Abstract
RNA-silencing mechanisms control many aspects of gene regulation including the detection and degradation of viral RNA through the action of, among others, Dicer-like and Argonaute (AGO) proteins. However, the extent to which RNA silencing restricts virus host range has been difficult to separate from other factors that can affect virus-plant compatibility. Here we show that Potato virus X (PVX) can infect Arabidopsis (Arabidopsis thaliana), which is normally a nonhost for PVX, if coinfected with a second virus, Pepper ringspot virus. Here we show that the pepper ringspot virus 12K protein functions as a suppressor of silencing that appears to enable PVX to infect Arabidopsis. We also show that PVX is able to infect Arabidopsis Dicer-like mutants, indicating that RNA silencing is responsible for Arabidopsis nonhost resistance to PVX. Furthermore, we find that restriction of PVX on Arabidopsis also depends on AGO2, suggesting that this AGO protein has evolved to specialize in antiviral defenses.
The outcome of a plant-virus interaction is determined by the balance between the ability of the virus to exploit host cellular mechanisms, and the multiple defense mechanisms employed against it by the host. As obligate parasites, viruses are unique in that their genetic material is transcribed, translated, and replicated entirely inside the host cell. As such, the inability of a virus to infect a given plant may be due to an incompatibility between viral proteins and the host translational machinery (Robaglia and Caranta, 2006), inhibition of viral replication (Ishibashi et al., 2007, 2009) and movement (Chisholm et al., 2000; Whitham et al., 2000; Cosson et al., 2010), or by detection and elimination by disease resistance (R) proteins (Moffett, 2009).
A major defense mechanism targeting foreign nucleic acids is based on RNA silencing (Ding and Voinnet, 2007). RNA silencing refers to a number of related cellular processes that employ small RNAs to regulate the expression of genetic material in a sequence-specific manner. Small RNAs are generated from dsRNA precursors by DICER-LIKE (DCL) RNase III-related enzymes and subsequently incorporated into RNA-induced silencing complexes (RISCs). The functions of RISCs are carried out in large part by the activity of RNAse H-like Argonaute (AGO) proteins. AGO proteins bind small RNAs, using them as guides to interact with homologous RNA molecules, and subsequently effect endonuclease activity, translational repression of mRNAs, or DNA methylation (Voinnet, 2009). The highly structured or dsRNA of viruses is targeted by DCL proteins to generate virus-derived small interfering RNAs (vsiRNAs). These vsiRNAs can be incorporated into virus-induced RISC complexes that target viral RNAs, thus making RNA silencing a doubly effective antiviral mechanism (Ding and Voinnet, 2007; Omarov et al., 2007). Furthermore, small RNAs can serve as primers for host RNA-dependent RNA polymerases to generate additional dsRNA targets for DCL enzymes to amplify the silencing signal (Voinnet, 2008; Vaistij and Jones, 2009; Garcia-Ruiz et al., 2010).
In response to the pressure exerted by RNA silencing, plant viruses have evolved viral suppressors of RNA silencing (VSRs; Diaz-Pendon and Ding, 2008; Alvarado and Scholthof, 2009). Mixed infection by two viruses can extend the host range, increase virus accumulation, and exacerbate viral symptoms (Latham and Wilson, 2008). In certain cases, this synergistic effect has been shown to be due to the action, in trans, of the VSR of one of the coinfecting viruses, likely because the VSRs of different viruses can target different RNA-silencing components (Wu et al., 2010). An example of such a synergism is illustrated by the helper component proteinase from potyviruses that enhances the accumulation and systemic spreading of Potato virus X (PVX) in tobacco (Nicotiana tabacum) and sweet potato (Ipomoea batatas; Pruss et al., 1997; Sonoda et al., 2000).
The Arabidopsis (Arabidopsis thaliana) genome encodes four DCL and 10 AGO proteins, suggesting a significant degree of diversification and specialization of RNA-silencing components (Vaucheret, 2008). DCL4 and DCL2 have been shown to play major, redundant but hierarchical, roles in silencing RNA viruses, while DCL3 also appears to play a minor role in antiviral defenses (Blevins et al., 2006; Bouche et al., 2006; Deleris et al., 2006; Fusaro et al., 2006; Donaire et al., 2008; Qu et al., 2008; Garcia-Ruiz et al., 2010). As such, mutation of dcl2 and dcl4 allows VSR-defective viruses to establish systemic infection and increases susceptibility to Tobacco rattle virus (TRV; Deleris et al., 2006; Garcia-Ruiz et al., 2010). With respect to the Arabidopsis AGO genes, hypomorphic ago1 mutants were found to be somewhat more susceptible to Cucumber mosaic virus (CMV; Morel et al., 2002) whereas infection of ago2 mutants with Turnip crinkle virus (TCV) and CMV was shown to result in more severe symptoms, but only a small and transient effect on virus accumulation (Harvey et al., 2011). At the same time, infection of ago1 and ago7 mutants with VSR-defective derivatives of TCV indicates complementary roles for AGO1 and AGO7 in antiviral defenses (Qu et al., 2008; Azevedo et al., 2010). Additionally, Ago4-like proteins have been implicated in virus defense, but only upon induction of an R-gene-mediated response (Bhattacharjee et al., 2009).
RNA silencing may also be an important mechanism of nonhost resistance against viruses, whereby a plant is resistant to all known isolates of a given pathogen. However, the extent to which this may be true has not been investigated. PVX is the type member of the genus Potexvirus (Martelli et al., 2007) and infects several solanaceous hosts, but Arabidopsis is considered a nonhost for PVX. However, an Arabidopsis-infecting strain of PVX (PVX 2.7) has been described (Cooper et al., 2003) and used in microarray expression studies (Whitham et al., 2003). We show here that PVX 2.7 can infect Arabidopsis because of the presence of a second virus, Pepper ringspot virus (PepRSV). PepRSV is a tobravirus (MacFarlane, 1999, 2010) and as expected, it encodes a VSR, which we demonstrate to be the PepRSV 12K protein. To demonstrate that PVX restriction in Arabidopsis is due to RNA-silencing mechanisms we show that PVX is able to infect a dcl2 dcl3 dcl4 triple mutant. Finally, we show that mutation of AGO2 alters the ability of PVX to infect Arabidopsis. This report demonstrates that RNA silencing may represent an important mechanism that restricts wild-type, non-host-adapted viruses, and that the AGO2 protein plays an important role in antiviral defenses.
RESULTS
Identification of a Virus Allowing PVX Infection of Arabidopsis
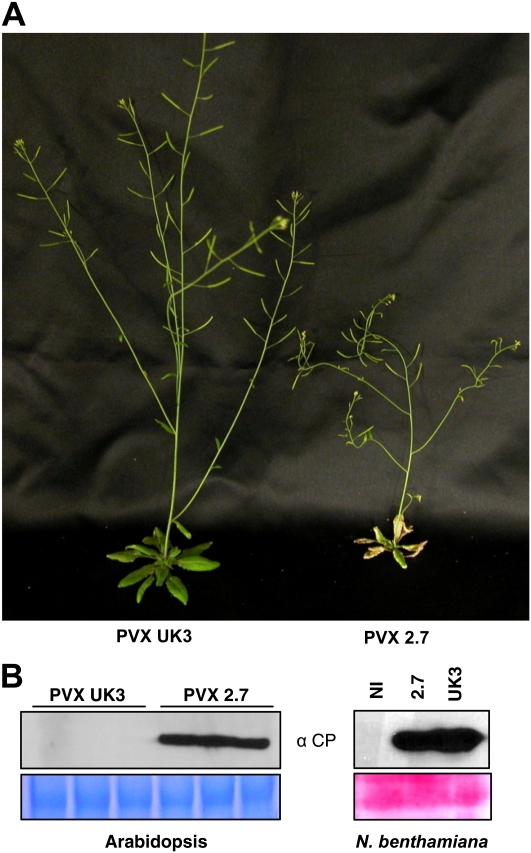
Sap derived from PVX 2.7-infected Nicotiana clevelandii leaves (Cooper et al., 2003) was used to rub inoculate Arabidopsis Columbia-0 (Col-0) plants and systemic disease symptoms were observed, including plant stunting, necrosis of rosette leaves, and curved stems (Fig. 1A). In contrast, Arabidopsis Col-0 plants infected with PVX strain UK3 showed no obvious viral symptoms. Five days post inoculation, high levels of accumulation of PVX coat protein (CP) were detected in protein extracts from leaves inoculated with PVX 2.7, but not in leaves inoculated with PVX UK3 (Fig. 1B). Both PVX strains infected Nicotiana benthamiana with equal efficiency as judged by PVX CP accumulation in inoculated leaves (Fig. 1B). Furthermore, sap preparations of both strains also elicited similar numbers of local lesions on Rx2 transgenic plants (see below, Supplemental Fig. S2), indicating that the two different inocula have similar viral infectious titres. At the same time however, rub inoculation of PVX 2.7 on Rx transgenic tobacco induced yellow ringspot lesions and vein necrosis whereas these plants are normally immune to PVX and show no visible lesions upon PVX infection (Supplemental Fig. S1; Bendahmane et al., 1999). Furthermore, in contrast to PVX UK3, PVX 2.7 induced necrotic lesions on cowpea (Vigna unguiculata; Supplemental Fig. S1), leading us to consider that PVX 2.7 might be contaminated with a second virus.
Figure 1.
Infection of Arabidopsis by PVX 2.7. A, Symptoms induced in wild-type Arabidopsis (Col-0) by the PVX 2.7 strain. Three leaves per plant were rub inoculated with sap made from either PVX UK3-infected tobacco (PVX UK3) or PVX 2.7-infected N. clevelandii (PVX 2.7). Photographs were taken 3 weeks after inoculation. B, Representative immunoblots of PVX CP accumulation in Arabidopsis and N. benthamiana. Total protein extracts were prepared from inoculated Arabidopsis or N. benthamiana leaves 5 or 6 d, respectively, after inoculation, and subjected to SDS-PAGE followed by anti-CP immunoblotting. Coomassie (left) or Ponceau (right) staining of the same extracts is shown to demonstrate equal loading. Each lane corresponds to the pool of the three inoculated leaves from a single plant for Arabidopsis and a single leaf for N. benthamiana. Three plants per treatment were tested in each Arabidopsis experiment and the experiment was repeated five times for Arabidopsis and twice for N. benthamiana.
To determine if another virus was present in the PVX 2.7 sap, we used an expanded version of a macroarray developed to identify viruses from infected plants (Agindotan and Perry, 2007) that consists of 70-mer DNA oligonucleotides specific to over 100 known viruses along with ribosomal plant RNA hybridization controls (K.L. Perry, unpublished data). The presence of specific viruses can be detected by hybridization with a labeled preparation of randomly primed, reverse-transcribed, and amplified cDNAs derived from infected plant RNAs (Agindotan and Perry, 2008). As expected, labeled cDNA derived from PVX 2.7-infected plants hybridized to PVX sequences present on the macroarray (Fig. 2). At the same time, a robust signal was detected corresponding to PepRSV (Fig. 2). PepRSV is a bipartite single-stranded positive-sense RNA virus, and a member of the genus Tobravirus (MacFarlane, 2010). To confirm the identity of PepRSV, total RNA from a PVX 2.7-infected Arabidopsis leaf was subjected to reverse transcription (RT)-PCR using generic tobravirus PCR primers that amplify an approximately 898-bp fragment from the tobravirus replicase open reading frame (Jones et al., 2008). The same PCR product was sequenced over a region of 668 nucleotides and BLAST analysis resulted in a single hit in GenBank, matching PepRSV (GenBank accession no. L23972) at 667 out of 668 nucleotides (Supplemental Fig. S3A). In contrast, the same sequence showed only 55% identity to TRV (Supplemental Fig. S3B).
Figure 2.
Detection of PepRSV. Total RNA from PVX 2.7-inoculated Arabidopsis leaves was extracted 5 d after inoculation, converted to labeled cDNA, and hybridized to the macroarray. Black arrowheads indicate the locations of the columns and rows with the spotted control oligonucleotides specific to plant ribosomal RNAs. Arrows indicate the boxed columns with the spotted PepRSV- and PVX-specific oligonucleotide probes.
To generated sap containing only PepRSV we employed Rx2 transgenic N. benthamiana plants, which are resistant to PVX (Bhattacharjee et al., 2009). These plants also display visible hypersensitive response (HR) lesions upon PVX infection and as such, can be used to ascertain the presence of PVX. PVX 2.7 was inoculated onto Rx2 N. benthamiana and sap was collected from systemic tissues. This sap was subsequently passaged to additional Rx2 plants. Rx2 plants inoculated with PVX UK3 showed only HR lesions on inoculated leaves. In contrast, plants inoculated with the PVX 2.7 sap passaged through Rx2 transgenic plants showed systemic viral symptoms, but no HR lesions (Supplemental Fig. S2). This result indicates that the latter sap contains only PepRSV and is referred to as such hereafter. To verify the presence of PepRSV in this inoculum, we performed RT-PCR on total RNA from N. benthamiana infected with PepRSV sap and amplified the open reading frames encoding the PepRSV-12K and -29K proteins (see below). Sequence analysis showed these to be 100% and 99% identical to corresponding sequences in the published PepRSV sequence (Supplemental Fig. S3). Given that sequence identity is low between tobravirus species (MacFarlane, 2008), we thus conclude that the contaminating virus in PVX 2.7 is indeed PepRSV.
To demonstrate that PepRSV is responsible for PVX accumulation in Arabidopsis, we inoculated plants with a mixture of PVX UK3- and PepRSV-containing saps. Five days post infection, inoculated leaves were collected and proteins extracted. Immunoblot analysis revealed high levels of PVX CP in the presence of PepRSV (Fig. 3). Moreover, although PepRSV by itself induces necrotic symptoms on Arabidopsis, coinfection of both viruses consistently induced more severe symptoms (Fig. 3), indicative of a synergistic interaction between the two viruses.
Figure 3.
Coinoculation of PVX and PepRSV on Arabidopsis. Three leaves per plant were rub inoculated with sap containing PVX UK3, sap containing PepRSV, or a mixture of both saps. A, Photographs were taken 3 weeks post infection. Enlargements of the singly or doubly infected rosettes are shown in the bottom section. B, Representative immunoblot analysis of the PVX CP accumulation in inoculated leaves. Total protein extracts were prepared from inoculated leaves 5 d after inoculation and subjected to SDS-PAGE followed by anti-CP immunoblotting. Coomassie staining of the same extracts is shown to demonstrate equal loading. Each lane corresponds to the pool of three inoculated leaves from the same plant. Three plants per treatment were tested in each experiment and the experiment was repeated five times.
The PepRSV 12K Protein Possesses VSR Activity
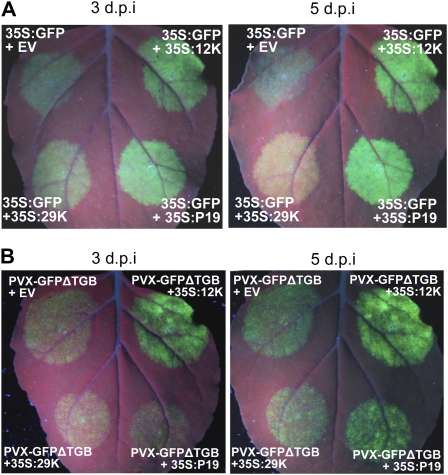
The apparent synergistic relationship between PepRSV and PVX suggests that coinfection involves the inhibition of RNA silencing. The PepRSV 12K protein is homologous to the TRV 16K protein that possesses VSR activity (Ghazala et al., 2008; Martin-Hernandez and Baulcombe, 2008; Martinez-Priego et al., 2008). To demonstrate VSR activity in the 12K protein we tested it in the standard Agrobacterium-mediated transient expression (agroinfiltration) assay that takes advantage of the fact that transiently expressed transgenes are subjected to RNA silencing, and that this silencing is suppressed by many VSRs (Voinnet et al., 2003). Transient expression of 35S-GFP by agroinfiltration in N. benthamiana leaves results in bright fluorescence visible at 3 d post infiltration (dpi). However, by 5 dpi, GFP expression is silenced and fluorescence only faintly visible (Fig. 4A). Coexpression of GFP with either PepRSV 12K or the well-characterized VSR, P19 (Voinnet et al., 2003), resulted in strong GFP fluorescence at 5 dpi, whereas the PepRSV 29K movement protein had no effect on GFP persistence (Fig. 4A). PVX encodes three genes required for viral movement, known as the triple gene block (TGB), including P25, which also possesses VSR activity (Voinnet et al., 2000; Bayne et al., 2005). We expressed 12K with a version of PVX that expresses GFP but lacks the TGB (PVX-GFPΔTGB), to determine whether the 12K protein could substitute for a lack of P25 as a VSR. Indeed, GFP fluorescence derived from PVX-GFPΔTGB was significantly augmented by coexpression with 12K, to a degree that was consistently higher than that afforded by coexpression with P19 (Fig. 4B).
Figure 4.
VSR activity of PepRSV-12K. A, Persistence of fluorescence from GFP upon coexpression with PepRSV-12K. GFP was transiently expressed via agroinfiltration from the 35S promoter in N. benthamiana, together with either 35S:PepRSV-12K:HA, 35S:P19, 35S:PepRSV-29K:HA, or pBin61 empty vector (EV). GFP signal was monitored by UV illumination at 3 and 5 d after infiltration. B, PepRSV-12K enhances PVX-GFPΔTGB in N. benthamiana. A derivative of PVX expressing GFP but lacking the TGB (PVX-GFPΔTGB) was expressed from the 35S promoter by agroinfiltration, together with either 35S:PepRSV-12K:HA, 35S:TBSV-P19, 35S:PepRSV-29K:HA, or pBIN61 empty vector (EV). GFP signal was monitored by UV illumination at 3 and 5 d after infiltration.
Role of RNA-Silencing Components in Arabidopsis Resistance to PVX
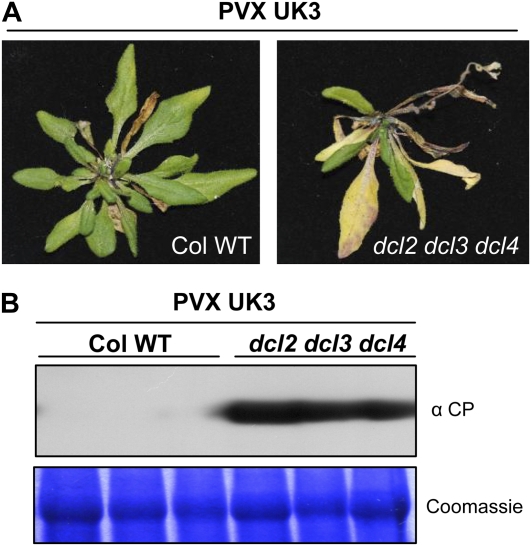
Given that PepRSV enables PVX to infect Arabidopsis, we hypothesized that this may be due to efficient elimination of PVX by RNA-silencing mechanisms rather than an intrinsic inability to replicate and move in this host. To test this, we infected an Arabidopsis triple dicer mutant, dcl2 dcl3 dcl4. Arabidopsis Dicer mutants allow enhanced accumulation of several Arabidopsis-infecting RNA viruses (Blevins et al., 2006; Bouche et al., 2006; Deleris et al., 2006; Fusaro et al., 2006; Donaire et al., 2008; Qu et al., 2008; Garcia-Ruiz et al., 2010), and the triple mutant has a dramatically reduced ability to produce vsiRNAs (Deleris et al., 2006). At 4 dpi we observed high levels of PVX CP accumulation in infected leaves and after 21 d, plants presented severe disease symptoms with systemic necrosis (Fig. 5).
Figure 5.
PVX infection of an Arabidopsis triple DICER mutant. A, Symptoms induced by PVX UK3 infection in wild type and dcl2 dcl3 dcl4 mutant Arabidopsis. Three leaves per plant were rub inoculated with sap containing PVX UK3. Photographs were taken 4 weeks after infection. B, Immunoblot analysis of PVX CP accumulation in inoculated leaves. Total protein extracts were prepared from inoculated leaves 6 d after inoculation and subjected to SDS-PAGE followed by anti-CP immunoblotting. Coomassie staining of the same extracts is shown to demonstrate equal loading. Each lane corresponds to a pool of the three inoculated leaves from a single plant. Four plants per treatment were tested in each experiment and the experiment was repeated five times.
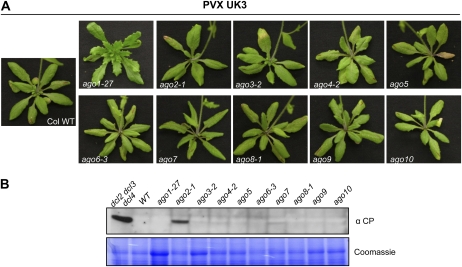
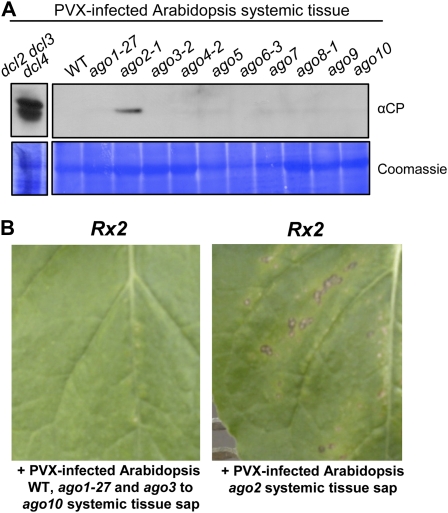
Hypomorphic ago1 and null ago7 mutants are more susceptible to VSR-defective TCV (Qu et al., 2008; Azevedo et al., 2010). To determine whether wild-type PVX is targeted by similar AGO proteins, PVX was inoculated onto Arabidopsis lines possessing the hypomorphic ago1-27 allele, mutant lines with null alleles in the AGO2, AGO3, AGO4, AGO5, AGO6, AGO7, AGO8, AGO9, and AGO10 genes, as well as the dcl2 dcl3 dcl4 mutant as a control. Twenty-eight days after inoculation (at least 18 plants for each genotype), no visible virus symptoms were apparent in the systemic tissues in any of the 10 ago mutants (Fig. 6A). However at 5 dpi, PVX CP accumulation was detected in the inoculated leaves of ago2 mutant plants (Fig. 6B). Despite a lack of visible systemic symptoms following PVX inoculation, PVX CP was detected by immunoblotting in the systemic tissues of eight out of 18 PVX-inoculated ago2 plants (compared to zero out of similar numbers of all other ago mutant genotypes), albeit at lower levels than in the dcl2 dcl3 dcl4 mutants (Fig. 7A). In addition, sap collected from systemic tissues of PVX-infected ago2 mutants induced HR lesions on Rx2 plants (2/4 times), indicating the presence of PVX in these plants (Fig. 7B). In contrast no HR lesions were induced with sap from PVX-inoculated wild-type Arabidopsis or any of the other ago mutants (Fig. 7B). This result indicates that AGO2 plays a major role in the antiviral defense against PVX in Arabidopsis.
Figure 6.
PVX infection in Arabidopsis ago mutants. A, Symptoms induced by PVX UK3 infection on wild type and ago1-27, ago2-1, ago3-2, ago4-2, ago5, ago6-3, ago7, ago8-1, ago9, and ago10 Arabidopsis mutants. Three leaves per plants were rub inoculated with sap made from PVX UK3-infected tobacco (PVX UK3). Photographs were taken 4 weeks after infection. B, Immunoblot analysis of PVX CP accumulation in inoculated leaves. Total protein extracts were prepared from inoculated leaves 5 d after inoculation and subjected to SDS-PAGE followed by anti-CP immunoblotting. Coomassie staining of the same extracts is shown to demonstrate equal loading. Each lane corresponds to a pool of the three inoculated leaves from a single plant. This result was obtained consistently in five experiments including at least three replicates for each genotype.
Figure 7.
Systemic movement of PVX in Arabidopsis RNA-silencing mutants. A, Immunoblot analysis of PVX CP accumulation in Arabidopsis systemic tissue. Total protein extracts were prepared from the systemic leaves of one Arabidopsis plant of each genotype 3 weeks after PVX inoculation, followed by SDS-PAGE and anti-CP immunoblotting. Coomassie staining of the same extracts is shown to demonstrate equal loading. The triple dicer mutant sample is taken from a separate experiment but similar amounts of tissue and exposure times were used. B, Detection of PVX by inoculation of Rx2 transgenic N. benthamiana. Sap was prepared from the systemic tissue of PVX UK3-infected wild type and ago mutant Arabidopsis. The left section indicates a representative result (lack of HR lesions) obtained from inoculation with sap from wild type, ago1-27, or mutants in ago3 to ago10. The right section depicts a representative result showing HR lesions, indicating the presence of PVX, observed upon inoculation with sap preparations from individual ago2-1 plants.
DISCUSSION
To our knowledge, these results provide the first demonstration that RNA silencing plays a major role in restricting a non-host-adapted virus. We show that synergy between PVX and PepRSV enable systemic infection of PVX in Arabidopsis. Both synergism and expansion of host range have been shown to occur upon virus coinfection and in certain cases VSRs, such as helper component proteinase, have been implicated in this phenomenon (Sonoda et al., 2000; Latham and Wilson, 2008). The PepRSV 12K Cys-rich protein is homologous to the TRV 16K protein, which has previously been shown to function as a VSR (Ghazala et al., 2008; Martin-Hernandez and Baulcombe, 2008; Martinez-Priego et al., 2008), and we find that PepRSV 12K functions efficiently as a VSR in N. benthamiana transient expression assays (Fig. 4).
We did not observe accumulation of a PVX-12K construct in either inoculated or systemic leaves of Arabidopsis (data not shown). This may not be surprising since, in addition to a cell-autonomous VSR function (Bayne et al., 2005), a critical property of the PVX VSR, the P25 protein, seems to be its ability to suppress a silencing signal, both locally and systemically (Voinnet et al., 2000; Himber et al., 2003; Bayne et al., 2005). The silencing signal involves the movement of siRNAs (Dunoyer et al., 2010; Molnar et al., 2010) and it is thought that such a mechanism allows vsiRNAs to accumulate in uninfected cells ahead of viral movement where they can target viral RNAs upon their entry into the cell. It is unclear how the P25 protein inhibits vsiRNA movement, although the use of this strategy may indicate that PVX is particularly sensitive to the effect of having mobile vsiRNAs present in uninfected cells prior to viral entry. If so, the presence of vsiRNAs in a cell prior to infection would be predicted to target PVX, via AGO2, before significant amounts of VSR protein can be expressed from the PVX genome. However, coinfection of Arabidopsis leaf cells by PepRSV appears to suppress silencing mechanisms, thereby allowing PVX to move into these cells. Likewise, a block in vsiRNA production in the triple dicer mutant, or a block in the appropriate vsiRNA effector AGO protein, would accomplish the same outcome.
The ability of PVX to infect the Arabidopsis triple dicer mutant suggests that the P25 protein may not fully function as an effective VSR in this host. Previous reports have shown that P25 is able to suppress silencing induced by RNA hairpins in both N. benthamiana and Arabidopsis (Dunoyer et al., 2004; Bayne et al., 2005). However the effect of P25 on the spread of silencing signals has not been investigated in Arabidopsis and the developmental phenotypes induced by transgenic expression of P25 in Arabidopsis are milder than for other VSRs (Dunoyer et al., 2004). Alternatively, the inability of PVX to infect Arabidopsis may be due to a combination of factors, with the inhibition or ablation of silencing allowing it to overcome a threshold required for systemic infection.
Several VSRs directly target AGO1 and either prevent it from loading siRNAs or induce its degradation (Zhang et al., 2006; Baumberger et al., 2007; Azevedo et al., 2010; Giner et al., 2010). The VSRs P1 and P38 exert their suppressor of silencing activity through direct interaction with AGO1 via their WG/GW motifs (Azevedo et al., 2010; Giner et al., 2010). Sequence analysis of tobravirus 16K homologs indicates the presence of a conserved GW motif in the N termini of these proteins (Ghazala et al., 2008). However, attempts to coimmunoprecipitate multiple AGO proteins with 12K and 16K did not reveal any interactions (data not shown). Recently, it was reported that the PVX P25 protein is able to interact with AGO1, AGO2, AGO3, and AGO4, as well as destabilizing AGO1 (Chiu et al., 2010). Using comparable methodology, we are similarly unable to detect interactions between P25 and a number of AGO proteins, including Arabidopsis and tobacco AGO2 (data not shown).
It is generally assumed that degradation of viral RNA by Dicer proteins alone is not sufficient to effect virus resistance and that their vsiRNA products must eventually be loaded into RISC complexes to target viral RNAs. The AGO1, AGO2, and AGO5 proteins can bind vsiRNAs (Zhang et al., 2006; Takeda et al., 2008; Harvey et al., 2011) but, of the 10 Arabidopsis AGO proteins, genetic analysis has implicated only AGO1, AGO2, and possibly AGO7 in virus resistance. The ago1-27 hypomorphic mutant shows a 5-fold greater accumulation of CMV RNA and both an ago1 hypomorphic mutant and an ago7 null mutant allow increased accumulation of a VSR-defective version of TCV (Morel et al., 2002; Qu et al., 2008; Azevedo et al., 2010). Infection of ago2 mutants with CMV and TCV resulted in more severe viral symptoms, but little difference in virus accumulation (Harvey et al., 2011). Surprisingly, we did not observe any difference in PVX accumulation on either ago1-27 or ago7 mutants (Fig. 6). Instead, we observed PVX accumulation in inoculated and systemic tissues only in the ago2 mutant (Figs. 6 and 7), indicating that AGO2 is essential for curtailing PVX on this host. Furthermore, our results show that AGO2 activity can absolutely limit virus infection rather than simply playing a secondary role to AGO1, as previously suggested (Harvey et al., 2011). Interestingly, AGO2 is placed in the same phylogenetic clade as AGO7 (Vaucheret, 2008) but unlike ago7, which is involved in tasi-RNA biogenesis, ago2 mutants show no obvious phenotype (Vaucheret, 2008). This may indicate that AGO2 has evolved to specialize in antiviral defenses.
We do not formally rule out a role for AGO1 in resistance to PVX as the protein encoded by ago1-27 may possess sufficient activity to target PVX, but not TCV or CMV. Indeed, although we find that ago2 is the only single mutant capable of allowing PVX infection, this infection is not as efficient as in the dcl2 dcl3 dcl4 mutant (Figs. 5–7), suggesting that at least one other AGO protein acts in PVX antiviral silencing. A supplemental or hierarchical role of another AGO protein(s) would not be surprising given the redundancy/complementarity seen with members of the DICER family (Deleris et al., 2006; Garcia-Ruiz et al., 2010). At the same time AGO2, which is regulated by an endogenous miRNA (Allen et al., 2005), is expressed at a 4.6-fold higher level in an ago1 hypomorphic mutant (Kurihara et al., 2009), and AGO2 protein levels are significantly up-regulated upon infection with TCV and CMV (Harvey et al., 2011). Thus, increased levels of AGO2 may in theory compensate for a lack of AGO1 with respect to PVX resistance.
A previous report has shown that the requirement for AGO1 versus AGO7 for resistance to a VSR-defective version of TCV differed depending on whether the virus contained a GFP insert (Qu et al., 2008). Likewise, although AGO1 has been shown to play an important role in defense against VSR-defective TCV in Arabidopsis (Qu et al., 2008; Azevedo et al., 2010), we find that down-regulation of NbAgo2 has a much greater effect than NbAgo1 on VSR-defective TBSV in N. benthamiana (Scholthof et al., 2011). This suggests that different AGO proteins may vary in their effectiveness in targeting different viral RNA genomes depending on as-yet-undefined characteristics of the viral RNA genome. Why certain AGO proteins might be more or less important for targeting different viruses is unclear. Recent reports have shown that the regions of a virus most effectively targeted by vsiRNAs do not necessarily correlate with those regions corresponding to the most abundant viRNAs (Szittya et al., 2010) and that AGO proteins show bias in the siRNAs they bind (Mi et al., 2008; Takeda et al., 2008; Ho et al., 2010). Thus, it is tempting to speculate that targeting by a given AGO protein may depend on a combination of the availability of the viral genome to RNA-silencing components, determined by RNA structure, together with the nucleotide composition of such regions.
CONCLUSION
Our results show that in addition to its well-defined role in the interaction between viruses and their hosts, RNA silencing is also an important mechanism of nonhost resistance against non-host-adapted viruses. Furthermore, our results indicate an important role for the AGO2 protein in antiviral defenses that will lay the groundwork for future studies on the interplay between different AGOs, DCLs, vsiRNAs, and viruses.
MATERIALS AND METHODS
Plant and Virus Materials
Nicotiana benthamiana, Nicotiana clevelandii, and tobacco (Nicotiana tabacum) were germinated and grown in a glasshouse maintained at 21°C. Samples of PVX 2.7-infected N. clevelandii leaves were generously provided by Bret Cooper (Cooper et al., 2003) and were used to produce sap for further virus infections. PVX strains were propagated in tobacco or N. clevelandii (UK3) and N. clevelandii (2.7), respectively. PepRSV was isolated by infecting Rx2 transgenic N. benthamiana (Bhattacharjee et al., 2009) with PVX 2.7 sap and subsequently propagated in N. clevelandii. Virus saps were prepared by grinding infected or systemic tissue in 0.1 m potassium phosphate buffer, pH 7.5. The concentration of PVX-containing inocula was normalized according to the immunoblot signal of PVX CP. Arabidopsis (Arabidopsis thaliana) wild type and mutants are of the Col-0 ecotype and were cultivated in growth chambers under long-day period (16 h/8 h). All Arabidopsis mutant lines were of the Col-0 ecotype and have been previously described, including the triple dicer mutant dcl2-1/dcl3-1/dcl4-2 (Deleris et al., 2006), ago1-27 (Morel et al., 2002), ago2-1, ago3-2, ago6-3, ago8-1 ago10-2 (Takeda et al., 2008), ago5, ago7, ago9 (Katiyar-Agarwal et al., 2007), and ago4-2 (Agorio and Vera, 2007). All virus infections of Arabidopsis were carried out by rubbing sap on carborundum-dusted leaves of 3-week-old plants with the side of pipette tip.
Immunoblot Analysis
Total proteins were extracted in 8 m urea solution, separated in a 10% SDS-polyacrylamide gel, and transferred to a polyvinylidene difluoride membrane by electroblotting. Each sample analyzed corresponds to a pool of inoculated leaves or systemic tissue of a single plant. For each infection, at least three plants were tested in more than five different experiments. PVX CP was probed with anti-PVX CP rabbit polyclonal antibodies (Agdia) and subsequently detected by a goat anti-rabbit Horseradish peroxidase-conjugated polyclonal antibody followed by chemiluminescence detection (ECL, GE Healthcare).
Cloning and Sequencing of PVX 2.7
Total RNA from PVX 2.7-infected Nicotiana glutinosa or Arabidopsis was extracted with Trizol reagent according to the manufacturer instructions (Invitrogen). cDNAs were obtained after RT using a primer targeting the 3′ untranslated region of PVX (5′-ATTTATATTATTCTAACAATCAAAC-3′) and PCR (Expand Hi Fidelity Taq DNA polymerase, Roche) with primers targeting overlapping fragments of PVX, based on the PVX UK3 sequence. PCR products were cloned into pGEMT (Promega) and sequenced and overlapping sequences were used to assemble a full-length sequence. At least three independent RT-PCRs were performed for both the Arabidopsis (PVX2.7a) and N. glutinosa (PVX2.7b) sequences, which have been deposited in GenBank as accession numbers HQ450387 and HQ450388.
Macroarray Hybridizations and Identification of PepRSV
cDNA were generated from PVX 2.7-infected Arabidopsis RNA extraction and used to probe the macroarray membrane as previously described (Agindotan and Perry, 2007, 2008). The identity of PepRSV was confirmed by RT-PCR from total RNA isolated from a PVX 2.7-infected Arabidopsis plant using generic tobravirus PCR primers Tobra-F3 and Tobra-R2, which amplify an approximately 898-bp fragment, as previously described (Jones et al., 2008). The latter PCR product was sequenced using the same primers and used to query sequence databases. The 668 bp that could be read from this sequencing have been deposited in GenBank as accession number JF906525. The identity of PepRSV was further confirmed by amplification of the PepRSV-12K and -29K genes from total Trizol-extracted (Invitrogen) RNA from PepRSV-infected N. benthamiana. cDNAs were obtained after RT using polydT primers and PCR (Expand Hi Fidelity Taq DNA polymerase, Roche) with the primer pairs PRSV12KXbaI.F (5′-CACTCTAGAACCATGACGAAGTGTGCTCTAC-3′) + PRSV12KBamHI.R (5′-CACGGATCCACACACCTCACTAAACAG-3′) or PRSV29KXbaI.F (5′-CACTCTAGAACCATGGAGAACGATAAGTCGT-3′) + PRSV29KBamHI.R (5′-CACGGATCCTTTTTTCTTTATGCTTG-3′). PCR products were subsequently cloned (see below) and sequenced. Sequences for the PepRSV-12K and -29K genes have been deposited in GenBank as accessions JF268315 and JF268316, respectively.
Plasmid Construction
Plasmids 35S-GFP, pBIN61, and 35S-P19 have been previously described (Voinnet et al., 2003). PVX-GFPΔTGB was constructed by overlapping PCR to delete the entire TGB from the binary vector pGr208 (PVX-GFP; Peart et al., 2002). To clone the PepRSV-12K and -29K genes into expression vectors, the PRSV12K and PRSV29K PCR products described above were digested with XbaI/BamHI and inserted into the same sites of the pBin61:HA plasmid (Moffett et al., 2002).
Transient Silencing Suppression Activity Assays
Wild-type N. benthamiana plants were infiltrated with Agrobacterium tumefaciens clones (agroinfiltration) as previously described (Moffett et al., 2002). Final concentrations for each Agrobacterium strain were: OD600 = 0.2. GFP expression was monitored under UV light using a handheld lamp (UVP BP-100AP).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers HQ450387, HQ450388, JF906525, JF268315, and JF268316.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Symptoms induced on Rx transgenic tobacco and cowpea by PVX UK3 and PVX 2.7.
Supplemental Figure S2. Isolation of PepRSV from PVX 2.7 sap using Rx2 transgenic plants.
Supplemental Figure S3. Sequence alignment of PepRSV sequences.
Supplementary Material
Acknowledgments
We thank Bret Cooper for initially sending us PVX 2.7-infected material and to Nadia Rodriguez for technical assistance. We are grateful to Hailing Jin, Yuichiro Watanabe, Olivier Voinnet, Lionel Navarro, and Hervé Vaucheret for mutant Arabidopsis lines and to Herman Scholthof for valuable discussion and critical reading of the manuscript.
References
- Agindotan B, Perry KL. (2007) Macroarray detection of plant RNA viruses using randomly primed and amplified complementary DNAs from infected plants. Phytopathology 97: 119–127 [DOI] [PubMed] [Google Scholar]
- Agindotan B, Perry KL. (2008) Macroarray detection of eleven potato-infecting viruses and potato spindle tuber viroid. Plant Dis 92: 730–740 [DOI] [PubMed] [Google Scholar]
- Agorio A, Vera P. (2007) ARGONAUTE4 is required for resistance to Pseudomonas syringae in Arabidopsis. Plant Cell 19: 3778–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Alvarado V, Scholthof HB. (2009) Plant responses against invasive nucleic acids: RNA silencing and its suppression by plant viral pathogens. Semin Cell Dev Biol 20: 1032–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo J, Garcia D, Pontier D, Ohnesorge S, Yu A, Garcia S, Braun L, Bergdoll M, Hakimi MA, Lagrange T, et al. (2010) Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes Dev 24: 904–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N, Tsai CH, Lie M, Havecker E, Baulcombe DC. (2007) The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr Biol 17: 1609–1614 [DOI] [PubMed] [Google Scholar]
- Bayne EH, Rakitina DV, Morozov SY, Baulcombe DC. (2005) Cell-to-cell movement of Potato Potexvirus X is dependent on suppression of RNA silencing. Plant J 44: 471–482 [DOI] [PubMed] [Google Scholar]
- Bendahmane A, Kanyuka K, Baulcombe DC. (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11: 781–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S, Zamora A, Azhar MT, Sacco MA, Lambert LH, Moffett P. (2009) Virus resistance induced by NB-LRR proteins involves Argonaute4-dependent translational control. Plant J 58: 940–951 [DOI] [PubMed] [Google Scholar]
- Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A, Park HS, Vazquez F, Robertson D, Meins F, Jr, Hohn T, et al. (2006) Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res 34: 6233–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche N, Lauressergues D, Gasciolli V, Vaucheret H. (2006) An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J 25: 3347–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Mahajan SK, Whitham SA, Yamamoto ML, Carrington JC. (2000) Cloning of the Arabidopsis RTM1 gene, which controls restriction of long-distance movement of tobacco etch virus. Proc Natl Acad Sci USA 97: 489–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu MH, Chen IH, Baulcombe DC, Tsai CH. (2010) The silencing suppressor P25 of Potato virus X interacts with Argonaute1 and mediates its degradation through the proteasome pathway. Mol Plant Pathol 11: 641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper B, Eckert D, Andon NL, Yates JR, Haynes PA. (2003) Investigative proteomics: identification of an unknown plant virus from infected plants using mass spectrometry. J Am Soc Mass Spectrom 14: 736–741 [DOI] [PubMed] [Google Scholar]
- Cosson P, Sofer L, Le QH, Leger V, Schurdi-Levraud V, Whitham SA, Yamamoto ML, Gopalan S, Le Gall O, Candresse T, et al. (2010) RTM3, which controls long-distance movement of potyviruses, is a member of a new plant gene family encoding a meprin and TRAF homology domain-containing protein. Plant Physiol 154: 222–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O. (2006) Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313: 68–71 [DOI] [PubMed] [Google Scholar]
- Diaz-Pendon JA, Ding S-W. (2008) Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu Rev Phytopathol 46: 303–326 [DOI] [PubMed] [Google Scholar]
- Ding SW, Voinnet O. (2007) Antiviral immunity directed by small RNAs. Cell 130: 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaire L, Barajas D, Martinez-Garcia B, Martinez-Priego L, Pagan I, Llave C. (2008) Structural and genetic requirements for the biogenesis of tobacco rattle virus-derived small interfering RNAs. J Virol 82: 5167–5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P, Brosnan CA, Schott G, Wang Y, Jay F, Alioua A, Himber C, Voinnet O. (2010) An endogenous, systemic RNAi pathway in plants. EMBO J 29: 1699–1712 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dunoyer P, Lecellier CH, Parizotto EA, Himber C, Voinnet O. (2004) Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16: 1235–1250 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fusaro AF, Matthew L, Smith NA, Curtin SJ, Dedic-Hagan J, Ellacott GA, Watson JM, Wang MB, Brosnan C, Carroll BJ, et al. (2006) RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep 7: 1168–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ruiz H, Takeda A, Chapman EJ, Sullivan CM, Fahlgren N, Brempelis KJ, Carrington JC. (2010) Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell 22: 481–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazala W, Waltermann A, Pilot R, Winter S, Varrelmann M. (2008) Functional characterization and subcellular localization of the 16K cysteine-rich suppressor of gene silencing protein of tobacco rattle virus. J Gen Virol 89: 1748–1758 [DOI] [PubMed] [Google Scholar]
- Giner A, Lakatos L, Garcia-Chapa M, Lopez-Moya JJ, Burgyan J. (2010) Viral protein inhibits RISC activity by argonaute binding through conserved WG/GW motifs. PLoS Pathog 6: e1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey JJ, Lewsey MG, Patel K, Westwood J, Heimstadt S, Carr JP, Baulcombe DC. (2011) An antiviral defense role of AGO2 in plants. PLoS ONE 6: e14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O. (2003) Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J 22: 4523–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T, Wang L, Huang L, Li Z, Pallett DW, Dalmay T, Ohshima K, Walsh JA, Wang H. (2010) Nucleotide bias of DCL and AGO in plant anti-virus gene silencing. Protein Cell 1: 847–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi K, Masuda K, Naito S, Meshi T, Ishikawa M. (2007) An inhibitor of viral RNA replication is encoded by a plant resistance gene. Proc Natl Acad Sci USA 104: 13833–13838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi K, Naito S, Meshi T, Ishikawa M. (2009) An inhibitory interaction between viral and cellular proteins underlies the resistance of tomato to nonadapted tobamoviruses. Proc Natl Acad Sci USA 106: 8778–8783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D, Farreyrol K, Clover GRG, Pearson MN. (2008) Development of a generic PCR detection method for tobraviruses. Australas Plant Pathol 37: 132–136 [Google Scholar]
- Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H. (2007) A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev 21: 3123–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Kaminuma E, Matsui A, Kawashima M, Tanaka M, Morosawa T, Ishida J, Mochizuki Y, Shinozaki K, Toyoda T, et al. (2009) Transcriptome analyses revealed diverse expression changes in ago1 and hyl1 Arabidopsis mutants. Plant Cell Physiol 50: 1715–1720 [DOI] [PubMed] [Google Scholar]
- Latham JR, Wilson AK. (2008) Transcomplementation and synergism in plants: implications for viral transgenes? Mol Plant Pathol 9: 85–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane SA. (1999) Molecular biology of the tobraviruses. J Gen Virol 80: 2799–2807 [DOI] [PubMed] [Google Scholar]
- MacFarlane SA. (2008) Tobravirus. Mahy BWJ, Van Regenmortel MHV, , Encyclopaedia of Virology, Ed 3 Elsevier, Oxford, pp 72–76 [Google Scholar]
- MacFarlane SA. (2010) Tobraviruses—plant pathogens and tools for biotechnology. Mol Plant Pathol 11: 577–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli GP, Adams MJ, Kreuze JF, Dolja VV. (2007) Family Flexiviridae: a case study in virion and genome plasticity. Annu Rev Phytopathol 45: 73–100 [DOI] [PubMed] [Google Scholar]
- Martin-Hernandez AM, Baulcombe DC. (2008) Tobacco rattle virus 16-kilodalton protein encodes a suppressor of RNA silencing that allows transient viral entry in meristems. J Virol 82: 4064–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Priego L, Donaire L, Barajas D, Llave C. (2008) Silencing suppressor activity of the Tobacco rattle virus-encoded 16-kDa protein and interference with endogenous small RNA-guided regulatory pathways. Virology 376: 346–356 [DOI] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. (2008) Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett P. (2009) Mechanisms of recognition in dominant R gene mediated resistance. Adv Virus Res 75C: 1–33 [DOI] [PubMed] [Google Scholar]
- Moffett P, Farnham G, Peart J, Baulcombe DC. (2002) Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J 21: 4511–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Melnyk CW, Bassett A, Hardcastle TJ, Dunn R, Baulcombe DC. (2010) Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328: 872–875 [DOI] [PubMed] [Google Scholar]
- Morel JB, Godon C, Mourrain P, Beclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H. (2002) Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omarov RT, Ciomperlik JJ, Scholthof HB. (2007) RNAi-associated ssRNA-specific ribonucleases in Tombusvirus P19 mutant-infected plants and evidence for a discrete siRNA-containing effector complex. Proc Natl Acad Sci USA 104: 1714–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart JR, Cook G, Feys BJ, Parker JE, Baulcombe DC. (2002) An EDS1 orthologue is required for N-mediated resistance against tobacco mosaic virus. Plant J 29: 569–579 [DOI] [PubMed] [Google Scholar]
- Pruss G, Ge X, Shi XM, Carrington JC, Bowman Vance V. (1997) Plant viral synergism: the potyviral genome encodes a broad-range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell 9: 859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Ye X, Morris TJ. (2008) Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc Natl Acad Sci USA 105: 14732–14737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robaglia C, Caranta C. (2006) Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci 11: 40–45 [DOI] [PubMed] [Google Scholar]
- Scholthof HB, Alvarado VY, Vega-Arreguin JC, Ciomperlik J, Odokonyero D, Brosseau C, Jaubert M, Zamora A, Moffett P. (2011) Identification of an ARGONAUTE for antiviral RNA silencing in Nicotiana benthamiana. Plant Physiol 156: 1548–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda S, Koiwa H, Kanda K, Kato H, Shimono M, Nishiguchi M. (2000) The helper component-proteinase of sweet potato feathery mottle virus facilitates systemic spread of Potato virus X in Ipomoea nil. Phytopathology 90: 944–950 [DOI] [PubMed] [Google Scholar]
- Szittya G, Moxon S, Pantaleo V, Toth G, Rusholme Pilcher RL, Moulton V, Burgyan J, Dalmay T. (2010) Structural and functional analysis of viral siRNAs. PLoS Pathog 6: e1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y. (2008) The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol 49: 493–500 [DOI] [PubMed] [Google Scholar]
- Vaistij FE, Jones L. (2009) Compromised virus-induced gene silencing in RDR6-deficient plants. Plant Physiol 149: 1399–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. (2008) Plant ARGONAUTES. Trends Plant Sci 13: 350–358 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2008) Use, tolerance and avoidance of amplified RNA silencing by plants. Trends Plant Sci 13: 317–328 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Lederer C, Baulcombe DC. (2000) A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103: 157–167 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Whitham SA, Anderberg RJ, Chisholm ST, Carrington JC. (2000) Arabidopsis RTM2 gene is necessary for specific restriction of tobacco etch virus and encodes an unusual small heat shock-like protein. Plant Cell 12: 569–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham SA, Quan S, Chang HS, Cooper B, Estes B, Zhu T, Wang X, Hou YM. (2003) Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J 33: 271–283 [DOI] [PubMed] [Google Scholar]
- Wu Q, Wang X, Ding SW. (2010) Viral suppressors of RNA-based viral immunity: host targets. Cell Host Microbe 8: 12–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH. (2006) Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev 20: 3255–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.