Abstract
The analysis of protein interactors in protein complexes can yield important insight into protein function and signal transduction. Thus, a reliable approach to distinguish true interactors from non-specific interacting proteins is of utmost importance for accurate data interpretation. Although stringent purification methods are critical, challenges still remain in the selection of criteria that will permit the objective differentiation of true members of the protein complex from non-specific background proteins. To address these challenges, we have developed a quantitative proteomic strategy combining stable isotope labeling with amino acids in cell culture (SILAC), affinity substrate trapping, and geLC-MS/MS protein quantitation. ATP hydrolysis-deficient vacuolar protein sorting-associated protein 4B (Vps4B) was used as the “bait” protein which served as a substrate trap since its lack of ATP hydrolysis enzymatic activity allows the stabilization of its transiently associated interacting proteins. A significant advantage of our approach is the use of our new in-house developed software program for SILAC-based mass spectrometry quantitation, which further facilitates the differentiation between the bait protein, endogenous bait-interacting proteins, and non-specific binding proteins based on their protein ratios. The strategy presented herein is applicable to the analysis of other protein complexes whose compositions are dependent upon the ATP hydrolysis activity of the bait protein used in affinity purification studies.
Keywords: SILAC, Vps4B, mass spectrometry, quantitative mass spectrometry, protein complex, substrate trapping, endogenous binding partners, saturable binding
INTRODUCTION
A commonly used approach to analyze protein complexes entails affinity purification followed by SDS-PAGE and mass spectrometry. Typically, four different classes of proteins can be identified in affinity purification studies: 1) bait protein; 2) proteins that specifically interact with the bait protein; 3) non-specific proteins associated with either the antibody or beads; and 4) proteins that cross-react with the antibody.1 Thus, an objective method is required for the differentiation of these classes of proteins. Although stringent purification procedures can be used, challenges remain in being able to reliably select the bait-interacting proteins from among the hundreds of proteins that will likely be identified.2 To meet this need, in the current study we have applied an unbiased and quantitative proteomic approach using a combination of stable isotope labeling, tetracycline-inducible bait protein expression, immunoprecipitation, and geLC-MS/MS mass spectrometry-based quantitation.
Among the strategies that have been applied in mass spectrometry-based proteomic analyses of protein complexes are tandem affinity purification3, 4, free-flow electrophoresis5, quantitative immunoprecipitation combined with knockdown (QUICK)1, native capillary isoelectric focusing6, and top-down mass spectrometry7. A comprehensive review of the methods combining affinity purification with quantitative mass spectrometry to study the in vivo protein interactions of protein complexes is beyond the scope of this study; however many thorough reviews have been published addressing this matter.8–10 Here we describe a substrate trapping strategy which allows the stabilization of endogenous, low abundance, transiently associated membrane protein complexes. Our approach relies upon the use of quantitative mass spectrometry to permit the application of stringent data filtering criteria for the selection of high quality protein interactor candidates.
To demonstrate the feasibility and robustness of our approach, we used tetracycline inducible HEK293 cells conditionally expressing an E235Q Vacuolar protein sorting associated protein 4B (Vps4B) mutant to generate samples for comparison in this study. Vps4B is an endosome-associated AAA+ (ATPase associated with a variety of cellular activities) type ATPase that uses its ATP-hydrolysis function to catalyze the dissociation and release from the membrane of endosomal sorting complex required for transport (ESCRT)-III proteins following the completion of endocytic and biosynthetic cargo sorting into multivesicular bodies.11–18 The assembly of Vps4B into a dodecamer is a critical regulatory step since Vps4B is an inactive monomer or dimer in the cytosol and assembles only transiently into its oligomeric and active state upon interacting with its endogenous substrates19–21. The dynamic nature of interactions between AAA+ ATPases such as Vps4B and their interacting proteins make it difficult to define the range of their endogenous protein substrates.
The well-characterized E235Q point mutation within the ATP binding domain of Vps4B abrogates ATP hydrolysis, consequently permitting this mutant to bind tightly to and “trap” its normal substrates.22–24 Therefore, Vps4B(E235Q) was used extensively as a “substrate trap” to permit the stabilization and detection of Vps4B-interacting membrane-associated proteins, including those that are only transiently associated with Vps4B and that are not detectable by classical immunoaffinity pull-down approaches.11–13, 25, 26
Here, we present our quantitative proteomic approach to detect endogenous transiently associated protein-protein interactions using Vps4B(E235Q) as a substrate trap. This approach combines stable isotope labeling with tetracycline-inducible bait protein expression, immunoprecipitation, and mass spectrometry-based quantitation to enable the objective analysis of affinity purified Vps4B protein complexes.
RESULTS and DISCUSSION
Biochemical characterization of tetracycline-inducible ATP hydrolysis deficient Vps4B(E235Q)-myc cells
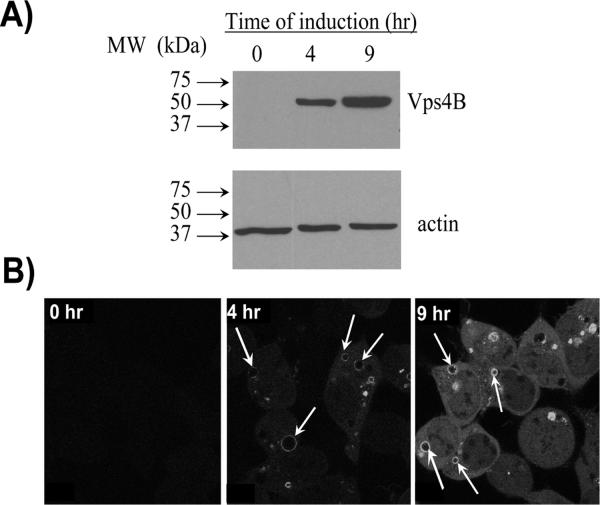
The in vitro mammalian experimental system we used to develop our substrate-trapping protein interaction analysis method consisted of HEK293 cells stably expressing Vps4B(E235Q) under the control of a tetracycline-regulated promoter.25 Abrogation of the ability of Vps4B to hydrolyze ATP by the introduction of an E235Q point mutation within the Walker B motif of its ATP binding domain is a well-characterized mutation that permits the irreversible binding of this Vps4B EQ mutant to its normal substrates.22, 23 The addition of tetracycline to stably-transfected HEK293 cells induced the expression of Vps4B(E235Q)-myc in a time-dependent manner (Figure 1A).
Figure 1. Tetracycline-induced Vps4B(E235Q) is specifically located on the endosomal membrane.
A) Fifty μg protein from soluble lysate of tetracycline-inducible Vps4B(E235Q)-myc HEK293 cells were separated by SDS-PAGE and analyzed by Western blot with a Vps4B antibody. Actin was used as a control for equal protein loading. Immunoblot is representative of three independent experiments. B) HEK293 cells stably expressing Vps4B(E235Q)-GFP for the indicated lengths of time were visualized directly by confocal microscopy with constant settings to permit the comparison of Vps4B(E235Q)-GFP expression levels. After 4 hr of tetracycline induction, Vps4B(E235Q)-GFP is specifically located on enlarged endosomal compartments (indicated by arrows). Mostly diffuse and cytosolic Vps4B(E235Q)-GFP localization can be observed after 9 hr of tetracycline induction.
Induction times of 4 hr and 9 hr were selected based on the observation that after 4 hr, Vps4B(E235Q)-GFP appeared entirely associated with enlarged endosomes, whereas at the 9 hr time-point, its binding sites had been saturated as indicated by accumulation of some of the protein in the cytoplasm (Figure 1B, middle panel 4 hr time point and indicated by arrows). These abnormally enlarged endosomal compartments, termed EQ compartments, have been shown to contain proteins characteristic of late endosomes and lysosomes, including CD63 and LAMP-2.25 Although the GFP labeling of the EQ compartments is more intense at the 9 hr timepoint than the 4 hr timepoint, our proteomic data suggest that the recruitment of Vps4B(E235Q)-GFP to the membranes of these EQ compartments is not associated with any additional recruitment of the endogenous binding partners of Vps4B.
A common adverse effect of protein over-expression is increased cell death.27, 28 Cell viability was not adversely affected by the level and duration of Vps4B(E235Q)-myc over-expression used in our studies; there was no statistically significant difference in the viability of HEK293 cells following 4 hr or 9 hr of tetracycline-induced expression of Vps4B(E235Q)-myc in comparison to the viability of HEK293 cells that were not induced. Thus, the induction time-points that were selected to analyze the Vps4B protein interaction complex permitted experimental analysis under conditions that did not trigger cell death pathways as a result of the level of Vps4B over-expression.
SILAC-based substrate trapping strategy to characterize transiently associated Vps4B protein complex components
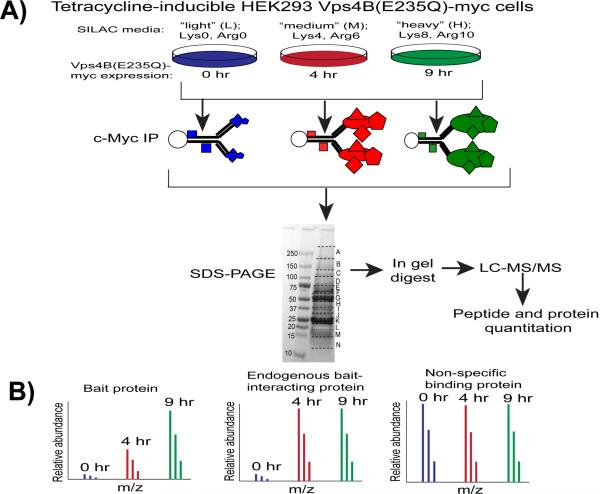
The identification of the interaction partners of AAA+ type ATPase proteins is complicated by the dynamic, ATP hydrolysis-dependent nature of their interactions with their protein substrates.23 ATP hydrolysis-deficient c-Myc-tagged Vps4B(E235Q)25 was used as the bait protein in the development of our proteomic method to detect protein interactions. Our rationale was that Vps4B(E235Q) would act as a substrate trap, thereby permitting the detection of its endogenous transiently associated interacting proteins. A schematic diagram of our workflow is illustrated in Figure 2A. Tetracycline-inducible Vps4B cells were first cultured in light (Lys0/Arg0), medium (Lys4/Arg6), or heavy (Lys8/Arg10) SILAC media individually. The expression of Vps4B was then induced by the addition of tetracycline for 0, 4, or 9 hr. To determine the endogenous binding partners of Vps4B, cell lysates isolated from 0 hr (light), 4 hr (medium) and 9 hr (heavy) induced cells were subjected to immuno-affinity purification with an anti-c-Myc antibody followed by quantitative LC-MS/MS (see Experimental Section).
Figure 2. A SILAC-based substrate trapping workflow to quantitatively characterize transient protein-protein interactions.
A) Tetracycline-inducible Vps4B(E235Q)-myc HEK293 cells were cultured in SILAC “light” (Lys0, Arg0), “medium” (Lys4, Arg6), or “heavy” (Lys8, Arg10) media and were harvested after 0 hr, 4 hr, or 9 hr of tetracycline induction, respectively. Three mg protein from each condition were combined and used for c-Myc immunoprecipitation. Immunoprecipitated proteins were separated by SDS-PAGE, followed by in-gel digestion and LC-MS/MS. Relative abundances of identified peptides were assessed by analyzing their MS1 peak intensities using IsoQuant, an in-house software program. B) The bait protein, endogenous bait-interacting proteins and non-specific binding proteins are readily distinguishable by their 4 hr/0 hr, 9 hr/0 hr, and 9 hr/4 hr SILAC-labeled peptide ratios.
Vps4B is an inactive monomer or dimer in the cytosol and assembles only transiently into its functional dodecameric state upon interacting with its endogenous substrates.19–21 Thus, the oligomerization of Vps4B monomers is an important regulatory step in the interaction between Vps4B and its endogenous interacting proteins. The immuno-affinity approach selected for this study permitted the analysis of the most thermodynamically-stable Vps4B protein interaction complex under near-physiological conditions, thereby avoiding some of the biases inherent in other protein-protein interaction approaches including TAP and cross-linking.29 For example, the use of a genetically-modified TAP tagged Vps4B chimeric construct would likely preclude the ability of Vps4B to oligomerize and bind to its endogenous interacting proteins.
The 4 hr/0 hr, 9 hr/0 hr, and 9 hr/4 hr relative abundance ratios of the quantified proteins were used to separate the proteins into three groups: bait protein, endogenous bait-interacting protein, and non-specific binding protein (Figure 2B). Reflecting the time-dependent increase in the tetracycline-induced expression of Vps4B(E235Q)-myc (bait protein), we expected that: 1) the relative abundance of Vps4B peptides would continuously increase from the 0 hr to the 4 hr time-point and from the 4 hr to the 9 hr time-point; 2) the relative abundance of endogenous bait-interacting proteins would increase from the 0 hr to the 4 hr time-point and would not increase further at the 9 hr time-point due to a saturation of binding sites on Vps4B(E235Q)-myc; and 3) the relative abundance of non-specific binding proteins would remain constant between all time-points.
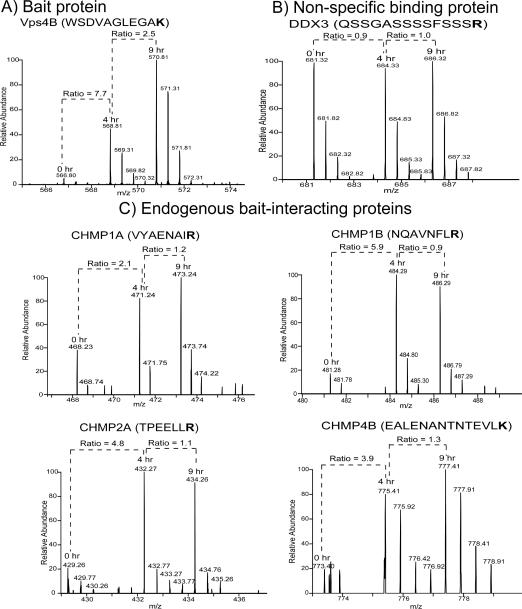
Bait Vps4B protein and its endogenous interacting proteins CHMPs are readily distinguished from non-specific binding proteins by their SILAC-labeled peptide ratios
Representative MS1 spectra of peptides from the bait protein, Vps4B; a non-specific binding protein, DEAD box protein 3 (DDX3); and endogenous bait-interacting proteins, CHMPs 1A, 1B, 2A and 4B, are shown in Figure 3. The relative abundance of the 4 hr Vps4B peptide is 7.7-fold higher than that of the 0 hr peptide, whereas the relative abundance of the 9 hr Vps4B peptide is 2.5-fold higher than that of the 4 hr peptide (Figure 3A). This trend reflects the tetracycline induction time-dependent increase in Vps4B expression and is consistent with the results from a traditional Vps4B immunoblotting analysis (Figure 1A). In contrast, the 4 hr/0 hr and 9 hr/4 hr peptide ratios from the non-specific binding protein DDX3 are equal to or close to 1.0: 0.9 and 1.0, respectively (Figure 3B). These ratios indicate that the relative abundance of this protein does not change irrespective of Vps4B expression.
Figure 3. Representative MS1 spectra of SILAC-labeled peptides from the bait protein, a non-specific interacting protein and endogenous bait-interacting proteins.
A) The bait protein, Vps4B, has relative abundance ratios (4 hr/0 hr and 9 hr/4 hr) > 2.0 which reflect its increased expression following 4 hr and 9 hr of tetracycline-induced expression. B) The relative abundance ratios of DEAD box protein 3 (DDX3) are ~1.0 indicating that DDX3 is a non-specific binding protein. C) CHMPs 1A, 1B, 2A and 4B are endogenous Vps4B-interacting proteins whose relative abundances increase following 4 hr induction of Vps4B expression. The 9 hr/4 hr ratios of these CHMP peptides (ratios ~1.0) suggest that the binding of CHMPs 1A, 1B, 2A and 4B to Vps4B can become saturated after 4 hr induction of Vps4B expression. Stable isotope-labeled amino acids are in bold.
The trends of the 0 hr, 4 hr, and 9 hr peptides from known Vps4B-interacting proteins CHMPs 1A, 1B, 2A and 4B (Figure 3C) suggest that after the 4 hr time-point, the pool of endogenous proteins that binds to Vps4B becomes depleted and their abundance does not increase further. The 4 hr/0 hr ratios of these peptides are all > 2.0, indicating that these peptides belong to proteins that interact specifically with Vps4B. CHMPs 1A, 1B, 2A and 4B are endosomal sorting complex required for transport (ESCRT)-III proteins that are known Vps4B interactors.15, 21, 25
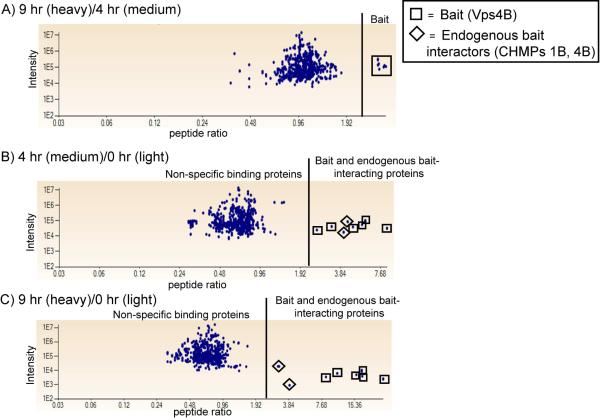
An in-house software program, IsoQuant, was used to calculate peptide and protein ratios based on MS1 peak intensity. The sampling of Vps4B interacting proteins reflects the abundance of endogenous proteins bound to Vps4B(E235Q)-myc. As shown in Figure 4, the distribution of peptides from the bait Vps4B protein (indicated by clear boxes) and its endogenous interacting proteins (indicated by clear diamonds) is distinct from that of the peptides belonging to non-specific binding proteins. The peptide identification and quantitation data for all three biological replicates is available as Supporting Information.
Figure 4. Peptide ratios of the bait protein and endogenous bait-interacting proteins have a distinct distribution from those of the non-specific binding proteins.
A) Peptides from the bait protein, Vps4B, are the only peptides in the representative dataset with 9 hr/4 hr (heavy/medium) ratios > 2.0, which reflects the time-dependent increase in the abundance of Vps4B. The relative abundance of Vps4B and its endogenous interacting proteins is increased following 4 hr (B) and 9 hr (C) expression of Vps4B, while the relative abundance of the non-specific binding proteins remains ≤ 1.0.
Figure 4A demonstrates that the peptides from Vps4B (labeled by clear boxes) are the only peptides in the representative dataset with 9 hr/4hr (heavy/medium) ratios > 2.0. The 4 hr/0 hr (medium/light) and 9 hr/0 hr (heavy/light) peptide ratios of Vps4B and its known endogenous interacting proteins, CHMPs 1B and 4B, are > 2.0 (Figures 4B and 4C, labeled by diamonds). CHMPs 1A and 2A were also identified with 4 hr/0 hr (medium/light) and 9 hr/0 hr (heavy/light) ratios > 2.0.
Using the criterion of having a 4 hr/0 hr (medium/light) ratio > 2.0, only four proteins other than Vps4B were identified as specific Vps4B protein interactors in the representative dataset: CHMPs 1A, 1B, 2A and 4B. These were the only ESCRT-III proteins quantifiably identified in our approach, suggesting that these are among the most abundant ESCRT-III Vps4B-interacting proteins.
CHMPs 1A, 1B, 2A and 4B were identified with 9 hr/4 hr ratios of ~ 1.0, whereas Vps4B had a 9 hr/4 hr ratio of ~ 2.0, which suggests that these CHMPs exhibit a saturable binding mechanism with Vps4B. CHMP4B was identified by our approach, but it is likely that our data are representative of the binding of both CHMPs 4A and 4B to Vps4B since the identified CHMP4B peptides share 70% sequence homology with CHMP4A.
The calculation of protein abundance ratios for CHMPs indicates that Vps4B(E235Q)-myc is expressed in the non-induced control (“light”-labeled) cells at a level not detected by Western blot (Figure 1A). This is most likely due to variability in the binding of Tet repressor homodimers to the Tet operator sequences in the inducible pcDNA6/TR expression vector that represses transcription of the Vps4B(E235Q)-myc gene in the stably transfected HEK293 cells.
The detection of Vps4B(E235Q)-myc binding proteins is directly proportional to their endogenous abundance. The distinct difference between the distribution of Vps4B, CHMP1B and CHMP4B peptides as compared to that of the peptides from non-specific binding proteins in Figures 4B and 4C demonstrates the robustness of our quantitative mass spectrometry-based proteomic approach. We expected that all non-specific binding proteins should have 4 hr/0 hr (medium/light) ratios ≤ 1.0. Tubulin (4 hr/0 hr = 0.91), 60S ribosomal protein L10a (4 hr/0 hr = 1.04), heterogeneous nuclear ribonuclear protein A2 (4 hr/0 hr = 0.92), heat shock 70 kDa protein (4 hr/0 hr = 1.01), histone H1.1 (4 hr/0 hr = 0.90), eukaryotic translation initiation factor 3 (4 hr/0 hr = 0.89) and DEAD box protein 3 (4 hr/0 hr = 0.88) were among the proteins classified as non-specific binding proteins. These proteins belong to the seven classes of proteins that bind non-specifically to sepharose and agarose and comprise the “bead proteome”: cytoskeletal/structural/motility proteins, DEAD box proteins, eukaryotic translation elongation and initiation factors, heat shock proteins, histones, hnRNP proteins, and ribosomal proteins.30
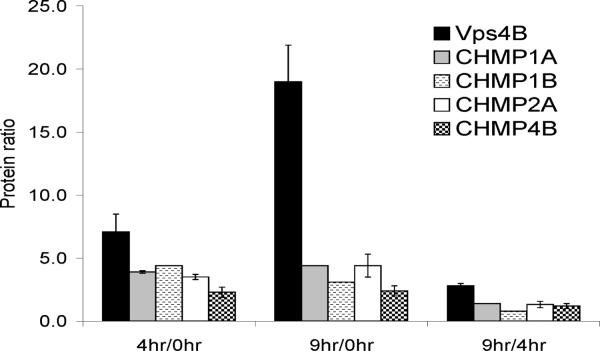
Figure 5 shows the average 4 hr/0 hr, 9 hr/0 hr and 9 hr/4hr protein ratios of Vps4B and its identified known interacting proteins across all three biological replicates. The 4hr/0hr protein ratios for Vps4B and its endogenous interacting proteins (CHMPs 1A, 1B, 2A and 4B) are all > 2.0. The 9hr/0hr ratio of Vps4B (19.0 ± 2.9) is indicative of its time-dependent, tetracycline-induced over-expression. The 9hr/4hr ratios of the CHMPs (~ 1.0) suggest that these proteins exhibit a saturable binding mechanism with Vps4B; their expression levels do not increase with the over-expression of Vps4B.
Figure 5. Average 4hr/0hr, 9hr/0hr, and 9hr/4hr protein ratios of bait and endogenous bait-interacting proteins (CHMPs).
The 4hr/0hr protein ratios for the bait protein (Vps4B) and its endogenous interacting proteins (CHMPs 1A, 1B, 2A and 4B) are all > 2.0. The 9hr/0hr ratio of Vps4B (19.0 ± 2.9) is indicative of the time-dependent, tetracycline-induced over-expression of this protein. The 9hr/4hr ratios of the CHMPs (~ 1.0) suggest that these proteins exhibit a saturable binding mechanism with Vps4B; their expression levels do not increase with the over-expression of Vps4B. n = 3 biological replicates. As CHMPs 1A, 1B and 4B were not identified in all three replicates, the s.d. of their protein ratios are calculated based on the biological replicate with the highest spectral count for each CHMP.
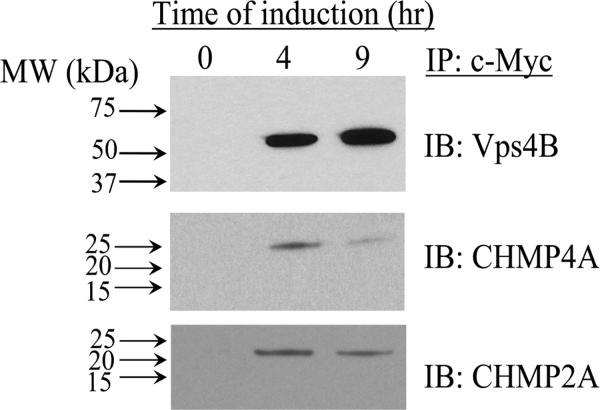
To confirm our mass spectrometric analysis, a separate Vps4B immunoaffinity purification was conducted. As shown in Figure 6, the Vps4B c-Myc IP immunoblotting analyses of Vps4B and two Vps4B interacting proteins, CHMPs 4A and 2A, confirm the mass spectrometry data from Figure 4. The amounts of immunoaffinity isolated CHMPs 4A and 2A present at 4 and 9 hr after the induction of Vps4B expression remain largely constant, further confirming that these CHMPs exhibit a saturable binding mechanism with Vps4B as determined by our quantitative SILAC analysis (Figure 3).
Figure 6. Validation of mass spectrometric results by immunoblotting of c-Myc IP eluant.
Mass spectrometry data was validated by immunoblotting of the c-Myc IP eluant from the 0, 4, and 9 hr induced HEK293 Vps4B(E235Q)-myc cells. Vps4B expression increased following 4 hr and 9 hr induction. The relative abundance of CHMP4A, a specific Vps4B-interacting protein, decreased slightly following 4 hr of expression of the bait protein, Vps4B. The relative abundance of CHMP2A, also a specific Vps4B-interacting protein, remained constant following 4 hr of Vps4B expression.
The use of the Vps4B “substrate trap” in our approach permitted the detection of normally transiently interacting ESCRT-III proteins that were not detected when wild-type Vps4B was used as the bait protein, which likely reflects the transient nature of their association with Vps4B. Thus, the ESCRT-III proteins identified in our approach comprise the most thermodynamically stable Vps4B protein interaction complex. The physical association of Vps4B with the ESCRT-III proteins is directly related to the hydrolysis of ATP by Vps4B. Cycles of Vps4B-mediated ATP hydrolysis induce conformational changes in ESCRT-III proteins that govern their ability to bind to Vps4B.31 Hence, in order to detect the ESCRT-III protein interactors of Vps4B in our studies, it was necessary to utilize an ATP hydrolysis-deficient Vps4B mutant (Vps4B(E235Q)).
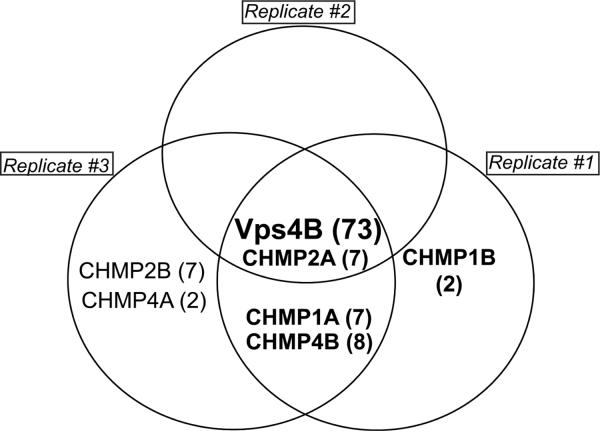
The reproducibility of the identification of the known Vps4B-interacting proteins in our study reflects the known heterogeneity with which these proteins bind to Vps4B. As indicated in Figure 7, CHMP2A is the most stable Vps4B-interacting protein among the ESCRT-III proteins quantified by our approach. This is in agreement with previously published work demonstrating that the biochemical function of CHMP2A is the recruitment of Vps4B32–34. Although other ESCRT-III components transiently interact with Vps4B, their functions are primarily the initiation of ESCRT-III assembly, membrane scission, and interaction with other proteins involved in multivesicular body biogenesis and autophagy.
Figure 7. CHMP2A is the most stable Vps4B interacting protein identified by our approach combining substrate trapping, SILAC, and quantitative proteomic analysis.
CHMP2A was identified in all three replicate IPs, suggesting that it is the most stable among the known Vps4B-interacting proteins identified in this study. The spectral count for each protein is in parentheses. Quantified proteins are in bold (Vps4B, CHMPs 1A, 1B, 2A and 4B).
CONCLUSIONS
In summary, we have demonstrated an objective and quantitative proteomic method to detect transient Vps4B protein interactions using substrate trapping. The use of Vps4B(E235Q) in the present studies allowed the stabilization and detection of protein interactors that are only transiently associated with Vps4B. Differential stable isotope labeling enabled us to definitively and quantitatively determine the true binding partners of Vps4B.
A significant advantage of our approach in comparison to traditional methods that are used to detect protein-protein interactions, such as yeast two-hybrid screens and co-immunoprecipitation of untagged endogenous target proteins followed by immunoblotting with antibodies against hypothesized protein binding partners, is the use of a conditional tetracycline-inducible ATP hydrolysis-deficient Vps4B mutant (E235Q) as our “bait” protein. This enhanced our ability to detect normally transient ATP-dependent Vps4B interactions since Vps4B dissociates from its interacting proteins following ATP hydrolysis. Furthermore, the tight tetracycline regulated expression of Vps4B reduced artifacts commonly associated with unrestricted over-expression of a bait protein.
The use of a quantitative mass spectrometry-based approach permitted the objective categorization of the identified immunoprecipitated proteins using their stable isotope ratios. The combination of substrate trapping, stable isotope labeling, tetracycline-inducible bait protein expression, immunoprecipitation, and mass spectrometry-based quantitation will have broad applicability for the objective and quantitative analysis of the protein interaction complexes of other AAA+ type ATPases such as torsin35–37, N-ethylmaleimide sensitive factor38, katanin39, ClpA40, 41 and mitochondrial respiratory chain complexes assembly protein YTA1242 – the ATP hydrolysis mutants (EQ mutation in Walker B ATPase motif) of which have been shown to result in high-affinity interactions with their respective protein targets.
EXPERIMENTAL SECTION
Materials
Dulbecco's modified Eagle's medium (DMEM), 0.25% trypsin, dialyzed fetal bovine serum, and penicillin/streptomycin were obtained from Invitrogen. SILAC DMEM (89985) and immobilized anti-c-myc agarose slurry (23620) were purchased from Thermo Fisher Scientific. Ammonium bicarbonate (NH4HCO3) was obtained from Sigma. Sequencing grade trypsin was purchased from Promega. All LC-MS/MS solvents (Water, Acetonitrile) were HPLC-grade (Baker) and were purchased from VWR Scientific.
Cell culture
Tetracycline-inducible HEK293 stable cell lines expressing c-myc or GFP-tagged Vps4B(E235Q) were generated as described previously25 using the T-REx tetracycline-regulated expression system (Invitrogen). Vps4B(E235Q)-GFP cells were maintained in DMEM supplemented with 10% tetracycline-free fetal bovine serum (Hyclone), 5 μg/mL blasticidin (Invitrogen), and 125 μg/mL zeocin (Invitrogen). For SILAC-based proteomics studies, Vps4B(E235Q)-myc cells were cultured in arginine- and lysine-deficient DMEM supplemented with 10% dialyzed fetal bovine serum, 1% penicillin/streptomycin, 5 μg/mL blasticidin, and 125 μg/mL zeocin. “Light”-labeled cells were cultured in supplemented SILAC medium containing 12C6-L-Lysine-2HCl (Lys0; 100 μg/mL) (Thermo Fisher Scientific) and 12C6-L-Arginine-HCl (Arg0; 100 μg/mL) (Thermo Fisher Scientific). “Medium”-labeled cells were cultured in supplemented SILAC medium to which D4-L-Lysine-2HCl (Lys4; 100 μg/mL) (Cambridge Isotope Laboratories) and 13C6-L-Arginine-HCl (Arg6; 100 μg/mL) (Cambridge Isotope Laboratories) were added. “Heavy”-labeled cells were cultured in supplemented SILAC medium containing 13C6,15N2-L-Lysine-2HCl (100 μg/mL) (Lys8; Sigma) and 13C6,15N4 LArginine-HCl (Arg10; 100 μg/mL) (Sigma). After seven cell doublings, the incorporation of the stable isotopes was 98% as determined by LC-MS/MS analysis of tryptic peptides isolated from the Lys4/Arg6 and Lys8/Arg10-labeled cells. To induce Vps4B(E235Q) expression, 0.75 μg/mL tetracycline (Sigma) was added for the indicated length of time. The cells were maintained at 37°C and 5% CO2 in a humidified incubator.
Immunoprecipitation and in-gel digestion
Vps4B(E235Q)-myc stable cells were grown to 80% confluence on 150 mm dishes prior to induction for 4 hr (SILAC “medium” cells) or 9 hr (SILAC “heavy” cells) with 0.75 μg/mL tetracycline. Cells cultured in SILAC “light” medium were not induced (0 hr, control). Cells were lysed (ball-bearing homogenizer, Industrial Tectonics) in immunoprecipitation (IP) buffer containing 20mM HEPES, pH 7.5, 150 mM NaCl, 1.0 % Triton X-100, 5 mM ATP, 2 mM MgCl2, 1 mM DTT, 1 mM PMSF, 1 mM ß-glycerophosphate, 1 mM sodium orthovanadate, 10 mM sodium pyrophosphate, and a protease inhibitor cocktail (Complete™, Roche Applied Science). Insoluble material was pelleted by centrifugation at 800 χ g for 5 min at 4°C. Protein concentration of the soluble cell lysate was determined by BCA protein assay (Thermo Fisher Scientific). Three mg protein from the SILAC light, medium and heavy cell lysates were combined and pre-cleared overnight with rotation at 4°C with Protein A agarose (50 μL agarose slurry/mg protein). Pre-cleared lysates were used for IP with immobilized anti-c-myc agarose slurry (1:100 (wt/wt) anti-c-myc:protein) overnight with rotation at 4°C. Agarose beads were washed three times with IP buffer. Three IPs were performed for three biological replicates. Immunoprecipitated proteins were eluted by incubation in 2× reducing sample buffer at 95°C for 5 min. Following separation by SDS-PAGE, proteins were enzymatically digested in-gel with trypsin as described previously.43 Extracted peptides were de-salted using in-house packed C18 STAGE Tips. De-salted peptides were stored at −80°C prior to LC-MS/MS analysis.
Immunoblot analysis
Protein in IP eluant was separated by SDS-PAGE using 4 – 20% Tris-Glycine gels (Bio-Rad). Protein was transferred to nitrocellulose membranes. Non-specific binding sites on the membranes were blocked for 1 hr at room temperature in PBS/0.1% Tween-20/5% milk, probed with primary antibodies overnight at 4°C and incubated with HRP-conjugated secondary antibodies (Thermo Fisher Scientific) for 1 hr at room temperature. Primary antibodies used were Vps4B25, CHMP4A and CHMP2A (Novus Biologicals), c-myc (Chemicon) and actin (Chemicon). Chemiluminescent detection was with Super Signal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific).
LC-MS/MS
Peptides were separated by nanoscale reverse-phase liquid chromatography using an Xtreme Simple nanoLC system (CVC/Micro-Tech). The analytical column was prepared by packing 1.7μm 200Å C18 material (Prospereon Life Sciences) into an integrated frit Pico Tip spray emitter (75 μm inner diameter, 10μm tip; New Objective) using a pressure injection cell (Next Advance). Peptides were manually injected into the sample loop and were loaded onto the column with 95 % solvent A (0.5% acetic acid in water). A 120 min LC gradient method from 5 – 35% B (80% acetonitrile, 0.5% acetic acid) with a post-split flow rate of 0.3 μL/min was used to elute the peptides into the mass spectrometer. The LTQ-Orbitrap mass spectrometer (Thermo Electron) was equipped with a nanospray ionization source. The spray voltage was 1.8 kV and the heated capillary temperature was 200 °C. MS1 data were acquired in the profile mode in the Orbitrap with a resolution of 60,000 at 400 m/z and the top five most intense ions in each MS1 scan were selected for collision induced dissociation in the linear ion trap. Dynamic exclusion was enabled with repeat count 1 and exclusion duration 60 sec. Other mass spectrometry data generation parameters were as follows: collision energy 35%, ion selection threshold for MS/MS 500 counts, isolation width 3, default charge state 3, charge state screening and rejection (1+ ions) enabled.
SILAC-labeled protein identification and quantitation
MS/MS spectra were searched against a UniProtKB human protein database (version Oct 5, 2010; 20,259 reviewed sequences; 75,498 non-reviewed sequences) using Bioworks 3.3.1 SP1 with the SEQUEST algorithm. Data were also searched using Mascot (Matrix Science). Search parameters included 50ppm peptide mass tolerance, 1.0Da fragment tolerance, static Cys +57.02510 (carbamidomethylation) modification and the following differential modifications: Met +15.99492 (oxidation), Lys +4.02511 (D4), Lys +8.01420 (13C6,15N2), Arg +6.02013 (13C6), and Arg +10.00827 (13C6,15N4). Fully tryptic peptides with up to two missed cleavages and charge-state dependent cross correlation scores ≥ 2.5, 3.0, and 3.5 for 2+, 3+ and 4+ peptides, respectively, were considered as positive identifications for further quantitative analyses.
An in-house developed quantitation method based on MS1 peak intensity, named IsoQuant (software is freely available for non-commercial use and can be downloaded from http://www.proteomeumb.org/MZw.html), was used for mass spectrometric data analysis.
Supplementary Material
ACKNOWLEDGMENT
This work was funded by the Maryland Cigarette Restitution Fund and National Institutes of Health Grants AG25323 to A.Y. and GM076686 to P.I.H. We thank Dr. Frank Yang of Micro-Tech Scientific for helpful insight regarding optimizing the performance of our liquid chromatography system.
Footnotes
Supporting Information Peptide quantitation data from three replicate IPs. The identified FIGL1 peptides share 100% sequence homology with Vps4B.
REFERENCES
- (1).Selbach M, Mann M. Nat Methods. 2006;3:981–983. doi: 10.1038/nmeth972. [DOI] [PubMed] [Google Scholar]
- (2).Moresco JJ, Carvalho PC, Yates JR., 3rd J Proteomics. 2010;73:2198–2204. doi: 10.1016/j.jprot.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Koch HB, Zhang R, Verdoodt B, Bailey A, Zhang CD, Yates JR, 3rd, Menssen A, Hermeking H. Cell Cycle. 2007;6:205–217. doi: 10.4161/cc.6.2.3742. [DOI] [PubMed] [Google Scholar]
- (4).Guerrero C, Tagwerker C, Kaiser P, Huang L. Mol Cell Proteomics. 2006;5:366–378. doi: 10.1074/mcp.M500303-MCP200. [DOI] [PubMed] [Google Scholar]
- (5).Gomes AV, Young GW, Wang Y, Zong C, Eghbali M, Drews O, Lu H, Stefani E, Ping P. Mol Cell Proteomics. 2009;8:302–315. doi: 10.1074/mcp.M800058-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Fonslow BR, Kang SA, Gestaut DR, Graczyk B, Davis TN, Sabatini DM, Yates JR., 3rd Anal Chem. 2010;82:6643–6651. doi: 10.1021/ac101235k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Lomeli SH, Peng IX, Yin S, Loo RR, Loo JA. J Am Soc Mass Spectrom. 2010;21:127–131. doi: 10.1016/j.jasms.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Ramisetty SR, Washburn MP. Crit Rev Biochem Mol Biol. 2011 doi: 10.3109/10409238.2011.567244. [DOI] [PubMed] [Google Scholar]
- (9).Kaake RM, Wang X, Huang L. Mol Cell Proteomics. 2010;9:1650–1665. doi: 10.1074/mcp.R110.000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Oeljeklaus S, Meyer HE, Warscheid B. FEBS Lett. 2009;583:1674–1683. doi: 10.1016/j.febslet.2009.04.018. [DOI] [PubMed] [Google Scholar]
- (11).Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. Dev Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- (12).Babst M, Wendland B, Estepa EJ, Emr SD. Embo J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Hurley JH, Emr SD. Annu Rev Biophys Biomol Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Shim S, Merrill SA, Hanson PI. Mol Biol Cell. 2008;19:2661–2672. doi: 10.1091/mbc.E07-12-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- (16).Tsang HT, Connell JW, Brown SE, Thompson A, Reid E, Sanderson CM. Genomics. 2006;88:333–346. doi: 10.1016/j.ygeno.2006.04.003. [DOI] [PubMed] [Google Scholar]
- (17).Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, Emr SD, Williams RL. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- (18).Kieffer C, Skalicky JJ, Morita E, De Domenico I, Ward DM, Kaplan J, Sundquist WI. Dev Cell. 2008;15:62–73. doi: 10.1016/j.devcel.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- (19).Azmi I, Davies B, Dimaano C, Payne J, Eckert D, Babst M, Katzmann DJ. J Cell Biol. 2006;172:705–717. doi: 10.1083/jcb.200508166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Gonciarz MD, Whitby FG, Eckert DM, Kieffer C, Heroux A, Sundquist WI, Hill CP. J Mol Biol. 2008;384:878–895. doi: 10.1016/j.jmb.2008.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Scott A, Chung HY, Gonciarz-Swiatek M, Hill GC, Whitby FG, Gaspar J, Holton JM, Viswanathan R, Ghaffarian S, Hill CP, Sundquist WI. Embo J. 2005;24:3658–3669. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Dalal S, Rosser MF, Cyr DM, Hanson PI. Mol Biol Cell. 2004;15:637–648. doi: 10.1091/mbc.E03-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Vale RD. J Cell Biol. 2000;150:F13–19. doi: 10.1083/jcb.150.1.f13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Finken-Eigen M, Rohricht RA, Kohrer K. Curr Genet. 1997;31:469–480. doi: 10.1007/s002940050232. [DOI] [PubMed] [Google Scholar]
- (25).Lin Y, Kimpler LA, Naismith TV, Lauer JM, Hanson PI. J Biol Chem. 2005;280:12799–12809. doi: 10.1074/jbc.M413968200. [DOI] [PubMed] [Google Scholar]
- (26).Shim S, Kimpler LA, Hanson PI. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- (27).Koskinen PJ, Alitalo K. Semin Cancer Biol. 1993;4:3–12. [PubMed] [Google Scholar]
- (28).Kuchino Y, Asai A, Kitanaka C. Prog Mol Subcell Biol. 1996;16:104–129. doi: 10.1007/978-3-642-79850-4_7. [DOI] [PubMed] [Google Scholar]
- (29).von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, Bork P. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- (30).Trinkle-Mulcahy L, Boulon S, Lam YW, Urcia R, Boisvert FM, Vandermoere F, Morrice NA, Swift S, Rothbauer U, Leonhardt H, Lamond A. J Cell Biol. 2008;183:223–239. doi: 10.1083/jcb.200805092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Hurley JH, Hanson PI. Nat Rev Mol Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Teis D, Saksena S, Emr SD. Dev Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- (33).Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Merrill SA, Hanson PI. J Biol Chem. 2010;285:35428–35438. doi: 10.1074/jbc.M110.126318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Jungwirth M, Dear ML, Brown P, Holbrook K, Goodchild R. Hum Mol Genet. 2010;19:888–900. doi: 10.1093/hmg/ddp557. [DOI] [PubMed] [Google Scholar]
- (36).Naismith TV, Dalal S, Hanson PI. J Biol Chem. 2009;284:27866–27874. doi: 10.1074/jbc.M109.020164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Naismith TV, Heuser JE, Breakefield XO, Hanson PI. Proc Natl Acad Sci U S A. 2004;101:7612–7617. doi: 10.1073/pnas.0308760101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Whiteheart SW, Rossnagel K, Buhrow SA, Brunner M, Jaenicke R, Rothman JE. J Cell Biol. 1994;126:945–954. doi: 10.1083/jcb.126.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Hartman JJ, Mahr J, McNally K, Okawa K, Iwamatsu A, Thomas S, Cheesman S, Heuser J, Vale RD, McNally FJ. Cell. 1998;93:277–287. doi: 10.1016/s0092-8674(00)81578-0. [DOI] [PubMed] [Google Scholar]
- (40).Pak M, Hoskins JR, Singh SK, Maurizi MR, Wickner S. J Biol Chem. 1999;274:19316–19322. doi: 10.1074/jbc.274.27.19316. [DOI] [PubMed] [Google Scholar]
- (41).Singh SK, Guo F, Maurizi MR. Biochemistry. 1999;38:14906–14915. doi: 10.1021/bi991615f. [DOI] [PubMed] [Google Scholar]
- (42).Arlt H, Tauer R, Feldmann H, Neupert W, Langer T. Cell. 1996;85:875–885. doi: 10.1016/s0092-8674(00)81271-4. [DOI] [PubMed] [Google Scholar]
- (43).Cripps D, Thomas SN, Jeng Y, Yang F, Davies P, Yang AJ. J Biol Chem. 2006;281:10825–10838. doi: 10.1074/jbc.M512786200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.