Abstract
White lupin (Lupinus albus) is a legume that is very efficient in accessing unavailable phosphorus (Pi). It develops short, densely clustered tertiary lateral roots (cluster/proteoid roots) in response to Pi limitation. In this report, we characterize two glycerophosphodiester phosphodiesterase (GPX-PDE) genes (GPX-PDE1 and GPX-PDE2) from white lupin and propose a role for these two GPX-PDEs in root hair growth and development and in a Pi stress-induced phospholipid degradation pathway in cluster roots. Both GPX-PDE1 and GPX-PDE2 are highly expressed in Pi-deficient cluster roots, particularly in root hairs, epidermal cells, and vascular bundles. Expression of both genes is a function of both Pi availability and photosynthate. GPX-PDE1 Pi deficiency-induced expression is attenuated as photosynthate is deprived, while that of GPX-PDE2 is strikingly enhanced. Yeast complementation assays and in vitro enzyme assays revealed that GPX-PDE1 shows catalytic activity with glycerophosphocholine while GPX-PDE2 shows highest activity with glycerophosphoinositol. Cell-free protein extracts from Pi-deficient cluster roots display GPX-PDE enzyme activity for both glycerophosphocholine and glycerophosphoinositol. Knockdown of expression of GPX-PDE through RNA interference resulted in impaired root hair development and density. We propose that white lupin GPX-PDE1 and GPX-PDE2 are involved in the acclimation to Pi limitation by enhancing glycerophosphodiester degradation and mediating root hair development.
Phosphorus (P) is an essential macronutrient required for plant growth and development (Bieleski, 1973). It is essential in cell constituents such as ATP, nucleic acids, and phospholipids. It also plays a pivotal role in energy conservation, metabolic regulation, and the signal transduction cascade (Marschner, 1995; Raghothama, 1999). Although P is abundant in many soils, its bioavailability is generally low due to fixation processes (Hinsinger, 2001); thus, plants have broad adaptive strategies to improve the acquisition, use, and recycling of inorganic phosphate (Pi) through the modification of gene expression and developmental processes (Williamson et al., 2001; Vance et al., 2003; Hammond et al., 2004; Lambers et al., 2006).
White lupin (Lupinus albus) is known for its extreme tolerance to low Pi availability and unusual root architecture (Gardner et al., 1983; Tadano and Sakai, 1991; Johnson et al., 1996; Gilbert et al., 2000; Neumann et al., 2000; Vance et al., 2003; Shen et al., 2005). In response to Pi deficiency, white lupin develops short, densely clustered lateral roots (cluster/proteoid roots). The formation of cluster roots having abundant root hairs results in a striking increase in surface area available for Pi uptake from the rhizosphere (Keerthisinghe et al., 1998; Neumann et al., 1999). Cluster roots also excrete large amounts of protons, citrate, and acid phosphatase (LaSAP) to the rhizosphere, consequently increasing Pi bioavailability (Dinkelaker et al., 1989; Neumann et al., 1999; Watt and Evans, 1999; Miller et al., 2001; Wasaki et al., 2003; Zhang et al., 2004). During the past few years, a number of Pi limitation-induced genes have been described in white lupin cluster roots, including a high-affinity Pi transporter gene (LaPT1; Liu et al., 2001), an acid phosphatase (LaSAP1; Wasaki et al., 1999; Miller et al., 2001), and a multidrug and toxin efflux gene (LaMATE; Uhde-Stone et al., 2005). Uhde-Stone et al. (2003) identified 35 ESTs or EST contigs that showed enhanced expression in cluster roots of Pi-deficient white lupin. Two of the most abundant ESTs encode deduced proteins containing a Pfam PF03009 motif characteristic of glycerophosphodiester phosphodiesterases (GPX-PDEs).
GPX-PDE enzymes catalyze the hydrolysis of deacylated phospholipid GPX to glycerol-3-phosphate and the corresponding alcohol. GPX-PDEs have been described in bacteria (Larson et al., 1983; Brzoska and Boos, 1988; Tommassen et al., 1991), yeast (Fisher et al., 2005; Santos-Beneit et al., 2009), and mammals (Zheng et al., 2000, 2003; Yanaka et al., 2003; Rao and Sockanathan, 2005; Corda et al., 2009). They appear to have important functions in Pi scavenging and carbon stress along with numerous other aspects of development. Until recently, much less was known about plant GPX-PDEs. Upon the development and use of arrays to evaluate gene expression, several studies have noted the enhanced expression of transcripts that annotate as GPX-PDEs (Uhde-Stone et al., 2003; Misson et al., 2005; Morcuende et al., 2007; Müller et al., 2007) Plant GPX-PDE enzyme was first identified in carrot (Daucus carota) cells. Both intracellular and extracellular GPX-PDE activities were induced in Pi-deficient cell cultures of carrot, Arabidopsis (Arabidopsis thaliana), and sycamore (Acer pseudoplatanus; Van der Rest et al., 2002). Enzyme assays performed with purified GPX-PDE enzyme from carrot cell walls showed a broad substrate range for glycerophosphodiesters such as glycerophosphocholine (GPC), glycerophosphoinositol (GPI), and glycerophosphoethanolamine. No functional genetic role for GPX-PDE was noted in these studies.
While this article was in review, Cheng et al. (2011) reported on two subfamilies of GPX-PDEs in Arabidopsis. They described a six-member gene family (A) of more typical GPX-PDEs and a seven-member family (B) of GPX-PDE-like genes. Recombinant forms of one member from each family were evaluated in some detail. At3g02040 representing more typical type A GPX-PDEs and At1g66970 representing type B GPX-PDE-like, as exemplified by SHAVEN3 (SHV3), hydrolyzed glycerolphosphoglycerol, GPC, and glycerophosphoethanolamine. Interestingly, expression of At3g02040 was induced by P insufficiency, while At1g66970 expression was not. At3g02040 appeared to be plastid localized, and a loss-of-function mutant of At3g02040 had a modestly reduced growth rate under Pi deficiency. Cheng et al. (2011) suggested that At3g02040 may be involved in Pi release from plastid phospholipids during P deficiency. Membrane phospholipid turnover and remodeling of the plasmalemma with galactolipids is well known in Pi-stressed plants (Andersson et al., 2003, 2005; Lee et al., 2003; Rietz et al., 2004, 2010; Misson et al., 2005; Morcuende et al., 2007; Nakamura et al., 2009).
Studies in white lupin (Liu et al., 2005; Tesfaye et al., 2007; Zhou et al., 2008) and Arabidopsis (Ciereszko et al., 2005; Jain et al., 2007; Hammond and White, 2008; Lei et al., 2011) provide evidence that sugar (Suc/photosynthate) modulates Pi deficiency-induced gene expression. Liu et al. (2005) and Tesfaye et al. (2007) demonstrated that the expression of several white lupin Pi-starved cluster root genes required photosynthate. Likewise, Zhou et al. (2008) showed that Suc was required for Pi deficiency-induced cluster root formation and Pi stress-induced gene expression. In Arabidopsis, the expression of 149 genes was shown to have a sugar-Pi availability interaction (Müller et al., 2007). Lei et al. (2011) showed that Suc transport in Arabidopsis was required for Pi deficiency-induced gene expression. It is noteworthy that microbial Pi stress-induced GPX-PDE gene expression is regulated in part by sugar availability (Kasahara et al., 1991; Santos-Beneit et al., 2009). Whether sugar plays a role in affecting plant GPX-PDE expression is currently unknown.
Increased proliferation of root hairs enhances plant acclimation to Pi deficiency by increasing root surface area and increasing exploitable soil volume (Bates and Lynch, 1996; Gahoonia and Nielsen, 1998; Ma et al., 2001). A root-hairless mutant line in barley (Hordeum vulgare) was less efficient in Pi uptake and showed attenuated acid phosphatase activity (Gahoonia et al., 2001). Arabidopsis mutants impaired in phospholipid metabolism displayed impaired root hair development and shorter root hairs (Böhme et al., 2004; Jones et al., 2006; Hayashi et al., 2008). Hayashi et al. (2008) presented a multialignment between SHV3 and other GPX-PDEs showing that conserved essential residues for GPX-PDE enzyme activity were not present in the SHV3 protein sequence. However, they did not report GPC-PDE activity. We question whether the white lupin GPX-PDEs may be involved in root hair morphological changes related to insufficient P.
In this study, we evaluated cluster root acclimation to Pi deficiency and the role that GPX-PDEs play in Pi deficiency and root hair development. Here, we report the following: (1) two white lupin GPX-PDEs (GPX-PDE1 and GPX-PDE2) are highly up-regulated by Pi limitation and are expressed in root hair, epidermal, and vascular cells; (2) GPX-PDE2 transcripts show rapid suppression in response to Pi resupply without a change in shoot P status, but GPX-PDE1 is less responsive; (3) GPX-PDEs are also affected by photosynthate deprivation, but in opposite manners; (4) GPX-PDE1 complements a yeast gde1Δ mutant revealing a GPC-PDE function, and recombinant GPX-PDE2 protein has highest activity with GPI but is also active with GPC; and (5) RNA interference (RNAi)-mediated knockdown of GPX-PDE1 and GPX-PDE2 impairs root hair development. Taken together, we propose a role for GPX-PDEs in root hair development and phospholipid degradation in Pi-stressed white lupin cluster roots.
RESULTS
Identification and Characterization of GPX-PDE1 and GPX-PDE2 Genes in White Lupin
We isolated and sequenced two full-length cDNA clones that have homology with GPX-PDE ESTs detected in a cluster root cDNA library of white lupin (Uhde-Stone et al., 2003). The identified cDNAs were designated as GPX-PDE1 and GPX-PDE2 according to their deduced amino acid sequences containing a Pfam PF03009 motif characteristic of GPX-PDE. GPX-PDE1 contains a 1,191-bp open reading frame encoding a 396-amino acid polypeptide with an estimated molecular mass of 45.8 kD. GPX-PDE2 contains a 1,125-bp open reading frame encoding a 374-amino acid polypeptide with an estimated molecular mass of 42.1 kD.
Multiple alignment of the white lupin GPX-PDE1 and GPX-PDE2 deduced amino acid sequences with known GPX-PDEs showed that both GPX-PDEs have the conserved sequence motif [H1RG(X)nE1XT(X)nE2XD1(X)nH2D2(X)nE3XK] comprising the active site residues for GPX-PDE activity (Supplemental Fig. S1A). Several residues in this motif (H1, R, E2, D1, H2, E3, K) have been identified as essential sites for GPX-PDE enzyme activity based on crystal structure and mutagenesis studies (Zheng et al., 2003; Shi et al., 2008; Corda et al., 2009). Based on signal prediction programs, the GPX-PDE1 deduced amino acid sequence contains a hydrophobic N-terminal region that was predicted to be a signal peptide with 25 amino acid residues (Supplemental Fig. S2) and targets GPX-PDE1 to the secretory system. One N-glycosylation site (-Asn-Xaa-Ser/Thr-) was predicted in both GPX-PDEs. Both GPX-PDE1 and GPX-PDE2 deduced amino acid sequences contain several predicted Ser/Thr/Tyr phosphorylation sites. Gene Ontogeny (GO) analysis of GPX-PDE1 indicated a GO biological process of GO.0006071, glycerol metabolism, GPX-PDE enzyme. InterPro (http://www.ebi.ac.uk/Tools/InterProScan/) analysis suggests that GPX-PDE2 is related to a cyclin-dependent kinase (e-10) similar to Pho85. However, we found neither a typical kinase domain nor kinase activity in GPX-PDE2. The highest predicted GO biological process for GPX-PDE2 function is GO.0007242, intracellular signaling cascade, GPX-PDE enzyme.
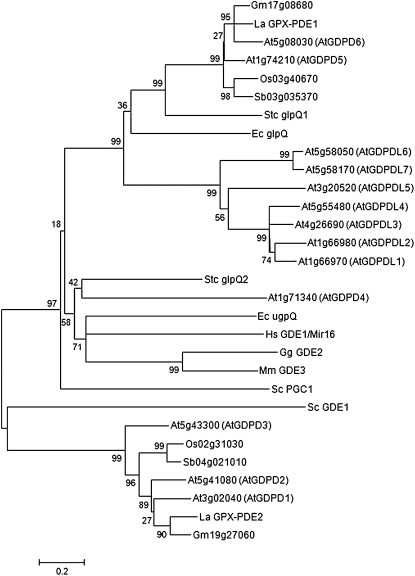
Using GPX-PDE1 and GPX-PDE2 deduced amino acid sequences to search in sequenced plant genomes including those of soybean (Glycine max), rice (Oryza sativa), Arabidopsis, and sorghum (Sorghum bicolor), we found that GPX-PDEs are widespread in various plant species (Supplemental Fig. S1B). The identities of GPX-PDE1 and GPX-PDE2 with the corresponding orthologs range from 68% to 79% and 57% to 75%, respectively. Interestingly, GPX-PDE1 and GPX-PDE2 share only 16% identity with each other, even though both of them contain the GPX-PDE domain and are assigned to the superfamily of phosphoinositide phospholipase C-like phosphodiesterases by the SCOP database (http://supfam.mrc-lmb.cam.ac.uk/SUPERFAMILY/). A phylogenetic analysis of these known GPX-PDEs from various species and putative orthologs is revealing. Figure 1 shows that the GPX-PDE family has diversified into distinct clades, and GPX-PDE1 and GPX-PDE2 belong to very different clades. GPX-PDE1 is closely related to glpQ and distantly related to another plant type B GPX-PDEL subgroup that contains Arabidopsis SHV3. In comparison, GPX-PDE2 sits in a highly divergent group that includes At3g02040, a putative plastid-targeted GPX-PDE (Cheng et al., 2011). These results show that although both GPX-PDE1 and GPX-PDE2 have the characteristics of GPX-PDE enzymes, they are highly divergent, may utilize different substrates, and have distinct catalytic functions.
Figure 1.
Phylogenetic tree of white lupin GPX-PDEs with related GPX-PDE proteins from various species. The phylogenetic tree of white lupin GPX-PDEs with proteins containing GPX-PDE domains was generated using MEGA4 from a ClustalX alignment (sequence alignment is available as Supplemental Data Set S1). Numbers represent bootstrap values obtained from 1,000 trials. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Evolutionary distances are displayed in units of the number of amino acid substitutions per site. Abbreviations for species are as follows: At, Arabidopsis thaliana; Ec, Escherichia coli; Gg, Gallus gallus; Gm, Glycine max; Hs, Homo sapiens; La, Lupinus albus; Mm, Mus musculus; Os, Oryza sativa; Sb, Sorghum bicolor; Sc, Saccharomyces cerevisiae; Stc, Streptomyces coelicolor.
The gene structure and 5′-upstream putative promoter regions for GPX-PDE1 and GPX-PDE2 are shown in Supplemental Figure S3. GPX-PDE1 is 3,272 bp and contains seven introns. The 1,751-bp 5′-upstream putative promoter region of GPX-PDE1 contains one P1BS element (GTATATTC; bp −116). GPX-PDE1 also contains two WRKY binding sites (W-box; TTGACC; bp −912 and −973) and a sequence (CACTTG; bp −1,001) that is similar to the G-box motif (CACGTG). By comparison, the GPX-PDE2 gene is 3,092 bp and contains six introns. The 1,772-bp 5′-upstream promoter region of GPX-PDE2 contains four P1BS elements (GNATATNC; bp −118, −131, −161, and −1,497). GPX-PDE2 also contains two W-boxes (TTGACT; bp −202 and −985) and a sequence (CACTTG; bp −1,010) that is similar to the G-box motif (CACGTG). The GPX-PDE2 promoter also contains four TATCCA elements (bp −158, −295, −1,350, −1,391).
Complementation of a gde1 Yeast Mutant with GPX-PDE1 and GPX-PDE2 and in Vitro GPX-PDE Enzyme Assays
To confirm that GPX-PDE1 and GPX-PDE2 encode functional GPX-PDE enzymes, we employed functional complementation of a yeast mutant by transformation with GPX-PDE1 and GPX-PDE2 and performed in vitro GPX-PDE enzyme assays. In Saccharomyces cerevisiae, an intracellular protein encoded by the GDE1 (YPL110c) gene has been characterized as a GPC-PDE (Fisher et al., 2005). Hydrolysis of GPC by Gde1 is required to utilize GPC as the sole phosphate source. A yeast mutant, gde1Δ, with a deletion of GDE1 results in the accumulation of GPC in the cell; consequently, the mutant cannot grow when GPC is supplied as the sole phosphate source (Fisher et al., 2005). The gde1Δ mutant cells were transformed with the GPX-PDE1 and GPX-PDE2 constructs, and growth was monitored by measuring the optical density at 600 nm. Table I shows that the gde1Δ mutant transformed with GPX-PDE1 gained wild-type growth characteristics when GPC was supplied as a sole phosphate source. In contrast, GPX-PDE2 transformants did not complement the gde1Δ mutant under the same conditions. The fact that GPX-PDE1 complements the function of yeast GDE1, which lacks GPC-PDE activity, provides strong evidence that GPX-PDE1 encodes an enzyme that can hydrolyze GPC to choline and glycerol-3-phosphate in eukaryotes. We further confirmed the specificity of GPX-PDE1 for GPC with recombinant enzyme activity. Although recombinant GPX-PDE1 activity was difficult to recover due to it being produced in inclusion bodies, when activity was recovered, GPC served as a substrate but not GPI (Supplemental Table S1).
Table I. Yeast complementation of GPX-PDE1 and GPX-PDE2.
Strains were grown in medium lacking phosphate (no-Pi medium), containing 200 μm KH2PO4 (low-Pi medium), and lacking phosphate but containing 200 μm GPC (no-Pi + GPC medium). For the gde1Δ mutant, medium also lacked uracil to maintain empty vector (EV) or vector containing GDX-PDE1 or GPX-PDE2. Cultures were grown for 3 d at 30°C. Data represent average growth (A600) ± se for two to four replicates.
| Strain | Medium |
||
| No Pi + GPC | Low Pi | No Pi | |
| Wild type | 1.21 ± 0.02 | 1.35 ± 0.25 | 0.03 ± 0.00 |
| gde1Δ + EV | 0.06 ± 0.03 | 1.74 ± 0.25 | 0.03 ± 0.01 |
| gde1Δ + GPX-PDE1 | 1.17 ± 0.07 | 1.79 ± 0.52 | 0.05 ± 0.01 |
| gde1Δ + GPX-PDE2 | 0.02 ± 0.01 | 1.93 ± 0.02 | 0.07 ± 0.05 |
The failure to complement the gde1Δ mutant with GPX-PDE2 suggested that GPX-PDE2 might cleave a substrate other than GPC. In vitro enzyme assays with purified recombinant GPX-PDE2 as a fusion protein with glutathione S-transferase (GST) from Escherichia coli were carried out to test whether the GPX-PDE2 protein could use GPC and GPI as substrates. Enzyme activity of purified recombinant GPX-PDE2 with GPI was strikingly high in the presence of 10 mm Mg2+. The enzyme requirement for Mg2+ could not be replaced by Ca2+ (Supplemental Table S1). By comparison, GPC activity with recombinant GPX-PDE2 was 73% lower than that of GPI. Free GST protein had no GPC-PDE or GPI-PDE activity. Based on these results, we conclude that white lupin GPX-PDE2 has both GPC and GPI activities but appears to favor GPI.
Preparing cell-free soluble protein and cell wall protein, we evaluated GPX-PDE enzyme activity in Pi-deficient and Pi-sufficient white lupin roots (Table II). In previous studies, Van der Rest et al. (2002) detected GPC-PDE activity in both extracellular and intracellular extracts of carrot cells, which was enhanced during P deficiency. Purified carrot cell GPC-PDE protein had a mass of 55 kD (Van der Rest et al., 2004). We prepared protein extracts from Pi-deficient zone 4 and 5 cluster roots and P-sufficient roots to enrich for GPX-PDE proteins. The 25% to 75% (NH4)2SO4 precipitated fraction from total soluble protein was resolubilized in extraction buffer. The cellular debris after the initial centrifugation of soluble protein was combined with the cell wall filtrate and then extracted in 200 mm CaCl2 to release cell wall proteins. The 25% to 75% (NH4)2SO4 precipitated protein fraction and the cell wall-extractable protein were used for enzyme assays and immunoblot analysis. The (NH4)2SO4-precipitable protein obtained from the soluble protein extract had both GPC-PDE and GPI-PDE activities (Table II). GPC-PDE activity in Pi-deficient cluster roots was 3- to 4-fold higher than that of Pi-sufficient roots. GPI-PDE activity was readily detectable in Pi-deficient cluster roots but was not detectable in Pi-sufficient roots. Enzyme activity for both GPC-PDE and GPI-PDE in cell wall extracts was not consistently detectable. However, when detected, both GPI-PDE and GPC-PDE served as substrates (data not shown). Control assays lacking Mg2+ and Ca2+ showed no GPX-PDE activity.
Table II. GPC-PDE and GPI-PDE enzyme activity in white lupin roots.
P ≤ 0.05 for treatments. Statistical significance was based on four biological replicates.
| Enzyme | 25% to 75% (NH4)2SO4 Cut |
|
| −Pi Cluster Roots | +Pi Normal Roots | |
| nmol min−1 mg−1 protein | ||
| GPC-PDE, Ca2+ | 35.2 ± 4.8 | 10.5 ± 2.2 |
| GPC-PDE, Mg2+ | 28.9 ± 3.6 | 6.6 ± 0.4 |
| GPI-PDE, Ca2+ | 4.5 ± 0.4 | 0 |
| GPI-PDE, Mg2+ | 3.4 ± 0.4 | 0 |
Immunodetection of GPX-PDEs
We raised enzyme-specific polyclonal antisera to recombinant forms of GPX-PDE1 and GPX-PDE2 (Supplemental Fig. S4). Antisera raised to GPX-PDE1 did not recognize recombinant GPX-PDE2; conversely, GPX-PDE2 antisera did not recognize recombinant GPX-PDE1 protein (Supplemental Fig. S4A). This is not unexpected, based upon their lack of deduced amino acid identity (Fig. 1; Supplemental Fig. S1A). Immunoblots of the 25% to 75% (NH4)2SO4 fraction of soluble protein and cell wall-extractable protein confirmed the in planta presence of both GPX-PDE1 and GPX-PDE2 (Supplemental Fig. S4B). Similar to the carrot cell GPC-PDE characterized by Van der Rest et al. (2004), white lupin GPX-PDE1 antiserum recognized two polypeptides of about 49 and 60 kD, somewhat larger than the 45-kD deduced protein. Although there was only a slight difference in the abundance of GPX-PDE1 60-kD protein in the 25% to 75% (NH4)2SO4 fraction of Pi-deficient plants as compared with Pi-sufficient plants, the abundance of GPX-PDE1 in cell wall protein of Pi deficiency-induced cluster roots was very abundant, while it was nearly nondetectable in Pi-sufficient plants. The GPX-PDE1 polypeptide migrating at 47 kD was more abundant in Pi-deficient plants. GPX-PDE2 antisera recognized polypeptides of varying mass in the 25% to 75% (NH4)2SO4 fraction of soluble protein as compared with the cell wall-extractable protein. The abundance of GPX-PDE2 protein was substantially greater in cell wall extracts of Pi-deficient plants as compared with Pi-sufficient plants. The trend was similar in the 25% to 75% (NH4)2SO4 fraction, but less striking. The GPX-PDE2 polypeptide detected in the 25% to 75% fraction was approximately 68 to 72 kD. This would coincide with a GPX-PDE2 dimer that was not effectively denatured by SDS. Van der Rest et al. (2002) noted that carrot cell native GPC-PDE enzyme was multimeric and was resistant to SDS denaturation. However, we did notice an inconsistency in cell wall protein abundance of GPX-PDE1 and GPX-PDE2 and GPX-PDE activities. We used several different buffer combinations to obtain proteins for enzyme assays but had difficulty consistently detecting GPC-PDE and GPI-PDE activities in cell wall protein extracts. In contrast, we consistently found GPX-PDE1 and GPX-PDE2 protein level differences between Pi-deficient and Pi-sufficient cell walls. Inconsistent detection of cell wall enzyme activity could reflect any number of difficulties, including but not limited to enzyme instability, loss of posttranslational modifications, and loss of accessory proteins needed for activity.
Transcript Expression of GPX-PDE1 and GPX-PDE2 in White Lupin Roots under Pi-Sufficient and Pi-Deficient Conditions
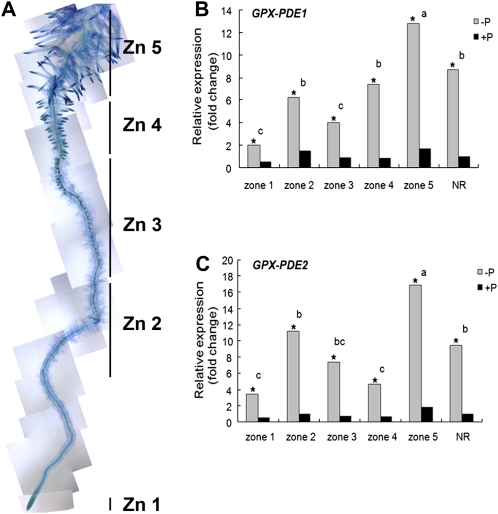
To understand the expression pattern of GPX-PDEs during cluster root development under both Pi-sufficient and Pi-deficient conditions, quantitative reverse transcription (qRT)-PCR and RNA blots were used to analyze transcripts from various developmental zones of cluster roots. As shown in Figure 2A, cluster roots from both Pi-sufficient and Pi-deficient plants were separated into five different zones: zone 1, root tip; zone 2, early meristem; zone 3, unemerged rootlets; zone 4, newly emerged and juvenile rootlets; and zone 5, mature cluster roots covered with abundant root hairs (Neumann et al., 1999; Sbabou et al., 2010). It should be noted that Pi-sufficient plants form cluster roots identical to Pi-sufficient plants but at a much lower density (Johnson et al., 1994, 1996). qRT-PCR of both GPX-PDE1 and GPX-PDE2 showed strikingly high transcript abundance in Pi-deficient roots as compared with Pi-sufficient roots during each developmental stage (Fig. 2, B and C). When comparing the qRT-PCR expression patterns among different zones, GPX-PDE1 and GPX-PDE2 showed greatest transcript abundance in cluster root zone 5 of Pi-deficient plants, where root hairs are most abundant (Supplemental Fig. S5). RNA-blot analysis of GPX-PDE1 and GPX-PDE2 reflected qRT-PCR results (data not shown).
Figure 2.
Cluster root development and qRT-PCR analysis of GPX-PDE expression in cluster roots of white lupin grown under Pi-sufficient and Pi-deficient conditions. A, Lupin cluster roots from Pi-deficient and Pi-sufficient plants can be separated into five zones (Zn): zone 1, root tip; zone 2, early meristems; zone 3, unemerged rootlets; zone 4, newly emerged and juvenile rootlets; zone 5, mature cluster roots covered with abundant root hairs (Neumann et al., 1999). Note that cluster roots can form on Pi-sufficient plants but constitute 10% or less of the root mass. However, on Pi-deficient plants, they constitute greater than 60% of the root mass (Johnson et al., 1994, 1996). B and C, Relative expression of white lupin GPX-PDE1 and GPX-PDE2 in the five developmental zones of Pi-deficient and Pi-sufficient cluster roots. qRT-PCR is shown for first-strand cDNA generated from Pi-sufficient (+P) and Pi-deficient (−P) lupin cluster root zones and normal roots (NR) using specific primer pairs for GPX-PDE1 and GPX-PDE2. Data are expressed as relative values based on the GPX-PDE1 or GPX-PDE2 expression level in +P normal roots and referenced as 1.0. Lowercase letters denote significant differences between zones (P ≤ 0.05). Asterisks denote significant differences between –P and +P (P ≤ 0.05). Statistical significance was based on three biological replicates.
The Response of GPX-PDE1 and GPX-PDE2 to Pi or Phosphite Resupply
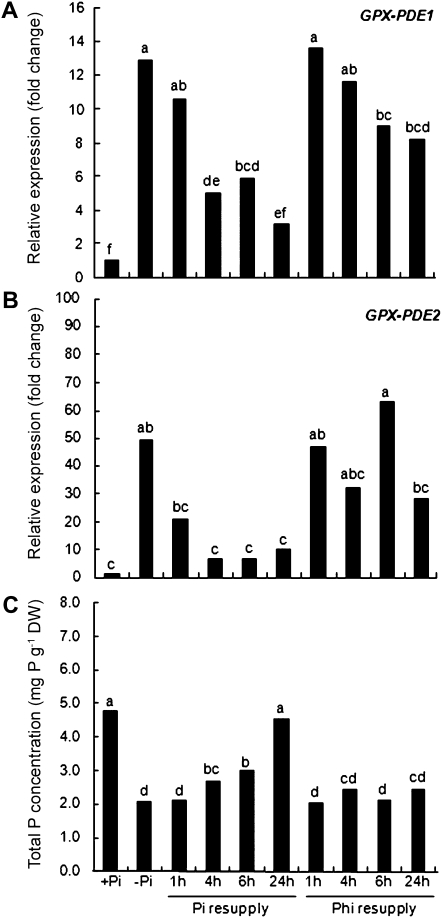
To better understand the effect of Pi on the expression of GPX-PDE1 and GPX-PDE2, 1 mm Pi or 1 mm phosphite, a nonmetabolized analog of Pi, was resupplied to Pi-deficient white lupin plants. The transcript abundance of both genes was monitored by qRT-PCR at 1, 4, 6, and 24 h after Pi or Phi resupply. As shown in Figure 3, when Pi-deficient plants were resupplied with Pi, GPX-PDE2 transcripts were reduced within 1 h, with further reduction by 4 h. However, GPX-PDE1 transcripts showed a slower response to Pi resupply, with reduction only after 4 h. By comparison, Phi, which cannot be metabolized, essentially mimicked the effect of Pi resupply after 24 h.
Figure 3.
The effect of Pi or Phi resupply on transcript abundance of Pi deficiency-induced GPX-PDE1 and GPX-PDE2. White lupin plants were grown to 14 d after emergence without Pi and then resupplied with 1 mm Pi. Control plants were grown for 14 d after emergence with +Pi (1 mm), −Pi (0 mm), or +Phi (1 mm). Cluster roots from control and treated Pi-resupplied plants excised at 1, 4, 6, and 24 h were used for RNA isolation and cDNA synthesis. A and B, GPX-PDE1 (A) and GPX-PDE2 (B) expression analyzed by qRT-PCR. C, Total P concentration in control and treated plants. Data are expressed as relative values based on the GPX-PDE1 or GPX-PDE2 expression level in +P normal roots and referenced as 1.0. Lowercase letters denote significant differences between treatments (P ≤ 0.05). Statistical significance was based on three biological replicates. DW, Dry weight.
The Effect of Photosynthate Deprivation by Dark Treatment on Transcriptional Levels of GPX-PDE1 and GPX-PDE2
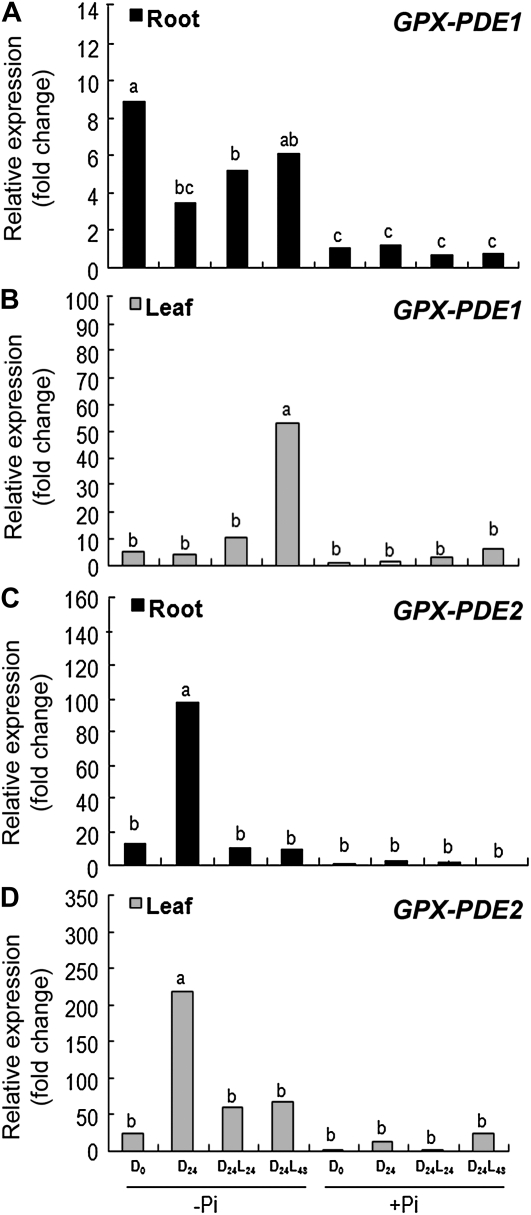
Sugar has been shown to be a signal that is involved in the Pi-sensing cascade in white lupin and affects cluster root formation. Sugars also change the transcription level of Pi starvation-induced genes (Liu et al., 2005, 2010a; Tesfaye et al., 2007; Zhou et al., 2008). Based on these results, we evaluated the effect of photosynthate deprivation through shoot dark treatment on the expression level of GPX-PDE1 and GPX-PDE2. The two genes displayed strikingly different responses to photosynthate deprivation (Fig. 4). When plants were deprived of light by dark treatment for 24 h, GPX-PDE1 transcripts in Pi-deficient roots were decreased by about 70% compared with under normal light/dark conditions. By comparison, the expression level of GPX-PDE1 in leaf tissue was not affected. We also observed that the effect of photosynthate deprivation on GPX-PDE1 expression was reversible by reexposure of dark-treated plants to continuous light for 24 to 48 h (Fig. 4, A and B). In contrast to GPX-PDE1, GPX-PDE2 showed the opposite response to photosynthate deprivation (Fig. 4, C and D). Transcripts of GPX-PDE2 accumulated to about 15-fold higher in photosynthate-deprived plants than normal light/dark plants, and the stimulation was displayed in both roots and leaves. Also, the dark effect could be reversed by reexposure to light. In summary, access to light/photosynthate can affect Pi stress-induced gene expression either positively or negatively.
Figure 4.
Effect of photosynthate deprivation on transcript abundance of Pi deficiency-induced GPX-PDE1 and GPX-PDE2. White lupin plants were grown to 14 d after emergence either with 1 mm Pi (+Pi) or without Pi (−Pi) under a 16/8-h photoperiod (D0). Plants were then placed in total darkness for 24 h (D24). After a 24-h dark treatment, plants were returned to continuous light for 24 h (D24L24) and 48 h (D24L48). Cluster roots and leaf tissue from the various treatments were harvested and used for RNA isolation and cDNA synthesis. qRT-PCR was used to assess transcript abundance. A and B, GPX-PDE1 transcript abundance in cluster roots (A) and leaves (B) in response to dark treatment and upon return to light. C and D, GPX-PDE2 transcript abundance in cluster roots (C) and leaves (D) in response to dark treatment and upon return to light. Data are expressed as relative values based on the GPX-PDE1 or GPX-PDE2 expression level in +P (D0) and referenced as 1.0. Lowercase letters denote significant differences between treatments (P ≤ 0.05). Statistical significance was based on three biological replicates.
Lupin GPX-PDE Promoter:GUS Reporters Are Highly Induced by Pi Deficiency
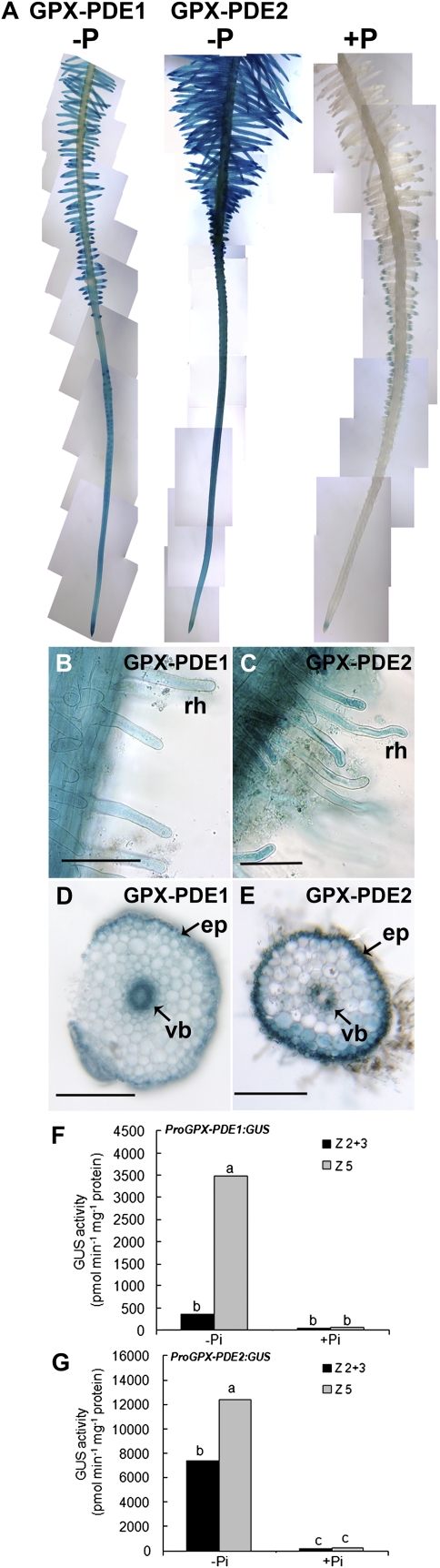
Translational fusions containing approximately 1,700 bp of GPX-PDE1 and GPX-PDE2 5′-upstream putative promoter (Pro) fragments were fused to the GUS reporter gene (ProGPX-PDE1:GUS or ProGPX-PDE2:GUS) and transformed into white lupin roots through the Agrobacterium rhizogenes-mediated hairy root transformation system. These composite transgenic plants were grown under Pi-deficient or Pi-sufficient conditions, and GUS staining activity was determined. We evaluated GUS staining on more than 50 root segments each representing an independent transgenic event. As seen in Figure 5, strikingly rapid and intense GUS staining (within 1–2 h) was observed in Pi-deficient transgenic plants carrying either the ProGPX-PDE1:GUS or ProGPX-PDE2:GUS construct. Furthermore, with both constructs, the GUS activity was activated along the entire root of P-deficient plants, appearing to show a higher activity in mature cluster roots with abundant root hairs (Fig. 5, A–C). That promoter activity was greatest in mature cluster root zone 5 was further supported by quantitative 4-methylumbelliferyl-β-d-glucuronide (MUG) assays (Fig. 5, F and G). Root hairs in Pi-deficient mature root zone 5 displayed strong GUS staining (Fig. 5, B and C). Cross sections of cluster rootlets also showed strong GUS staining in epidermal cells and vascular bundle cells in Pi-deficient transgenic roots transformed with either ProGPX-PDE1:GUS or ProGPX-PDE2:GUS (Fig. 5, D and E). Little to no staining was seen in Pi-sufficient plants (Fig. 5A).
Figure 5.
GPX-PDE1 and GPX-PDE2 reporter gene activity in Pi-deficient and Pi-sufficient cluster roots of white lupin. The 1,681- and 1,743-bp 5′-upstream promoter regions of GPX-PDE1 and GPX-PDE2, respectively, were fused to the GUS coding sequence and transformed into white lupin roots through A. rhizogenes-mediated hairy root transformation. Transgenic roots from Pi-sufficient and Pi-deficient roots were harvested 5 weeks after transformation and stained for GUS activity. Six roots per plant from a total of 12 transgenic plants were evaluated. Transgenic root tissue was incubated in GUS solution for 2 h at 37°C. Tissue from zone 5 and pooled zone 2 and 3 tissue was used for MUG enzyme assays. Lupin cluster roots are separated into five zones as described in Figure 2. A, GUS staining occurs throughout the entire root of both GPX-PDE1 and GPX-PDE2, with strong staining in the mature cluster root zone of –Pi plants; cluster roots of +Pi transgenic plants showed very low GUS staining. B and C, Root hairs (rh) from rootlets of the mature cluster root zone 5 of GPX-PDE1 (B) and GPX-PDE2 (C) also showed strong GUS staining. Bars = 0.05 mm. D and E, Cross sections of GPX-PDE1 (D) and GPX-PDE2 (E) rootlets from zone 5 displayed strong GUS staining in the epidermal cells (ep) and the vascular bundle (vb) in Pi-deficient transgenic roots. Bars = 0.5 mm. F and G, MUG assays confirmed the high promoter:reporter activity response to –Pi for GPX-PDE1 (F) and GPX-PDE2 (G). Z2+3, Cluster root zone 2 and 3 tissue pooled; Z5, cluster root zone 5 tissue. Lowercase letters denote significant differences between zones (P ≤ 0.05). Statistical significance was based on four biological replicates.
The MYB transcription factor PHR1, which binds to the P1BS element, is characterized as playing an important role in the regulation of Pi sensing (Rubio et al., 2001; Bari et al., 2006; Nilsson et al., 2007; Pant et al., 2008). Sequence analysis showed that GPX-PDE1 contains one P1BS element and GPX-PDE2 contains four P1BS repeats in the 2-kb upstream region from the translation start of each gene (Supplemental Fig. S6). To evaluate whether the P1BS elements of GPX-PDE1 and GPX-PDE2 play a role in the Pi-sensing mechanism, approximately 1,700 bp of the 5′-upstream promoters with mutated P1BS elements (GNATATNC to GNCCATNC) of mProGPX-PDE1:GUS or mProGPX-PDE2:GUS was used to transform white lupin roots. The comparable nonmutated promoters (ProGPX-PDE1:GUS and ProGPX-PDE2:GUS) were used as controls. Mutation of the single P1BS element in the promoter region of GPX-PDE1 abolished strong GUS activity in Pi-deficient roots (Supplemental Fig. S6, A and B). Similarly, strong GUS activity in Pi-deficient roots was also abolished by mutating all four P1BS elements in the promoter region of GPX-PDE2 (Supplemental Fig. S6, A and B). The negative effect of P1BS mutagenesis on the response of lupin GPX-PDEs to Pi starvation was further confirmed by MUG assays (Supplemental Fig. S6, C and D).
White Lupin GPX-PDE1 Is Associated with the Endoplasmic Reticulum
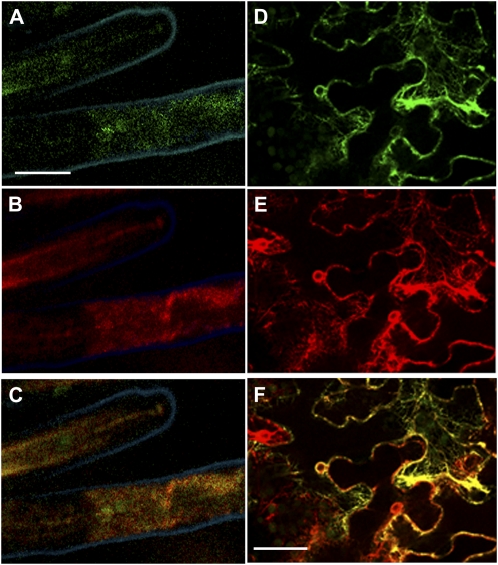
GPX-PDE1, which cleaves GPC, has a signal peptide of 25 amino acids, which predicts that the protein is targeted to the secretory system. To assess the subcellular localization of GPX-PDE1, we prepared a GPX-PDE1:EGFP translational fusion and used it to transform white lupin and Medicago truncatula. In earlier studies, we have shown that promoters from white lupin Pi deficiency-induced genes drive appropriate expression in M. truncatula roots (Liu et al., 2005; Zinn et al., 2009). The GFP protein signal was found in root hairs (Fig. 6). Counterstaining of the tissue samples with ER-Tracker Red and evaluation by confocal microscopy showed that GFP fluorescence colocalized with ER-Tracker Red when the images were merged (Fig. 6, A–C). Similarly, in a separate system using Nicotiana benthamiana epidermal cells transformed with constructs for both GPX-PDE1:GFP and HDEL:RFP (for red fluorescent protein), the GFP and RFP fluorescence colocalized to the endoplasmic reticulum (ER) membrane network and nuclear envelope (Fig. 6, D–F). Thus, two different approaches indicate that GPX-PDE1 is found associated with the ER.
Figure 6.
Subcellular localization of GPX-PDE1 protein in root hairs. A to C, Confocal laser scanning micrographs of root hair cells of transgenic M. truncatula roots expressing a fusion of GPX-PDE1, including the transit sequence, and EGFP driven by the GPX-PDE1 promoter (GPD-PDE1:GFP) stained with ER-Tracker Dye. A, GPX-PDE1:GFP fluorescence. B, ER-Tracker Red fluorescence. C, Merged images of A and B showing colocalization of GPX-PDE1 and ER-Tracker Red as orange-yellow color. Bar = 0.5 mm. D to F, Confocal laser scanning micrographs of N. benthamiana leaf epidermal cells infiltrated with A. tumefaciens carrying both GPX-PDE1:GFP and HDEL:RFP. D, GPX-PDE1:GFP fluorescence. E, HDEL:RFP fluorescence. F, Merged images of D and E. Images show colocalization of GPX-PDE1:GFP and HDEL:RFP as orange-yellow color. Bar = 25 μm.
Silencing of GPX-PDE1 or GPX-PDE2 in Composite Plants of White Lupin Impairs Root Hair Development
To investigate the role of GPX-PDEs in the acclimation to Pi deficiency in white lupin, we generated transgenic hairy roots of white lupin with reduced GPX-PDE1 or GPX-PDE2 expression by A. rhizogenes-mediated RNAi. A 631-bp fragment of the GPX-PDE1 mRNA sequence (bp 631–1,261) and a 510-bp fragment of the GPX-PDE2 mRNA sequence (bp 678–1,187) were PCR amplified and cloned into the Gateway RNAi vector pK7GWIWG2-DsRED containing the red fluorescent marker DsRED1 and were further transformed into white lupin. In parallel, roots transformed with a human myosin gene were used as a control to exclude the effects due to the formation of hairy roots. All the transgenic plants were grown under Pi-deficient conditions and harvested after 5 weeks. The DsRED fluorescence was used to screen for transformed roots. Each transformed root represented an independent transgenic event. Previous work from our laboratory (Uhde-Stone et al., 2005; Sbabou et al., 2010) showed that white lupin roots transformed with A. rhizogenes respond similarly to nontransformed roots during Pi deficiency. A total of 48 RNAi composite plants for GPX-PDE1, 54 RNAi composite plants for GPX-PDE2, and 30 myosin RNAi composite plants showing high DsRED fluorescence in hairy roots were analyzed in two independent experiments.
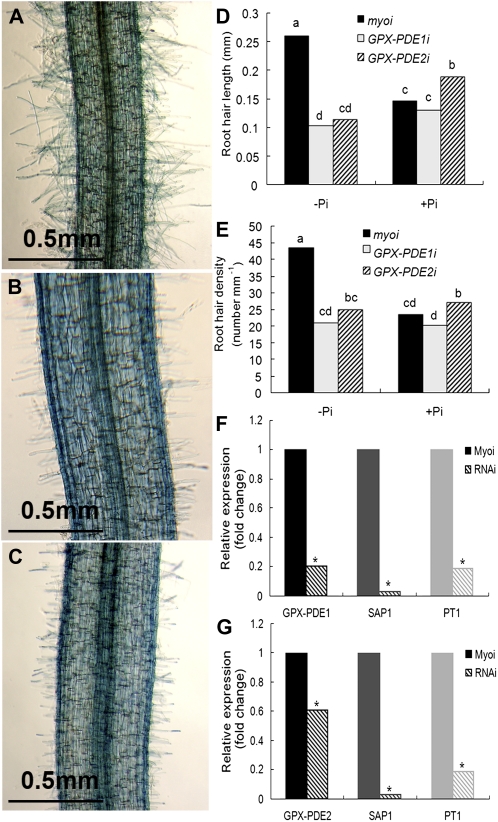
Root morphology changes evaluated were lateral root formation, root length, and root hair formation. In roots having strongly reduced expression of GPX-PDE1 or GPX-PDE2, we observed that the number of root hairs and root hair length were reduced as compared with the abundant and long root hairs in control transgenic cluster roots (Fig. 7, A–C). The frequency of impaired root hair development in GPX-PDE1 and GPX-PDE2 RNAi roots was 77% and 50%, respectively (Fig. 7, D and E). There was no difference in lateral root morphology between GPX-PDE1 and GPX-PDE2 RNAi roots and control roots. We used qRT-PCR to assess whether transgenic roots containing GPX-PDE RNAi constructs had altered transcript levels. As shown in Figure 7, F and G, knockdown roots had significant reductions of both GPX-PDE1 and GPX-PDE2 transcripts in corresponding RNAi roots as compared with control roots.
Figure 7.
Effect of GPX-PDE1 or GPX-PDE2 RNAi silencing on root hair development and expression of Pi starvation-induced genes. Reduced expression of GPX-PDE1 and GPX-PDE2 was achieved by subcloning a 631-bp mRNA sequence of GPX-PDE1 and a 510-bp mRNA sequence of GPX-PDE2 into an RNAi vector containing DsRED as a reporter and subsequent transformation to white lupin roots using A. rhizogenes. The human myosin gene was used as a control. Transgenic plants were grown under Pi-deficient conditions and harvested after 5 weeks. The DsRED reporter was used to screen transformed roots. A total of 48, 54, and 30 RNAi plants were evaluated for GPX-PDE1, GPX-PDE2, and myosin, respectively. A to C, Control RNAi (A), GPX-PDE1 RNAi (B), and GPX-PDE2 RNAi (C) showing root hair density and length from a zone 5 mature rootlet. D and E, Root hair length (D) and root hair density (E) of GPX-PDE1 and GPX-PDE2 RNAi zone 5 mature cluster rootlets show a significant reduction relative to the control myosin RNAi root hair length and density grown under Pi-deficient conditions. No significant differences were evident when grown under Pi-sufficient conditions. Lowercase letters denote significant differences between genes (P ≤ 0.05). Statistical significance was based on 11, 10, and eight biological replicates for myosin, GPX-PDE1, and GPX-PDE2 RNAi transgenic plants, respectively. F and G, qRT-PCR confirmed the reduction of the GPX-PDE1 (F) and GPX-PDE2 (G) transcript in the corresponding RNAi roots (patterned columns) relative to the control (solid columns). The transcript abundance of two other Pi deficiency-induced genes (SAP1 and PT1) was also reduced in GPX-PDE1 and GPX-PDE2 RNAi roots. Asterisks denote significant differences in expression levels relative to the control myosin RNAi gene (P ≤ 0.05).
To investigate whether knockdown of GPX-PDE1 and GPX-PDE2 transcripts affected the expression of other Pi starvation-induced genes in white lupin, GPX-PDE1 and GPX-PDE2 RNAi roots were used to evaluate the expression level of white lupin Pi stress-induced LaSAP1 and LaPT1. The knockdown of either GPX-PDE1 or GPX-PDE2 clearly resulted in reduced transcripts of Pi starvation-induced LaSAP1 and LaPT1 (Fig. 7, F and G). These data suggest that GPX-PDEs play a role, directly or indirectly, in mediating the expression of other Pi deficiency-induced genes in white lupin cluster roots.
DISCUSSION
Two Divergent GPX-PDEs Are Induced in White Lupin Cluster Roots during P Deficiency
In this report, we extend our understanding of plant biochemical and molecular response of cluster roots to Pi deprivation through molecular characterization of two very different GPX-PDEs, demonstrate a role for GPX-PDEs in root hair development, and suggest that GPX-PDEs may be involved in a Pi stress-induced phospholipid degradation/signaling pathway in white lupin cluster roots. GPX-PDE1 and GPX-PDE2 RNA abundance, promoter::GUS reporter gene studies, enzyme activities, and immunodetection studies reflect that Pi deficiency induces enhanced expression of two very distinct GPX-PDEs in white lupin cluster roots. Moreover, white lupin roots having reduced expression of GPX-PDE1 and GPX-PDE2 through RNAi-induced silencing have impaired root hair development. The white lupin GPX-PDE proteins share only 16% identity, resulting in the two being immunologically non-cross-reactive. Antibodies to GPX-PDE1 and GPX-PDE2 recognized their respective polypeptides in both cluster root soluble protein and cell wall protein extracts. The proteins were more highly abundant in cell walls of Pi-stressed cluster roots but only slightly so in the 25% to 75% soluble protein fraction. Unlike GPC-PDE activity, we found GPI-PDE activity only in cluster roots of Pi-stressed roots. Recombinant GPX-PDE1:GFP data show that white lupin GPX-PDE1 appears to be located in the ER membrane system. By comparison, white lupin GPX-PDE2 has no detectable targeting sequence. Van der Rest et al. (2004) purified a Pi stress-induced GPC-PDE enzyme from carrot cells that appears to be quite similar in sequence and size to white lupin GPX-PDE1. They found the native enzyme to have a molecular mass of 280 kD composed of polypeptides ranging in mass from 55 to 58 kD. Based upon organelle isolation, the carrot protein appeared to be targeted to the vacuole. They isolated an Arabidopsis cDNA (At1g74210) homologous to the carrot cell protein. Antibodies produced to recombinant At1g74210 recognized a carrot protein of 55 to 60 kD. They noted that carrot GPX-PDE was probably glycosylated and/or phosphorylated, which leads to an SDS gel product of 55 to 58 kD, which is larger than the 47-kD deduced protein size. The white lupin GPX-PDE1 protein is 71% identical to At1g74210 and has very similar attributes to the carrot cell protein on gels but is found in the ER membrane system. We did not, however, purify the lupin enzyme from cluster roots to assess the size of the native enzyme.
Very recently, Cheng et al. (2011) reported on a Pi deficiency-induced Arabidopsis GPX-PDE (At3g02040) that has moderate similarity (64%) to white lupin GPX-PDE2. However, unlike white lupin GPX-PDE2, which has no signal sequence at the N terminus, At3g02040 appears to have a 53-amino acid plastid-targeting sequence. The recombinant form of At3g02040 GPX-PDE has an approximate molecular mass of 40 kD, a broad substrate specificity, and greatest activity with Mg2+. Although not tested by Cheng et al. (2011), processing of At3g02040 would lead to a mass of 34 kD. The deduced molecular mass of white lupin GPX-PDE2 is 42 kD, but in planta it can be immunodetected as a 49- and a 70-kD protein from cluster roots. White lupin GPX-PDE2, although strikingly different in amino acid sequence from GPX-PDE1, also has high-probability glycosylation and phosphorylation sites, suggesting that posttranslational modification may result in a larger molecular mass. The substrate preference of the recombinant white lupin GPX-PDE2 protein has attributes similar to the Arabidopsis At3g02040 GPX-PDE but has highest activity with GPI as the substrate. Lack of complementation of the yeast GPC deletion mutant gde1Δ by GPX-PDE2 may be due to either its lower GPC activity or processing errors in yeast. Our data along with those of Van der Rest et al. (2004) and Cheng et al. (2011) provide good evidence that plant GPX-PDEs have variable substrate specificities, are highly expressed in roots, and appear to be important in acclimation to P deficiency.
We did notice an inconsistency in cell wall protein abundance of GPX-PDE1 and GPX-PDE2 and GPX-PDE activity. Although we used several different buffer combinations to obtain cell wall proteins for enzyme assays, we had difficulty in consistently detecting GPC-PDE and GPI-PDE activities in cell wall protein extracts. However, consistent differences were found in GPX-PDE1 and GPX-PDE2 protein between Pi-deficient and Pi-sufficient cell walls. Inconsistent detection of cell wall enzyme activity could reflect any number of difficulties, including but not limited to enzyme instability, loss of posttranslational modifications, and loss of accessory proteins needed for activity.
The ER Is Involved in Acclimation to Stress
Our confocal microscopy analysis of transgenic root hairs and epidermal cells expressing GPX-PDE1:EGFP translational fusions showed the protein to be located in the ER membrane network. It is well known that the ER plays a central role in phospholipid metabolism and signaling during stress in eukaryotes (Loewen et al., 2004; Sriburi et al., 2007; Gaspar et al., 2008; Xu et al., 2008; Young et al., 2010). In plants, several gene products involved in acclimation to Pi stress and phospholipid metabolism have been localized to the ER. The Arabidopsis GPX-PDE-like Root Hair Defective3 (RDH3) encodes a GTP-binding protein, and RDH4 encodes a phosphatidylinositol-4-phosphate phosphatase and is required for root hair formation and growth (Zheng et al., 2004; Thole et al., 2008). Stenzel et al. (2008) have shown that type B phosphatidylinositol-4-phosphate-5′-kinase is expressed in root hairs and is essential for root hair formation. Recently Lee et al. (2010) and Eastmond et al. (2010) have demonstrated that Arabidopsis Phosphatidic Acid Phosphohydrolyase1 and -2 (PHA1 and PHA2) and Phospholipase A2 (PA2), respectively, are localized to the ER. Mutations in PHA1 and -2 as well as PA2 impair Pi stress-induced modifications in membrane galactolipid synthesis (Nakamura et al., 2009; Eastmond et al., 2010). Moreover, two Arabidopsis gene products that transduce the low-Pi-availability signal, PDR2 and LPR1 (Ticconi et al., 2009), are localized to the ER of root stem cells. As a whole, it appears that a portion of phospholipid metabolism in roots is associated with ER trafficking and plays a significant role in acclimation to Pi limitation and cell development. Other aspects of phospholipid degradation, particularly as related to membrane remodeling in response to Pi deficiency, have been previously noted in Arabidopsis, maize (Zea mays), oat (Avena sativa), and common bean (Phaseolus vulgaris; Gniazdowska et al., 1999; Andersson et al., 2003, 2005; Jouhet et al., 2004; Nakamura et al., 2005; Cruz-Ramírez et al., 2006; Li et al., 2006; Calderon-Vazquez et al., 2008; Gaude et al., 2008).
Photosynthate (Sugar) Availability Modulates Root Gene Expression during Pi Deficiency
Sugars, derived from photosynthesis, have been implicated in coordinating the plant response to Pi limitation at both physiological and molecular levels (Liu et al., 2005; Müller et al., 2007; Tesfaye et al., 2007; Hammond and White, 2008; Zhou et al., 2008; Lei et al., 2011). Our results have shown that photosynthate deprivation by darkening leaves affects the expression of both GPX-PDE1 and GPX-PDE2 but in an opposite fashion. These results confirm reports of Liu et al. (2005, 2010a) and Müller et al. (2007) showing that many Pi deficiency-induced genes in white lupin cluster roots and Arabidopsis require sugars and photosynthate for expression. Surprisingly, however, darkening of Pi-deficient plants greatly enhanced GPX-PDE2 transcript expression in both cluster roots and leaves. There are few, if any, reports demonstrating that removal of photosynthetic capacity stimulates Pi deficiency-induced genes. However, Müller et al. (2007) noted that in leaves, At5g41080, an ortholog of white lupin GPX-PDE2, displayed an interaction between Pi and Suc. In addition, Lei et al. (2011), utilizing the Arabidopsis sucrose transporter2 mutant, have shown that Suc and Pi deficiency interaction can affect the expression of numerous Pi response genes either cooperatively or antagonistically. Also noteworthy is that the consumption of substrates for GPX-PDE, such as GPC and GPI, show an initial increase followed by a decrease in sycamore cells subjected to Suc starvation (Roby et al., 1987; Aubert et al., 1996). Further support for carbon starvation in the regulation of GPX-PDE expression is gleaned from studies of the ugp operon in microbial species (Kasahara et al., 1991; Su et al., 1991; Santos-Beneit et al., 2009). Interestingly, the promoter of lupin GPX-PDE2 contains four random repeats of the TATCCA element that has been shown to be an important component of the sugar starvation complex in rice (Lu et al., 2002). The TATCCA element binds with three MYB transcription factors (OsMYBS1, OsMYBS2, and OsMYBS3) that regulate carbon starvation-induced genes. In our white lupin Pi-induced cluster root cDNA library, we have found ESTs having high similarity (91%) to the OsMYBSs. Although we have not mutated the TATCCA elements, we postulate that this element may participate in the carbon starvation induction of GPX-PDE2.
Suppression of GPX-PDE Transcripts Impairs Root Hair Formation
Gahoonia and Nielsen (1998) report that greater than 60% of Pi may be acquired by root hairs, and their density becomes greater under Pi-deficient conditions (Bates and Lynch, 1996; Ma et al., 2001). Root hairs are highly abundant in mature cluster roots under Pi stress (Gardner et al., 1983; Dinkelaker et al., 1989; Watt and Evans, 1999). The fact that the two lupin GPX-PDE promoter:GUS reporter genes drive high expression in root epidermal and root hair cells suggests that the genes may have a role in root hair function. In addition, impaired root hair development and growth in white lupin GPX-PDE1 and GPX-PDE2 RNAi knockdown roots provides strong support to the interpretation that these genes play a role in root hair biology. We evaluated the phenotype of Arabidopsis mutants corresponding to GPX-PDE1 and GPX-PDE2 but did not detect a phenotype. This is probably due to the fact that Arabidopsis has multiple paralogs of GPX-PDEs (Cheng et al., 2011). In Arabidopsis, the type B GPX-PDE-like shv3 mutant displays shorter root hairs (Jones et al., 2006; Hayashi et al., 2008). SHV3 encodes a protein consisting of two tandemly repeated GPX-PDE domains and a GPI anchor (Hayashi et al., 2008), but the authors did not find GPX-PDE enzyme activity. SHV3:reporter gene activity occurred in root hairs coinciding with root hair initiation. GUS activity was also high in the vascular bundle of roots. Mutants in at least four other genes unrelated to GPX-PDEs but involved in phospholipid signaling have impaired root hair development (Ohashi et al., 2003; Böhme et al., 2004; Kusano et al., 2008; Thole et al., 2008). Overexpression of the transcription factor GLABRA2 promotes ectopic root hair initiation by activating the expression of the PHOSPHOLIPASE Dζ1 gene (PLDζ1), which in turn modulates phospholipid signaling (Ohashi et al., 2003). Suppression of AtPLDζ1 results in abnormal root hair development. Mutation of Arabidopsis phosphatidylinositol phosphate-5-kinase (PIP5K3) blocks the synthesis of phosphatidylinositol 4,5-bisphosphate, which functions as a site-specific signal to promote cytoskeleton reorganization and membrane trafficking. T-DNA insertions in PIP5K3 resulted in very short root hairs, while overexpression of the gene caused longer and multiple root hairs on a single trichoblast (Kusano et al., 2008; Stenzel et al., 2008). The ROOT HAIR DEFECTIVE4 (RHD4) gene encodes a phosphatidylinositol-4-phosphate phosphatase (PIPP) enzyme. Mutations in PIPP (rhd4-1) accumulate high amounts of phosphatidylinositol-4-phosphate, resulting in fewer and much shorter root hairs (Thole et al., 2008). Moreover, the Arabidopsis COW1 gene, which encodes a phosphatidylinositol transfer protein, is essential for root hair tip growth (Böhme et al., 2004). Interestingly, knockout mutants of an Arabidopsis PLDε had reduced root growth but when overexpressed increased root and root hair growth (Hong et al., 2009). Thus, several Arabidopsis phospholipid mutants demonstrate a critical role for phospholipids in root and root hair development. Our data for white lupin GPX-PDEs extend this understanding by showing that GPX-PDE1 and GPX-PDE2 or a product of their reaction also appear to be involved in root hair growth and development.
Potential Role of White Lupin GPX-PDEs in Phospholipid Ester Metabolism and Pi Recycling
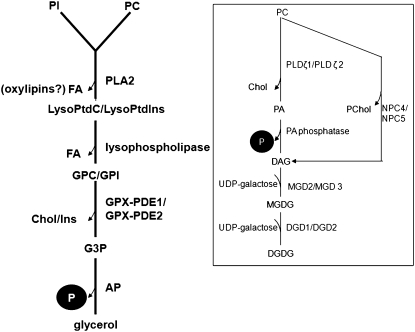
It is well established that under Pi stress, plant membrane synthesis is redirected toward the production of galactolipids/sulfolipids to stabilize membranes as phospholipids are degraded for Pi recycling (Yu et al., 2002; Andersson et al., 2003; Hammond et al., 2004; Misson et al., 2005; Morcuende et al., 2007; Calderon-Vazquez et al., 2008). The degradation pathway for phosphatidylcholine (PC) in Pi-deficient plants is fairly well characterized (Fig. 8). PC can be degraded directly to diacylglycerol (DAG) by nonspecific phospholipase C (Andersson et al., 2005; Nakamura et al., 2005; Gaude et al., 2008). Alternatively, PC may be degraded by phospholipase D (PLDζ1/PLDζ2) to phosphatidic acid, a signal-transducing compound (Hong et al., 2010), followed by cleavage to DAG by phosphatidic acid phosphohydrolyase (Cruz-Ramírez et al., 2006; Li et al., 2006; Nakamura et al., 2009; Eastmond et al., 2010). The galactolipid precursors monogalactosyl/digalactosyl diacylglycerol are synthesized from the condensation of UDP-Gal and DAG by monogalactosyl and digalactosyl diacylglycerol synthase (Shimojima et al., 1997; Kelly and Dörmann 2002; Moellering and Benning, 2011). By comparison, the sulfolipid precursors are synthesized through the formation of UDP-sulfoquinovose and subsequent condensation with DAG through the action of UDP-sulfoquinovose synthase and sulfoquinovosyl transferase (Yu et al., 2002; Andersson et al., 2003, 2005). Most of the genes involved with PC degradation and galactolipid/sulfolipid precursor synthesis have been shown to be expressed in several plant species subjected to Pi stress (Yu et al., 2002; Andersson et al., 2003, 2005; Nakamura et al., 2005; Hong et al., 2010; Moellering and Benning, 2011).
Figure 8.
A novel pathway for the degradation of phospholipids and Pi recycling via a GPX-PDE-mediated pathway during phosphate limitation. Under Pi limitation, plant phospholipids (PC and PI) decrease. The degradation of PC mediated by either nonspecific phospholipase C (NPC4 and NPC5) or phospholipase D (PLDζ1 and PLDζ2) with the subsequent release of Pi is well documented (pathway in box). We propose an alternative phospholipid degradation and Pi recycling pathway that catabolizes PC and PI. PC and PI are deacylated into GPC and GPI by Pi deficiency-induced phospholipase A2 (PLA2) and lysophospholipase. GPC and GPI are further hydrolyzed by Pi deficiency-induced GPX-PDE1 and GPX-PDE2 to glycerol-3-phosphate (G3P) and choline (Chol) or inositol (Ins). G3P is subsequently catalyzed by Pi deficiency-induced acid phosphatase (AP) to release Pi and glycerol. DGDG, Digalactosyldiacylglycerol; DGD, DGDG synthase; FA, fatty acid; LysoPtdC, lysophosphatidylcholine; LysoPtdIns, lysophosphatidylinositol; MGDG, monogalactosyldiacylglycerol; MGD, MGDG synthase; PA, phosphatidic acid; PChol, phosphocholine.
In currently proposed models, GPX-PDEs do not play a direct role in Pi deficiency-induced phospholipid degradation with subsequent synthesis of galactolipid/sulfolipid precursors (Gaude et al., 2008; Cheng et al., 2011). However, high expression of GPX-PDEs occurs during Pi deficiency not only in white lupin but also in Arabidopsis (Misson et al., 2005; Morcuende et al., 2007; Cheng et al., 2011), maize (Calderon-Vazquez et al., 2008), oat (Andersson et al., 2005), and potato (Solanum tuberosum; Hammond et al., 2009). Van der Rest et al. (2004) and Cheng et al. (2011) demonstrated that GPX-PDEs have enhanced expression under Pi deficiency. They proposed a role for GPX-PDE in hydrolyzing deacylated phospholipids to release Pi. We have built upon the findings above to show that Pi stress-induced cluster roots of white lupin have separate unique GPX-PDEs that can cleave GPC and GPI. Degradation of GPC and GPI can provide a source of choline, inositol, and recycled Pi during Pi stress. While the role of GPX-PDEs in the degradation of phosphatidylinositol (PI) and PC needs further clarification, we propose an alternative pathway for Pi deficiency degradation of PI and PC through a phospholipase A2/lysophospholipase/GPX-PDE pathway to release inositol, choline, and Pi (Fig. 8). This pathway could provide a novel route to provide inositol phosphates as well as jasmonates for signaling (Drissner et al., 2007). We recently initiated a next-generation sequencing project and obtained 100 million 72-bp reads from Pi-stressed cluster roots and leaves (S. Yang and C.P. Vance, unpublished data). It is noteworthy that we have found several Pi stress-induced transcripts involved in this proposed alternative pathway to be up-regulated in Pi-stressed cluster roots, including (1) phospholipase A2, which can cleave PI and PC to lysophosphatidylcholine/lysophosphatidylinositol and fatty acids, which in turn may be precursors for oxylipin molecules (Wang, 2004; Drissner et al., 2007); (2) lipoxygenase 2/3 and allene oxide synthase, the first steps in jasmonate synthesis; (3) lysophospholipases, which degrade lysophosphatidylcholine/lysophosphatidylinositol to GPC and GPI; and (4) phosphatidylinositol-4-phosphate-5′-kinase and phosphatidylinositol phosphate synthase, key steps in inositol phosphate synthesis. Our data suggest that the GPX-PDE pathway for PI and PC degradation to choline and inositol is induced during Pi stress (Fig. 8). Because inositol phosphates and jasmonate can serve as signal transducers (Berridge, 1993; DeWald et al., 2001; Wang, 2004; Kusano et al., 2008; Perera et al., 2008; Thole et al., 2008; Gfeller et al., 2010), we question whether an alternative pathway for phospholipid metabolism involving GPX-PDEs may play a role in Pi stress signal transduction and Pi recycling.
MATERIALS AND METHODS
Plant Material and Growth Conditions
White lupin (Lupinus albus var Ultra) was grown in a growth chamber at 20°C/15°C under a 16-h photoperiod (300 μmol photons m−2 s−1) as described by Gilbert et al. (2000). The Pi-sufficient and Pi-deficient solutions contained 1 mm Ca(H2PO4)2 and 1 mm CaSO4, respectively. Plants were harvested at 14 d after emergence. P concentrations in plant tissues were measured using ARL 3560 inductively coupled plasma-atomic emission spectroscopy. To determine the effects of Pi and Phi addition, −Pi plants at 14 d after emergence were treated with 0.5 mm Ca(H2PO4)2 or 1 mm Phi. Plants were harvested at 1, 4, 6, and 24 h. Phi was prepared by neutralizing a 100 mm solution of phosphonic acid with KOH as described previously (Carswell et al., 1996).
Quantitative Real-Time PCR, RT-PCR, and RNA Gel-Blot Analysis
Total RNA was isolated and treated with DNase I, and cDNA was synthesized to perform quantitative real-time RT-PCR using the SYBR Green (QuantiFast SYBR Green PCR kit; Qiagen) method in the StepOne Plus system (Applied Biosystems) with the appropriate primers (Supplemental Table S2). Relative quantitative results were calculated by normalization to the lupin tubulin gene. At least three biological replicates were performed per condition. Genomic contamination of cDNA was tested by the amplification of GPX-PDE2 using primers spanning intron 6 (274 bp), and no additional product was observed. RT-PCR analysis was performed on cDNA samples synthesized as described for real-time PCR with the appropriate primers (Supplemental Table S2). The lupin tubulin gene was used as a quantitative control.
For RNA gel-blot analysis, 20 μg of RNA was electrophoretically separated on a denaturing formaldehyde agarose gel, and the resulting blots were hybridized overnight with [α-32P]dCTP-labeled DNA probes and washed under high-stringency conditions using the formamide protocol according to the manufacturer’s instructions. The lupin ubiquitin gene was used as a quantitative control.
Screening of Lupin Genomic Libraries
A partial genomic library of white lupin constructed in the DASH II vector was made to isolate GPX-PDE clones as described previously (Liu et al., 2001). The cDNA insert representing specific GPX-PDE1 and GPX-PDE2 proteins was labeled and hybridized to plaque lifts. Single plaques were purified, and the inserts were subcloned into pBluescript SK− vector (Stratagene) for restriction mapping and sequencing.
Plant Protein Extraction
Lupin cluster roots were homogenized with 50 mm Tris (pH 8.0) containing 250 mm Suc, 3 mm dithiothreitol, and 1 mm phenylmethylsulfonyl fluoride (extraction buffer). The majority of the cell debris was filtered out from the soluble fraction by gentle vacuum filtration through two layers of Miracloth, then the filtrate was further clarified by centrifugation (15,000g, 20 min, 4°C). Soluble proteins were fractionated by (NH4)2SO4 precipitation, and the 25% to 75% fraction pellet was resuspended in 2.5 mL of extraction buffer and then desalted by elution through a PD-10 column (Pharmacia). Root debris from filtration through the Miracloth was combined with the pellet from the first centrifugation and extracted with 200 mm CaCl2 for 3 to 4 h at 4°C with stirring to elute salt-extractable cell wall-associated proteins using a protocol adapted from Van der Rest et al. (2004). Suspended debris was centrifuged at 15,000g for 20 min at 4°C. Following centrifugation, the supernatant was concentrated in a Macrosep 30K Ω centrifugal device (Pall Life Sciences) and desalted by elution through a PD-10 column (Pharmacia) equilibrated in 50 mm Tris, pH 7.4, containing 0.5% β-mercaptoethanol, 100 mm Suc, and 15% glycerol. The protein extracts were used to perform enzyme assays and protein immunoblot analysis.
Enzyme Assay
GPX-PDE activity was quantitated by a directly enzyme-coupled spectrophotometric assay measuring the amount of glycerol-3-phosphate generated by the GPX-PDE reaction adapted from Larson et al. (1983). The assay mixture contained 0.5 m hydrazine-Gly buffer (pH 9.0), 1 mm NAD, 5 units of glycerol-3-phosphate dehydrogenase, either 10 mm CaCl2 or MgCl2, 0.5 mm substrate (glycerophosphocholine or glycerophosphorylinositol), and 75 to 100 μL (235–360 μg) of 25% to 75% root extract, 150 μL (45–80 μg) of cell wall extract, or 25 μL (5–8 μg for GPX-PDE1 and 17–18 μg for GPX-PDE2) of partially purified fusion protein in a total volume of 0.5 mL. The rate of NAD reduction was measured by recording the increase in A340 (molar absorbance coefficient of 6,300 m−1 cm−1). Glycerophosphorylinositol was kindly provided by Jana Patton-Vogt (Department of Biological Sciences, Duquesne University).
Antisera Production and Protein Gel-Blot Analysis
The partial coding sequence without the predicted signal peptide (120 bp downstream of the translation start) of GPX-PDE1 and the whole coding sequence of GPX-PDE2 were amplified by PCR using appropriate primers (Supplemental Table S2) and were subsequently cloned into the pGEX-2T expression vector (Pharmacia). The constructed pGEX-2T plasmid containing either GPX-PDE1 or GPX-PDE2 was then transformed into Escherichia coli DH5α to express proteins by induction with 0.2 mm isopropyl-β-d-thiogalactopyranoside. The DH5α strain proved to be as good a host as the BL21 strain for this construct, since the pGEX-2T vector itself encodes the repressor protein and therefore works well in many E. coli strains. Freshly transformed colonies were used to inoculate liquid cultures for both proteins and grown at 37°C for 7 h followed by induction with isopropyl-β-d-thiogalactopyranoside and overnight growth at 16°C to 23°C prior to cell harvest. This lower temperature growth period was necessary for the production of small amounts of soluble GPX-PDE1, which could not be achieved when growth continued under induction at 30°C. GPX-PDE2 colonies always produced large amounts of soluble GPX-PDE2 protein. The resulting GST fusion proteins were partially purified using Glutathione Sepharose 4B (Pharmacia). Protein concentrations were determined using the Bio-Rad protein assay kit. Soluble fusion proteins of GPX-PDE1 and GPX-PDE2 were used to perform in vitro GPX-PDE enzyme assays as described for enzyme assay methods. Anti-GPX-PDE1 and anti-GPX-PDE2 polyclonal antisera were generated by injection of rabbits (New Zealand White) using purified fusion proteins.
Fractionated proteins prepared as mentioned above were used for immunoblot analysis according to Miller et al. (2001). Forty micrograms of protein for each sample was separated on a 10% SDS-PAGE gel and then electrophoretically transferred to nitrocellulose for exposure to antiserum. Rabbit anti-GPX-PDE1 (1:60,000 dilution) and anti-GPX-PDE2 (1:15,000 dilution) sera were used as the primary antibodies, and goat anti-rabbit IgG (H+L) horseradish peroxidase conjugate (Bio-Rad) at 1:20,000 dilution was used as the secondary antibody. Blots were developed using the Western Lightning Ultra kit (Perkin-Elmer).
Plasmid Construction
GPX-PDE1 and GPX-PDE2 promoter variants including the full-length promoters (1,681 and 1,743 bp, respectively) and the promoters with P1BS mutated were amplified by PCR from corresponding genomic clones with appropriate primers and cloned into the transformation vector pBI101.2 (Clontech). For construction of a translational GPX-PDE1:EGFP fusion driven by the GPX-PDE1 promoter, plasmid pBI101.2-EGFP was constructed as described by Uhde-Stone et al. (2005). The promoter region of 1,681 bp upstream of the start ATG of GPX-PDE1 and the protein-coding regions of the GPX-PDE1 gene were amplified from corresponding genomic and cDNA clones, separately, with the appropriate primers. These two fragments were further cloned in frame into the pBI101.2-EGFP vector.
To create constructs for RNAi-mediated silencing of GPX-PDE1 and GPX-PDE2, fragments corresponding to the coding regions of the GPX-PDE1 and GPX-PDE2 genes were amplified by PCR from cDNA clones using the appropriate primers. The PCR products were cloned into the pENTR/D-TOPO entry vector following the manufacturer’s instructions (Invitrogen) and then recombined into the Gateway-compatible destination vector pK7GWIWG2 (II)-DsRED to produce the GPX-PDE1 or GPX-PDE2 RNAi construct. The pK7GWIWG2 (II)-DsRED vector with a fragment of the human myosin gene was used as a control and was kindly provided by J. Stephen Gantt (Department of Plant Biology, University of Minnesota).
All the resulting constructs were introduced into Agrobacterium rhizogenes A4TC24 (Quandt et al., 1993) by electroporation and used for white lupin transformation. All primer sequences used for plasmid construction are listed in Supplemental Table S2.
Complementation of Yeast Mutants
Both whole coding sequence and partial coding sequence without the predicted signal peptide (132 bp after the transcription start) of GPX-PDE1 and the whole coding sequence of GPX-PDE2 were amplified from corresponding cDNA clones by PCR using the appropriate primers (Supplemental Table S2) and were subsequently cloned into the yeast expression vector pCu416CUP1 (Labbé and Thiele, 1999). Wild-type yeast (strain JPV203) and a gde1Δ deletion mutant (JPV125) were then transformed with these plasmids using a standard lithium acetate-based technique. A gde1Δ mutant strain was transformed to uracil prototrophy with the indicated plasmids. Transformants and wild-type cells were grown in low-Pi medium overnight. For the growth of all strains containing plasmids, the medium also lacked uracil. Overnight cultures were used to inoculate fresh cultures at A600 = 0.01 into the following media, each containing 50 μm CuSO4 to induce the expression of plasmid-borne genes under the control of the CUP1 promoter: (1) no-Pi medium; (2) low-Pi medium; and (3) no-Pi + GPC medium. Cultures were grown at 30°C on a rotating wheel. Growth was monitored by measuring turbidity at 600 nm on a Biomate 3 Thermo Spectronic spectrophotometer. The yeast medium used in this study was synthetic complete medium or –uracil dropout medium, made as described previously (Patton et al., 1995). Saccharomyces cerevisiae strains used in this study are listed in Supplemental Table S3.
A. rhizogenes-Mediated Transformation of White Lupin and Medicago truncatula
White lupin (ABC1082) seeds were surface sterilized with 95% ethanol for 10 min followed by rinsing with sterile water. Then, seeds were sterilized with 30% bleach for 10 min and rinsed with sterilized water. Seeds were germinated in the dark for 2 d on 0.6% agar plates at room temperature and then transformed with A. rhizogenes as described previously by Uhde-Stone et al. (2005) with the following modifications: when emerging radicles reached a length of approximately 2 cm on the plates, seedlings were transplanted to prewetted vermiculite and kept in the dark at room temperature for 2 d followed by 2 d in the dark at 4°C. After the cold treatment, the primary root tip (approximately 0.5–1 cm) was excised and the cut end was inoculated with the A4TC24 containing the plasmid of interest. One hour after inoculation, seedlings were placed in an aeroponic system (EZ Clone Enterprise) filled with deionized water for the first week and with 1 mm Pi nutrient solution for the subsequent 1 week. Plants were then supplied with either 0 mm Pi (−P) or 1 mm Pi (+P) and were harvested 3 to 4 weeks later. M. truncatula A17 ENOD11 (A17 containing a ProENOD11-GUS construct; Journet et al., 2001) seeds were used for the transformation of GFP reporter constructs.
Microscopy
Localization of GPX-PDE1:GFP in transgenic roots was performed using a Nikon D-eclipse C1si laser scanning confocal microscope. GFP unmixing from autofluorescence was performed using the Clontech spectra for eGFP. ER-Tracker Red (587/615 nm) stain was used at a 1 μm working solution (Cole et al., 2000). Tissue was stained for 30 to 60 min. Nicotiana benthamiana leaf epidermal cells were cotransformed with 35S:GPX-PDE1:GFP and an ER-targeted marker protein, 35S:HDEL:RFP (Ticconi et al., 2009). The Agrobacterium tumefaciens strains (EHA105) carrying p19, GPX-PDE1:GFP, and HDEL:RFP were coinfiltrated into the abaxial surface of fully expanded N. benthamiana leaves (Liu et al., 2010b). Equal volumes of p19 and GPX-PDE1:GFP were combined with HDEL:RFP suspension (5:1, v/v) prior to infiltration. All subsequent light microscopy used a Nikon SMZ800 stereoscope and a Nikon Labophot light microscope attached to a Nikon DXM1200 digital camera. GUS activity (Jefferson, 1989) measurements were performed as described by Uhde-Stone et al. (2005).
Phylogenetic Analysis
Amino acid sequences of GPX-PDEs were retrieved from public databases. Accession numbers for protein sequences used for alignment are listed below. Sequences were aligned using ClustalX (Thompson et al., 1997), and the phylogenetic analysis was performed with the MEGA4 package developed by Tamura et al. (2007) using the neighbor-joining method (Saitou and Nei, 1987) with 1,000 trials to obtain bootstrap values. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965). All positions containing gaps and missing data were eliminated from the data set. The alignment used for the phylogenetic analysis is available as Supplemental Data Set S1.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Lupinus albus GPX-PDE1 (GU647221 and GU647223) and GPX-PDE2 (GU647222 and GU647224); Escherichia coli glpQ (NP_754666) and ugpQ (YP_002331155); Homo sapiens GDE1/Mir16 (NP_057725); and Thermoanaerobacter tengcongensis TtGDPD (2PZ0B). Sequences used in the analysis of GPX-PDE homology among various plant species (Supplemental Fig. S2) can be found in Phytozome (http://www.phytozome.net/). The accession numbers of GPX-PDEs used in the phylogeny are as follows: Streptomyces coelicolor Stc_glpQ1 (NP_733538) and Stc_glpQ2 (NP_626231); Homo sapiens Hs_GDE1/Mir16 (NP_057725); Gallus gallus Gg_GDE2 (AAX89036); Mus musculus Mm_GDE3 (NP_076097); Escherichia coli Ec_glpQ (NP_754666) and Ec_ugpQ (YP_002331155); Saccharomyces cerevisiae Sc_GDE1 (NP_015215) and Sc_PGC1 (NP_015118); Arabidopsis thaliana At4g26690 (NP_567755), At5g55480 (NP_200359), At1g66970 (NP_176869), At3g20520 (NP_188688), At5g58170 (NP_200613), At5g58050 (NP_200625), At1g66980 (NP_176870), At1g74210 (NP_177561), At5g08030 (NP_196420), At5g41080 (NP_198924), At3g02040 (NP_566159), At5g43300 (NP_199144), and At1g71340 (NP_177290); Glycine max Gm17g08680 and Gm19g27060; Oryza sativa Os03g40670 and Os02g31030; and Sorghum bicolor Sb04g021010 and Sb03g035370.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of the conserved catalytic domain of white lupin GPX-PDEs with the orthologs of various species, and the amino acid identities of the corresponding plant orthologs.
Supplemental Figure S2. Analysis of deduced amino acid sequences of GPX-PDE1 and GPX-PDE2.
Supplemental Figure S3. Gene structures and 5′ upstream cis-element analysis of GPX-PDE1 and GPX-PDE2.
Supplemental Figure S4. Immunological detection of GPX-PDE1 and GPX-PDE2 in cluster roots of white lupin grown under Pi-sufficient and Pi-deficient conditions.
Supplemental Figure S5. Cluster root development in white lupin grown under Pi-deficient conditions.
Supplemental Figure S6. GUS reporter gene activity driven by the P1BS-mutated promoter of GPX-PDE1 or GPX-PDE2 in Pi-deficient and Pi-sufficient cluster roots of white lupin.
Supplemental Table S1. In vitro enzyme activity for recombinantly produced GPX-PDE1 and GPX-PDE2.
Supplemental Table S2. Oligonucleotides used in this study.
Supplemental Table S3. Yeast strains used in this study.
Supplemental Data Set S1. Text file of the alignment used for the phylogenetic analysis in Figure 1.
Supplementary Material
Acknowledgments
We thank Dr. Mark Sanders (College of Biological Sciences Imaging Center, University of Minnesota) and Dr. Qi Xie (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for their help with the GFP localization.
References
- Andersson MX, Larsson KE, Tjellström H, Liljenberg C, Sandelius AS. (2005) Phosphate-limited oat: the plasma membrane and the tonoplast as major targets for phospholipid-to-glycolipid replacement and stimulation of phospholipases in the plasma membrane. J Biol Chem 280: 27578–27586 [DOI] [PubMed] [Google Scholar]
- Andersson MX, Stridh MH, Larsson KE, Liljenberg C, Sandelius AS. (2003) Phosphate-deficient oat replaces a major portion of the plasma membrane phospholipids with the galactolipid digalactosyldiacylglycerol. FEBS Lett 537: 128–132 [DOI] [PubMed] [Google Scholar]
- Aubert S, Gout E, Bligny R, Marty-Mazars D, Barrieu F, Alabouvette J, Marty F, Douce R. (1996) Ultrastructural and biochemical characterization of autophagy in higher plant cells subjected to carbon deprivation: control by the supply of mitochondria with respiratory substrates. J Cell Biol 133: 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible W-R. (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates TR, Lynch JP. (1996) Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ 19: 529–538 [Google Scholar]
- Berridge MJ. (1993) Inositol trisphosphate and calcium signalling. Nature 361: 315–325 [DOI] [PubMed] [Google Scholar]
- Bieleski RL. (1973) Phosphate pools, phosphate transport and phosphate availability. Annu Rev Plant Physiol 24: 225–252 [Google Scholar]
- Böhme K, Li Y, Charlot F, Grierson C, Marrocco K, Okada K, Laloue M, Nogué F. (2004) The Arabidopsis COW1 gene encodes a phosphatidylinositol transfer protein essential for root hair tip growth. Plant J 40: 686–698 [DOI] [PubMed] [Google Scholar]
- Brzoska P, Boos W. (1988) Characteristics of a ugp-encoded and phoB-dependent glycerophosphoryl diester phosphodiesterase which is physically dependent on the ugp transport system of Escherichia coli. J Bacteriol 170: 4125–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon-Vazquez C, Ibarra-Laclette E, Caballero-Perez J, Herrera-Estrella L. (2008) Transcript profiling of Zea mays roots reveals gene responses to phosphate deficiency at the plant- and species-specific levels. J Exp Bot 59: 2479–2497 [DOI] [PubMed] [Google Scholar]
- Carswell C, Grant BR, Theodorou ME, Harris J, Niere JO, Plaxton WC. (1996) The fungicide phosphonate disrupts the phosphate-starvation response in Brassica nigra seedlings. Plant Physiol 110: 105–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Zhou W, El Sheery NI, Peters C, Li M, Wang X, Huang J. (2011) Characterization of the Arabidopsis glycerophosphodiester phosphodiesterase (GDPD) family reveals a role of the plastid-localized AtGDPD1 in maintaining cellular phosphate homeostasis under phosphate starvation. Plant J 66: 781–795 [DOI] [PubMed] [Google Scholar]
- Ciereszko I, Johansson H, Kleczkowski LA. (2005) Interactive effects of phosphate deficiency, sucrose and light/dark conditions on gene expression of UDP-glucose pyrophosphorylase in Arabidopsis. J Plant Physiol 162: 343–353 [DOI] [PubMed] [Google Scholar]
- Cole L, Davies D, Hyde GJ, Ashford AE. (2000) ER-Tracker dye and BODIPY-brefeldin A differentiate the endoplasmic reticulum and Golgi bodies from the tubular-vacuole system in living hyphae of Pisolithus tinctorius. J Microsc 197: 239–249 [DOI] [PubMed] [Google Scholar]
- Corda D, Kudo T, Zizza P, Iurisci C, Kawai E, Kato N, Yanaka N, Mariggiò S. (2009) The developmentally regulated osteoblast phosphodiesterase GDE3 is glycerophosphoinositol-specific and modulates cell growth. J Biol Chem 284: 24848–24856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ramírez A, Oropeza-Aburto A, Razo-Hernández F, Ramírez-Chávez E, Herrera-Estrella L. (2006) Phospholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proc Natl Acad Sci USA 103: 6765–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWald DB, Torabinejad J, Jones CA, Shope JC, Cangelosi AR, Thompson JE, Prestwich GD, Hama H. (2001) Rapid accumulation of phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate correlates with calcium mobilization in salt-stressed Arabidopsis. Plant Physiol 126: 759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkelaker B, Römheld V, Marschner H. (1989) Citric acid excretion and precipitation of calcium citrate in the rhizosphere of white lupin (Lupinus albus L.). Plant Cell Environ 12: 285–292 [Google Scholar]
- Drissner D, Kunze G, Callewaert N, Gehrig P, Tamasloukht M, Boller T, Felix G, Amrhein N, Bucher M. (2007) Lyso-phosphatidylcholine is a signal in the arbuscular mycorrhizal symbiosis. Science 318: 265–268 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Quettier A-L, Kroon JTM, Craddock C, Adams N, Slabas AR. (2010) Phosphatidic acid phosphohydrolase 1 and 2 regulate phospholipid synthesis at the endoplasmic reticulum in Arabidopsis. Plant Cell 22: 2796–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher E, Almaguer C, Holic R, Griac P, Patton-Vogt J. (2005) Glycerophosphocholine-dependent growth requires Gde1p (YPL110c) and Git1p in Saccharomyces cerevisiae. J Biol Chem 280: 36110–36117 [DOI] [PubMed] [Google Scholar]
- Gahoonia TS, Nielsen NE. (1998) Direct evidence on participation of root hairs in phosphorus (32P) uptake from soil. Plant Soil 198: 147–152 [Google Scholar]
- Gahoonia TS, Nielsen NE, Joshi PA, Jahoor AA. (2001) A root hairless barley mutant for elucidating genetic of root hairs and phosphorus uptake. Plant Soil 235: 211–219 [Google Scholar]
- Gardner WK, Barber DA, Parbery DG. (1983) The acquisition of phosphorus by Lupinus albus L. III. The probable mechanism by which phosphorus movement in the soil/root interface is enhanced. Plant Soil 70: 107–124 [Google Scholar]
- Gaspar ML, Jesch SA, Viswanatha R, Antosh AL, Brown WJ, Kohlwein SD, Henry SA. (2008) A block in endoplasmic reticulum-to-Golgi trafficking inhibits phospholipid synthesis and induces neutral lipid accumulation. J Biol Chem 283: 25735–25751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaude N, Nakamura Y, Scheible WR, Ohta H, Dörmann P. (2008) Phospholipase C5 (NPC5) is involved in galactolipid accumulation during phosphate limitation in leaves of Arabidopsis. Plant J 56: 28–39 [DOI] [PubMed] [Google Scholar]
- Gfeller A, Dubugnon L, Liechti R, Farmer EF. (2010) Jasmonate biochemical pathway. Sci Signal 3: cm3. [DOI] [PubMed] [Google Scholar]
- Gilbert GA, Knight JD, Vance CP, Allan DL. (2000) Proteoid root development of phosphorus deficient lupin is mimicked by auxin and phosphonate. Ann Bot (Lond) 85: 921–928 [Google Scholar]
- Gniazdowska A, Szal B, Rychter AM. (1999) The effect of phosphate deficiency on membrane phospholipid composition of bean (Phaseolus vulgaris L.) roots. Acta Physiol Plant 21: 263–269 [Google Scholar]
- Hammond JP, Broadley MR, Bowen HC, Hayden R, Spracklen WP, White PJ. (2009) A molecular diagnostic for phosphorus deficiency in potatoes. Proceedings of the International Plant Nutrition Colloquium XVI, University of California Davis; http://www.escholarship.org/uc/item/9w97m6mz (May 27, 2009) [Google Scholar]
- Hammond JP, Broadley MR, White PJ. (2004) Genetic responses to phosphorus deficiency. Ann Bot (Lond) 94: 323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, White PJ. (2008) Sucrose transport in the phloem: integrating root responses to phosphorus starvation. J Exp Bot 59: 93–109 [DOI] [PubMed] [Google Scholar]
- Hayashi S, Ishii T, Matsunaga T, Tominaga R, Kuromori T, Wada T, Shinozaki K, Hirayama T. (2008) The glycerophosphoryl diester phosphodiesterase-like proteins SHV3 and its homologs play important roles in cell wall organization. Plant Cell Physiol 49: 1522–1535 [DOI] [PubMed] [Google Scholar]
- Hinsinger P. (2001) Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: a review. Plant Soil 237: 173–195 [Google Scholar]
- Hong Y, Devaiah SP, Bahn SC, Thamasandra BN, Li M, Welti R, Wang X. (2009) Phospholipase D epsilon and phosphatidic acid enhance Arabidopsis nitrogen signaling and growth. Plant J 58: 376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Zhang W, Wang X. (2010) Lipid signaling in plant response to hyperosmotic stress. Plant Cell Environ 33: 627–635 [DOI] [PubMed] [Google Scholar]
- Jain A, Poling MD, Karthikeyan AS, Blakeslee JJ, Peer WA, Titapiwatanakun B, Murphy AS, Raghothama KG. (2007) Differential effects of sucrose and auxin on localized phosphate deficiency-induced modulation of different traits of root system architecture in Arabidopsis. Plant Physiol 144: 232–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA. (1989) The GUS reporter gene system. Nature 342: 837–838 [DOI] [PubMed] [Google Scholar]
- Johnson JF, Allan DL, Vance CP. (1994) Phosphorus stress-induced proteoid roots show altered metabolism in Lupinus albus. Plant Physiol 104: 657–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JF, Vance CP, Allan DL. (1996) Phosphorus deficiency in Lupinus albus: altered lateral root development and enhanced expression of phosphoenolpyruvate carboxylase. Plant Physiol 112: 31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MA, Raymond MJ, Smirnoff N. (2006) Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J 45: 83–100 [DOI] [PubMed] [Google Scholar]
- Jouhet J, Maréchal E, Baldan B, Bligny R, Joyard J, Block MA. (2004) Phosphate deprivation induces transfer of DGDG galactolipid from chloroplast to mitochondria. J Cell Biol 167: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet EP, El-Gachtouli N, Vernoud V, de Billy F, Pichon M, Dedieu A, Arnould C, Morandi D, Barker DG, Gianinazzi-Pearson V. (2001) Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol Plant Microbe Interact 14: 737–748 [DOI] [PubMed] [Google Scholar]
- Kasahara M, Makino K, Amemura M, Nakata A, Shinagawa H. (1991) Dual regulation of the ugp operon by phosphate and carbon starvation at two interspaced promoters. J Bacteriol 173: 549–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keerthisinghe G, Hocking PJ, Ryan PR, Delhaize E. (1998) Effect of phosphorus supply on the formation and function of proteoid roots of white lupin (Lupinus albus L.). Plant Cell Environ 21: 467–478 [Google Scholar]
- Kelly AA, Dörmann P. (2002) DGD2, an Arabidopsis gene encoding a UDP-galactose-dependent digalactosyldiacylglycerol synthase is expressed during growth under phosphate-limiting conditions. J Biol Chem 277: 1166–1173 [DOI] [PubMed] [Google Scholar]
- Kusano H, Testerink C, Vermeer JEM, Tsuge T, Shimada H, Oka A, Munnik T, Aoyama T. (2008) The Arabidopsis phosphatidylinositol phosphate 5-kinase PIP5K3 is a key regulator of root hair tip growth. Plant Cell 20: 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé S, Thiele DJ. (1999) Copper ion inducible and repressible promoter systems in yeast. Methods Enzymol 306: 145–153 [DOI] [PubMed] [Google Scholar]
- Lambers H, Shane MW, Cramer MD, Pearse SJ, Veneklaas EJ. (2006) Root structure and functioning for efficient acquisition of phosphorus: matching morphological and physiological traits. Ann Bot (Lond) 98: 693–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson TJ, Ehrmann M, Boos W. (1983) Periplasmic glycerophosphodiester phosphodiesterase of Escherichia coli, a new enzyme of the glp regulon. J Biol Chem 258: 5428–5432 [PubMed] [Google Scholar]
- Lee HY, Bahn SC, Kang Y-M, Lee KH, Kim HJ, Noh EK, Palta JP, Shin JS, Ryu SB. (2003) Secretory low molecular weight phospholipase A2 plays important roles in cell elongation and shoot gravitropism in Arabidopsis. Plant Cell 15: 1990–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee OR, Kim SJ, Kim HJ, Hong JK, Ryu SB, Lee SH, Ganguly A, Choa H-T. (2010) Phospholipase A2 is required for PIN-FORMED protein trafficking to the plasma membrane in the Arabidopsis root. Plant Cell 22: 812–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Liu Y, Zhang B, Zhao Y, Wang X, Zhou Y, Raghothama KG, Liu D. (2011) Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in Arabidopsis. Plant Physiol 156: 1116–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Qin C, Welti R, Wang X. (2006) Double knockouts of phospholipases Dζ1 and Dζ2 in Arabidopsis affect root elongation during phosphate-limited growth but do not affect root hair patterning. Plant Physiol 140: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Uhde-Stone C, Li A, Vance CP, Allan DL. (2001) A phosphate transporter with enhanced expression in proteoid roots of white lupin (Lupinus albus L.). Plant Soil 237: 257–266 [Google Scholar]
- Liu JQ, Allan DL, Vance CP. (2010a) Systemic signaling and local sensing of phosphate in common bean: cross-talk between photosynthate and microRNA399. Mol Plant 3: 428–437 [DOI] [PubMed] [Google Scholar]
- Liu JQ, Samac DA, Bucciarelli B, Allan DL, Vance CP. (2005) Signaling of phosphorus deficiency-induced gene expression in white lupin requires sugar and phloem transport. Plant J 41: 257–268 [DOI] [PubMed] [Google Scholar]
- Liu L, Zhang Y, Tang S, Zhao Q, Zhang Z, Zhang H, Dong L, Guo H, Xie Q. (2010b) An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J 61: 893–903 [DOI] [PubMed] [Google Scholar]
- Loewen CJR, Gaspar ML, Jesch SA, Delon C, Ktistakis NT, Henry SA, Levine TP. (2004) Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science 304: 1644–1647 [DOI] [PubMed] [Google Scholar]
- Lu CA, Ho TH, Ho SL, Yu SM. (2002) Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of α-amylase gene expression. Plant Cell 14: 1963–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Bielenberg DG, Brown KM, Lynch JP. (2001) Regulation of root hair density by phosphorus availability in Arabidopsis thaliana. Plant Cell Environ 24: 459–467 [Google Scholar]
- Marschner H. (1995) Mineral Nutrition of Higher Plants, Ed 2. Academic Press, San Diego [Google Scholar]
- Miller SS, Liu J, Allan DL, Menzhuber CJ, Fedorova M, Vance CP. (2001) Molecular control of acid phosphatase secretion into the rhizosphere of proteoid roots from phosphorus-stressed white lupin. Plant Physiol 127: 594–606 [PMC free article] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering ER, Benning C. (2011) Galactoglycerolipid metabolism under stress: a time for remodeling. Trends Plant Sci 16: 98–107 [DOI] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, Bläsing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, et al. (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30: 85–112 [DOI] [PubMed] [Google Scholar]
- Müller R, Morant M, Jarmer H, Nilsson L, Nielsen TH. (2007) Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol 143: 156–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Awai K, Masuda T, Yoshioka Y, Takamiya KI, Ohta H. (2005) A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. J Biol Chem 280: 7469–7476 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Koizumi R, Shui G, Shimojima M, Wenk MR, Ito T, Ohta H. (2009) Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. Proc Natl Acad Sci USA 106: 20978–20983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Massonneau A, Langlade N, Dinkelaker C, Romheld V, Martinoia E. (2000) Physiological aspects of cluster root function and development in phosphorus-deficient white lupin (Lupinus albus L.). Ann Bot (Lond) 85: 909–919 [Google Scholar]
- Neumann G, Massonneau A, Martinoia E, Römheld V. (1999) Physiological adaptation to phosphorus deficiency during proteoid root development in white lupin. Planta 208: 373–382 [Google Scholar]
- Nilsson L, Müller R, Nielsen TH. (2007) Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ 30: 1499–1512 [DOI] [PubMed] [Google Scholar]
- Ohashi Y, Oka A, Rodrigues-Pousada R, Possenti M, Ruberti I, Morelli G, Aoyama T. (2003) Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science 300: 1427–1430 [DOI] [PubMed] [Google Scholar]
- Pant BD, Buhtz A, Kehr J, Scheible WR. (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53: 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton JL, Pessoa-Brandao L, Henry SA. (1995) Production and reutilization of an extracellular phosphatidylinositol catabolite, glycerophosphoinositol, by Saccharomyces cerevisiae. J Bacteriol 177: 3379–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera IY, Hung C-Y, Moore CD, Stevenson-Paulik J, Boss WF. (2008) Transgenic Arabidopsis plants expressing the type 1 inositol 5-phosphatase exhibit increased drought tolerance and altered abscisic acid signaling. Plant Cell 20: 2876–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt H-J, Puehler A, Broer I. (1993) Transgenic root nodules of Vicia hirsuta: a fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol Plant Microbe Interact 6: 699–706 [Google Scholar]
- Raghothama KG. (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Rao M, Sockanathan S. (2005) Transmembrane protein GDE2 induces motor neuron differentiation in vivo. Science 309: 2212–2215 [DOI] [PubMed] [Google Scholar]
- Rietz S, Dermendjiev G, Oppermann E, Tafesse FG, Effendi Y, Holk A, Parker JE, Teige M, Scherer GFE. (2010) Roles of Arabidopsis patatin-related phospholipases a in root development are related to auxin responses and phosphate deficiency. Mol Plant 3: 524–538 [DOI] [PubMed] [Google Scholar]
- Rietz S, Holk A, Scherer GFE. (2004) Expression of the patatin-related phospholipase A gene AtPLA IIA in Arabidopsis thaliana is up-regulated by salicylic acid, wounding, ethylene, and iron and phosphate deficiency. Planta 219: 743–753 [DOI] [PubMed] [Google Scholar]
- Roby C, Martin J-B, Bligny R, Douce R. (1987) Biochemical changes during sucrose deprivation in higher plant cells: phosphorus-31 nuclear magnetic resonance studies. J Biol Chem 262: 5000–5007 [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Santos-Beneit F, Rodríguez-García A, Apel AK, Martín JF. (2009) Phosphate and carbon source regulation of two PhoP-dependent glycerophosphodiester phosphodiesterase genes of Streptomyces coelicolor. Microbiology 155: 1800–1811 [DOI] [PubMed] [Google Scholar]
- Sbabou L, Bucciarelli B, Miller S, Liu J, Berhada F, Filali-Maltouf A, Allan D, Vance C. (2010) Molecular analysis of SCARECROW genes expressed in white lupin cluster roots. J Exp Bot 61: 1351–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Li H, Neumann G, Zhang F. (2005) Nutrient uptake, cluster root formation and exudation of protons and citrate in Lupinus albus as affected by localized supply of phosphorus in a split-root system. Plant Sci 168: 837–845 [Google Scholar]
- Shi L, Liu JF, An XM, Liang DC. (2008) Crystal structure of glycerophosphodiester phosphodiesterase (GDPD) from Thermoanaerobacter tengcongensis, a metal ion-dependent enzyme: insight into the catalytic mechanism. Proteins 72: 280–288 [DOI] [PubMed] [Google Scholar]
- Shimojima M, Ohta H, Iwamatsu A, Masuda T, Shioi Y, Takamiya K. (1997) Cloning of the gene for monogalactosyldiacylglycerol synthase and its evolutionary origin. Proc Natl Acad Sci USA 94: 333–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriburi R, Bommiasamy H, Buldak GL, Robbins GR, Frank M, Jackowski S, Brewer JW. (2007) Coordinate regulation of phospholipid biosynthesis and secretory pathway gene expression in XBP-1(S)-induced endoplasmic reticulum biogenesis. J Biol Chem 282: 7024–7034 [DOI] [PubMed] [Google Scholar]
- Stenzel I, Ischebeck T, König S, Hołubowska A, Sporysz M, Hause B, Heilmann I. (2008) The type B phosphatidylinositol-4-phosphate 5-kinase 3 is essential for root hair formation in Arabidopsis thaliana. Plant Cell 20: 124–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T-Z, Schweizer HP, Oxender DL. (1991) Carbon-starvation induction of the ugp operon, encoding the binding protein-dependent sn-glycerol-3-phosphate transport system in Escherichia coli. Mol Gen Genet 230: 28–32 [DOI] [PubMed] [Google Scholar]
- Tadano T, Sakai H. (1991) Secretion of acid phosphatase by the roots of several crop species under phosphorus-deficient conditions. Soil Sci Plant Nutr 37: 129–140 [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tesfaye M, Liu J, Allan DL, Vance CP. (2007) Genomic and genetic control of phosphate stress in legumes. Plant Physiol 144: 954–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thole JM, Vermeer JE, Zhang Y, Gadella TW, Jr, Nielsen E. (2008) Root hair defective4 encodes a phosphatidylinositol-4-phosphate phosphatase required for proper root hair development in Arabidopsis thaliana. Plant Cell 20: 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. (1997) The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticconi CA, Lucero RD, Sakhonwasee S, Adamson AW, Creff A, Nussaume L, Desnos T, Abel S. (2009) ER-resident proteins PDR2 and LPR1 mediate the developmental response of root meristems to phosphate availability. Proc Natl Acad Sci USA 106: 14174–14179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommassen J, Eiglmeier K, Cole ST, Overduin P, Larson TJ, Boos W. (1991) Characterization of two genes, glpQ and ugpQ, encoding glycerophosphoryl diester phosphodiesterases of Escherichia coli. Mol Gen Genet 226: 321–327 [DOI] [PubMed] [Google Scholar]
- Uhde-Stone C, Liu J, Zinn KE, Allan DL, Vance CP. (2005) Transgenic proteoid roots of white lupin: a vehicle for characterizing and silencing root genes involved in adaptation to P stress. Plant J 44: 840–853 [DOI] [PubMed] [Google Scholar]
- Uhde-Stone C, Zinn KE, Ramirez-Yáñez M, Li A, Vance CP, Allan DL. (2003) Nylon filter arrays reveal differential gene expression in proteoid roots of white lupin in response to phosphorus deficiency. Plant Physiol 131: 1064–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL. (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable source. New Phytol 157: 423–447 [DOI] [PubMed] [Google Scholar]
- Van der Rest B, Boisson AM, Gout E, Bligny R, Douce R. (2002) Glycerophosphocholine metabolism in higher plant cells: evidence of a new glyceryl-phosphodiester phosphodiesterase. Plant Physiol 130: 244–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Rest B, Rolland N, Boisson AM, Ferro M, Bligny R, Douce R. (2004) Identification and characterization of plant glycerophosphodiester phosphodiesterase. Biochem J 379: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. (2004) Lipid signaling. Curr Opin Plant Biol 7: 329–336 [DOI] [PubMed] [Google Scholar]
- Wasaki J, Omura M, Ando M, Shinano T, Osaki M, Tadano T. (1999) Secreting portion of acid phosphatase in roots of lupin (Lupinus albus L.) and a key signal for the secretion from the roots. Soil Sci Plant Nutr 45: 937–945 [Google Scholar]
- Wasaki J, Yamamura T, Shinano T, Osaki M. (2003) Secreted acid phosphatase is expressed in cluster roots of lupin in response to phosphorus deficiency. Plant Soil 248: 129–136 [Google Scholar]
- Watt M, Evans JR. (1999) Linking development and determinacy with organic acid efflux from proteoid roots of white lupin grown with low phosphorus and ambient or elevated atmospheric CO2 concentration. Plant Physiol 120: 705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson LC, Ribrioux SPCP, Fitter AH, Leyser HMO. (2001) Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol 126: 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Fan J, Cornish AJ, Benning C. (2008) Lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis requires the extraplastidic TGD4 protein. Plant Cell 20: 2190–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaka N, Imai Y, Kawai E, Akatsuka H, Wakimoto K, Nogusa Y, Kato N, Chiba H, Kotani E, Omori K, et al. (2003) Novel membrane protein containing glycerophosphodiester phosphodiesterase motif is transiently expressed during osteoblast differentiation. J Biol Chem 278: 43595–43602 [DOI] [PubMed] [Google Scholar]
- Young BP, Shin JJH, Orij R, Chao JT, Li SC, Guan XL, Khong A, Jan E, Wenk MR, Prinz WA, et al. (2010) Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science 329: 1085–1088 [DOI] [PubMed] [Google Scholar]
- Yu B, Xu C, Benning C. (2002) Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc Natl Acad Sci USA 99: 5732–5737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WH, Ryan PR, Tyerman SD. (2004) Citrate-permeable channels in the plasma membrane of cluster roots from white lupin. Plant Physiol 136: 3771–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Berrie CP, Corda D, Farquhar MG. (2003) GDE1/MIR16 is a glycerophosphoinositol phosphodiesterase regulated by stimulation of G protein-coupled receptors. Proc Natl Acad Sci USA 100: 1745–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Chen D, Farquhar MG. (2000) MIR16, a putative membrane glycerophosphodiester phosphodiesterase, interacts with RGS16. Proc Natl Acad Sci USA 97: 3999–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Kunst L, Hawes C, Moore I. (2004) A GFP-based assay reveals a role for RHD3 in transport between the endoplasmic reticulum and Golgi apparatus. Plant J 37: 398–414 [DOI] [PubMed] [Google Scholar]
- Zhou K, Yamagishi M, Osaki M, Masuda K. (2008) Sugar signalling mediates cluster root formation and phosphorus starvation-induced gene expression in white lupin. J Exp Bot 59: 2749–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn KE, Liu J, Allan DL, Vance CP. (2009) White lupin (Lupinus albus) response to phosphorus stress: evidence for complex regulation of LaSAP1. Plant Soil 322: 1–15 [Google Scholar]
- Zuckerkandl E, Pauling L. (1965) Evolutionary divergence and convergence in proteins. Bryson V, Vogel HJ, eds, Evolving Genes and Proteins. Academic Press, New York, pp 97–166 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.