Abstract
The ICP34.5 protein of herpes simplex virus type 1 is a neurovirulence factor that plays critical roles in viral replication and anti-host responses. One of its functions is to recruit protein phosphatase 1 (PP1) that leads to the dephosphorylation of the α subunit of translation initiation factor eIF2 (eIF2α), which is inactivated by infection-induced phosphorylation. As PP1 is a protein phosphatase with a wide range of substrates, the question remains to be answered how ICP34.5 directs PP1 to specifically dephosphorylate eIF2α. Here we report that ICP34.5 not only binds PP1 but also associates with eIF2α by in vitro and in vivo assays. The binding site of eIF2α is identified at amino acids 233–248 of ICP34.5, which falls in the highly homologous region with human gene growth arrest and DNA damage 34. The interaction between ICP34.5 and eIF2α is independent of the phosphorylation status of eIF2α at serine 51. Deletion mutation of this region results in the failure of dephosphorylation of eIF2α by PP1 and, consequently, interrupts viral protein synthesis and replication. Our data illustrated that the binding between viral protein ICP34.5 and the host eIF2α is crucial for the specific dephosphorylation of eIF2α by PP1. We propose that herpes simplex virus protein ICP34.5 bridges PP1 and eIF2α via their binding motifs and thereby facilitates the protein synthesis and viral replication.
Keywords: Herpesvirus, Protein Synthesis, Protein-Protein Interactions, Translation Initiation Factors, Translation Regulation, ICP34.5, Dephosphorylation, eIF2α, Protein Phosphatase 1
Introduction
Viral infection can activate a series of host immune responses. One of the essential responses is the interruption of the viral protein synthesis, which is executed by double-stranded RNA-dependent protein kinase (PKR) (1). PKR is induced and activated upon viral infection and leads to the phosphorylation of the α subunit of translation initiation factor eIF2 (eIF2α)2 at serine 51 (2). The phosphorylation of eIF2α globally inhibits the synthesis of viral proteins and cellular proteins (3), thus halting the viral replication.
Many viruses have evolved strategies to counteract the antiviral response of PKR (4). For example, PKR activity can be inhibited by HIV-encoded Tar (5), hepatitis C virus NS5A (6), influenza virus NS1 (7), and so forth. In addition to affecting PKR activation as mentioned above, HSV-1 adopted mechanisms not only inhibiting PKR by Us11 (8) but also reversing the biochemical reaction catalyzed by PKR with its neurovirulent factor ICP34.5 (9).
ICP34.5 is encoded by the γ134.5 gene of HSV-1 and HSV-2. The HSV-1(F) ICP34.5 consists of 263 amino acids and can be divided into three domains: an amino-terminal domain, a linker region of ATP (Ala-Thr-Pro) repeats, and a carboxyl-terminal domain (9, 10). The function of the amino-terminal domain is implicated in the control of TBK1-mediated signaling (11) and also related to autophagy (12). The linker region, with a varying number of the Ala-Thr-Pro repeats, may affect the protein localization (13). The carboxyl-terminal domain is a stretch of 84 amino acids containing a consensus binding motif (R/KVXF) for protein phosphatase 1 (PP1) followed by an Ala-Arg-rich motif and is highly homologous to the carboxyl-terminal region of mammalian growth arrest and DNA damage protein 34 (GADD34) (10, 14–17). Substitution of the C-terminal sequences from mouse GADD34 homologue MyD116 rescues ICP34.5 function in cells infected with mutant HSV-1 (18), demonstrating that the C-terminal regions of ICP34.5 and GADD34 are functionally interchangeable regarding the dephosphorylation of eIF2α.
Previous studies illustrated that the carboxyl terminus of ICP34.5 is essential in the dephosphorylation of eIF2α (9). Mutation at PP1 binding motif of ICP34.5 completely abolished the dephosphorylation of eIF2α in vitro and rendered the reduced replication of virus (19), whereas the truncation of 10 amino acid residues from the C-terminal of ICP34.5 or substitution of the conserved Arg residues to acidic residues also elicited the dephosphorylation and protein synthesis (20). Thus, the PP1 binding motif of ICP34.5 is prerequisite but not sufficient for the dephosphorylation of eIF2α. It remains to be investigated how the ICP34.5-mediated dephosphorylation is achieved. In this study, we demonstrated the association of ICP34.5 with cellular eIF2α in vitro and in vivo. With the bimolecular fluorescence complementation (BiFC) approach, we showed that the complex formation of PP1 and eIF2α occurs only in the presence of ICP34.5. This interaction between ICP34.5 and eIF2α is independent of the phosphorylation status of eIF2α. Furthermore, we identified that the carboxyl terminus (233 to 248 aa) of ICP34.5 is responsible for the binding with eIF2α and that ICP34.5 with a deletion in the 233–248 aa region failed to rescue the eIF2α dephosphorylation and viral replication. Taken together, this study suggests that ICP34.5 bridges PP1 and eIF2α via their respective binding motifs, thereby facilitating the specific dephosphorylation of eIF2α and the protein synthesis. This study may lead to a common mechanism in controlling protein synthesis by dephosphorylation of eIF2α, which is inactivated by other PKR-like kinases.
EXPERIMENTAL PROCEDURES
Cells and Viruses
Vero, HeLa, and HEK293T cells were from the American Type Culture Collection. Cells were propagated in DMEM supplemented with 10% fetal bovine serum for HeLa and HEK293T cells or 10% newborn calf serum for Vero cells. HSV-1(F) is a prototype HSV-1 strain, and R3616 is a mutation virus with a 1-kb fragment including the coding region of the γ134.5 gene deleted (21).
Plasmids
Plasmids pcDNA3-YN, pcDNA3-YC, pcDNA3-YN-PP1, pcDNA3-YC-ICP34.5 (WT), pcDNA3-YC-ICP34.5 (V193E,F195L), pHIV-ICP34.5 (WT), and pHIV-ICP34.5 (V193E, F195L) have been described previously (20). For the BiFC assay, the expression vectors of ICP34.5 variants WT-N, WT-C, ΔC15, ΔC30, and ΔC45 were obtained by PCR and cloned into the EcoRI and XhoI sites of pcDNA3, with the nucleotide encoding the carboxyl-terminal fragment containing residues 155–238 (YC) of YFP fused to these mutants, respectively. To construct pcDNA3-YN-eIF2α and pcDNA3-YC-eIF2α, the nucleotide encoding residues 1–154 (YN) of YFP or 155–238 (YC) of YFP was fused to the eIF2α coding region. To obtain pcDNA3-YN-pDlim2, the amino-terminal fragment containing residues 1–154 (YN) of YFP was fused to the pDlim2 coding region. The ICP34.5 (Δ233–248) or eIF2α mutants were obtained by overlap PCR and were cloned into the EcoRI and XhoI sites of pcDNA3 with a YC or YN fragment. For the coimmunoprecipitation assay, YN-pDlim2, YN-eIF2α, and YN-PP1 in pcDNA3 were isolated by BamHI and XhoI digestion and then cloned into the BamHI and XhoI sites of the pCMVtag2B vector. GFP-PP1 and GFP-ICP34.5 (WT) were obtained by PCR and cloned into the XhoI and BamHI sites of the pEGFP-C1 vector. To construct pHIV-ICP34.5 (Δ233–248), ICP34.5 (Δ233–248) was cloned into the EcoRI and XhoI sites of pHIV-vFlip. Nucleotide sequences of the constructions were confirmed by DNA sequencing.
BiFC Assay
BiFC analysis enables direct visualization of protein interactions in living cells (22, 23). The YN-eIF2α and YC-ICP34.5 variants were cotransfected into HeLa cells using polyethyleneimine. At 36-hour post-transfection, cells were cultured at room temperature for 1–3 h for fluorophore maturation. The fluorescence was then examined by fluorescence microscopy, and the cells were subjected to FACS analysis.
Immunoblotting and Coimmunoprecipitation Analysis
To detect the protein expression, cells were harvested, washed with phosphate-buffered saline, and lysed in ice-cold radioimmune precipitation assay buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 5 mm EDTA, 1 mm phenylmethylsulonyl fluoride, 1 mm benzamidine, 2 μg/ml leupeptin, 1 μg/ml pepstatin, 1 mm Na3VO4, 1 mm NaF). After centrifugation, supernatants were subjected to electrophoresis and transferred to PVDF membranes, blocked with 5% nonfat milk and then probed with antibodies against ICP34.5, eIF2α (Cell Signaling Technology, Inc.), phosphorylated eIF2α Ser-51 (Sigma), HSV-1 (Dako, Inc.), or α-tubulin (Sigma). To examine the protein interactions, HEK293T or HeLa cells were transfected with the FLAG-tagged eIF2α and ICP34.5 variants. At 24–36 h post-transfection, as indicated, HEK293T cells were harvested and lysed in 50 mm Tris-HCl (pH 7.4) containing 150 mm NaCl, 5 mm EDTA, 1.0% Triton X-100, and protease inhibitor mixtures. Lysates were incubated, respectively, with anti-FLAG antibody (Sigma) or anti-ICP34.5 antibody plus protein A/G-agarose beads (Santa Cruz Biotechnology) for 3 h at 4 °C. The beads were then washed four times with WASH buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 5 mm EDTA, 0.1% Triton X-100, protease inhibitor mixtures), and protein A/G-agarose beads were subjected to electrophoresis and immunoblotting analysis. Furthermore, to examine the interaction between ICP34.5 and endogenous eIF2α, HEK293T cells were transfected with GFP-tagged PP1 or ICP34.5. At 36 h post-transfection, cells were harvested and lysed in 50 mm Tris-HCl (pH 7.4) containing 150 mm NaCl, 5 mm EDTA, 1.0% Triton X-100, and protease inhibitor mixtures. Then lysates were incubated, respectively, with anti-GFP antibody (Abmart) or anti-eIF2α antibody (Cell Signaling Technology, Inc.) plus protein A/G-agarose beads (Santa Cruz Biotechnology) for 3 h at 4 °C. Then the beads were washed four times with WASH buffer, and protein A/G-agarose beads were subjected to electrophoresis and immunoblotting analysis. To detect the interaction between ICP34.5 encoded by virus and endogenous eIF2α, HeLa cells were infected with R3616 or HSV-1(F). At 18 h post-infection, cells were harvested and lysed in radioimmune precipitation assay buffer. Then lysates were incubated with anti-ICP34.5 antibody plus protein A/G-agarose beads for 2 h at 4 °C. The beads were then washed four times with WASH buffer, and the beads were subjected to electrophoresis and immunoblotting analysis.
Viral Infection Assays
HeLa cells were transfected with pHIV-GFP, pHIV-ICP34.5 (WT), pHIV-ICP34.5 (V193E, F195L), and pHIV-ICP34.5 (Δ233–248). To detect the viral protein synthesis and eIF2α dephosphorylation, at 24 h post-transfection, cells were infected with R3616 at 10 pfu per cell. At 24 h post-infection, cells were harvested to determine the viral protein synthesis and eIF2α dephosphorylation by Western blot analysis. To detect the viral replication, at 24-hour post transfection, cells were infected with R3616 at 0.05 pfu/cell Also at 24 h post-infection, virus yields were determined on Vero cells.
RESULTS
ICP34.5 Mediates the Association of PP1 and eIF2α
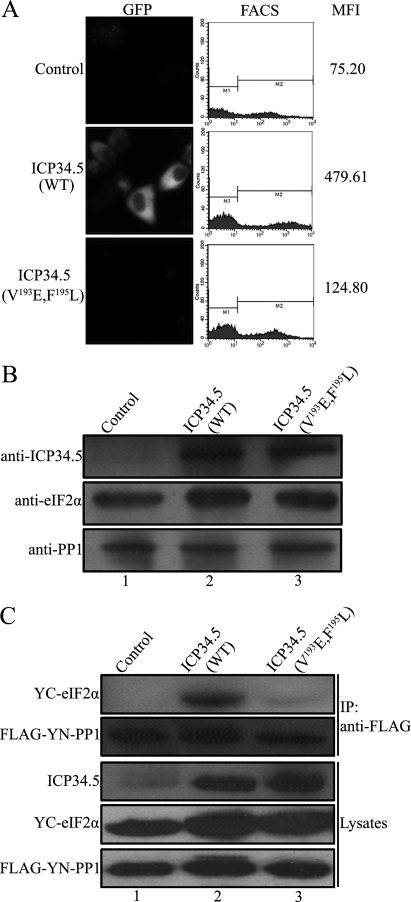
The substrate specificity of the PP1 catalytic subunit is normally determined by the regulatory subunit (24). ICP34.5 has been identified as a regulatory subunit of PP1 that contains a consensus PP1 binding motif (19). Mutation of this motif in ICP34.5 is functionally deleterious (25). Nevertheless, C-terminal mutations, although bearing an intact PP1 binding motif, failed to rescue the protein synthesis, suggesting that certain functional determinants exist in the C-terminal region (25). Because HSV-1 ICP34.5 plays a role as a PP1 regulatory subunit to determine the specific dephosphorylation of eIF2α, we examined the association of PP1 and eIF2α in the presence or absence of ICP34.5. We first investigated the interaction of PP1 and eIF2α by BiFC assay, an approach that allows the visualization of protein interactions in living cells (26). PP1 or eIF2α was fused to the YN or YC fragment of YFP, and an interaction between the two proteins facilitated association between the fragments to produce a bimolecular fluorescent complex (23). Cells were cotransfected with the constructs YN-PP1 and YC-eIF2α, along with plasmids encoding the wild-type ICP34.5 (WT) or ICP34.5 mutation (V193E, F195L) that failed to bind PP1, respectively, or a control plasmid. Then the cells were examined by fluorescence microscopy and subjected to flow cytometry analysis. The mean fluorescence intensity reflects the binding efficiency of the associated YN and YC fragments (23). As shown in Fig. 1A, high fluorescence was detected in the cells cotransfected with ICP34.5 (WT) (center panel), with a mean fluorescence intensity (MFI) of 479.61, reflecting a close contact between PP1 and eIF2α. In contrast, little fluorescence was seen in the presence of mutated ICP34.5 (V193E, F195L) (bottom panel) or the control plasmid, with much lower MFI scores, indicating a very rare interaction. The expression levels of the indicated proteins were monitored by Western blot analysis (Fig. 1B), validating the proteins in the BiFC analysis.
FIGURE 1.
ICP34.5 mediates the association of PP1 and eIF2α. A, fluorescence visualization and FACS analysis. HeLa cells were transfected with plasmids encoding YN-PP1 and YC-eIF2α, along with a plasmid encoding ICP34.5 (WT), ICP34.5 (V193E, F195L), or a control plasmid. Then cells were subjected to fluorescence microscopy (left column), and the FACS analysis profile of each transfectant was plotted in the center column, with the mean YFP fluorescence intensity (MFI) in the right column. B, the protein expression of each transfected component was verified by Western blot analysis with the indicated antibodies. C, coimmunoprecipitation (IP) of eIF2α with PP1. HEK293T cells were cotransfected with plasmids encoding FLAG-tagged YN-PP1 and YC-eIF2α together with pHIV-vFlip, pHIV-ICP34.5 (WT), or pHIV-ICP34.5 (V193E, F195L). At 48 h post-transfection, cell lysates were collected and incubated with anti-FLAG antibody and protein A/G-agarose for 2 h at 4 °C with gentle agitation. The immobilized proteins were resolved by Western blot analysis using antibodies against eIF2α and FLAG, respectively.
We confirmed the role of ICP34.5 in the interaction between PP1 and eIF2α by coimmunoprecipitation. Cells were cotransfected plasmids encoding Flag-tagged PP1 and eIF2α together with the control plasmid ICP34.5 (WT) or ICP34.5 (V193E, F195L), respectively, and PP1 were immunoprecipitated with anti-FLAG antibody. As shown in Fig. 1C, eIF2α was coimmunoprecipitated with PP1 in cell lysates containing wild-type ICP34.5 but not PP1 binding-defect ICP34.5 (V193E, F195L), even though the three components were expressed evenly as analyzed by Western blot analysis (three bottom panels). Collectively, the above in vivo and in vitro results indicate that PP1 associates with eIF2α only when ICP34.5 presents, and PP1 binding capability of ICP34.5 is essential in this process. Because PP1 has a variety of substrates, its specificity for eIF2α is to be achieved by ICP34.5. We thus hypothesized that ICP34.5 may bind to eIF2α so as to bring the substrate to PP1 specifically.
ICP34.5 Associates with eIF2α
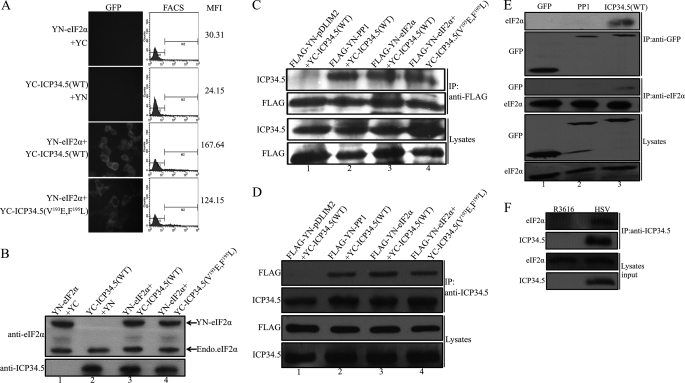
We next tested the association between ICP34.5 and eIF2α by BiFC assay. The nonspecific binding between the YN and YC fragments were excluded, as negative binding was observed in the combination of YN-eIF2α with YC or YC-ICP34.5 (WT) with YN (Fig. 2A, two top panels). The binding between ICP34.5 (WT) and eIF2α was positively indicated with an MFI score of 167.64 (Fig. 2A, third panel). Intriguingly, positive interaction was obtained in ICP34.5 (V193E, F195L) and eIF2α (Fig. 2A, lower panel), with MFI much higher than the two control groups, suggesting that the PP1 binding capability of ICP34.5 is not required for the interaction between eIF2α and ICP34.5. The expression level of YN-eIF2α is comparable with the endogenous eIF2α (Fig. 2B), thus the effect of endogenous protein in BiFC assay is negligible as reported (23).
FIGURE 2.
ICP34.5 associates with eIF2α. A, HeLa cells were cotransfected with pcDNA3-YN-eIF2α and pcDNA3-YC, pcDNA3-YC-ICP34.5 (WT) and pcDNA3-YN, pcDNA3-YN-eIF2α and pcDNA3-YC-ICP34.5 (WT), and pcDNA3-YN-eIF2α and pcDNA3-YC-ICP34.5 (V193E, F195L), respectively. At 36 h post-transfection, cells were subjected to fluorescence microscopy and FACS analysis. The FACS analysis profile of each transfectant was plotted in the center column, with the mean YFP fluorescence intensity (MFI) in the right column. The MFI scores indicate the interaction between the proteins attached to the N-terminal or the C-terminal of the YFP fragments, as described under “Experimental Procedures.” The protein expression levels in the BiFC assay were verified with anti-eIF2α and anti-ICP34.5 antibodies (B). C, coimmunoprecipitation (IP) of ICP34.5 with eIF2α. HEK293T cells were cotransfected with the indicated combinations: pTag2B-YN-PDlim2 and pcDNA3-YC-ICP34.5 (WT) as the negative control, pTag2B-YN-PP1 and pcDNA3-YC-ICP34.5 (WT) as the positive control, pTag2B-YN-eIF2α and pcDNA3-YC-ICP34.5 (WT), and pTag2B-YN-eIF2α and pcDNA3-YC-ICP34.5 (V193E, F195L). At 48 h post-transfection, cell lysates were prepared and incubated with anti-FLAG antibody and protein A/G-agarose for 2 h at 4 °C. Precipitated proteins were resolved by Western blot analysis using antibodies against ICP34.5 and FLAG, respectively (two top panels). Protein expression were also inspected in lysates by Western blot analysis (two bottom panels). On the other hand, coimmunoprecipitation with anti-ICP34.5 antibody to precipitate eIF2α was performed with the same combination of the binding partners, and the Western blot analyses are displayed in D. E, detection of the binding of endogenous eIF2α with ICP34.5. HEK293T cells were transfected with plasmids encoding GFP, GFP-tagged-PP1, or GFP-tagged ICP34.5 (WT), respectively. 36 h post-transfection, cell lysates were obtained and divided evenly into two potions. One of them was incubated with anti-eIF2α antibody; and the other was incubated with anti-GFP antibody to perform the immunoprecipitation as described in C. Immobilized proteins as well as cell lysates were resolved by Western blot analysis using antibodies as indicated. F. HeLa cells were infected with R3616 or HSV-1(F). At 18 h post-infection, cell lysates were prepared and incubated with anti-ICP34.5 antibody plus protein A/G-agarose beads for 2 h at 4 °C. Samples from cell lysates and immunoprecipitates were analyzed with antibodies against eIF2α and ICP34.5, respectively.
The coimmunoprecipitation assay was also carried out to detect the interaction between ICP34.5 and eIF2α. The cells were cotransfected with combinations of plasmids encoding eIF2α and ICP34.5 or its variant (V193E, F195L) or ICP34.5 with an unrelated protein as a negative control (Fig. 2C, lane 1) or PP1 plus ICP34.5 as a positive control (lane 2). When PP1 or eIF2α was precipitated with anti-FLAG antibodies, although ICP34.5 (WT) was expectedly pulled down with PP1 (Fig. 2C, lane 2), ICP34.5 WT and the mutation (V193E, F195L) were both coprecipitated with eIF2α (lane 3 and 4). All proteins were expressed comparably in each transfectant, as shown in Fig. 2C (lysates panels). Alternatively, when ICP34.5 was used as an immobilized phase, the FLAG-tagged eIF2α was coimmunoprecipitated and detected in the pellet (Fig. 2D, lanes 3 and 4), with similar negative and positive controls in lanes 1 and 2.
To exclude the effect of the ectopic expression of eIF2α as in the above assays, we confirmed the binding ICP34.5 with endogenous eIF2α by coimmunoprecipitation. In this experiment, we used plasmids encoding fusion proteins GFP-PP1 and GFP-ICP34.5 and then immunoprecipitated them with anti-GFP antibody. As show in Fig. 2E, the endogenous eIF2α was detected only in lane 3 with ICP34.5 but not in the GFP control (lane 1) or GFP-PP1 (lane 2) in the coimmunoprecipitation sample by anti-GFP antibody (Fig. 2E, two top panels). On the other hand, detectable ICP34.5 was coimmunoprecipitated with the endogenous eIF2α by anti-eIF2α antibody (Fig. 2E, two center rows, lane 3).
We further attempted to detect this interaction in live virus-infected cells. As shown in Fig. 2F, coimmunoprecipitation with ICP34.5 antibody resolved a component of eIF2α in the HSV-infected cell lysate (right lane), whereas eIF2α from the infected HSV cell lysate was detected with the immobilized ICP34.5 (right lane) but not in the R3616-infected sample (left lane). All protein expression levels were shown to be comparable (Fig. 2F, lower panels).
Together, the above coimmunoprecipitation data witnessed the interaction between eIF2α and ICP34.5 seen by BiFC assay (Fig. 2A) and demonstrated the molecular tangency of eIF2α with viral protein ICP34.5. It is further revealed that the PP1 binding motif of ICP34.5 is not mandatory to its association with eIF2α (Fig. 2, A, C, and D), suggesting that HSV ICP34.5 contains two independent motifs, one for binding PP1 (RVxF), and the other for eIF2α.
The Phosphorylation State of eIF2α Does Not Affect Its Association with ICP34.5
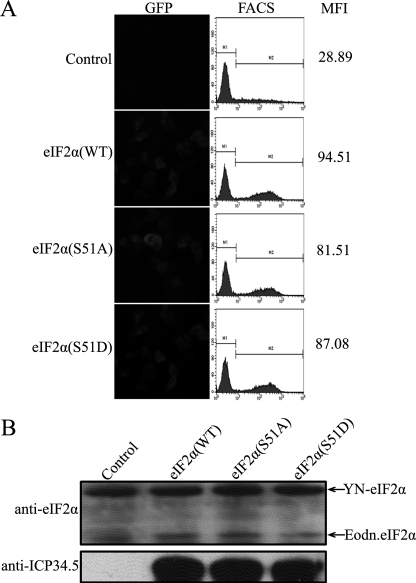
As illustrated previously, eIF2α is phosphorylated upon viral infection by PKR, and ICP34.5 mediated the dephosphorylation of eIF2α. We then asked whether ICP34.5 is selective for the phosphorylated eIF2α. Serine 51 is the phosphorylation site of eIF2α by PKR or other PKR-like kinases to halt protein synthesis. We thus substituted Ser 51 with an aspartic acid (S51D) or an alanine (S51A) residue to mimic the phosphorylated eIF2α or dephosphorylated eIF2α. In the BiFC assay, except for the control sample (Fig. 3A, top panel), ICP34.5 exhibits similar binding features with wild-type eIF2α (Fig. 3A, second panel from top), S51A (third panel from top), and S51D (bottom panel). The protein expression of ICP34.5 and eIF2α variants was comparable, as shown in Fig. 3B. The FACS analyses and MFI scores of samples with the eIF2α wild type, S51A, and S51D (Fig. 3A, 94.51, 81.51, and 87.08, respectively) are much higher than the control (28.89), indicating that ICP34.5 associates with eIF2α regardless of the phosphorylation state of eIF2α at Ser-51.
FIGURE 3.
Effect of eIF2α phosphorylation on its association with ICP34.5. A, HeLa cells were transfected with pcDNA3-YC and pcDNA3-YN-eIF2α as a control or pcDNA3-YC-ICP34.5 (WT) together with plasmids encoding YN fused to eIF2α (WT), eIF2α (S51A), or eIF2α (S51D), respectively. At 36 h post-transfection, cells were subjected to fluorescence microscopy and FACS analysis, and a FACS profile of each transfectant was plotted in the center column, with the mean YFP fluorescence intensity (MFI) in the right column. B, the protein expression levels in the BiFC assay were verified with anti-eIF2α and anti-ICP34.5 antibodies.
Identification of the eIF2α Binding Region of ICP34.5
As described above, ICP34.5 associates eIF2α without compromising its PP1 binding motif. There must be such a region responsible for binding eIF2α. Indeed, the dephosphorylation of eIF2α is interrupted even when the PP1 binding feature is intact (20, 25). We therefore set up to identify the region of ICP34.5 responsible for eIF2α interaction.
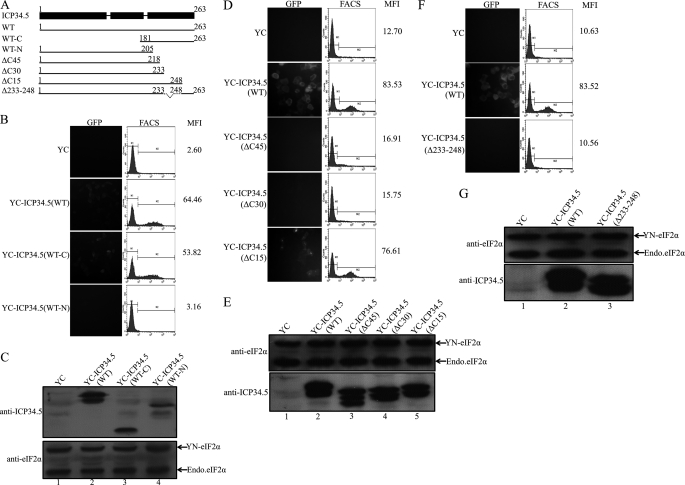
As mentioned previously, ICP34.5 is divided into three domains: an amino-terminal domain, 10 repeats of three amino acids (Ala-Thr-Asp), and a carboxyl-terminal domain. We first constructed two truncation mutants: WT-N (a deletion of C-terminal amino acids 205–263) and WT-C (a deletion of N-terminal aa 1–180), as depicted in Fig. 4A. Then we detected the binding ability of mutants with eIF2α in a BiFC assay. As shown in Fig. 4B, we can see that there is little interaction between eIF2α and the ICP34.5WT-N mutant, with a score of 3.16. However, there is a distinct binding between eIF2α and the ICP34.5WT-C mutant, with a score of 53.82. This result clearly excludes the involvement of the N-terminal peptide and addressed that the C-terminal stretch of peptides contains the eIF2α binding domain.
FIGURE 4.
Identification of the ICP34.5 amino acids responsible for the association with eIF2α. A, schematic diagrams of HSV-1(F) ICP34.5 and its mutations. ICP34.5 consists of an amino-terminal domain, the ATP repeat domain, and a carboxyl-terminal effector domain, as depicted by the filled bar at the top. The solid lines represent the wild-type ICP34.5 and deletion mutants with the amino acid numbered, and the designated names are listed in the left column. BiFC assays were performed as described previously. HeLa cells were transfected with plasmid pcDNA3-YN-eIF2α together with one of the following ICP34.5 constructs: pcDNA3-YC-ICP34.5 (WT-C) or pcDNA3-YC-ICP34.5 (WT-N) (B and C), the plasmids encoding YC fused to ICP34.5ΔC45/ΔC30/ΔC15 mutants (D and E), and the plasmid encoding YC-ICP34.5 (ΔC233–248) fusion protein (F and G). Each independent experiment contains the transfectants of YN-eIF2α plus YC as the negative control and YN-eIF2α plus YC-ICP34.5 (WT) as the positive control. At 36 h post-transfection, cells were subjected to fluorescence microscopy and FACS analysis. The FACS profile of each transfectant was plotted in the center column, with the mean YFP fluorescence intensity (MFI) in the right column (B, D, and F). The protein expression in BiFC assay was confirmed with anti-eIF2α and anti-ICP34.5 antibodies, respectively (C, E, and G).
To investigate which region of carboxyl-terminal domain is required for eIF2α binding, we constructed a series of plasmids encoding YC-fused ICP34.5, as listed in Fig. 4A, ΔC45 (a deletion of amino acids 218–263), ΔC30 (a deletion of amino acids 233–263), and ΔC15 (a deletion of amino acids 248–263). The BiFC assay narrowed down the binding region, as shown in Fig. 4D. The fluorescence and the mean values of YFP intensity in the upper panel indicate the interaction between eIF2α and ICP34.5 (WT) or mutants. Valid interactions occurred between eIF2α and ICP34.5 WT, with a score of 83.53 (Fig. 4D, second row) and ICP34.5 (ΔC15), with a score of 76.61 (A, bottom row), whereas ICP34.5 (ΔC45) and ICP34.5 (ΔC30) (as indicated in A) exhibited no binding as that in the control (A, top row). The sharp contrast between ΔC30 and ΔC15 in the binding of eIF2α indicates that the region covering aa 233–248 may be responsible for the interaction with eIF2α.
Thereafter, an ICP34.5 mutation Δ233–248 (a deletion of amino acids 233–248) was constructed, and the binding of eIF2α was analyzed by BiFC assay. As predicted, deletion of amino acids 233–248 completely abolished its interaction with eIF2α, with an MFI score of 10.56 (Fig. 4F, bottom row), the same as the negative control (top row), in comparison with the positive eIF2α binding by wild-type ICP34.5 with a score of 83.52. Taken together, these results indicate that the peptide stretch from 233–248 of ICP34.5 is responsible for eIF2α binding. Sequence alignment indicates the high homology with human and mouse GADD34, and secondary structure prediction implies a coil-to-helix structure along this region. It is likely that ICP34.5 binds to eIF2α via a helix structure. Further studies on the structure-function relationship of the conserved amino acids are ongoing. The crystal structure of the complex is also investigated to obtain detailed information of the interaction between ICP34.5 and eIF2α.
Interaction of ICP34.5 with eIF2α Is Required for Viral Replication
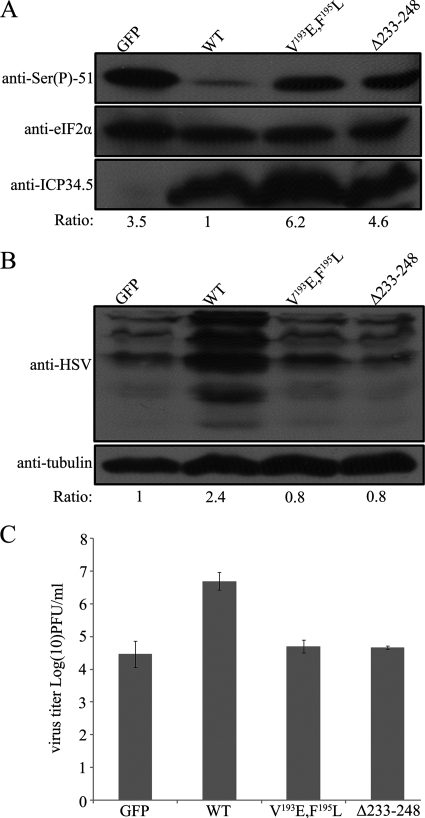
The C-terminal region (233–248) of ICP34.5 is identified to bind to eIF2α. The next question is whether this region is relevant to viral replication. We therefore tested the rescue of viral protein synthesis and replication in HeLa cells infected with the ICP34.5-negative virus R3616 by GFP as a negative control or pretransfected ICP34.5 (WT), ICP34.5 (V193E, F195L), or ICP34.5 (Δ233–248), respectively. As shown in Fig. 5A, the infection-induced phosphorylation of eIF2α was obvious in the control group, with a ratio of phosphorylated to total eIF2α of 3.5. The expression of ICP34.5 (WT) significantly rescued the PP1 dephosphorylation of eIF2α to a ratio of 1.0, as we defined. However, no rescue of the dephosphorylation was seen in the presence of the ICP34.5 mutation with a PP1 binding defect (V193E, F195L) nor the mutation with the deletion of aa 233–248, which was designated to bind eIF2α. Correspondingly, the lack of the active form of eIF2α rendered a significant deficiency in viral protein synthesis (Fig. 5B). Viral protein synthesis in cells transfected with ICP34.5 (WT) was nearly 3-fold that of cells with ICP34.5 (Δ233–248). Collectively, the binding between eIF2α and ICP34.5 is essential for eIF2α dephosphorylation and viral protein synthesis.
FIGURE 5.
Effect of eIF2α interaction on the function of ICP34. 5 in viral infection. A, effect on the endogenous eIF2α dephosphorylation. HeLa cells were pretransfected with plasmid encoding GFP, ICP34.5 (WT), ICP34.5 (V193E, F195L), or ICP34.5 (Δ233–248), respectively. At 24 h post-transfection, cells were infected with R3616 (multiplicity of infection = 10) for 24 h and harvested for electrophoresis and Western blot analysis. Phosphorylation of eIF2α was detected with anti-Ser(P)51-eIF2α antibody (top panel), and the total endogenous eIF2α was displayed by anti-eIF2α antibody (center panel). The expression of ICP34.5 or its variants were inspected using anti-ICP34.5 antibody (bottom panel). The phospho-eIF2α and total eIF2α were quantitated by densitometer, and the phosphorylation of eIF2α was scored as a ratio of phospho-eIF2α to total eIF2α, with the lowest phosphorylation level (WT) designated as score = 1 (bottom panel). B, Then the viral protein synthesis was detected with anti-HSV antibody, and tubulin was blotted with anti-α-tubulin antibody as an internal control. The level of the protein synthesis was quantitated by densitometer and presented as a ratio to tubulin. The ratio in the control group was defined as 1. C, viral replication. HeLa cells were pretransfected as described above followed by infection with R3616 (multiplicity of infection = 0.05). At 24 h post-infection, virus yields were titrated on Vero cells as described under “Experimental Procedures,” and viral replication was presented as a log value in the vertical axis. Data is mean ± S.D. of three independent experiments.
Deficient Protein synthesis results in compromised virus replication. Thereby, virus replication was examined in the presence of described ICP34.5 and its variants as described above. HeLa cells belong to an interferon competent cell line. Upon infection, HSV-1 exhibited nearly 1000 fold higher replication than its ICP34.5 deleted virus, R3616, as previously described. Here we pre-transfected cells with plasmids encoding GFP control, wild type ICP34.5, PP1 binding defective variant, and Δ233–248, respectively, as indicated in Fig. 5C, and challenged with R3616. At 24 h post infection, virus yields were determined. As shown in Fig. 5C, R3616 replicated to a titer of 6 × 104 pfu/ml, and the expression of ICP34.5 (WT) efficiently rescued viral replication to nearly 1 × 107pfu/ml, to the extent of a wild type HSV-1 (previous data, and data not shown). However, the protein deficient mutations of ICP34.5, V193E, F195L and Δ233–248 were hardly restore the virus replication (4.6 × 104pfu/ml) (Fig. 5C). Therefore, for the efficient replication of HSV-1, not only the PP1 binding motif is required for the determinant factor ICP34.5, but also the site 233–248 responsible for interaction with eIF2α is indispensable. These two components of ICP34.5 are interdependent to achieve the dephosphorylation of eIF2α, and thus protein synthesis and viral replication.
DISCUSSION
ICP34.5 of herpes simplex viruses is a virulent factor with multiple cellular functions. As reported for GADD34, ICP34.5 contains a consensus binding site for PP1. The PP1 binding motif is required for the formation of a high-molecular-weight complex that contains the dephosphorylation activity toward eIF2α (27). On the other hand, ICP34.5 truncation mutations or replacement of the conserved amino acids completely abolishes the dephosphorylation of eIF2α. Even the PP1 binding motif remains intact. We illustrated that the enzyme-substrate interaction, in this case the PP1-eIF2α interaction, occurs in the presence of ICP34.5 (Fig. 1) as well as GADD34 (data not shown) and suggested that ICP34.5 brings PP1 and eIF2α into the vicinity for dephosphorylation. The heterotrimer of PP1-ICP34.5-eIF2α may be part of the components in the high-molecular-weight complex, as mentioned earlier (25).
The C-terminal region of ICP34.5 was identified previously to contain a PP1 binding motif at aa 192–196 (19). Virus bearing a mutated ICP34.5 at this motif was rendered by reduced replication similar to that of the deletion mutation R3616 (25). Nevertheless, the C-terminal truncation of ICP34.5 with an intact PP1 binding motif also displayed deficient replication. An alternative site that indirectly binds PP1 was suggested, using ICP34.5 human homologue GADD34, and it was proposed that GADD34 has another site for the binding of PP1 inhibitor-1 for function (28). Here we addressed that the GADD34-homologous region of ICP34.5 contains a stretch of peptide that facilitates the recruitment of eIF2α to PP1. This binding is independent of the PP1 binding motif, as the mutation failed to bind PP1 does not affect the association of ICP34.5 and eIF2α (Fig. 2). Inversely, ICP34.5 lacking the eIF2α binding site still binds PP1 in in vivo and in vitro assays (data not shown).
This work identified a region (aa 233–248) of ICP34.5 responsible for the association with eIF2α. This region is highly homologous to GADD34. Prediction of the secondary structure displayed a coil structure right next to the helix region made with the arginine-rich domain (20). Further effort on crystal structure of ICP34.5 would provide molecular details in the dephosphorylation module, consisting of ICP34.5, PP1, and eIF2α.
In HSV-infected cells, ICP34.5 counteracts PKR by mediating eIF2α activity (9). ICP34.5 perhaps plays a scaffold role for PP1 and its substrate eIF2α that is of great importance to the dephosphorylation reaction. In this regard, aa 233–248 of ICP34.5 are considered to be another functional determinant in HSV infection in addition to its PP1 binding motif. The effect of this region on the physiological measures is 3-fold. 1) Viral protein synthesis in cells infected with R3616 can be rescued by wild-type ICP34.5, by the mutation with a deletion of aa 233–248, or by the mutated PP1 binding motif. 2) The dephosphorylation of eIF2α exhibits great correlation with the protein synthesis. 3) The above defect in protein synthesis is rendered by the deficient viral replication.
In summary, we present the new finding that ICP34.5 associates with eIF2α and identify the corresponding region that locates apart from the PP1 binding motif at the C-terminal effector domain. We also addressed that the interaction of ICP34.5 with eIF2α is independent of the binding with PP1, leading to the hypothetic model of ICP34.5 as a scaffold to bridge PP1 and eIF2α. Amino acids 233–248 of ICP34.5 are essential for the function of ICP34.5 in viral replication. Further studies on resolving the three-dimensional structure of the complex will provide detailed information on the mechanism.
Acknowledgment
We thank Dr. Shirish Shenolikar (Duke University) for discussions and providing the GADD34 construct.
This work was supported by National Natural Science Foundation of China Grants 30670080 and 30870128 (to Y. C.), Ministry of Science and Technology of China Grants 2007CB914800 and 2006DFB32420, and National Institute of Allergy and Infectious Diseases Grant AI092230 (to B. H.).
- eIF2α
- eIF2 subunit α
- HSV
- herpes simplex virus
- PP1
- protein phosphatase 1
- GADD34
- growth arrest and DNA damage protein 34
- BiFC
- bimolecular fluorescence complementation
- MFI
- mean fluorescence intensity
- aa
- amino acid(s)
- pfu
- plaque-forming unit(s).
REFERENCES
- 1. Clemens M. J., Elia A. (1997) J. Interferon Cytokine Res. 17, 503–524 [DOI] [PubMed] [Google Scholar]
- 2. Gale M., Jr., Katze M. G. (1998) Pharmacol. Ther. 78, 29–46 [DOI] [PubMed] [Google Scholar]
- 3. de Haro C., Méndez R., Santoyo J. (1996) FASEB J. 10, 1378–1387 [DOI] [PubMed] [Google Scholar]
- 4. Schneider R. J., Mohr I. (2003) Trends Biochem. Sci. 28, 130–136 [DOI] [PubMed] [Google Scholar]
- 5. Gunnery S., Rice A. P., Robertson H. D., Mathews M. B. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 8687–8691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gale M., Jr., Blakely C. M., Kwieciszewski B., Tan S. L., Dossett M., Tang N. M., Korth M. J., Polyak S. J., Gretch D. R., Katze M. G. (1998) Mol. Cell. Biol. 18, 5208–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Salvatore M., Basler C. F., Parisien J. P., Horvath C. M., Bourmakina S., Zheng H., Muster T., Palese P., García-Sastre A. (2002) J. Virol. 76, 1206–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khoo D., Perez C., Mohr I. (2002) J. Virol. 76, 11971–11981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. He B., Gross M., Roizman B. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 843–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chou J., Roizman B. (1990) J. Virol. 64, 1014–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Verpooten D., Ma Y., Hou S., Yan Z., He B. (2009) J. Biol. Chem. 284, 1097–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Orvedahl A., Alexander D., Tallóczy Z., Sun Q., Wei Y., Zhang W., Burns D., Leib D. A., Levine B. (2007) Cell Host Microbe 1, 23–35 [DOI] [PubMed] [Google Scholar]
- 13. Mao H., Rosenthal K. S. (2002) J. Biol. Chem. 277, 11423–11431 [DOI] [PubMed] [Google Scholar]
- 14. Chou J., Roizman B. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 5247–5251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hollander M. C., Zhan Q., Bae I., Fornace A. J., Jr. (1997) J. Biol. Chem. 272, 13731–13737 [DOI] [PubMed] [Google Scholar]
- 16. Lord K. A., Hoffman-Liebermann B., Liebermann D. A. (1990) Nucleic Acids Res. 18, 2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhan Q., Lord K. A., Alamo I., Jr., Hollander M. C., Carrier F., Ron D., Kohn K. W., Hoffman B., Liebermann D. A., Fornace A. J., Jr. (1994) Mol. Cell. Biol. 14, 2361–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. He B., Chou J., Liebermann D. A., Hoffman B., Roizman B. (1996) J. Virol. 70, 84–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. He B., Gross M., Roizman B. (1998) J. Biol. Chem. 273, 20737–20743 [DOI] [PubMed] [Google Scholar]
- 20. Zhang C., Tang J., Xie J., Zhang H., Li Y., Zhang J., Verpooten D., He B., Cao Y. (2008) FEBS Lett. 582, 171–176 [DOI] [PubMed] [Google Scholar]
- 21. Chou J., Kern E. R., Whitley R. J., Roizman B. (1990) Science 250, 1262–1266 [DOI] [PubMed] [Google Scholar]
- 22. Hu C. D., Chinenov Y., Kerppola T. K. (2002) Mol. Cell 9, 789–798 [DOI] [PubMed] [Google Scholar]
- 23. Kerppola T. K. (2006) Nat. Protoc. 1, 1278–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Depaoli-Roach A. A., Park I. K., Cerovsky V., Csortos C., Durbin S. D., Kuntz M. J., Sitikov A., Tang P. M., Verin A., Zolnierowicz S. (1994) Adv. Enzyme Regul. 34, 199–224 [DOI] [PubMed] [Google Scholar]
- 25. Cheng G., Gross M., Brett M. E., He B. (2001) J. Virol. 75, 3666–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hu C. D., Kerppola T. K. (2003) Nat. Biotechnol. 21, 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cheng G., Yang K., He B. (2003) J. Virol. 77, 10154–10161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brush M. H., Weiser D. C., Shenolikar S. (2003) Mol. Cell. Biol. 23, 1292–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]