Abstract
Little is known about genes that control growth and development under low carbon (C) availability. The Arabidopsis (Arabidopsis thaliana) EXORDIUM-LIKE1 (EXL1) gene (At1g35140) was identified as a brassinosteroid-regulated gene in a previous study. We show here that the EXL1 protein is required for adaptation to C- and energy-limiting growth conditions. In-depth analysis of EXL1 transcript levels under various environmental conditions indicated that EXL1 expression is controlled by the C and energy status. Sugar starvation, extended night, and anoxia stress induced EXL1 gene expression. The C status also determined EXL1 protein levels. These results suggested that EXL1 is involved in the C-starvation response. Phenotypic changes of an exl1 loss-of-function mutant became evident only under corresponding experimental conditions. The mutant showed diminished biomass production in a short-day/low-light growth regime, impaired survival during extended night, and impaired survival of anoxia stress. Basic metabolic processes and signaling pathways are presumed to be barely impaired in exl1, because the mutant showed wild-type levels of major sugars, and transcript levels of only a few genes such as QUA-QUINE STARCH were altered. Our data suggest that EXL1 is part of a regulatory pathway that controls growth and development when C and energy supply is poor.
The coordination of carbon (C) supply and growth in varying environmental conditions is a major challenge for plants. The rate of CO2 assimilation, the accumulation of storage carbohydrates during the day, the use of assimilates for growth, and the rate of carbohydrate degradation at night represent processes that require coordinated regulation (Paul and Pellny, 2003; Smith and Stitt, 2007). The diurnal C balance is of pivotal importance for the maintenance of growth, because even transient periods of carbohydrate deficiency cause growth inhibition by temporary inhibition of carbohydrate utilization (Gibon et al., 2004). Short-term undesirable conditions such as moisture deficit and cold temperatures reduce photosynthetic C supply, and biotic damage to leaves or defoliation can interrupt photosynthetic activity.
Plants evolved mechanisms that ensure the coordination of growth with metabolism. These mechanisms imply both long-distance and tissue- or cell type-specific signaling mechanisms (Rolland et al., 2006). Glc, Fru, and Suc are the main products of photosynthesis. Sugar-dependent feedback mechanisms control photosynthesis (Paul and Pellny, 2003). Sink tissues modify local and long-distance sugar signaling by the level of sugar import and the action of Suc-cleaving enzymes (Koch, 2004). The C and energy status controls metabolic pathways by posttranslational regulation of enzymes (Huber and Hardin, 2004). In addition, sugar availability controls transcription, translation, and protein stability of transcription factors (Smeekens et al., 2010) and modulates global gene expression patterns (Ho et al., 2001; Contento et al., 2004; Lee et al., 2004; Thimm et al., 2004; Bläsing et al., 2005; Usadel et al., 2008).
Although sugar regulation is necessarily far more complex in multicellular organisms, yeast is a useful model for sugar sensing and signaling at the cellular level (Rolland et al., 2006). Three major Glc sensing and signaling pathways have been identified in yeast. Glc is the preferred C source for yeast. The metabolism of other C substrates is repressed if Glc is available. This phenomenon is referred to as “Glc repression.” It requires the activity of the glycolytic enzyme and sensor Hexokinase2 (Hxk2). In response to Glc, Hxk2 is transported into the nucleus, where it interacts with the transcription factor Multicopy inhibitor of GAL expression1 (Mig1) and generates a repressor complex. This complex is responsible for the repression of genes involved in the transport and metabolism of alternative C substrates, gluconeogenesis, and respiration (Moreno et al., 2005). The activity of Mig1 is also controlled by a key protein kinase, Sucrose nonfermenting1 (Snf1). Upon Glc depletion, Snf1 is activated and controls many processes through the phosphorylation of transcription factors and metabolic enzymes. The general effect of the Snf1 kinase in yeast is to switch the cellular metabolism from fermentation to respiration (Zhang et al., 2010). Phosphorylation of Mig1 by Snf1 is essential for the nuclear export of Mig1 and the derepression of genes that are repressed in the presence of high Glc concentrations (Ahuatzi et al., 2007). Fermentative Glc metabolism requires a high metabolic flux through glycolysis. The expression of hexose transporters (Hxts) is up-regulated through the action of the Hxt homologs Snf3 and Restores glucose transport2 (Rgt2; Rolland et al., 2006). Snf3 and Rgt2 function as sensors for extracellular Glc (Gancedo, 2008). The third Glc signaling pathway in yeast is based on Glc activation of cAMP synthesis. Activation of cAMP synthesis by the adenylate cyclase Cyr1 requires Glc phosphorylation by either Glucokinase1 (Glk1) or one of the two hexokinases (Hxk1 or Hxk2) and Glc activation of the G-protein-coupled receptor Gpr1. The cAMP-dependent protein kinases A (PKAs; Tpk1–Tpk3) control a large number of proteins, including transcriptional regulators such as Rgt1, Msn2, and Msn4. The PKAs stimulate glycolytic activity, the mobilization of reserve carbohydrates, and ribosome biogenesis. Furthermore, the cAMP-PKA pathway mediates cell growth and represses the expression of stress resistance genes (Rolland et al., 2006; Gancedo, 2008).
Experimental evidence suggests the presence of at least three systems for Glc sensing and additional systems for Suc sensing in plants. The AtHXK1/GIN2 hexokinase has Glc signaling functions that are uncoupled from metabolic activities (Jang et al., 1997; Zhou et al., 1998; Moore et al., 2003). It is mainly associated with mitochondria, possibly as part of a glycolytic metabolon, but also found in the nucleus (Yanagisawa et al., 2003). The Arabidopsis (Arabidopsis thaliana) genome encodes two further hexokinases and three hexokinase-like (HKL) proteins. The HKL1 protein lacks glucokinase activity and plays a specific role (Karve and Moore, 2009). In contrast to yeast and animals, G-protein signaling is poorly characterized in plants. AtRGS1, an unusual hybrid seven-transmembrane domain protein with a C-terminal RGS box with GTPase-accelerating activity (Chen et al., 2003), potentially acts as an extracellular receptor for Glc and mediates Glc regulation of a limited set of genes (Grigston et al., 2008). The Suc transporter-like protein SUT2 resembles the yeast Snf3 and Rgt2 proteins (Barker et al., 2000), but a role as extracellular Glc or Suc sensor has not been demonstrated. Evidence for additional sugar sensing mechanisms in plants comes from experiments with Glc and Suc analogs and mutants with defects in sugar responses.
Evolutionarily conserved protein kinases termed Snf1 in yeast, AMPK in mammals, and Snf1-Related Protein Kinase1 (SnRK1) in plants are major components of energy homeostasis maintenance in eukaryotes (Baena-González and Sheen, 2008; Zhang et al., 2010). As mentioned above, Snf1 in yeast is activated upon Glc depletion and controls many processes through the regulation of transcription factors and metabolic enzymes. SnRK1 in plants represents a master regulator during C and energy deprivation (Baena-González and Sheen, 2008; Halford and Hey, 2009; Jossier et al., 2009). SnRK1.1/KIN10-mediated gene expression is positively correlated with that regulated by C deprivation and negatively correlated with that controlled by sugar (Baena-González et al., 2007). Plant SnRK1 kinases are also involved in the posttranslational regulation of key enzymes and thus may directly regulate major biosynthetic pathways (Sugden et al., 1999). The activity of SnRK1 is inhibited by proteins such as PRL1 (Bhalerao et al., 1999) and the sugar-induced signaling molecule trehalose-6-phosphate (Zhang et al., 2009). AtSnAK1/GRIK2 and AtSnAK2/GRIK1 act as upstream kinases and activate SnRK1 (Hey et al., 2007; Shen et al., 2009). The calcineurin B-like-interacting protein kinase CIPK15 is an upstream regulator of SnRK1A in rice (Oryza sativa) and links oxygen-deficiency signals to the SnRK1-dependent sugar sensing cascade (Lu et al., 2007; Lee et al., 2009).
Several mutants impaired in phytohormone biosynthesis or signaling also show altered sugar responses. For example, abscisic acid-deficient or abscisic acid-insensitive mutants are Glc insensitive (Arenas-Huertero et al., 2000; Huijser et al., 2000; Laby et al., 2000; Cheng et al., 2002). Ethylene overproduction mutants and mutants with constitutive ethylene signaling show reduced responses to high levels of Glc or Suc, whereas ethylene-insensitive mutants are hypersensitive to sugar (Zhou et al., 1998; Cheng et al., 2002).
In this study, we characterize the Arabidopsis gene EXORDIUM-LIKE1 (EXL1). EXL1 was identified as a BR-induced gene (Schröder et al., 2009). EXL1 encodes a putative protein of 309 amino acids that is the closest homolog to the previously characterized EXO protein (67% identity and 79% similarity; Farrar et al., 2003; Coll-Garcia et al., 2004; Schröder et al., 2009). Structure prediction tools such as PSIPRED (McGuffin et al., 2000) suggest that the positions of helices and strands are highly conserved in EXO and EXL1. The major structural feature of the proteins is the so-called PHOSPHATE-INDUCED PROTEIN1 conserved region (see Pfam entry PF04674 [Finn et al., 2008]). Proteins harboring this region were identified in evolutionarily distant green plants (Schröder et al., 2009). The molecular function of the region is unknown. The EXO and EXL1 primary sequences do not show similarities to other known protein domains. In silico analyses and experimental evidence indicate an extracellular localization of the proteins. EXO, EXL1, and other members of the protein family carry an N-terminal signal peptide. Sequence analysis tools such as TargetP (Emanuelsson et al., 2000) and WoLF PSORT (Horton et al., 2007) predict that the proteins enter the secretory pathway (Schröder et al., 2009). Several proteomics approaches identified EXO, EXL1, and other members of the protein family as part of the cell wall proteome (Borderies et al., 2003; Bayer et al., 2006; Feiz et al., 2006; Jamet et al., 2006), and EXO:HA (for hemagglutinin) and EXO:GFP fusion proteins were detected in the apoplast (Schröder et al., 2009).
Here, we show that EXL1 plays an essential role under specific environmental conditions. EXL1 is largely irrelevant under favorable light conditions and nonlimiting C supply but controls growth and development under low C and energy availability. Our evidence suggests that EXL1 is part of a regulatory mechanism that serves to balance the C status with development and growth.
RESULTS
Identification of Mutants
An exl1 knockout mutant (GABI-Kat line 592A10; Rosso et al., 2003) was identified. The mutant carries a T-DNA insertion in the coding region (126 bp upstream of the stop codon). Northern-blot and reverse transcription (RT)-PCR analyses did not detect EXL1 transcript (data not shown). Introduction of a 35S::EXL1 construct into the exl1 mutant (exl1/35S::EXL1) resulted in strong EXL1 expression in the mutant and normalized the phenotypic changes (see below). Thus, the exl1 phenotype was caused by the T-DNA insertion into the EXL1 coding sequence. Furthermore, a knockon mutant termed exl1-D (SALK line 027172; Alonso et al., 2003) was identified. The exl1-D mutant carries a T-DNA insertion at position −467 upstream of the translation start codon and showed a several-fold increase in EXL1 transcript levels in comparison with the wild type.
EXL1 Expression Is Induced by C Starvation
Expression of EXL1 and EXO is induced by BR (Schröder et al., 2009), and the encoded proteins are highly similar. Thus, we assumed that EXL1 and EXO play similar roles in the control of vegetative growth and tested biomass production and development of the exl1 mutant in a greenhouse. In contrast to EXO, the loss of EXL1 was largely irrelevant for growth and development under standard conditions.
Therefore, we used Web-based platforms such as Genevestigator (Hruz et al., 2008) and the Arabidopsis eFP Browser (Winter et al., 2007) to identify experimental conditions that have a strong impact on EXL1 expression. The data indicated that EXL1 expression is controlled by the C status. Relevant Affymetrix ATH1 microarray profiles were downloaded from the Gene Expression Omnibus (Edgar et al., 2002) and analyzed using the ROBIN software that is based on functions of the R/BioConductor project (Lohse et al., 2010). In addition, plants were grown in synthetic medium supplemented with different sugar levels or in soil under different light regimes. EXL1 transcript and protein levels were analyzed by means of quantitative RT-PCR and western-blot analysis, respectively.
The expression of EXL1 showed the following profile. First, EXL1 transcript levels decreased slightly during the light period and increased slightly during the night in wild-type plants (Fig. 1). This diurnal cycling was intensified in the pgm mutant (Fig. 1A, top right panel). The pgm mutant does not form transitory starch (Caspar et al., 1985) and is unable to maintain appropriate carbohydrate and energy levels during the night. Consequently, pgm plants show poor growth and symptoms of C starvation in the dark period (Gibon et al., 2006). Second, wild-type plants showed strong EXL1 expression in extended night (Fig. 1, A, bottom left panel, and C). Third, EXL1 mRNA levels decreased in wild-type seedlings grown in liquid culture upon sugar supply (Fig. 1A, bottom right panel). Fourth, wild-type plants grown in an atmosphere with 50 μL L−1 CO2 showed higher EXL1 expression than plants grown in 350 μL L−1 CO2 (Fig. 1A, bottom right panel). Fifth, addition of increasing levels of Suc to synthetic growth medium suppressed EXL1 expression (data not shown). Sixth, overexpression of SnRK1.1 (also known as KIN10 or AKIN10) resulted in elevated EXL1 mRNA levels (Baena-González et al., 2007). In summary, low C availability and SnRK1.1/KIN10 action result in elevated EXL1 transcript levels.
Figure 1.
EXL1 expression in response to C availability. A, Expression profiles from publicly available microarray data (Bläsing et al., 2005; Gibon et al., 2006; Osuna et al., 2007; Usadel et al., 2008). The data were normalized using RMA and log2 transformed. The means of two or three replicates are shown. The se is shown if three replicates were available. Expression of EXO is shown for comparison. FN, Full nutrition; C-starv, C starvation. B, Quantitative RT-PCR analysis of EXL1 transcript levels in wild-type (Col-0) shoots. Plants were grown in soil under the low-total-irradiance regime (4 h of light [60 μmol m−2 s−1]/20 h of dark). RNA was extracted from shoots of 28-d-old plants. eIF1α cycle threshold values were subtracted from respective cycle threshold values of the gene of interest. Subsequently, differences were subtracted from an arbitrary value (i.e. 40). Higher numbers indicate higher transcript levels. A difference of 1 unit indicates a 2-fold change. Error bars indicate se of the gene of interest in three technical replicates. The data shown are from one experiment representative of three independent biological replicates. eN, End of night; eD, end of day. C, Quantitative RT-PCR analysis of EXL1 transcript levels in wild-type (Col-0) shoots. Plants were grown in soil under short-day conditions (8 h of light [140 μmol m−2 s−1]/16 h of dark). RNA was extracted from 35-d-old plants. Technical details are as given for B. [See online article for color version of this figure.]
In addition to mRNA synthesis and stability, translation efficiency and protein stability could determine the EXL1 protein level. EXL1:HA transcript and protein levels were assessed in plants carrying the pEXL1::EXL1:HA and 35S::EXL1:HA constructs. The pEXL1::EXL1:HA construct conferred expression of an HA-tagged EXL1 protein driven by a genomic EXL1 fragment that is located upstream of the EXL1 translation start codon (positions −1 to −928). The sequence of several full-length cDNAs and promoter analysis tools such as PPDB (Yamamoto and Obokata, 2008) indicate a length for the 5′ untranslated region of EXL1 of about 92 bp and a putative TATA box at positions −110 to −121. Thus, the cloned promoter sequence in the pEXL::EXL1:HA construct spans about 830 bp. The 35S::EXL1:HA construct resulted in the expression of an identical EXL1:HA protein under control of the cauliflower mosaic virus 35S promoter.
High levels of EXL1:HA transcript were detected in shoots of the pEXL1::EXL1:HA transgenic lines when plants were grown in the presence of 0.2% (w/v) Suc. Accordingly, decreasing transcript levels were detected when plants were grown in the presence of 0.5%, 1%, 3%, or 5% Suc (Fig. 2A). EXL1:HA expression was less repressed by sugar in roots (Fig. 2B). EXL1:HA expression driven by the EXL1 promoter resulted in corresponding EXL1:HA protein levels (Fig. 3). The 35S promoter caused strong EXL1:HA expression under all conditions (Fig. 2) and largely prevented the modulation of EXL1:HA transcript (Fig. 2) and EXL1:HA protein levels (data not shown) by the sugar supply and light regime. These data indicate that transcriptional regulation of EXL1 confers an adjustment of EXL1 protein levels to the C status.
Figure 2.
EXL1:HA expression in response to different Suc levels and light regimes. A, Relative EXL1:HA transcript levels in shoots. Plants carrying the pEXL1::EXL:HA and 35S::EXL1:HA constructs were grown in synthetic medium supplemented with 0.2%, 0.5%, 1%, 3%, and 5% (w/v) Suc. RNA was extracted from 14-d-old plants. Technical details are as given for Figure 1B. The data shown are from one experiment representative of three independent biological replicates. B, Relative EXL1:HA transcript levels in roots. RNA was extracted from the same plants as in A. C, Relative EXL1:HA transcript levels in shoots. Plants were grown in soil under light-limited conditions (4 h of light [60 μmol m−2 s−1]/20 h of dark) and subjected to extended night. RNA was extracted from 35-d-old plants. eD, End of day; eN, end of night. D, Relative EXL1:HA transcript levels in shoots. Plants were grown in soil under long-day conditions (16 h of light/8 h of dark) and subjected to extended night. RNA was extracted from 28-d-old plants.
Figure 3.
Western-blot analysis of EXL1:HA protein levels. Plants carrying the pEXL1::EXL:HA construct were grown under different conditions. The EXL1:HA fusion protein was detected using a monoclonal anti-HA antibody and an enhanced chemiluminescence detection system. Similar results were obtained for independent transgenic lines. A, Plants were grown in synthetic medium supplemented with 0.2%, 0.5%, 1%, 3%, and 5% (w/v) Suc. Protein was extracted from the same shoot material as used for transcript analysis (Fig. 2A). B, Plants were grown in long-day conditions as described in the legend of Figure 2D. eD, End of day; eN, end of night. C, Plants were grown under light-limited conditions as described in the legend of Figure 2C.
EXL1 Controls Growth and Development during Low C Availability
Plants were grown in half-concentrated Murashige and Skoog (MS) medium supplemented with different sugar quantities. Sugar responses were tested both under standard growth conditions (16 h of light [145 μmol m−2 s−1]/8 h of dark) and in continuous darkness.
The exl1 mutant developed like the wild type when grown in the presence of 1% (w/v) Suc in a diurnal cycle. Biomass production and leaf size were similar. Next, plants were grown under standard conditions in the presence of high sugar levels (3%–6% [w/v] Suc). High sugar levels similarly impaired the development of the mutant and the wild type. No differences between the genotypes were observed. In contrast, loss of EXL1 caused slightly reduced leaf growth and a significant 27% reduction in biomass in the presence of 0.2% Suc (controls in Figs. 4 and 5).
Figure 4.
Biomass production in response to Suc and BR. Plants were grown in synthetic medium supplemented with 0.2% or 1% (w/v) Suc and brassinosteroids (50 nm castasterone [CS] or 50 nm brassinolide [BL]). Fresh weight was determined after 16 d. Results are means ± se (n = 5 plates per genotype and treatment containing 10 plants each). Biomass of exl1 plants was significantly reduced in the control treatment with 0.2% Suc (denoted with an asterisk; t test, P < 0.01). Percentage differences in BR treatments with 0.2% Suc supply were significantly larger in exl1 in comparison with the wild type (t test, P < 0.05).
Figure 5.
Leaf growth in response to Suc and BR. Plants were grown in synthetic medium supplemented with 0.2% (w/v) Suc and brassinosteroids (50 nm brassinolide [BL] or 50 nm 24-epibrassinolide [EBL]). Length and width of primary leaves were determined after 16 d. Results are means ± se (n = 5 plates per genotype and treatment containing 10 plants each). Mutant values denoted with an asterisk are significantly different from those of their wild type (t test, P < 0.05). Percentage differences of leaf length in BR treatments were significantly larger in exl1 in comparison with the wild type (t test, P < 0.05).
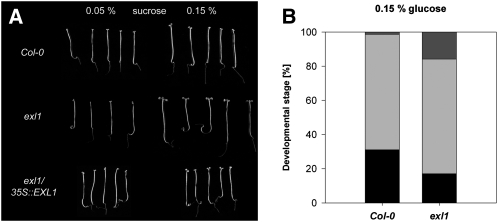
Arabidopsis seedlings can develop leaf- and flower-like organs in the dark, but skotomorphogenesis critically depends on the sugar availability (Roldán et al., 1999; Baier et al., 2004; Li et al., 2007). Similar to photomorphogenic development, skotomorphogenic development of exl1 was identical with the wild type when grown in medium supplemented with 1% Suc or 1% Glc. Nearly all plants developed true leaves after 3 weeks. Sporadically, plants developed another pair of true leaves (data not shown). There were also no clear differences between the genotypes in the presence of 0.05% Suc (Fig. 6A), presumably because very low sugar levels prohibited growth. Supply of 0.15% Suc or Glc, however, caused accelerated development of the exl1 mutant. Wild-type plants typically developed elongated hypocotyls and opened cotyledons but hardly produced true leaves. In contrast, approximately 15% of the exl1 plants developed an internodium and true leaves (Fig. 6). Introduction of the 35S::EXL1 construct into the exl1 mutant suppressed true leaf formation (Fig. 6A). Thus, the loss of EXL1 resulted in enhanced dark development in the presence of low sugar levels.
Figure 6.
Dark development phenotypes. A, Seedlings were grown in half-concentrated MS medium supplemented with 0.05% or 0.15% (w/v) Suc for 3 weeks in the dark. Only the largest seedlings of a representative experiment are shown. B, Seedlings were grown in half-concentrated MS medium supplemented with 0.15% (w/v) Glc for 3 weeks in the dark. Development of plants was grouped into three categories: elongated hypocotyls with closed cotyledons (black bars), opened cotyledons (gray bars), and formation of true leaves (dark gray bars). The experiment was carried out independently three times. The observed responses from one representative experiment are shown.
EXL1 Limits BR-Promoted Growth during Low C Availability
BR treatments were used to enforce growth. Growth of wild-type and exl1 plants was tested in half-concentrated MS medium supplemented with 0.2% or 1% (w/v) Suc and 0 nm BR (control), 50 nm castasterone, or 50 nm brassinolide. Exogenous BR stimulated growth at both Suc levels, but the relative BR effect on biomass production was more pronounced at 0.2% Suc. Application of 50 nm castasterone or 50 nm brassinolide resulted in a 1.7- to 2-fold increase of biomass in wild-type plants and a 2.8- to 3.3-fold increase of biomass in exl1, respectively (Fig. 4). In contrast, the biomass of exl1 was similar to that of the wild type at 1% Suc (Fig. 4).
Several studies have shown that BR promotes leaf growth. For example, enhancement of BR responses by means of DWF4, BRI1, or BAK1 overexpression or application of synthetic BR resulted in leaves that are characterized by longer petioles and an increased leaf index (Choe et al., 2001; Wang et al., 2001; Nam and Li, 2002). To investigate the influence of EXL1 on leaf growth, plants were grown in synthetic medium supplemented with 0.2% Suc and 0 nm BR (control), 50 nm 24-epibrassinolide, or 50 nm brassinolide. Leaf length and leaf width were determined after 16 d. In the absence of exogenous BR, leaf size of exl1 plants was slightly smaller in comparison with the wild type, but leaf indices did not differ significantly (Fig. 5). Application of 50 nm 24-epibrassinolide or 50 nm brassinolide resulted in a significant increase of the leaf index in all genotypes. The BR-promoted growth response was more pronounced in plants with impaired EXL1 expression. For example, the presence of 50 nm brassinolide caused a 1.7-fold increase of leaf length in the wild type and a 2.4-fold increase in exl1 (Fig. 5).
EXL1 Is Required for Growth under Light-Limited Conditions
In order to examine the role of EXL1 in the adaptation to low C under physiological conditions, plants were grown in soil under short-day conditions with low light (4 h of light [60 μmol m−2 s−1]/20 h of darkness). The low photosynthetically active radiation and long dark period resulted in a low total irradiance that limited growth. Several independent experiments revealed that the exl1 mutant showed less growth in comparison with the wild type (Fig. 7A). Fresh weight and dry weight of exl1 were reduced by approximately 50% (Fig. 7B). Thus, EXL1 is necessary for growth under light-limited conditions. The exl1-D mutant did not show improved growth and biomass production, suggesting that wild-type EXL1 expression is sufficient under these conditions.
Figure 7.
Phenotype of the exl1 mutant under growth-limiting light conditions. A, Wild-type, exl1, exl1-D, and complemented exl1 plants were raised for 2 weeks in standard conditions and then grown for 2 weeks under light-limited conditions (4 h of light [60 μmol s−1 m−2]/20 h of dark). B, Fresh weight (FW) and dry weight (DW) of plants shown in A. Results are means ± se (n = 6 individuals per genotype). Mutant values denoted with an asterisk are significantly different from those of their wild type (t test, P < 0.01). [See online article for color version of this figure.]
Reduced growth of exl1 could be due to defects in the primary metabolism of major carbohydrates. Altered levels of starch, Glc, Fru, or Suc are indicative of such defects. Starch levels of wild-type, exl1-D, and exl1 plants were measured based on the enzymatic hydrolysis and photometric determination of Glc. Plant material of three independent short-day/low-light experiments was analyzed. No significant differences were detected between the genotypes. Differences between independent experiments actually exceeded differences between the genotypes (wild-type, 2.5–3.8 μmol hexose equivalents mg−1 chlorophyll [means, end of night] and 8.9–12.9 μmol hexose equivalents mg−1 chlorophyll [means, end of day]; exl1-D, 2.6–3.7 and 8.8–10.5 μmol mg−1 chlorophyll; exl1, 2.2–3.5 and 11.5–11.6 μmol mg−1 chlorophyll; similar results were obtained when starch levels were related to dry weight). Similarly, Glc, Fru, and Suc levels did not differ significantly (data not shown). This suggests that major metabolic pathways are functional in exl1, although it cannot be excluded that the plasticity of metabolic networks compensates for perturbations.
Wild-type and exl1 plants were also analyzed by means of gene expression profiling. Eight profiles were established using Affymetrix ATH1 microarrays. First, wild-type and exl1 plants were grown in an 8-h-day (140 μmol m−2 s−1)/16-h-night regime. These conditions allowed largely normal growth of exl1. Second, wild-type and exl1 plants were grown under the low-total-irradiance regime mentioned above (4 h of light [60 μmol m−2 s−1]/20 h of darkness). These conditions caused significantly less growth of exl1 in comparison with the wild-type. For each light regime and genotype, plant material was harvested at the beginning and the end of the light period.
The activity of most genes was virtually identical under the 8-h-light/16-h-dark growth regime. Only nine and 14 genes showed at least a 2-fold change in the comparison of exl1 versus the wild type at the end of the night and the end of the day, respectively (Supplemental File S1). A single gene, QUA-QUINE STARCH (QQS; At3g30720), showed consistent up-regulation in exl1 (7.3- and 5.9-fold change; Supplemental File S1). The wild-type and exl1 expression profiles were more different under the low-total-irradiance regime. Twenty-six and 40 genes showed at least a 2-fold change in exl1 compared with the wild type at the end of the night and the end of the day, respectively. The QQS gene again showed stronger expression in exl1 (2.8- and 3.4-fold change; Supplemental File S1). QQS transcript levels were studied in more detail by quantitative RT-PCR under light-limited conditions (4 h of low light; Fig. 8A), in a short-day diurnal cycle (8 h of light/16 h of dark), and in extended night (more than 16 h of dark; Fig. 8B). The diurnal regulation and induction in extended night of QQS in wild-type plants were unaltered in exl1, but QQS transcript levels were higher at all times in exl1 compared with the wild type. QQS encodes a protein of 59 amino acids. The precise function is unknown, but it was suggested to play a role in the control of starch metabolism (Li et al., 2009).
Figure 8.
QQS transcript levels in wild-type and exl1 plants. A, Wild-type and exl1 plants were grown in soil under the low-total-irradiance regime (4 h of light [60 μmol m−2 s−1]/20 h of dark). For the wild type, the same cDNAs were used as for EXL1 analysis (Fig. 1B). eN, End of night; eD, end of day. B, QQS expression in a diurnal cycle (end of night to 16 h of dark) and in extended night in wild-type and exl1 plants. eN corresponds to 16 h of dark. The same wild-type cDNAs were used as for EXL1 analysis (Fig. 1C).
Transcript levels of sugar-responsive genes (Price et al., 2004; Osuna et al., 2007), of genes that show diurnal changes (Schaffer et al., 2001; Bläsing et al., 2005; Michael et al., 2008), and of phytohormone-responsive genes (Goda et al., 2008) were barely changed in exl1 in comparison with the wild type. The marginal changes in the expression profiles indicate that exl1 does not suffer from a severe disturbance of sugar responses, from incorrect phasing of transcription under circadian conditions, or from defects in the phytohormone signaling network.
EXL1 Warrants the Survival of Extended Night Periods and Confers Anoxia Tolerance
To examine the role of EXL1 in prolonged darkness, wild-type and exl1 plants were grown in soil for 3 weeks in a controlled-growth chamber. Subsequently, the light was switched off. Survival rates were determined by transferring 15 plants per genotype and point in time into an illuminated greenhouse after different periods of extended night. Five independent experiments were performed. Survival rates of the exl1 mutant were significantly lower in comparison with the wild type after 11 to 15 d in darkness (Fig. 9A).
Figure 9.
Survival rates after extended night and anoxic stress. A, Plants were raised under short-day conditions (8 h of light/16 h of dark) and subjected to different periods of continuous darkness. After retransfer to a greenhouse with long-day conditions, plants were grown for 1 week prior to determining survival rates. Five independent experiments were analyzed. Results are mean survival rates ± se (n = 15 plants per genotype, time, and experiment). Mutant values denoted with an asterisk are significantly different from those of their wild type (t test, P < 0.01). B, Seven-day-old plants were subjected to anoxia for 8 and 10 h. After the treatment, plants were transferred to ambient air, and survival rates were determined after a 10-d recovery period. Results are mean survival rates ± se (n = 4 plates containing at least 50 seedlings each). Mutant values denoted with an asterisk are significantly different from those of their wild type (t test, P < 0.01).
Several experiments in the Genevestigator Stimulus database (Hruz et al., 2008) indicated that EXL1 transcript levels were elevated under oxygen deficiency in comparison with normoxia. The most obvious effect of an oxygen shortage is an energy deficit, and genes that are under the control of the oxygen level may mediate the adaptation to an altered energy status. Anoxia tolerance of wild-type, exl1, and exl1-D plants was tested as described by Licausi et al. (2010). Plants were established in half-concentrated MS medium supplemented with 1% (w/v) Suc and subjected to temporary anoxia stress. Survival rates were negatively correlated with the length of the anoxic treatment and positively correlated with EXL1 expression. Short anoxic treatments (6 h) allowed the survival of all plants (data not shown). However, 8- and 10-h anoxic treatments revealed differences between the genotypes. The exl1 mutant showed significantly reduced survival rates, and the exl1-D mutant showed significantly higher survival rates in comparison with the wild type (Fig. 9B). The anoxia tolerance of exl1-D seedlings was not due to stronger growth (Supplemental File S2). Neither genotype survived longer anoxic treatments (more than 11 h).
DISCUSSION
EXL1 Transcript Accumulates during Low C and Low Energy Availability
Several lines of evidence indicate that EXL1 expression is induced under low C and low energy availability (Figs. 1 and 2). For example, extended night, sugar starvation in synthetic medium, and diminished oxygen supply promoted EXL1 expression. Diurnal changes of EXL1 expression were strongly pronounced in the starch-deficient pgm mutant (Fig. 1A), and EXL1 expression was under the control of SnRK1.1/KIN10 (Baena-González et al., 2007). The increase of the EXL1 mRNA level was associated with an increase in the protein level (Fig. 3).
EXL1 Adapts Growth and Development to Low C Supply
Plants are able to avoid C deficiency under fluctuating environmental conditions because growth and development are synchronized with the supply and utilization of C (Gibon et al., 2004; Hummel et al., 2010). C signaling may ultimately determine the activity of biosynthetic and catabolic pathways (Stitt et al., 2007).
Sugar responses were assessed by plant cultivation on medium supplemented with different levels of sugar. Growth and development of exl1 under a diurnal light regime and in constant darkness were similar to the wild type in the presence, for example, of 1% (w/v) Suc. Phenotypic changes only became apparent when C supply was suboptimal. The mutant produced less biomass and slightly smaller leaves under a diurnal light regime in medium supplemented with 0.2% Suc (Figs. 4 and 5). Skotomorphogenesis is a sensitive indicator of the effects of sugar on development, because C availability determines activity of the shoot apical meristem (Roldán et al., 1999; Baier et al., 2004). Dark development of exl1 was accelerated in medium supplemented with 0.15% Glc or Suc (Fig. 6). The molecular basis of this result is unknown and could be manifold. Altered sugar transport, enhanced sugar responses, and cell wall changes were demonstrated to accelerate leaf and internodium formation in darkness (Baier et al., 2004; Li et al., 2007).
The conditional phenotype suggests that EXL1 controls growth and development only during low C availability. This hypothesis was tested further by the application of synthetic BR. The primary function of BR is growth promotion of nearly all plant organs (Müssig, 2005). BR-promoted growth depends on the C supply (Fig. 4; data not shown). The exl1 mutant showed a wild-type response to Suc and BR in the presence of 1% Suc. In contrast, BR-dependent biomass production and leaf growth were much stronger in exl1 at 0.2% Suc (Figs. 4 and 5). The EXL1 protein could limit BR-dependent growth under low C availability. Alternatively, enhanced growth of the exl1 mutant could be due to higher sensitivity to BR in low-C conditions. The molecular processes that link sugar responses and BR signaling have not been resolved so far. BR presumably coordinates and integrates diverse processes required for growth, partly via interactions with other phytohormones and signaling pathways. BR has been implicated in the control of C metabolism and C allocation. For example, BR modifies enzyme activities (e.g. acid invertases, Rubisco, and cytosolic β-amylase) and controls sink strength and source efficiency (Goetz et al., 2000; Schlüter et al., 2002; Lisso et al., 2006; Wu et al., 2008).
EXL1 Is Essential for Growth during Low Total Irradiance, Survival of Extended Night, and Survival of Anoxia Stress
The physiological role of EXL1 was tested in three additional experimental setups that are characterized by low C and low energy availability.
First, plants were grown in soil under short-day conditions with low light (4 h of light [60 μmol m−2 s−1]/20 h of dark). Plants can adapt to limited light, although this adaptation requires comprehensive reprogramming of the central metabolism and decrease of the growth rate (Gibon et al., 2004, 2009). Growth of the exl1 mutant was clearly impaired under these conditions in comparison with the wild type, resulting in approximately 50% less biomass production (Fig. 7). Therefore, the adaptation to light-limited conditions depends on EXL1. In contrast, growth was largely normal under short-day (8 h of light [140 μmol m−2 s−1]/16 h of dark), long-day (16 h of light [180 μmol m−2 s−1]/8 h of dark), and high-light (16 h of light [700 μmol m−2 s−1]/8 h of dark) conditions (data not shown).
Second, plants were subjected to extended night. Plants switch each day between high C availability in the light and a negative C balance at night. Starch turnover is regulated in wild-type plants, and only a small amount of starch is left at the end of the night (Smith et al., 2004; Usadel et al., 2008). Abrupt periods of extended night cause massive changes in global gene expression and of metabolite levels and eventually provoke an acute C limitation (Smith and Stitt, 2007). The exl1 mutant is impaired in the adaptation to prolonged darkness, because survival rates were significantly smaller in comparison with the wild type (Fig. 9A).
Third, plants were subjected to anoxia stress. Plant responses to too little oxygen serve to reduce oxygen consumption in order to maintain a minimal internal oxygen concentration that is required for oxidative phosphorylation (i.e. activity of cytochrome oxidase) and sustainment of several metabolic pathways that require molecular oxygen (Geigenberger, 2003). Decreasing internal oxygen is sensed in plants and leads to an inhibition of respiration, a decrease of the ATP/ADP ratio, and the prioritization of metabolic pathways that conserve energy. Several components that control anaerobic responses have been identified (Licausi et al., 2010; Mustroph et al., 2010). EXL1 represents another factor that is essential for survival of anoxia (Fig. 9B).
CONCLUSION
The phenotypic changes of exl1 in soil and in synthetic medium only became evident when C and energy supply was suboptimal. Low sugar supply in synthetic medium, limited light and elongated dark periods in soil, and oxygen deficiency require comprehensive adaptations of metabolism and growth. EXL1 is essential for these adaptations. The mode of action of EXL1 is as yet unknown. The presumed extracellular localization of EXL1 suggests a specific role that is distinct from known sugar signaling pathways.
MATERIALS AND METHODS
Screen for Mutants and Establishment of Transgenic Lines
The SALK 027172 line (Alonso et al., 2003) carries a T-DNA insertion in the EXL1 promoter and was named exl1-D. The GABI-Kat line GK 592A10 (Rosso et al., 2003) carries a T-DNA insertion in the EXL1 coding sequence and was named exl1. DNA insertion sites were confirmed by sequencing. Homozygosity of both T-DNA insertion lines was confirmed by PCR on genomic DNA using T-DNA border-specific and gene-specific primers.
Several constructs were established using Gateway-compatible vectors. The EXL1 coding sequence was amplified using the primers EXL1ga_fw (5′-CACCATGGCTTCTTTTGTGATGGG-3′) and EXL1ga_ rev (5′-AAACAGAGTCGAGCAAGAATCTG-3′). The PCR fragments were cloned into the pENTR/D-TOPO (Invitrogen) entry vector. Sequencing confirmed full sequence identity of the PCR product with the ecotype Columbia (Col-0) genomic sequence. The EXL1 coding sequence was inserted into the pH7WG2 vector for expression under control of the 35S promoter (Karimi et al., 2002) and into the pGWB14 vector (Nakagawa et al., 2007) for fusion of three copies of the HA epitope to the C terminus of EXL1. The resulting constructs were termed 35S::EXL1 and 35S::EXL1:HA, respectively. A genomic EXL1 fragment comprising 928 bp upstream of the translation start codon and the coding sequence was amplified using the primers pEXL1ga_fw (5′-CACCTTGATTCGGTTTTTCGGATT-3′) and EXL1ga_rev. Cloning into the pGWB13 vector (Nakagawa et al., 2007) resulted in the pEXL1::EXL1:HA construct. All constructs were transformed into Arabidopsis (Arabidopsis thaliana) plants using the floral dip method.
Growth Conditions
Seeds for growth experiments were derived from plants grown in parallel in a greenhouse. Plants were grown in half-concentrated MS medium supplemented with different Glc or Suc concentrations and solidified with 0.8% (w/v) agar. After 2 to 3 d in a cold room (4°C), plants were transferred into a growth chamber with a long-day light regime (16 h of light [140 μmol m−2 s−1, 22°C]/8 h of dark [22°C]) and grown in a randomized manner. Alternatively, plants were established in soil. Seeds were allowed to germinate and grow for 2 weeks in controlled-growth chambers (7 d of 16 h of light [140 μmol m−2 s−1, 20°C, 75% relative humidity]/8 h of dark [6°C, 75% relative humidity]; thereafter, 7 d of 8 h of light [140 μmol m−2 s−1, 20°C, 60% relative humidity]/16 h of dark [16°C, 75% relative humidity]). Subsequently, plants were either left in short-day conditions or transferred to long-day conditions (Schröder et al., 2009), or plants were grown in a controlled-growth chamber under low total irradiance (4 h of light [60 μmol m−2 s−1, 20°C, 60% relative humidity]/20 h of dark [16°C, 75% relative humidity]). All genotypes were grown in the same chamber at the same time in a randomized manner. Anaerobic responses were tested as described by Licausi et al. (2010).
Gene Expression Analysis and Protein Extraction
Gene expression analyses were performed as described (Schröder et al., 2009). Primer sequences for quantitative RT-PCR were as follows: EXL1_fw (5′-TGGATCGGCTTGTACTGGAG-3′) and EXL1_rev (5′-GGGTCAAACAAAGCCGGTAA-3′; E = 1.982 ± 0.003 [mean ± se]); EXL1:HA_fw (identical to EXL1_fw) and EXL1:HA_rev (5′-TCCTGCATAGTCCGGGACGTCA-3′; E = 1.983 ± 0.005); QQS_fw (5′-AGCCATTGAAGAAACCTCCTTTCG-3′) and QQS_rev (5′-ATGGCTGACCGTGTGAGTCTTG-3′; E = 1.977 ± 0.002); and eIF1α_fw (5′-TTGACAGGCGTTCTGGTAAGG-3′) and eIF1α_rev (5′-CAGCGTCACCATTCTTCAAAAA-3′; At5g60390). Average PCR efficiencies (E) were computed for each primer pair across all analyzed samples with the LinRegPCR software (Ramakers et al., 2003). Protein for western-blot analysis was isolated using ice-cold extraction buffer (125 mm Tris [pH 8.8], 1% SDS, 10% glycerol, 50 mm Na2SO4, and Complete Protease Inhibitor Cocktail [Roche]).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AY084831.1 (EXL1) and EU805808.1 (QQS).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental File S1. Affymetrix_ATH1_profiles_SUPPLEMENT.xls.
Supplemental File S2. Biomass_exl1-D_SUPPLEMENT.xls.
Supplementary Material
Acknowledgments
We thank Justyna Sikocinska for implementation of sterile culture experiments and analyses of sugar levels. We thank Francesco Licausi and Joost van Dongen for assistance with the anoxia treatments.
References
- Ahuatzi D, Riera A, Peláez R, Herrero P, Moreno F. (2007) Hxk2 regulates the phosphorylation state of Mig1 and therefore its nucleocytoplasmic distribution. J Biol Chem 282: 4485–4493 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, León P. (2000) Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev 14: 2085–2096 [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J. (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Sheen J. (2008) Convergent energy and stress signaling. Trends Plant Sci 13: 474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier M, Hemmann G, Holman R, Corke F, Card R, Smith C, Rook F, Bevan MW. (2004) Characterization of mutants in Arabidopsis showing increased sugar-specific gene expression, growth, and developmental responses. Plant Physiol 134: 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker L, Kühn C, Weise A, Schulz A, Gebhardt C, Hirner B, Hellmann H, Schulze W, Ward JM, Frommer WB. (2000) SUT2, a putative sucrose sensor in sieve elements. Plant Cell 12: 1153–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer EM, Bottrill AR, Walshaw J, Vigouroux M, Naldrett MJ, Thomas CL, Maule AJ. (2006) Arabidopsis cell wall proteome defined using multidimensional protein identification technology. Proteomics 6: 301–311 [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Salchert K, Bakó L, Okrész L, Szabados L, Muranaka T, Machida Y, Schell J, Koncz C. (1999) Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc Natl Acad Sci USA 96: 5322–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bläsing OE, Gibon Y, Günther M, Höhne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible W-R, Stitt M. (2005) Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17: 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borderies G, Jamet E, Lafitte C, Rossignol M, Jauneau A, Boudart G, Monsarrat B, Esquerré-Tugayé M-T, Boudet A, Pont-Lezica R. (2003) Proteomics of loosely bound cell wall proteins of Arabidopsis thaliana cell suspension cultures: a critical analysis. Electrophoresis 24: 3421–3432 [DOI] [PubMed] [Google Scholar]
- Caspar T, Huber SC, Somerville C. (1985) Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol 79: 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-G, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP. (2003) A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 301: 1728–1731 [DOI] [PubMed] [Google Scholar]
- Cheng W-H, Endo A, Zhou L, Penney J, Chen H-C, Arroyo A, Leon P, Nambara E, Asami T, Seo M, et al. (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14: 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann KA. (2001) Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J 26: 573–582 [DOI] [PubMed] [Google Scholar]
- Coll-Garcia D, Mazuch J, Altmann T, Müssig C. (2004) EXORDIUM regulates brassinosteroid-responsive genes. FEBS Lett 563: 82–86 [DOI] [PubMed] [Google Scholar]
- Contento AL, Kim S-J, Bassham DC. (2004) Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol 135: 2330–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Farrar K, Evans IM, Topping JF, Souter MA, Nielsen JE, Lindsey K. (2003) EXORDIUM: a gene expressed in proliferating cells and with a role in meristem function, identified by promoter trapping in Arabidopsis. Plant J 33: 61–73 [DOI] [PubMed] [Google Scholar]
- Feiz L, Irshad M, Pont-Lezica RF, Canut H, Jamet E. (2006) Evaluation of cell wall preparations for proteomics: a new procedure for purifying cell walls from Arabidopsis hypocotyls. Plant Methods 2: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz H-R, Ceric G, Forslund K, Eddy SR, Sonnhammer ELL, et al. (2008) The Pfam protein families database. Nucleic Acids Res (Database issue) 36: D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo JM. (2008) The early steps of glucose signalling in yeast. FEMS Microbiol Rev 32: 673–704 [DOI] [PubMed] [Google Scholar]
- Geigenberger P. (2003) Response of plant metabolism to too little oxygen. Curr Opin Plant Biol 6: 247–256 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Bläsing OE, Palacios-Rojas N, Pankovic D, Hendriks JHM, Fisahn J, Höhne M, Günther M, Stitt M. (2004) Adjustment of diurnal starch turnover to short days: depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post-translational activation of ADP-glucose pyrophosphorylase in the following light period. Plant J 39: 847–862 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Pyl E-T, Sulpice R, Lunn JE, Höhne M, Günther M, Stitt M. (2009) Adjustment of growth, starch turnover, protein content and central metabolism to a decrease of the carbon supply when Arabidopsis is grown in very short photoperiods. Plant Cell Environ 32: 859–874 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Usadel B, Blaesing OE, Kamlage B, Hoehne M, Trethewey R, Stitt M. (2006) Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol 7: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K, Li W, Ogawa M, Yamauchi Y, Preston J, Aoki K, et al. (2008) The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J 55: 526–542 [DOI] [PubMed] [Google Scholar]
- Goetz M, Godt DE, Roitsch T. (2000) Tissue-specific induction of the mRNA for an extracellular invertase isoenzyme of tomato by brassinosteroids suggests a role for steroid hormones in assimilate partitioning. Plant J 22: 515–522 [DOI] [PubMed] [Google Scholar]
- Grigston JC, Osuna D, Scheible W-R, Liu C, Stitt M, Jones AM. (2008) D-Glucose sensing by a plasma membrane regulator of G signaling protein, AtRGS1. FEBS Lett 582: 3577–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford NG, Hey SJ. (2009) Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem J 419: 247–259 [DOI] [PubMed] [Google Scholar]
- Hey S, Mayerhofer H, Halford NG, Dickinson JR. (2007) DNA sequences from Arabidopsis, which encode protein kinases and function as upstream regulators of Snf1 in yeast. J Biol Chem 282: 10472–10479 [DOI] [PubMed] [Google Scholar]
- Ho S-L, Chao Y-C, Tong W-F, Yu S-M. (2001) Sugar coordinately and differentially regulates growth- and stress-related gene expression via a complex signal transduction network and multiple control mechanisms. Plant Physiol 125: 877–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park K-J, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res (Web Server issue) 35: W585–W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber SC, Hardin SC. (2004) Numerous posttranslational modifications provide opportunities for the intricate regulation of metabolic enzymes at multiple levels. Curr Opin Plant Biol 7: 318–322 [DOI] [PubMed] [Google Scholar]
- Huijser C, Kortstee A, Pego J, Weisbeek P, Wisman E, Smeekens S. (2000) The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: involvement of abscisic acid in sugar responses. Plant J 23: 577–585 [DOI] [PubMed] [Google Scholar]
- Hummel I, Pantin F, Sulpice R, Piques M, Rolland G, Dauzat M, Christophe A, Pervent M, Bouteillé M, Stitt M, et al. (2010) Arabidopsis plants acclimate to water deficit at low cost through changes of carbon usage: an integrated perspective using growth, metabolite, enzyme, and gene expression analysis. Plant Physiol 154: 357–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator V3: a reference expression database for the meta-analysis of transcriptomics. Adv Bioinformat 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamet E, Canut H, Boudart G, Pont-Lezica RF. (2006) Cell wall proteins: a new insight through proteomics. Trends Plant Sci 11: 33–39 [DOI] [PubMed] [Google Scholar]
- Jang J-C, León P, Zhou L, Sheen J. (1997) Hexokinase as a sugar sensor in higher plants. Plant Cell 9: 5–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossier M, Bouly J-P, Meimoun P, Arjmand A, Lessard P, Hawley S, Grahame Hardie D, Thomas M. (2009) SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana. Plant J 59: 316–328 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Karve A, Moore BD. (2009) Function of Arabidopsis hexokinase-like1 as a negative regulator of plant growth. J Exp Bot 60: 4137–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch K. (2004) Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol 7: 235–246 [DOI] [PubMed] [Google Scholar]
- Laby RJ, Kincaid MS, Kim D, Gibson SI. (2000) The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J 23: 587–596 [DOI] [PubMed] [Google Scholar]
- Lee E-J, Koizumi N, Sano H. (2004) Identification of genes that are up-regulated in concert during sugar depletion in Arabidopsis. Plant Cell Environ 27: 337–345 [Google Scholar]
- Lee K-W, Chen P-W, Lu C-A, Chen S, Ho T-H, Yu S-M. (2009) Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci Signal 2: ra61. [DOI] [PubMed] [Google Scholar]
- Li L, Foster CM, Gan Q, Nettleton D, James MG, Myers AM, Wurtele ES. (2009) Identification of the novel protein QQS as a component of the starch metabolic network in Arabidopsis leaves. Plant J 58: 485–498 [DOI] [PubMed] [Google Scholar]
- Li Y, Smith C, Corke F, Zheng L, Merali Z, Ryden P, Derbyshire P, Waldron K, Bevan MW. (2007) Signaling from an altered cell wall to the nucleus mediates sugar-responsive growth and development in Arabidopsis thaliana. Plant Cell 19: 2500–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F, van Dongen JT, Giuntoli B, Novi G, Santaniello A, Geigenberger P, Perata P. (2010) HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J 62: 302–315 [DOI] [PubMed] [Google Scholar]
- Lisso J, Altmann T, Müssig C. (2006) Metabolic changes in fruits of the tomato dx mutant. Phytochemistry 67: 2232–2238 [DOI] [PubMed] [Google Scholar]
- Lohse M, Nunes-Nesi A, Krüger P, Nagel A, Hannemann J, Giorgi FM, Childs L, Osorio S, Walther D, Selbig J, et al. (2010) Robin: an intuitive wizard application for R-based expression microarray quality assessment and analysis. Plant Physiol 153: 642–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C-A, Lin C-C, Lee K-W, Chen J-L, Huang L-F, Ho S-L, Liu H-J, Hsing Y-I, Yu S-M. (2007) The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. Plant Cell 19: 2484–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. (2000) The PSIPRED protein structure prediction server. Bioinformatics 16: 404–405 [DOI] [PubMed] [Google Scholar]
- Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM, et al. (2008) Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet 4: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng W-H, Liu Y-X, Hwang I, Jones T, Sheen J. (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332–336 [DOI] [PubMed] [Google Scholar]
- Moreno F, Ahuatzi D, Riera A, Palomino CA, Herrero P. (2005) Glucose sensing through the Hxk2-dependent signalling pathway. Biochem Soc Trans 33: 265–268 [DOI] [PubMed] [Google Scholar]
- Müssig C. (2005) Brassinosteroid-promoted growth. Plant Biol (Stuttg) 7: 110–117 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Lee SC, Oosumi T, Zanetti ME, Yang H, Ma K, Yaghoubi-Masihi A, Fukao T, Bailey-Serres J. (2010) Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol 152: 1484–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nam HK, Li J. (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signalling. Cell 26: 203–212 [DOI] [PubMed] [Google Scholar]
- Osuna D, Usadel B, Morcuende R, Gibon Y, Bläsing OE, Höhne M, Günter M, Kamlage B, Trethewey R, Scheible W-R, et al. (2007) Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J 49: 463–491 [DOI] [PubMed] [Google Scholar]
- Paul MJ, Pellny TK. (2003) Carbon metabolite feedback regulation of leaf photosynthesis and development. J Exp Bot 54: 539–547 [DOI] [PubMed] [Google Scholar]
- Price J, Laxmi A, St Martin SK, Jang J-C. (2004) Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16: 2128–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM. (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Roldán M, Gómez-Mena C, Ruiz-García L, Salinas J, Martínez-Zapater JM. (1999) Sucrose availability on the aerial part of the plant promotes morphogenesis and flowering of Arabidopsis in the dark. Plant J 20: 581–590 [DOI] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57: 675–709 [DOI] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E. (2001) Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13: 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter U, Köpke D, Altmann T, Müssig C. (2002) Analysis of carbohydrate metabolism of CPD-antisense plants and the brassinosteroid-deficient cbb1 mutant. Plant Cell Environ 25: 783–791 [Google Scholar]
- Schröder F, Lisso J, Lange P, Müssig C. (2009) The extracellular EXO protein mediates cell expansion in Arabidopsis leaves. BMC Plant Biol 9: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Reyes MI, Hanley-Bowdoin L. (2009) Arabidopsis protein kinases GRIK1 and GRIK2 specifically activate SnRK1 by phosphorylating its activation loop. Plant Physiol 150: 996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S, Ma J, Hanson J, Rolland F. (2010) Sugar signals and molecular networks controlling plant growth. Curr Opin Plant Biol 13: 274–279 [DOI] [PubMed] [Google Scholar]
- Smith AM, Stitt M. (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30: 1126–1149 [DOI] [PubMed] [Google Scholar]
- Smith SM, Fulton DC, Chia T, Thorneycroft D, Chapple A, Dunstan H, Hylton C, Zeeman SC, Smith AM. (2004) Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol 136: 2687–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Gibon Y, Lunn JE, Piques M. (2007) Multilevel genomics analysis of carbon signalling during low carbon availability: coordinating the supply and utilisation of carbon in a fluctuating environment. Funct Plant Biol 34: 526–549 [DOI] [PubMed] [Google Scholar]
- Sugden C, Donaghy PG, Halford NG, Hardie DG. (1999) Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol 120: 257–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Usadel B, Bläsing OE, Gibon Y, Retzlaff K, Höhne M, Günther M, Stitt M. (2008) Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol 146: 1834–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-Y, Seto H, Fujioka S, Yoshida S, Chory J. (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CY, Trieu A, Radhakrishnan P, Kwok SF, Harris S, Zhang K, Wang J, Wan J, Zhai H, Takatsuto S, et al. (2008) Brassinosteroids regulate grain filling in rice. Plant Cell 20: 2130–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YY, Obokata J. (2008) PPDB: a plant promoter database. Nucleic Acids Res (Database issue) 36: D977–D981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Yoo SD, Sheen J. (2003) Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425: 521–525 [DOI] [PubMed] [Google Scholar]
- Zhang J, Vemuri G, Nielsen J. (2010) Systems biology of energy homeostasis in yeast. Curr Opin Microbiol 13: 382–388 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Primavesi LF, Jhurreea D, Andralojc PJ, Mitchell RA, Powers SJ, Schluepmann H, Delatte T, Wingler A, Paul MJ. (2009) Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol 149: 1860–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Jang J-C, Jones TL, Sheen J. (1998) Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc Natl Acad Sci USA 95: 10294–10299 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.