Abstract
The transition from etiolated to green seedlings involves a shift from hypocotyl growth-promoting conditions to growth restraint. These changes occur through a complex light-driven process involving multiple and tightly coordinated hormonal signaling pathways. Nitric oxide (NO) has been lately characterized as a regulator of plant development interacting with hormone signaling. Here, we show that Arabidopsis (Arabidopsis thaliana) NO-deficient mutant hypocotyls are longer than those from wild-type seedlings under red light but not under blue or far-red light. Accordingly, exogenous treatment with the NO donor sodium nitroprusside and mutant plants with increased endogenous NO levels resulted in reduced hypocotyl length. In addition to increased hypocotyl elongation, NO deficiency led to increased anthocyanin levels and reduced PHYB content under red light, all processes governed by phytochrome-interacting factors (PIFs). NO-deficient plants accordingly showed an enhanced expression of PIF3, PIF1, and PIF4. Moreover, exogenous NO increased the levels of the gibberellin (GA)-regulated DELLA proteins and shortened hypocotyls, likely through the negative regulation of the GA Insensitive Dwarf1 (GID1)-Sleepy1 (SLY1) module. Consequently, NO-deficient seedlings displayed up-regulation of SLY1, defective DELLA accumulation, and altered GA sensitivity, thus resulting in defective deetiolation under red light. Accumulation of NO in wild-type seedlings undergoing red light-triggered deetiolation and elevated levels of NO in the GA-deficient ga1-3 mutant in darkness suggest a mutual NO-GA antagonism in controlling photomorphogenesis. PHYB-dependent NO production promotes photomorphogenesis by a GID1-GA-SLY1-mediated mechanism based on the coordinated repression of growth-promoting PIF genes and the increase in the content of DELLA proteins.
Light controls plant development through a complex mechanism that is dependent on the quality, intensity, timing, and duration of the incoming light. After embryonic development, germinated seeds can follow two main developmental programs, depending on whether early postgermination occurs in darkness or light. The most common situation in nature is that germinated seeds grow initially skotomorphogenically until cotyledons emerge from the ground. The deetiolation program starts with the loss of the apical hook and the opening of cotyledons to display maximal surface for photosynthesis (Alabadí and Blázquez, 2009). The dark-to-light transition is accompanied by an extensive transcriptional reprogramming that is mostly controlled by the function of two kinds of photoreceptors, phytochromes and cryptochromes, for red/far-red and UV/blue light, respectively (Jiao et al., 2007). The common understanding suggests that photomorphogenesis is the default program in plant development in such a way that skotomorphogenesis is the result of the efficient repression of the photomorphogenic program. Repression is based on the action of CONSTITUTIVE PHOTOMORPHOGENIC1 E3 ubiquitin ligase (Osterlund et al., 1999), which targets several photomorphogenesis-promoting transcription factors for degradation, including LAF1, HFR1, HY5, and HYH (Holm et al., 2002; Seo et al., 2003; Jang et al., 2005; Yang et al., 2005), but not others driving etiolated growth, such as the phytochrome-interacting factors PIF1, PIF3, and PIF4 (Castillon et al., 2007; Leivar et al., 2008b). To avoid skotomorphogenic growth in light, PIFs are rapidly phosphorylated and degraded by light-activated phytochromes (Shen et al., 2005, 2007; Al-Sady et al., 2006). The balance between the activities of both groups of transcription factors is thus determined by the transition from skotomorphogenesis to photomorphogenesis (Huq, 2006; Leivar et al., 2009; Shin et al., 2009). However, light is not the only factor controlling this developmental transition. Plant development is tightly subject to hormonal control, and this regulatory level represents an integration point between endogenous cues and stress-related environmental factors (Nemhauser, 2008; Alabadí and Blázquez, 2009; Wolters and Jürgens, 2009). Among major hormonal growth regulators, it is well known that GA signaling exerts an essential role in repressing photomorphogenesis under darkness (Alabadí et al., 2008; de Lucas et al., 2008; Feng et al., 2008). Repression is achieved through the GA-induced degradation of DELLA proteins, which are indeed negative modulators of PIF function (Achard et al., 2007; Alabadí et al., 2008; de Lucas et al., 2008; Feng et al., 2008), thus leading to hypocotyl elongation. The function of the GAs and its receptor GA Insensitive Dwarf1 (GID1) in DELLAs represents a regulatory module able to control plant growth under fluctuating environmental conditions (Harberd et al., 2009). This signaling module could allow growth arrest when plants face adverse environmental conditions, and it might function potentially as a link between stress-triggered responses and development (Achard et al., 2006, 2008; Navarro et al., 2008).
Many stress factors share the activation of rapid responses in the plant, characterized by the production of reactive oxygen and nitrogen species. Some of these molecules have been characterized as potential primary modulators because of their ubiquity and strong reactivity. Among them, nitric oxide (NO) has been extensively characterized as a regulator of stress-related responses (Hong et al., 2008; Neill et al., 2008) and developmental processes as well (He et al., 2004; Prado et al., 2004; Hu et al., 2005; López-Bucio et al., 2006). NO often exerts its regulatory activity in tight coordination with other regulators, including salicylic acid (Mur et al., 2008), jasmonic acid (Saito et al., 2009), abscisic acid (Saito et al., 2009), and ethylene (Ederli et al., 2006; Wang et al., 2009). It has also been reported that NO may function as a modulator of auxins in root development (Pagnussat et al., 2003; Guo et al., 2008; Lanteri et al., 2008). Conversely, NO production is activated by different hormones, including abscisic acid (Guo et al., 2003; Bright et al., 2006), cytokinins (Tun et al., 2008), auxins (Kolbert et al., 2008), jasmonic acid (Huang et al., 2004), and salicylic acid (Zottini et al., 2007; Hao et al., 2010). We have recently demonstrated that production of NO through two biosynthetic pathways involving nitrate reductase (NR/NIA) and nitric oxide-associated 1 (NOA1) activities is essential for the correct development of Arabidopsis (Arabidopsis thaliana), affecting a wide range of processes including seed production, germination, vegetative growth, and the control of stomatal closure (Lozano-Juste and León, 2010). NO’s effect on seed germination is exerted in tight coordination with abscisic acid (Lozano-Juste and León, 2010), but the participation of other hormones such as GAs is also very likely. In fact, there is evidence about the interaction between GAs and NO in regulating germination (Beligni et al., 2002; Bethke et al., 2007). Interestingly, there are also some reports proposing that NO controls other aspects of development where GAs have an essential role, including hypocotyl elongation, the acquisition of photomorphogenic traits (Beligni and Lamattina, 2000; Tonón et al., 2010), and the growth and reorientation of pollen tubes (Prado et al., 2004). However, the underlying mechanism explaining how NO controls plant growth in connection with hormones remains unknown. Here, we show that NO counteracts GA signaling and thus promotes photomorphogenesis through a regulatory mechanism affecting DELLA protein accumulation positively and PIF expression negatively.
RESULTS
Increased Hypocotyl Elongation in Light-Grown NO-Deficient nia1,2noa1-2 Mutant
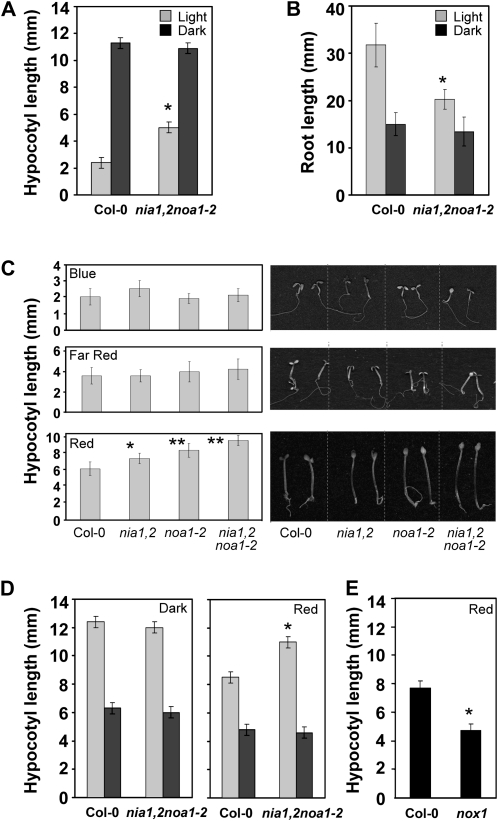
Despite the delayed vegetative growth characteristic of nia1,2noa1-2 seedlings (Lozano-Juste and León, 2010), we observed that their hypocotyls were unusually long, reaching a size that doubled that of wild-type seedlings 5 d after germination under white light (Fig. 1A). Enhanced hypocotyl elongation caused by NO deficiency was not evident under every growth condition; for instance, nia1,2noa1-2 seedlings were as long as the wild type in darkness (Fig. 1A). Moreover, it was not the result of a negative effect of NO on the growth of the whole seedling, since the nia1,2noa1-2 mutant displayed the opposite pattern in root elongation, with shorter roots than wild-type seedlings in light but no significant differences in darkness (Fig. 1B). The differential effect on hypocotyl elongation and root growth in light but not in darkness suggests that the nia1,2noa1-2 mutant is fully able to repress photomorphogenesis in darkness, thus indicating that NO is not required for it. However, it displayed a defect in light-induced deetiolation, suggesting that NO is necessary for the repression of growth by light. To investigate whether NO would act in a specific branch of the light signaling pathway, we tested the ability of different light qualities to repress hypocotyl elongation in different NO-deficient mutant backgrounds. Figure 1C shows that blue and far-red light were able to repress hypocotyl elongation in all NO-deficient genotypes to the same extent observed in wild-type seedlings. However, under red light conditions, NO-deficient seedlings showed longer hypocotyls than wild-type seedlings, the effect being mild in the backgrounds with partial NO deficiency and additive in the triple mutant (Fig. 1C). Further characterization of the reduced inhibition of hypocotyl elongation of NO-deficient seedlings by red light showed that the differential effect was already significant by 4 d after germination. Up to 5 d after germination, when wild-type hypocotyl growth started to slow, the triple nia1,2noa1-2 hypocotyls were still growing at the maximal rate (Supplemental Fig. S1A). The differential effect was also dependent on the fluence rate of red light, with significant differences in the inhibition of hypocotyl elongation of noa1-2 and nia1,2noa1-2 mutants at fluence rates as low as 1 μmol m−2 s−1 (Supplemental Fig. S1B). At 10 μmol m−2 s−1 red light, a 40% inhibition in wild-type hypocotyl length was observed, whereas only 25% was observed for nia1nia2 and less than 5% for noa1-2 and the triple mutant (Supplemental Fig. S1B). Importantly, differential hypocotyl elongation of nia1,2noa1-2 versus accession Columbia (Col-0) seems to be due to reduced NO content, since both exogenous application of NO by the NO donor sodium nitroprusside (SNP) and the nox1 mutant plant with increased NO content (He et al., 2004) resulted in shorter hypocotyls (Fig. 1, D and E), clearly showing that NO controls hypocotyl elongation.
Figure 1.
Hypocotyl and root elongation of NO-deficient plants under different light conditions. A, and B, Hypocotyl and root length, respectively, of the wild type and the nia1,2noa1-2 mutant under white light (Light) or darkness (Dark). Hypocotyl and root length were measured in 5- and 10-d-old seedlings, respectively. C, Hypocotyl length was measured in 5-d-old seedlings grown under 16.5 μmol m−2 s−1 blue light, 5 μmol m−2 s−1 far-red light, and 20 μmol m−2 s−1 red light. D, Hypocotyl length in untreated (light gray bars) and NO-treated (250 μm SNP; dark gray bars) Col-0 and nia1,2noa1-2 seedlings. E, Length of the hypocotyls of Col-0 and NO-overproducer nox1 seedlings. Values are means ± se of three independent experiments (at least 20 seedlings per experiment were measured). Asterisks represent statistically significant differential values at * P < 0.005 and ** P < 0.001 when comparing mutant versus the wild type under the same treatment conditions.
PHYB Levels and Signaling Are Altered in a NO-Deficient Mutant
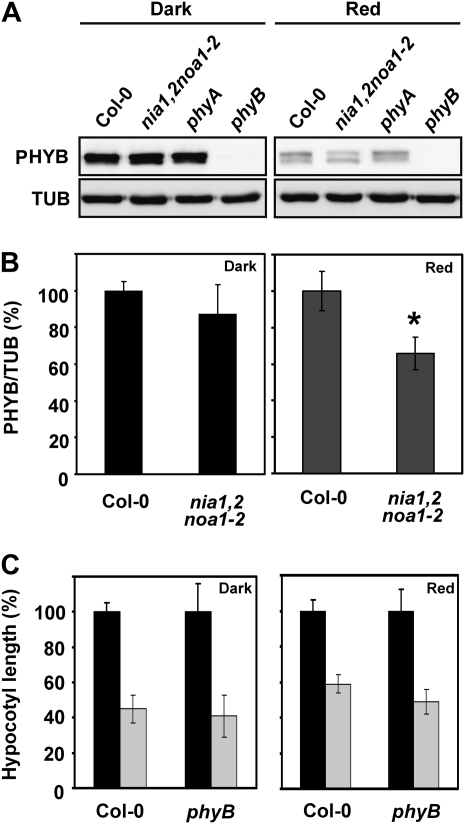
The reduced inhibition of hypocotyl elongation observed for NO-deficient seedlings only under red light conditions suggests that mutant seedlings are partially blind to red light, which may be caused by changes in the level or activity of the PHYB photoreceptor or by interference with PHYB signaling. If the nia1,2noa1-2 mutant had altered PHYB levels or was impaired in PHYB function, it would be expected to phenocopy most if not every phenotype displayed by phyB mutant seedlings. Table I summarizes some of the traits characteristic of the phyB mutant phenotype and the correspondence in nia1,2noa1-2 seedlings. Both mutants share overlapping traits, such as elongated hypocotyls under red light, pale green leaves with reduced chlorophyll content, low transpiration rates, accumulation of anthocyanins, and reduced branching. However, the elongated petioles, long root hairs, elongated stems, and early-flowering phenotype under a short-day photoperiod, which are characteristic of the phyB mutant, were not evident in nia1,2noa1-2 mutant plants. In fact, the triple mutant was not impaired in PHYB production, since its level in the NO-deficient mutant did not differ significantly from that in wild-type seedlings in darkness (Fig. 2, A and B). However, a significantly reduced content of PHYB was detected in nia1,2noa1-2 when compared with wild-type seedlings under red light (Fig. 2, A and B). Nevertheless, the phyB mutant was not altered in NO-triggered inhibition of hypocotyl elongation (Fig. 2C). All these observations suggest that NO-deficient plants might be altered in red light signaling but not in PHYB protein activity and that NO acts downstream of PHYB in this process.
Table I. Comparative phenotypic analysis of phyB and nia1,2noa1-2 mutants.
Traits analyzed in the cited references were compared with those reported for the nia1,2noa1-2 mutant (Lozano-Juste and León, 2010).
| Trait | phyB | nia1,2noa1-2 | Reference |
| Elongated hypocotyls | Yes | Yes | Reed et al. (1993) |
| Long petioles | Yes | Yes | Reed et al. (1993) |
| Pale leaves in light | Yes | Yes | Reed et al. (1993) |
| Elongated stems | Yes | No | Reed et al. (1993) |
| Reduced root elongation | Yes | Yes | Correll and Kiss (2005) |
| Long root hairs | Yes | No | Reed et al. (1993) |
| Early flowering | Yes | No | Reed et al. (1993); Guo et al. (1998) |
| Lower transpiration rate | Yes | Yes | Boccalandro et al. (2009) |
| Reduced branching | Yes | Yes | Reed et al. (1993) |
| Reduced seed germination | Yes | Yes | Shinomura et al. (1994) |
Figure 2.
PHYB protein levels in wild-type Col-0, NO-deficient nia1,2noa1-2, phyA, and phyB mutants grown in darkness or red light. A, Western blots with anti-PHYB antibodies of protein samples from representative hypocotyls of the indicated genotypes grown for 3 d under darkness or 7 μmol m−2 s−1 red light. Western blots with anti-tubulin (TUB) are shown as loading controls. B, Quantification of PHYB levels normalized to TUB and expressed relative to the levels of Col-0. Values are means of three independent experiments ± sd. C, Relative hypocotyl length of untreated (black bars) and NO-treated (250 μm SNP; gray bars) Col-0 and phyB seedlings under dark and red light conditions. Values are means ± se of three independent experiments (at least 20 seedlings per experiment were measured). Asterisks represent statistically significant differential values at P < 0.05 when comparing mutant versus the wild type under the same treatment conditions.
PIFs Are Targets of the NO-Triggered Inhibition of Hypocotyl Growth
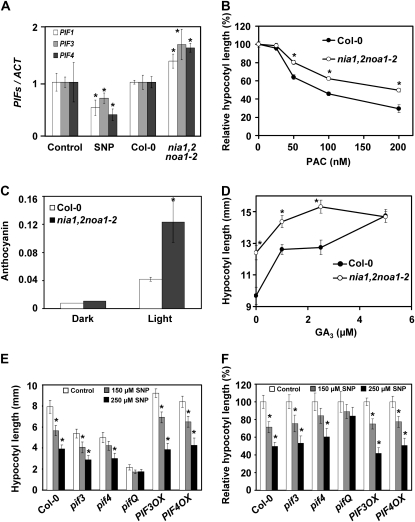
Since nia1,2noa1-2 seedlings contained around 3-fold more anthocyanins than wild-type seedlings in light (Fig. 3C), and given that many of the genes encoding the enzymes of flavonol and anthocyanin biosynthesis are targets of the PIF3 transcription factor (Shin et al., 2007), NO might interfere with PHYB signaling through PIF3. As previously reported, PIF3, together with PIF4, negatively regulates PHYB content under red light conditions (Al-Sady et al., 2008; Leivar et al., 2008a), which is fully in agreement with the reduced levels of PHYB detected in nia1,2noa1-2 seedlings only under red light (Fig. 2). Supporting this hypothesis, we have found that nia1,2noa1-2 hypocotyls accumulate more PIF3 transcript than wild-type seedlings under red light (Fig. 3A), and this increase was also detected for other bHLH members of the PIF family such as PIF1 and PIF4 (Fig. 3A). Accordingly, the levels of these three PIF transcripts were significantly reduced upon treatment of wild-type seedlings with SNP (Fig. 3A). Furthermore, as expected for plants with enhanced PIF function (de Lucas et al., 2008; Feng et al., 2008), nia1,2noa1-2 seedlings had an altered response to both GA3 and the GA biosynthetic inhibitor paclobutrazol (PAC). Figure 3B shows that wild-type and nia1,2noa1-2 hypocotyls treated with 200 nm PAC were 70% and 50% shorter, respectively, than those from untreated seedlings. At lower PAC concentrations, nia1,2noa1-2 hypocotyls were always hyposensitive to PAC (Fig. 3C). Altered sensitivity to GAs was also observed in the nia1,2noa1-2 mutant. Whereas nia1,2noa1-2 hypocotyls reached their maximum length at GA3 concentrations around 1 μm, wild-type hypocotyls increased elongation to at least 5 μm GA3 (Fig. 3D). On the other hand, the hypocotyls of seedlings overexpressing PIF3 were significantly more sensitive (P < 0.05) than wild-type seedlings to NO-mediated inhibition of hypocotyl elongation (Fig. 3F). By contrast, a quadruple pif1pif3pif4pif5 mutant (pifQ) was almost completely insensitive to NO-triggered hypocotyl shortening (Fig. 3, E and F). All these data strongly suggest that there is an enhanced PIF function in NO-deficient mutants and also that PIFs are targets of the NO-triggered inhibition of hypocotyl growth.
Figure 3.
Functional connection between NO, GAs, and PIF proteins. A, PIF1, PIF3, and PIF4 transcript levels in Col-0 or nia1,2noa1-2 hypocotyls and on wild-type hypocotyls either treated with 1 mm SNP (NO donor) for 2 h or untreated as a control. B, Relative length of Col-0 and nia1,2noa1-2 hypocotyls (mean ± se) in seedlings exposed to the indicated concentrations of PAC. C, Anthocyanin levels in Col-0 and nia1,2noa1-2 seedlings under dark or white light conditions expressed in arbitrary units of A530. D, Length of Col-0 and nia1,2noa1-2 hypocotyls (mean ± se) treated with the indicated concentrations of GA3. E and F, Total and relative length, respectively, of control untreated and SNP-treated hypocotyls in the indicated genotypes and concentrations. All the experiments were performed with seedlings grown under 20 μmol m−2 s−1 red light unless otherwise mentioned. Values are means of three biological replicates ± se. For hypocotyl length, at least 20 seedlings per independent experiment were measured. Asterisks represent statistically significant differential values with at least P < 0.05 when comparing mutant versus the wild type for the same treatment conditions (A–D) or untreated versus treated samples in each genotype (E and F).
NO Promotes DELLA Protein Accumulation
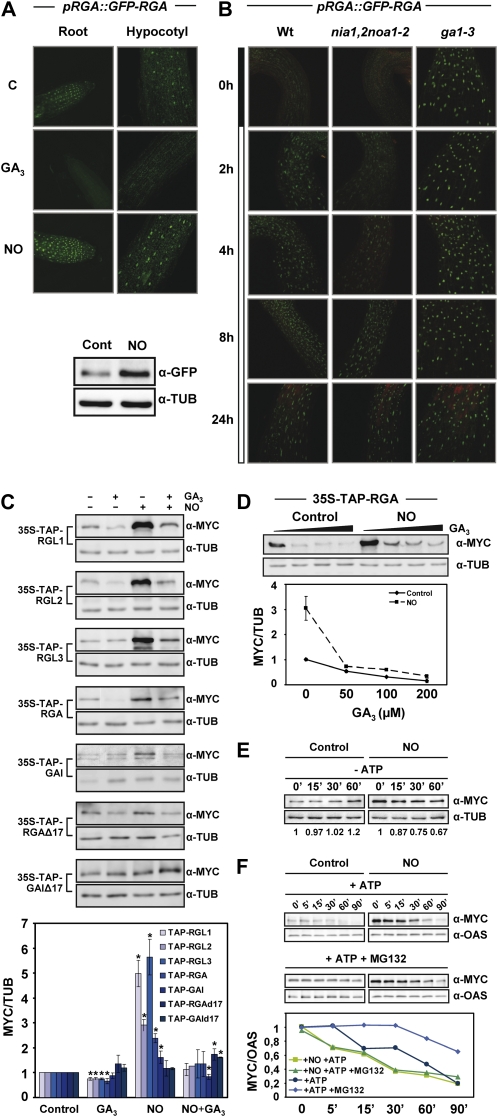
PIF activity is controlled by its association with DELLA proteins (de Lucas et al., 2008; Feng et al., 2008), whose levels are regulated by GAs through proteasome-mediated degradation (Dill et al., 2004: Achard et al., 2007; Harberd et al., 2009). The altered sensitivity of nia1,2noa1-2 seedlings to PAC and GAs could also be explained by reduced levels of DELLAs in the NO-deficient seedlings. To test the possible effect of NO on DELLA accumulation, transgenic seedlings expressing a GFP-tagged version of the DELLA protein RGA under its endogenous promoter (pRGA::GFP-RGA) were treated with SNP. Figure 4A shows that in accordance with GA promoting DELLA degradation (Dill et al., 2004; Achard and Genschik, 2009), GFP-RGA fluorescence was absent both in roots and hypocotyls from GA-treated seedlings but, in turn, GFP-RGA-associated fluorescence, at nuclei, increased in NO-exposed seedlings (Fig. 4A). The increased amount of GFP-RGA protein was also detected in whole seedlings by western blot with anti-GFP antibodies (Fig. 4A). To check whether endogenous NO controls RGA levels, we generated pRGA::GFP-RGA transgenic plants in the nia1,2noa1-2 background. Figure 4B shows GFP-RGA fluorescence in different backgrounds after transferring seedlings from darkness to light. Fluorescence was detected as soon as 2 h after the shift in hypocotyls of wild-type background, when almost no fluorescence was detected in hypocotyls of the NO-deficient background plants (Fig. 4B). By 4 h after the shift, GFP-RGA started to accumulate in the nia1,2noa1-2 background, although its level was significantly lower than in the wild-type background, suggesting that NO is necessary for the timely light-dependent GFP-RGA accumulation. By contrast, hypocotyls of the ga1-3 background showed enhanced GFP-RGA fluorescence even in darkness (Fig. 4B), due to the lack of GAs in this mutant (Achard et al., 2007). Moreover, we have further checked that this effect was not specific for RGA, and it is extended to all DELLAs. By using transgenic lines expressing tandem affinity purification (TAP)-tagged versions of the five DELLAs, we have analyzed the levels of DELLA proteins after treatment with the NO donor or GA3. Western blots with anti-MYC antibody of whole seedling extracts showed increased protein levels of every DELLA in seedlings exposed to NO and decreased levels in GA-treated seedlings (Fig. 4C). Additionally, pretreatment with the NO donor and further application of GAs did not prevent GA-induced degradation of DELLAs (Fig. 4C), suggesting that NO is not modifying DELLA proteins, turning them resistant to proteolysis. We confirmed this idea by following TAP-RGA degradation upon exposure to increasing concentrations of GAs in NO-treated and untreated seedlings. Figure 4D shows that TAP-RGA was efficiently degraded in NO-treated seedlings to levels similar to those detected in untreated control seedlings. In fact, the degradation of TAP-RGA by GA3 seemed to be more efficient in NO-treated seedlings. Then, we explored whether active degradation of DELLAs in NO-treated seedlings is also mediated by the ATP-consuming proteasome by a previously reported cell-free assay system (Wang et al., 2009). Figure 4E shows no significant degradation of TAP-RGA proteins in the absence of exogenously added ATP in untreated seedlings, but degradation was observed in NO-treated seedlings, indicating that TAP-RGA in these extracts could be degraded by a proteasome-independent mechanism. However, both untreated and NO-treated TAP-RGAs were progressively degraded with time in ATP-treated extracts, the latter being more susceptible to degradation (Fig. 4F). Moreover, the proteasome inhibitor MG132 retarded TAP-RGA degradation in untreated controls but only to a limited extent in NO-treated samples (Fig. 4F), suggesting that in NO-treated samples, proteasome-independent hydrolysis also occurred. This alternative degradation mechanism could also explain the more efficient degradation of TAP-RGA in NO-treated seedlings upon GA3 treatment (Fig. 4D) or in the absence of ATP (Fig. 4E).
Figure 4.
Effect of NO on DELLA protein accumulation. A, GFP-RGA in pRGA::GFP-RGA roots and hypocotyls, either untreated (control [C]) or treated for 2 h with 50 μm GA3 or 250 μm SNP as a source of NO, visualized by confocal microscopy. GFP-RGA levels and the loading control tubulin (TUB) were analyzed by western blot. B, GFP-RGA protein in hypocotyls of pRGA::GFP-RGA in wild-type, nia1,2noa1-2, and ga1-3 backgrounds at different times after the shift from darkness to red light, as indicated in the bar at left. C, TAP-tagged versions of every DELLA protein were used to analyze the levels of each protein in seedlings treated (+) or not (−) with 50 μm GA3 and/or 250 μm SNP (NO) for 2.5 h. TAP-DELLAs were detected with anti-MYC antibodies, and the levels of tubulin are shown as a loading control. The values normalized with tubulin and relative to control untreated samples were quantified, and the values shown in the bottom panel correspond to means of three independent experiments ± sd. Asterisks represent statistically significant differential values with at least P < 0.05 when comparing treated versus untreated controls in each genotype. D, GA-induced degradation of RGA in untreated (Control) and NO-treated (NO; as described in C) 35S-TAP-RGA seedlings exposed to increasing GA3 concentrations (0, 50, 100, and 200 μm). Values normalized to tubulin were quantified and are shown in the bottom panel as means of three independent experiments ± se. E and F, Cell-free degradation assay of RGA in the absence (E) or presence (F) of ATP and the proteasome inhibitor MG132. Protein samples were incubated at room temperature for the indicated times and treatment conditions and detected with anti-MYC antibodies. Tubulin or O-acetyl-Ser(thiol)lyase 1 (OAS) was detected as a loading control (D–F). ATP and MG132 were used at 10 mm and 100 μm, respectively. The protein levels detected on western blots in E and F were quantified and normalized to TUB or OAS content.
We have also checked that increased tagged DELLA levels are not the result of an enhancing effect of NO on 35S-driven expression, as 35S::TAP-GFP seedlings showed similar levels of GFP in NO-treated and control untreated seedlings (Supplemental Fig. S2). Moreover, 35S::TAP-GAIΔ17 and 35S::TAP-RGAΔ17 seedlings, expressing truncated versions of GAI and RGA resistant to GA-induced proteolysis, showed unaltered protein levels upon treatment (Fig. 4C). Increased DELLA levels upon NO treatment in 35S::TAP-DELLA transgenic plants were then not likely due to transcriptional activation of DELLA genes, because the expression of these genes is under the control of the constitutive 35S promoter. Nevertheless, to check whether the enhanced accumulation of GFP-RGA in pRGA::GFP-RGA lines was due to the transcriptional control of RGA by NO, quantitative reverse transcription (RT)-PCR with RNA extracted from hypocotyls of wild-type seedlings exposed to the NO donor SNP was performed. We did not find significant changes in the transcript levels of DELLA genes in response to NO (Supplemental Fig. S3). Moreover, DELLA transcript levels were not altered in the NO-deficient nia1,2noa1-2 mutant compared with wild-type seedlings (Supplemental Fig. S3). Only RGL1 transcripts were significantly changed in the NO-deficient mutant when compared with Col-0, and it was not down-regulated, as expected for a putative role of NO as a gene activator (Supplemental Fig. S3).
Alternatively, the control of endogenous GA levels by NO might explain the accumulation of DELLAs in NO-treated seedlings. Regarding this, we have quantified the levels of GA biosynthesis and metabolism gene transcripts. Only the GA20ox3 gene was down-regulated by NO treatment and up-regulated in the NO-deficient nia1,2noa1-2 mutant (Supplemental Fig. S4). These data suggest that NO might reduce the biosynthesis of active GAs through specific effects on GA20ox3. However, this effect should be restricted to hypocotyl, because no other GA-related phenotype was observed in NO-deficient seedlings.
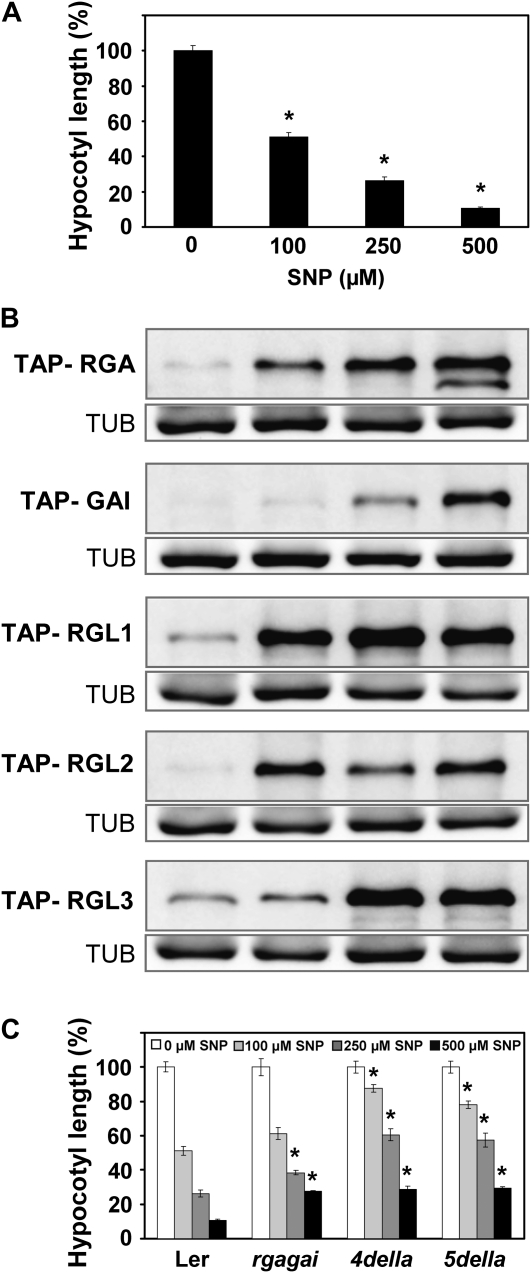
NO Induces DELLA Accumulation by Repressing SLY1
Wild-type hypocotyls grown in red light under increasing NO concentrations were progressively shortened. At 0.5 mm SNP, hypocotyl length was below 10% of those from untreated seedlings (Fig. 5A). Shortening of hypocotyls with increasing SNP concentrations correlated well with the accumulation of all DELLAs, with GAI being the less sensitive to NO (Fig. 5B). Additionally, Figure 5C shows that the inhibition of hypocotyl elongation exerted by NO under red light conditions was partially dependent on DELLA function, as revealed by the slightly reduced response to NO detected in the rgagai double mutant and the more pronounced insensitivity detected in the quadruple mutant 4della, lacking all DELLAs but RGL3 (Achard et al., 2006), and the 5della global knockout mutant (Fig. 5C). In the concentration range from 0.1 to 0.5 mm SNP tested, the length of 4della and 5della hypocotyls was always 20% to 35% longer than wild-type hypocotyls (Fig. 5C). These data suggest that NO inhibits hypocotyl growth mostly through DELLA function. However, even at the lower NO donor concentration tested, around one-quarter of the effect was exerted via DELLA-independent mechanisms. These data point to DELLAs as the main but not only target for NO action in controlling growth, which is in agreement with the proposed regulation of PIF1, PIF3, and PIF4 expression by NO indicated above.
Figure 5.
Effect of NO on hypocotyl elongation and DELLA content under red light. A and C, Hypocotyl length of wild-type Ler, rga-24gai-t6, and quadruple (4della) and quintuple (5della) DELLA mutants was measured after growing seedlings in the indicated SNP concentrations for 3 d. Values of hypocotyl length are means ± se of three independent experiments (at least 20 seedlings per experiment were measured). Asterisks represent statistically significant differential values with at least P < 0.05 when comparing hypocotyls of treated versus untreated wild-type seedlings (A) or mutant versus wild-type hypocotyls from different genotypes under the same treatment conditions (C). B, TAP-DELLA accumulation under the SNP concentrations shown in A was detected with anti-MYC antibodies, and the loading controls of tubulin (TUB) are included.
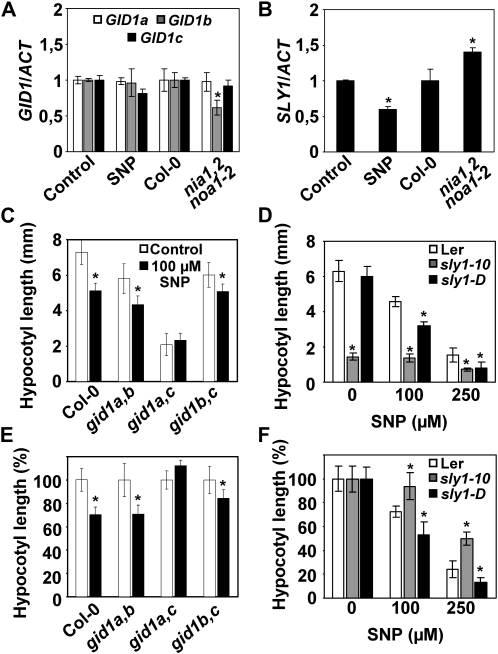
DELLA levels are controlled by GAs through a mechanism dependent on GA perception by GID1, ubiquitination by the ubiquitin ligase Sleepy1 (SLY1), and further degradation by the proteasome. Whereas transcripts of genes coding for GID1s were not significantly affected by exogenous NO and only GID1b was down-regulated in nia1,2noa1-2 seedlings (Fig. 6A), SLY1 expression was significantly down-regulated by NO and up-regulated in the NO-deficient nia1,2noa1-2 mutant (Fig. 6B). In agreement with an important role of SLY1 in NO-mediated inhibition of hypocotyl elongation, the loss- and gain-of-function mutants of SLY1, sly1-10 and sly1-D, were less and more sensitive to NO, respectively, than wild-type seedlings (Fig. 6, D and F). Again, the insensitivity of sly1-10 mutant to NO was not complete, indicating the existence of another pathway for NO regulation of hypocotyl elongation. Interestingly, although no transcriptional regulation of GID1 genes by NO was observed (Fig. 6A), the double gid1a,c mutant hypocotyls were less sensitive to NO-triggered shortening (Fig. 6, C and E), thus suggesting that NO action requires the activity of both GID1a and GID1c receptors.
Figure 6.
Functional connection between NO and the GA signaling components GID1s and SLY1. A and B, GID1s and SLY1 transcript levels normalized to actin in hypocotyls of untreated (Control) and 1 mm SNP-treated wild-type seedlings as well as in the nia1,2noa1-2 mutants and its wild-type background, Col-0. Values are means ± se of three experiments. C and E, Total and relative hypocotyl length, respectively, of different combinations of double gid1 mutants in untreated (white bars) and 100 μm SNP-treated (black bars) seedlings. D and F, Total and relative hypocotyl length of Col-0, the loss-of-function mutant sly1-10, and the gain-of-function mutant sly1-D at the indicated SNP concentrations. For C to F, values of hypocotyl length are means ± se of three independent experiments (at least 20 seedlings per experiment were measured). Asterisks represent statistically significant differential values at P < 0.05 when comparing mutant versus wild-type hypocotyls under the same treatment conditions (A, B, D, and F) or treated versus untreated hypocotyls in each genotype (C and E).
NO Production Is Negatively Regulated by GAs
The proposed role of NO on photomorphogenesis would require physiological support to have a functional significance in the deetiolation process. We have tested whether the transition from darkness to light is accompanied by the endogenous production of NO in Arabidopsis. Endogenous NO content analysis in dark-grown hypocotyls shifted to red light has been performed by using 3-amino,4-aminomethyl-2',7'-difluorofluorescein diacetate (DAF-FM DA) fluorescein that specifically detects NO. Figure 7 shows that NO-associated fluorescence was very low in dark-grown wild-type hypocotyls. However, by 1 h after the shift to red light, NO-associated fluorescence was raised along the hypocotyls (Fig. 7A). However, the increased NO-associated fluorescence upon transition to red light was not detected either in the nia1,2noa1-2 or in the phyB mutant hypocotyls (Fig. 7A). Interestingly, the GA-deficient mutant ga1-3 contained NO levels in darkness significantly higher than those detected in wild-type plants (Fig. 7A), suggesting that GAs exert a negative control on NO production in hypocotyls. In addition, no increase in NO was detected in the ga1-3 mutant upon the shift to red light (Fig. 7A). As further support for the negative regulation exerted by GAs on NO production, both the wild type and the ga1-3 mutant displayed reduced NO-associated fluorescence in GA3-treated seedlings compared with untreated controls (Fig. 7B). By contrast, the GA-insensitive gai-1D mutant did not change its NO content upon treatment with exogenously supplied GAs (Fig. 7B).
Figure 7.
NO levels in the wild-type Col-0 and Ler as well as in nia1,2noa1-2, phyB, ga1-3, and gai-1D mutant plants. A, Endogenous NO was visualized with the cell-permeable DAF-FM DA fluorescein in seedlings grown in darkness for 4 d and 1 h after the shift to red light (20 μmol m−2 s−1). B, NO-related fluorescence in Ler, ga1-3, and gai-1D in untreated (Control) and 50 μm GA3-treated seedlings for 2.5 h. Values are shown relative to untreated wild-type levels. The right panel of A and B show the quantification of three independent experiments as mean values of fluorescence (arbitrary units [a.u.]) ± se. Asterisks represent statistically significant differential values at P < 0.05 when comparing controls in darkness (A) or untreated (B).
DISCUSSION
Oxygen and nitrogen reactive species have important roles in plant development that are characteristic of developmental transitions (Gapper and Dolan, 2006; Tsukagoshi et al., 2010). These molecules have well-documented roles regulating the adaptation to changes in temperature (Allakhverdiev et al., 2008) or light (Li et al., 2009), conditions that regulate developmental processes like seed germination and seedling establishment (Bailly et al., 2008), hypocotyl elongation (Stavang et al., 2009), flowering (Ye et al., 2000), leaf senescence (Jing et al., 2008), and tuber formation (Agrawal et al., 2008). Among reactive species produced in plants in response to changes in their environment, NO has been the focus of increasing interest in recent years, since it participates in both defensive (Romero-Puertas et al., 2004; Wendehenne et al., 2004; Hong et al., 2008) and developmental (He et al., 2004; López-Bucio et al., 2006) processes. It has been reported that the addition of a NO donor to etiolated Arabidopsis seedlings promotes deetiolation and inhibits hypocotyl elongation (Beligni and Lamattina, 2000). In addition, a severe heat shock treatment (45°C) leads to a NO burst and the concomitant hypocotyl growth inhibition (Lee et al., 2008). Furthermore, the hot 5-1 mutant, impaired in S-nitrosoglutathione reductase, which metabolizes the NO adduct S-nitrosoglutathione, was markedly susceptible to hot temperatures, accumulated increased concentrations of NO, and presented a deetiolated phenotype in darkness (Lee et al., 2008), the latter being dependent on the increased NO accumulation of the mutant (Lee et al., 2008). Accordingly, we have found that nia1,2noa1-2 seedlings severely impaired in NO biosynthesis displayed elongated hypocotyls under red light conditions, in accordance with the previously reported data for a mutant impaired in the regulation of nitrate reductase gene expression (Lin and Cheng, 1997). Moreover, as presented in this study, a mutant that accumulates increased NO levels presented shorter hypocotyls than the wild type under the same conditions. All this evidence points to a positive role of NO in promoting photomorphogenesis.
However, nothing is known to date about the molecular mechanism by which NO is regulating photomorphogenesis. The observation that the triple mutant only showed elongated hypocotyls under red light but not under far-red or blue light conditions, and because PHYB is the main photoreceptor mediating deetiolation in red light (de Lucas et al., 2008), we first focused on PHYB as the target of NO. After comparing phyB mutant phenotypes with those of nia1,2noa1-2 plants and, importantly, once we noticed that nia1,2noa1-2 seedlings were able to produce wild-type levels of PHYB protein in darkness and also that the phyB mutant responds to NO like wild-type plants, it seemed to us that PHYB-dependent signaling and not its function was impaired in nia1,2noa1-2 seedlings. This hypothesis agrees with previously reported data supporting a role for the NO second messenger cGMP in controlling the phytochrome signaling involved in some processes such as chloroplast development (Bowler et al., 1994). It is noteworthy to mention that by comparing the effect of NO on hypocotyl elongation under dark and red light conditions, we observed essentially similar responses (Supplemental Fig. S5). This suggests that NO in darkness can modulate the same signaling components than in red light. There are several essential downstream components of PHYB in red light signaling, but the interaction of PHYB with the PIF transcription factors may be one of the earliest. The fact that nia1,2noa1-2 presented longer hypocotyls and contained more anthocyanins than wild-type seedlings pointed to an enhanced expression of PIF genes, and particularly of PIF3, in this mutant. In addition, the reduced accumulation of PHYB protein in the nia1,2noa1-2 mutant under red light conditions fits well with an enhanced expression of PIF3 in this mutant, given the previously described negative regulation of PHYB levels by PIF3 and PIF4 (Leivar et al., 2008a). Therefore, it is reasonable that the reduction in PHYB protein levels of the nia1,2noa1-2 mutant in red light contributes significantly to the observed hypocotyl phenotype. We demonstrated that PIF3 together with PIF1 and PIF4 were down-regulated by NO and up-regulated in NO-deficient seedlings. Interestingly, an interaction between HY5, HYH, and PIF4 regulating the light activation of NR2/NIA2 expression in Arabidopsis has been described (Jonassen et al., 2009). These data suggest the existence of a regulatory loop between NR/NIA and PIFs with potential as a mechanism to integrate light- and NO-related factors controlling plant development.
Although it has been previously proposed that NO regulates hypocotyl elongation (Beligni and Lamattina, 2000; Tonón et al., 2010), no mechanism has been reported, to our knowledge, for explaining the control of hypocotyl growth by NO. We have identified PIFs as important regulators of hypocotyl elongation phenotypes observed in NO-deficient seedlings. Furthermore, we found that the pifQ mutant is almost insensitive to the inhibition of hypocotyl elongation by NO. However, neither pif3 nor pif4 single mutants exhibited that phenotype, likely due to redundancy of PIF members in the NO control of hypocotyl length. Yet, the transactivating activity of PIFs is finely regulated through heterodimerization (Hornitschek et al., 2009) and/or interaction with other proteins, such as DELLAs, inhibiting their activity (de Lucas et al., 2008; Feng et al., 2008). The negative regulation of PIF function by NO (Fig. 3A) is accompanied by an increase in the accumulation of DELLA proteins (Fig. 4). It is well known that PIF3 and PIF4 physically interact with members of the DELLA family such as RGA, resulting in a reduction of the PIF3- and PIF4-triggered activation of their targets (de Lucas et al., 2008; Feng et al., 2008; Gallego-Bartolomé et al., 2010). After the dark-to-light transition, NO production leads to repressed PIF expression and enhanced DELLA accumulation, thus resulting in a very limited pool of transactivating PIFs. So, how is DELLA accumulation promoted by NO? First, we checked that NO-treated seedlings could degrade DELLA proteins in response to exogenously applied GAs (Fig. 4). Surprisingly, it seemed that NO-treated seedlings degraded DELLAs more efficiently in response to GAs than control-treated seedlings. This mechanism, far from being unusual, operates in other conditions where exaggerated DELLA accumulation is achieved due to the altered expression of important genes (Silverstone et al., 2007; Willige et al., 2007; Richter et al., 2010) or to inhibition of GA synthesis by PAC (Muangprom et al., 2005). After cell-free assays, we can propose that this is due to a proteasome-independent mechanism (Fig. 4). Then, we explored the possibility of NO regulating the transcription of genes coding for DELLAs. However, NO did not affect DELLA gene expression either in NO-deficient mutants or after NO treatment of wild-type plants (Supplemental Fig. S3). Eventually, NO might be controlling GA biosynthesis or catabolism. After an extensive analysis of the expression of GA20oxidase and GA3oxidase biosynthetic genes as well as GA2oxidase catabolic genes, only GA20ox3 fulfills the criteria to be considered a potential target of NO in controlling GA production, as it was down-regulated upon exogenous NO treatment and up-regulated in the NO-deficient triple mutant (Supplemental Fig. S4). Nevertheless, we anticipate that changes in GA biosynthetic or catabolic gene expression in NO-treated or NO-deficient plants, if any, should be restricted to an organ, tissue, or limited number of cells, but certainly not to the whole seedling. nia1,2noa1-2 plants did not show any general phenotype characteristic of GA overproducers; on the contrary, it had small shoots and produced seeds with low germination potential and increased dormancy (Lozano-Juste and León, 2010). An increase in GA levels specifically in the elongation zone of the hypocotyls might explain the phenotype of long hypocotyls of NO-deficient seedlings in red light, but this is technically difficult, and new methodologies or sensor tools should be developed in the future to accomplish this purpose.
Despite potential fine regulatory functions of NO on GA levels, NO certainly regulates GA signaling through the control of DELLA abundance and function. Our data support a role for NO inducing the accumulation of DELLAs (Fig. 4) despite finding no transcriptional induction of DELLA genes (Supplemental Fig. S3). Because DELLA protein levels are regulated through ubiquitin-proteasome-dependent degradation by the GID1-SCFSLY1 complex, we next focused our attention on this module as a possible target of NO action. First, GID1a,b and GID1c genes were not transcriptionally regulated by NO. However, the gid1a,c mutant, affected in the GID1 receptor isoforms, which are more involved in hypocotyl elongation (Griffiths et al., 2006; Stavang et al., 2009), showed a NO-resistant phenotype. Second, down-regulation of SLY1 expression by NO and the up-regulation detected in the nia1,2noa1-2 hypocotyls might explain the control of DELLA accumulation by NO. Moreover, the loss-of-function mutant sly1-10 was partially insensitive in the inhibition of hypocotyl elongation by NO under red light (Fig. 6, D and F). In contrast to the short-hypocotyl phenotype of sly1-10, we have not found an altered hypocotyl length in sly1-D mutant seedlings under red light (Fig. 6, D and F), thus suggesting that the previously reported enhanced DELLA degradation by SLY1-D protein in roots (Fu et al., 2004) is not functional in the regulation of hypocotyl length under red light conditions. Additionally, sly1-D mutant seedlings responded unexpectedly stronger than wild-type seedlings to NO-triggered hypocotyl shortening (Fig. 6, D and F). It has been proposed that the SLY1-D protein interacts more efficiently than SLY1 with DELLAs and also that the interaction is increased by DELLA phosphorylation (Fu et al., 2004). In addition to the transcriptional regulation of the SLY1 gene by NO described in this work, we cannot rule out the interference of NO in SLY1-DELLA interaction or even NO-mediated modifications of any of the components of the GA perception and signaling complex.
The fact that mutants in DELLA proteins, the GID1 receptor, or the SLY1 F-box behave as partially insensitive to NO strongly suggest that the GID1-DELLA-SLY1 complex is a target of NO in the control of hypocotyl length. Interestingly, the sly1-10 mutant responded differentially to NO-triggered hypocotyl shortening under dark and red light conditions (Fig. 6, D and F; Supplemental Fig. S5E). The wild-type response to NO of the sly1-10 mutant in darkness suggests that transcriptional control of SLY1 by NO is mainly functional in light. This also points to PIFs as important targets of NO in darkness. Moreover, the pifQ mutant is almost completely insensitive to NO, thus suggesting that PIF proteins should have a pivotal role in the negative regulation exerted by NO on the basic GA signaling module. Furthermore, negative functional interaction between NO and GAs in controlling photomorphogenesis is somehow potentiated by the reciprocal negative effect of GAs on NO production, as demonstrated by the enhanced NO content, which can be reverted by GA3 application, in the GA-deficient ga1-3 mutant.
A model integrating the putative functions of NO, GAs, DELLA, and PIF proteins in controlling photomorphogenesis is depicted in Figure 8. The dark-to-light transition leads to increased levels of NO and decreases of GAs, which in turn would lead to a rapid increase in DELLA content as a result of less GID1-GA-DELLA-SLY1 functional interactions. The large pool of DELLA protein would associate with transcription factors, such as PIFs, whose transcription has been nevertheless repressed by NO. DELLAs acting as efficient scavengers of the free form of those transcription factors would lead to the arrest of hypocotyl growth, among other processes characteristics of photomorphogenesis. In the nia1,2noa1-2 mutant, which is severely impaired in NO biosynthesis, the dark-to-light transition leads to a deficient production of NO and, therefore, to a loss of NO-mediated repression of PIFs. Simultaneously, ubiquitination by SLY1 and further degradation of DELLAs may be enhanced under low-NO conditions, thus leading to a decrease in DELLA content. As a result, the enlarged pool of free PIFs and/or their enhanced performance in nia1,2noa1-2 seedlings would be responsible for a partially etiolated phenotype under light conditions.
Figure 8.
Scheme integrating NO and GA antagonist functions in the control of light-regulated photomorphogenesis through the balance between DELLAs and PIFs.
MATERIALS AND METHODS
Plant Material and Treatments
Arabidopsis (Arabidopsis thaliana) accession Col-0 was the wild-type control of nia1nia2, noa1-2, nia1,2noa1-2, gid1a,b, gid1a,c, gid1b,c, pif3-3, pif4-2, and pifQ mutants. The single ga1-3, gai-1D, sly1-10, and sly1-D, double rga-24gai-t6, quadruple 4della, and quintuple 5della mutants and the transgenic plants 35S::TAP-RGA, 35S::TAP-GAI, 35S::TAP-RGL1, 35S::TAP-RGL2, 35S::TAP-RGL3, 35S::TAP-RGAΔ17, 35S::TAP-GAIΔ17, and pRGA::GFP-RGA were used along with the Landsberg erecta (Ler) wild-type background accession. Seeds were surface sterilized with 30% bleach and 0.01% Tween 20, washed extensively with milliQ sterile water, and sown in Murashige and Skoog medium supplemented with 0.8% agar and 1% Suc. After 3 d of stratification at 4°C, germination was synchronized by 3 h of illumination with white light and subsequent incubation in the dark for 22 h before transfer to the different growth conditions. White light-grown seedlings were grown at 19°C to 23°C under fluorescent white light (fluence rate of 70 μmol m−2 s−1) with a 16-h-light/8-h-dark photoperiod. For different light quality treatments, seedlings were grown under continuous blue, far-red, or red light, provided by light-emitting diodes (Percival Science), at the indicated fluence rates (blue light, 16.5 μmol m−2 s−1; far-red light, 5 μmol m−2 s−1; red light, 20 μmol m−2 s−1).
NO treatments were performed by photochemically mediated release of NO gas from a solution of SNP (Fluka) always contained in internal vessels separated from Murashige and Skoog medium, as reported previously (Bethke et al., 2006; Lozano-Juste and León, 2010). For gene expression or protein analysis, pulse treatments (2 h) of SNP were done. To test the SNP effect on hypocotyl growth, SNP treatment was performed upon seeds synchronized for germination and then maintained until the end point of the experiment. Freshly prepared GA3 (Duchefa) and PAC (Duchefa) were added at the indicated concentrations.
Seedlings were manipulated in darkness under dim (below 0.05 μmol m−2 s−1) green safelight when required.
For the generation of pRGA::GFP-RGA in the nia1,2noa1-2 background, pRGA::GFP-RGA plants were crossed with nia1,2noa1-2 and F2 and F3 progeny were PCR genotyped for nia1, nia2, and noa1-2 mutations as reported previously (Lozano-Juste and León, 2010) and selected with kanamycin for the pRGA::GFP-RGA construct.
Hypocotyl Length Measurements
Seedlings were harvested from petri dishes, laid on acetate sheets, and scanned at 600 dots per inch. The resulting images were used for measuring hypocotyl length by using ImageJ software. Values of hypocotyl length are means ± se of three independent experiments (at least 20 seedlings per experiment were measured).
Protein Extraction and Western Blot
Protein extraction and quantification were performed as reported previously (Lozano-Juste et al., 2011). Samples (15 μg) were separated on 8% polyacrylamide minigels (Bio-Rad; http://www.bio-rad.com) and transferred, when required, onto nitrocellulose membranes (GE Healthcare Spain; http://www.gehealthcare.com/eses/index.html).
GFP-tagged proteins were detected with a 1:8,000 dilution of a monoclonal anti-GFP antibody (Clontech), and TAP-tagged proteins were detected with a 1:10,000 dilution of anti-C-MYC tag-peroxidase conjugate antibody (Sigma). PHYB protein levels were detected with a 1:2,500 dilution of anti-PHYB monoclonal antibody from the laboratory of Akira Nagatani. Loading controls were performed with anti-tubulin antibodies (Sigma) or anti-OAS A1 antibodies from the laboratory of Cecilia Gotor. The secondary antibody was anti-mouse (1:10,000) or anti-rabbit (1:10,000) coupled to horseradish peroxidase, and further detection was performed with the ECL or the ECL Advance kit (GE Healthcare Spain; http://www.gehealthcare.com/eses/index.html). Images were captured with LAS3000 (Fuji) and quantified by using Image Gauge software (Fuji) where indicated.
Quantitative Real-Time PCR
Total RNA was isolated from hypocotyls of 5-d-old seedlings and further analyzed by quantitative RT-PCR techniques as described previously (Castillo and León, 2008). Transcript levels of PIF1, PIF3, PIF4, RGA, GAI, RGL1, RGL2, and RGL3 genes coding for PIF and DELLA proteins, as well as GA biosynthetic and catabolic genes, were analyzed by quantitative RT-PCR using specific primers as reported previously (Frigerio et al., 2006; Alabadí et al., 2008). GID1a, -1b, -1c, and SLY1 were analyzed with primers as follows: qGID1a-F, 5′-GTGACGGTTAGAGACCGCGA-3′; qGID1a-R, 5′-TCCCTCGGGTAAAAACGCTT-3′; qGID1b-F, 5′-TTACGGTCAAGGAACTCGGC-3′; qGID1b-R, 5′-TCGCCCTGACGGTTCTTTC-3′; qGID1c-F, 5′-CGGCTCAAATCTTCGATCTGG-3′; qGID1c-R, 5′-TTGGCATTTGCAGGGACTTTC-3′; qSLY1-F, 5′-GGGCAGAACCAGCTCAGATC-3′; qSLY1-R, 5′-TCTTCGGAAGCCACCAAGC-3′.
Anthocyanin Extraction and Quantification
Samples of 100 mg fresh weight of 5-d-old seedlings grown in darkness or white light were harvested, anthocyanin extracted, and quantified on acidic methanol buffer overnight at 4°C as reported previously (Francis, 1982). Anthocyanin content is expressed as A530 mg−1 fresh weight.
NO and GFP Detection by Fluorescence and Confocal Microscopy
The endogenous levels of NO in hypocotyls were determined by seedling staining with 15 μm DAF-FM DA for 1 h as described (Guo et al., 2003). NO-associated fluorescence was detected under UV illumination with a Nikon Eclipse fluorescence microscope using unchanged parameters for every measurement. Fluorescence intensity was quantified as reported previously (Lozano-Juste and León, 2010).
Fluorescence from pRGA::GFP-RGA was detected using a TCS SL confocal laser scanning microscope (Leica) with a 40× oil-immersion objective lens. For GFP and chloroplast autofluorescence, samples were excited with an argon laser at 488 nm. The fluorescence emission was collected between 497 and 537 nm for GFP (rendered in green) and between 579 and 647 nm band pass for chloroplast autofluorescence (rendered in red). TCS SL average projections of the Z-stack reconstructions (10 slices, 3 μm each) were taken and are presented in Figure 4. Ten seedlings were analyzed per experiment in three independent experiments.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Time and fluence rate dependence effect of red light on the elongation of wild-type and NO-deficient hypocotyls.
Supplemental Figure S2. Similar levels of GFP in NO-treated and control untreated 35S-TAP-GFP seedlings.
Supplemental Figure S3. Relative transcript levels of genes coding for DELLA proteins in wild-type and NO-deficient hypocotyls.
Supplemental Figure S4. Biosynthetic pathway of gibberellins in Arabidopsis and levels of the biosynthetic and catabolic gene transcripts.
Supplemental Figure S5. Control experiments corresponding to Figures 1, 3, 5, and 6 performed in darkness.
Supplementary Material
Acknowledgments
We are especially grateful to Miguel A. Blázquez, David Alabadí, and Oscar Lorenzo for helpful discussions and suggestions and also for comments on the manuscript and to Javier Gallego-Bartolomé and Verónica Arana for experimental advice. We thank the following colleagues for sharing biological material: Tai-Pin Sun for transgenic pRGA::GFP-RGA, sly1-10, and sly1-D mutants; X.W. Deng for transgenic lines overexpressing the TAP-tagged version of DELLAs and the 5della mutant; Peter Quail for pif3, pif4, and pifQ mutants; Giltsu Choi for PIF3OX and Christian Fankhauser for PIF4OX transgenic lines; Steve Thomas for gid1 double mutants; David Alabadí for pRGA:GFP-RGA/ga1-3; Vicente Rubio for 35S::TAP-GFP seeds; and Akira Nagatani and Cecilia Gotor for the generous gifts of anti-PHYB and anti-OAS A1 antibodies, respectively. We also thank Marisol Gascón for technical assistance with confocal microscopy.
References
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP. (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Achard P, Genschik P. (2009) Releasing the brakes of plant growth: how GAs shut down DELLA proteins. J Exp Bot 60: 1085–1092 [DOI] [PubMed] [Google Scholar]
- Achard P, Liao L, Jiang C, Desnos T, Bartlett J, Fu X, Harberd NP. (2007) DELLAs contribute to plant photomorphogenesis. Plant Physiol 143: 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P, Renou J-P, Berthomé R, Harberd NP, Genschik P. (2008) Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr Biol 18: 656–660 [DOI] [PubMed] [Google Scholar]
- Agrawal L, Chakraborty S, Jaiswal DK, Gupta S, Datta A, Chakraborty N. (2008) Comparative proteomics of tuber induction, development and maturation reveal the complexity of tuberization process in potato (Solanum tuberosum L.). J Proteome Res 7: 3803–3817 [DOI] [PubMed] [Google Scholar]
- Alabadí D, Blázquez MA. (2009) Molecular interactions between light and hormone signaling to control plant growth. Plant Mol Biol 69: 409–417 [DOI] [PubMed] [Google Scholar]
- Alabadí D, Gallego-Bartolomé J, Orlando L, García-Cárcel L, Rubio V, Martínez C, Frigerio M, Iglesias-Pedraz JM, Espinosa A, Deng XW, et al. (2008) Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J 53: 324–335 [DOI] [PubMed] [Google Scholar]
- Allakhverdiev SI, Kreslavski VD, Klimov VV, Los DA, Carpentier R, Mohanty P. (2008) Heat stress: an overview of molecular responses in photosynthesis. Photosynth Res 98: 541–550 [DOI] [PubMed] [Google Scholar]
- Al-Sady B, Kikis EA, Monte E, Quail PH. (2008) Mechanistic duality of transcription factor function in phytochrome signaling. Proc Natl Acad Sci USA 105: 2232–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH. (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Bailly C, El-Maarouf-Bouteau H, Corbineau F. (2008) From intracellular signaling networks to cell death: the dual role of reactive oxygen species in seed physiology. C R Biol 331: 806–814 [DOI] [PubMed] [Google Scholar]
- Beligni MV, Fath A, Bethke PC, Lamattina L, Jones RL. (2002) Nitric oxide acts as an antioxidant and delays programmed cell death in barley aleurone layers. Plant Physiol 129: 1642–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beligni MV, Lamattina L. (2000) Nitric oxide stimulates seed germination and de-etiolation, and inhibits hypocotyl elongation, three light-inducible responses in plants. Planta 210: 215–221 [DOI] [PubMed] [Google Scholar]
- Bethke PC, Libourel IG, Aoyama N, Chung YY, Still DW, Jones RL. (2007) The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol 143: 1173–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke PC, Libourel IGL, Jones RL. (2006) Nitric oxide reduces seed dormancy in Arabidopsis. J Exp Bot 57: 517–526 [DOI] [PubMed] [Google Scholar]
- Boccalandro HE, Rugnone ML, Moreno JE, Ploschuk EL, Serna L, Yanovsky MJ, Casal JJ. (2009) Phytochrome B enhances photosynthesis at the expense of water-use efficiency in Arabidopsis. Plant Physiol 150: 1083–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Yamagata H, Neuhaus G, Chua NH. (1994) Phytochrome signal transduction pathways are regulated by reciprocal control mechanisms. Genes Dev 8: 2188–2202 [DOI] [PubMed] [Google Scholar]
- Bright J, Desikan R, Hancock JT, Weir IS, Neill SJ. (2006) ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J 45: 113–122 [DOI] [PubMed] [Google Scholar]
- Castillo MC, León J. (2008) Expression of the beta-oxidation gene 3-ketoacyl-CoA thiolase 2 (KAT2) is required for the timely onset of natural and dark-induced leaf senescence in Arabidopsis. J Exp Bot 59: 2171–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E. (2007) Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci 12: 514–521 [DOI] [PubMed] [Google Scholar]
- Correll MJ, Kiss JZ. (2005) The roles of phytochromes in elongation and gravitropism of roots. Plant Cell Physiol 46: 317–323 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun TP. (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ederli L, Morettini R, Borgogni A, Wasternack C, Miersch O, Reale L, Ferranti F, Tosti N, Pasqualini S. (2006) Interaction between nitric oxide and ethylene in the induction of alternative oxidase in ozone-treated tobacco plants. Plant Physiol 142: 595–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis FJ. (1982) Analysis of anthocyanins. Markakis P, , Anthocyanins as Food Colors. Academic Press, New York, pp 181–207 [Google Scholar]
- Frigerio M, Alabadí D, Pérez-Gómez J, García-Cárcel L, Phillips AL, Hedden P, Blázquez MA. (2006) Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol 142: 553–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Richards DE, Fleck B, Xie D, Burton N, Harberd NP. (2004) The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16: 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Minguet EG, Marín JA, Prat S, Blázquez MA, Alabadí D. (2010) Transcriptional diversification and functional conservation between DELLA proteins in Arabidopsis. Mol Biol Evol 27: 1247–1256 [DOI] [PubMed] [Google Scholar]
- Gapper C, Dolan L. (2006) Control of plant development by reactive oxygen species. Plant Physiol 141: 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, et al. (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo FQ, Okamoto M, Crawford NM. (2003) Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302: 100–103 [DOI] [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C. (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Guo K, Xia K, Yang ZM. (2008) Regulation of tomato lateral root development by carbon monoxide and involvement in auxin and nitric oxide. J Exp Bot 59: 3443–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao F, Zhao S, Dong H, Zhang H, Sun L, Miao C. (2010) Nia1 and Nia2 are involved in exogenous salicylic acid-induced nitric oxide generation and stomatal closure in Arabidopsis. J Integr Plant Biol 52: 298–307 [DOI] [PubMed] [Google Scholar]
- Harberd NP, Belfield E, Yasumura Y. (2009) The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: how an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 21: 1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Tang RH, Hao Y, Stevens RD, Cook CW, Ahn SM, Jing L, Yang Z, Chen L, Guo F, et al. (2004) Nitric oxide represses the Arabidopsis floral transition. Science 305: 1968–1971 [DOI] [PubMed] [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW. (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JK, Yun BW, Kang JG, Raja MU, Kwon E, Sorhagen K, Chu C, Wang Y, Loake GJ. (2008) Nitric oxide function and signalling in plant disease resistance. J Exp Bot 59: 147–154 [DOI] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. (2009) Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J 28: 3893–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Neill SJ, Tang Z, Cai W. (2005) Nitric oxide mediates gravitropic bending in soybean roots. Plant Physiol 137: 663–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Stettmaier K, Michel C, Hutzler P, Mueller MJ, Durner J. (2004) Nitric oxide is induced by wounding and influences jasmonic acid signaling in Arabidopsis thaliana. Planta 218: 938–946 [DOI] [PubMed] [Google Scholar]
- Huq E. (2006) Degradation of negative regulators: a common theme in hormone and light signaling networks? Trends Plant Sci 11: 4–7 [DOI] [PubMed] [Google Scholar]
- Jang IC, Yang JY, Seo HS, Chua NH. (2005) HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev 19: 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW. (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Jing HC, Hebeler R, Oeljeklaus S, Sitek B, Stühler K, Meyer HE, Sturre MJ, Hille J, Warscheid B, Dijkwel PP. (2008) Early leaf senescence is associated with an altered cellular redox balance in Arabidopsis cpr5/old1 mutants. Plant Biol (Stuttg) (Suppl 1) 10: 85–98 [DOI] [PubMed] [Google Scholar]
- Jonassen EM, Sandsmark BA, Lillo C. (2009) Unique status of NIA2 in nitrate assimilation: NIA2 expression is promoted by HY5/HYH and inhibited by PIF4. Plant Signal Behav 4: 1084–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbert Z, Bartha B, Erdei L. (2008) Exogenous auxin-induced NO synthesis is nitrate reductase-associated in Arabidopsis thaliana root primordia. J Plant Physiol 165: 967–975 [DOI] [PubMed] [Google Scholar]
- Lanteri ML, Laxalt AM, Lamattina L. (2008) Nitric oxide triggers phosphatidic acid accumulation via phospholipase D during auxin-induced adventitious root formation in cucumber. Plant Physiol 147: 188–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee U, Wie C, Fernandez BO, Feelisch M, Vierling E. (2008) Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant Cell 20: 786–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Al-Sady B, Carle C, Storer A, Alonso JM, Ecker JR, Quail PH. (2008a) The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH. (2008b) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, Quail PH. (2009) Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wakao S, Fischer BB, Niyogi KK. (2009) Sensing and responding to excess light. Annu Rev Plant Biol 60: 239–260 [DOI] [PubMed] [Google Scholar]
- Lin Y, Cheng CL. (1997) A chlorate-resistant mutant defective in the regulation of nitrate reductase gene expression in Arabidopsis defines a new HY locus. Plant Cell 9: 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J, Acevedo-Hernández G, Ramírez-Chávez E, Molina-Torres J, Herrera-Estrella L. (2006) Novel signals for plant development. Curr Opin Plant Biol 9: 523–529 [DOI] [PubMed] [Google Scholar]
- Lozano-Juste J, Colom-Moreno R, León J. (March 4, 2011) In vivo protein tyrosine nitration in Arabidopsis thaliana. J Exp Bot http://dx.doi.org/10.1093/jxb/err042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Juste J, León J. (2010) Enhanced abscisic acid-mediated responses in nia1nia2noa1-2 triple mutant impaired in NIA/NR- and AtNOA1-dependent nitric oxide biosynthesis in Arabidopsis. Plant Physiol 152: 891–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muangprom A, Thomas SG, Sun TP, Osborn TC. (2005) A novel dwarfing mutation in a green revolution gene from Brassica rapa. Plant Physiol 137: 931–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur LA, Laarhoven LJ, Harren FJ, Hall MA, Smith AR. (2008) Nitric oxide interacts with salicylate to regulate biphasic ethylene production during the hypersensitive response. Plant Physiol 148: 1537–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP, Jones JD. (2008) DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol 18: 650–655 [DOI] [PubMed] [Google Scholar]
- Neill S, Barros R, Bright J, Desikan R, Hancock J, Harrison J, Morris P, Ribeiro D, Wilson I. (2008) Nitric oxide, stomatal closure, and abiotic stress. J Exp Bot 59: 165–176 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL. (2008) Dawning of a new era: photomorphogenesis as an integrated molecular network. Curr Opin Plant Biol 11: 4–8 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Ang LH, Deng XW. (1999) The role of COP1 in repression of Arabidopsis photomorphogenic development. Trends Cell Biol 9: 113–118 [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Lanteri ML, Lamattina L. (2003) Nitric oxide and cyclic GMP are messengers in the indole acetic acid-induced adventitious rooting process. Plant Physiol 132: 1241–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado AM, Porterfield DM, Feijó JA. (2004) Nitric oxide is involved in growth regulation and re-orientation of pollen tubes. Development 131: 2707–2714 [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R, Behringer C, Müller IK, Schwechheimer C. (2010) The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and phytochrome-interacting factors. Genes Dev 24: 2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Puertas MC, Perazzolli M, Zago ED, Delledonne M. (2004) Nitric oxide signalling functions in plant-pathogen interactions. Cell Microbiol 6: 795–803 [DOI] [PubMed] [Google Scholar]
- Saito N, Nakamura Y, Mori IC, Murata Y. (2009) Nitric oxide functions in both methyl jasmonate signaling and abscisic acid signaling in Arabidopsis guard cells. Plant Signal Behav 4: 119–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH. (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999 [DOI] [PubMed] [Google Scholar]
- Shen H, Moon J, Huq E. (2005) PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J 44: 1023–1035 [DOI] [PubMed] [Google Scholar]
- Shen Y, Khanna R, Carle CM, Quail PH. (2007) Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol 145: 1043–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G. (2009) Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci USA 106: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Park E, Choi G. (2007) PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J 49: 981–994 [DOI] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Chory J, Furuya M. (1994) The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome B and secondarily by phytochrome A. Plant Physiol 104: 363–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Tseng TS, Swain SM, Dill A, Jeong SY, Olszewski NE, Sun TP. (2007) Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol 143: 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavang JA, Gallego-Bartolomé J, Gómez MD, Yoshida S, Asami T, Olsen JE, García-Martínez JL, Alabadí D, Blázquez MA. (2009) Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J 60: 589–601 [DOI] [PubMed] [Google Scholar]
- Tonón C, Terrile MC, Iglesias MJ, Lamattina L, Casalongué C. (2010) Extracellular ATP, nitric oxide and superoxide act coordinately to regulate hypocotyl growth in etiolated Arabidopsis seedlings. J Plant Physiol 167: 540–546 [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN. (2010) Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- Tun NN, Livaja M, Kieber JJ, Scherer GF. (2008) Zeatin-induced nitric oxide (NO) biosynthesis in Arabidopsis thaliana mutants of NO biosynthesis and of two-component signaling genes. New Phytol 178: 515–531 [DOI] [PubMed] [Google Scholar]
- Wang H, Liang X, Wan Q, Wang X, Bi Y. (2009) Ethylene and nitric oxide are involved in maintaining ion homeostasis in Arabidopsis callus under salt stress. Planta 230: 293–307 [DOI] [PubMed] [Google Scholar]
- Wendehenne D, Durner J, Klessig DF. (2004) Nitric oxide: a new player in plant signalling and defence responses. Curr Opin Plant Biol 7: 449–455 [DOI] [PubMed] [Google Scholar]
- Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EM, Maier A, Schwechheimer C. (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters H, Jürgens G. (2009) Survival of the flexible: hormonal growth control and adaptation in plant development. Nat Rev Genet 10: 305–317 [DOI] [PubMed] [Google Scholar]
- Yang J, Lin R, Sullivan J, Hoecker U, Liu B, Xu L, Deng XW, Wang H. (2005) Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17: 804–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, Rodriguez R, Tran A, Hoang H, de los Santos D, Brown S, Vellanoweth RL. (2000) The developmental transition to flowering represses ascorbate peroxidase activity and induces enzymatic lipid peroxidation in leaf tissue in Arabidopsis thaliana. Plant Sci 158: 115–127 [DOI] [PubMed] [Google Scholar]
- Zottini M, Costa A, De Michele R, Ruzzene M, Carimi F, Lo Schiavo F. (2007) Salicylic acid activates nitric oxide synthesis in Arabidopsis. J Exp Bot 58: 1397–1405 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.