Abstract
Interactions between bacteria and the viruses that infect them (i.e., phages) have profound effects on biological processes, but despite their importance, little is known on the general structure of infection and resistance between most phages and bacteria. For example, are bacteria–phage communities characterized by complex patterns of overlapping exploitation networks, do they conform to a more ordered general pattern across all communities, or are they idiosyncratic and hard to predict from one ecosystem to the next? To answer these questions, we collect and present a detailed metaanalysis of 38 laboratory-verified studies of host–phage interactions representing almost 12,000 distinct experimental infection assays across a broad spectrum of taxa, habitat, and mode of selection. In so doing, we present evidence that currently available host–phage infection networks are statistically different from random networks and that they possess a characteristic nested structure. This nested structure is typified by the finding that hard to infect bacteria are infected by generalist phages (and not specialist phages) and that easy to infect bacteria are infected by generalist and specialist phages. Moreover, we find that currently available host–phage infection networks do not typically possess a modular structure. We explore possible underlying mechanisms and significance of the observed nested host–phage interaction structure. In addition, given that most of the available host–phage infection networks examined here are composed of taxa separated by short phylogenetic distances, we propose that the lack of modularity is a scale-dependent effect, and then, we describe experimental studies to test whether modular patterns exist at macroevolutionary scales.
Keywords: bacteriophage, complex networks, ecology, evolution, nestedness
Bacteria and their viruses (phages) make up two of the most abundant and genetically diverse groups of organisms (1–3). The extent of this diversity has become increasingly apparent with the advent of community genomics. Microbial DNA isolated from oceans, lakes, soils, and human guts has revealed tremendous taxonomic diversity in a broad range of environmental habitats and conditions (4–11). The ongoing discovery of new taxonomic diversity has, thus far, outpaced gains in understanding the function of specific microbes and their most basic ecology of who interacts with whom. One of the starkest examples of this disparity is the lack of an efficient (bioinformatic or otherwise) approach for determining which viruses can infect which bacteria. Although it is well-known that individual phages do not infect all bacteria, we have little understanding of what the precise host range for any given phage is or whether there are universal patterns or principles governing the set of viruses able to infect a given bacterium and the set of bacteria that a given virus can infect. This deficit is unfortunate given that phage–bacterial interactions are important for both human health and ecosystem function (12–16).
Phages have multifaceted effects on their hosts: they can lyse host cells, thereby releasing new virons, transfer genes between hosts, and form lysogens that can modify host function (17–19). In some cases, phages can transfer genes for pathogenicity between pathogenic and labile strains (e.g., for both Vibrio cholerae and Shigella), facilitating the spread of bacterial infections (20–22). Phages also alter ecosystem functions by the high levels of bacterial mortality that they cause. Bacteria lysed by phage will release their contents, which consequently are scavenged by other bacteria rather than being incorporated into bactivorous eukaryotes (23, 24). This weakened connection early in the food chain can have effects that ripple throughout the ecosystem. Information on a general pattern of infection by phages on hosts could improve predictions of microbial population dynamics, ecosystem functioning, and microbial community assembly (25, 26).
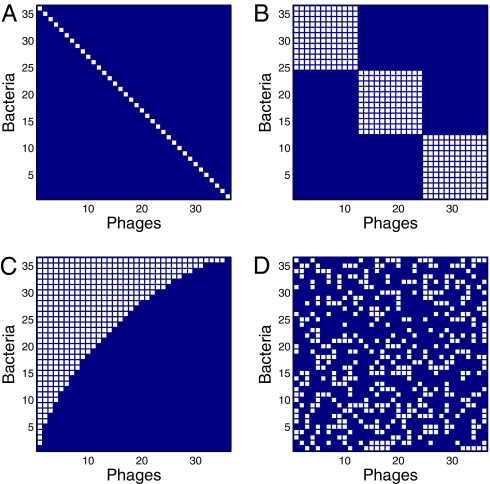
What is our expectation for the general pattern of host–phage infection networks? Host–phage infection networks have, in the past, been measured by performing pair-wise infections of hosts by phages isolated from natural ecological communities, evolution experiments, or strain collections. The results of such pair-wise infections can be represented as a network or a matrix, where the rows indicate host isolates, the columns indicate phage isolates, and the cells within the matrix describe whether each combination results in a successful infection. We consider different classes of host–phage interaction networks as alternative hypotheses for an expected pattern (Fig. 1). First, phages may infect a unique host or a limited number of closely related hosts, leading to nearly diagonal matrices (Fig. 1A) or block-like matrices that exhibit high degrees of modularity (Fig. 1B). These patterns should occur if host–viral interactions are the result of coevolutionary processes that lead to specialization. Second, diversification of hosts and phages may result in nested matrices in which the most specialist phages infect those hosts that are most susceptible to infection rather than infecting those hosts that are most resistant to infection (Fig. 1C). The nested pattern is the predicted outcome of a prominent theory of gene-for-gene coevolution, where phages evolve so as to broaden host ranges and bacteria evolve so as to increase the number of phages to which they are resistant (27, 28). We should note that these two patterns and hypotheses for the forms of coevolution are not mutually exclusive and in fact, could be scale-dependent. Nested patterns could form within modules if, for instance, microevolutionary changes result in nestedness; however, genetic differences between species or genera that accumulate over macroevolutionary time may limit the exchange of viruses between these phylogenetic groups and create an overall modular structure. Finally, we consider a null model to be that matrices of host–phage infection are statistically indistinguishable from random matrices (Fig. 1D).
Fig. 1.
Schematic of expected host–phage interaction matrices (white cells denote infection). (A) Host–phage interactions are unique (i.e., only one phage infects a given host, and only one host is infected by a given phage). (B) Host–phage interactions are modular (i.e., blocks of phages can infect blocks of bacteria, but cross-block infections are not present). (C) Host–phage interactions are nested (i.e., the generalist phage infects the most sensitive and the most resistant bacteria, whereas the specialist phage infects the host that is infected by the most viruses). (D) Host–phage interactions are random and lack any particular structure. For B–D, a connectance of 0.33 was used so that the expected total number of interactions was the same in each case.
Contrary to this null expectation, we show that currently available host–phage interaction matrices are, as a whole, statistically distinguishable from random matrices and possess a characteristic nested structure. We reach this conclusion by performing a metaanalysis on the patterns of host–phage infection matrices collected by a comprehensive search of the literature and supplementing these matrices with an experimental analysis of host–phage infection. The data that we assemble consist of 38 matrices of host–phage infection assays representing the cumulative study of 1,009 bacterial isolates, 502 phage isolates, and almost 12,000 separate attempts to infect a bacteria host with a phage strain (27, 29–64) (SI Appendix, Tables S1 and S2 have more information on the examined studies). This work is an attempt to subject host–phage infection assays to a unified analysis. In doing so, we find a general pattern of host–phage interactions. We discuss biophysical, ecological, and evolutionary mechanisms that could lead to this nested (and not modular) pattern as well as future studies to explore how such a pattern may change as a function of phylogenetic scale.
Results
Compiling a Large-Scale Host–Phage Interaction Dataset.
We compiled a set of 37 studies with direct laboratory evidence of host–phage interactions using an extensive literature search supplemented by an experimental study of an evolved Escherichia coli and phage λ-system (SI Appendix, Tables S1 and S2 have complete details of all studies) (27, 29–64). The method of evaluating infection ability in assembling a host–phage infection matrix varies; however, the most commonly used approach is that of spot assays, in which a single virus type is combined with a population of bacteria cells from a single strain. Infection is considered to have occurred given evidence that the phage has infected and lysed (part of) the bacterial population. Hence, the result of each study is a matrix of the infection ability for each phage on each host. The studies included in the host–phage infection assays analyzed here were isolated from one of three sources: co-occurring isolates within natural communities taken directly from the environment and then cultured, coevolutionary laboratory experiments where a single bacterial clone and a single phage clone were allowed to coevolve for a fixed amount of time and then, their evolved progenitors examined, and laboratory stocks of phages and hosts that were artificially combined. Some of the matrices used were composed of bacteria and phage acquired from two separate isolation strategies. For these studies, we classified the matrix by which isolation strategy represented the majority of matrix cells and made a note of the other sources (SI Appendix, Table S2). The criterion by which we searched and cataloged these studies is explained in more detail in SI Appendix, SI Materials and Methods. Overall, we identified and analyzed a wide range of infection networks for organisms that varied in their phylogenic position, traits, and habitats. For example, the bacterial hosts included Gram-positives and -negatives, heterotrophs, and phototrophs as well as pathogens and nonpathogens.
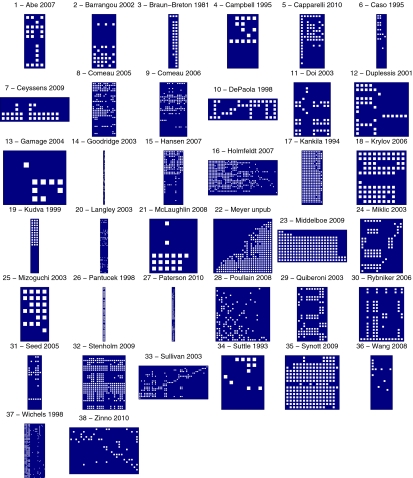
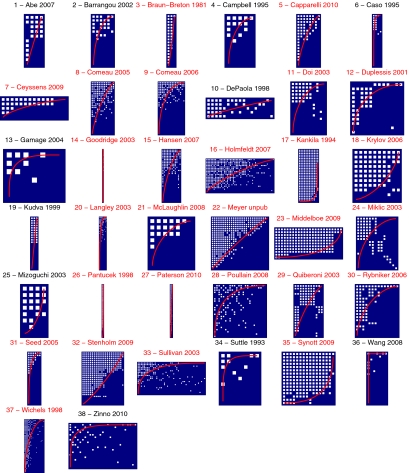
Some of the assays include graded information about infection (for example, whether a phage simply inhibits bacterial growth or forms regions of complete bacterial mortality like plaques). In other studies, replicate phage populations were used to deduce whether phages always or only sometimes cause plaques. Details of the criteria for the interactions can be found in the original works (27, 29–64), and the experimental methods for the experimental study of host–phage infection can be found in Materials and Methods. Because graded information about infection was not uniformly available in all studies, assays were standardized using hand-curated extraction of original data into a single matrix of ones and zeros with H rows (one for every bacterial host) and P columns (one for every phage), where a 1-valued cell represents evidence for infection (either full or partial) and a 0-valued cell represents no evidence for infection (Fig. 2 shows a visual depiction of all host–phage interaction matrices).
Fig. 2.
Matrix representation of the compiled studies. The rows represent the hosts, and the columns represent the phages. White cells indicate the recorded infections. Note the diversity in the size of these matrices.
Host–Phage Infection Statistics Do Not Vary with Study Type or Show Significant Cross-Correlations.
We calculated a variety of global properties of these matrices: number of hosts (H), number of phages (P), number of interactions (I), number of species (S = H + P), size (M = HP), connectance (C = I/M), mean number of interactions across host species (LH = I/H), and mean number of interactions across phage species (LP = I/P) (SI Appendix, Tables S1, S2, and S3 show values of each property within each of the 38 studies). Importantly, on a per-study basis, we find that the average number of phages infecting a given host is 4.88 (median = 3.04), whereas the average number of hosts that a phage can infect is 10.91 (median = 6.13). Both results are inconsistent with the hypothesis that phages only infect one host and that hosts are only infected by one phage (Fig. 1A).
We first sought to establish whether the source type (natural communities taken directly from the environment and then cultured, coevolutionary laboratory experiments where a single bacterial clone and a single phage clone were allowed to coevolve for a fixed amount of time and then, their evolved progenitors examined, and laboratory stocks of phages and hosts that were artificially combined) had any influence on basic characteristics of the matrices. We performed a principal component analysis (SI Appendix, SI Materials and Methods, SI Appendix, Table S4, and SI Appendix, Fig. S1) using these eight global properties. Despite the significant variation in global properties, we find no statistically significant distinction between the three different types of studies. For example, the distributions of type-specific matrices do not cluster into three groups. We apply a Jaccard clustering validity index (65) and find that the degree of clustering validity is 0.26 (indicating poor separation of labeled classes into distinct clusters), which is not significantly different from random (P = 0.33) (SI Appendix, SI Materials and Methods and SI Appendix, Figs. S3 and S4).
Not only do we not find evidence for clustering, we also do not find evidence for significant and biologically meaningful correlations among the global properties of all matrices when grouped together. For example, previous work on the analysis of bipartite networks within plant and pollinator systems found inverse relationships between the total number of species in the network and the fraction of interactions that actually occurred (66, 67). We do not find this relationship here. SI Appendix, Fig. S2 plots connectance (C) vs. number of species (S). The observed slope is small and nonsignificant (SI Appendix, Table S5). Moreover, the other correlations between connectance and the size of host–phage infection matrices are not significant (Materials and Methods has details and SI Appendix, Table S5 shows the correlation values).
Host–Phage Infection Assays Are Typically Nested and Not Modular.
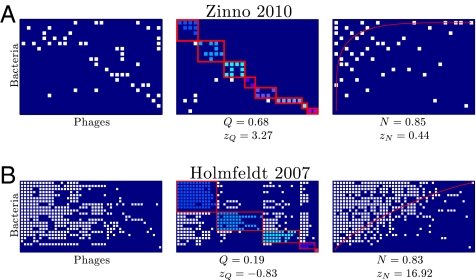
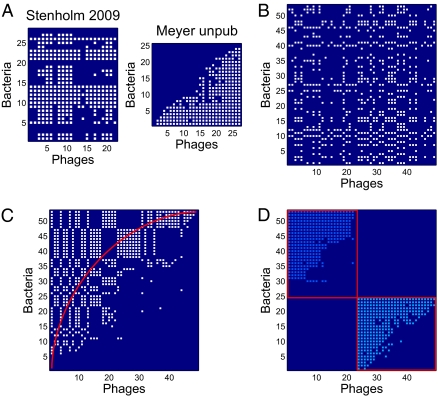
We measured higher-order properties of the host–phage interaction matrices, specifically modularity and nestedness. In this context, modularity is determined by the occurrence of groups of phages that infect groups of hosts significantly more often than they infect other hosts in the system. Modularity is typically found in biological systems in which groups of organisms preferentially interact with organisms within the group (e.g., plant–pollinator network) (66, 67) and is thought to be an important feature underlying the maintenance of biodiversity (68). Likewise, nestedness is determined by the extent to which phages that infect the most hosts tend to infect bacteria that are infected by the fewest phages (69, 70). Nestedness has been used to characterize species interactions because it is predicted to affect important properties of communities such as stability and extinction potential (67, 71). Both modularity and nestedness may emerge because of coevolutionary adaptation of hosts and phages (28, 72). The individual host–phage infection studies collected here were not subjected to a network analysis with one exception (27). Hence, we examined each study to see if previously unrealized patterns existed within each host–phage interaction network (Fig. 3 and SI Appendix, Fig. S5 have an example of how network properties are extracted from two matrices, Datasets S1 and S2 shows data corresponding to each matrix, and Materials and Methods has additional details on how to calculate modularity and nestedness).
Fig. 3.
Two example matrices were resorted to maximize modularity and nestedness. (A and B) The matrix in Left is the original data, the matrix in Center is the output from the modularity algorithm (102), and the matrix in Right is the output from the modified nestedness algorithm (103, 104). Colors represent different communities within the maximal modular configuration. (A) An example of a matrix with significantly elevated modularity and insignificant nestedness. (B) An example of a matrix with insignificant modularity and significantly elevated nestedness.
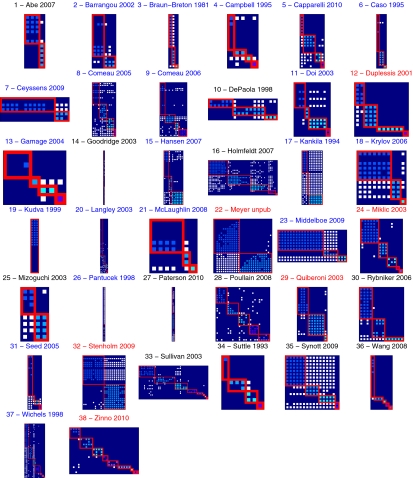
For the 38 matrices shown in Fig. 2, the maximally modular relabeling of each matrix is displayed in Fig. 4 and the maximally nested resorting of each matrix is displayed in Fig. 5. To evaluate the statistical significance of the modularity and nestedness values of observed host–phage matrices, we have to compare the observed values to those values of random matrices. We generate random matrices that have the same size and number of interactions as the original data (SI Appendix, SI Materials and Methods). In that way, we constrain our null model to have exactly the same global properties as detailed in SI Appendix, Table S1 for each study, whereas the nestedness and modularity will vary between realizations.
Fig. 4.
Modularity sorts of the collected studies. Blue labels (20/38) represent studies statistically antimodular, and red labels (6/38) represent studies statistically modular.
Fig. 5.
Nestedness sorts of the collected studies. Red line represents the isocline. Blue labels (0/38) represent studies statistically antinested, and red labels (27/38) represent studies statistically nested.
The titles of the study in Fig. 4 (the maximally modular configuration) are red if they are significantly modular, blue if they are significantly antimodular, and black if they are nonsignificantly modular. The majority of studies are significantly antimodular (where we used a P value = 0.05 and 105 random matrices as our null). Our findings stand in contrast to expectations that groups of phages adsorb to nonoverlapping groups of hosts, which would be expected if groups of phages had specialized on groups of hosts within the study systems. The titles of each study in Fig. 5 (the maximally nested configuration) are red if they are significantly nested, blue if they are significantly antinested, and black if they are nonsignificantly nested. The majority of studies are significantly nested (P < 0.05), where we used 105 random matrices as our null. Overall, we find 27 of 38 studies to be significantly nested, and when broken down by type, we find significant nestedness in 13 of 19 ecological, 7 of 10 experimental, and 7 of 9 artificial studies. Our findings corroborate, in one case, an earlier effort to characterize nestedness by Poullain et al. (27) using a different nestedness metric. It is also apparent that some matrices are almost perfectly nested [e.g., matrices in the works of Ceyssens et al. (35), McLaughlin and King (49), and Seed and Dennis (57)]. In some cases, like the work of Middelboe et al. (50), the data came from a mix of ecological and experimental studies in that the bacteria were derived from environmental and experimentally evolved isolates, whereas the phages were wild from the same environment as the host. Does the finding of a strongly nested matrix mean, in this case, that in vitro evolution mimics selection in nature, suggesting that there exists robust principles underlying the emergence of nestedness?
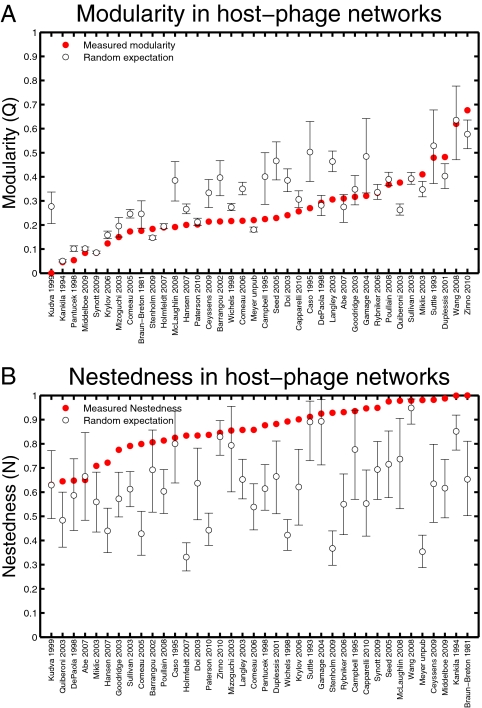
Hence, given the number of studies, we ask what evidence is there that host–phage matrices are, as a whole, nested and not modular. We rank all 38 matrices from lowest to largest modularity and lowest to largest nestedness (Fig. 6 A and B). It is evident that matrices tend to be more nested than their random counterparts but not more modular (and apparently, antimodular) than their random counterparts. How often do we expect to find 27 significantly nested matrices in a sample of 38 random matrices if each of the significantly nested matrices has a P < 0.05? Combinatorically, such a result is highly improbable and given by a binomial distribution with resulting P << 10−10. Likewise, the finding of an excess of antimodular matrices (20 of 38) compared with a small number of modular matrices (6 of 38) is a highly improbable result. Moreover, most of the significantly modular matrices have low values of modularity, suggesting that, although modularity may be deemed significant in a few cases, it is not a driving mechanism underlying the structure of most of these matrices and may be incidental to other patterns. Together, these results imply that currently available host–phage infection networks are typically nested and not modular.
Fig. 6.
Statistical distribution of modularity and nestedness for random matrices compared with that of the original data. (A) Sorted comparison of modularity of the collected studies vs. random networks. (B) Sorted comparison of nestedness of the collected studies vs. random networks. In both cases, error bars denote 95% confidence intervals based on 105 randomizations.
Previously Overlooked Nested Patterns Uncovered.
An additional power of subjecting host–phage infection networks to a unified analysis is that, by doing so, we can extract meaningful biological information about the organization of a system that may not have been possible given the original placement of hosts and phages in matrix format. For example, the work by Zinno et al. (64) mentions variability in phage infection; however, Zinno et al. (64) make no mention of the fact that there are evidently groups of phages that preferentially infect groups of hosts (Fig. 3A). Such block-like variability suggests that resistance mechanisms are less haphazard than they seem when network characteristics are not analyzed. Similarly, the work by Holmfeldt et al. (44) highlighted the variability and possibly unique signature of infection for each host and phage. However, reordering hosts according to the number of infecting phages while also reordering phages based on the number of hosts that they can infect leads to a nested pattern, suggesting that specific forms of infection rules may underlie infection variability (Fig. 3B). To what extent is our finding of nestedness novel? As a reminder, nestedness is a property of a host–phage infection matrix as calculated for a given row and column ordering. Hence, we calculated nestedness for all of the matrices in the format as they were first reported in the literature and then compared these results to the nestedness calculated from our reshuffled matrices. We found that, in 35 of 37 cases of the previously published studies, the reshuffled matrix had a nestedness value higher than that of the original publication, whereas in 2 of 37 studies, the nestedness was equal (47, 50) (SI Appendix, Fig. S6). Hence, our results suggest that, by and large, prior efforts did not identify the extent to which their matrices were nested or whether such nestedness was significant.
Addressing Sample Composition Biases as Potential Drivers of Network Structure.
We report a set of analyses to quantify the extent to which potential biases might impact our results. One potential bias in our study derives from the methods some researchers used for phage isolation. Phages require a bacterial host to reproduce, and therefore, the bacterial host(s) chosen by the researcher can affect the form of the interaction matrix. For instance, if researchers used a single host to isolate phages and included this host in the matrix, then their matrix will necessarily possess a full row of positive infections, thereby introducing the first element of a perfectly nested matrix. We found only six studies that used such an approach (46, 47, 49, 50, 56, 58). To determine if phage isolation strategy biased our results to nestedness, we reanalyzed all six of these matrices after removing the isolation host(s). We found no significant difference between the nestedness and modularity for each of these six matrices with or without the excluded host (SI Appendix, Table S6).
Another potential bias is that studies included zero rows and columns, which implies that there are hosts that no phages infect and phages that do not infect hosts, respectively. Note that inclusion of zero rows and columns has the potential to bias the structure to a nested pattern. However, such zero rows and columns may be biologically meaningful if hosts or phages have evolved resistance that leads to noninteraction between particular sets of strains. Nonetheless, we performed the entire analysis again by generating alternative matrices such that hosts and phages were only included if they had had at least one nonzero element in their row or column, respectively. Then, we recalculated nestedness for the modified matrices and compared it to the nestedness of appropriately resized null matrices. We found that 26 of 38 studies were nested compared with 27 of 38 using the original analysis (SI Appendix, Fig. S7). Moreover, although the quantitative value of nestedness did decrease in one case, that particular study (39) was, in fact, still highly nested and marginally significant at a P = 0.067 level. We also recalculated modularity for the modified matrices and found that 9 of 38 are modular compared with 6 of 38 in the original analysis (SI Appendix, Fig. S8). Hence, although there are minor changes in the number of significantly nested and modular networks, our finding that matrices have a characteristic nested structure is robust to either of these sources of bias.
Finally, we ask whether there are certain characteristics of matrices that defy the general pattern of nestedness and if it is possible to learn from these outliers? Interestingly, the three matrices with the most significant modular structures (40, 55, 64) were determined for a single bacterial species, Streptococcus thermophilus, and its phages. This finding seems robust, because different laboratories performed the studies and the microbes were isolated from three separate continents. Additionally, we did not find an example where a matrix that included S. thermophilus did not have the modular structure. We examined bacteria from the same taxonomic order (Lactobacillales) and isolated from the same environment (dairy products), but these bacteria lacked a modular structure. The consistent modularity observed for this species suggests that species-specific traits may have strong deterministic effects on the form that their interactions with parasites take. We are unsure of which traits produce the modular interactions; however, additional research may help reveal if and what resistance mechanisms determine the shape of microbial interaction networks.
Possible Scale Dependence of Host–Phage Interactions: From Nestedness to Modularity?
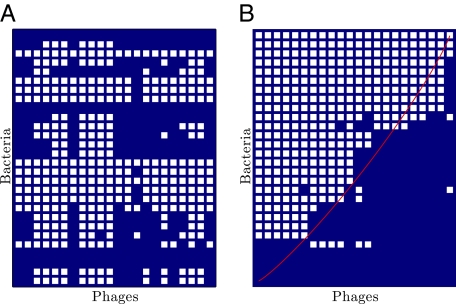
The data that we analyzed included almost 12,000 separate attempts to infect a host isolate with a phage isolate. Although the scale of the current data is beyond the scope of any individual project, it still pales compared with the number of possible interactions in a community at local or regional levels. Scaling up to larger assays presents technical challenges aside from increasing the depth of sampling. Studying many host strains beyond the species (or genus) level often requires distinct culture conditions, a prerequisite for studies that many laboratories cannot or do not want to reach. Here, we present an analysis of what such a hypothesized study may reveal. Consider an experiment in which the hosts from two groups of experiments were combined in a large cross-infection assay with the phages from the same two groups of experiments. If the original matrix sizes were H1 • P1 and H2 • P2, then the final matrix size is (H1 + H2) • (P1 + P2). A total of H1P2 + H2P1 new experiments would need to be performed. If the hosts were of sufficiently distant types (e.g., E. coli and Synecoccocus), we should expect that nearly all of the new cross-infection experiments would lead to no additional infections. Hence, if the original matrices were nested, then the new matrix would have two modules, each of which was nested (Fig. 7 has the results of such a numerical experiment). In other words, we predict that, at larger, possibly macroevolutionary scales, host–phage interaction matrices should be typified by a modular structure, even if there is nested structure at smaller scales.
Fig. 7.
Union of two nested matrices indicates possible host–phage interaction structure at larger, possibly macroevolutionary scales. In this figure, we selected two of the most nested studies and performed a union while presuming that there were no cross-infections of hosts by phages of the other study. In this case, E. coli and cyanobacteria were the host types. (A) Depiction of the original matrices. (B) Randomization of the union matrix. (C) Nested sort of the union matrix. (D) Modularity sort of the union matrix with a nested sort of each module.
Discussion
Summary of Major Results.
We have established a unified approach to analyzing host–phage infection matrices. In so doing, we find that a compilation of 38 empirical studies of host–phage interaction networks is nested on average and not modular (Figs. 4 and 5). In most cases, our finding of higher-order structure such as nestedness within an individual study was not previously observed, in that prior analyses of host–phage interaction matrices usually did not attempt to estimate the network characteristics examined here. We found that host–phage interaction networks are not perfectly nested and that interactions that defy perfect nestedness are typical throughout nearly all of the data. Additionally, we found no significant difference in nestedness or modularity based on taxa, sources, or isolation method. This dataset, although far larger than any individual study, is limited to (largely) microevolutionary scales, an issue that we addressed in Results and will return to later in Discussion. Considering the large range of taxa, habitats, and sampling techniques used to construct the matrices, the repeated sampling of a nested pattern of host–phage infections is salient, although the process driving the nestedness is not obvious. It could result from multiple mechanisms or a single principle. Here, we examine three hypotheses to explain the nestedness pattern based on biochemical, ecological, and evolutionary principles. Note that these hypotheses are not mutually exclusive and that we have only limited ability to test them given our comparative approach. However, each of these hypotheses can be tested with additional laboratory-based or field experiments.
Mechanisms Responsible for Nestedness: Biophysical, Ecological, and Evolutionary.
Phage and bacterial infection matrices at microevolutionary scales may be constrained to a nested shape by the nature of their molecular interactions. Phages infect bacteria by using specialized proteins that target and bind to molecules on the outer membranes of bacteria (receptor molecules). Nested infection matrices have been shown for T-phages, which infect strains of E. coli, to be the result of the interactions of the phage proteins and receptor molecules (73). T-phages bind to the lipopolysaccharide (LPS) chains on the cell surface. Mutant E. coli has been observed with shortened LPS chains that confer resistance to some but not all T-phages. There are T-phages that are able to infect these mutants, because they require fewer segments of the LPS molecule to bind. If phage–bacterial molecular interactions are dominated by single traits and variation in these traits is constrained along a single hierarchical dimension such as LPS, then one should expect the nested pattern to arise. There are other examples of traits with physical characteristics that behave similarly: bacteria that evolve a thicker and thicker protective coating (74), phages that evolve increased host range by continually reducing tail length (73), bacteria that reduce their number of receptors, and phages that target fewer receptors (75). Although there are many examples of this type of one-dimensional interaction, the problem with this finding being a universal explanation for the form of bacterial–phage interactions are that host–phage interactions are governed by hundreds of other genes (76), bacteria can use multiple strategies for resistance (74), and phages have complex mechanism to evade bacteria defenses (74, 77). Moreover, a recent discovery of an adaptive immune system, where bacteria acquire targeted sequences to prevent phage infection and phages evolve to evade such immunity, suggests a complex interaction space (78). Given the diversity of host–phage interactions, it seems unlikely that the molecular details alone would constrain the form of their relationship (79). Instead, we turn to the potential guiding forces of community assembly and coevolution to explain this reoccurring pattern.
The nested pattern may be common, because the processes of microbial community assembly select for species with nested relationships. One could imagine that communities may settle into this pattern if this interaction structure is more stable than others (67, 71), noting that the stability of host–phage interaction structures may depend on ecological factors such as resource availability (80). Cohesive interaction structures such as nested patterns have been shown to be more stable than other structures for mutualistic networks (81, 82). The regularity of the interactions and redundancies make these communities less susceptible to the random removal of nodes. However, these networks are thought to be susceptible to invasion by new species that violate the nested pattern, suggesting that migration of a species would perturb the nestedness. Furthermore, the spatiotemporal complexity of microbial and viral communities suggests that prior theoretical efforts that consider community addition as a process in which invasions occur infrequently may not be widely applicable. Moreover, community assembly models rarely invoke the influence of evolutionary change at similar time scales as ecological change—an issue highly relevant to the study of microbial and viral communities.
Indeed, there may be an evolutionary explanation for nestedness. Most attempts to characterize the form of coevolution with host–phage experiments to date have shown a form of antagonistic evolution called expanded host range (or gene for gene) coevolution (52, 83, 84). Under this model, bacteria evolve ever-increasing resistance to more and more phage genotypes, and phages evolve broader host ranges. If one were to sample a community of bacteria and phages coevolving under this model, they would uncover a diversity of phages and bacteria that exhibit a nested interaction pattern. At any time point, the most-derived bacteria should exist, which is either completely resistant or depending on the timing, sensitive to the most-derived phage. Given that selection by phage may be slow to alleviate the more sensitive ancestral variants or that there may be a tradeoff between resistance and competitiveness, there will exist a diversity of bacteria with ever decreasing sets of phages to which they are resistant. Similarly, the most-derived phages will have the broadest host range, and by the same logic as for the bacteria, its ancestors are likely to persist in the community and display ever-decreasing host ranges. The nested pattern could be a product of taking a snapshot of a dynamically evolving community. Although the majority of experimental results observed in artificial laboratory settings support this hypothesis, there is a single laboratory experiment (85) and models of bacterial host–parasite coevolution that suggest that other forms of coevolution are possible when there are bottom-up costs for modifications to resistance (86, 87). Furthermore, if coevolution provided the only explanation, then the artificially assembled matrices would not have the nested pattern.
Dispelling and Recognizing Potential Biases.
Three sources of sampling bias challenge the generality of our findings. First, the taxa sampled may poorly represent microbial diversity given that they are subject to both human and methodological biases. If, for instance, only taxa associated with humans were selected or all taxa were cultured similarly, then our results would only be relevant for a small group of microbes. Indeed, the majority of microbial studies were performed on the family Enterobacteriaceae, which lives within human digestive systems; however, the spectrum of bacteria that we examined is much broader and includes both heterotrophic and photosynthetic species. Further, gram-negative and -positive bacteria examined here were isolated from six continents and many disparate environments from the extreme conditions of hot springs, the rich resource conditions of sewage, depauperate marine environments, and the complex matrix of soil to the simplified laboratory environment. Although this study cannot feasibly test the full microbial diversity of the globe, it does include examples from much of it (SI Appendix, Tables S1 and S2).
Second, as previously discussed, the number of hosts used to isolate phages and the inclusion of noninteracting hosts and phages have the potential to alter the nestedness of a matrix. Ideally, the same number of hosts studied in the matrix would be used to isolate phages, or if only a subset of hosts was used, then these hosts would not be included in the matrix. This finding is important to ensure that the pattern of infection is independent of how the parasites were isolated. We found that these biases were not a problem by (i) testing matrices that were created by isolating phages on a single host and (ii) removing hosts and phages that were not interacting. We found that whether the matrices were significantly nested was not affected by including the isolation host in the matrix or by removing noninteracting hosts and phages, which is strong support that the isolation method did not enrich for nestedness.
The last category of bias, phylogenetic, is likely to mean that our results define a pattern at relatively narrow taxonomic scales. The majority of our studies was of closely related genotypes and species. As described in Results, we anticipate that more complex patterns of infection may form at larger phylogenetic scales that likely include increasing compartmentalization. Hence, we hypothesize that a multiscale view of host–phage infection networks will reveal nestedness at small scales and modularity at large scales. Our finding of nested interaction matrices is still relevant for characterizing patterns at short phylogenetic distances; they are, arguably, the most relevant for many ecological and evolutionary scenarios, because they likely share the richest connections.
Prospective View.
Whatever the limitations of this dataset, it is important to point out that viewing host–phage interaction networks through a unifying lens will likely unveil other commonalities of microbial and viral communities. By way of analogy, over 25 y ago, the study of food webs was radically altered by the compilation of many small food webs that were subject to a unified analysis (88–91). The key finding of the earliest food web studies was that the members of a community could be ranked, and that larger species would eat a random fraction of those species smaller than them. From this stage, there were two ways forward. First, by studying larger food webs, the original pattern was refined such that species ranking was found to be correlated with body size (but not equivalent to body size); therefore, individuals eat prey that are smaller, although they are a part of a well-defined size class (92, 93). Second, the topology of food webs was then used as a target and basis for dynamic models of community behavior (i.e., what mechanisms can explain the patterns and how do the patterns influence community function) (94). We hope and envision that a similar process unfolds here in that the finding of a general pattern in the current dataset will stimulate the collection of more and larger host–phage infection networks to continue to provide a fuller picture of who infects whom across an entire community. In so doing, we caution that data completeness can alter the observed patterns of connectivity and refer readers to a number of recent papers that address this topic (95–99).
What do we expect to find when analyzing ever larger host–phage interaction networks collected from an ecological community, evolution experiments, or culture collections? We hypothesize that host–phage interaction matrices are likely characterized by modularity at larger taxonomic scales even if there is structure (e.g., nestedness) at small taxonomic scales (Fig. 7). What would such a multiscale phenomenon inform us about the structure and function of microbiological communities? First, it would suggest the existence of diversifying coevolutionary-induced selection that gave rise to (largely) independent host–phage communities. The molecular basis of such diversification could then be explored. Second, cross-infection assays or similar laboratory-based strategies (100) that test whether phages can infect or at least transmit their genes between phylogenetically divergent hosts have the potential to provide significant advances in understanding patterns of global gene transfer. Such phages (and the bacteria that they infect) may be critical to understanding the direct transfer of genes on a global scale. Instead of phages acting locally (in a taxonomic sense) to shuttle genes between closely related bacteria, a few rare links would permit greater cross-talk between bacterial taxa. Quantifying the frequency of such events may represent the small-world links that connect distant microbial populations (101), and it is in need of experimental testing.
Furthermore, infections of distantly related groups by the same phages would imply that the bacteria are in indirect competition with one another, even if they do not seem to compete directly for the same set of carbon and nutrient sources. Although whole genome-based approaches to infer host range and phage susceptibility may help provide candidates for such rare links, they are not the only solution. Rather, we suggest that the continued use of laboratory-based assays to catalog the life history traits of culturable host–phage pairs is essential if we are to improve our understanding of the population dynamics of host–phage communities in the wild. Of course, many (if not most) bacteria and phages are not currently culturable. Hence, in parallel, we recommend attention be given to the development of inverse methods to catalog the life history traits of phages based on community infection assays in those circumstances in which culturing is impossible or yet intractable.
Materials and Methods
Network Statistics.
Modularity is estimated by reshuffling the rows and columns of the matrix to find groupings of highly interconnected phages and bacteria, labeling these groups and assessing matrix-wide the ratio of the number of within to outside group connections. This calculation is done using a heuristic called the BRIM algorithm (102) to efficiently find the configuration that maximizes this ratio. We ported the BRIM algorithm to MATLAB from the original code in Octave and used the adaptive BRIM algorithm for all calculations here. By this definition, a perfectly modular matrix is comprised of clusters of completely isolated groups, and modularity declines as the number of cross-group connections increases. Nestedness is estimated by reordering the rows and columns (103, 104) to determine whether phages that infect fewer hosts are only able to infect a subset of bacteria that are susceptible to many phages. This reordering tries to maximize the position of ones in the matrix such that they clusters above a nullcline (Fig. 1C shows a perfectly nested matrix). The value for nestedness depends on how frequently ones fall above rather than below this nullcline. Complete details are provided in SI Appendix, SI Materials and Methods.
Host–Phage Infection Assay.
Matrix 22 is the only dataset not previously published. We constructed the matrix by coevolving an obligately lytic phage-λ strain with its host E. coli. The E. coli studied was of strain REL606, a derivative of E. coli B acquired from Richard Lenski (Michigan State University, Lansing, MI) and described in ref. 105, and phages were of strain cI21 (λvir) provided by Donald Court (National Cancer Institute, Frederick, MD). The phages and bacteria were cocultured in 50-mL Erlenmeyer flasks with 10 mL liquid medium, shaken at 120 rpm, and incubated at 37 °C (New Brunswick Innova 4300 Incubator Shaker). This flask was incubated, and the cycle of transfer and incubation was continued one more time. Three 24-h incubations were long enough for the bacteria to evolve resistance and the phages to counter it; however, it was not long enough for a second round of coevolution. We randomly selected 150 bacteria and 150 phage isolates. We determined which of the 150 bacteria isolates were resistant to the 150 phage isolates. To do this task, we performed spot plate assays. All bacterial–phage combinations were replicated five separate times, and a total of 28,125 spots were assayed. To make this processes more efficient, we placed up to 96 separate phage stocks onto a single dish (150 mm radius). Phage stock replicates were never placed on the same plate to reduce the signal of any stochastic plating effects. The five replicates were combined, and a phage was only determined to be able to infect a bacterium if three of five replicates were given ones. Lastly, phages or bacteria that had identical infection or resistance profiles as their ancestors were removed from the matrix. Complete details are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank the anonymous reviewers for comments and suggestions that improved the manuscript. J.S.W. acknowledges the support of the James S. McDonnell Foundation and Defense Advanced Projects Research Agency Grant HR0011-09-1-0055. J.S.W. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Author Summary on page 11309.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101595108/-/DCSupplemental.
References
- 1.Edwards RA, Rohwer F. Viral metagenomics. Nat Rev Microbiol. 2005;3:504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- 2.Torsvik V, Øvreås L, Thingstad TF. Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science. 2002;296:1064–1066. doi: 10.1126/science.1071698. [DOI] [PubMed] [Google Scholar]
- 3.Fraser C, Alm EJ, Polz MF, Spratt BG, Hanage WP. The bacterial species challenge: Making sense of genetic and ecological diversity. Science. 2009;323:741–746. doi: 10.1126/science.1159388. [DOI] [PubMed] [Google Scholar]
- 4.Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 5.Huse SM, et al. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angly FE, et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yooseph S, et al. The Sorcerer II Global Ocean Sampling expedition: Expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox-Foster DLL, et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318:283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- 9.Tyson GW, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 10.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 12.Levin BR, Bull JJ. Population and evolutionary dynamics of phage therapy. Nat Rev Microbiol. 2004;2:166–173. doi: 10.1038/nrmicro822. [DOI] [PubMed] [Google Scholar]
- 13.Rohwer F, Thurber RV. Viruses manipulate the marine environment. Nature. 2009;459:207–212. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- 14.Faruque SM, et al. Self-limiting nature of seasonal cholera epidemics: Role of host-mediated amplification of phage. Proc Natl Acad Sci USA. 2005;102:6119–6124. doi: 10.1073/pnas.0502069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faruque SM, et al. Seasonal epidemics of cholera inversely correlate with the prevalence of environmental cholera phages. Proc Natl Acad Sci USA. 2005;102:1702–1707. doi: 10.1073/pnas.0408992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodridge L, Abedon ST. Bacteriophage biocontrol and bioprocessing: Application of phage therapy to industry. SIM News. 2003;53:254–262. [Google Scholar]
- 17.Weinbauer MG. Ecology of prokaryotic viruses. FEMS Microbiol Rev. 2004;28:127–181. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Wommack KE, Colwell RR. Virioplankton: Viruses in aquatic ecosystems. Microbiol Mol Biol Rev. 2000;64:69–114. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abedon ST. Bacteriophage Ecology: Population Growth, Evolution and Impact of Bacterial Viruses. Cambridge, UK: Cambridge University Press; 2008. [Google Scholar]
- 20.Boyd EF, Davis BM, Hochhut B. Bacteriophage-bacteriophage interactions in the evolution of pathogenic bacteria. Trends Microbiol. 2001;9:137–144. doi: 10.1016/s0966-842x(01)01960-6. [DOI] [PubMed] [Google Scholar]
- 21.Silander OK, et al. Widespread genetic exchange among terrestrial bacteriophages. Proc Natl Acad Sci USA. 2005;102:19009–19014. doi: 10.1073/pnas.0503074102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sano E, Carlson S, Wegley L, Rohwer F. Movement of viruses between biomes. Appl Environ Microbiol. 2004;70:5842–5846. doi: 10.1128/AEM.70.10.5842-5846.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuhrman JA, Noble RT. Viruses and protists cause similar bacterial mortality in coastal seawater. Limnol Oceanogr. 1995;40:1236–1242. [Google Scholar]
- 24.Gobler CJ, Hutchins DA, Fisher NS, Cosper EM, Sañudo-Wilhelmy SA. Release and bioavailability of C, N, P, Se, and Fe following viral lysis of a marine chrysophyte. Limnol Oceanogr. 1997;42:1492–1504. [Google Scholar]
- 25.Bohannan BJM, Lenski RE. Linking genetic change to community evolution: Insights from studies of bacteria and bacteriophage. Ecol Lett. 2000;3:362–377. [Google Scholar]
- 26.Thingstad TF. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol Oceanogr. 2000;45:1320–1328. [Google Scholar]
- 27.Poullain V, Gandon S, Brockhurst MA, Buckling A, Hochberg ME. The evolution of specificity in evolving and coevolving antagonistic interactions between a bacteria and its phage. Evolution. 2008;62:1–11. doi: 10.1111/j.1558-5646.2007.00260.x. [DOI] [PubMed] [Google Scholar]
- 28.Agrawal A, Lively CM. Infection genetics: Gene-for-gene versus matching-alleles models and all points in between. Evol Ecol Res. 2002;4:79–90. [Google Scholar]
- 29.Abe M, Izumoji Y, Tanji Y. Phenotypic transformation including host-range transition through superinfection of T-even phages. FEMS Microbiol Lett. 2007;269:145–152. doi: 10.1111/j.1574-6968.2006.00615.x. [DOI] [PubMed] [Google Scholar]
- 30.Barrangou R, Yoon SS, Breidt F, Jr., Fleming HP, Klaenhammer TR. Characterization of six Leuconostoc fallax bacteriophages isolated from an industrial sauerkraut fermentation. Appl Environ Microbiol. 2002;68:5452–5458. doi: 10.1128/AEM.68.11.5452-5458.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braun-Breton C, Hofnung M. In vivo and in vitro functional alterations of the bacteriophage lambda receptor in lamB missense mutants of Escherichia coli K-12. J Bacteriol. 1981;148:845–852. doi: 10.1128/jb.148.3.845-852.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell JIA, Albrechtsen M, Sørensen J. Large Pseudomonas phages isolated from barley rhizosphere. FEMS Microbiol Ecol. 2006;18:63–74. [Google Scholar]
- 33.Capparelli R, et al. Bacteriophage therapy of Salmonella enterica: A fresh appraisal of bacteriophage therapy. J Infect Dis. 2010;201:52–61. doi: 10.1086/648478. [DOI] [PubMed] [Google Scholar]
- 34.Caso JL, et al. Isolation and characterization of temperate and virulent bacteriophages of Lactobacillus plantarum. J Dairy Sci. 1995;78:741–750. [Google Scholar]
- 35.Ceyssens P-J, et al. Survey of Pseudomonas aeruginosa and its phages: De novo peptide sequencing as a novel tool to assess the diversity of worldwide collected viruses. Environ Microbiol. 2009;11:1303–1313. doi: 10.1111/j.1462-2920.2008.01862.x. [DOI] [PubMed] [Google Scholar]
- 36.Comeau AM, Buenaventura E, Suttle CA. A persistent, productive, and seasonally dynamic vibriophage population within Pacific oysters (Crassostrea gigas) Appl Environ Microbiol. 2005;71:5324–5331. doi: 10.1128/AEM.71.9.5324-5331.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Comeau AM, Chan AM, Suttle CA. Genetic richness of vibriophages isolated in a coastal environment. Environ Microbiol. 2006;8:1164–1176. doi: 10.1111/j.1462-2920.2006.01006.x. [DOI] [PubMed] [Google Scholar]
- 38.DePaola A, Motes ML, Chan AM, Suttle CA. Phages infecting Vibrio vulnificus are abundant and diverse in oysters (Crassostrea virginica) collected from the Gulf of Mexico. Appl Environ Microbiol. 1998;64:346–351. doi: 10.1128/aem.64.1.346-351.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doi K, et al. A comparative study and phage typing of silage-making Lactobacillus bacteriophages. J Biosci Bioeng. 2003;95:518–525. doi: 10.1016/s1389-1723(03)80054-3. [DOI] [PubMed] [Google Scholar]
- 40.Duplessis M, Moineau S. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol Microbiol. 2001;41:325–336. doi: 10.1046/j.1365-2958.2001.02521.x. [DOI] [PubMed] [Google Scholar]
- 41.Gamage SD, Patton AK, Hanson JF, Weiss AA. Diversity and host range of Shiga toxin-encoding phage. Infect Immun. 2004;72:7131–7139. doi: 10.1128/IAI.72.12.7131-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodridge L, Gallaccio A, Griffiths MW. Morphological, host range, and genetic characterization of two coliphages. Appl Environ Microbiol. 2003;69:5364–5371. doi: 10.1128/AEM.69.9.5364-5371.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hansen VM, Rosenquist H, Baggesen DL, Brown S, Christensen BB. Characterization of Campylobacter phages including analysis of host range by selected Campylobacter Penner serotypes. BMC Microbiol. 2007;7:90. doi: 10.1186/1471-2180-7-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holmfeldt K, Middelboe M, Nybroe O, Riemann L. Large variabilities in host strain susceptibility and phage host range govern interactions between lytic marine phages and their Flavobacterium hosts. Appl Environ Microbiol. 2007;73:6730–6739. doi: 10.1128/AEM.01399-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kankila J, Lindstrom K. Host range, morphology and DNA restriction patterns of bacteriophage isolates infecting Rhizobium leguminosarum bv. trifolii. Soil Biol Biochem. 1994;26:429–437. [Google Scholar]
- 46.Krylov VN, et al. Ambivalent bacteriophages of different species active on Escherichia coli K12 and Salmonella sp. strains. Russ J Genet. 2006;42:106–114. [PubMed] [Google Scholar]
- 47.Kudva IT, Jelacic S, Tarr PI, Youderian P, Hovde CJ. Biocontrol of Escherichia coli O157 with O157-specific bacteriophages. Appl Environ Microbiol. 1999;65:3767–3773. doi: 10.1128/aem.65.9.3767-3773.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langley R, Kenna DT, Vandamme P, Ure R, Govan JR. Lysogeny and bacteriophage host range within the Burkholderia cepacia complex. J Med Microbiol. 2003;52:483–490. doi: 10.1099/jmm.0.05099-0. [DOI] [PubMed] [Google Scholar]
- 49.McLaughlin MR, King RA. Characterization of Salmonella bacteriophages isolated from swine lagoon effluent. Curr Microbiol. 2008;56:208–213. doi: 10.1007/s00284-007-9057-9. [DOI] [PubMed] [Google Scholar]
- 50.Middelboe M, Holmfeldt K, Riemann L, Nybroe O, Haaber J. Bacteriophages drive strain diversification in a marine Flavobacterium: Implications for phage resistance and physiological properties. Environ Microbiol. 2009;11:1971–1982. doi: 10.1111/j.1462-2920.2009.01920.x. [DOI] [PubMed] [Google Scholar]
- 51.Miklič A, Rogelj I. Characterization of lactococcal bacteriophages isolated from Slovenian dairies. Int J Food Sci Technol. 2003;38:305–311. [Google Scholar]
- 52.Mizoguchi K, et al. Coevolution of bacteriophage PP01 and Escherichia coli O157:H7 in continuous culture. Appl Environ Microbiol. 2003;69:170–176. doi: 10.1128/AEM.69.1.170-176.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pantůček R, et al. The polyvalent staphylococcal phage ϕ 812: Its host-range mutants and related phages. Virology. 1998;246:241–252. doi: 10.1006/viro.1998.9203. [DOI] [PubMed] [Google Scholar]
- 54.Paterson S, et al. Antagonistic coevolution accelerates molecular evolution. Nature. 2010;464:275–278. doi: 10.1038/nature08798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quiberoni A, et al. Comparative analysis of Streptococcus thermophilus bacteriophages isolated from a yogurt industrial plant. Food Microbiol. 2003;20:461–469. [Google Scholar]
- 56.Rybniker J, Kramme S, Small PL. Host range of 14 mycobacteriophages in Mycobacterium ulcerans and seven other mycobacteria including Mycobacterium tuberculosis—application for identification and susceptibility testing. J Med Microbiol. 2006;55:37–42. doi: 10.1099/jmm.0.46238-0. [DOI] [PubMed] [Google Scholar]
- 57.Seed KD, Dennis JJ. Isolation and characterization of bacteriophages of the Burkholderia cepacia complex. FEMS Microbiol Lett. 2005;251:273–280. doi: 10.1016/j.femsle.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 58.Stenholm AR, Dalsgaard I, Middelboe M. Isolation and characterization of bacteriophages infecting the fish pathogen Flavobacterium psychrophilum. Appl Environ Microbiol. 2008;74:4070–4078. doi: 10.1128/AEM.00428-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sullivan MB, Waterbury JB, Chisholm SW. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature. 2003;424:1047–1051. doi: 10.1038/nature01929. [DOI] [PubMed] [Google Scholar]
- 60.Suttle CA, Chan A. Marine cyanophages infecting oceanic and coastal strains of Synechococcus: Abundance, morphology, cross-infectivity and growth characteristics. Mar Ecol Prog Ser. 1993;92:99–109. [Google Scholar]
- 61.Synnott AJ, et al. Isolation from sewage influent and characterization of novel Staphylococcus aureus bacteriophages with wide host ranges and potent lytic capabilities. Appl Environ Microbiol. 2009;75:4483–4490. doi: 10.1128/AEM.02641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang K, Chen F. Prevalence of highly host-specific cyanophages in the estuarine environment. Environ Microbiol. 2008;10:300–312. doi: 10.1111/j.1462-2920.2007.01452.x. [DOI] [PubMed] [Google Scholar]
- 63.Wichels A, et al. Bacteriophage diversity in the North Sea. Appl Environ Microbiol. 1998;64:4128–4133. doi: 10.1128/aem.64.11.4128-4133.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zinno P, Janzen T, Bennedsen M, Ercolini D, Mauriello G. Characterization of Streptococcus thermophilus lytic bacteriophages from mozzarella cheese plants. Int J Food Microbiol. 2010;138:137–144. doi: 10.1016/j.ijfoodmicro.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 65.Jaccard P. The distribution of flora in the alpine zone. New Phytol. 1912;11:37–50. [Google Scholar]
- 66.Memmott J. The structure of a plant-pollinator food web. Ecol Lett. 1999;2:276–280. doi: 10.1046/j.1461-0248.1999.00087.x. [DOI] [PubMed] [Google Scholar]
- 67.Bascompte J, Jordano P, Melián CJ, Olesen JM. The nested assembly of plant-animal mutualistic networks. Proc Natl Acad Sci USA. 2003;100:9383–9387. doi: 10.1073/pnas.1633576100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thompson JN. The Geographic Mosaic of Coevolution. Chicago: University of Chicago Press; 2005. [Google Scholar]
- 69.Ulrich W, Gotelli NJ. Null model analysis of species nestedness patterns. Ecology. 2007;88:1824–1831. doi: 10.1890/06-1208.1. [DOI] [PubMed] [Google Scholar]
- 70.Almeida-Neto M, Guimarães PR, Lewinsohn TM. On nestedness analyses: Rethinking matrix temperature and anti-nestedness. Oikos. 2007;116:716–722. [Google Scholar]
- 71.Bastolla U, et al. The architecture of mutualistic networks minimizes competition and increases biodiversity. Nature. 2009;458:1018–1020. doi: 10.1038/nature07950. [DOI] [PubMed] [Google Scholar]
- 72.Sasaki A. Host-parasite coevolution in a multilocus gene-for-gene system. Proc Biol Sci. 2000;267:2183–2188. doi: 10.1098/rspb.2000.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Forde SE, et al. Understanding the limits to generalizability of experimental evolutionary models. Nature. 2008;455:220–223. doi: 10.1038/nature07152. [DOI] [PubMed] [Google Scholar]
- 74.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 75.Hyman P, Abedon ST. Bacteriophage host range and bacterial resistance. In: Laskin AI, Sariaslani S, Gadd GM, editors. Advanced Applied Microbiology. Vol. 70. San Diego: Academic Press; 2010. pp. 217–248. [DOI] [PubMed] [Google Scholar]
- 76.Maynard ND, et al. A forward-genetic screen and dynamic analysis of lambda phage host-dependencies reveals an extensive interaction network and a new anti-viral strategy. PLoS Genet. 2010;6:e1001017. doi: 10.1371/journal.pgen.1001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 78.Karginov FV, Hannon GJ. The CRISPR system: Small RNA-guided defense in bacteria and archaea. Mol Cell. 2010;37:7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gudelj I, et al. An integrative approach to understanding microbial diversity: From intracellular mechanisms to community structure. Ecol Lett. 2010;13:1073–1084. doi: 10.1111/j.1461-0248.2010.01507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Poisot T, Lepennetier G, Martinez E, Ramsayer J, Hochberg ME. Resource availability affects the structure of a natural bacteria-bacteriophage community. Biol Lett. 2011;7:201–204. doi: 10.1098/rsbl.2010.0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bascompte J, Jordano P. The structure of plant-animal mutualistic networks. In: Pascual M, Dunne J, editors. Ecological Networks: Linking Structure to Dynamics in Food Webs. Oxford: Oxford University Press; 2006. pp. 143–159. [Google Scholar]
- 82.Bascompte J. Disentangling the web of life. Science. 2009;325:416–419. doi: 10.1126/science.1170749. [DOI] [PubMed] [Google Scholar]
- 83.Lenski RE, Levin BR. Constraints on the coevolution of bacteria and virulent phage: A model, some experiments, and predictions for natural communities. Am Nat. 1985;125:585–602. [Google Scholar]
- 84.Buckling A, Rainey PB. Antagonistic coevolution between a bacterium and a bacteriophage. Proc Biol Sci. 2002;269:931–936. doi: 10.1098/rspb.2001.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gómez P, Buckling A. Bacteria-phage antagonistic coevolution in soil. Science. 2011;332:106–109. doi: 10.1126/science.1198767. [DOI] [PubMed] [Google Scholar]
- 86.Weitz JS, Hartman H, Levin SA. Coevolutionary arms races between bacteria and bacteriophage. Proc Natl Acad Sci USA. 2005;102:9535–9540. doi: 10.1073/pnas.0504062102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Spanakis E, Horne MT. Co-adaptation of Escherichia coli and coliphage λ vir in continuous culture. J Gen Microbiol. 1987;133:353–360. doi: 10.1099/00221287-133-2-353. [DOI] [PubMed] [Google Scholar]
- 88.Cohen JE, Newman CM. A stochastic theory of community food webs I. Models and aggregated data. Proc R Soc Lond B Biol Sci. 1985;224:421–448. [Google Scholar]
- 89.Cohen JE, Newman CM, Briand F. A stochastic theory of community food webs II. Individual webs. Proc R Soc Lond B Biol Sci. 1985;224:449–461. [Google Scholar]
- 90.Pimm SL, Lawton JH, Cohen JE. Food web patterns and their consequences. Nature. 1991;350:669–674. [Google Scholar]
- 91.Cohen JE, Briand F, Newman CM. Community Food Webs: Data and Theory. Berlin: Springer; 1990. [Google Scholar]
- 92.Allesina S, Alonso D, Pascual M. A general model for food web structure. Science. 2008;320:658–661. doi: 10.1126/science.1156269. [DOI] [PubMed] [Google Scholar]
- 93.Williams RJ, Martinez ND. Simple rules yield complex food webs. Nature. 2000;404:180–183. doi: 10.1038/35004572. [DOI] [PubMed] [Google Scholar]
- 94.Pascual M, Dunne JA, editors. Ecological Networks: Linking Structure to Dynamics in Food Webs. New York: Oxford University Press; 2005. [Google Scholar]
- 95.Mestres J, Gregori-Puigjané E, Valverde S, Solé RV. Data completeness—the Achilles heel of drug-target networks. Nat Biotechnol. 2008;26:983–984. doi: 10.1038/nbt0908-983. [DOI] [PubMed] [Google Scholar]
- 96.Olesen JM, et al. From Broadstone to Zackenberg: Space, time and hierarchies in ecological networks. Adv Ecol Res. 2010;42:1–69. [Google Scholar]
- 97.Waser NM, Ollerton J. Plant-Pollinator Interactions: From Specialization to Generalization. Chicago: University of Chicago; 2006. [Google Scholar]
- 98.Fortunato S, Barthélemy M. Resolution limit in community detection. Proc Natl Acad Sci USA. 2007;104:36–41. doi: 10.1073/pnas.0605965104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Genini J, Morellato LPC, Guimarães PR, Jr., Olesen JM. Cheaters in mutualism networks. Biol Lett. 2010;6:494–497. doi: 10.1098/rsbl.2009.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chiura HX. Generalized gene transfer by virus-like particles from marine bacteria. Aquat Microb Ecol. 1997;13:75–83. [Google Scholar]
- 101.Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 102.Barber MJ. Modularity and community detection in bipartite networks. Phys Rev E. 2007;76:066102. doi: 10.1103/PhysRevE.76.066102. [DOI] [PubMed] [Google Scholar]
- 103.Atmar W, Patterson BD. The measure of order and disorder in the distribution of species in fragmented habitat. Oecologia. 1993;96:373–382. doi: 10.1007/BF00317508. [DOI] [PubMed] [Google Scholar]
- 104.Rodriguez-Girones MA, Santamaria L. A new algorithm to calculate the nestedness temperature of presence-absence matrices. J Biogeogr. 2006;33:924–935. [Google Scholar]
- 105.Daegelen P, Studier FW, Lenski RE, Cure S, Kim JF. Tracing ancestors and relatives of Escherichia coli B, and the derivation of B strains REL606 and BL21(DE3) J Mol Biol. 2009;394:634–643. doi: 10.1016/j.jmb.2009.09.022. [DOI] [PubMed] [Google Scholar]