Abstract
Plant phosphate transporters (PTs) are active in the uptake of inorganic phosphate (Pi) from the soil and its translocation within the plant. Here, we report on the biological properties and physiological roles of OsPht1;8 (OsPT8), one of the PTs belonging to the Pht1 family in rice (Oryza sativa). Expression of a β-glucuronidase and green fluorescent protein reporter gene driven by the OsPT8 promoter showed that OsPT8 is expressed in various tissue organs from roots to seeds independent of Pi supply. OsPT8 was able to complement a yeast Pi-uptake mutant and increase Pi accumulation of Xenopus laevis oocytes when supplied with micromolar 33Pi concentrations at their external solution, indicating that it has a high affinity for Pi transport. Overexpression of OsPT8 resulted in excessive Pi in both roots and shoots and Pi toxic symptoms under the high-Pi supply condition. In contrast, knockdown of OsPT8 by RNA interference decreased Pi uptake and plant growth under both high- and low-Pi conditions. Moreover, OsPT8 suppression resulted in an increase of phosphorus content in the panicle axis and in a decrease of phosphorus content in unfilled grain hulls, accompanied by lower seed-setting rate. Altogether, our data suggest that OsPT8 is involved in Pi homeostasis in rice and is critical for plant growth and development.
Phosphorus (P) is one of the major macronutrients for plant growth and development. The acquisition process of inorganic phosphate (Pi) by plant roots is accomplished through its active absorption via the Pi transporters into the epidermal and cortical cells of the root. Once in the root cortical cells, Pi must eventually be loaded into the apoplastic space of the xylem and transported to the shoot mediated by Pi transporters (PTs; Marschner, 1995).
A number of plant PT genes have been identified in plants based on amino acid homology with a yeast (Saccharomyces cerevisiae) PT (PHO84) and functionally characterized using yeast mutants lacking endogenous high-affinity PTs or plant suspension cells (Raghothama, 1999; Rausch and Bucher, 2002; Rae et al., 2003). Recently, the Xenopus laevis oocyte expression system was also successfully used for detecting the kinetic properties of plant PTs for Pi transport (Ai et al., 2009; Preuss et al., 2010, 2011). The identified plant PT genes were classified into four families, Pht1 to Pht4. The varied subcellular localizations of the PT genes from the four families (Pht1, plasma membrane; Pht2, chloroplast; Pht3, mitochondria; Pht4, Golgi apparatus) suggest their diverse biological functions for plant growth and development (Rausch and Bucher, 2002). Among all the known PTs, members belonging to the Pht1 family, which are presumed as high-affinity PTs, are studied more intensively (Paszkowski, 2006; Bucher, 2007).
In Arabidopsis (Arabidopsis thaliana), only two of the nine Pht1 Pi transporters have been functionally characterized (Misson et al., 2004; Shin et al., 2004; Catarecha et al., 2007). AtPht1;1 and AtPht1;4 play significant roles in Pi acquisition from both low- and high-Pi environments. Thirteen putative high-affinity Pi transporter genes belonging to the Pht1 family (OsPT1–OsPT13) have been identified in the rice (Oryza sativa) genome (Goff et al., 2002). Two of them, OsPT11 and OsPT13, were exclusively induced in roots by inoculation with arbuscular mycorrhiza fungi (Paszkowski et al., 2002; Glassop et al., 2005; Güimil et al., 2005). In our previous report, we demonstrated that two Pi starvation-responsive Pht1 members in rice, OsPT2 and OsPT6, have different functions and kinetic properties in Pi uptake and translocation (Ai et al., 2009). OsPT6 is broadly involved in Pi uptake and translocation through the plants. However, OsPT2, unlike other Pht1 members, is a low-affinity Pi transporter that might mainly play roles during the Pi translocation process (Ai et al., 2009). Overexpession of OsPT2 can cause overaccumulation of shoot Pi in rice and thus a Pi toxicity phenotype (Liu et al., 2010).
Components in the Pi starvation signaling pathways under the regulation of AtPHR1 were continuously unveiled during the past decades in Arabidopsis (Schachtman and Shin, 2007). AtPHR1, a transcription factor with a MYB domain, is a key regulator in the Pi signaling pathway (Rubio et al., 2001). Overexpression of AtPHR1 leads to increased concentrations of Pi in the shoot tissues together with induction of a range of Pi starvation-induced genes that encode Pi transporters, phosphatases, and RNases (Nilsson et al., 2007). The functional ortholog gene of AtPHR1 in rice (designated as OsPHR2) was also identified, and it was found that overexpression of OsPHR2 results in excessive accumulation of Pi in shoots and up-regulation of some OsPht1 genes under the Pi-sufficient condition (Zhou et al., 2008; Liu et al., 2010). Wang et al. (2009) and Liu et al. (2010) reported that suppression of OsSPX1 resulted in Pi accumulation in shoots, similar to that found in OsPT2 and OsPHR2 overexpressor and the ospho2 mutant. Subsequently, a new regulatory mechanism for Pi starvation signaling in plants has been proposed in which OsSPX1 suppresses OsPHR2 function in the expression of OsPT2 and Pi homeostasis in rice shoots via a negative feedback regulation (Liu et al., 2010).
Although the functions and regulatory mechanisms of plant Pht1 genes have been widely studied and elucidated, a large amount of work is needed to dissect the biological roles of each member. In this study, we comprehensively investigated the effects of OsPT8 on Pi acquisition and plant growth and development. Our results showed that OsPT8 is a high-affinity PT. Both enhanced and suppressed expression of this gene inhibited rice growth. Pi uptake were significantly enhanced in OsPT8-overexpressing plants and decreased in OsPT8 knockdown mutants. These data demonstrate that OsPT8 plays a critical role in Pi homeostasis in rice.
RESULTS
Expression Response of the OsPT8 Gene to Pi Starvation
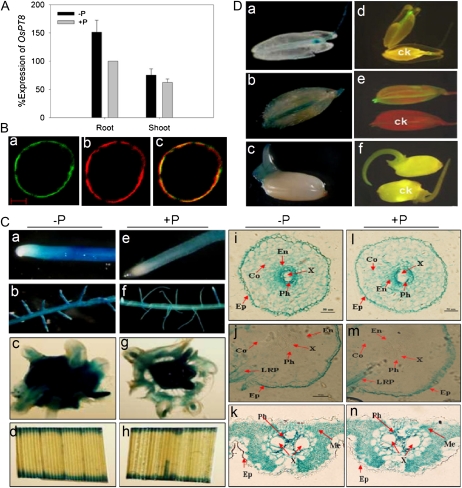
To examine the expression response of OsPT8 to Pi supply status, both reverse transcription (RT)-PCR (data not shown) and quantitative real-time RT-PCR (qRT-PCR) analyses were used to detect the expression patterns of OsPT8. Our results showed that the expression of OsPT8 was abundantly expressed in roots and moderately in shoots grown in Pi-sufficient solution. We detected with several biological replicates that OsPT8 expression in roots was up-regulated distinctly under Pi deprivation, while its expression in shoots was relative stable and not apparently affected by Pi supply status (Fig. 1A).
Figure 1.
Expression pattern in response to Pi availability, and subcellular and tissue localization analysis of OsPT8. A, Detection of the expression level of OsPT8 in wild-type plants by analysis. Total RNA was extracted from rice plants grown for 21 d in the nutrient solution with 0.3 mm Pi (HP; +P) or 0.015 mm Pi (LP; −P). PCR were performed with specific primers for OsPT8 (Supplemental Tables S1 and S2). The expression of OsActin was used as an internal control. B, Expression of cauliflower mosaic virus 35S promoter::OsPT8::GFP fusion genes in rice protoplast. a, Confocal image of the protoplast under the GFP channel showing the plasma membrane localization of 35S::OsPT8::GFP. b, The red fluorescence reflects the position of the plasma membrane, as indicated by the plasma membrane-specific dye 1007PM-rk. c, The merged image of a and b. Bar = 100 nm. C, GUS staining observation of transgenic rice plants harboring the OsPT8 promoter::GUS fusion. Expression is shown in different tissues of rice supplied with 0.3 mm Pi (HP; +P [e–h]) or 0.015 mm Pi (LP; −P [a–d]) for 21 d. a and e, Root tip. b and f, Lateral root branching zone. c and g, Hand-cut cross sections of the root-shoot junction. d and h, Leaf blade. i and l, Root tip cross sections of Pi-deficient (i) and Pi-sufficient (l) plants showing GUS activity in the epidermis (Ep), cortex (Co), endodermis (En), phloem (Ph), and xylem (X). j and m, Cross sections of the lateral root branching zone of Pi-deficient (j) and Pi-sufficient (m) plants showing GUS activity only in the phloem, xylem, and lateral root primordium (LRP). k and n, Cross sections of leaf blade from Pi-deficient (k) and Pi-sufficient (n) plants. Leaf cell types showing GUS expression include the phloem, xylem, and mesophyll (Me) cells. D, Tissue localization of OsPT8 promoter::GUS and OsPT8 promoter::GFP expression in stamens (a and d), caryopsis (b and e), and 3-d-old germinated seeds (c and f) of Pi-sufficient plants. ck, Stamens (d), caryopsis (e), and germinated seeds (f) of wild-type plants.
Subcellular and Tissue Localization of OsPT8
Plant Pht1 members were predicted to be localized to the plasma membrane. To verify its subcellular localization, we constructed C-terminal GFP fusions driven by the cauliflower mosaic virus 35S promoter and transfected the derived expression vector into rice protoplast. As expected, the fused protein was restricted to the plasma membrane by microscopic observation (Fig. 1B), confirming the potential transport activity of OsPT8.
For histochemical analysis, the 2,184-bp promoter and 5′ untranslated region of OsPT8 (pOsPT8) was amplified and fused to a GUS and GFP reporter gene. Subsequently, the constructs were transformed into rice (cv Nipponbare). The transgenic plants carrying pOsPT8::GUS/GFP were cultured in nutrient solution with either high Pi (HP; 300 μm Pi) or low Pi (LP; 15 μm Pi) and stained for GUS activity. Strong GUS activity was detected in root tips, lateral roots, leaves, stamens, caryopses, and germinated seeds under both HP and LP conditions (Fig. 1C). The GUS activity was slightly enhanced by Pi starvation, which was consistent with the expression patterns determined by RT-PCR and qRT-PCR (Supplemental Fig. S2). The localization indicated by the GFP reporter gene (Supplemental Fig. S1) was consistent with that observed in pOsPT8::GUS plants.
Functional Assay of OsPT8 in Yeast and Oocyte Cells
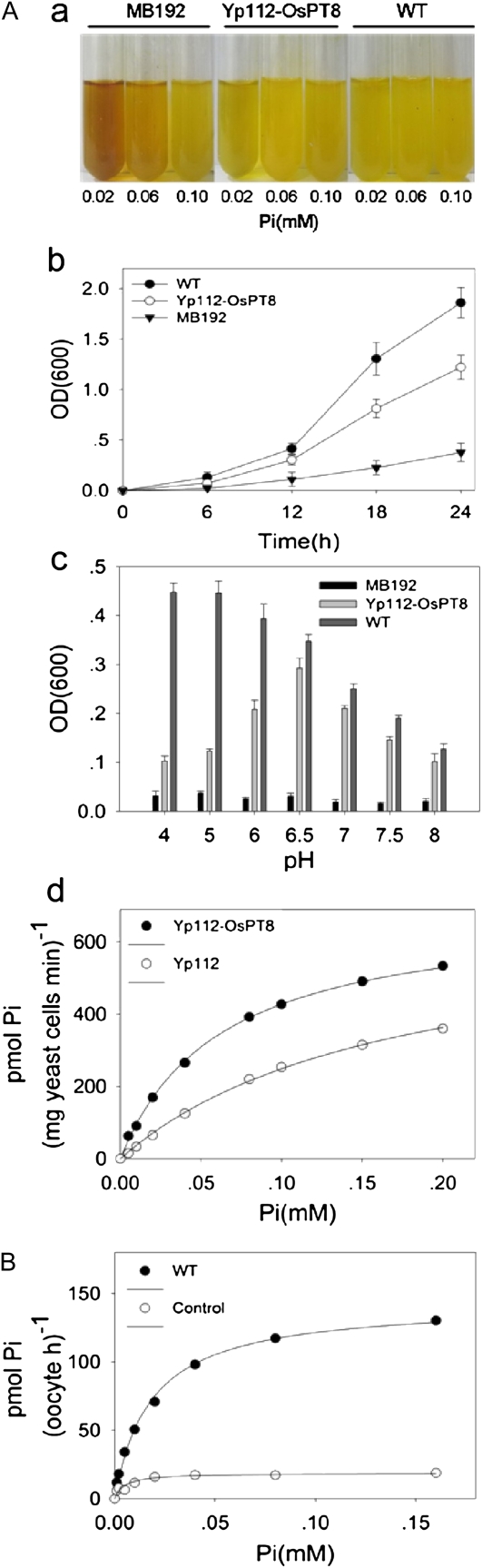
For evaluation of the Pi transport activity of OsPT8, we isolated the coding sequence of OsPT8 and cloned it into the yeast expression vector p112A1NE. The construct was then transferred to a yeast mutant strain, MB192, which is defective in high-affinity Pi transport. The mutant cells expressing with OsPT8 (Yp112-OsPT8) could partially restore their growth at 20 μm Pi and grew well at 60 μm Pi in comparison with the cells of both wild-type plants and the mutant MB192 (Fig. 2A, a and b). There was no obvious diversity in growth at 100 μm Pi concentration. Both the wild type and the MB192 yeast strain expressing OsPT8 grew much faster at pH 4 to 6 when compared with pH 7 and 8 (Fig. 2Ac). The pH optimum for the yeast mutant cells carrying Yp112-OsPT8 was 6.5, whereas in comparison, for the wild type it was pH 4 to 5. The data indicate the different proton (H+) dependence or the change in affinity or structure of OsPT8 and the endogenous high-affinity yeast PTs at various external pH values.
Figure 2.
Functional expression of OsPT8 in yeast and oocytes. A, Functional characterization of OsPT8 in a yeast mutant. a, Staining test for acid phosphatase activity in the yeast strain MB192 (control), Yp112-OsPT8, which contains OsPT8 in MB192, and the wild type (WT). The culture medium contains 0.02, 0.06, and 0.10 mm Pi, respectively. b, Growth curves of the wild type, MB192, and MB192 transformed with Yp112-OsPT8 generated from a 24-h culture under 60 μm Pi. c, Effects of different pH levels in the culture medium on the growth of the three yeast strains: Yp112-OsPT8, MB192, and the wild type. d, Velocity of 33Pi transport by Yp112-OsPT8 as a function of Pi concentration. The nonlinear regression of Pi uptake of strain Yp112-OsPT8 versus the external concentration at pH 6.5 was used to estimate the apparent Km value for Pi uptake. OD(600), Optical density at 600 nm. B, Functional characterization of OsPT8 in Xenopus oocytes. The absorption of 33Pi by oocytes injected with OsPT8 mRNA and water is shown.
To determine the kinetic properties of OsPT8, Pi uptake experiments using 33Pi were performed using the transformed yeast MB192. To avoid the effects of other endogenous PTs, particularly the low-affinity PTs in MB192, we show the Pi transport dynamics of the strains transformed with both OsPT8 and empty vector up to 0.2 mm Pi supply. The OsPT8 mediated 33Pi uptake velocities after subtracting the Pi transport with an empty vector following the Michaelis-Menten kinetics equation (Fig. 2Ad). The apparent mean Km value for Pi transport of OsPT8 was 23 μm Pi, as determined by three independent experiments (Fig. 2Ad).
To confirm that OsPT8 is a putative high-affinity Pi transporter, we further assayed its function in Pi transport in a Xenopus oocyte expression system following the method described by Ai et al. (2009). The oocytes injected with mRNA encoding OsPT8 were compared with water-injected controls in Pi uptake assays. The 33Pi uptake velocities of OsPT8 in the oocytes, after subtracting the Pi transport of the water-injected controls from its concentration in the external bath solution, showed typical Michaelis-Menten kinetics (Fig. 2B). The predicted mean Km resulting from three oocyte expression experiments was 27 μm for Pi transport of OsPT8. Both the yeast and oocyte expression systems suggest that OsPT8 has a high Pi affinity, mediating Pi uptake in the micromolar range (Fig. 2).
Transgenic Rice Showing Growth Inhibition by Overexpression or Suppression of OsPT8
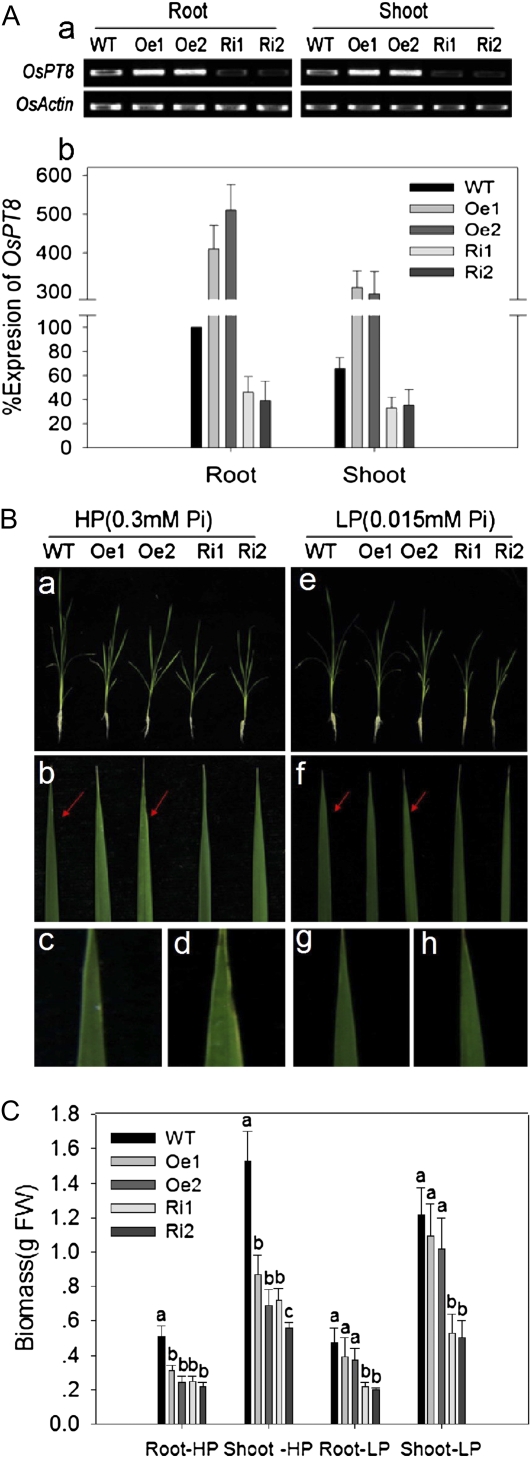
To further characterize the role of OsPT8 for maintaining Pi homeostasis in planta, transgenic lines were generated by introducing the OsPT8 overexpression construct or the OsPT8 RNA interference construct into the japonica cv Nipponbare. Overexpression and knockdown efficiency of OsPT8 in the transgenic plants were confirmed by RT-PCR and qRT-PCR analyses. Two independent transgenic lines, OsPT8-Oe detected by Southern blot (Supplemental Fig. S3) and OsPT8-Ri, were selected for further experimental analysis. Under the Pi-sufficient condition, the OsPT8-Oe plants showed enhanced amounts of the transcript compared with the wild-type plants, whereas the transcript abundance decreased significantly in OsPT8-Ri (Fig. 3A, a and b).
Figure 3.
Expression of OsPT8 in transgenic plants, and characterization of the wild-type and transgenic plants. A, Expression of OsPT8 in transgenic plants. Detection of the transcript abundance of OsPT8 in wild-type (WT) and transgenic plants is shown by RT-PCR (a) and qRT-PCR (b). Ten-day-old seedlings were transferred to nutrient solution containing 0.3 mm Pi (HP) or 0.015 mm Pi (LP) for 21 d. RNA was extracted from the roots and shoots of the seedlings. Oe1, Oe2, Ri1, and Ri2 represent independent OsPT8-overexpressing and RNA interference lines. The expression of OsActin was used as an internal control. Relative expression levels are shown in percentage as compared with the wild type as 100% expression. B, Characterization of wild-type and transgenic plants. Plants were grown in nutrient solution to which 0.3 mm Pi (HP) or 0.015 mm Pi (LP) was added for 21 d. a and e, Seedlings of wild-type and transgenic plants. b to d and f to h, Mature leaves of wild-type and transgenic plants. Red arrows indicate the sites of the mature leaves for phenotype (with or without Pi toxicity symptoms: chlorosis and necrosis) observation of wild-type and OsPT8-Oe plants. c, d, g, and h, Enlarged images for the corresponding sites indicated by the red arrows in b and f. C, Root and shoot biomass of wild-type and transgenic plants under HP and LP conditions. Biomass measurements were obtained from the roots and shoots of 21-d-old seedlings of wild-type and transgenic plants grown in nutrient solution to which 0.3 mm Pi (HP) or 0.015 mm Pi (LP) was added. Five plants per line were measured. FW, Fresh weight.
A stunted growth of 30-d-old OsPT8-Oe and -Ri plants under either HP (300 μm) or LP (15 μm) supply was observed. Under the HP condition, the root and shoot biomass of the 30-d-old OsPT8-Oe and -Ri plants were significantly lower than those of wild-type plants (Fig. 3C), whereas the root-shoot ratio remained the same regardless of the altered expression of OsPT8 (data not shown). It is noteworthy that under HP supply, OsPT8-Oe plants showed Pi toxic symptoms (necrotic leaves; Fig. 3B, b and d). Under the LP condition, no significant difference in biomass was found between wild-type and OsPT8-Oe plants, while the root and shoot biomass of OsPT8-Ri plants was only about 40% to 50% of the wild-type value (Fig. 3C).
Alteration of Pi Uptake and Translocation in OsPT8 Transgenic Plants
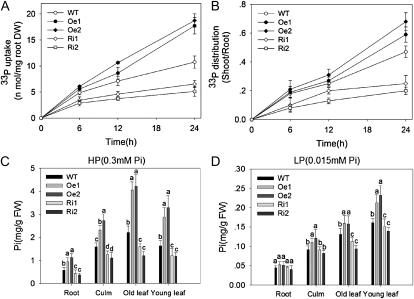
To determine the function of OsPT8 in Pi uptake and translocation, Pi uptake was first monitored during a 24-h period in 30-d-old transgenic materials supplied with 300 μm Pi (HP condition supplemented with 33P). The OsPT8-Oe plants exhibited a significant increase in Pi uptake as compared with the wild-type plants. In contrast, Pi uptake was impaired in the OsPT8-Ri plants (Fig. 4A). These results suggested that OsPT8 plays a crucial role in Pi uptake. To examine the Pi translocation in OsPT8-Oe and OsPT8-Ri plants, the shoot-root ratio of 33P was measured under the same Pi supply as that used in the Pi uptake experiment. The shoot-root ratio of 33P was moderately increased (especially after 24 h of Pi uptake) in OsPT8-Oe plants, whereas it was significantly decreased in OsPT8-Ri plants (Fig. 4B), indicating that OsPT8 is also required for Pi translocation within plants.
Figure 4.
Pi uptake in OsPT8-overexpressing and RNA interference lines. A, Pi uptake activity of wild-type (WT) and transgenic plants. DW, Dry weight. B, Shoot-root ratios of the 33P taken up by wild-type and transgenic plants. Error bars represent sd (n = 3). C and D, Pi contents in root, culm, old leaf, and young leaf were measured in 21-d-old seedlings of wild-type and transgenic plants grown in nutrient solution to which 0.3 mm Pi (HP) or 0.015 mm Pi (LP) was added. Five plants per line were measured. Error bars represent sd (n = 5). FW, Fresh weight.
Pi concentration in roots, stems, old leaves, and young leaves was also measured under HP and LP conditions. Under the HP condition, Pi concentration in all these tissues was elevated in OsPT8-Oe plants and decreased in OsPT8-Ri plants (Fig. 4C). Such effects of altered OsPT8 expression on Pi concentration in different organs became less significant between OsPT8-Oe/-Ri and wild-type plants under the LP condition (Fig. 4D). However, the total amount of Pi accumulated in the whole plant was significantly decreased by OsPT8-Ri, confirming that OsPT8 could play its high-affinity Pi transporter role in rice.
Function of OsPT8 in Pi Translocation from Vegetative Organs to Reproductive Organs in Rice
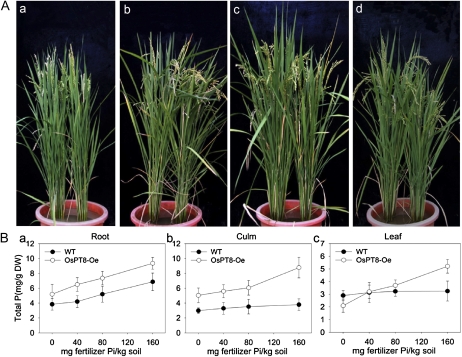
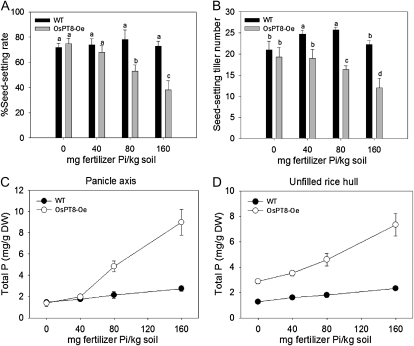
For phenotypic observation during the entire plant growth period, soil pot experiments were carried out in a greenhouse. Four levels of available Pi extracted by the Bray I method before planting in soil were designed for the experiment: 6.5 mg kg−1 (no fertilizer Pi was added), 13.5 mg kg−1 (40 mg fertilizer Pi kg−1 soil), 22.5 mg kg−1 (80 mg fertilizer Pi kg−1 soil), and 35.1 mg kg−1 (160 mg fertilizer Pi kg−1 soil). We observed that all three independent lines of OsPT8-Oe plants showed stunted growth at 80 and 160 mg fertilizer Pi kg−1 soil. We measured the growth parameters, including plant height, panicle length, maximum tiller number, seed-setting tiller number, and seed-setting rate, as well as total P concentration in root, culm, leaves, panicle axis, and unfilled rice hull (Figs. 5 and 6; Supplemental Figs. S5–S7) at the harvest stage for one of the three OsPT8-Oe lines. At high P level, most parameters of OsPT8-Oe plants were suppressed as compared with wild-type plants, especially maximum tiller number (1.4-fold lower), seed-setting tiller number (1.9-fold lower), and seed-setting rate (2-fold lower; Fig. 6, A and B; Supplemental Fig. S5, A–C). The toxicity phenotypes (suppressed physiological parameters) were gradually recovered with decreasing Pi supply from moderate to low levels. In general, the total P concentration increased in root, culm, leaves, panicle axis, and unfilled rice hull with the increasing P level in the soil from 0 to 160 mg fertilizer Pi kg−1 (Fig. 5B). The OsPT8-Oe plants contained 2.3- and 3.2-fold higher total P concentration in the culms and panicle axis, respectively, than wild-type plants grown at high P level. Overexpression of OsPT8 resulted in an increase of the total P concentration in unfilled rice hulls by 2- to 3-fold at both high-P and low-P levels (Fig. 6, C and D). In addition, we found that 17 of 20 independent OsPT8-overexpressing lines in a field experiment showed the same phenotype change as that observed in the pot experiment. All four lines that we analyzed showed significant growth suppression and increased total P concentration in their reproductive organs grown in HP acid red soil containing 20.3 mg available Pi kg−1 soil (Supplemental Table S5). The data suggested that OsPT8 might contribute to Pi translocation from the vegetative organs to the reproductive organs in rice.
Figure 5.
Growth performances of the wild-type and OsPT8-overexpressing lines at different Pi levels in a pot experiment. A, Growth performance of wild-type and OsPT8-overexpressing plants at 0 mg fertilizer Pi kg−1 soil (a), 40 mg fertilizer Pi kg−1 soil (b), 80 mg fertilizer Pi kg−1 soil (c), and 160 mg fertilizer Pi kg−1 soil (d). Two seedlings were grown in each pot. The left one is the wild-type plant and the right one is the OsPT8-Oe plant. The photographs are representatives of five independent biological replicates. B, Total P concentration in root, culm, and leaf of wild-type (WT) and OsPT8-overexpressing plants at different Pi levels in the above pot experiment. Error bars represent sd (n = 5). DW, Dry weight.
Figure 6.
Physiological parameters of wild-type and OsPT8-overexpressing plants. A and B, Percentage seed-setting rate per panicle and seed-setting tiller number of wild-type (WT) and OsPT8-overespressing plants at different Pi levels in the pot experiment (Fig. 5A). C and D, Total P concentration in panicle axis and unfilled rice hull of wild-type and OsPT8-overexpressing plants at different Pi levels in the pot experiment (Fig. 5A). DW, Dry weight.
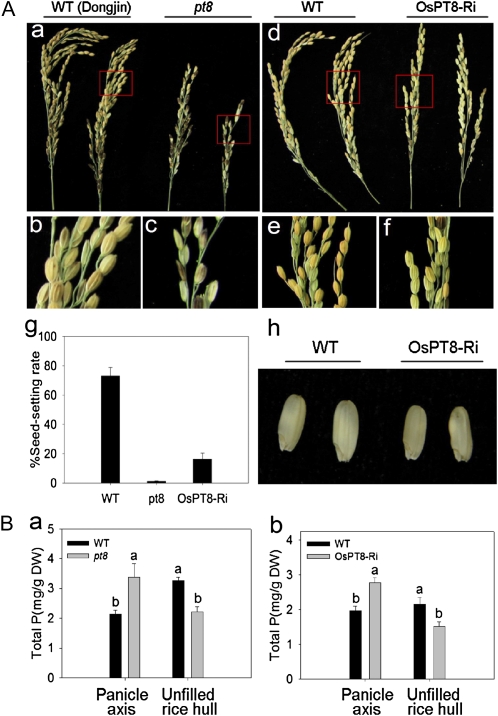
To confirm the effect of OsPT8 on Pi transport from vegetative organs to reproductive organs, OsPT8-Ri plants as well as an OsPT8 T-DNA insertion mutant (pt8) were used for further study. Field experiments were carried out in HP acid red soil (available Pi of 20.3 mg kg−1 soil) in our experimental station. Most growth and developmental parameters, especially seed-setting rate, of OsPT8-Ri and pt8 were decreased in comparison with wild-type plants. We acquired no and very few filled grains in the pt8 homozygous mutant and OsPT8-Ri, respectively (Fig. 7A, a–g), and the grain size of OsPT8-Ri lines was smaller and thinner compared with that of wild-type plants (Fig. 7Ah). The pt8 and OsPT8-Ri mutants accumulated about 30% higher P than their respective wild types in the panicle axis (Fig. 7Ba). In contrast, the total P of the unfilled rice hull in the mutants was decreased to about only 70% of that in wild-type plants (Fig. 7Bb). These data imply that suppression of OsPT8 expression impaired the translocation of Pi from the panicle axis to the grains.
Figure 7.
Panicle-filling performance and total P concentration in panicle axis and unfilled rice hull of the wild-type (WT) and OsPT8 knockdown plants. A, Seed-setting rate and seed size of OsPT8 T-DNA insertion mutant and -Ri plants. A, a to f, Phenotypes of seed-setting performance in pt8 and OsPT8-Ri plants. g, Percentage seed-setting rate per panicle in pt8 and OsPT8-Ri at high P level (available Pi of 20.3 mg kg−1 soil) in the field experiment. h, Decreased size of the brown rice seed of OsPT8-Ri plants. B, Total P concentration in panicle axis and unfilled rice hull of the wild type, pt8 (cv Dongjin), and the OsPT8-Ri line (cv Nipponbare) at sufficient Pi levels in the field experiment. pt8 represent the homozygous OsPT8 T-DNA insertion mutant. Error bars represent sd (n = 5). DW, Dry weight.
DISCUSSION
Plants acquire Pi by its active uptake into the epidermal and cortical cells of the root via the H+/Pi symporters of the Pht1 family (Raghothama, 2000; Poirier and Bucher, 2002). In rice, 13 Pht1 members (named as OsPT1–OsPT13) have been isolated (Goff et al., 2002; Paszkowski et al., 2002); however, only two of them (OsPT2 and OsPT6), with the most abundant transcripts among the Pi starvation-regulated Pht1 genes in rice roots, were functionally characterized (Ai et al., 2009). In this work, we provide direct evidence that OsPT8 is a high-affinity Pi transporter and plays important roles in both the acquisition of Pi from the external environment and the translocation of Pi within plants.
OsPT8 Is a High-Affinity Pi Transporter
It has been well recognized that plant PT functions in a H+/Pi cotransport manner with a stoichiometry of 2 to 4 H+/Pi (Rausch and Bucher, 2002). Based on this property, its transport activity could be monitored by using heterologous expression systems, such as plant cells (Leggewie et al., 1997; Mitsukawa et al., 1997; Rae et al., 2003), yeast mutants defective in high-affinity Pi transport (Daram et al., 1998; Liu et al., 1998; Rausch et al., 2001; Harrison et al., 2002), and Xenopus oocytes (Ai et al., 2009; Preuss et al., 2010, 2011). It has been suggested that PT expression in a plant cell has significant interference from the endogenous PT, and its expression in yeast cells might display altered transport properties owing to the existence of a native PT system (Preuss et al., 2011). Therefore, Km values derived from yeast cells were generally much higher than those from plant Pi uptake analyses. Nevertheless, we noticed that the Km values of Pht1 members of plant PTs reported previously ranged from several micromolar (3 μm for AtPT1 in tobacco [Nicotiana tabacum] suspension cells [Mitsukawa et al., 1997] and 9 μm for HvPT1 in barley [Hordeum vulgare] endosperm cells [Rae et al., 2003]) to about 60 to 200 μm obtained in a yeast expression system (64 μm for StPT3 [Rausch et al., 2001], 97 μm for OsPT6 [Ai et al., 2009], and 192 μm for MtPT1 [Liu et al., 1998]). Several plant PT members belonging to the Pht1 family have also been reported as low-affinity PTs. Using a yeast expression system, Harrison et al. (2002) detected that the Km value of MtPT4 from Medicago was 490 to 670 μm, and they claimed MtPT4 as a relatively low-affinity PT. Recently, using a Xenopus oocyte expression system, Ai et al. (2009) and Preuss et al. (2010) characterized that OsPht1;2 from rice and HvPht1;6 from barley function at the millimolar Pi range.
In this study, we detected that OsPT8 has Km values of 23 and 27 μm, respectively, as indicated by yeast mutant complementation and Xenopus oocyte injection experiments (Fig. 2). The Km values are lower than those obtained by a yeast expression system for several other Pht1 members in dicots, which were defined as high-affinity Pi transporters previously (Leggewie et al., 1997; Daram et al., 1998; Liu et al., 1998; Rausch et al., 2001). Preuss et al. (2011) utilized a two-electrode voltage clamp for determining the electrophysiological properties of HvPht1; 1 by the Xenopus oocyte expression system and obtained an even lower Km value (1.9 μm) than that of OsPT8. One possible explanation for the variation in the Km value is technical differences; the other is that the affinity of plant Pht1 members themselves fluctuates, suggesting the functional diversification among the high-affinity PTs of the Pht1 family. Moreover, knockdown of OsPT8 expression caused large decreases of root and shoot biomass and total Pi uptake under the 15 μm Pi supply condition (Fig. 3C), also providing evidence for an in planta role of OsPT8 as a high-affinity PT in rice. Consequently, we still suggest that OsPT8 belongs to the high-affinity PTs of the Pht1 family.
OsPT8 in Planta Functions in Pi Homeostasis
Histochemical analysis by fusing an OsPT8 putative promoter to a GUS reporter gene revealed that it is expressed abundantly in the root epidermis (Fig. 1C, a, b, e, f, i, and l). That expression is very similar to the expression pattern of Pht1;1-4 in Arabidopsis (Mudge et al., 2002; Misson et al., 2004; Shin et al., 2004), indicating that OsPT8 is likely to be involved in Pi uptake from the soil solution. High expression levels of OsPT8 were also detected in root-shoot junctions and leaves (Fig. 1C, c and g), indicating that it may be involved in the translocation of Pi from root to shoot in rice and play a role in the redistribution of Pi to young organs during leaf senescence (Rausch et al., 2004). In addition, we found higher GUS activity in the growing points of germinated seeds, anther, rice hull, and awn (Fig. 1D), suggesting that this PT may be involved in rice pollination, grouting, and Pi release and remobilization. Based on their similar spatial expression patterns, OsPT8 and AtPht1;5 in Arabidopsis are likely to perform a similar role during these processes (Mudge et al., 2002). Altogether, the histochemical analysis led us to assume that OsPT8 plays an important role at all developmental stages involving Pi homeostasis in rice.
Recent reports have suggested that altered expression of several genes causes an accumulation of excessive Pi in the shoots and thus a Pi toxic phenotype (necrotic leaf tip and stunted growth) in rice. These genes include OsPHR2, OsmiR399 (microRNA399), OsPHO2, OsSPX1, and OsPT2 (Ai et al., 2009; Wang et al., 2009; Liu et al., 2010). Overexpression of OsPHR2 or OsmiR399 and repression of OsPHO2 resulted in excessive accumulation of Pi in plant shoots but not in roots (Wang et al., 2009; Liu et al., 2010; Hu et al., 2011) The Pi overaccumulation, caused by the overexpression of OsPT2 and the repression of OsSPX1, was found in shoots and roots but to a different degree (more than 2-fold higher in OsPT2-overexpressing plant roots and 50%–60% higher in OsSPX1 knockdown plant roots; Wang et al., 2009; Liu et al., 2010). Our data here showed that the Pi concentration in the roots, culms, old leaves, and young leaves of OsPT8-Oe plants all increased about 2-fold over that in the wild type. In contrast, the Pi uptake rate and concentration in all plant parts investigated of OsPT8-Ri plants decreased as compared with the wild type (Fig. 4). These results suggest that OsPHR2 and its reciprocal downstream regulation system (OsPHR2-OsIPS1-OsmiR399-OsPHO2) play a role in Pi uptake and translocation in roots, while constitutively enhanced expression of OsPTs and the negative effect of OsSPX1 exert an enhanced Pi uptake and translocation in whole plant tissues.
OsPT8 Is Involved in Pi Translocation from Vegetative Organs to Reproductive Organs in Rice
Pi plays a very important role in the process of fecundation and grouting in rice (Marschner, 1995). Rice plants can accumulate abundant Pi in leaves at the early developmental stage and transport the stored Pi in the leaves to the panicle at the late developmental stage (Marschner, 1995). The soil pot experiments in this study showed that OsPT8 was involved in the translocation of Pi from the panicle axis to the rice hull. The total P concentrations in the panicle axis and in the rice hull were measured in transgenic and wild-type plants grown at different levels of Pi in the soil. The total P concentration in panicle axis of OsPT8-Oe plants remained at the same level as in wild-type plants grown at 0 and 40 mg Pi kg−1 soil (Fig. 6C). However, in 80 and 160 mg Pi kg−1 soil treatments, the total P concentration in the panicle axis was dramatically enhanced in the OsPT8-Oe plants as compared with the wild-type plants (Fig. 6C), and the enhancement correlates well with the increased Pi supply. Furthermore, the rice hull accumulated about 3-fold higher P in the OsPT8-Oe plants than in the wild-type plants irrespective of soil Pi availability (Fig. 6D). Since the seed-setting rate of OsPT8-Ri plants was significantly lower than that of the wild type and no filled seeds were harvested for the OsPT8 T-DNA insertion mutant, we measured P content in the panicle axis and unfilled rice hull in OsPT8-Ri and the T-DNA insertion mutant. The suppression of OsPT8 expression resulted in an increase of P concentration in the panicle axis but a decrease of P concentration in the unfilled rice hull (Fig. 7B, a and b). The impaired translocation of Pi from panicle axis to unfilled rice hull implies that OsPT8 is involved in Pi distribution in rice grains. OsPT8 overexpression or OsPT8 suppression also affected P concentration in the rice hulls and brown rice of the normal filled grains (Supplemental Fig. S6). Taken together, we conclude that OsPT8 not only takes part in the uptake and translocation Pi but also affects the rice grouting process.
Altered Expression of OsPT8 Affects the Expression of Other Pht1 Members in Rice
Translational products of several Pht1 ortholog genes in one plant share the same final destiny (plasma membrane) for their protein trafficking process. Deciphering whether a potential interaction or sensing mechanism exists between these PT genes and/or their protein products is of importance and interest. It has been revealed in the yeast Pi-responsive signal transduction (PHO) pathway that the cells starved for Pi can activate feedback loops that regulate high- and low-affinity Pi transport. Therefore, the interplay of positive and negative feedback loops leads to bistability in Pi transporter usage: individual cells express predominantly either low- or high-affinity transporters, both of which can yield similar Pi uptake capacity. In this study, we attempted to investigate the transcriptional expression alteration of PT genes caused by overexpression or suppression of OsPT8.
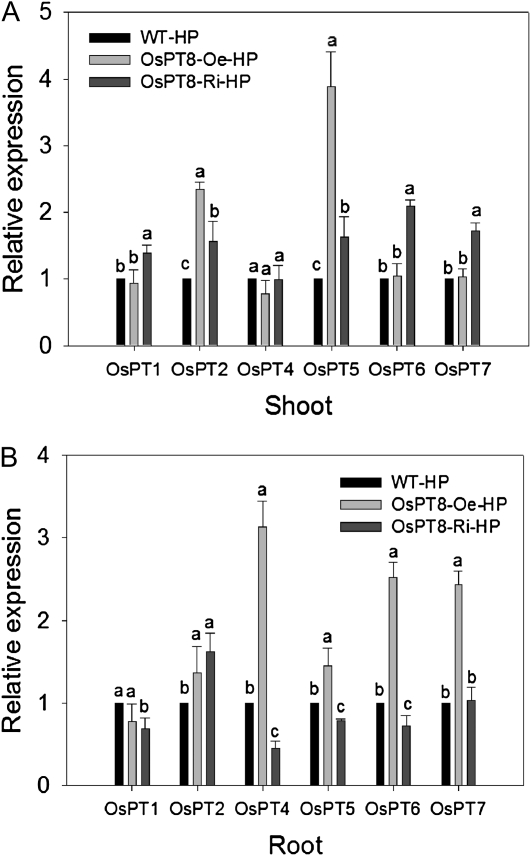
Under the Pi-sufficient condition, a set of PT genes were transcriptionally up- or down-regulated upon overexpression or suppression of OsPT8. It could be questioned whether the altered phenotype and Pi uptake and translocation capacity in OsPT8-Oe and -Ri plants were attributed to, at least partially, its indirect effect on the expression of other Pht1 members. This possibility was investigated by an analysis of the expression of other Pht1 genes (Fig. 8).
Figure 8.
Expression of six members of the rice Pht family in wild-type (WT), OsPT8-Oe, and OsPT8-Ri plants under the HP condition. Effects of OsPT8 overexpression on transcript levels of Pht family members are shown in shoots (A) and roots (B) when grown at HP (0.3 mm Pi) compared with the wild type.
In shoots, the expression levels of OsPT2 and OsPT5 were significantly elevated in OsPT8-Oe plants, which prompted us to assume that the manifestation of OsPT8 function on Pi translocation might require the assistance of OsPT2 and OsPT5 and that they might function as trimers or even polymers. Interestingly, the expression of OsPT2 and OsPT5 as well as OsPT6 and OsPT7 was also increased in OsPT8-Ri plants, indicating a possible functional complementation to OsPT8 by other PTs during Pi translocation. It should be noted that the expression level of OsPT2 in rice shoots is much lower than that of other PTs (Ai et al., 2009; Liu et al., 2010; Hu et al., 2011), suggesting that its contribution could be marginal in OsPT8-Oe and/or -Ri plants. Moreover, since increased expression of OsPT2, OsPT5, OsPT6, and OsPT7 in the shoots of OsPT8-Ri plants (Fig. 8A) did not restore the impaired performance of OsPT8-Ri plants in Pi translocation (Fig. 4B), it could be reasoned that OsPT8 plays a different role from OsPT2, OsPT5, OsPT6, and OsPT7 or that they share only partially functional overlap with OsPT8 with regard to Pi translocation. The expression of OsPT1 and OsPT4 is not significantly changed in shoots of either OsPT8-Oe or -Ri plants, suggesting a probable functional diversification.
In roots, as in the case of OsPT5 in shoots, the transcript abundance of OsPT4, OsPT6, and OsPT7 was enhanced dramatically in OsPT8-Oe plants (Fig. 8B). Thus, it could be assumed that some of the effects on the dynamics of Pi uptake (e.g. increased Pi uptake) may not be the result of OsPT8 overexpression alone but also the indirect effect of altered OsPT4, OsPT6, and OsPT7 expression caused by OsPT8 overexpression, again indicating their potential functional complementation to OsPT8 in Pi uptake. All these results suggested that the Pi transporters might function in a complex combinational way in Pi uptake and translocation.
MATERIALS AND METHODS
RT-PCR and qRT-PCR
Total RNA was extracted from root and shoot tissues using TRIzol reagent (Invitrogen; http://www.invitrogen.com/) according to the manufacturer’s instructions. RT-PCR was performed using gene-specific primers for OsPT8 (accession no. AF536968) and OsActin (accession no. AB047313). qRT-PCR of the two genes was performed using the MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad; http://www.bio-rad.com), and the products were labeled using the SYBR Green master mix (SYBR Premix Ex Tag TM II; TaKaRa Bio; http://www.takara-bio.com) according to the manufacturer’s instructions. All of the primers used for RT-PCR and qRT-PCR are listed in Supplemental Tables S1 and S2.
Transient Expression of OsPTs in Rice Protoplast and Fluorescence Microscopy Imaging
Rice (Oryza sativa) protoplast preparation and transfection followed previously described procedures (Miao and Jiang, 2007) with some modifications. Briefly, 0.2 mL of protoplast suspension (approximately 2 × 105 cells) was transfected with DNA for various constructs (10 μg each). For plasma membrane indication, 10 μg of plasma membrane marker was used in combination with the OsPT8::GFP fusion vectors (Nelson et al., 2007). After transfection, cells were cultured in protoplast medium (R2S + 0.4 m mannitol) overnight (approximately 12 h). Observations were made on a Nikon Eclipse 90i microscope, and images were captured with a SPOT camera. Excitation and emission filters (Ex460-500/DM505/BA510-560 and Ex516/10/DM575/BA590; Nikon) were used for GFP and a monomeric red fluorescent protein (Campbell et al., 2002). Protoplasts were observed under a 60× objective.
Construction of an OsPT8 Promoter Fusion with GUS and GFP, Overexpression and RNA Interference Vectors, and Generation of Transgenic Plants
The putative OsPT8 promoter was amplified from upstream of its coding regions with rice (cv Nipponbare) genomic DNA using the specific primers listed in Supplemental Table S3. The PCR product was digested with AscI and PacI and ligated into the pS1aGUS-3 and pS1aGFP-8 vectors (kindly provided by Dr. Delhaize Schünmann, Commonwealth Scientific and Industrial Research Organization Plant Industry; http://www.pi.csiro.au). For OsPT8 overexpression, the 1,626-bp open reading frame of OsPT8 was amplified using the specific primers listed in Supplemental Table S4 from the Nipponbare cDNA clone (GenBank accession no. J033028K24). The PCR product was digested with HpaI and XhoI and ligated into the pS1aG-4 vector (kindly provided by Dr. Delhaize), driven by a maize (Zea mays) ubiquitin promoter, with a nopaline synthase terminator. For the OsPT8 RNA interference construct, a 221-bp fragment of the OsPT8 coding sequence was amplified using the specific primers listed in Supplemental Table S4 from the Nipponbare cDNA clone (GenBank accession no. J033028K24). The PCR product was cleaved with BamHI, KpnI, SacI, and SpeI and ligated into pTCK303 expression vector (Ai et al.,2009). The above constructs were transferred to Agrobacterium tumefaciens strain EHA105 by electroporation and then transformed into rice as described previously (Upadhyaya et al., 2000).
Southern-Blot Analysis
The independent transgenic lines with overexpression of OsPT8, namely OsPT8-Oe1 and OsPT8-Oe2, were determined by Southern-blot analysis. Genomic DNA was extracted from leaves of wild-type and T1 transgenic plants using the SDS method, and 8 μg of genomic DNA was digested with the restriction enzyme HindIII overnight at 37°C. The digested DNA was separated on a 0.8% (w/v) agarose gel, transferred to a Hybond-N+ nylon membrane, and hybridized with the coding sequence of the hygromycin-resistant gene used as the hybridization probe following the procedures described previously (Zhou et al., 2008).>
Hydroponic and Pot Experiments
Rice seeds were surface sterilized in a 30% (v/v) hydrogen peroxide solution for 30 min, washed, and germinated for 3 d at 25°C in the dark (Li et al., 2006). The 10-d-old seedlings were transferred to nutrient solution containing 1.25 mm NH4NO3, 0.35 mm K2SO4, 1 mm CaCl2·2H2O, 1 mm MgSO4·7H2O, 0.5 mm Na2SiO3·9H2O, 20 μm Fe-EDTA, 20 μm H3BO3, 9 μm MnCl2·4H2O, 0.32 μm CuSO4·5H2O, 0.77 μm ZnSO4·7H2O, and 0.39 μm Na2MoO4·2H2O, pH 5.5, supplemented with 0.3 mm Pi (HP) or 0.015 mm Pi (LP). The hydroponic experiments were carried out in a growth room with a 16-h-light (30°C)/8-h-dark (22°C) photoperiod, and the relative humidity was controlled at approximately 70%. The solution was refreshed every 3 d.
The soil pot experiment was performed with four replications in a greenhouse using the soil collected from an experimental farm of Nanjing Agricultural University. The acid soil (pH 5.0, soil:water = 1:1) contained 6.5 mg Pi kg−1 extracted by the Bray I method (Bray and Kurtz, 1945). One wild-type plant and one OsPT8-Oe plant were grown in each pot containing 16 kg of air-dried soil. The Pi supply levels to the plants were 0, 40, 80, and 160 mg fertilizer Pi kg−1 soil.
Histochemical Localization of GUS Expression
The histochemical analysis was performed as described previously (Ai et al., 2009). The stained tissues were photographed using an Olympus MVX10 stereomicroscope with a color CCD camera (http://www.olympus-global.com). For the experiments of subcellular expression patterns, the stained tissues were rinsed and fixed in formalin:acetic acid:70% ethanol (1:1:18) for 24 h, embedded in paraffin, and then sectioned. The sections (15 μm thick) were transferred onto a slide and visualized with an Olympus BX51T stereomicroscope with a color CCD camera (Olympus).
Functional Complementation Assay of OsPT8 in Yeast
The yeast mutant MB192 defective in the high-affinity Pi transporter gene PHO84 (Bun-Ya et al., 1991) was used for functional complementation assay of OsPT8, following the protocol described previously (Ai et al., 2009). The yeast expression vector carrying the OsPT8 open reading frame was transformed into MB192. The MB192-OsPT8 and control cells were grown to the logarithmic phase and then subjected to yeast nitrogen base liquid medium containing different Pi concentrations (20, 60, and 100 μm) evenly. Bromcresol purple was used as pH indicator in the medium. To substantiate the pH dependence of Pi uptake, different extracellular pH values from 4 to 8 were used at a fixed amount of 80 μm K2HPO4.
For 33P uptake experiments in yeast, about 1-mg fresh yeast cell samples were used following the previously described method (Ai et al., 2009).
Functional Assay of OsPT8 in Oocytes
The full-length cDNA of OsPT8 was amplified and subcloned into the EcoRV and SpeI sites of the oocyte expression vector pT7TS (Cleaver et al., 1996). The plasmid was linearized using XbaI (TaKaRa) and used as a template for the synthesis of capped copy RNA using a Message Machine T7 kit (Ambion). The stage V to VI defolliculated oocytes from Xenopus laevis were isolated and maintained as described previously (Tong et al., 2005). For gene expression in oocytes, 50 ng of mRNA was injected, whereas 50 ng of water was injected as a control. After injection, all oocytes were incubated at 18°C in ND-96 (pH 7.2). The solution was supplemented with 5 mg L−1 doxycyclin. Pi transport experiments were performed 3 d after injection.
The oocyte was exposed for 3 h to ND-96 solution containing nine different Pi (from 1 to 160 μm NaH2PO4, pH 7.2) and 33P (H3PO4; specific activity of 0.5 μCi per 0.1 μmol of Pi, about 1.11 × 104 dpm nmol−1) levels. At the end of the uptake period, oocytes were washed five times in ice-cold ND-96. The oocyte was placed in a scintillation vial and lysed in 250 μL of 10% SDS. 33P activities of individual oocytes were counted using a Beckman LS6500 scintillation counter. Seven to 10 oocytes were used for each Pi uptake experiment.
pt8 Mutant Identification
A T-DNA insertion mutant line was requested from RiceGE (the Rice Functional Genomic Express Database) in Korea (http://signal.salk.edu/cgi-bin/RiceGE). Based on the insertional information, two primers flanking the T-DNA borders and one primer specifically for T-DNA were used to confirm the insertional site (Supplemental Fig. S4). To determine the expression of OsPT8 in the mutant, RT-PCR analysis was performed using primers designed from the gene sequence (Supplemental Fig. S4).
Radioactive 33P Uptake Assay in Transgenic Plants
Seedlings of OsPT8-Oe, OsPT8-Ri, and wild-type plants that had been subjected to HP treatment for 3 weeks were incubated for 6, 12, and 24 h in 250 mL of the same nutrient solution as described above containing 16 μCi of H333PO4. After incubation, the seedlings were washed thoroughly in sterile distilled water, and the roots and shoots were dried and weighed separately. The tissues were dried at 70°C for 2 d and then wet digested in a mixture of H2SO4 and hydrogen peroxide. The radioactivity of these solutions was measured with a Beckman LS6500 scintillation counter.
Measurement of Pi Concentration and Total P Concentration in Plants
For the measurement of unassimilated Pi concentration in the plants, about 0.5-g fresh samples were used following the previously described method (Zhou et al., 2008). For the measurement of total P concentration in the plant, about 0.05-g dry samples were used following the method described by Chen et al. (2007).
Sequence data from this article can be found in the Rice Genome Initiative/GenBank data libraries under accession numbers AF536961 to AF536968 (OsPT1–OsPT8).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Tissue localization analysis of OsPT8.
Supplemental Figure S2. GUS activity analysis of the transgenic rice carrying the OsPT8 promoter::GUS construct, and cis-regulatory element analysis of the putative OsPT8 promoter.
Supplemental Figure S3. Phenotype and Southern-blot analysis of the OsPT8-Oe plant.
Supplemental Figure S4. pt8 mutant identification.
Supplemental Figure S5. Physiological parameters of wild-type and OsPT8-Oe plants.
Supplemental Figure S6. Total P concentration in rice hull and brown rice of OsPT8 expression-altered plants.
Supplemental Table S1. Primers used to amplify the Actin and OsPT8 cDNA for RT-PCR.
Supplemental Table S2. Primers used to amplify the Actin and OsPT8 cDNA for real-time PCR.
Supplemental Table S3. Primers used to amplify the sequences of OsPT8 promoters immediately upstream of their translation start for the genes.
Supplemental Table S4. Primers used to amplify the sequences of OsPT8 cDNA for the construction of overexpression and RNA interference vectors.
Supplemental Table S5. Growth performance of wild-type and OsPT8-Oe plants from four independent lines in a field experiment.
Supplementary Material
Acknowledgments
We thank Dr. Penghui Ai and Ms. Hongye Qu from Nanjing Agricultural University for technical assistance and Prof. Uzi Kafkafi from Hebrew University of Jerusalem for critical reading of the manuscript.
References
- Ai PH, Sun SB, Zhao JN, Fan XR, Xin WJ, Guo Q, Yu L, Shen QR, Wu P, Miller AJ, et al. (2009) Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J 57: 798–809 [DOI] [PubMed] [Google Scholar]
- Bray RH, Kurtz LT. (1945) Determination of total, organic, and available forms of phosphorus in soils. Soil Sci 59: 39–45 [Google Scholar]
- Bucher M. (2007) Functional biology of plant phosphate uptake at root and mycorrhiza interfaces. New Phytol 173: 11–26 [DOI] [PubMed] [Google Scholar]
- Bun-Ya M, Nishimura M, Harashima S, Oshima Y. (1991) The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol Cell Biol 11: 3229–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RE, Tour OP, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. (2002) A monomeric red fluorescent protein. Proc Natl Acad Sci USA 99: 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catarecha P, Segura MD, Franco-Zorrilla JM, García-Ponce B, Lanza M, Solano R, Paz-Ares J, Leyva A. (2007) A mutant of the Arabidopsis phosphate transporter PHT1;1 displays enhanced arsenic accumulation. Plant Cell 19: 1123–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AQ, Hu J, Sun SB, Xu GH. (2007) Conservation and divergence of both phosphate- and mycorrhiza-regulated physiological responses and expression patterns of phosphate transporters in solanaceous species. New Phytol 173: 817–831 [DOI] [PubMed] [Google Scholar]
- Cleaver OB, Patterson KD, Krieg PA. (1996) Overexpression of the tinman-related genes XNkx-2.5 and XNkx-2.3 in Xenopus embryos results in myocardial hyperplasia. Development 122: 3549–3556 [DOI] [PubMed] [Google Scholar]
- Daram P, Brunner S, Persson BL, Amrhein N, Bucher M. (1998) Functional analysis and cell-specific expression of a phosphate transporter from tomato. Planta 206: 225–233 [DOI] [PubMed] [Google Scholar]
- Glassop D, Smith SE, Smith FW. (2005) Cereal phosphate transporters associated with the mycorrhizal pathway of phosphate uptake into roots. Planta 222: 688–698 [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Güimil S, Chang HS, Zhu T, Sesma A, Osbourn A, Roux C, Ioannidis V, Oakeley EJ, Docquier M, Descombes P, et al. (2005) Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc Natl Acad Sci USA 102: 8066–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MJ, Dewbre GR, Liu JY. (2002) A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14: 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Zhu C, Li F, Tang J, Wang Y, Lin A, Liu L, Che R, Chu C. (2011) LEAF TIP NECROSIS1 plays a pivotal role in the regulation of multiple phosphate starvation responses in rice. Plant Physiol 156: 1101–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggewie G, Willmitzer L, Riesmeier JW. (1997) Two cDNAs from potato are able to complement a phosphate uptake-deficient yeast mutant: identification of phosphate transporters from higher plants. Plant Cell 9: 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BZ, Xin WJ, Sun SB, Shen QR, Xu GH. (2006) Physiological and molecular responses of nitrogen-starved rice plants to re-supply of different nitrogen sources. Plant Soil 287: 145–159 [Google Scholar]
- Liu F, Wang ZY, Ren HY, Shen C, Li Y, Ling HQ, Wu C, Lian XM, Wu P. (2010) OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J 62: 508–517 [DOI] [PubMed] [Google Scholar]
- Liu H, Trieu AT, Blaylock LA, Harrison MJ. (1998) Cloning and characterization of two phosphate transporters from Medicago truncatula roots: regulation in response to phosphate and to colonization by arbuscular mycorrhizal (AM) fungi. Mol Plant Microbe Interact 11: 14–22 [DOI] [PubMed] [Google Scholar]
- Marschner H. (1995) Mineral Nutrition of Higher Plants. Academic Press, London [Google Scholar]
- Miao YS, Jiang LW. (2007) Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nat Protoc 2: 2348–2353 [DOI] [PubMed] [Google Scholar]
- Misson J, Thibaud MC, Bechtold N, Raghothama K, Nussaume L. (2004) Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol Biol 55: 727–741 [DOI] [PubMed] [Google Scholar]
- Mitsukawa N, Okumura S, Shirano Y, Sato S, Kato T, Harashima S, Shibata D. (1997) Overexpression of an Arabidopsis thaliana high-affinity phosphate transporter gene in tobacco cultured cells enhances cell growth under phosphate-limited conditions. Proc Natl Acad Sci USA 94: 7098–7102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW. (2002) Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J 31: 341–353 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nilsson L, Müller R, Nielsen TH. (2007) Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ 30: 1499–1512 [DOI] [PubMed] [Google Scholar]
- Paszkowski U. (2006) A journey through signaling in arbuscular mycorrhizal symbioses 2006. New Phytol 172: 35–46 [DOI] [PubMed] [Google Scholar]
- Paszkowski U, Kroken S, Roux C, Briggs SP. (2002) Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 99: 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Bucher M. (2002) Phosphate transport and homeostasis in Arabidopsis. The Arabidopsis Book 1: e0009, doi/10.1199/tab.0009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss CP, Huang CY, Gilliham M, Tyerman SD. (2010) Channel-like characteristics of the low-affinity barley phosphate transporter PHT1;6 when expressed in Xenopus oocytes. Plant Physiol 152: 1431–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss CP, Huang CY, Tyerman SD. (2011) Proton-coupled high-affinity phosphate transport revealed from heterologous characterization in Xenopus of barley-root plasma membrane transporter, HvPHT1;1. Plant Cell Environ 34: 681–689 [DOI] [PubMed] [Google Scholar]
- Rae AL, Cybinski DH, Jarmey JM, Smith FW. (2003) Characterization of two phosphate transporters from barley: evidence for diverse function and kinetic properties among members of the Pht1 family. Plant Mol Biol 53: 27–36 [DOI] [PubMed] [Google Scholar]
- Raghothama KG. (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Raghothama KG. (2000) Phosphate transport and signaling. Curr Opin Plant Biol 3: 182–187 [PubMed] [Google Scholar]
- Rausch C, Bucher M. (2002) Molecular mechanisms of phosphate transport in plants. Planta 216: 23–37 [DOI] [PubMed] [Google Scholar]
- Rausch C, Daram P, Brunner S, Jansa J, Laloi M, Leggewie G, Amrhein N, Bucher M. (2001) A phosphate transporter expressed in arbuscule-containing cells in potato. Nature 414: 462–470 [DOI] [PubMed] [Google Scholar]
- Rausch C, Zimmermann P, Amrhein N, Bucher M. (2004) Expression analysis suggests novel roles for the plastidic phosphate transporter Pht2;1 in auto- and heterotrophic tissues in potato and Arabidopsis. Plant J 39: 13–28 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman DP, Shin R. (2007) Nutrient sensing and signaling: NPKS. Annu Rev Plant Biol 58: 47–69 [DOI] [PubMed] [Google Scholar]
- Shin H, Shin HS, Dewbre GR, Harrison MJ. (2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J 39: 629–642 [DOI] [PubMed] [Google Scholar]
- Tong YP, Zhou JJ, Li ZS, Miller AJ. (2005) A two-component high-affinity nitrate uptake system in barley. Plant J 41: 442–450 [DOI] [PubMed] [Google Scholar]
- Upadhyaya NM, Surin B, Ramm K, Gaudron J, Schunmann PHD, Taylor W, Waterhouse PM, Wang MB. (2000) Agrobacterium-mediated transformation of Australian rice cultivars Jarrah and Amaroo using modified promoters and selectable markers. Aust J Plant Physiol 27: 201–210 [Google Scholar]
- Wang C, Ying S, Huang H, Li K, Wu P, Shou HX. (2009) Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J 57: 895–904 [DOI] [PubMed] [Google Scholar]
- Zhou J, Jiao FC, Wu ZC, Li Y, Wang X, He X, Zhong W, Wu P. (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146: 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.