Abstract
Neutrophils play critical roles in modulating the immune response. We present a robust methodology for rapidly isolating neutrophils directly from whole blood and develop ‘on-chip’ processing for mRNA and protein isolation for genomics and proteomics. We validate this device with an ex vivo stimulation experiment and by comparison with standard bulk isolation methodologies. Lastly, we implement this tool as part of a near patient blood processing system within a multi-center clinical study of the immune response to severe trauma and burn injury. The preliminary results from a small cohort of patients in our study and healthy controls show a unique time-dependent gene expression pattern clearly demonstrating the ability of this tool to discriminate temporal transcriptional events of neutrophils within a clinical setting.
Introduction
Neutrophils, which are the most common type of blood leukocyte (white blood cell), are important for protection against infections and modulation of inflammatory responses1. Although neutrophils were once thought of as terminally-differentiated cells with few roles outside of phagocytosis and granule content release, it is clear from the literature that protein synthesis and gene regulation play an important role in neutrophil function and innate immune signaling2,3. Furthermore, it is well recognized that neutrophils contribute to both chronic and acute inflammation, and are a critical part of a complex temporal pattern of activation of the immune system after injury and as such, an expanded role for neutrophils in adaptive immunity is now being suggested1.
In a clinical setting, the peripheral blood is an easily accessible tissue, and there is great interest in using leukocyte transcript profiling to understand disease processes4,5. Laudanski et al. recently demonstrated that differential genomic changes can be observed in distinct blood leukocyte subpopulations in response to the same in vivo stimulus6. However, the genomic changes seen in the total leukocyte population were blunted in comparison to individual cell types as would be expected. This observation has led to the practice of enriching leukocytes into more homogeneous subpopulations to allow for clearer functional interpretation of gene expression patterns6. Unfortunately, the fractionation of leukocytes into several subpopulations is technically challenging and extremely time consuming.
To date, the investigation of neutrophils by genomic and proteomic technologies has been hampered by three major challenges. First, standard methods for neutrophil isolation require multi-step density gradient separation, which involves several hours (>2) of processing time and uses milliliter volumes of blood (typically 4–8 ml)7. Second, neutrophils are especially sensitive to external perturbations and can be easily activated during the isolation process8. Third, neutrophils contain 10–20 times less mRNA per cell than monocytes9 thus requiring increased cell numbers for an equivalent quantity of nucleic acids. We thus sought to develop a cost-effective, facile method to isolate a highly enriched population of neutrophils from whole blood that could be readily utilized in a clinical setting.
We have previously created microfluidic tools that use affinity capture to isolate cellular subpopulations directly from whole blood in HIV infected patients10, and in metastatic cancer patients11. While these studies demonstrate the potential for microfluidic sample processing within a specialized laboratory setting, the tools have not been applied in a multi-center clinical program due to the high technical skills required to use the microfluidic devices. Additionally, molecular analysis of the captured cell populations in these studies was limited to a narrow set of known candidate genes due to a lack of cell purity, and sensitivity for proteomics analysis has been challenged by the total protein content available. As a result, microfluidic blood processing tools have not yet been combined with types of genome-wide microarray or proteomic analyses that would be fundamental to identifying biomarkers and understanding the biological basis of disease at a molecular level.
In this article, we report a microfluidic tool that captures highly enriched (>95%) neutrophils directly from 150 μL of whole blood within five minutes in sufficient quantity and purity for genome-wide microarray and mass-spectrometry based proteomic analysis. We show that the genomic and proteomic samples resulting from microfluidic cell isolation are of sufficiently high quality to discriminate between subtle differences in neutrophil activation states. Moreover, we implemented the use of this microfluidic neutrophil-capture cassette by non-specialized staff in a multi-center clinical program and we observe gene expression changes in neutrophils that are highly regulated following trauma injury. We anticipate that this device will have wide range of applications for furthering the biological and therapeutic investigations of neutrophils, and that this approach will be transferrable to other cell types in the peripheral blood.
Results
Characterization of cell capture
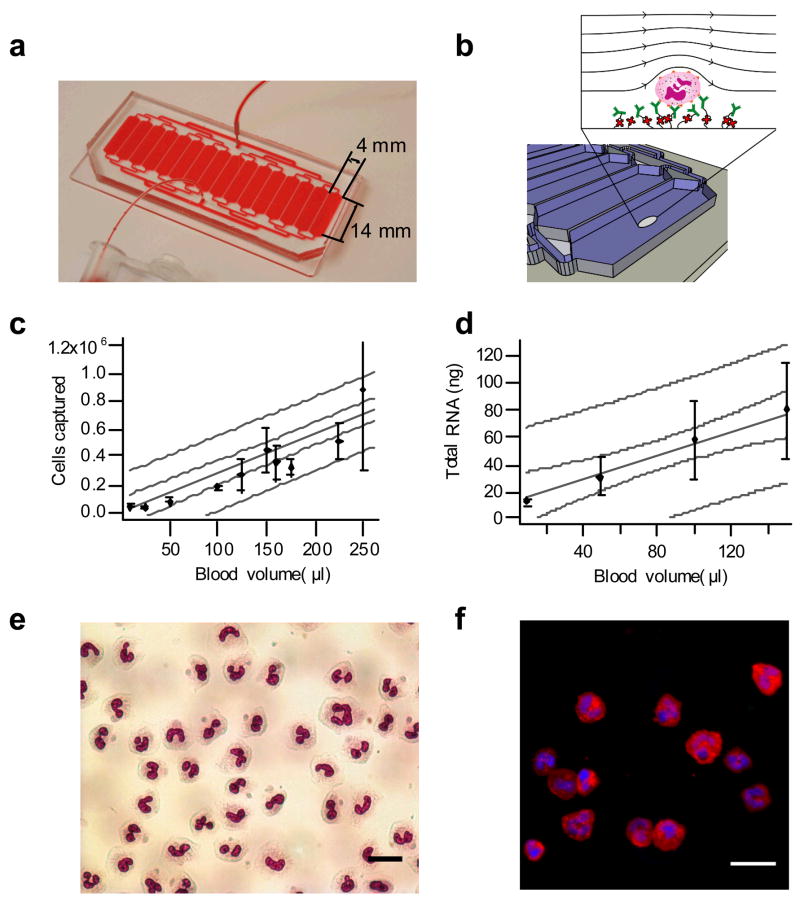
We designed a device that could capture neutrophils directly from 150 μL of whole blood. We chose anti-CD66b as the capture antibody, which captures all polymorphonuclear leukocytes bearing CEACAM-8 (carcinoembryonic antigen-related cell adhesion molecule 8), an adhesion molecule expressed on neutrophils and eosinophils. Neutrophils typically outnumber eosinophils by a factor of 30, and potential eosinophil contamination of ~3% was deemed acceptable for the studies described below. We determined the optimal shear stress of 0.45 dynes cm−2 for capturing neutrophils at a high density on the device surface, and a chamber height of 50 μm was chosen based on previous studies10. We then determined the surface area of the chambers that would capture sufficient numbers of cells for carrying out downstream genomic and proteomic studies. We finalized the design (Fig. 1a,b) which maximizes the width of a device on a 38 × 75 mm microscope slide. The final device consists of discrete chambers that divide the flow in order to obtain a more uniform flow distribution across the width of the device. At an optimal flow rate of 30 μl min−1, the number of neutrophils captured by the device is proportional to the volume of starting whole blood over a range of 10–250 μl (Fig. 1c,d), with a captured neutrophil purity of > 96% as evaluated by on-device Wright-Giemsa and immunofluorescent staining (Fig. 1e,f, Table 1; phenotyping of the remaining 4% population is given in Supplementary Table 1). To test the effects of severe inflammation on the ability to capture cells, we examined samples from five individual severely burned patients and found no change in the obtained cell purity (Table 1 and Supplementary Table 1).
Figure 1.
Summary of microfluidic device characterization. (a) Microfluidic chip design and (b) schematic of the surface functionalization of antibodies to the device. Green biotinylated α-CD66b monoclonal antibodies bind to red Neutravidin molecules that are covalently linked to the surface. Whole blood flows through each parallel capture channel and cells with CD66b antigen are specifically bound to the surface. Chip loading for cells captured (c) and RNA (d) with a linear fit (grey solid line), 95% confidence limits(grey dashed line), and 95% prediction bands (grey dotted line). The R value for the fits for (e) and (f) are 0.95 and 0.98, respectively. (e) Wright-Giemsa stain of burn blood captured from burn patient 10 days post injury showing mixture of fully segmented neutrophils and band forms (scale bar 20 μm).(f) Immunofluorescence of healthy volunteer stained with DAPI (blue), CD14-FITC (green), and CD16b-PE (red) (scale bar 25 μm);
Table 1.
Performance characteristics of microfluidic capture device
| Healthy normal volunteers (n=12) | Burn patients (n=13)a | ex vivo stimulation samples (n=12) | Burn and trauma (n=308)b | |
|---|---|---|---|---|
| Cells | ||||
| On-chip purity | 95% ± 1% (n=10) | 95% ± 3% (n=8) | 98 ± 2% (n=6) | N/D |
| On-chip viability | >99% (n=3) | >99% (n=3) | >99% (n=3) | N/D |
| Cells captured | 450K ± 160K | 890K ± 500K | 200K ± 100K | N/D |
| RNA | ||||
| RNA quantity (ng) | 200 ± 60 | 80 ± 20 | 210 ± 90 | 82 ± 45 |
| RNA RIN score | 8.2 ± 0.6 | 9.3 ± 0.2 | 7.8 ± 0.2 | 8.5 ± 0.6 |
| Proteins (TFE Lysis) | ||||
| Total protein (BCA) | 25 ± 14 μg | N/D | N/D | |
| Peptides | 2100 ± 400 | N/D | 3800 ± 100 | N/D |
| Proteins identified | 510 ± 80 | N/D | 860 ± 70 | N/D |
Massachusetts General Hospital burn unit, see supplementary methods for patient inclusion data
Combined Inflammation and Host Response to Injury program clinical sampling sites (Supplementary Table 7)
Not determined
RNA and protein extraction
For genomic analyses, captured cells were lysed in situ on the device with guanadinium-based chaotropes and total RNA was extracted and analyzed for nucleic acid quantity and quality. Total RNA recovered was linear over the range of capture (Fig. 1d), RNA quantity (0.33 ± 0.15 pg cell−1) was similar to other studies12, and RNA quality as measured on an Agilent Bioanalyzer 2100 was excellent with a RNA Integrity Number (RIN) range of 7.4–9.9 (Table 1, Supplementary Fig. 1). The correlations of mRNA abundance between multiple samples isolated by the microfluidic cassettes were 0.98 ± 0.02 (n = 12) demonstrating high reproducibility of the sampling.
For mass spectrometry (MS)-based proteomic analysis, we optimized the reagent and lysis conditions (Supplementary Table 2) to maximize the number of peptides and proteins identified for samples with low total protein content13. We observed that the chips can consistently capture ~4 × 105 neutrophils, which are sufficient for proteomic analysis with an average of ~2430 peptides and ~860 proteins identified per chip using a single reverse-phase LC separation coupled with mass spectrometry (LC-MS) (Table 1). Importantly, chip-to-chip correlations based on peptide intensities were extremely high (0.96 ± 0.02) for replicate samples. Protein distribution based on cellular components was nearly identical to Ficoll-dextran isolated neutrophils, suggesting that there was no differential adsorption of cellular proteins to the surface of the device (Supplementary Fig. 2).
Ex vivo stimulation studies
We next assessed whether the relatively small number of isolated neutrophils captured by the microfluidics cassettes would permit the potential discriminatory capabilities of genomic and proteomic analyses in response to various stimuli. We compared the genome-wide expression profile and protein abundance profile in neutrophils following ex vivo activation with either Escherichia coli lipopolysaccharide (LPS) or lymphokine stimulation using granulocyte-macrophage colony-stimulating factor (GM-CSF) and interferon-gamma (INF-γ) (abbr. GM+I). In both protocols, whole blood was stimulated ex vivo to allow leukocyte and plasma protein interactions14. In order to limit the effects of inter-subject biological variability, multiple cassette isolations from a single biological sample were performed for each experiment arm. Neither capture purity nor cell viability (Trypan blue exclusion) of the isolated cells was affected by the stimulation (Table 1).
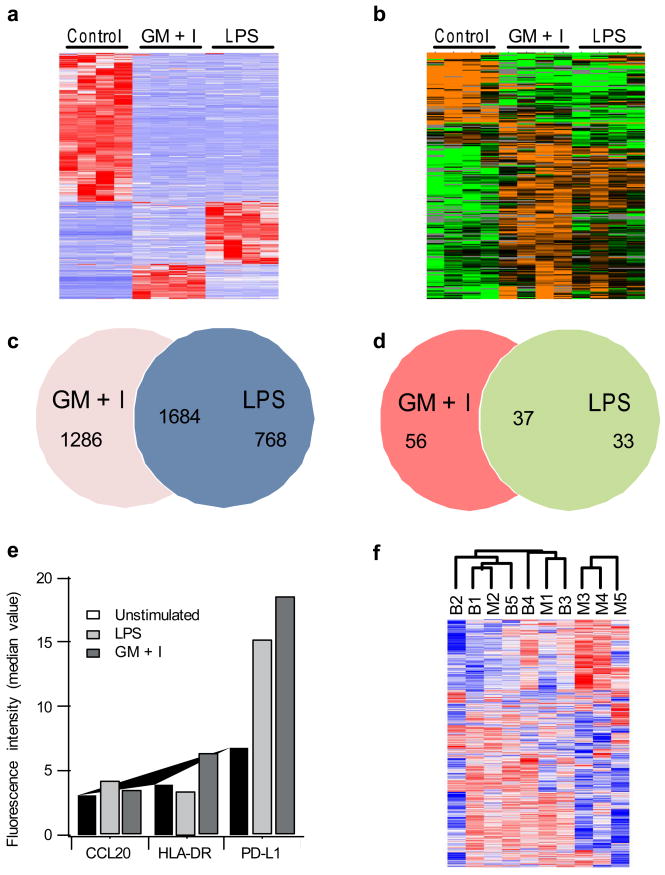
Unsupervised hierarchical clustering of the genomic and proteomic data from the cassette extractions showed patterns of neutrophil RNA expression and protein abundance distinct among the two stimulation protocols and from unstimulated neutrophils (Fig. 2). Samples cluster according to stimulation condition, thus verifying the ability of the cassette to isolate cells with varying activation states and subsequently resolve details of the distinct activation pathways. Correlations between multiple extractions within a single sample were excellent for both the genomic data (0.98 ± 0.02) and the proteomics data (0.96 ± 0.02) demonstrating a high level of sampling reproducibility. Overall, 12–14% of the approximately twenty-one thousand genes measured on the microarray were significantly perturbed (fold changes > 2, and false discovery rate < 0.01) following either stimulation (LPS or GM+I), and 12–15% among the six hundred consistently detectable proteins were also significantly changed (fold changes > 2, and false discovery rate < 0.05) (Supplementary Table 3). 62% of the significant genes overlapped between LPS and GM+I (Fig. 2c); among these, most genes (1684 of 1690) showed the same trend of changes after either stimulation. Similarly 45% of the significant proteins overlap and all showed the same trend of changes between the two stimulations (Fig. 2d).
Figure 2.
Genomic and protemic characterization of neutrophil lysates. Unsupervised cluster analysis for PMN validation studies for (a) microarray data and (b) LC-MS proteomics data. Red bars indicate upregulated genes, blue bars downregulated genes, orange bars upregulated proteins, and green bars downregulated proteins. Venn diagram of significant gene expression changes (c) and protein abundance changes (d) following ex vivo stimulation. 1684 genes overlapped between the two lists and showed the same directions of changes, while 6 genes showed opposite changes. For the proteins 37 proteins overlapped between the two lists and showed the same directions of changes, and none showed opposite changes. (e) Flow cytometry validation of ex vivo stimulation results, showing the mean fluorescence signal measured in CD66b+ granulocytes for unstimulated blood (white bars), LPS stimulated blood (blue checks), and GM+I stimulated blood (red stripes) (f) Unsupervised hierarchical clustering of genes (1690 probesets with SD>1) from five different healthy subjects isolated using microfluidics (M) or bulk Ficoll-dextran (B) methods. Note that there are no significant genes differentially expressed between the microfluidics and bulk isolation at FDR < 5%.
In addition, we examined a number of the genes found in the microarray studies using flow cytometry. While neutrophils are traditionally considered phagocytic cells, recent studies have shown their capability for antigen presentation with HLA-DR, and chemokine signaling with CCL20 associated with activation and migration of regulatory T cells and Th17 lymphocytes 15–17. CCL20 is upregulated in the LPS-stimulated sample with respect to the GM+I sample while HLA-DR shows the opposite trend (Fig. 2e). These results examining surface proteins are consistent with the microarray results, confirming that the changes in HLA-DR and CCL20 are likely not due to artifact or a contaminating cell population.
We next examined the metabolic and cell signaling pathways from the gene expression and protein identification data in Fig. 2 using the Ingenuity Pathway Analysis system. A complete list of the statistically significant functional pathways identified is provided in Supplementary Table 4. Cytokine signaling pathways were upregulated in the stimulated samples as compared to the unstimulated samples. Also, antigen presentation was significantly upregulated for the GM+I stimulation when compared to both unstimulated and LPS stimulated samples. Thus, microfluidic cassette isolated neutrophils yield genomic data that can be discriminated at the functional level as well as the gene level.
Inter-subject reproducibility
To further ensure the reliability of the microfluidic method, we performed parallel neutrophil isolations using both microfluidics and Ficoll-dextran methodologies from five different healthy volunteers and processed the cell lysates for microarray analysis. Correlations of the microarray data between subjects were 0.95 ± 0.01 and 0.96 ± 0.01 for the microfluidic samples and Ficoll-dextran isolations, respectively, and are consistent with subject-to-subject gene expression variability seen in total leukocyte preparations 18. To test possible differences at the gene level, we first performed unsupervised hierarchical clustering of the expression data. The microfluidic and Ficoll-dextran isolated neutrophils do not display any distinct clustering according to methods of neutrophil isolation (Fig. 2f and Supplementary Table 5). Furthermore, no probesets were identified as significantly different in expression between the two isolation methods at a false discovery rate (FDR) < 0.05, suggesting that the variance due to baseline gene expression and sample processing is on the order of or larger than the differences between isolation methodologies. While the number of probe sets where the average expression differs by more than 2-fold between microfluidic and Ficoll-dextran isolation may suggest a difference between the two isolation methods, we cannot observe these probe sets with statistical certainty even at a FDR < 0.25 (Supplementary Table 6). Furthermore, it should be noted that the differences between isolation methodologies are small compared to changes due to activation, where > 3,700 genes change following activation by either LPS or GM+I at a FDR < 0.001 and a fold-change > 2.
Clinical implementation
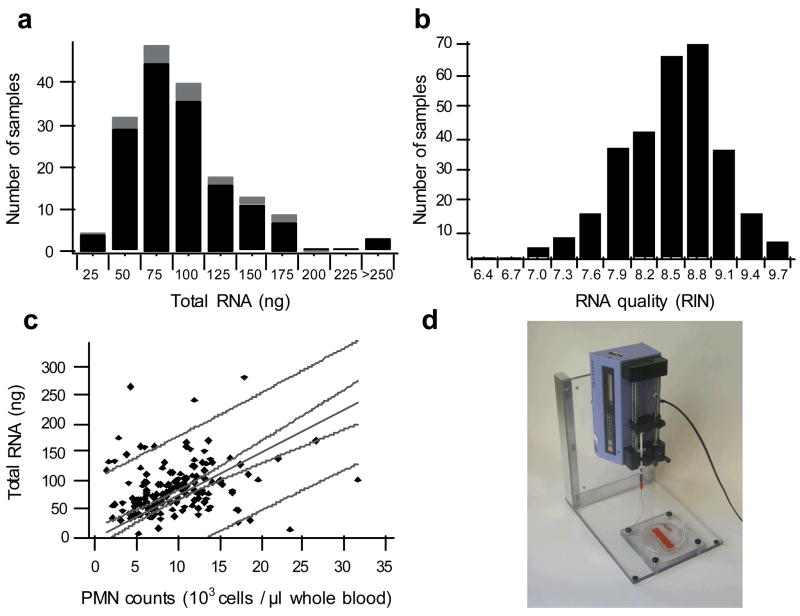
To demonstrate the utility of this technology in the clinical setting, we implemented microfluidic neutrophil isolation and processing methods in the genomic sampling arm of a multi-center study looking at genomic changes in severe trauma. After providing hands-on training, we supplied preassembled microfluidic devices as part of single use kits to clinical staff or research technicians at six clinical sites listed in Supplementary Table 7. Using the device loading data (Fig. 1) and complete blood counts from severely burned and trauma patients, we calculated the blood requirements (150 μl) needed to yield >20 ng of total cellular RNA in 99% of the clinical samples, a quantity of RNA that is more than sufficient to achieve reliable amplification for microarray expression studies using currently available amplification systems 19. In order to reduce the biological effects of sample processing, we developed a protocol for obtaining the cell lysate from whole blood within 20 minutes20. This processing time is shorter than the 30 minute transcriptional time lag seen in vitro following TLR activation with LPS 21 and in vivo with LPS-stimulated volunteers where gene expression showed marked differences within a 2 hour period 5. We have provided over 1000 cassettes to process blood from both severely burned or trauma patients. The total RNA extracted from the samples processed at the clinical sites meets targets in quantity and quality in 98% of the samples. Total RNA extraction quantities correlated in a linear fashion with clinical neutrophil counts based on CBC analysis (Fig. 3c). Additionally, there were no significant effects in the quantity and quality data based on cassette production batches (Supplementary Fig. 3).
Figure 3.
Summary of RNA extractions from cell lysates collected at six different clinical sites. (a) Histogram of the total RNA isolated from the trauma samples (black) and burn samples (gray). (b) Histogram of the RNA RIN quality score from both groups in panel a; RNA is scored on a scale of one to ten (higher is better), and any sample that scores four or higher is processed for microarray expression analysis. (c) Correlation of the total extracted RNA with clinical PMN counts taken from a complete blood count with five part differential; the solid line is a linear fit (R=0.23) through the origin with 95% confidence limits (grey dashed line), and 95% prediction bands (grey dotted line). (d) Syringe pump unit used at the clinical sites for sample processing.
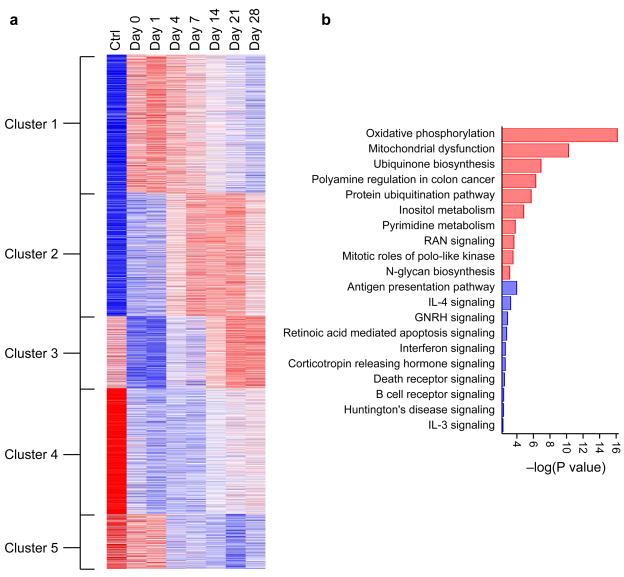
Because the RNA met internal standards for quantity and quality, 187 samples were processed for microarray analysis. In Fig. 4, we present the results from 26 trauma subjects at seven time points over first 28 days post injury. Control data were derived from ten healthy normal subjects and used as a baseline for data clustering. Remarkably, the expression level of 25% of the genes (7818/34834) is significantly changed over the time course at an FDR adjusted probability level of < 0.001 (Supplementary Table 8). An unsupervised hierarchical clustering shows five distinct groups as follows: (1) genes that show early upregulation in the first day and resolution to baseline around day 14; (2) genes that show delayed upregulation 4–7 days post trauma, with peak expression between 7 and 21 days; (3) genes that are down-regulated early in day 1 and resolve by day 21 (mirror of cluster 1); (4) genes that are initially down-regulated and remain so throughout the sampling period (28 days); and (5) genes that down-regulate after day 4 without recovery to baseline. A complete list of the statistically significant functional pathways identified from these genes is provided in Supplementary Table 9. These data demonstrate that we are able to discern complex transcriptional regulation patterns that occur over a 28-day period post-injury. This uniform performance demonstrates the potential of this device for on-site and near-site clinical sample genomics.
Figure 4.
Summary of the microarray results for a subset of the clinical samples from Figure 3. For the preliminary analysis shown here, we chose transcripts with a statistical significance of ≤ 0.001 (Q-value) corresponding to 8719 genes. (a) Unsupervised K-means clustering of these 8719 genes identified from the 187 microarrays in the time-course clinical data leads to five distinct clusters (from top to bottom): (1) Early up-regulation with resolution; (2) late up-regulation with a peak signal at 7–21 days; (3) Early down-regulation with resolution at 14–21 days; (4) Early down-regulation without recovery; and (5) late down-regulation without recovery. (b) Bar graph of the ten most statistically significant up-regulated pathways (red) and down-regulated pathways from the genes in (a).
Discussion
To date, several key genomic and proteomic studies have focused on aspects of neutrophil biology, such as development 22, apoptosis23, and response to pathogens24,25. Of these basic studies, our ex vivo stimulation data compare well with previous microarray data looking at LPS stimulation of enriched neutrophils 24–26. We identify all of the genes in the study by Fessler et al. and Zhang et al., with 87% of the relative change in expression as with our study. We see 78% of the genes in the study by Malcom et al., with 84% of the genes changing in the same direction. There is another microarray study by Kobayashi et al. that examined the response of purified neutrophils to GM-CSF at 18 hours 27. Our GM+I data identifies 99.9% of the gene transcripts in this study, with 65% of the genes showing the same trend in relative gene expression. These are remarkable observations given that we are using different microarray platforms, different experimental conditions, and different cell numbers. This verifies the expression changes that we observe are originating from neutrophil signals, and cross-signals from possible contaminants are generally small if not absent 16.
Further, no statistically significant difference was seen when comparing gene expression of neutrophils derived from different isolation methodologies, suggesting that minimal bias is introduced with the microfluidic isolation method. We can, however, measure large, statistically significant gene responses between model adaptive and innate stimuli in a small population of cells, which is important for downstream “-omics” analyses.
To our knowledge, the genomic data from neutrophils isolated from trauma patients using the microfluidic device described in this paper represents the single largest microarray study of neutrophils published to date, both in terms of numbers of subjects (26 patients, 10 controls) and number of microarrays (187). Pathway analysis shows that there is a general increase in metabolic pathways for this patient subset, while there is a general down-regulation of cell-death and chemokine signaling pathways (Fig. 4b). Current analysis is ongoing to determine the biological significance of these results and the correlation of gene expression with clinical trajectories over time.
While microarray technology for gene expression is well established, mass spectrometry based proteomics is being rapidly developed to quantitatively examine a large number of proteins in cells28, providing important information on post-transcriptional and post-translational regulation of cells 29. In our ex vivo stimulation experiment, we observed a strong concordance between gene expression and protein abundance changes for the 15 proteins that were significantly changed and were known to interact with LPS as identified through the Ingenuity Pathway Analysis system. Among these 15 genes, 12 genes show the same direction of changes between mRNA and protein abundances (Supplementary Table 10). This observation is consistent with recent report that mRNA and protein data often agree well on specific biological pathways 29. In contrast, we examined all the gene expression profiles for a set of proteins significantly perturbed by LPS, to identify changes of opposite directions at the gene vs. protein level (Supplementary Table 11). Several examples are provided. Nucleolin (NCL), a protein involved in the synthesis and maturation of ribosomes, is 2.4 fold increased at mRNA level but 29 times decreased at protein level, which corresponds to a trend observed previously 30, suggesting a selective degradation of this protein after LPS stimulation. In addition, the endogenous amount of lipocalin 2 (LCN2), a secretive protein important to antibacterial innate immune response 31 and known to be induced by LPS 32, was decreased by 3-fold, despite of a 6 fold increase of mRNA. The integrative analysis of genes and proteins of neutrophils isolated from patient samples using this microfluidics device will likely provide unique insights into the regulation of protein expression in the clinical setting.
In this article, we described the design and application of a new microfluidic device that rapidly and reproducibly separates neutrophils from whole blood for both microarray and mass spectrometry based proteomic analyses. We demonstrated the scalability of the device, its applicability in a clinical setting, and its versatility in processing samples for multiple downstream analyses.
This neutrophil separation device should be useful for monitoring diseases which are in some part characterized by inflammation, and will enable new patient populations where blood sampling volumes are limited (e.g. neonates and small children). With minor modifications this device can also be used to isolate other cell subpopulations as well as capture cells from other clinical samples, such as bronchoalveolar lavage, urine, and cerebrospinal fluid. As a result, this device should open up new applications of clinical genomics to disease and therapeutic monitoring.
Methods
All studies involving human samples were approved by the appropriate human use committees at all institutions within the NIH-funded “Inflammation and Host Response to Injury” Large Scale Collaborative Research Program. Patient enrollment criteria and patient demographics are given in the supplementary information. Unless otherwise stated, blood was collected into EDTA Vacutainer collection tubes (Becton Dickinson). All samples were run on the microfluidic device within one hour of blood draw.
Microfluidic device design and fabrication
Microfluidic devices in this study were fabricated following standard rapid-prototyping methods 33. Devices were functionalized with antibody (AbD Serotec CB66b, clone 80H3) in batches of ~100, and an internal quality control program was put in place for devices used for clinical sampling. Blood was pumped through the devices using a syringe pump (Harvard Apparatus), followed by a washing step using nuclease-free phosphate-buffered saline (PBS) (Ambion). Cell purity was assessed en bloc by immunofluorescent staining and Wright-Giemsa staining. Cell lysis for either genomics or proteomics was performed in situ. Guanadinium isothiocyanate buffer and buffered 2,2,2-trifluoroethanol were the lytic reagents for genomics and proteomics, respectively.
Ficoll-dextran isolation of neutrophils
Peripheral blood neutrophils were isolated by a modified Ficoll-dextran gradient as previously described by Nauseef et al7. Briefly, 4 mL whole blood was diluted with PBS with 2% fetal calf serum & 100 U ml−1 polymixin B, layered on top of Ficoll Paque Plus (Amersham Pharmacia Biotech) and centrifuged at 500 x g for 20 minutes at room temperature. The subsequent neutrophil/erythrocyte suspension was mixed with 6% dextran solution and allowed to sediment in a CO2 incubator at 37°C for 30 minutes. The remaining erythrocytes were lysed with ammonium chloride buffer (Qiagen). Following PBS wash, the neutrophil pellet was lysed in RLT buffer (Qiagen) and was stored at −80°C.
Ex vivo stimulation
Freshly drawn peripheral blood was anti-coagulated with sodium heparin and was directly processed for cell isolation by microfluidics or stimulated with LPS (50 ng ml−1) or GM-CSF (20 ng ml−1) and IFN-γ (100 IU ml−1) for 16 hours on a rocker at 37 °C in a 5% CO2 atmosphere before cell isolation. Leukocytes were also isolated using density gradient techniques from both unstimulated and stimulated samples and then tested for expression of surface HLA-DR and intra-cellular CCL20 in CD66b-identified PMNs by flow cytometry. Data are expressed as median fluorescence intensity.
Genomic analysis
RNA extraction followed a modified commercial protocol (QIAGEN RNeasy Plus) yielding purified total RNA that was analyzed on an Agilent Bioanalyzer 2100 system. cDNA was synthesized using Ovation BiotinRNA Amplification and Labeling System (NuGEN Technologies) from 20 ng oftotal RNA as starting material. For the ex vivo experiments, the labeled cDNA was hybridizedonto GeneChip™ Human Genome U133 Plus 2.0 Arrays (Affymetrix), while for the clinical study, the labeled cDNA was hybridized onto a custom-designed 6.9 million feature Affymetrix human exon-junction array with on average 100+ unique probes targeting 34,834 human genes. Chips were washed and scanned as recommended by the Ovation System User Guide (version 1.0).
Low level analysis was performed using dChip using the perfect match only option 34. Significance analysis of microarrays35 was applied on the basis of 1,000 permutations, to identify probesets significantly perturbed by LPS and GM+I (with false discovery rate (FDR) of < 0.01), gene expression significantly different between isolation methods (FDR < 0.05), and genes significantly changed by trauma injury over the 28 day time course (FDR < 0.001) as described previously5. Ingenuity Pathways Analysis was used to identify pathways and functional modules 5.
Proteomic Analysis
Enriched neutrophil lysates were digested using trypsin and aliquots of peptide samples were analyzed by reversed phase capillary LC-MS and –MS/MS using a LTQ-Orbitrap instrument (Thermo Scientific). The LC-MS datasets were analyzed using a custom software package36. Peptides were identified based on the accurate mass and time (AMT) tag strategy against a neutrophil AMT tag database pre-established by extensive LC-MS/MS profiling37 with a false discovery adjusted probability for peptide identifications < 0.0538. The peptide MS intensity values for each feature, was normalized, re-scaled and rolled up to protein level abundance using the tool DAnTE39. ANOVA was used to compare proteins under different conditions and adjusted for FDR < 0.05.
Supplementary Material
Acknowledgments
We thank O. Hurtado, K. Eken, K. Richter, and A. Gupta for microfabrication support. We thank C. Vanderburg for help with nucleic acid analysis and use of the Harvard NeuroDiscovery Ceneter Agilent Bioanalyzer 2100. We thank the UF technical staff (A. Abouhamze, C. Tannahill, and R. Ungaro) for managing clinical implementation of the microfluidic devices. K.T.K. was supported with a US National Institutes of Health (NIH) training grant T32 GM-007035-32. These studies were supported by the US NIH, “Inflammation and the Host Response to Injury Large Scale Collaborative Project”, U54 GM-062119, and BioMEMS Resource Center, P41 EB-002503, and Proteomics Research Resource for Integrative Biology, RR018522. The proteomics work was performed in the Environmental Molecular Sciences Laboratory, a U.S. Department of Energy (DOE) Office of Biological and Environmental Research national scientific user facility on the PNNL campus. PNNL is multi-program national laboratory operated by Battelle for the DOE under Contract No. DE-AC05-76RLO 1830.
The magnitude of the clinical and genomic and proteomic data reported here required the efforts of many individuals at participating institutions. In particular, we wish to acknowledge the supportive research environment created and sustained by the participants in the Glue Grant Program:
Henry V. Baker, Ph.D.1, Ulysses G.J. Balis, M.D.2, Timothy R. Billiar, M.D.3, Steven E. Calvano, Ph.D.5, Irshad H. Chaudry, Ph.D.6, J. Perren Cobb, M.D.4, Joseph Cuschieri, M.D.7, Celeste C. Finnerty, Ph.D.9, Richard L. Gamelli10, M.D., Nicole S. Gibran, M.D.7, Brian G. Harbrecht, M.D.11, Douglas L. Hayden, M.A.12, Laura Hennessy, R.N.7, David N. Herndon, M.D.9, Marc G. Jeschke, M.D., Ph.D. 9, Jeffrey L. Johnson, M.D.13, Matthew B. Klein, M.D.7, James A. Lederer, Ph.D.14, Stephen F. Lowry, M.D.2, Ronald V. Maier, M.D.7, John A. Mannick, M.D.14, Philip H. Mason, Ph.D.12, Grace P. McDonald-Smith, M.Ed.12, Joseph P. Minei, M.D.15, Ernest E. Moore, M.D.13, Avery B. Nathens, M.D., Ph.D., M.P.H.16, Grant E. O’Keefe, M.D., M.P.H.7, Laurence G. Rahme, Ph.D.12, Daniel G. Remick, M.D.17, David A. Schoenfeld, Ph.D.12, Michael B. Shapiro, M.D.19, Geoffrey M. Silver, M.D.10, Jason Sperry, M.D., Ph.D.3, John D. Storey, Ph.D.20, Robert Tibshirani, Ph.D.8, H. Shaw Warren, M.D.12, Michael A. West, M.D., PhD.18, and Bram Wispelwey, M.S.12
1 University of Florida, Gainesville, FL 32610, 2 University of Michigan School of Medicine, Ann Arbor, MI 48109, 3 University of Pittsburgh Medical Center, Pittsburgh, PA 15213, 4 Washington University School of Medicine, St. Louis, MO 63110, 5 University of Medicine and Dentistry of New Jersey, New Brunswick, NJ 08903, 6 University of Alabama Medical School, Birmingham, AL 35294, 7 University of Washington, Seattle, WA 98195, 8 Stanford University, Palo Alto, CA 94304, 9 University of Texas Medical Branch, Galveston, TX 77550, 10 Loyola University School of Medicine, Maywood, IL 60153, 11 University of Louisville, Louisville, KY 40292, 12 Massachusetts General Hospital, Boston, MA 02114,13 University of Colorado Health Sciences Center, Denver, CO 80204, 14 Brigham & Women’s Hospital, Boston, MA 02115, 15 University of Texas Southwestern Medical School, Dallas, TX 75390, 16 St. Michael’s Hospital, Toronto, Ontario, CA M5B 1W8, 17 Boston University School of Medicine, Boston, MA 02118, 18 San Francisco General Hospital, San Francisco, CA 04110, 19 Northwestern University, Chicago, IL 60611, 20 Princeton University, Princeton, NJ 08544
Footnotes
Author Contributions
KTK performed and analyzed experiments. WX and WJQ preformed microarray and proteomics analyses. KTK, WX, JW, MNM, WXu, AR, EAW, LLM, DI, BHB, RWD, MT designed genomic experiments. KTK, WJQ, DGC, RDS designed proteomic experiments. JG, SPF, AER, RGT, aided in clinical sample studies at MGH. KTK, CMG, LMM, WX, MNM, JW, WJQ, BOP, DGC, AER, PEB, MT designed, conducted, and analyzed the ex vivo stimulation experiment. KTK, CMG, WX, MNM, LLM wrote the manuscript. All authors contributed to the final editing of the manuscript.
References
- 1.Nathan C. Neutrophils and immunity: challenges and opportunities. Nat Rev Immunol. 2006;6:173–82. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- 2.Cassatella MA, Gasperini S, Russo MP. Cytokine expression and release by neutrophils. Ann N Y Acad Sci. 1997;832:233–42. doi: 10.1111/j.1749-6632.1997.tb46251.x. [DOI] [PubMed] [Google Scholar]
- 3.McDonald PP, Bald A, Cassatella MA. Activation of the NF-kappaB pathway by inflammatory stimuli in human neutrophils. Blood. 1997;89:3421–33. [PubMed] [Google Scholar]
- 4.Burczynski ME, Dorner AJ. Transcriptional profiling of peripheral blood cells in clinical pharmacogenomic studies. Pharmacogenomics. 2006;7:187–202. doi: 10.2217/14622416.7.2.187. [DOI] [PubMed] [Google Scholar]
- 5.Calvano SE, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–7. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 6.Laudanski K, et al. Cell-specific expression and pathway analyses reveal alterations in trauma-related human T cell and monocyte pathways. Proc Natl Acad Sci U S A. 2006;103:15564–9. doi: 10.1073/pnas.0607028103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nauseef WM. Isolation of human neutrophils from venous blood. Methods Mol Biol. 2007;412:15–20. doi: 10.1007/978-1-59745-467-4_2. [DOI] [PubMed] [Google Scholar]
- 8.Elghetany MT, Davis BH. Impact of preanalytical variables on granulocytic surface antigen expression: a review. Cytometry B Clin Cytom. 2005;65:1–5. doi: 10.1002/cyto.b.20051. [DOI] [PubMed] [Google Scholar]
- 9.Cassatella MA. The production of cytokines by polymorphonuclear neutrophils. Immunol Today. 1995;16:21–6. doi: 10.1016/0167-5699(95)80066-2. [DOI] [PubMed] [Google Scholar]
- 10.Cheng X, et al. A microchip approach for practical label-free CD4+ T-cell counting of HIV-infected subjects in resource-poor settings. J Acquir Immune Defic Syndr. 2007;45:257–61. doi: 10.1097/QAI.0b013e3180500303. [DOI] [PubMed] [Google Scholar]
- 11.Nagrath S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–9. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyons PA, et al. Microarray analysis of human leucocyte subsets: the advantages of positive selection and rapid purification. BMC Genomics. 2007;8:64. doi: 10.1186/1471-2164-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, et al. Development and evaluation of a micro- and nanoscale proteomic sample preparation method. J Proteome Res. 2005;4:2397–403. doi: 10.1021/pr050160f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeForge LE, Kenney JS, Jones ML, Warren JS, Remick DG. Biphasic Production of IL-8 in Lipopolysaccharide (LPS)-Stimulated Human Whole Blood. J Immunol. 1992;148:2133–2141. [PubMed] [Google Scholar]
- 15.Pelletier M, et al. Evidence for a cross-talk between human neutrophils and Th17 cells. Blood. 2009 doi: 10.1182/blood-2009-04-216085. [DOI] [PubMed] [Google Scholar]
- 16.De AK, et al. Selective activation of peripheral blood T cell subsets by endotoxin infusion in healthy human subjects corresponds to differential chemokine activation. J Immunol. 2005;175:6155–62. doi: 10.4049/jimmunol.175.9.6155. [DOI] [PubMed] [Google Scholar]
- 17.Gosselin EJ, Wardwell K, Rigby WF, Guyre PM. Induction of MHC class II on human polymorphonuclear neutrophils by granulocyte/macrophage colony-stimulating factor, IFN-gamma, and IL-3. J Immunol. 1993;151:1482–90. [PubMed] [Google Scholar]
- 18.Cobb JP, et al. Application of genome-wide expression analysis to human health and disease. Proc Natl Acad Sci U S A. 2005;102:4801–6. doi: 10.1073/pnas.0409768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh R, et al. Microarray-based comparison of three amplification methods for nanogram amounts of total RNA. Am J Physiol Cell Physiol. 2005;288:C1179–89. doi: 10.1152/ajpcell.00258.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotz K, Cheng X, Toner M. Cell capture using a microfluidic device. J Vis Exp. 2007;320 doi: 10.3791/320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Covert MW, Leung TH, Gaston JE, Baltimore D. Achieving stability of lipopolysaccharide-induced NF-kappaB activation. Science. 2005;309:1854–7. doi: 10.1126/science.1112304. [DOI] [PubMed] [Google Scholar]
- 22.Theilgaard-Monch K, et al. The transcriptional program of terminal granulocytic differentiation. Blood. 2005;105:1785–96. doi: 10.1182/blood-2004-08-3346. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi SD, Voyich JM, Braughton KR, DeLeo FR. Down-regulation of proinflammatory capacity during apoptosis in human polymorphonuclear leukocytes. J Immunol. 2003;170:3357–68. doi: 10.4049/jimmunol.170.6.3357. [DOI] [PubMed] [Google Scholar]
- 24.Fessler MB, Malcolm KC, Duncan MW, Worthen GS. A genomic and proteomic analysis of activation of the human neutrophil by lipopolysaccharide and its mediation by p38 mitogen-activated protein kinase. J Biol Chem. 2002;277:31291–302. doi: 10.1074/jbc.M200755200. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, et al. Gene expression in mature neutrophils: early responses to inflammatory stimuli. J Leukoc Biol. 2004;75:358–72. doi: 10.1189/jlb.0903412. [DOI] [PubMed] [Google Scholar]
- 26.Malcolm KC, Arndt PG, Manos EJ, Jones DA, Worthen GS. Microarray analysis of lipopolysaccharide-treated human neutrophils. Am J Physiol Lung Cell Mol Physiol. 2003;284:L663–70. doi: 10.1152/ajplung.00094.2002. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi SD, Voyich JM, Whitney AR, DeLeo FR. Spontaneous neutrophil apoptosis and regulation of cell survival by granulocyte macrophage-colony stimulating factor. J Leukoc Biol. 2005;78:1408–18. doi: 10.1189/jlb.0605289. [DOI] [PubMed] [Google Scholar]
- 28.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1:252–62. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 29.de Godoy LM, et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–4. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X, et al. Proteomic analysis of macrophages stimulated by lipopolysaccharide: Lipopolysaccharide inhibits the cleavage of nucleophosmin. Electrophoresis. 2006;27:1659–68. doi: 10.1002/elps.200500736. [DOI] [PubMed] [Google Scholar]
- 31.Berger T, et al. Lipocalin 2-deficient mice exhibit increased sensitivity to Escherichia coli infection but not to ischemia-reperfusion injury. Proc Natl Acad Sci U S A. 2006;103:1834–9. doi: 10.1073/pnas.0510847103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meheus LA, et al. Identification by microsequencing of lipopolysaccharide-induced proteins secreted by mouse macrophages. J Immunol. 1993;151:1535–47. [PubMed] [Google Scholar]
- 33.Kotz K, Cheng X, Toner M. PDMS device fabrication and surface modification. J Vis Exp. 2007;319 doi: 10.3791/319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–6. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zimmer JS, Monroe ME, Qian WJ, Smith RD. Advances in proteomics data analysis and display using an accurate mass and time tag approach. Mass Spectrom Rev. 2006;25:450–82. doi: 10.1002/mas.20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qian WJ, Jacobs JM, Liu T, Camp DG, 2nd, Smith RD. Advances and challenges in liquid chromatography-mass spectrometry-based proteomics profiling for clinical applications. Mol Cell Proteomics. 2006;5:1727–44. doi: 10.1074/mcp.M600162-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petyuk VA, et al. Spatial mapping of protein abundances in the mouse brain by voxelation integrated with high-throughput liquid chromatography-mass spectrometry. Genome Res. 2007;17:328–36. doi: 10.1101/gr.5799207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polpitiya AD, et al. DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics. 2008;24:1556–8. doi: 10.1093/bioinformatics/btn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.