the intracellular concentration of H2O2 was induced (i.e. 0.46 ± 0.05 nmol g–1 [wet weight] ACSC) when 1 mm SA was added to the culture medium at LL (Fig. 5G).
Figure 5.
Representative micrographs illustrating the production of H2O2 in ACSC after treatment with SA and HL for 45 min. H2O2 was detected with DCFH-DA by the bright green fluorescence. The top panels are bright-field images of ACSC at 50 μE m–2 s–1 (A), ACSC at 50 μE m–2 s–1 in the presence of 1 mm SA (B), and ACSC at 1,800 μE m–2 s–1 (C). The middle panels show the chlorophyll autofluorescence of ACSC at 50 μE m–2 s–1 (D), the green fluorescence of DCFH-DA in ACSC at 50 μE m–2 s–1 in the presence of 1 mm SA (E), and the combination of the autofluorescence of chlorophyll and the green fluorescence of DCFH-DA in ACSC at 1,800 μE m–2 s–1 (F). The bottom panel (G) shows the intracellular concentration of H2O2 (nmol g–1 wet weight of cell culture) in ACSC. Bar = 100 μm.
D1 Photodamage in ACSC under HL Stress
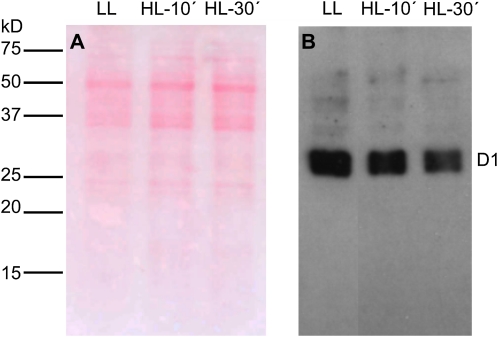
In order to test experimentally that 1O2 was the predominant ROS responsible for the observed transcriptional defense responses, we also attempted to detect its production indirectly following the photodamage of the D1 protein of the PSII reaction center in ACSC under HL stress. The progress of the in vivo photodamage of D1 was detected by western blot (Fig. 6), and the decrease in the intensity of the D1 band indicates that 1O2 must be responsible for the D1 photodamage. The intensity of the immunodetected band of the D1 protein decreased to about 70% and 50% of the initial value after 10 and 30 min of HL treatment, respectively (Fig. 6B). Other ROS have also been proposed to be responsible for the photodamage of the D1 protein; however, the concentration of such ROS or the polypeptide pattern of the degradation products of the D1 protein did not match with those found under our experimental conditions (see “Discussion”).
Figure 6.
Western-blot analysis of the D1 protein of PSII in ACSC under HL stress. Each lane was loaded with 10 μg of protein. Arabidopsis cells were exposed to LL (50 μE m–2 s–1) and HL (1,800 μE m–2 s–1) stress conditions for 10 and 30 min. A, Ponceau red staining of the nitrocellulose membrane is shown to visualize the ACSC protein pattern. B, Western blot showing the photodamage of the D1 protein. [See online article for color version of this figure.]
Selection of Key Mutants and Hormone-Treated Plants for Meta-Analysis
Prior to meta-analysis, the 418 up-regulated transcripts identified in ACSC under HL stress were subjected to comparative analysis using the biclustering tool of Genevestigator (Hruz et al., 2008). As expected, the comparative analysis yielded a very high correlation between the up-regulated transcripts in our study and those found in the flu mutant family of Arabidopsis (data not shown). Unexpectedly, a high correlation was also observed with the (apo)carotenoid biosynthesis pathway-deficient mutants aba1 and max4 of Arabidopsis. Therefore, all the above mutants were selected for further analysis, together with the O2·−-producing mutant over-tAPX. Additionally, microarray data from experiments with wild-type plants of Arabidopsis treated with hormones such as ABA, the ethylene intermediate 1-aminocyclopropane-1-carboxylic acid (ACC), SA, and methyl jasmonate (MeJA) were also selected for meta-analysis based on our functional enrichment analysis explained above.
Meta-Analysis
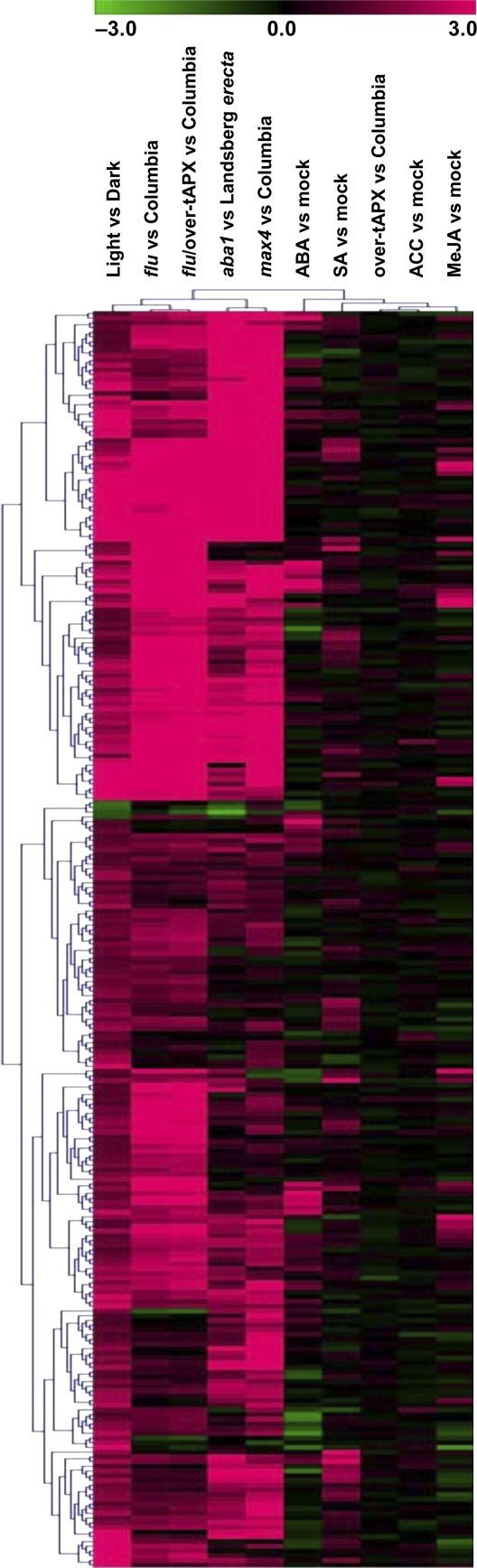
In order to gain insight into the coregulation patterns on transcript expression profiles, we performed a hierarchical clustering analysis of transcripts differentially expressed in response to HL as compared with several flu and hormone-deficient mutants and hormone-treated plants of Arabidopsis. The list of these 305 transcripts, which were selected based on the signal log2 ratios (up-regulation when 1 or greater, down-regulation when –1 or less), is given in Supplemental Table S4, together with their corresponding coregulated transcripts. An image of the clustering analysis is represented in Figure 7.
Figure 7.
Overall picture of the hierarchical clustering analysis on transcripts differentially expressed in ACSC under HL stress. Data were clustered together with available expression data from hormone treatment experiments within the AtGenExpress database and the aba1, max4, flu, flu/over-tAPX, and over-tAPX mutants within the Gene Expression Omnibus. Gene subclusters of interest are discussed in the text.
As summarized in Supplemental Figure S5A, 214 out of the 297 (72%) up-regulated transcripts selected from ACSC at HL were also up-regulated in the flu mutant. A similar percentage was also observed by comparison with the flu/over-tAPX mutant, as 217 out of the 297 (73%) up-regulated transcripts were found to be coregulated. Indeed, Pearson’s correlation was observed to be high between the ACSC at HL and the two flu mutants (Supplemental Table S5). In contrast, no significant correlation was observed when ACSC at HL was compared with over-tAPX (Supplemental Table S5). Surprisingly, a cluster of coregulated transcripts was also observed when compared with the aba1 and max4 mutants (Supplemental Fig. S5B). Two hundred six out of the 297 (69%) up-regulated transcripts selected in the HL cultures were found to be coregulated in max4. Similarly, 183 out of 297 (62%) up-regulated transcripts were also up-regulated in aba1. Pearson’s correlation was also observed to be high between the ACSC at HL and the aba1 and max4 mutants (Supplemental Table S5). When the clustering analysis was carried out by comparison with the hormone-treated plants, approximately only 15% of the 297 up-regulated transcripts were found to be coregulated in the treatments with ABA (42 out of 297) and SA (45 out of 297). A lower percentage was even observed when compared with the MeJA- and ACC-treated plants. No significant coregulation was found between the HL cultures and Arabidopsis plants exposed to hormones (Supplemental Table S5). Further analysis showed that only 22 out of the 297 selected transcripts were specifically up-regulated in ACSC at HL (Table II). Within this list, two transcripts (At5g39580 and At1g14540) encoding peroxidases were identified.
Table II. Early HL-responsive transcripts specifically up-regulated in ACSC after 30 min of treatment.
Fold induction is expressed as log2 ratio. Adjusted P < 0.05.
| Gene Identifier | Light Versus Dark | aba1 Versus Landsberg erecta | max4 Versus Columbia | Description |
| At3g57750 | 1.01 | −0.31 | 0.36 | Protein kinase, putative |
| At2g46780 | 1.03 | 0.38 | 0.46 | RNA-binding protein |
| At1g68765 | 1.04 | 0.67 | 0.18 | Receptor-binding protein |
| At5g60270 | 1.24 | 0.14 | −0.6 | Lectin protein kinase family protein |
| At1g27820 | 1.27 | 0.59 | 0.46 | CCR4-NOT transcription complex protein, putative |
| At5g23130 | 1.28 | 0.53 | 0.42 | Peptidoglycan-binding LysM domain-containing protein |
| At1g04490 | 1.37 | 0.13 | 0.82 | Protein of unknown function |
| At1g80450 | 1.42 | 0.25 | −0.19 | VQ motif-containing protein |
| At1g55700 | 1.43 | 0.08 | 0.05 | DC1 domain-containing protein |
| At1g49780 | 1.47 | −0.07 | −0.09 | U-box domain-containing protein |
| At5g40460 | 1.49 | −0.45 | 0.59 | Protein of unknown function |
| At4g01950 | 1.63 | −0.29 | 0.77 | Glycerol-3-phosphate acyltransferase |
| At5g43520 | 1.68 | −0.03 | 0.32 | DC1 domain-containing protein |
| At5g39580 | 1.85 | 0.04 | 0.1 | Peroxidase, putative |
| At5g46295 | 1.87 | 0.33 | 0.66 | Protein of unknown function |
| At2g20960 | 1.99 | 0.09 | 0.66 | Protein of unknown function |
| At4g37780 | 2.07 | −0.05 | 0.16 | MYB87 |
| At5g37490 | 2.12 | 0.43 | 0.25 | U-box domain-containing protein |
| At2g37880 | 2.58 | −0.07 | −0.10 | Protein of unknown function |
| At1g14540 | 2.81 | 0.01 | 0.08 | Anionic peroxidase, putative |
| At4g20000 | 3.04 | 0.19 | −0.09 | VQ motif-containing protein |
| At1g77640 | 3.56 | 0.26 | 0.37 | AP2 domain-containing transcription factor, putative |
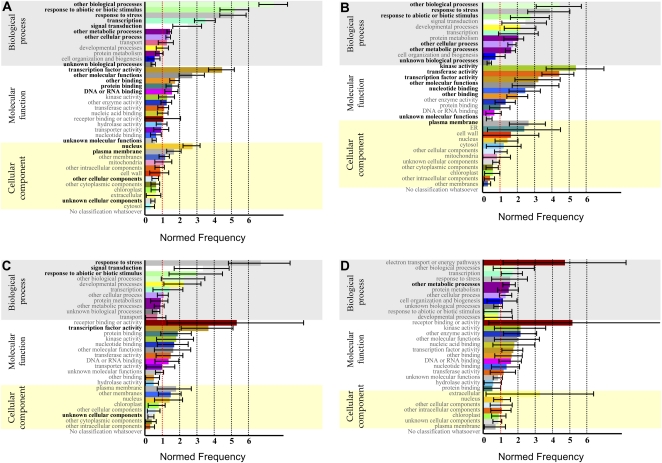
The 305 transcripts differentially expressed in HL cultures were also classified according to the Classification SuperViewer bioinformatic tool. Prior to analysis, the transcripts were assigned to four different categories: A, coregulation in flu, flu/over-tAPX, aba1, and max4; B, coregulation in flu and flu/over-tAPX; C, coregulation in aba1 and max4 (Table III); and D, specific coregulation in HL cultures (Table II). As shown in Figure 8, A to C, the functional gene enrichment was mainly associated with the response to abiotic or biotic stimulus for categories A to C. Interestingly, most of the stress-related transcripts were associated with the response to ABA stimulus and water deprivation. Analysis of transcripts specifically up-regulated in HL cultures (i.e. not responsive to hormonal treatments or selected mutants) showed that most of the induced transcripts belong to a major category: electron transport or energy pathways (Fig. 8D).
Table III. Early HL-responsive transcripts of ACSC at 30 min showing specific coregulation with transcripts up-regulated in aba1 and max4.
Fold induction is expressed as log2 ratio. Adjusted P < 0.05.
| Gene Identifier | Light Versus Dark | aba1 Versus Landsberg erecta | max4 Versus Columbia | Description |
| At2g43290 | 1.07 | 1.05 | 2.83 | Calcium ion-binding protein |
| At1g67880 | 1.07 | 1.8 | 2.52 | Glycosyl transferase family 17 protein |
| At1g14870/At1g14880 | 1.1 | 2.45 | 2.36 | Protein of unknown function |
| At3g45660/At3g45650 | 1.15 | 1.27 | 1.89 | NAXT1 (nitrate excretion transporter 1) |
| At1g71400 | 1.17 | 1.28 | 2.31 | RLP12 (receptor-like protein 12) |
| At1g27100 | 1.20 | 1.05 | 3.27 | Protein of unknown function |
| At1g19025 | 1.22 | 1.12 | 1.09 | DNA cross-link repair protein-related |
| At2g01300 | 1.28 | 2.53 | 2.20 | Protein of unknown function |
| At2g45680 | 1.32 | 2.11 | 3.13 | TCP family transcription factor, putative |
| At3g20590/At3g20600 | 1.32 | 3.24 | 2.96 | NDR1 (non-race-specific disease resistance 1); signal transducer |
| At5g10750 | 1.33 | 1.38 | 2.27 | Protein of unknown function |
| At2g41010 | 1.45 | 2.04 | 2.32 | Calmodulin-binding protein of 25 kD |
| At2g25250 | 1.48 | 1.19 | 3.13 | Protein of unknown function |
| At1g70740 | 1.67 | 1.8 | 1.78 | Protein kinase family protein |
| At3g05320 | 1.77 | 2.20 | 1.66 | Protein of unknown function |
| At5g58120 | 1.86 | 1.87 | 3.58 | Disease resistance protein (TIR-NBS-LRR class), putative |
| At3g50060 | 1.95 | 3.31 | 2.92 | MYB77 |
| At5g19240 | 2.06 | 3.19 | 3.06 | Protein of unknown function |
| At5g64900 | 2.09 | 1.67 | 1.74 | Elicitor peptide 1 precursor |
| At3g54810 | 2.18 | 2.08 | 2.48 | BME3/BME3-ZF transcription factor |
| At2g31880 | 2.19 | 2.75 | 2.75 | Leu-rich-repeat transmembrane protein kinase, putative |
| At1g20823 | 2.32 | 3.36 | 3.38 | Zinc finger (C3HC4-type RING finger) family protein |
| At5g44060 | 2.47 | 1.02 | 2.63 | Protein of unknown function |
| At1g12610 | 2.8 | 5.18 | 3.89 | DDF1 transcription factor |
| At5g52020 | 3.31 | 2.78 | 2.94 | AP2 domain-containing protein |
| At5g64905 | 5.02 | 1.53 | 1.10 | Elicitor peptide 3 precursor |
Figure 8.
Functional classification of differentially expressed transcripts in ACSC under HL stress. The 305 transcripts differentially expressed were classified based on the Classification SuperViewer bioinformatic tool at the Bio-Array Resource Web site (http://bar.utoronto.ca/ntools/cgi-bin/ntools_classification_superviewer.cgi). A, Coregulation of aba1, max4, flu, and flu/over-tAPX. B, Coregulation of flu and flu/over-tAPX. C, Coregulation of aba1 and max4. D, Light- versus dark-specific responses in ACSC under HL stress.
DISCUSSION
Functioning of Chloroplasts in ACSC
The functioning of chloroplasts in ACSC was first investigated before the cells were subjected to HL stress. The oxygen evolution rate was observed to increase gradually during growth, and it is worth noting that no electron acceptors were added to ACSC during the measurement of the Hill reaction. Thus, the water-to-ferredoxin (and from there to NADP+) reaction provides evidence to support that the photosynthetic electron transport chain of thylakoid membranes in ACSC was active. However, other physiological parameters indicated that chloroplasts were not completely mature. In particular, the maximum value for the Fv/Fm ratio (approximately 0.6) was lower than the one (0.83) determined for healthy and mature chloroplasts of green leaves (Björkman and Demmig, 1987), suggesting the presence of nonreducing QB PSII complexes in the thylakoid membranes due to slow activation and/or slow development of the oxygen-evolving complex (Lebkuecher et al., 1999). Additionally, the chlorophyll a/b ratio was higher (approximately 3.8) than the same ratio (3.5) for mature chloroplasts of many higher plants. These results were in accordance with the study by Doyle et al. (2010), where chloroplasts from ACSC and Arabidopsis leaves were compared, and the former were found both to be smaller and less regular on average and to contain a lower quantity of grana. Despite all this, chloroplasts in Arabidopsis cultures are able to sense environmental changes and to activate nucleus-encoded genes in a manner similar to that described in whole plants exposed to environmental stresses (Piñas-Fernández and Strand, 2008). For example, Oswald et al. (2001) observed changes in the transcriptional expression of some nucleus-encoded photosynthetic genes when the photosynthetic electron transport chain was inhibited by 3-(3,4-dichlorophenyl)-1,1-dimethylurea, and Doyle et al. (2010) proposed an interplay between light, chloroplast, ROS, and nuclear protein synthesis during the apoptosis-like programmed cell death of the Arabidopsis culture. All these results, together with the results discussed below, show that the chloroplast-to-nucleus communication is functional in ACSC under stress conditions.
ROS Production in ACSC
Our study has proven that chloroplasts in ACSC sense HL stress and initiate a signaling cascade that leads to the up- and down-regulation of a set of approximately 449 transcripts with functions associated with different types of cellular defense responses. HL stress is responsible for the photosynthetic photoinhibition and concomitant enhancement of ROS production in chloroplasts, where 1O2 is proposed to be the major ROS generated under excess light conditions (Triantaphylidès et al., 2008; Triantaphylidès and Havaux, 2009). The oxygen evolution rate measured in ACSC clearly indicated that PSII was active in chloroplast thylakoids, although some PSII complexes were unable to reduce QB. This means that 1O2 can be generated by the radical pair mechanism (Takahashi et al., 1987) in both types of PSII, because either the acceptor side of PSII is overreduced by HL or PSII is simply nonactive. In addition, O2·−H2O2 production is also possible if PSI electron acceptors remain reduced in the chloroplast stroma (Fryer et al., 2003).
In our attempt to determine experimentally what type of ROS was produced in ACSC, we made use of fluorescence probes that react with 1O2 or H2O2. SOSG has been successfully used to detect 1O2 in leaves of wild-type and flu mutant plants under certain experimental conditions (Flors et al., 2006). Our results showed that the HL treatment also induced green fluorescence emission in ACSC at HL when the cell cultures were incubated with SOSG, indicating that there was a chemical reaction between the fluorescence probe and 1O2. A similar result was observed when bromoxynil was added to block electron transport on the acceptor side of PSII (Fufezan et al., 2002).
Briefly, 1O2 production has also been observed at LL (30 μE m–2 s–1 or less) or under illumination with widely spaced, single-turnover flashes (Keren et al., 1997; Vass and Cser, 2009). In our study, a basal 1O2 production is presumed during ACSC growth at LL (50 μE m–2 s–1) based on the inherent pigment-binding properties of the PSII reaction center. In fact, a very faint green fluorescence is observed in ACSC at LL when the cell culture is incubated with SOSG. However, no significant changes in the transcriptional profile of ACSC were observed after 1 h in the dark, a considerable period of time during which the basal 1O2 production must stop, and consequently, no conclusions were drawn about a signaling role of 1O2 at LL in ACSC.
When DCFH-DA was used instead, green fluorescence emission was not observed during the first 30 min of HL treatment, but it was observed in a few Arabidopsis cells after 45 min, indicating that the induction of H2O2 production required a longer incubation time. This latter result was further supported when the intracellular concentration of H2O2 was measured and no significant changes in the concentration of H2O2 were determined between ACSC at LL and HL conditions.
Further evidence supporting that 1O2 was the major ROS produced under our experimental conditions came from the fact that the D1 protein was partially damaged during the HL treatment. Photodamage of the D1 protein is known to take place under aerobic conditions, when the triplet state of the primary oxidant, P680, is quenched by molecular oxygen and, subsequently, 1O2 reacts with D1 (Aro et al., 1993; Mishra et al., 1994). Photodamage of the D1 protein is also present in preparations of PSII treated with H2O2 or O2·−. However, the H2O2 concentrations required to induce D1 degradation were found to be in the millimolar range (Miyao et al., 1995). Since the intracellular concentration of H2O2 is well below the millimolar range and does not vary during the HL treatment, it is very unlikely that H2O2 can be responsible for the D1 photodamage under our experimental conditions. Additionally, O2·− is known to trigger the degradation of the D1 protein following a pathway that differs from the acceptor-side damage of PSII at HL (Hideg et al., 1995). In this pathway, D1 degradation appears in parallel with the specific C-terminal fragments of D1 in the 17- to 19-kD region. A close inspection of the western-blot analysis of ACSC at HL did not reveal the formation of C-terminal fragments of D1 in that region.
All this supports the idea that 1O2 was generated within the PSII reaction center and was the major ROS responsible for the early transcriptional defense responses triggered in ACSC under HL stress.
Comparison between the Transcriptional Profiling of ACSC at HL and the flu Mutant
In our search for transcripts up-regulated by ROS, we found a significant number of transcripts described as specifically up-regulated by 1O2, all of them present in the flu mutant (Gadjev et al., 2006). On the contrary, our search for transcripts specifically up-regulated by O2·−H2O2 only rendered four (Table I). This finding is in agreement with the results presented by op den Camp et al. (2003), who also reported that O2·−H2O2 did not interfere with the early stress responses of the flu mutant after the dark/light transition. Despite the substantial differences in the experimental approach between using the flu mutant or ACSC at HL to investigate the 1O2-mediated stress responses of plant cells, we found that the cellular transcriptional responses in both studies were similar in many respects, but not in all.
The localization of PSII and protochlorophyllide in thylakoids of the flu mutant (Meskauskiene et al., 2001; Przybyla et al., 2008) suggests that the first steps in the 1O2-mediated signaling cascade are equivalent in both systems. This argument finds some support from the fact that several transcripts involved in either the biosynthesis or the signaling pathway of two phytohormones, ET and JA, are up-regulated in both the flu mutant (Danon et al., 2005; Kim et al., 2008) and ACSC under HL stress. In the first instance, the up-regulated transcripts involved in the biosynthesis or signaling pathway of ET closely coincide with those reported in the flu mutant (Danon et al., 2005), although there were a few more ERF transcripts (ERF4, ERF11, and ERF13) up-regulated in ACSC under HL stress. ERF1 was also induced, suggesting a direct induction by ET/JA (Lorenzo et al., 2003), while other ERF transcripts could also be induced by other phytohormones or stress conditions (Wang et al., 2002). With regard to JA, At1g17420, encoding a chloroplast-located lipoxygenase 3 with a key role in the biosynthesis pathway of oxylipins, was up-regulated in ACSC. There were also other transcripts encoding proteins with functions related to JA signaling, such as (1) At5g45110, the product of which is an NPR1-like protein, a type of transcriptional regulator proposed to be involved in the redox-dependent transmission of oxylipin signals (Böttcher and Pollmann, 2009); (2) At1g19180 (JAZ1) and At1g17380 (JAZ5), two members of the recently identified JAZ family of negative regulators that respond to JA stimulus (Chini et al., 2007); and (3) At3g50260 (CEJ1), which encodes a transcription factor whose expression is cooperatively regulated by ET and JA (Nakano et al., 2006). Additionally, At1g02920, At1g02930, At1g69920, and At1g74590 were all up-regulated; these four transcripts encode glutathione S-transferase enzymes belonging to the phi and tau classes, where the glutathione S-transferase tau family is known to catalyze the reaction between the oxylipin 12-oxo-phytodienoic acid and reduced glutathione (Böttcher and Pollmann, 2009).
In contrast to ET and JA, the set of early up-regulated transcripts of ACSC at HL did not include any transcript that could suggest that SA biosynthesis was triggered. We found a short list of transcripts, most of them encoding MYB and WRKY transcription factors and the BON association protein 1, that responded to SA stimulus but also to other hormones or stress stimuli. Specifically, there is no indication for the early up-regulated transcript At3g48090 encoding the enhanced disease susceptibility protein 1 (EDS1), a lipase implicated in the release of polyunsaturated fatty acids needed for the biosynthesis of oxylipins (Ochsenbein et al., 2006). EDS1 is required for SA accumulation in the flu mutant during the 1O2-mediated stress response. In our transcriptional analysis, there are three early up-regulated transcripts (At1g30370, At1g56670, At5g50890) encoding lipases in ACSC. At5g50890 encodes a pathogen-inducible lipase protein belonging to subgroup III (Jakab et al., 2003), as does At1g30370, but there is no evidence to support the contention that either can regulate the biosynthesis or accumulation of SA in the absence of EDS1. This represents a significant difference with respect to the flu mutant, and it means that the induced resistance response in ACSC at HL mainly relies on pathways mediated by ET and JA but not by SA, whose cellular accumulation is usually associated with systemic acquired resistance in plants (Durrant and Dong, 2004).
While medium light irradiance (80–100 μE m–2 s–1) was enough to induce the 1O2-mediated signaling cascade in the flu mutant during the dark/light shift (op den Camp et al., 2003), we required HL irradiance (1,800 μE m–2 s–1) to observe a similar transcriptional defense response in ACSC. Such a difference in the light irradiance has important implications. Specifically, the SA-mediated systemic acquired resistance pathway does not operate at high irradiance because SA does not accumulate (Zeier et al., 2004; Bechtold et al., 2005), explaining why the EDS1-dependent signaling is not activated in ACSC under HL stress. Instead, there are three up-regulated transcripts, At3g20600, At2g35980, and At5g06320, encoding NDR1 (for non-race-specific resistance 1) or NDR1-like proteins, known to mediate EDS1-independent systemic acquired resistance (Bechtold et al., 2005) and to play a role in hypersensitive response-like cell death.
The induced transcriptional stress response of ACSC under HL stress includes approximately 80 transcription factors (zinc finger, WRKY, MYB, HSF, NPR, and RAV), the expression of which is mediated by several types of ROS (Mittler et al., 2004). A few transcription factors of the above families, together with other transcription factors belonging to the ERF, AP2, and DREB families, are up-regulated in ACSC. Some of these transcription factors are known to be specifically up-regulated in the flu mutant (Gadjev et al., 2006), reconfirming the view that 1O2 is the major ROS in our study. However, the set of up-regulated transcription factors includes others that are not well represented in the flu mutant. Five of these belong to the NAC family, and one is a DRE/CRT transcription factor. The NAC transcription factor named ATAF1 (At1g01720) and DRE/CRT are characterized by their high transcriptional activation in response to water deficit stress (Sakuma et al., 2006; Lu et al., 2007; Wu et al., 2009). At present, the reason why these types of transcription factors are up-regulated in ACSC is not clear, where presumably water is not a limiting factor. Recently, it has been suggested that plants are more sensitive to drought when ROS-scavenging mechanisms are deficient in chloroplasts (Miller et al., 2010).
In addition to the above set of transcription factors, there are other key components in ROS signaling pathways and calcium regulation with significant changes in transcript expression, for example, protein kinases (Ser/Thr kinase, OXI1, mitogen-activated protein kinase family) and calmodulin-binding proteins. This shows that the generalized model of ROS signaling transduction pathway proposed by Mittler et al. (2004) is also induced by 1O2 and provides an insight into the complexity of the signaling cascade mediated by this type of ROS (Kim et al., 2008). However, as stated above, it is worth noting that in our experimental conditions, there were very few transcripts that were either specifically up-regulated by H2O2 and O2·− or encoded enzymes involved with ROS synthesis or scavenging, suggesting some signaling cross-talk and an antagonistic interaction between 1O2 and the other types of ROS. This cross-talk is present in the flu/over-tAPX mutant, where antagonism by H2O2 is shown by an enhanced expression of early-activated transcripts by 1O2 (Laloi et al., 2007). Intriguingly, the number of clustered transcripts between ACSC at HL and the flu/over-tAPX mutant was found to be largest when compared with other members of the flu mutant family. Besides, several transcription factors of the NAC family up-regulated in ACSC under HL stress are also up-regulated in flu/over-tAPX (ATAF1, At3g49530, and At5g63790).
Coregulation of the aba1 and max4 Mutants with ACSC at HL
Further meta-analysis was performed by comparison with wild-type plants exposed to hormones including ABA, ACC, SA, and MeJA. These hormones are known to be involved in the regulation of stress responses to abiotic and biotic stimuli and cell death. Indeed, they were present in the GO biological process terms found to be significantly overrepresented in ACSC under HL stress. However, no significant coregulation was observed between the HL cultures and the hormone-treated Arabidopsis plants in the clustering analysis. Only 15% of the up-regulated transcripts identified in ACSC were found to be coregulated in plants exposed to ABA and SA. In contrast, a significant and unexpected coregulation of early HL stress-induced transcripts was found with the aba1 and max4 mutants. Zeaxanthin epoxidase, the product of the ABA1 gene of Arabidopsis, catalyzes the epoxidation of zeaxanthin to antheraxanthin and violaxanthin, generating the epoxycarotenoid precursor of the ABA biosynthetic pathway (Koornneef et al., 1982; Ishitani et al., 1997; Niyogi et al., 1998; Xiong et al., 2001). Therefore, the aba1 mutant accumulates zeaxanthin. In addition, the Arabidopsis MAX4 (AtCCD8) gene encodes a plastid-targeted carotenoid cleavage dioxygenase involved in the production of strigolactones (Sorefan et al., 2003). These compounds belong to the carotenoid-derived terpenoid lactones, previously shown to be involved in shoot branching, mycorrhizal interactions, and seed germination of the Striga plant parasite (Akiyama et al., 2005; Humphrey and Beale, 2006; Hayward et al., 2009). The corresponding max4 mutant is thus deficient in the production of strigolactones.
In our analysis, we have identified a cluster of HL-responsive stress resistance transcripts that are coregulated in aba1 and max4 mutants (Table III): At3g20590 (NDR1) encodes a putative signal transducer of unknown molecular function; At5g64900 encodes a putative 92-amino acid protein that is the precursor of AtPep1, a 23-amino acid peptide that activates transcription of the defensive gene defensin (PDF1.2) and activates the synthesis of H2O2, both being components of the innate immune response (Huffaker et al., 2006); At5g64905 (PROPEP3) encodes an elicitor peptide 3 precursor paralog of PROPEP1 in Arabidopsis; At5g58120 encodes a putative disease resistance protein, intrinsic to membrane (TIR-NBS-LRR class), involved in signal transduction, defense response, apoptosis, and innate immune response (Meyers et al., 2003); At1g71400 encodes a CLAVATA2-related gene, located in the endomembrane system and also involved in stress response (Wang et al., 2010); At2g31880 (EVR_SOBIR1) encodes a putative Leu-rich-repeat transmembrane protein kinase involved in the regulation of cell death and innate immunity (Gao et al., 2009); and finally, At5g52020 encodes a member of the DREB subfamily A-4 of the ERF/AP2 transcription factor family that could be the direct or indirect target of downy mildew effector proteins that promote disease susceptibility (Huibers et al., 2009). Consequently, HL stress triggers a common and specific signaling pathway with aba1 and max4 mutants that controls stress resistance.
Arabidopsis leaves exposed to HL have been shown to activate the biosynthesis of ABA and to trigger an ABA-mediated signaling network responsible for the maintenance of the photochemical quenching required for dissipation of excess energy excitation (Galvez-Valdivieso et al., 2009; Galvez-Valdivieso and Mullineaux, 2010). This new role for ABA is different from its well-documented roles in, for example, dehydration stress, stomatal response, and pathogen defense and is consistent with a role in the compromised nonphotochemical quenching observed in aba1, where the nonphotochemical quenching induction is more rapid but has lower amplitude than in wild-type Arabidopsis (Pogson et al., 1998). In aba1, the chlorophyll a/b ratio is higher and the Fv/Fm is lower than those respective ratios in wild-type Arabidopsis (Lokstein et al., 2002). In addition, aba1 accumulates monomeric, instead of trimeric, light-harvesting complexes (LHC) and exhibits a delayed-greening virescent phenotype (Pogson et al., 1998). Recently, Johnson et al. (2010) have demonstrated that changes in the xanthophyll content of several mutant lines of Arabidopsis or the oligomeric state of LHC significantly affect the chlorophyll fluorescence lifetime and the dynamic range between the light-harvesting and photoprotective states of LHC. These variations could favor 1O2 production in LHC and might also account for the transcriptional responses observed in aba1. In our study, ACSC showed several of the above physiological features described for aba1, such as a high chlorophyll a/b ratio, a low Fv/Fm ratio, and slow greening during growth. To the best of our knowledge, no relationship has been established between aba1 and enhanced 1O2 production in thylakoids of this mutant. Only two studies have demonstrated that aba1 is less tolerant to heat stress or salinity when these types of stresses are combined with HL (Cramer, 2002; Larkindale et al., 2005); however, neither of these studies indicates what type of ROS is produced under their experimental conditions. When the transcript expression profile of aba1 was compared with the set of transcripts specifically up-regulated with 1O2, H2O2, or O2·− (Gadjev et al., 2006), we found that about 40 transcripts specifically up-regulated by 1O2 were also included in the list of transcripts up-regulated in aba1 (i.e. aba1 versus Landsberg erecta), but very few transcripts specifically up-regulated by O2·−H2O2 shared expression with the list of up-regulated transcripts in aba1.
On the other hand, strigolactones have been proposed to have a positive effect on the gene expression of several photosynthetic genes, particularly associated with PSI and PSII and the enzyme Rubisco (Mayzlish-Gati et al., 2010). In the tomato (Solanum lycopersicum) mutant named Slort1, deficient in strigolactone biosynthesis, a reduced level of chlorophyll was detected in leaves relative to the wild type (Koltai et al., 2010). All this suggests that strigolactones are inducers of photosynthetic genes and that their absence provokes alterations in the photosynthetic apparatus. However, no information is available yet on whether their absence can also be responsible for an enhancement of oxidative stress in plant cells. As above, the comparison between the transcripts specifically up-regulated with 1O2, H2O2, or O2·−the transcripts overexpressed in max4 showed that about 70 transcripts specifically up-regulated by 1O2 were also listed in the set of transcripts up-regulated in max4 (i.e. max4 versus mock), but very few transcripts associated with H2O2 or O2·− were retrieved. The fact that a significant set of transcripts in aba1 and max4 are included in the list of transcripts specifically up-regulated by 1O2 creates a new opportunity to study and confirm the production of 1O2 in these two mutants, both intriguingly defective in the biosynthesis of two plant hormones that have carotenoids, the most efficient quenchers of 1O2, as the initial substrates.
CONCLUSION
Although there are several problems affecting the physiological and genetic quality of plant cell cultures (Cassells and Curry, 2001), ACSC is a valuable cellular system for studying the activation of transcriptional defense responses under adverse stimuli. We conclude that chloroplasts of ACSC are functional organelles able to sense HL stress and initiate defense responses mediated by 1O2-responsive transcripts, which closely correspond with the up-regulated transcripts of the flu mutant family of Arabidopsis. The set of up-regulated transcripts also included a remarkable number of transcription factors, most associated with the ROS signaling cascade but others associated with water deficit stress conditions. Furthermore, HL is responsible for the activation of a stress resistance signaling pathway similar to those observed in the aba1 and max4 mutants.
MATERIALS AND METHODS
Growth Conditions for ACSC
ACSC was kindly provided by the Institut de Biochimie et Physiologie Moléculaire des Plantes (Montpellier, France). The culture was maintained in 200 mL of liquid growth medium (Jouannea and Peaudlen, 1967; Axelos et al., 1992) by gentle agitation at 120 rpm and 24ºC under continuous illumination (50 μE m–2 s–1) in an incubator shaker (model innova TM 44/44R; New Brunswick Scientific). Cells were subcultured with a 1/20th dilution every 7 d.
Functioning of Chloroplasts: Respiratory and Photosynthetic Parameters
In order to determine the functioning of chloroplasts in ACSC, the following parameters were measured: oxygen consumption rate, mitochondrial respiration, Fv/Fm, and chlorophyll a/b ratio. Oxygen evolution rates were measured polarographically using the Chlorolab 2 system (Hansatech Instruments). A volume of 1 mL of ACSC at a cell density of 100 mg mL–1 was placed in the electrode chamber and incubated for a few minutes at 20ºC with constant magnetic stirring. Mitochondrial respiration was always monitored in the dark, whereas overall consumption or production of oxygen, depending on the growth stage, was measured at a light irradiance of 300 μE m–2 s–1. The difference between the light and dark measurements yielded the photosynthetic oxygen evolution. The ratio Fv/Fm, where Fv stands for the variable fluorescence and Fm for the maximum fluorescence, was measured using the PAM-2000 Portable Chlorophyll Fluorometer (Heinz Walz). The chlorophyll a fluorescence induction was monitored by special fiber optics adjusted to the DW2/2 electrode chamber of the Chlorolab 2 system. The chlorophyll a/b ratio was determined spectrophotometrically (Porra et al., 1989).
HL Stress Conditions
ACSC was grown for 9 d, until they reached a cell density of approximately 150 to 200 mg mL–1. A volume of 200 mL was then transferred to a glass vessel, immersed in a water bath to maintain temperature at 24ºC during treatment, and stirred in front of a slide projector irradiating light at 1,800 μE m–2 s–1. The culture was previously incubated in complete dark for 1 h before switching on the light. Control culture was subjected to the same procedure, but the light was kept at 50 μE m–2 s–1. Aliquots of 10 mL were first collected at 10 and 30 min after HL treatment, then filtered and frozen in liquid nitrogen, and finally stored at –80°C until further analysis.
Target Transcripts to Evaluate Early ROS-Mediated Responses in ACSC under HL Stress
Target transcripts to monitor the ROS-mediated responses of ACSC under HL stress were selected from a list containing ROS markers specifically up- or down-regulated by H2O2, O2·–1O2, or general ROS (op den Camp et al., 2003; Gadjev et al., 2006). The selected transcripts were as follows: (1) At5g64870, a nodulin-like protein transcript (NOD) that specifically responds to 1O2; (2) At4g10500, a 2-oxoglutarate-Fe(II) oxygenase family transcript (2-OXO) that specifically responds to H2O2; and (3) At2g43510, a defensin-like family transcript (DEF) that responds to general abiotic stress. The profilin family transcript (PROF) At2g19760 was selected as a housekeeping transcript for internal reference (Laloi et al., 2007). The primers designed to amplify the selected transcripts are shown in Supplemental Table S6.
RNA Isolation and Reverse Transcription-PCR Analysis
Total RNA was extracted with the acid guanidinium isothiocyanate-phenol-chloroform method using Trizol reagent (Ambion) as described by Chomczynski and Sacchi (2006). RNA was treated with TURBO DNase (Ambion) to eliminate traces of contaminating genomic DNA. DNA-free RNA was reverse transcribed using the PrimeScript first-strand cDNA synthesis kit from Takara Bio. Reverse transcription (RT)-PCR analyses were performed on the ABI PRISM 7000 sequence detector system (Applied Biosystems) using the SYBR Premix Ex Taq kit (Takara Bio) and the specific primers for the selected transcripts indicated above. The thermal profile of the RT-PCR consisted of an initial cycle at 95ºC for 30 s, followed by 40 cycles of 5 s at 95ºC, and a final extension at 60ºC for 20 s. Transcripts were amplified on a 96-well format plate by using three technical replicates of samples obtained from at least three biological replicates. Relative quantification of mRNA expression was then calculated using the mathematical method described by Livak and Schmittgen (2001). Expression levels were normalized using the housekeeping transcript PROF.
Microarray Experiments
Transcriptomic analyses were performed using Affymetrix GeneChip Arabidopsis (Arabidopsis thaliana) genome ATH1 arrays. The quality of total, DNA-free RNA was first verified by using the 2100 Bioanalyzer (Agilent Technologies). All samples had 260:280 ratios greater than 1.8 and clear 18S and 28S ribosomal RNA bands. Retrotranscription was performed using the SuperScript Choice System for cDNA synthesis kit (Invitrogen) according to the manufacturer’s instructions. Biotin-labeled complementary RNA was produced by in vitro transcription using the GeneChip 3′ IVT Express Kit (Affymetrix). The biotin-labeled complementary RNA was then degraded by alkaline digestion and used for hybridization with ATH1 arrays. Technical steps such as target hybridization, washing, staining, and scanning of the arrays were performed sequentially as described in the Affymetrix GeneChip expression analysis technical manual using the Affymetrix one-cycle target labeling and control reagents, an Affymetrix GeneChip Hybridization Oven 640, an Affymetrix Fluidics Station 450, and an Affymetrix GeneChip Scanner 3000 7G. The Affymetrix GeneChip Operating Software was used to automate the control of GeneChip Fluidics Stations and Scanners and also to perform the transcript expression data analysis. Experiments were performed from at least three biological replicates. PCA was used as described by Johnson and Wichern (1998) in order to explore the variability between the replicates.
Microarray Data Analysis
Data from microarrays were standardized using quantile normalization and the robust multiarray average method (Bolstad et al., 2003). Differential transcript expression was carried out using the Limma (Smyth, 2004) package from Bioconductor (http://www.bioconductor.org/). Multiple testing adjustment of P values was done according to Benjamini and Hochberg (1995). Significantly overrepresented or underrepresented GO biological process terms were obtained using FatiGO and FatiScan from the Babelomics suite (http://babelomics.bioinfo.cipf.es/) as described previously (Al-Shahrour et al., 2004, 2007). Multiple testing adjustment of P values was then carried out according to the false discovery rate method (Benjamini and Hochberg, 1995; Benjamini and Yekutieli, 2001). GO terms were annotated from Ensembl 56 (http://www.ensembl.org; The Arabidopsis Information Resource 9).
Validation of Microarray Experiments
In order to validate the microarray experiments, 10 of the most up-regulated transcripts after the HL treatment were selected from the list included in Supplemental Table S1. The selected transcripts and their corresponding primers are shown in Supplemental Table S6. In addition, the transcripts NOD, DEF, and 2-OXO, used previously to evaluate the early ROS-mediated responses in ACSC, were also added to the validation.
Detection of 1O2 and H2O2 in Cultures under HL Stress by Fluorescence Microscopy
The detection of 1O2 was performed using the SOSG reagent (Molecular Probes, Invitrogen) as described previously by Flors et al. (2006) with minor modifications. SOSG was added to 9-d-old cultures to give a final concentration of 5 μm. Following the SOSG addition, ACSC was kept in the dark for 10 min and subsequently exposed to HL for 30 min as described above. After the HL treatment, an aliquot of 500 μL of ACSC was washed four times with 5 mL of 2.7 mm KCl, 147.3 mm NaCl, and 0.01 m sodium phosphate (PBS), pH 7.4, following centrifugation at 120g for 3 min at 4°C. The fluorescence emission was monitored in a confocal microscope (model DM IRB; Leica Microsystems) following excitation at 488 nm with an argon laser and a RSP500 excitation beam splitter. The fluorescence emission was collected at 500 to 575 nm.
The detection of H2O2 was carried out following a similar procedure but using the fluorescence probe DCFH-DA (Rhee et al., 2010). Briefly, an aliquot of 500 μL of ACSC, previously exposed to HL stress for 45 min, was washed four times with 5 mL of 0.01 m PBS, pH 7.4, following centrifugation at 120g for 3 min at 4°C, acclimatized to 10 mm Tris-HCl, pH 7.4, and finally incubated with 5 μm DCFH-DA in the former buffer for 15 min at 25°C in the dark on a rotating plate shaker. Cells were then washed four times with 0.01 m PBS, pH 7.4, and visualized through an Eclipse E800 microscope (Nikon Instruments) with an excitation band from 450 to 490 nm and a 520-nm long-pass filter.
Control experiments included ACSC treated at LL for 30 min, ACSC treated with 20 μm bromoxynil at LL to induce the photoproduction of 1O2 in chloroplasts, and ACSC treated with 1 mm SA at LL to induce intracellular production of H2O2.
Spectrophotometric Detection of H2O2 in ACSC
The concentration of H2O2 was measured using a spectrophotometric assay based on the ferrous oxidation in the presence of xylenol orange method (De Michele et al., 2009). A volume of 1 mL of Arabidopsis cells previously treated at HL for 30 min was centrifuged at 120g for 3 min at 4°C, then washed twice with 5 mL of 0.01 m PBS, pH 7.4, following centrifugation in the same conditions, and finally suspended in 1 mL of 0.01 m PBS, pH 7.4. The suspended cells were mixed with 200 mg of acid-washed glass beads, shaken vigorously for 30 min at 4°C, and centrifuged at 18,000g for 15 min at 4°C. An aliquot of 500 μL of the supernatant was then added to an equal volume of assay reagent [500 μm (NH4)2Fe(SO4)2, 50 mm H2SO4, 200 μm xylenol orange, and 200 mm sorbitol] and incubated for 45 min in the dark. The H2O2-mediated oxidation of Fe2+ to Fe3+ was determined by measuring the A560 of the Fe3+-xylenol orange complex. A calibration curve obtained by measuring the A560 of H2O2 standards allowed the conversion of the absorbance values into concentration estimates. All reactions were carried out at least in duplicate, and the values were expressed as nmol H2O2 g−1 fresh weight of Arabidopsis cells.
Indirect Detection of 1O2 in Cultures under HL Stress by Western-Blot Analysis
Approximately 100 mg of HL-treated cultures was disrupted with an electric homogenizer in Nonidet P-40 lysis buffer consisting of 20 mm Tris-HCl, pH 8.0, 137 mm NaCl, 10% (w/v) glycerol, 1% (w/v) Nonidet P-40, 2 mm EDTA, 1 μg mL–1 leupeptin, 1 μg mL–1 aprotinin, 1 μg mL–1 pepstatin A, and 1 mm phenylmethanesulfonyl fluoride. The samples were sonicated for 10 s at a setting of 1 in a Virsonic 300 sonicator (Virtis Co.). The final homogenate was centrifuged at 12,000g for 3 min at 4°C. Protein concentration of the supernatant was determined using the bicinchoninic acid protein assay kit (Pierce) and bovine serum albumin (BSA) as the protein standard (Smith et al., 1985). Protein samples of about 10 μg were subjected to 15% (w/v) SDS-PAGE and transferred overnight to nitrocellulose membranes. Nitrocellulose membranes were stained with Ponceau S solution (Applichem) for 1 min to visualize the protein transfer and then destained with water. The membranes were blocked with 5% (w/v) BSA, 0.6% (w/v) Tween 20 in 150 mm NaCl, and 50 mm Tris-HCl, pH 7.4 (TBS; blocking buffer). For detection of the D1 protein (or PsbA), membranes were incubated overnight at 4°C with a polyclonal anti-PsbA (C-terminal) antibody (Agrisera) using a dilution of 1:5,000 in TBS. After extensive washing in 0.6% (w/v) Tween 20 TBS, the immunocomplexed membranes were probed for 1 h at 25°C with an anti-rabbit, peroxidase-linked secondary antibody using a dilution of 1:10,000 in 0.3% (w/v) Tween 20 and 0.1% (w/v) BSA TBS. Probed membranes were washed with 0.3% (w/v) Tween 20 TBS, and immunoreactive proteins were visualized by means of luminol-enhanced chemiluminescence reagents (Immuno-Star HRP substrate kit; Bio-Rad) and scanned using a Fluor-S MultiImager system (Bio-Rad). Western-blot bands were integrated using the ImageMaster 2D platinum software (GE Healthcare Bio-Sciences).
Meta-Analysis
Data from microarray experiments were clustered together with expression data obtained from key mutants and hormone treatments in Arabidopsis previously selected on the basis of our own microarray data analysis and a comparative analysis using Genevestigator (Hruz et al., 2008). CEL.file data from mutants flu, flu/over-tAPX, over-tAPX, aba1, and max4 were obtained from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), and CEL.file data from plants treated with ABA, ACC, SA, and MeJA were provided by the AtGenExpress project (http://Arabidopsis.org/portals/expression/microarray/ATGenExpress.jsp). Only transcripts presenting signal log2 ratios of 1 or greater (induction) or –1 or less (repression) were considered for analysis. The hierarchical clustering analysis was then carried out using the TIGR Multiarray Experiment Viewer (version 4.4) software provided by The Institute for Genomic Research (Saeed et al., 2003). Data were normalized using the robust multiarray average method. The selected transcripts were also assigned to specific functional categories using the Classification SuperViewer tool available at the Bio-Array Resource Web site (http://bar.utoronto.ca/ntools/cgi-bin/ntools_classification_superviewer.cgi). A correlation analysis to evaluate the linear relationship between the different treatments was performed (Quinn and Keough, 2002).
The raw microarrays data from the HL-treated ACSC have been deposited in the Gene Expression Omnibus database under accession number GSE22671.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Oxygen evolution rates of ACSC after the HL treatment.
Supplemental Figure S2. PCA plot of the microarray experiments: ACSC under control (50 μE m–2 s–1), 1-h dark, and HL (1,800 μE m–2 s–1) conditions.
Supplemental Figure S3. FatiScan overrepresented biological processes in ACSC under HL stress.
Supplemental Figure S4. Validation of the microarray experiments in ACSC under HL stress by RT-PCR.
Supplemental Figure S5. Venn diagrams representing the number of up-regulated transcripts in ACSC at HL that are coregulated in the 1O2-producing mutants flu and flu/over-tAPX (A) and the (apo)carotenoid-deficient mutants aba1 and max4 (B).
Supplemental Table S1. Set of transcripts down- and up-regulated in Arabidopsis cell cultures under 30 min of HL stress (adjusted P < 0.05).
Supplemental Table S2. GO biological process terms significantly overrepresented in the list of 418 up-regulated transcripts in ACSC under HL stress.
Supplemental Table S3. Transcription factors up-regulated in ACSC under HL stress.
Supplemental Table S4. List of transcripts up- and down-regulated in ACSC under HL together with their resulting coregulated transcripts when compared with key selected Arabidopsis plants.
Supplemental Table S5. Pearson’s correlation between different treatments.
Supplemental Table S6. Transcripts and their corresponding primers designed for monitoring ROS-mediated responses in ACSC under HL stress (first four rows) and RT-PCR validation of DNA microarray experiments in ACSC.
Note Added in Proof
Although the western-blot analysis indicates that 1O2 is produced in the PSII reaction center of ACSC at HL, exogenous 1O2 production by the SOSG endoperoxide product might enhance the fluorescence emission collected at 500 to 575 nm (Gollmer et al., 2011).
Supplementary Material
Acknowledgments
We are very grateful to the Institut de Biochimie et Physiologie Moléculaire des Plantes for providing the cell culture. We also thank Dr. M. Balsera, Dr. J.B. Barroso, and Dr. F. Cabello-Hurtado for their critical comments and reading and Prof. R. Martínez-Carrasco, Dr. R. Valderrama, and Mr. J.J. Martín for their valuable technical assistance.
References
- Akiyama K, Matsuzaki K, Hayashi H. (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827 [DOI] [PubMed] [Google Scholar]
- Al-Shahrour F, Arbiza L, Dopazo H, Huerta J, Mínguez P, Montaner D, Dopazo J. (2007) From genes to functional classes in the study of biological systems. BMC Bioinformatics 8: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shahrour F, Díaz-Uriarte R, Dopazo J. (2004) FatiGO: a Web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics 20: 578–580 [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Physiol Plant Mol Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Aro EM, Virgin I, Andersson B. (1993) Photoinhibition of photosystem II: inactivation, protein damage and turnover. Biochim Biophys Acta 1143: 113–134 [DOI] [PubMed] [Google Scholar]
- Asada K. (2006) Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol 141: 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelos M, Curie C, Mazzolini L, Bardet C, Lescure B. (1992) A protocol for transient gene-expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant Physiol Biochem 30: 123–128 [Google Scholar]
- Bechtold U, Karpinski S, Mullineaux PM. (2005) The influence of the light environment and photosynthesis on oxidative signalling responses in plant-biotrophic pathogen interactions. Plant Cell Environ 28: 1046–1055 [Google Scholar]
- Bell E, Takeda S, Dolan L. (2009) Reactive oxygen species in growth and development. del Río LA, Puppo A, , Reactive Oxygen Species in Plant Signaling. Springer, Dordrecht, The Netherlands, pp 55–71 [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol 57: 289–300 [Google Scholar]
- Benjamini Y, Yekutieli D. (2001) The control of the false discovery rate in multiple testing under dependency. Ann Stat 29: 1165–1188 [Google Scholar]
- Björkman O, Demmig B. (1987) Photon yield of O2 evolution and chlorophyll fluorescence characteristics at 77 K among vascular plants of diverse origins. Planta 170: 489–504 [DOI] [PubMed] [Google Scholar]
- Bolstad B, Irizarry R, Astrand M, Speed T. (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Böttcher C, Pollmann S. (2009) Plant oxylipins: plant responses to 12-oxo-phytodienoic acid are governed by its specific structural and functional properties. FEBS J 276: 4693–4704 [DOI] [PubMed] [Google Scholar]
- Cassells AC, Curry RF. (2001) Oxidative stress and physiological, epigenetic and genetic variability in plant tissue culture: implications for micropropagators and genetic engineers. Plant Cell Tissue Organ Cult 64: 145–157 [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. (2006) The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat Protoc 1: 581–585 [DOI] [PubMed] [Google Scholar]
- Cramer GR. (2002) Response of abscisic acid mutants of Arabidopsis to salinity. Funct Plant Biol 29: 561–567 [DOI] [PubMed] [Google Scholar]
- Danon A, Miersch O, Felix G, op den Camp RGL, Apel K. (2005) Concurrent activation of cell death-regulating signaling pathways by singlet oxygen in Arabidopsis thaliana. Plant J 41: 68–80 [DOI] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R. (2005) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Michele R, Vurro E, Rigo C, Costa A, Elviri L, Di Valentin M, Careri M, Zottini M, Sanità di Toppi L, Lo Schiavo F. (2009) Nitric oxide is involved in cadmium-induced programmed cell death in Arabidopsis suspension cultures. Plant Physiol 150: 217–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SM, Diamond M, McCabe PF. (2010) Chloroplast and reactive oxygen species involvement in apoptotic-like programmed cell death in Arabidopsis suspension cultures. J Exp Bot 61: 473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Dong X. (2004) Systemic acquired resistance (2004). Annu Rev Phytopathol 42: 185–209 [DOI] [PubMed] [Google Scholar]
- Flors C, Fryer MJ, Waring J, Reeder B, Bechtold U, Mullineaux PM, Nonell S, Wilson MT, Baker NR. (2006) Imaging the production of singlet oxygen in vivo using a new fluorescent sensor, Singlet Oxygen Sensor Green. J Exp Bot 57: 1725–1734 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. (2005a) Oxidant and antioxidant signalling in plants: a re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ 28: 1056–1071 [Google Scholar]
- Foyer CH, Noctor G. (2005b) Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17: 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer MJ, Ball L, Oxborough K, Karpinski S, Mullineaux PM, Baker NR. (2003) Control of ascorbate peroxidase 2 expression by hydrogen peroxide and leaf water status during excess light stress reveals a functional organisation of Arabidopsis leaves. Plant J 33: 691–705 [DOI] [PubMed] [Google Scholar]
- Fufezan C, Rutherford AW, Krieger-Liszkay A. (2002) Singlet oxygen production in herbicide-treated photosystem II. FEBS Lett 532: 407–410 [DOI] [PubMed] [Google Scholar]
- Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inze D, Mittler R, van Breusegem F. (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141: 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez-Valdivieso G, Fryer MJ, Lawson T, Slattery K, Truman W, Smirnoff N, Asami T, Davies WJ, Jones AM, Baker NR, et al. (2009) The high light response in Arabidopsis involves ABA signaling between vascular and bundle sheath cells. Plant Cell 21: 2143–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez-Valdivieso G, Mullineaux PM. (2010) The role of reactive oxygen species in signalling from chloroplasts to the nucleus. Physiol Plant 138: 430–439 [DOI] [PubMed] [Google Scholar]
- Gao M, Wang X, Wang D, Xu F, Ding X, Zhang Z, Bi D, Cheng YT, Chen S, Li X, et al. (2009) Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6: 34–44 [DOI] [PubMed] [Google Scholar]
- Gechev TS, van Breusegem F, Stone JM, Denev I, Laloi C. (2006) Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays 28: 1091–1101 [DOI] [PubMed] [Google Scholar]
- Gollmer A, Arnbjerg J, Blaikie FH, Pedersen BW, Breitenbach T, Daasbjerg K, Glasius M, Ogilby PR. (2011) Singlet oxygen sensor green(®): photochemical behavior in solution and in a mammalian cell. Photochem Photobiol 87: 671–679 [DOI] [PubMed] [Google Scholar]
- Halliwell B. (2006) Reactive species and antioxidants: redox biology is a fundamental theme of aerobic life. Plant Physiol 141: 312–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A, Stirnberg P, Beveridge C, Leyser O. (2009) Interactions between auxin and strigolactone in shoot branching control. Plant Physiol 151: 400–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideg E, Spetea C, Vass I. (1995) Superoxide radicals are not the main promoters of acceptor-side-induced photoinhibitory damage in spinach thylakoids. Photosynth Res 46: 399–407 [DOI] [PubMed] [Google Scholar]
- Hideg E, Vass I. (1995) Singlet oxygen is not produced in photosystem I under photoinhibitory conditions. Photochem Photobiol 62: 949–952 [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendrop F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Pearce G, Ryan CA. (2006) An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci USA 103: 10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibers RP, de Jong M, Dekter RW, Van den Ackerveken G. (2009) Disease-specific expression of host genes during downy mildew infection of Arabidopsis. Mol Plant Microbe Interact 22: 1104–1115 [DOI] [PubMed] [Google Scholar]
- Humphrey AJ, Beale MH. (2006) Strigol: biogenesis and physiological activity. Phytochemistry 67: 636–640 [DOI] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Stevenson B, Zhu JK. (1997) Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis thaliana: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 9: 1935–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakab G, Manrique A, Zimmerli L, Metraux JP, Mauch-Mani B. (2003) Molecular characterization of a novel lipase-like pathogen-inducible gene family of Arabidopsis. Plant Physiol 132: 2230–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MP, Zia A, Horton P, Ruban AV. (2010) Effect of xanthophyll composition on the chlorophyll excited state lifetime in plant leaves and isolated LHCII. Chem Phys 373: 23–32 [Google Scholar]
- Johnson RA, Wichern DW. (1998) Applied Multivariate Statistical Analysis, Ed 2 Prentice Hall, Upper Saddle River, NJ [Google Scholar]
- Jouannea JP, Peaudlen C. (1967) Growth and synthesis of proteins in cell suspensions of a kinetin dependent tobacco. Physiol Plant 20: 834–850 [Google Scholar]
- Keren N, Berg A, van Kan PJM, Levanon H, Ohad I. (1997) Mechanism of photosystem II photoinactivation and D1 protein degradation at low light: the role of back electron flow. Proc Natl Acad Sci USA 94: 1579–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelwal A, Elvitigala T, Ghosh B, Quatrano RS. (2008) Arabidopsis transcriptome reveals control circuits regulating redox homeostasis and the role of an AP2 transcription factor. Plant Physiol 148: 2050–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, Meskauskiene R, Apel K, Laloi C. (2008) No single way to understand singlet oxygen signalling in plants. EMBO Rep 9: 435–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltai H, LekKala SP, Bhattacharya C, Mayzlish-Gati E, Resnick N, Wininger S, Dor E, Yoneyama K, Yoneyama K, Hershenhorn J, et al. (2010) A tomato strigolactone-impaired mutant displays aberrant shoot morphology and plant interactions. J Exp Bot 61: 1739–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Dellaert LWM, van der Veen JH. (1982) EMS- and radiation-induced mutation frequencies at individual loci in Arabidopsis thaliana (L.) Heynh. Mutat Res 93: 109–123 [DOI] [PubMed] [Google Scholar]
- Krieger-Liszkay A, Fufezan C, Trebst A. (2008) Singlet oxygen production in photosystem II and related protection mechanism. Photosynth Res 98: 551–564 [DOI] [PubMed] [Google Scholar]
- Laloi C, Stachowiak M, Pers-Kamczyc E, Warzych E, Murgia I, Apel K. (2007) Cross-talk between singlet oxygen- and hydrogen peroxide-dependent suspension of stress responses in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 672–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale J, Hall JD, Knight MR, Vierling E. (2005) Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol 138: 882–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebkuecher JG, Haldeman KA, Harris CE, Holz SL, Joudah SA, Minton DA. (1999) Development of photosystem II activity during irradiance of etiolated Helianthus (Asteraceae) seedlings. Am J Bot 86: 1087–1092 [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lokstein H, Tian L, Polle JEW, DellaPenna D. (2002) Xanthophyll biosynthetic mutants of Arabidopsis thaliana: altered nonphotochemical quenching of chlorophyll fluorescence is due to changes in photosystem II antenna size and stability. Biochim Biophys Acta 1553: 309–319 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R. (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PL, Chen NZ, An R, Su Z, Qi BS, Ren F, Chen J, Wang XC. (2007) A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol Biol 63: 289–305 [DOI] [PubMed] [Google Scholar]
- Mayzlish-Gati E, LekKala SP, Resnick N, Wininger S, Bhattacharya C, Lemcoff JH, Kapulnik Y, Koltai H. (2010) Strigolactones are positive regulators of light-harvesting genes in tomato. J Exp Bot 61: 3129–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JAH. (2003) Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol Biol 53: 423–442 [DOI] [PubMed] [Google Scholar]
- Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K. (2001) FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 98: 12826–12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33: 453–467 [DOI] [PubMed] [Google Scholar]
- Mishra NP, Francke C, van Gorkom HJ, Ghanotakis DF. (1994) Destructive role of singlet oxygen during aerobic illumination of the photosystem II core complex. Biochim Biophys Acta 1186: 81–90 [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, van Breusegem F. (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9: 490–498 [DOI] [PubMed] [Google Scholar]
- Miyao M, Ikeuchi M, Yamamoto N, Ono T. (1995) Specific degradation of the D1 protein of photosystem II by treatment with hydrogen peroxide in darkness: implications for the mechanism of degradation of the D1 protein under illumination. Biochemistry 34: 10019–10026 [DOI] [PubMed] [Google Scholar]
- Mullineaux PM, Baker NR. (2010) Oxidative stress: antagonistic signaling for acclimation or cell death? Plant Physiol 154: 521–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Ohtsuki N, Tsujimoto Y, Fujimura T, Shinshi H. (2006) Identification of genes of the plant-specific transcription-factor families cooperatively regulated by ethylene and jasmonate in Arabidopsis thaliana. J Plant Res 119: 407–413 [DOI] [PubMed] [Google Scholar]
- Niyogi KK. (1999) Photoprotection revisited: genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol 50: 333–359 [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Grossman AR, Björkman O. (1998) Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10: 1121–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenbein C, Przybyla D, Danon A, Landgraf F, Gobel C, Imboden A, Feussner I, Apel K. (2006) The role of EDS1 (enhanced disease susceptibility) during singlet oxygen-mediated stress responses of Arabidopsis. Plant J 47: 445–456 [DOI] [PubMed] [Google Scholar]
- op den Camp RG, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg E, Göbel C, Feussner I, et al. (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15: 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald O, Martin T, Dominy PJ, Graham IA. (2001) Plastid redox state and sugars: interactive regulators of nuclear-encoded photosynthetic gene expression. Proc Natl Acad Sci USA 98: 2047–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñas-Fernández A, Strand Å. (2008) Retrograde signaling and plant stress: plastid signals initiate cellular stress responses. Curr Opin Plant Biol 11: 509–513 [DOI] [PubMed] [Google Scholar]
- Pogson BJ, Niyogi KK, Bjorkman O, DellaPenna D. (1998) Altered xanthophyll compositions adversely affect chlorophyll accumulation and nonphotochemical quenching in Arabidopsis mutants. Proc Natl Acad Sci USA 95: 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Przybyla D, Gobel C, Imboden A, Hamberg M, Feussner I, Apel K. (2008) Enzymatic, but not non-enzymatic, 1O2-mediated peroxidation of polyunsaturated fatty acids forms part of the EXECUTER1-dependent stress response program in the flu mutant of Arabidopsis thaliana. Plant J 54: 236–248 [DOI] [PubMed] [Google Scholar]
- Quinn GP, Keough MJ. (2002) Experimental Design and Data Analysis for Biologists. Cambridge University Press, Cambridge, UK [Google Scholar]
- Rhee SG, Chang T-S, Jeong W, Kang D. (2010) Methods for detection and measurement of hydrogen peroxide inside and outside of cells. Mol Cells 29: 539–549 [DOI] [PubMed] [Google Scholar]
- Rinalducci S, Pedersen JZ, Zolla L. (2004) Formation of radicals from singlet oxygen produced during photoinhibition of isolated light-harvesting proteins of photosystem II. Biochim Biophys Acta 1608: 63–73 [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K. (2006) Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci USA 103: 18822–18827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. (1985) Measurement of protein using bicinchoninic acid. Anal Biochem 150: 76–85 [DOI] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3 [DOI] [PubMed] [Google Scholar]
- Sorefan K, Booker J, Haurogne K, Goussot M, Bainbridge K, Foo E, Chatfield S, Ward S, Beveridge C, Rameau C, et al. (2003) MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev 17: 1469–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Hansson O, Mathis P, Satoh K. (1987) Primary radical pair in the photosystem II reaction center. Biochim Biophys Acta 893: 49–59 [Google Scholar]
- Triantaphylidès C, Havaux M. (2009) Singlet oxygen in plants: production, detoxification and signalling. Trends Plant Sci 14: 219–228 [DOI] [PubMed] [Google Scholar]
- Triantaphylidès C, Krischke M, Hoeberichts FA, Ksas B, Gresser G, Havaux M, van Breusegem F, Mueller MJ. (2008) Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol 148: 960–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ülker B, Somssich IE. (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7: 491–498 [DOI] [PubMed] [Google Scholar]
- van Breusegem F, Bailey-Serres J, Mittler R. (2008) Unraveling the tapestry of networks involving reactive oxygen species in plants. Plant Physiol 147: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vass I, Cser K. (2009) Janus-faced charge recombinations in photosystem II photoinhibition. Trends Plant Sci 14: 200–205 [DOI] [PubMed] [Google Scholar]
- Wang GD, Long YC, Thomma BPHJ, de Wit PJGM, Angenent GC, Fiers M. (2010) Functional analyses of the CLAVATA2-like proteins and their domains that contribute to CLAVATA2 specificity. Plant Physiol 152: 320–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KLC, Li H, Ecker JR. (2002) Ethylene biosynthesis and signaling networks. Plant Cell 14: S131–S151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YR, Deng ZY, Lai JB, Zhang YY, Yang CP, Yin BJ, Zhao QZ, Zhang L, Li Y, Yang CW, et al. (2009) Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res 19: 1279–1290 [DOI] [PubMed] [Google Scholar]
- Xiong L, Gong Z, Rock C, Subramanian S, Guo Y, Xu W, Galbraith D, Zhu JK. (2001) Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev Cell 6: 771–781 [DOI] [PubMed] [Google Scholar]
- Zeier J, Pink B, Mueller MJ, Berger S. (2004) Light conditions influence specific defence responses in incompatible plant-pathogen interactions: uncoupling systemic resistance from salicylic acid and PR-1 accumulation. Planta 219: 673–683 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.