Abstract
The intracellular localization and shape of the nucleus plays a central role in cellular and developmental processes. In fibroblasts, nuclear movement and shape are controlled by a specific perinuclear actin network made of contractile actin filament bundles called transmembrane actin-associated nuclear (TAN) lines that form a structure called the actin cap. The identification of regulatory proteins associated with this specific actin cytoskeletal dynamic is a priority for understanding actin-based changes in nuclear shape and position in normal and pathological situations. Here, we first identify a unique family of actin regulators, the refilin proteins (RefilinA and RefilinB), that stabilize specifically perinuclear actin filament bundles. We next identify the actin-binding filamin A (FLNA) protein as the downstream effector of refilins. Refilins act as molecular switches to convert FLNA from an actin branching protein into one that bundles. In NIH 3T3 fibroblasts, the RefilinB/FLNA complex organizes the perinuclear actin filament bundles forming the actin cap. Finally, we demonstrate that in epithelial normal murine mammary gland (NmuMG) cells, the RefilinB/FLNA complex controls formation of a new perinuclear actin network that accompanies nuclear shape changes during the epithelial–mesenchymal transition (EMT). Our studies open perspectives for further functional analyses of this unique actin-based network and shed light on FLNA function during development and in human syndromes associated with FLNA mutations.
Keywords: cfm1, cfm2, neural progenitors
Physical connections between perinuclear actin bundles and the nuclear envelope are essential for nuclear movement that controls cell migration and mammalian developmental processes. Defects in perinuclear actin organization are associated with many disease states (1). In fibroblasts, cell shape, nuclear shape, and movement are controlled by specific perinuclear actin networks of actin cables anchored to the nuclear membrane, called transmembrane actin-associated nuclear (TAN) lines (2) or actin cap (3). Despite extensive work on cytoskeleton anchorage at the nuclear envelope, it remains to be determined how actin bundle dynamics are regulated at the nuclear surface. Here we identify a unique family of F-actin bundling proteins that we call refilin (RefilinA and RefilinB) (for REgulator of FILamin proteIN), which function to organize perinuclear actin networks in fibroblasts and in epithelial cells during epithelial–mesenchymal transition (EMT). Refilins bind to the actin-binding filamins. Filamins (FLNA, FLNB, and FLNC) are a family of actin binding and scaffolding proteins that integrate cellular architecture and signaling and are essential for normal fetal development (4–6). Mammalian FLNA is composed of an amino-terminal actin binding domain followed by 24 repeats of which the last repeat mediates dimerization. Two flexible hinge regions, H1 and H2, separate repeats 15 and 16 and 23 and 24, respectively (7). Dimerization of FLNA forms V-shaped molecules that cross-link actin filaments into orthogonal networks (7). FLNA-null murine embryos die with severe vascular, cardiac, and brain morphogenic defects (8, 9). In humans, pathogenic mutations in FLNA cause a wide range of developmental malformations in the heart, skeleton, and brain (10–12). A comprehensive model of FLNA functions is still difficult to formulate and it remains unclear how different mutations in the same protein can cause such a broad spectrum of diseases. Here we provide evidence that refilin converts FLNA from an F-actin branching protein into an F-actin bundler and that the refilin/FLNA complex functions to organize an actin cap in fibroblasts and a unique perinuclear actin network in epithelial cells during EMT mediated by TGF-beta (TGF-β). EMT is a biological process that plays crucial roles in the differentiation of multiple tissues and organs (13, 14). These findings open unique perspectives for understanding FLNA function during embryonic development and in human syndromes associated with FLNA mutations.
Results
Refilin Promotes Formation of Perinuclear Actin Bundles in Astrocytoma U373A Cells.
Refilin proteins belong to the FAM101 family of genes conserved in mammals (FAM101A–RefilinA and FAM101B–RefilinB) that are widely expressed during early embryonic development in the mouse (15). We identified refilins because their transcripts are up-regulated during cell differentiation switches. RefilinA mRNA is up-regulated during the commitment of multipotent neural precursor cells into glial progenitor cells. RefilinB mRNA is up-regulated during EMT mediated by TGF-β. Refilins are hydrophilic proteins and enriched in proline with a secondary structure predicted to be composed of β structures and coiled domains lacking α helices. Refilins are characterized by a conserved N-terminal sequence harboring a DSG(X)2–4S motif that mediates degradation of short-lived proteins (16) and is also found in nuclear transcription factors involved in TGF-β–dependent EMT signaling (17).
To gain insight into the function of refilin proteins, we first compared the cellular localization of GFP and RefilinA/B–GFP fusion proteins in U373A cells that do not express endogenous refilins (Figs. S1A and S2 and Movies S1 and S2). Refilin–GFP constructs localized along actin fibers and promoted formation of parallel actin filament bundles organized into a crescentic shell on top of the nucleus. The height of the nuclei was reduced in RefilinB–GFP-expressing cells that had perinuclear actin bundles compared with control cells (Fig. S1 A, xz and B). The perinuclear actin filaments were immunostained with myosin II antibody, indicating that they are contractile actin bundles (Fig. S1C). Cells with high expression of refilin–GFP are also characterized by star-shaped actin superstructures that are connected to the perinuclear actin bundles (Figs. S1A, arrowheads and S2). Electron microscope observation revealed that these superstructures are enriched in F-actin from which actin stress fibers project (Fig. S3). Similar star-shaped actin superstructures are occasionally observed in NIH 3T3 cells expressing endogenous RefilinB (Fig. 4C, arrowhead).
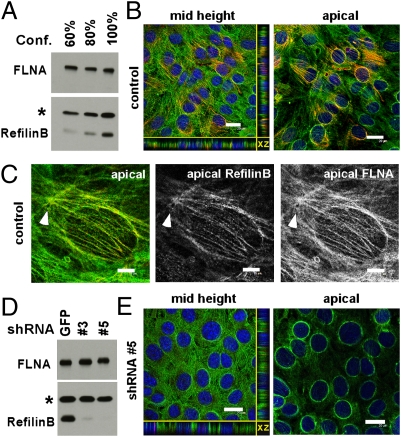
Fig. 4.
RefilinB promotes the recruitment of FLNA on perinuclear actin cap in NIH 3T3 cells. (A) Western blot analysis for RefilinB and FLNA expression in NIH 3T3 cell extracts from exponentially growing cells (lane 1), cell reaching confluence (lane 2), and confluent cells (lane 3). Asterisk indicates position of a cross-reacting protein used as an internal loading control. (B and C) NIH 3T3 cells were fixed with methanol and double labeled with guinea pig RefilinB (red) and mouse FLNA (green) antibodies. Low (B) and high (C) magnification observations are shown. In C, merged and individual RefilinB and FLNA staining at the apical surface are shown. See also Movie S5. Arrowhead points to superstructure (see text for detail). (D) Western blot analysis for RefilinB and FLNA expression in NIH 3T3 cells stably transfected with recombinant lentivirus expressing shRNA against GFP as a control (GFP) or two different shRNA against RefilinB (#3 and #5). (E) Stably transfected NIH 3T3 cells with recombinant lentivirus expressing RefilinB–shRNA #5 were fixed with methanol and double labeled with RefilinB (red) and FLNA (green) antibodies. (B and E, Left) Merged sections at the midheight and profile view (xz), nuclei are stained with Hoechst (blue). (Right) Merged staining at the apical surface. [Scale bars, 20 μm (B and E); 5 μm (C).]
Refilins Dimerize and Interact with Filamins.
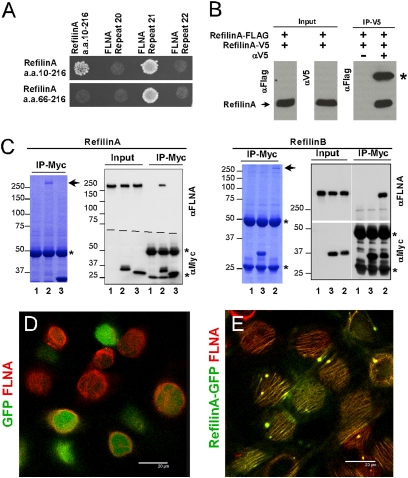
To understand the cellular mechanism through which refilins promote perinuclear actin bundles, we searched for partner proteins by yeast two-hybrid screening (Materials and Methods). Using a RefilinA bait encompassing residues 11–216 of the human protein, 400 interacting colonies were obtained (mating efficiency 6 × 106), 88% of which were identified as being nearly full-length RefilinA, strongly suggestive of a homodimerizing capability of this protein. Upon screening with a second truncated RefilinA bait comprising residues 66–216 (mating efficiency 5.3 × 106), no RefilinA interacting clones were found, indicating that the RefilinA homodimerizing domain resides within the N terminus of the protein. Of the 96 interacting clones in this experiment, 33 (34%) encoded sequences derived from the C-terminal region of FLNA, extending from repeat 15 to the C terminus of the protein. C-terminal domains of FLNB and FLNC were also identified as interactants. RefilinA dimerization was confirmed by yeast two-hybrid–directed matings (Fig. 1A) and the dimerization interface was similarly shown to be distinct from the FLNA interaction domain (Fig. 1A). The ability of RefilinA to homodimerize was confirmed by coimmunoprecipitation (co-IP) of expressed tagged proteins (Fig. 1B). A physical interaction between refilins and the full-length FLNA was also confirmed by coimmunoprecipitation of Myc-tagged RefilinA or RefilinB expressed in human astrocytoma U373 cells (Fig. 1C). Coomassie blue staining of the immunoprecipitates shows an abundant protein of ∼280 kDa (Fig. 1C, blue panels, arrows). Mass spectroscopy sequence analysis of peptides derived from a trypsin digest of this 280-kDa protein revealed it to be a mixture of FLNA and FLNB. Western blot analysis with anti–FLNA-specific antibodies confirmed the identity of the 280-kDa protein as FLNA (Fig. 1C, black/white panels).
Fig. 1.
Refilin targets FLNA in vivo. (A) Directed yeast two-hybrid mating of pGBAD-BD RefilinA (amino acids 11–216) and pGBAD-BD RefilinA (amino acids 66–216) with pACT2B RefilinA (amino acids 11–216), pACT2B FLNA repeat 21 and pACT2B FLNA repeat 20 and 22 used as negative controls. (B) HEK293 cells were cotransfected with RefilinA (amino acids 11–216)-FLAG and RefilinA-V5. Total cell extracts (input) and immunoprecipitates (IP–V5) were analyzed by Western blot using mouse anti-V5 and anti-FLAG antibodies. Asterisk indicates the heavy chain IgG of mouse anti-V5. (C) RefilinA (Left) or RefilinB (Right) coimmunoprecipitate with FLNA. Untransfected U373 MG cells (lane 1) or cells transfected with Myc-tagged RefilinA/B (lane 2) or control Myc-tagged protein corresponding to the 1–220 N-terminal amino acid sequence of ATAD3A protein (lane 3) were used for immunoprecipitation with anti-Myc antibodies. Total cell extracts (input) and immunoprecipitates (IP-Myc) were resolved on SDS/PAGE. Immunoprecipitates were analyzed by Coomassie blue staining. Arrows indicate the position of FLNA. Total cell extracts and immunoprecipitates were analyzed by Western blot using anti-Myc or anti-FLNA antibodies. Asterisk indicates the position of heavy and light IgG chains. (D and E) U373A cells, infected with recombinant adenovirus expressing GFP–control (D) or GFP–RefilinB (E) were fixed and immunostained with mouse anti-FLNA antibody (red). Merged confocal microscopy sections at the apical surface are shown. (Scale bar, 20 μm.) See also Movies S3 and S4.
In U373A cells, the binding of RefilinB or RefilinA to FLNA promotes a relocalization of FLNA from diffuse cytoplasmic staining (in control cells expressiong GFP) onto newly formed perinuclear parallel actin bundles and actin superstructures (Fig. 1 D and E and Movies S3 and S4 and Fig. S2). The refilin/FLNA complex concentrated at the periphery of the actin superstructure but not at their center (Fig. S2 C and D). To confirm a strict cooperativity between refilins and FLNA for perinuclear parallel actin localization and stabilization, we expressed RefilinA in melanoma M2 cells that do not express detectable level of FLNA and in their A7 cells’ counterpart that express FLNA through stable transfection of the FLNA gene (18). In M2 cells, perinuclear actin stress fibers are never observed and RefilinA expression does not stimulate their formation (Fig. S4A). In A7 cells, RefilinA protein promotes formation of perinuclear parallel actin stress fibers (Fig. S4A) and specifically relocalizes FLNA on perinuclear parallel actin stress fibers (Fig. S4B).
FilaminA Mediates Refilin-Dependent Actin Bundling.
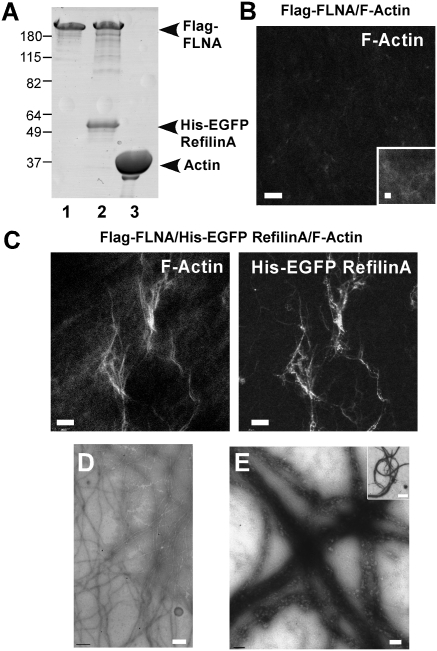
To determine the impact of refilins bound to FLNA on F-actin organization, G-actin was polymerized in the presence of Flag–FLNA or a Flag–FLNA/His–EGFP–refilin complex purified from insect sf9 cells (Fig. 2A). Results obtained with His–EGFP–RefilinA are presented and were reproduced with His–EGFP–RefilinB. Fluorescence microscopy of the samples after costaining F-actin with fluorescent phalloidin (Fig. 2 B and C) and electron microscopy (Fig. 2 D and E) revealed homogenous actin filament networks formed with FLNA in the absence of refilins (Fig. 2 B and D), whereas large F-actin bundles formed in the Flag–FLNA/His–EGFP–refilin samples even at a low ratio of FLNA to actin (1:100) (Fig. 2 C and E). Deletion of a short sequence within RefilinA that was subsequently shown to be critical to the function of the RefilinA/FLNA complex (BD2 domain, see below) totally abrogated the bundling activity of the FLNA/ΔBD2 RefilinB complex both in vitro and in cells (Fig. S5). Altogether, these results indicate that refilins function as molecular switches that convert FLNA from an actin branching protein into one that bundles actin into fibers.
Fig. 2.
RefilinA binding to FLNA promotes actin bundles in vitro. (A) Coomassie blue stain of 10% SDS/PAGE of purified Flag–FLNA (lane 1), Flag–FLNA/His–EGFP–RefilinA complex (lane 2), and actin (lane 3). (B and C) Actin (12 μM) was mixed with 0.12 μM of Flag–FLNA (B) or Flag–FLNA/His–EGFP–RefilinA complex (C), polymerized by the addition of 2 mM MgCl2 and 0.1 M KCl in the presence of Alexa-Fluor 568 phalloidin. Polymerized actin was observed by confocal microscopy. Only Flag–FLNA/His–EGFP–RefilinA complex induces actin bundles. (Scale bar, 200 μm.) (B, Inset) High magnification. (Scale bar, 2 μm.) (D and E) Transmission electron microscopy of negatively stained actin filaments cross-linked by FLNA (D) or FLNA–RefilinA complex (E). (Scale bar, 0.2 μm.) (Inset) Low magnification of bundles. (Scale bar, 1 μm.) Specimens in D and E were diluted to 2 μM F-actin and stained with 2% uranyl acetate.
Mechanism of Refilin–FLNA Interaction.
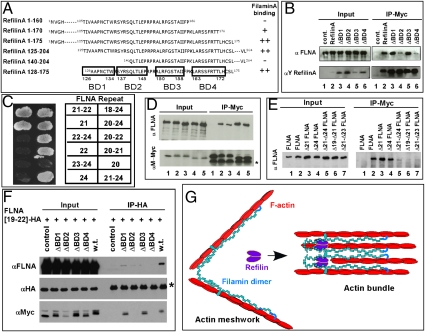
Using N- and C-terminal deletion RefilinA mutants, the minimum RefilinA region required to interact with FLNA was mapped to residues 128–175 (Fig. 3A). Comparison of the 128–175 RefilinA sequence to other FLNA binding partners identified four potential FLNA binding domains (BD1–BD4) (Fig. 3A and Fig. S6). These domains were sequentially deleted from the full-length protein and the mutants (ΔBD1–ΔBD4) were studied, compared with wild-type RefilinA, for binding FLNA in U373 MG cells (Fig. 3B). The ΔBD1 and ΔBD3 deletion mutants coimmunoprecipitated with FLNA to the same extent as the wild-type protein (Fig. 3B, lanes 2, 3, and 5). FLNA binding by the ΔBD2 and ΔBD4 deletion mutants was maintained but dramatically decreased (Fig. 3B, lanes 4 and 6). These results suggest the presence of at least two noncontiguous regions of RefilinA that are necessary for binding to FLNA.
Fig. 3.
Characterization of the RefilinA–FLNA interaction. (A) Sequences of C-terminal and N-terminal Myc-tagged RefilinA deletion mutants used to map the minimal domain required for interaction with FLNA. Interaction of various N-terminal and C-terminal RefilinA–Myc deletion mutants with endogenous FLNA was monitored by coimmunoprecipitation of the myc tag after transfection in U373 MG cells. Region 128–175 is involved in FLNA binding. Within this region, four putative binding domains were identified after sequence alignment with other FLNA binding proteins. The putative FLNA binding domains BD1–4 are boxed. (B) U373 MG cells were transfected with a control plasmid (lane 1) or recombinant plasmids expressing Myc-tagged wild-type RefilinA and RefilinA deletion mutants (ΔBD1–4) (lanes 2–6). Total cell extracts (input) and immunoprecipitates (IP-Myc) were analyzed by Western blot using mouse anti- FLNA and chicken anti-RefilinA antibodies. (C) Directed yeast two-hybrid mating of pACT2B RefilinA (amino acids 11–216) with truncated FLNA variants cloned into pGBAD-BD in yeast strain AH109. (D) M2 cells were cotransfected with either GFP-tagged FLNA (lanes 1, 2, and 4) or GFP-tagged Δ21-FLNA mutant (lanes 3 and 5) and Myc-tagged control protein (lane 1), RefilinA–Myc (lanes 2 and 3) or RefilinB–Myc (lanes 4 and 5). (E) M2 cells were cotransfected with either Myc-tagged control protein (lane 1) or Myc-tagged RefilinA (lanes 2–9) and either GFP-tagged FLNA (lanes 1 and 2) or one of the following GFP-tagged FLNA mutants: Δ21-FLNA (lane 3), Δ24-FLNA (lane 4), Δ21/Δ24-FLNA (lane 5), Δ19/Δ21-FLNA (lane 6), and Δ21/Δ23-FLNA (lane 7). Myc-tagged proteins were immunoprecipitated with mouse anti-Myc antibody. Total cell extracts (input) and immunoprecipitates (IP-Myc) were analyzed by Western blot using mouse anti-FLNA and mouse anti-Myc antibodies as indicated. (D) Asterisk indicates positions of IgG light chains. (F) U373MG cells were cotransfected with FLNA[19–22]–HA and Myc-tagged control protein, ΔBD11–4–RefilinA–Myc mutants or wild-type RefilinA–Myc as indicated. FLNA[19–22]–HA was immunoprecipitated with rat anti-HA antibody. Total cell extracts (input) and immunoprecipitates (IP-HA) were analyzed by Western blot using mouse anti-FLNA, mouse anti-HA, and mouse anti-Myc antibodies. Asterisk indicates positions of rat heavy IgG chains present in the immunoprecipitates that migrate just above FLNA[19–22]–HA construct and cross-react with the mouse secondary antibody used to reveal the Western blot. (G) Proposed schematic representation of RefilinA dimer as a molecular switch that converts FLNA from an actin branching protein into one that bundles. See text for details.
To map the refilin binding domains on FLNA, various yeast clones expressing FLNA truncated C-terminal constructs were mated to clones expressing RefilinA (residues 11–216) using the yeast two-hybrid vector system (Fig. 3C). Bait and prey vectors were reversed for these experiments relative to those used previously (Fig. 1A). Repeat 21 on FLNA was found to be critical for RefilinA binding. These results were confirmed by coimmunoprecipitation of RefilinA–Myc with different HA-tagged FLNA constructs coexpressed in M2 cells that lack endogenous FLNA (Fig. S7A). To determine whether FLNA repeat 21 is the only refilin binding site we next compared binding of refilins to the full-length FLNA and Δ21–FLNA mutant lacking repeat 21. Deletion of domain 21 was not sufficient to inhibit interaction of RefilinA or RefilinB with FLNA, as assessed by co-IP (Fig. 3D, lanes 3 and 5), suggesting that another refilin binding domain is present on FLNA. To identify this second refilin binding domain, we deleted additional domains on full-length FLNA protein (Fig. 3E). Sole deletion of domain 24 or 21 did not inhibit the interaction (Fig. 3E, lanes 3 and 4). However, if both domain 21 and domain 24 were deleted, the interaction of the Δ21/Δ24–FLNA mutant with RefilinA was drastically reduced (Fig. 3E, lane 5). If domain 21 and adjacent domains 19 or 23 were then deleted while keeping the dimerization domain 24 intact, the interaction of these FLNA mutants with refilin was also drastically decreased (Fig. 3E, lanes 6 and 7). We conclude that repeat 21 provides a template for the interaction of refilin with FLNA monomer and that a second refilin binding domain, which requires FLNA dimerization, strengthens this interaction. This model is consistent with the presence of two putative FLNA binding domains on refilin and possible cooperative binding of refilin to FLNA.
We next asked whether refilin dimer promotes the formation of multimolecular FLNA complexes to bundle F-actin. To answer this question, we cotransfected a construct encoding only repeats 19–22 fused to HA tag (FLNA[19–22]–HA) with either a Myc tagged control protein, RefilinA–Myc or one of the RefilinA–Myc deletion mutants (ΔBD1–ΔBD4) in U373 cells. FLNA[19–22]–HA was immunoprecipitated and immunoprecipitates were analyzed by Western blot for their RefilinA and FLNA contents (Fig. 3F). Results showed that FLNA[19–22]–HA, which is unable to dimerize or to bind actin, can bind to endogenous FLNA only when coexpressed with wild-type RefilinA. Note that in this experiment, the truncated FLNA[19–22]–HA will be in competition with the endogenous full-length FLNA dimer for binding refilin. This explains the limited amount of refilin and FLNA recovered within the immunoprecipitates. It is also noteworthy that ΔBD1 and ΔBD3 refilin mutants complex with FLNA[19–22]–HA but could not coimmunoprecipitate endogenous FLNA, indicating that BD1 and BD3 domains are necessary to nucleate a functional refilin–FLNA complex. The capacity of RefilinA to dimerize FLNA was confirmed with repeats 18–23 of FLNA fused to V5 or FLAG tags (Fig. S7B).
Altogether, these results support a model where the RefilinA dimer may function as a zipper to promote formation of a multimolecular FLNA complex on F-actin and to convert FLNA from an actin branching protein into one that bundles actin into fibers (Fig. 3G).
RefilinB/FLNA Complex Stabilizes Perinuclear Actin Cap in NIH 3T3 Cells.
The perinuclear actin cytoskeleton organization mediated by refilin in U373A cells resembles in many aspects the “perinuclear actin cap” previously described in mouse 3T3 fibroblasts (3). To investigate whether the refilin/FLNA complex contributes to actin cap formation in 3T3 fibroblasts, we first analyzed the protein expression in NIH 3T3 cells. Western blot analysis showed specific RefilinB expression that increased as cells in culture reached confluence (Fig. 4A). The level of expression of RefilinA was below the detection limit. The specificity of the RefilinB band was confirmed by its down-regulation in stably transfected NIH 3T3 cells with two different shRNAs targeted to RefilinB (Fig. 4D).
Double indirect immunofluorescence analyses on cells fixed with methanol showed heterogeneous RefilinB immunoreactivity among cells. Cells showing the highest RefilinB immunostaining also demonstrated apical FLNA staining (Fig. 4 B and C). RefilinB immunoreactivity colocalized with FLNA on parallel perinuclear filamentous structures organized into a crescentric shell above the nucleus (Fig. 4C and Movie S5). With NIH 3T3 cells fixed with PFA and stained with phalloidin, we confirmed the specific localization of FLNA on parallel perinuclear actin bundles forming the actin cap (Fig. S8A). In NIH 3T3 cells stably transfected with shRNA targeted against RefilinB, perinuclear actin bundles that previously immunostained with RefilinB and FLNA in untransfected cells disappeared (Fig. 4E), and cells showed a disrupted cap organization (Fig. S8B). Cells with down-regulated RefilinB were also characterized by increased nuclear height (see reconstituted profile views in Fig. 4 B and E and Fig. S8) and a more flattened nuclear morphology. To exclude possible off-target effects of RefilinB shRNA, we demonstrated that ectopic expression of RefilinA–Myc can sponsor actin cap formation in RefilinB null NIH 3T3 cells (Fig. S9). We conclude that the RefilinB/FLNA complex is a key regulator of actin cap dynamics in fibroblast cells.
RefilinB/FLNA Complex Mediates Organization of a Perinuclear Actin Network During EMT.
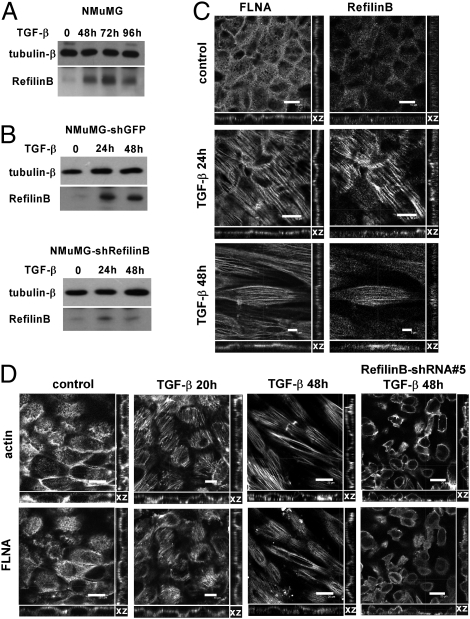
In mouse epithelial NMuMG cells, RefilinB expression increases in response to TGF-β stimulation (Fig. 5A). The specificity of the RefilinB band was confirmed by its down-regulation in stably transfected NMuMG cell with RefilinB shRNA #5 (Fig. 5B). Here again, both RefilinA protein and mRNA were below the detection limit. In NMuMG cells stimulated with TGF-β and fixed with methanol, RefilinB colocalized with FLNA exclusively on a new actin network that formed at the apical surface above the nuclei during early stages of the EMT process (Fig. 5C). In NMuMG cells fixed with PFA, staining with phalloidin and anti-FLNA antibodies confirmed the specific recruitment of FLNA on the new apical perinuclear actin network that forms during EMT (Fig. 5D and Movies S6–S8). In nonstimulated cells, F-actin organizes the cortical actin cytoskeleton and FLNA shows a diffuse cytoplasmic staining (Fig. 5D, control and Movie S6). Following TGF-β stimulation, a population of FLNA is specifically relocalized on perinuclear actin filament bundles that form at the apical surface (Fig. 5D, TGF-β 20 h and Movie S7). Later, in cells that have acquired fibroblastic morphology with elongated nuclei, FLNA immunoreactivity remains present on perinuclear actin bundles aligned with the overall nucleus-cell orientation (Fig. 5D, TGF-β 48 h and Movie S8). Remarkably, in those cells, FLNA immunoreactivity was excluded from basal actin stress fibers (Movies S7 and S8).
Fig. 5.
FLNA relocalizes on actin cap in NMuMG cells during EMT. (A and B) Western blot analysis for RefilinB expression in NMuMG cells (A) and in stably transfected NMuMG cell lines with recombinant lentivirus expressing control shRNA (NMuMG–shGFP) (B, Upper) or shRNA #5 against RefilinB (NMuMG–shRefilinB) (B, Lower). Cells were stimulated with 2 ng/mL TGF-β for the indicated times. β-Tubulin was used as loading control. (C) RefilinB colocalizes with FLNA in NMuMG cells stimulated with TGF-β. NMuMG cells not stimulated (control) or stimulated with TGF-β for 24 h or 48 h were fixed with methanol and double labeled with mouse FLNA antibody and guinea pig RefilinB antibody. Reconstituted profile view (xz) and individual RefilinB and FLNA staining at the apical surface are shown. [Scale bar, 10 μm (control and TGF-β 24 h) and 5 μm (TGF-β 48 h).] (D) Stably transfected NMuMG cells with recombinant lentivirus expressing GFP–control shRNA or RefilinB–shRNA #5 (far Right column) were not stimulated (control) or stimulated with TGF-β for 20 h or 48 h (as indicated), fixed with PFA, and double labeled with Alexa 546-phalloidin and FLNA antibody. Individual FLNA and F-actin staining at the apical surface and reconstituted profile view (xz) are shown. (Scale bar, 20 μm.) See also Movies S6–S9.
In the NMuMG cell line stably expressing RefilinB shRNA #5, down-regulation of RefilinB expression was associated with diffuse FLNA staining, disorganized apical actin network, and a delay in nuclear and cell shape remodeling in response to TGF-β stimulation (Fig. 5D, far Right column and Movie S9). Down-regulation of RefilinB had no impact on basal actin reorganization (Movie S9). Altogether, these results demonstrate a specific function of the RefilinB/FLNA complex in the regulation of a unique perinuclear actin network that forms during EMT and that contributes to nuclear cell-shape remodeling.
Discussion
Here we identify a unique actin regulatory family, refilins, which specifically promote actin filament bundles to organize apical perinuclear actin networks in mesenchymal cells. Refilins function through binding to the actin binding protein FLNA and conversion of FLNA from an F-actin branching protein into an F-actin bundler. Detailed analyses of the mechanisms of the refilin–FLNA interaction identified the presence of at least two FLNA binding domains on RefilinA (BD2 and BD4). Sequence comparison revealed conserved amino acids between RefilinA BD2 and the FLNA binding domains on other proteins that bind to FLNA repeat 21 (Fig. S6). Specific activation of the actin bundling activity of FLNA by refilin could be mediated by additional contributions from the BD4 domain within refilin to trigger conformational changes to the FLNA molecule. Determination of the structure of a three-domain fragment of human FLNA (repeats 19–21) revealed interactions between repeats that could function as an autoinhibitory mode of regulation of FLNA function (19). We propose that refilins, through binding to repeat 21 and possibly adjacent repeats, may reorganize the overall conformation of the FLNA 19–21 domain. This would, in turn, change the orientation of the FLNA rod regions, as previously discussed (19), and convert the high-angle F-actin branching (7, 20) into low-angle structures for actin bundling (Fig. 3G). It is possible that when complexed to actin, larger refilin/FLNA complexes may also form, that would explain the formation of a larger actin-rich superstructure in cells overexpressing refilins (Figs. S1A and S2). Further studies are thus required to refine the specific molecular organization of the refilin/FLNA complex on F-actin.
In NIH 3T3 fibroblast cells, the perinuclear actin bundles stabilized by the refilin/FLNA complex most likely correspond to the “TAN lines” or perinuclear actin cap, previously described in mouse 3T3 fibroblasts, and which connect actin cables that form on the dorsal surface of nuclei to the outer nuclear membrane (2, 3). The proteins involved in this connection are referred to as the LINC complex (1). In epithelial NMuMG cells stimulated by TGF-β, RefilinB, and FLNA also contributed to organize perinuclear actin bundles that accompany EMT. The identification of a unique perinuclear actin network organization that is specifically regulated by the refilin/FLNA complex during a well-characterized cell process, the EMT, opens a unique perspective for further functional analysis of this subcellular structure. In U373A and NIH 3T3 cells, the induction or inhibition of the actin cap was associated with discrete changes in nuclear and cellular shapes. In NMuMG cells stably expressing RefilinB shRNA, a delay in nuclear shape remodeling was observed but this did not affect cell viability. This suggests that the regulation of perinuclear actin bundle dynamics by the refilin/FLNA complex is probably not fundamental for cell viability in culture and is also consistent with the absence of a demonstrable phenotype in cultured FLNA-null fibroblasts (8, 9). In contrast, loss of function of FLNA in mice and humans is associated with severe cardiac, skeletal, vascular, and brain morphogenic defects during embryonic development (9, 11), and gain-of-function mutations that alter its avidity for actin lead to still further developmental anomalies (10, 12). Loss of FLNA shows similarities with loss of LINC complex proteins that structure the TAN lines in this respect (21). One LINC protein, LaminA, is dispensable for survival and proliferation in cell culture but its deletion or mutation are responsible for developmental defects and a broad spectrum of diseases (21, 22). We propose that alterations in FLNA function can induce dysregulation of perinuclear actin dynamics in specific developmental processes, such as EMT, leading to developmental defects or pathologies. In mice, maximal expression of FLNA in cardiac tissues occurs during expression of EMT programs associated with cardiac and vascular morphogenesis (23). Defective EMT programming may therefore explain the major cardiac and vascular defects seen in FLNA-null mouse embryos (8, 9). Similarly EMT programs are also important in development of human tissues, notably cardiac morphogenesis (24) and because several phenotypically discrete syndromes associated with FLNA mutations have defective morphogenesis of the cardiac valves as features of their phenotype (10, 12, 25), disruption of the FLNA–refilin-mediated formation of the actin cap could lie at the center of the pathophysiology of these disorders. It is also possible that altered perinuclear actin dynamics could contribute to abnormal neuronal migration, as seen in patients with FLNA mutations associated with periventricular heterotopia (11, 26). The possibility is supported by a recent study showing that nuclear migration within the neuroepithelium, mediated in part by LINC proteins, is essential for radial neuronal migration in mice (27). Finally, the refilin binding domains of FLNA (including repeat 21) are highly conserved in other filamin isoforms. We have also confirmed a physical interaction of FLNB and FLNC with refilin by yeast two hybrid and co-IP. Hence, refilin very likely regulates the function of all cellular filamins. It would now be interesting to evaluate whether mutant filamin proteins found across the full range of human syndromes caused by mutations in filamins have altered responses to binding to refilins.
Materials and Methods
Materials.
Antibodies, plasmids, ShRNA, and vector constructions are described in SI Materials and Methods.
Cell Culture.
NIH 3T3, U373A (a clone obtained by xenograft of the human astrocytoma U373 MG selected for its tumorigenic properties), and M2 and A7 cells (18) were grown in Dulbecco’s modified Eagle’s medium (DMEM containing 4.5 g/L glucose) plus 10% FCS. NMuMG cells were grown in DMEM + 4.5 g/L glucose, 10% FCS, and 0.1% insulin. For EMT induction, NMuMG medium was supplemented with TGF-β1 2 ng/mL + EGF 2 ng/mL.
Cloning of Refilins.
Rat FAM101A and FAM101B genes were cloned by RT-PCR on rat oligodendrocyte progenitor cell extracts. Sequences were confirmed by DNA sequencing. Nucleotide and protein sequences correspond to reference sequences Ensembl ENSRNOP00000001337 and ENSRNOT00000008982. The human RefilinA correspond to reference Ensembl ENSP00000374377.
Additional Methods.
Yeast two-hybrid screening and directed yeast two-hybrid mating, production and transduction of adenovirus and lentiviral vectors, purification of FLNA–RefilinA complex expressed in sf9 insect cells, visual assay for F-actin bundling, indirect immunofluorescence, and immunoprecipitation procedures are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by Institut National du Cancer INCA-PL 114 (to J.B.), Association contre le Cancer ARC-SFI20101201517 (to J.B.), Ligue National contre le Cancer (O.G.), National Institutes of Health Grant HL-56252 (to J.H.H.), and the Harvard University Science and Engineering Committee Seed Fund for Interdisciplinary Science (F.N.). Work in S.P.R.’s laboratory is supported by the Marsden Fund of New Zealand and Curekids New Zealand. We thank colleagues from Institut National de la Santé et de la Recherche Médicale U 880 for mass spectrometry analyses, N. Bertacchi for technical assistance, Dr D. Markie for reagents, and Dr. J. LaMarre and C. Benaud for stimulating discussions.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104211108/-/DCSupplemental.
References
- 1.Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khatau SB, et al. A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci USA. 2009;106:19017–19022. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou AX, Hartwig JH, Akyurek LM. Filamins in cell signaling, transcription and organ development. Trends Cell Biol. 2010;20:113–123. doi: 10.1016/j.tcb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Feng Y, Walsh CA. The many faces of filamin: A versatile molecular scaffold for cell motility and signalling. Nat Cell Biol. 2004;6:1034–1038. doi: 10.1038/ncb1104-1034. [DOI] [PubMed] [Google Scholar]
- 6.Stossel TP, et al. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura F, Osborn TM, Hartemink CA, Hartwig JH, Stossel TP. Structural basis of filamin A functions. J Cell Biol. 2007;179:1011–1025. doi: 10.1083/jcb.200707073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng Y, et al. Filamin A (FLNA) is required for cell-cell contact in vascular development and cardiac morphogenesis. Proc Natl Acad Sci USA. 2006;103:19836–19841. doi: 10.1073/pnas.0609628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hart AW, et al. Cardiac malformations and midline skeletal defects in mice lacking filamin A. Hum Mol Genet. 2006;15:2457–2467. doi: 10.1093/hmg/ddl168. [DOI] [PubMed] [Google Scholar]
- 10.Kyndt F, et al. Mutations in the gene encoding filamin A as a cause for familial cardiac valvular dystrophy. Circulation. 2007;115:40–49. doi: 10.1161/CIRCULATIONAHA.106.622621. [DOI] [PubMed] [Google Scholar]
- 11.Fox JW, et al. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21:1315–1325. doi: 10.1016/s0896-6273(00)80651-0. [DOI] [PubMed] [Google Scholar]
- 12.Robertson SP, et al. OPD-spectrum Disorders Clinical Collaborative Group Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat Genet. 33:487–491. doi: 10.1038/ng1119. [DOI] [PubMed] [Google Scholar]
- 13.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Hirano M, et al. cfm is a novel gene uniquely expressed in developing forebrain and midbrain, but its null mutant exhibits no obvious phenotype. Gene Expr Patterns. 2005;5:439–444. doi: 10.1016/j.modgep.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 16.Busino L, et al. Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature. 2003;426:87–91. doi: 10.1038/nature02082. [DOI] [PubMed] [Google Scholar]
- 17.Zhou BP, et al. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004;6:931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 18.Cunningham CC, et al. Actin-binding protein requirement for cortical stability and efficient locomotion. Science. 1992;255:325–327. doi: 10.1126/science.1549777. [DOI] [PubMed] [Google Scholar]
- 19.Lad Y, et al. Structure of three tandem filamin domains reveals auto-inhibition of ligand binding. EMBO J. 2007;26:3993–4004. doi: 10.1038/sj.emboj.7601827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartwig JH, Tyler J, Stossel TP. Actin-binding protein promotes the bipolar and perpendicular branching of actin filaments. J Cell Biol. 1980;87:841–848. doi: 10.1083/jcb.87.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Folker ES, Ostlund C, Luxton GW, Worman HJ, Gundersen GG. Lamin A variants that cause striated muscle disease are defective in anchoring transmembrane actin-associated nuclear lines for nuclear movement. Proc Natl Acad Sci USA. 2011;108:131–136. doi: 10.1073/pnas.1000824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parnaik VK, Manju K. Laminopathies: Multiple disorders arising from defects in nuclear architecture. J Biosci. 2006;31:405–421. doi: 10.1007/BF02704113. [DOI] [PubMed] [Google Scholar]
- 23.Norris RA, et al. Expression of the familial cardiac valvular dystrophy gene, filamin-A, during heart morphogenesis. Dev Dyn. 2010;239:2118–2127. doi: 10.1002/dvdy.22346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Person AD, Klewer SE, Runyan RB. Cell biology of cardiac cushion development. Int Rev Cytol. 2005;243:287–335. doi: 10.1016/S0074-7696(05)43005-3. [DOI] [PubMed] [Google Scholar]
- 25.Robertson SP, et al. Frontometaphyseal dysplasia: Mutations in FLNA and phenotypic diversity. Am J Med Genet A. 2006;140:1726–1736. doi: 10.1002/ajmg.a.31322. [DOI] [PubMed] [Google Scholar]
- 26.Sheen VL, et al. Mutations in the X-linked filamin 1 gene cause periventricular nodular heterotopia in males as well as in females. Hum Mol Genet. 2001;10:1775–1783. doi: 10.1093/hmg/10.17.1775. [DOI] [PubMed] [Google Scholar]
- 27.Taverna E, Huttner WB. Neural progenitor nuclei IN motion. Neuron. 2010;67:906–914. doi: 10.1016/j.neuron.2010.08.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.