Abstract
The brown alga Ectocarpus siliculosus has a haploid–diploid life cycle that involves an alternation between two distinct generations, the sporophyte and the gametophyte. We describe a mutant, ouroboros (oro), in which the sporophyte generation is converted into a functional, gamete-producing gametophyte. The life history of the mutant thus consists of a continuous reiteration of the gametophyte generation. The oro mutant exhibited morphological features typical of the gametophyte generation and accumulated transcripts of gametophyte generation marker genes. Genetic analysis showed that oro behaved as a single, recessive, Mendelian locus that was unlinked to the IMMEDIATE UPRIGHT locus, which has been shown to be necessary for full expression of the sporophyte developmental program. The data presented here indicate that ORO is a master regulator of the gametophyte-to-sporophyte life cycle transition and, moreover, that oro represents a unique class of homeotic mutation that results in switching between two developmental programs that operate at the level of the whole organism.
Keywords: brown algae, development, phaeophyceae, stramenopiles
Many organisms have life histories that involve alternation between two multicellular generations, a sporophyte and a gametophyte (1). A considerable amount of theoretical work has been carried out to understand the evolutionary and ecological significance of such complex life cycles (1). Haploid–diploid life cycles (i.e., cycles in which there is an alternation between a diploid sporophyte and a haploid gametophyte generation) pose a particular problem because it is necessary to explain why they do not evolve towards either haploid or diploid life cycles, both of which have theoretical advantages under some conditions. Possible advantages specific to haploid–diploid life cycles include the potential of the two generations to exploit alternative ecological niches, avoidance of biotic aggressors (because of differential susceptibility of the two generations), and a reduced cost of sex (one meiosis/fertilization every two generations). However, very little experimental work has been carried out to test these various hypotheses (1, 2). One of the limiting factors in this regard is that the mechanisms that regulate life cycle progression at the molecular level remain poorly understood.
Variant life cycles could potentially provide insight into underlying regulatory mechanisms. In a normal haploid–diploid life cycle, the two generations are demarcated by meiosis, which produces the spores that will become the gametophyte generation, and gamete fusion, which produces the zygote that will become the sporophyte generation. However, many species exhibit various forms of apomixis, in which the life cycle progresses without the occurrence of meiosis or fertilization (3, 4). Apomictic life cycles can involve both apospory (formation of a gametophyte from a sporophyte without meiosis) and apogamy (formation of a sporophyte by parthenogenesis of a gametophyte cell). Genetic analysis of apomictic systems is complicated by a number of factors (5), but the existence of these systems does suggest that life cycle regulation may be amenable to genetic analysis. This contention has been supported by experimental work on the non-apomictic species Arabidopsis thaliana and on non-apomictic strains of Physcomitrella patens, in which phenomena associated with apomixis, such as parthenogenesis, have been observed in mutant strains.
Arabidopsis strains carrying mutations in homologs of PRC2 polycomb complex component genes or in the RETINOBLASTOMA-RELATED gene, a regulator of the PRC2 complex, exhibit fertilization-independent development of the endosperm and, in some cases (FERTILIZATION INDEPENDENT ENDOSPERM1/MEDEA, FERTILIZATION INDEPENDENT ENDOSPERM2, and MULTICOPY SUPPRESSOR OF IRA 1 [MSI1] mutants), parthenogenetic development of embryo-like structures (6–11). These embryo-like structures usually abort after undergoing a few cell divisions, making it difficult to determine definitively whether the structure formed corresponds to a bona fide embryo. However, strong evidence for a switch from the gametophyte to the sporophyte developmental program has been obtained for the MSI1 mutant by analyzing the expression of sporophyte generation marker genes in the embryo-like structure (12). Together, these observations suggest that the PRC2 complex acts to prevent transitions from the gametophyte to the sporophyte developmental program in the Arabidopsis gametophyte, acting as part of a developmental switch for this life cycle transition. Similar observations have been made in the moss P. patens, where mutations in either the PHYSCOMITRELLA PATENS CURLY LEAF (PpCLF) or the PHYSCOMITRELLA PATENS FERTILIZATION INDEPENDENT ENDOSPERM genes (both of which are predicted to encode components of a PRC2 complex) result in fertilization-independent production of a sporophyte-like body on side branches of the gametophytic protonema filaments (13, 14). In both studies, sporophyte marker genes were used to verify that the structures produced corresponded to the sporophyte generation. As in Arabidopsis, sporophyte development arrests prematurely. Arrest occurs partly because an active PRC2 complex seems to be needed for a later transition within the sporophyte, from the vegetative to the reproductive phase (production of the sporangium). However, even restoration of PpCLF expression at this later stage was not sufficient to produce a functional sporophyte, suggesting either that the sporophyte generation also depends on a factor supplied by the underlying gametophore (on which it normally grows) or that the haploid nature of the structure prevents full sporophyte development. Similar factors may explain the abortion of fertilization-independent embryos in Arabidopsis mutants. In this respect, brown macroalgae represent interesting alternative model systems to study the alternation of generations in haploid–diploid life cycles because they can exhibit a remarkable plasticity with regard to ploidy and because, in species with haploid–diploid life cycles, the two generations of the life cycle often develop completely independently, after the release of single-cell propagules into the surrounding seawater.
The filamentous alga Ectocarpus siliculosus (Dillwyn) Lyngbye is an emerging model for the brown algae (15), and a number of molecular and genetic tools have recently been made available, including a complete genome sequence (16, 17). Ectocarpus has a haploid–diploid life cycle involving alternation between two independent heteromorphic generations, the sporophyte and the gametophyte (Fig. S1A). In addition, unfused gametes can develop parthenogenetically to produce partheno-sporophytes. One experimental advantage of this parthenogenesis is that mutations affecting sporophyte development are directly expressed phenotypically in haploid partheno-sporophytes, facilitating mutant screens (18). A spontaneous mutation (immediate upright; imm), in which the sporophyte generation exhibits several characteristics of the gametophyte, including an asymmetric initial cell division and absence of a basal system of prostrate filaments, has recently been described (18). Here we characterize a second Ectocarpus life cycle mutant, ouroboros (oro), which exhibits homeotic conversion of the sporophyte generation into a fully functional gametophyte. The oro mutation represents a unique class of homeotic mutation, causing switching between two different developmental programs that operate at the level of the whole organism.
Results
ouroboros Mutant Parthenotes Closely Resemble Wild-Type Gametophytes.
The oro mutant was isolated after UV mutagenesis of freshly released gametes of Ectocarpus strain Ec 32. In contrast to the situation in wild-type Ectocarpus, where gametes germinate parthenogenetically (i.e., in the absence of gamete fusion) to produce partheno-sporophytes (Fig. S1A), parthenotes of the oro mutant exhibited both morphological and functional features typical of gametophytes. The first cell division was asymmetrical, rather than being symmetrical as in the wild-type partheno-sporophyte (18), resulting in cells with rhizoid and upright filament identities (Fig. 1 A–C). Mutant individuals produced richly branched thalli, again typical of the gametophyte, which lacked the prostrate basal system found in the sporophyte (Fig. 1 D–F). Plurilocular sexual organs formed after 3 wk in culture and resembled gametangia rather than sporangia, having an elongated shape and never occurring at the tips of the upright filaments (Fig. 1 G–I). Furthermore, oro parthenotes never produced unilocular sporangia, a feature that has been observed only during the sporophyte generation.
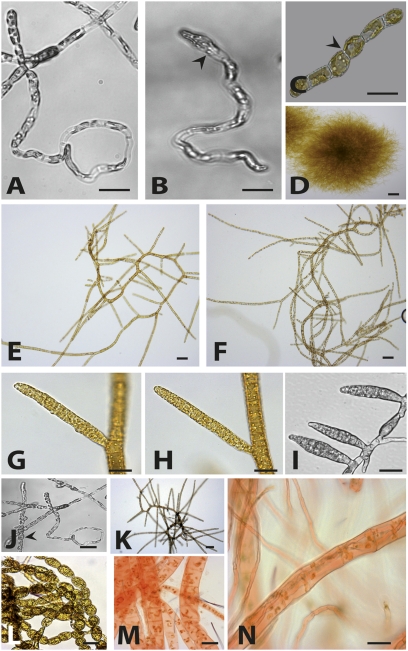
Fig. 1.
oro mutant parthenotes closely resemble wild-type gametophytes. Representative images are shown. (A) Wild-type gametophyte germling (4 d old). (B) oro parthenote germling (4 d old). Note the asymmetrical first cell division (arrowhead). (C) Wild-type partheno-sporophyte germling (2 d old). Note the symmetrical first cell division (arrowhead). (D) Adult wild-type sporophyte (3 wk old). (E) Adult wild-type gametophyte (3 wk old). (F) Adult oro parthenote (3 wk old). (G) Plurilocular gametangium of a wild-type gametophyte. (H) Plurilocular gametangium of an oro parthenote. (I) Plurilocular sporangia of a wild-type partheno-sporophyte. (J) Round cells present during the early development of a few oro individuals (arrowhead). (K) oro individuals that had round cells during early development reverted to gametophyte morphology. (L) Wild-type partheno-sporophytes stained with Congo red (no red staining). (M) Wild-type gametophytes stained with Congo red (showing the characteristic red color). (N) oro individuals are Congo red positive. (Scale bars: 10 μm.)
Many oro parthenotes were indistinguishable from wild-type gametophytes throughout their development, but minor differences were observed for some oro individuals during early development. For example, wild-type sporophytes produce a base consisting of round cells strongly adhering to the substratum, whereas gametophytes tend to float off into the medium (18). Some oro individuals exhibited weak adherence to the substratum and produced a small number of round cells during the early stages of development (Fig. 1J). However, these sporophyte-like features were only observed transiently. When 45 parthenotes that exhibited this intermediate phenotype were isolated and their development was monitored, all developed gametophyte morphology within 6 d (Fig. 1K). Zoids released by the plurilocular organs of the oro mutant germinated to produce individuals that reiterated the oro phenotype (Fig. S1B). Transmission of the phenotype by this asexual pathway occurred stably for at least 36 asexual generations, and the phenotype was not affected by cultivation at 5, 10, or 20 °C or under different daylength regimes.
To further investigate the nature of the oro parthenotes, we developed a simple staining technique to distinguish between the sporophyte and gametophyte generations. The carbohydrate binding dye Congo red, which is used to stain xylan fibers in algal cell walls (19), stained the gametophyte, but not the sporophyte, of wild-type Ectocarpus (Fig. 1 L and M). The oro parthenotes were stained by Congo red, which again indicates that these individuals are gametophytes (Fig. 1N). Wild-type sporophytes and gametophytes also differ in their response to unidirectional light during early development, with gametophytes exhibiting a significantly more marked response to this stimulus (18). Approximately 90% of the oro parthenotes germinated away from unidirectional light (Fig. S2). This result was not significantly different from that obtained for a wild-type gametophyte (χ2 = 1.46, P > 0.05, df = 1, n = 305) but was significantly different from that obtained with wild-type partheno-sporophytes (χ2 = 29.60, P < 0.001, df 1, n = 1,030). An analysis of marker genes whose transcripts have been shown to exhibit significantly different levels of abundance during the two generations of the life cycle (18) showed that oro parthenotes accumulated transcripts that had been shown to be significantly more abundant in the gametophyte generation and, conversely, had reduced levels of transcripts that had been shown to be significantly more abundant in the sporophyte (Fig. 2).
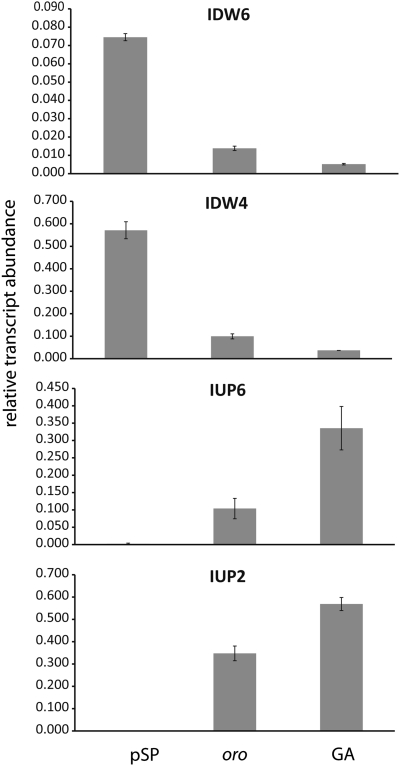
Fig. 2.
qRT-PCR analysis of the abundances of gene transcripts in the oro mutant compared with wild-type gametophyte (GA) and partheno-sporophyte (pSP). The abundance of transcripts of four genes that have been shown to be differentially expressed in the gametophyte and sporophyte generations (18) was assayed in oro individuals. Data are means of three independent biological replicates ± SE. Transcript abundance was significantly different in the oro and GA samples compared with the wild-type pSP sample for all of the genes tested (ANOVA, P < 0.05). (From top to bottom) IDW6, Esi0351_0007; IDW4, Esi0085_0048; IUP6, Esi0031_0170; IUP2, Esi0416_0001.
oro Is a Single-Locus, Recessive Mutation That Causes Conversion of the Sporophyte into a Functional Gametophyte.
Zoids released from the plurilocular sexual organs of the male oro mutant fused with female gametes from the wild-type strain Ec 25 in crossing experiments to produce zygotes, indicating that the oro zoids were gametes and not spores (Fig. S3A). This result demonstrated that the oro parthenotes not only exhibit gametophyte morphology but are actually functional gametophytes.
Three of the zygotes from the cross between the oro mutant and Ec 25 were raised to maturity. All three individuals exhibited a typical sporophyte pattern of growth throughout their development and produced unilocular sporangia, indicating that the oro mutation was recessive (Fig. S3B). Sporophyte progeny were also obtained when the oro mutant was crossed with another, more distantly genetically related strain, Ec 568 (20).
To analyze the genetic inheritance of the oro locus, 14 unilocular sporangia were isolated individually from 3 diploid sporophytes derived from the cross between the oro mutant and the female strain Ec 25. These sporangia, which each contained >100 meiospores (derived from a single meiosis followed by at least five mitotic divisions), produced 14 meiotic “families” of gametophytes, all of which exhibited a germination pattern typical of wild-type gametophytes. Therefore, there was no evidence that the oro mutation had a phenotypic effect during the gametophyte generation, which was to be expected if the effect of the mutation is to convert the sporophyte into a gametophyte. For each of the families derived from a single unilocular sporangium, between 19 and 27 gametophytes were sub-isolated. Gametes produced by these gametophytes were allowed to germinate parthenogenetically, and the pattern of growth of the parthenotes was monitored (Table S1). Overall, 162 gametophytes produced parthenotes that developed as partheno-sporophytes (as expected for wild-type individuals), and 177 produced parthenotes with the oro phenotype. These figures are consistent with a 1:1 segregation ratio and Mendelian inheritance of a single-locus recessive mutation (ANOVA, F1,26 = 1.07, P = 0.3). The sex of several individuals each from 12 of the unilocular-sporangium-derived families that exhibited an oro phenotype was determined in test crosses with male and female tester strains: 33 of the oro plants tested were females, and 48 were males (Table S2). These data indicated that the ORO locus was not linked to the sex locus (ANOVA, F1,22 = 3.21, P = 0.09).
Diploid Individuals That Are Homozygous for the oro Mutation Exhibit a Gametophyte Phenotype.
To determine whether diploid sporophytes (produced by the fusion of two gametes) were also converted to gametophytes in the presence of the oro mutation, a female oro strain (generated by the segregation analysis described above) was back-crossed with the male oro strain. The diploid progeny of this cross exhibited a similar phenotype to oro parthenotes except that the conversion to gametophyte morphology was slightly less marked. Many round cells (typical of sporophytes) could be observed during early development, but, as had been observed with the parthenotes, gametophyte morphology was adopted as growth progressed. Crosses (n = 11) with female (Ec 25 and Ec 410) and male (Ec 400 and Ec 32) tester strains demonstrated that the oro homozygous individuals produced functional gametes and that these gametes behaved as males, despite the presence of both the male and female haplotypes of the sex locus.
Genetic Interactions Between the oro and imm Mutations.
Individuals carrying both the oro and the imm mutations were constructed by crossing imm strain Ec 419 and oro strain Ec 494. All 14 of the zygotes isolated from this cross developed as wild-type sporophytes, indicating that complementation had occurred. Sixteen unilocular sporangia were isolated from one of the sporophytes (Ec 560), and between 9–20 gametophytes were sub-isolated from each of the meiotic families derived each from a single unilocular sporangium. Parthenotes that developed from gametes produced by these gametophytes exhibited one of four different patterns of early development (Table S3 and Fig. 3 A–E): (a) individuals morphologically identical to the wild-type partheno-sporophyte; (b) immediate development of erect filaments and production of a straight, rather than wavy, rhizoid (the phenotype of the imm mutant; ref. 18); (c) parthenotes with the phenotype described above for the oro mutant, i.e., gametophyte-like but with the formation of a small number of round cells during early development in some individuals; and (d) parthenotes that more strongly resembled the gametophyte during early development (absence of round cells) except that some individuals possessed a straight rather than wavy rhizoid. For the phenotypic classes c and d, all individuals rapidly adopted a typical gametophyte morphology as they developed (Fig. 3F). At maturity, parthenotes of classes a and b were functional sporophytes (producing unilocular sporangia and plurilocular sporangia releasing mitospores and not gametes), whereas parthenotes of classes c and d were functional gametophytes, producing gametes that were able to fuse with gametes of the opposite sex. These phenotypic data indicated that the four classes corresponded to the following genotypes: (a) IMM ORO, (b) imm ORO, (c) IMM oro, and (d) imm oro. The four different phenotypic groups occurred in equal proportions (ANOVA, F3,60 = 0.83, P = 0.5). Note that the three mutant classes exhibited gametophyte-like phenotypic features to an increasing degree as follows: imm < oro < imm oro. All of the phenotypic classes were stable for at least eight asexual generations (asexual propagation being via mitospores in the case of classes a and b and via parthenogenetic development of gametes in the case of classes c and d).
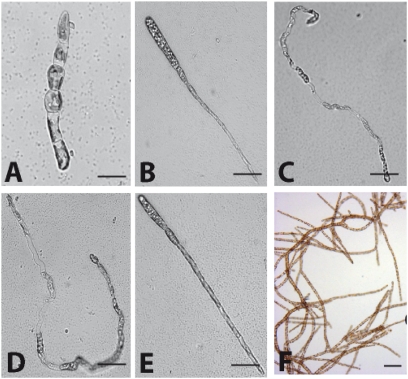
Fig. 3.
Segregation analysis of the oro and imm loci. (A) Germling showing a wild-type partheno-sporophyte phenotype. (B) Germling showing an imm phenotype. (C) Germling showing an oro phenotype. (D and E) Germlings showing imm oro double-mutant phenotypes. (F) imm-like germlings from the imm oro mutants develop into gametophytes. See text for details. (Scale bars: 10 μm.)
To confirm that parthenotes of class d corresponded to imm oro double mutants, one individual from this class was crossed with a wild-type strain (Ec 32). Seven sporophytes with wild-type phenotypes were derived from this cross, and 15–25 gametophytes were produced from each sporophyte. Phenotypic analysis of the parthenotes derived from these gametophytes confirmed that the original individual carried both the imm and oro mutations (Table S4).
Comparison of mRNA Abundances in Life Cycle Mutants with the Wild-Type Sporophyte and Gametophyte.
An expressed-sequence-tag-based microarray carrying probes corresponding to 10,600 of the 16,256 genes identified in the Ectocarpus genome (16, 21) was used to carry out a broad analysis of changes in steady-state mRNA abundances in the life cycle mutants (ArrayExpress accession no. E-MTAB-485). Transcript abundances were compared in parthenotes of all four possible classes of segregant in the population derived from the cross between the imm and oro mutants: (a) IMM ORO (wild-type partheno-sporophyte), (b) imm ORO, (c) IMM oro, and (d) imm oro individuals, using bulked segregants to minimize variance due to unlinked loci. A fifth sample corresponding to the wild-type gametophyte was also analyzed to provide a reference for the gametophyte generation of the life cycle. We carried out pairwise comparisons of each of the three mutant samples and the gametophyte sample with the wild-type partheno-sporophyte sample using the Significance Analysis of Microarrays (SAM) method (22). In each case, the genes for which we detected significant changes in mRNA abundance were assigned to a broad range of Gene Ontology (GO) categories (Dataset S1). This result indicated that both the imm and the oro mutations lead to complex modifications of cellular mRNA levels, involving changes in the expression levels of genes with roles in many different cellular processes. A GO slim analysis (http://www.geneontology.org) was carried out to look for general trends across the four different comparisons. Only one GO slim category, “protein metabolic process” (GO:0019538), was represented solely in samples that contained functional gametophytes (oro mutant, imm oro double mutant, and wild-type gametophyte).
Fig. 4 illustrates the overlaps between the lists of differentially expressed genes identified in the four comparisons (imm, oro, and imm oro mutant strains and wild-type gametophyte with partheno-sporophyte). In all four comparisons, more genes were identified as being up-regulated than down-regulated (between 3- and 62-fold difference). This result suggests that the transition from the sporophyte to the gametophyte stage involves primarily gene induction rather than gene repression.
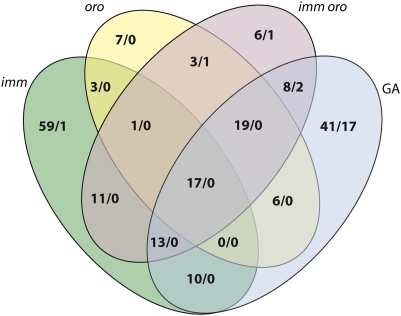
Fig. 4.
Venn diagram showing the overlap between the sets of genes that had significantly different transcript abundances in the mutant (imm, oro, imm oro) strains and in the wild-type gametophyte (GA) compared with the wild-type partheno-sporophyte. The two numbers separated by a slash indicate the number of genes whose transcripts were significantly more (to the left) or less (to the right) abundant in the indicated sample, compared with the wild-type partheno-sporophyte.
To carry out a broad cluster analysis, a set of 4,046 genes that exhibited significant changes in transcript abundance across the five samples analyzed in the microarray experiment were identified by applying a one-way ANOVA test. The 4,046 genes could be grouped into four clusters (Fig. S4A). The two largest clusters corresponded to genes whose transcripts tended to exhibit abundances in the mutant samples that were intermediate between those in the wild-type sporophyte and gametophyte samples, with preferential expression either in the sporophyte (cluster 1) or in the gametophyte (cluster 2). These expression patterns were consistent with intermediate phenotypes exhibited by the single- and double-mutant lines. A GO slim analysis indicated that GO categories associated with primary metabolism, including carbohydrate and lipid metabolism, and related functions such as transport were overrepresented in cluster 1 and underrepresented in cluster 2. In contrast, GO categories associated with protein synthesis and modification and other basic cellular processes, such as cell cycle, DNA replication, and developmental processes, showed the opposite trend (Fig. S4B). These trends indicate that there may be differences in the allocation of cellular resources in the two generations, perhaps associated with adaptation to different ecological niches or growth habits.
Quantitative RT-PCR (qRT-PCR) Analysis of Gene Expression.
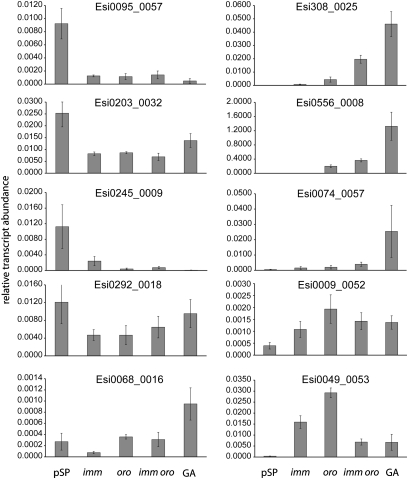
In an effort to better understand the molecular mechanisms underlying the alternation between the sporophyte and gametophyte generations, we searched the microarray data for genes that were differentially expressed in mutant and wild-type samples and that were predicted to encode proteins with potentially interesting functions (regulatory proteins, for example). Nine genes were selected, to which we added the Ectocarpus ARGONAUT1 gene because there is accumulating evidence that small RNA pathways play important roles at key stages of the life cycle (23, 24). Transcript abundances for these 10 genes were then assayed by using qRT-PCR in five samples corresponding to those analyzed in the microarray experiment (Fig. 5 and Table S5). Transcripts of 9 of the genes analyzed exhibited either increases (Esi0308_0025, Esi0556_0008, Esi0074_0057, Esi0049_0053, Esi0009_0052) or decreases (Esi0095_0057, Esi0203_0032, Esi0245_0009, Esi0292_0018) in abundance in both the wild-type gametophyte sample and in some or all of the mutant samples, suggesting that these are life cycle-regulated genes that are influenced by the imm and/or the oro loci. The proteins encoded by these genes include a putative transcription factor (Esi0095_0057) and the ARGONAUT1 protein (Esi0203_0032). Interestingly, the gradual increases in the abundances of the Esi0308_0025 (predicted to encode a serine/threonine kinase) and Esi0074_0057 (predicted to encode a Homeodomain-like protein) transcripts when the imm, oro, imm oro, and wild-type gametophyte samples were compared correlated with the increasingly gametophyte-like nature of the individuals in each of these samples that was observed during the morphological analysis described above. Moreover, for Esi0308_0025, there was a synergistic effect of the imm and oro mutations resulting in a greater increase in transcript abundance in the imm oro double mutant than with either of the individual mutations alone. The Esi0556_0008 transcript increased in abundance only in the samples that contained functional gametophytes (wild-type gametophyte, oro mutant, and imm oro double mutant), suggesting that this gene may be important for the transition from sporophyte to gametophyte function rather than, for example, for a morphological feature of the gametophyte that is not directly related to life cycle function. Esi0556_0008 is predicted to encode an extracellular structural protein. For Esi0068_0016, there was no obvious correlation between transcript levels in the mutant lines and in the wild-type gametophyte. This result was consistent with the data from the microarray analysis, which suggested that the mutations do not exactly reproduce the phenotype of the wild-type gametophyte, even in the case of the imm oro double mutant, which is a functional gametophyte with morphological characteristics that are almost identical to those of the wild-type gametophyte.
Fig. 5.
qRT-PCR analysis of the abundance of gene transcripts in wild-type partheno-sporophyte, imm, oro, imm oro, and wild-type gametophytes. Genes were selected either from the microarray analysis or based on their predicted function (see text for details). Data are means of three independent biological replicates ± SE.
Discussion
Mutation at the ORO Locus Results in Conversion of the Sporophyte Generation into a Functional Gametophyte.
oro parthenotes exhibited several features typical of young wild-type gametophytes, including an asymmetric first cell division, production of highly ramified filaments composed exclusively of cylindrical cells, production of sexual structures resembling plurilocular gametangia, positive staining with Congo red, the presence of transcripts of genes that are normally expressed during the gametophyte generation, and production of functional gametes (Fig. 1 and Fig. S2). Together, these results indicate that parthenotes derived from oro gametes take on a gametophyte fate and that ORO functions either to activate the sporophyte program at the appropriate stage of the life cycle or to repress the gametophyte developmental program during the sporophyte generation. Transcriptomic analysis indicated that the switch from the gametophyte to the sporophyte generation involved predominantly gene repression, providing support for the latter hypothesis.
The global, organism-level developmental modification observed in the oro mutant indicates that this gene is a master regulator, influencing a broad range of processes associated with the gametophyte developmental program. This hypothesis was supported by the microarray analysis, which identified a significant number of genes whose transcript abundances were modified in the absence of a functional ORO locus.
Interactions Between the ORO and IMM Loci.
The imm mutation results in partial conversion of the sporophyte into a gametophyte (18), whereas individuals carrying the oro mutation are functional gametophytes that are almost indistinguishable from wild-type gametophytes. The phenotype of the imm oro double mutant closely resembled that of the oro mutant alone, indicating that ORO is at least partially epistatic to IMM. There was partial overlap between the sets of genes that exhibited significant changes in transcript abundance compared with the wild-type sporophyte reference for the imm and oro mutants (16% and 37% of the gene sets for each mutant, respectively), consistent with a partial epistatic relationship between the two genes. Like oro, the imm mutation resulted in modifications of the transcript abundances of a large number of genes, suggesting that both genes control the expression of many downstream genes (ref. 18 and this study). However, a proportion of the genes that exhibited significant changes in transcript abundance in the wild-type gametophyte compared with the wild-type sporophyte were not represented in the gene lists for the imm and oro single mutants and the imm oro double mutant (70%, 68%, and 56%, respectively), suggesting that even the imm oro double mutant does not completely recapitulate the gametophyte developmental program.
oro Mutation and Life Cycle Variability.
The isolation of the oro mutant has allowed previous observations about the Ectocarpus life cycle and sexuality to be independently confirmed. For example, several lines of evidence indicated that life cycle generation can be uncoupled from ploidy. Haploid, diploid, or tetraploid partheno-sporophytes (25, 26) and haploid, diploid, and aneuploid gametophytes (26–28) have been observed in culture. Here we have shown that zygotes derived from a cross between male and female oro mutants give rise to functional, diploid gametophytes. Moreover, crosses carried out using this strain and male and female tester lines indicated that the male haplotype of the sex locus was dominant over the female haplotype. Again, this finding confirms Müller's earlier work on sexually heterozygous gametophytes constructed through laboratory crosses of non-mutant strains (27, 28).
In some species of brown algae, gametes develop parthenogenetically to produce partheno-gametophytes rather than partheno-sporophytes (e.g., Myriotrichia clavaeformis; ref. 29). Hence, the modified life cycle observed in individuals carrying the oro mutation is the “normal” situation for some brown algal species. However, unlike in the oro mutant, their zygotes always develop into sporophytes. Once the genetic lesion that corresponds to the oro mutation has been identified, it will be interesting to compare the structure of this locus in species that produce either partheno-sporophytes or partheno-gametophytes.
Changes in life cycle structure have been associated with several important transitions during evolution. In land plants, for example, adaptation to the terrestrial environment appears to have been linked with increasing dominance of the sporophytic stage of the life cycle. The brown algae include organisms with remarkably varied life cycles, ranging from isomorphic, haploid–diploid cycles to diploid cycles in which there is only one multicellular generation. They therefore represent an ideal group both for understanding the adaptive benefits of particular life cycle structures and for investigating the general role played by life cycle modifications in evolutionary processes.
Materials and Methods
Biological Material.
Cultivation, crossing, raising of sporophytes from zygotes, isolation of meiotic families, and sexing of gametophyte strains were carried out as described (20, 30). See SI Materials and Methods and Table S6 for details.
UV Mutagenesis.
Gametes from Ec 32m strain were irradiated with a UV (254 nm) lamp for 45 min immediately after release from plurilocular gametangia. Irradiated gametes were allowed to settle in the dark at 13 °C for 4 h. Petri dishes were then transferred to a culture chamber at 13 °C and cultivated as described above.
Microarray Analysis of mRNA Abundances.
Preparation of material, hybridization, and analysis of microarray data were performed essentially as described by Dittami et al. (21). Lists of genes with significant differences in transcript abundance were established by pairwise analyses using the SAM method (22) in the TIGR MeV package (Version 3.1). See SI Materials and Methods for details.
qRT-PCR Analysis of Transcript Abundance.
qRT-PCR validation was carried out essentially as described by Peters et al. (18). Transcript abundance was estimated based on the analysis of three biological replicates, normalized against the EF1α reference gene. See SI Materials and Methods and Table S7 for details.
Supplementary Material
Acknowledgments
We thank Delphine Scornet, Guillaume Hatte, and Laurence Dartevelle for technical assistance. This work was supported by Centre National de la Recherche Scientifique, Université Pierre et Marie Curie, Groupement d'Interet Scientifique Génomique Marine, the Interreg program France (Channel)-England (project Marinexus), and Agence Nationale de la Recherche (Project Bi-cycle). A.A. was supported by a European Erasmus Mundus Program Fellowship.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the ArrayExpress database, http://www.ebi.ac.uk/arrayexpress/ (accession no. E-MTAB-485).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102274108/-/DCSupplemental.
References
- 1.Coelho SM, et al. Complex life cycles of multicellular eukaryotes: New approaches based on the use of model organisms. Gene. 2007;406:152–170. doi: 10.1016/j.gene.2007.07.025. [DOI] [PubMed] [Google Scholar]
- 2.Thornber CS. Functional properties of the isomorphic biphasic algal life cycle. Integr Comp Biol. 2006;46:605–614. doi: 10.1093/icb/icl018. [DOI] [PubMed] [Google Scholar]
- 3.Bicknell RA, Koltunow AM. Understanding apomixis: Recent advances and remaining conundrums. Plant Cell. 2004;16(Suppl):S228–S245. doi: 10.1105/tpc.017921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koltunow AM, Grossniklaus U. Apomixis: A developmental perspective. Annu Rev Plant Biol. 2003;54:547–574. doi: 10.1146/annurev.arplant.54.110901.160842. [DOI] [PubMed] [Google Scholar]
- 5.Ozias-Akins P, van Dijk PJ. Mendelian genetics of apomixis in plants. Annu Rev Genet. 2007;41:509–537. doi: 10.1146/annurev.genet.40.110405.090511. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhury AM, et al. Fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1997;94:4223–4228. doi: 10.1073/pnas.94.8.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guitton AE, et al. Identification of new members of Fertilisation Independent Seed Polycomb Group pathway involved in the control of seed development in Arabidopsis thaliana. Development. 2004;131:2971–2981. doi: 10.1242/dev.01168. [DOI] [PubMed] [Google Scholar]
- 8.Köhler C, et al. Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 2003;22:4804–4814. doi: 10.1093/emboj/cdg444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohad N, et al. A mutation that allows endosperm development without fertilization. Proc Natl Acad Sci USA. 1996;93:5319–5324. doi: 10.1073/pnas.93.11.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohad N, et al. Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell. 1999;11:407–416. doi: 10.1105/tpc.11.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebel C, Mariconti L, Gruissem W. Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature. 2004;429:776–780. doi: 10.1038/nature02637. [DOI] [PubMed] [Google Scholar]
- 12.Guitton AE, Berger F. Loss of function of MULTICOPY SUPPRESSOR OF IRA 1 produces nonviable parthenogenetic embryos in Arabidopsis. Curr Biol. 2005;15:750–754. doi: 10.1016/j.cub.2005.02.066. [DOI] [PubMed] [Google Scholar]
- 13.Okano Y, et al. A polycomb repressive complex 2 gene regulates apogamy and gives evolutionary insights into early land plant evolution. Proc Natl Acad Sci USA. 2009;106:16321–16326. doi: 10.1073/pnas.0906997106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mosquna A, et al. Regulation of stem cell maintenance by the Polycomb protein FIE has been conserved during land plant evolution. Development. 2009;136:2433–2444. doi: 10.1242/dev.035048. [DOI] [PubMed] [Google Scholar]
- 15.Charrier B, et al. Development and physiology of the brown alga Ectocarpus siliculosus: Two centuries of research. New Phytol. 2008;177:319–332. doi: 10.1111/j.1469-8137.2007.02304.x. [DOI] [PubMed] [Google Scholar]
- 16.Cock JM, et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature. 2010;465:617–621. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]
- 17.Cock JM, Coelho SM, Brownlee C, Taylor AR. The Ectocarpus genome sequence: insights into brown algal biology and the evolutionary diversity of the eukaryotes. New Phytol. 2010;188:1–4. doi: 10.1111/j.1469-8137.2010.03454.x. [DOI] [PubMed] [Google Scholar]
- 18.Peters AF, et al. Life-cycle-generation-specific developmental processes are modified in the immediate upright mutant of the brown alga Ectocarpus siliculosus. Development. 2008;135:1503–1512. doi: 10.1242/dev.016303. [DOI] [PubMed] [Google Scholar]
- 19.Yamagishi T, Hishinuma T, Kataoka H. Novel sporophyte-like plants are regenerated from protoplasts fused between sporophytic and gametophytic protoplasts of Bryopsis plumosa. Planta. 2004;219:253–260. doi: 10.1007/s00425-004-1230-9. [DOI] [PubMed] [Google Scholar]
- 20.Peters AF, et al. Genetic diversity of Ectocarpus (Ectocarpales, Phaeophyceae) in Peru and northern Chile, the area of origin of the genome-sequenced strain. New Phytol. 2010;188:30–41. doi: 10.1111/j.1469-8137.2010.03303.x. [DOI] [PubMed] [Google Scholar]
- 21.Dittami SM, et al. Global expression analysis of the brown alga Ectocarpus siliculosus (Phaeophyceae) reveals large-scale reprogramming of the transcriptome in response to abiotic stress. Genome Biol. 2009;10:R66. doi: 10.1186/gb-2009-10-6-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wuest SE, et al. Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr Biol. 2010;20:506–512. doi: 10.1016/j.cub.2010.01.051. [DOI] [PubMed] [Google Scholar]
- 24.Khurana JS, Theurkauf W. piRNAs, transposon silencing, and Drosophila germline development. J Cell Biol. 2010;191:905–913. doi: 10.1083/jcb.201006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bothwell JH, Marie D, Peters AF, Cock JM, Coelho SM. Role of endoreduplication and apomeiosis during parthenogenetic reproduction in the model brown alga Ectocarpus. New Phytol. 2010;188:111–121. doi: 10.1111/j.1469-8137.2010.03357.x. [DOI] [PubMed] [Google Scholar]
- 26.Müller DG. Generationswechsel, Kernphasenwechsel und Sexualität der Braunalge Ectocarpus siliculosus im Kulturversuch. Planta. 1967;75:39–54. doi: 10.1007/BF00380838. [DOI] [PubMed] [Google Scholar]
- 27.Müller DG. Sex expression in aneuploid gametophytes of the brown alga Ectocarpus siliculosus (Dillw.) Lyngb. Arch. Protistenk. Bd. 1975;117:297–302. [Google Scholar]
- 28.Müller DG. Diploide, heterozygote gametophyten bei der braunalge Ectocarpus siliculosus. Naturwissenschaften. 1970;57:357–358. [Google Scholar]
- 29.Peters AF, Marie D, Scornet D, Kloareg B, Cock JM. Proposal of Ectocarpus siliculosus (Ectocarpales, Phaeophyceae) as a model organism for brown algal genetics and genomics. J Phycol. 2004;40:1079–1088. [Google Scholar]
- 30.Peters AF, Scornet D, Müller DG, Kloareg B, Cock JM. Inheritance of organelles in artificial hybrids of the isogamous multicellular chromist alga Ectocarpus siliculosus (Phaeophyceae) Eur J Phycol. 2004;39:235–242. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.