Abstract
Fluorescent proteins have revolutionized modern biology with their ability to report the presence of tagged proteins in living systems. Although several fluorescent proteins have been described in which the excitation and emission properties can be modulated by external triggers, no fluorescent proteins have been described that can be activated from a silent dark state to a bright fluorescent state directly by the activity of an enzyme. We have developed a version of GFP in which fluorescence is completely quenched by appendage of a hydrophobic quenching peptide that tetramerizes GFP and prevents maturation of the chromophore. The fluorescence can be fully restored by catalytic removal of the quenching peptide, making it a robust reporter of proteolysis. We have demonstrated the utility of this uniquely dark state of GFP as a genetically encoded apoptosis reporter that monitors the function of caspases, which catalyze the fate-determining step in programmed cell death. Caspase Activatable-GFP (CA-GFP) can be activated both in vitro and in vivo, resulting in up to a 45-fold increase in fluorescent signal in bacteria and a 3-fold increase in mammalian cells. We used CA-GFP successfully to monitor real-time apoptosis in mammalian cells. This dark state of GFP may ultimately serve as a useful platform for probes of other enzymatic processes.
Keywords: Apoptosis, Cell Death, Fluorescence, Protease, Protein Chemistry
Introduction
The ever-growing palate of fluorescent proteins has arguably become the most widely used set of tools in cell and developmental biology. These fluorescent proteins act as markers for visualization of cellular processes, localization, gene expression patterns, and protein function. A spectrum of fluorescent proteins are now available, further expanding the multi-factorial imaging possibilities (for review see Ref. 1). The variously colored, split and photo-activatable fluorescent proteins (for review see Ref. 2) are excellent tools for precise labeling and tracking of a variety of targets in living systems, but there is still a need for tools that can report on the functional state of the protein they interrogate. Many proteins are only activated by a post-translational event, so genetically encoded fluorophores that undergo a change in fluorescent properties in direct response to enzymatic action would meet this need. A dark-to-bright fluorescent reporter would be especially useful for reporting enzymatic activity, as the low intrinsic background would allow robust detection of even relatively rare events. Such a reporter could enable the same level of real-time, noninvasive, longitudinal studies of enzymatic activity as parent fluorescent proteins have enabled on expression and localization.
In an effort to develop and characterize a dark-to-bright activatable GFP, we sought to apply our reporter to a complex biological pathway that would allow thorough characterization of the quenching mechanism and kinetics of activation both in vitro and in vivo. We ultimately selected apoptosis, the process of programmed cell death. Apoptosis is essential for embryonic patterning and vertebrate development at all stages and is causally involved in up to half of human diseases lacking suitable treatment (3). Thus, methods for monitoring apoptosis longitudinally would provide a full temporal understanding of the contributions of apoptosis to tissue formation, remodeling, response to drug treatment, and disease progression.
Apoptosis can be activated through an intrinsic caspase-9/apoptosome-dependent pathway or through an extrinsic, caspase-8/DISC complex-dependent pathway. Caspase-8 and -9 are apical initiators in the proteolytic cascade cleaving and thereby activating the downstream executioners, caspase-3 and -7. Cleavage of caspase-3 or -7 removes a prodomain and cuts an intersubunit linker, generating a small and a large subunit from each half of the dimer. Activated executioner caspases then cleave a select group of substrates (4), sentencing the cell to death. Caspase activation is the fate-determining step in the irreversible onset of apoptotic cell death, so they offer the most accurate and sensitive enzymatic indicator of apoptosis. Caspases show exquisite specificity for cleaving after aspartic acid residues in well-defined recognition sequences. The recognition specificities of the executioner caspases have been extensively mapped in vitro (5–9) and in vivo (4, 10). The canonical and widely used recognition sequence comprising the amino acids DEVD remains an appropriate and selective sensor of executioner caspase activity (11).
Executioner caspase activity can be quantified using fluorogenic peptide substrates containing a caspase-recognition sequence linked to a synthetic fluorophore, whose fluorescence is completely quenched prior to cleavage (12). The synthetic nature of these peptide reporters clearly precludes their expression in living systems. A great deal of effort to date has focused on developing genetically encoded fluorescent apoptosis reporters based on cleavage by caspases. Because of the pervasive need for monitoring apoptotic cell death, several clever genetically encoded apoptosis reporters have been developed.
The largest class of genetically encoded apoptosis reporters uses fluorescence resonance energy transfer (FRET)2 pairs separated by caspase cleavage sites. These reporters monitor a change in the FRET intensity upon caspase cleavage (13–15). The multimodality reporter uses copies of the caspase-recognition sequence to link a fluorescent protein, a bioluminescent protein, and a positron emission tomography reporter, which can each be more readily detected after caspase cleavage (16). The Apoliner and ApoAlertTM reporters use nuclear localization sequences to target a fluorescent protein to the nucleus following caspase cleavage (17, 18). Apoptosis in these cells can be visualized based on changes in fluorescent protein localization. Another reporter relies on destruction of green fluorescent protein (GFP) using a tag for ubiquitin-dependent degradation, which can be proteolytically removed by active caspase (19). The continued development of apoptosis reporters suggests that none to date have fully optimized brightness, sensitivity, and reporting mechanism.
An ideal apoptosis reporter would be genetically encodable, yet function in vitro, have very low background and high signal, consist of a single activatable fluorophore requiring no cofactors or additional biological processes, and be amenable to use in whole organisms, cells, microscopy, and flow cytometry. The caspase activatable-GFP we have developed meets all of these criteria. The dark-to-bright transition in direct response to enzymatic activation constitutes a new class of fluorescent reporter.
The goals of this work are to develop a version of GFP in which fluorescence is fully quenched by a short peptide and characterize the mechanism of quenching and gain of fluorescence. A thorough understanding of the mechanism of quenching will not only enable use in the study of apoptosis but will also pave the way for other applications, including exploration of other enzymatic processes.
EXPERIMENTAL PROCEDURES
Cloning
The gene for dark GFP (S65T) (referred to as GFP from this point forward) was created by PCR amplifying the GFP gene using a reverse primer that also encoded the 27 amino acid quenching peptide derived from the transmembrane region of influenza M2. This PCR product was ligated into the XhoI and NdeI sites of pET21b (Novagen). A sequence encoding the linker (LEVLFQGP) was then inserted between the EGFP and M2 genes using the QuikChange mutagenesis (Stratagene) approach. The newly inserted linker sequence was then mutated to encode the caspase-7 cleavage recognition site (DEVDFQGP). The final sequence of CA-GFP protein is GFP with the fusion of DEVDFQGPCNDSSDPLVVAASIIGILHLILWILDRL at the C terminus. This construct was used for expression and purification of CA-GFP.
The CA-GFP gene was then amplified by PCR and ligated into NdeI and XhoI of the pBB75 vector (a gift from Adrien Batchelor), which has a p15 origin of replication, different than the pET family of vectors which have ColE1 origins of replication. This plasmid was used for co-expression of CA-GFP with caspase-7.
The mammalian expression vectors pCA-GFP and pGFP-STOP were constructed by insertion of the CA-GFP or GFP, obtained by PCR into the NheI and EcoRI sites of pmKate2-C (Clontech, Palo Alto, CA, USA). CA-GFP from pCA-GFP was isolated and ligated into the NheI and EcoRI sites of pT-CD8-IRES-SCN2B (a kind gift from Dr. Alfred George, Vanderbilt University) to produce pT-CA-GFP-IRES-SCN2B. The resulting plasmid was cut with SalI and NotI and the coding sequence for mLumin (20) (mKate2-S158A) was inserted, generating pT-CA-GFP-IRES-mLumin. All constructs were confirmed by sequencing.
Lysate-based GFP Fluorescence Assay
CA-GFP was co-transformed with either wild-type caspase-7 or the C186A caspase-7 mutant into the BL21(DE3) strain of Escherichia coli. 50-ml cultures were inoculated from a 5 ml dense overnight culture, allowed to grow to an A600 of 0.6 at 37 °C and induced with 1 mm IPTG for 18 h at 25 °C. One ml of the culture was centrifuged using a tabletop centrifuge, the media was decanted and the pellet resuspended in 400 μl of a lysis buffer (0.5 mg/ml lysozyme, 2 units DNase). The suspension was lysed by freeze-thaw and the lysate was then centrifuged using a tabletop centrifuge. 100 μl of the supernatant was analyzed on a Molecular Devices Spectramax M5 spectrophotometer, measuring fluorescence (Ex. 475 nm/Em. 512 nm) in a costar 96-well flat bottom black plate.
Immunoblotting of Bacterial Cultures
Aliquots of the cultured and lysed CA-GFP/capase-7 expressing E. coli samples described above were reserved for immunoblotting analysis. SDS-loading buffer was added to lysate samples and the samples were boiled for 10 min. Three identical SDS-PAGE mini-gels were run. One gel was stained with Coomassie dye and imaged with a Gel Doc work station (Syngene). A second gel was transferred to a nitrocellulose membrane and immunoblotted using rat monoclonal anti-mouse caspase-7 antibody (Sigma) that is specific for the caspase-7 large subunit. The third gel was was also transferred to a nitrocellulose membrane and and immunoblotted using a mouse IgG monoclonal anti-GFP primary antibody (Abgent). Gels immunoblotted with either anti-caspase-7 or anti-GFP primary antibodies were treated with anti-mouse IgG alkaline phosphatase produced in goat (Sigma) and visualized using 1-StepTM NBT/BCIP (Thermo Scientific).
Fluorescence Microscopy
To prepare samples of E. coli for imaging, 50 ml LB cultures of CA-GFP, CA-GFP co-transformed with inactive caspase-7, and CAGFP co-transformed with wild-type caspase-7 were inoculated from dense 5 ml overnight cultures and grown to an A600 of 0.6. They were induced with 1 mm IPTG and grown 16–18 h at 25 °C. From these cultures 1-ml aliquots were spun down for 1 min at 13,000 rpm in an Eppendorf tabletop centrifuge. The media was decanted and the pellet was resuspended in PBS. Slides were prepared from these samples for DIC and fluorescence images taken using a Nikon Spot/E600 fluorescent microscope. All fluorescence images used the same exposure time of 150 ms.
Flow Cytometry
Flow cytometric analysis was performed on a Becton Dickinson (BD) LSR II configured with a 488 nm blue laser and a 530 ± 30 nm bandpass filter. Samples were prepared by harvesting 300 μl of cultures expressing CA-GFP with or without wild-type or C186A caspase-7, induced overnight as described above. Cells were washed twice with PBS and resuspended at a final concentration of 5 × 107 cells/ml. With BD FACS Diva Software, measurements were recorded for 10,000 cells analyzed at a rate of 200 events s−1. A gate was set to define a target population of fluorescent cells by including only events with a higher GFP intensity than cells expressing CA-GFP alone (uncleaved CA-GFP).
Cell Culture and Transfection
NIH 3T3 cells were obtained from ATCC and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mm glutaMAX and 10% fetal bovine serum (Atlanta Biologicals). Cultures were maintained in a humidified atmosphere of 5% CO2 in air at 37 °C. For transfection, cells were plated in 12-well plates or 35-mm dishes (70–80% confluence), cultured for 12 h and transfected with 1.5 μg of plasmid DNA per 35 mm well using 6 μl Fugene HD (Roche) according to the manufacturer's instructions. For induction of apoptosis, cells were incubated with 1 μm staurosporine (STS, Ascent Scientific NJ) for a period of time as indicated in “Results.” Controls were treated with vehicle alone (0.05% DMSO).
Immunoblotting of Mammalian Cultures
NIH 3T3 cells were transiently transfected with the indicated expression vectors and treated with 1 μm staurosporine (STS) or left untreated. Cells were harvested at the indicated time points and solubilized in loading buffer (62.5 mm Tris-HCl pH 6.8, 2% SDS, 41.7 mm dithiothreitol, 10% glycerol, 0.01% bromphenol blue). After being boiled for 5 min, proteins were resolved on a 5–20% gradient gel or 12% homogenous SDS-PAGE gel and electrotransferred to PVDF membranes for immunoblotting using enhanced chemiluminescence detection. Primary antibodies against GFP (1:5000), caspase-3 (1:1000), and β-tubulin (1:5000) were applied. Mouse monoclonal anti-GFP (Clone 3F8.2) and anti-β-tubulin (Clone AA2) antibodies were purchased from Millipore, rabbit polyclonal anti-caspase-3 antibody from Cell Signaling (San Diego, CA) and HRP-conjugated goat anti-mouse IgG and goat anti-rabbit IgG from Jackson ImmunoResearch Labs (West Grove, PA).
Live Cell Imaging
Epifluorescence was performed on an inverted wide-field fluorescence microscope (TE2000; Nikon, Japan) equipped with a cooled CCD camera (Qimaging, Canada) and a Plan Fluor 10×/0.30 objective using Elements imaging software (Nikon). GFP fluorescence was captured using a 515/30 filter. Time-lapse, confocal imaging was carried out on a Zeiss LSM 510 Meta Confocal Microscope (Zeiss, Germany) over a 6-h period. GFP and mLumin (20) were excited at 488 nm and 543 nm, respectively, and fluorescence detected at 520/30 nm (for GFP) and 585LP nm (for mLumin) using a 40× (1.3 N.A.) oil immersion objective lens. The live cells were maintained in 12.5 mm Hepes buffered DMEM and kept at 37 °C with a 5% CO2 atmosphere using heaters and a CO2 regulator. Images from the confocal microscope were manually focused and captured every 20 min using LSM 510 software (version 4.2; Zeiss). All fluorescence images were corrected by subtracting the background and analyzed using ImageJ and Adobe Photoshop 9.0. The ratio of green fluorescence intensity at each time point (IGt) was compared with the red fluorescence intensity (IRt) corrected for the initial intensity green:red ratio, IGt/IRt − IG0/IR0. The ratio was calculated based on the average pixel value for the whole cell, wherein IGt and IRt are the fluorescence intensities of GFP and mLumin at the indicated time point. As cells changed their morphology during imaging, the whole cell region was determined separately in each image.
Statistical Analysis for Live Cell Imaging
Manual cell counting was performed with ImageJ software using the Cell Counter plugin. More than 1000 cells were counted for both treatment and control groups. Statistics were calculated using Excel. Statistical significance was assessed by an unpaired Student two-tailed t test. Values were considered statistically significant at p < 0.05.
Protein Expression and Purification
All GFP-based expression constructs described were transformed into E. coli strain BL21(DE3). Cleaved CA-GFP was obtained by co-expression of CA-GFP and wild-type full-length caspase-7 lacking the His6 tag. Flasks containing 1 liter of 2×YT media were inoculated with 1 ml of dense culture and grown at 37 °C to an A600 of 0.6. The cultures were then induced using 1 mm isopropyl β-d-1-thioglalctopyranoside (IPTG) and the temperature reduced to 25 °C for three hours of expression. Cells were harvested by centrifugation and disrupted by microfluidization. Clarified lysates were prepared by centrifugation at 15,000 × g for 45 min. The GFP-based proteins were then purified using Co2+-affinity liquid chromatography (HiTrap Chelating HP, GE). The column was washed with a buffer composed of 50 mm imidazole, 300 mm NaCl, and 50 mm NaH2PO4 pH 8.0. Proteins eluted using a buffer of 300 mm imidazole, 300 mm NaCl, and 50 mm NaH2PO4 pH 8.0 and stored at 4 °C. Protein purity was assessed by gel electrophoresis. Proteins were estimated to be at least 95% pure.
Wild-type caspase-7 was expressed from a plasmid comprising pET23b with full-length caspase-7 gene (21) (gift of Guy Salvesen). This plasmid was mutagenized by QuikChange (Stratagene) to introduce the C186A mutation. Wild-type or C186A caspase-7 were expressed in E. coli strain BL21(DE3). Protein expression was induced with 1 mm IPTG and allowed to proceed for 12–18 h at 14 °C. Wild-type caspase-7 was purified by affinity chromatography on a Ni-NTA superflow column (Qiagen) developed with a step gradient of 250 mm imidazole. Eluted caspases were further purified using ion exchange chromatography on a 5 ml High-Q column (Bio-Rad) with a linear gradient from 50 mm to 750 mm NaCl. The purity of all mutants was assessed by SDS-PAGE gel stained with Coomassie Blue (Bio-Rad) and found to be >95% pure.
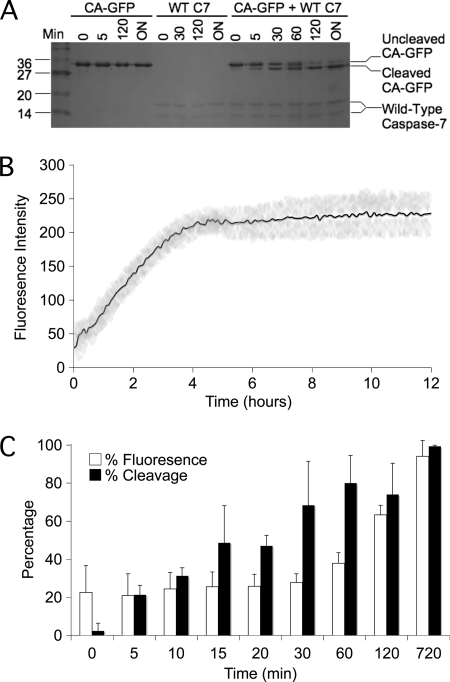
In Vitro CA-GFP Cleavage and Fluorescence Assay
Samples containing 10 mm CA-GFP with or without 9 mm wild-type caspase-7 were prepared at time zero to assess the ability of caspase-7 to digest CA-GFP directly. At initiation of the reaction 100 μl aliquots of the digest and controls were added to a costar 96-well black plate, and the fluorescence was measured every 5 min (Ex. 475 nm/Em. 512 nm) for 15 h at 27 °C. The remainder of the sample was incubated at 27 °C in 15-μl aliquots to which SDS loading buffer was added at time points of 0, 5, 10, 15, 20, 30, 60, 90, 120 min, and at 15 h. These samples were then run on an SDS-PAGE gel to determine the ratio of cleaved to uncleaved CA-GFP. The percent of cleaved product was determined by quantification using GeneTools software (product version 4.00) on gels imaged with a Gel Doc work station (Syngene).
Absorbance of GFP Chromophore
To measure the absorbance spectra of the GFP chromophore, 100 ml of purified CA-GFP and GFP were buffer exchanged into a buffer containing 10 mm NaH2PO4, pH 7 using Millipore Ultra-free 5K NMWL membrane concentrators and diluted to a concentration of 10 μm. The absorbance spectra were collected on a Molecular Devices Spectramax M5 spectrophotometer measuring absorbance from 350–550 nm in a Costar UV, flatbottom 96-well plate (product number 3635).
Mass Spectrometry
Purified CA-GFP and GFP samples were run on a one-dimensional SDS-PAGE gel and Coomassie stained. The bands of interest were excised and cut into 1 × 1 mm pieces and incubated in water for 1 h. The water was removed and 250 mm ammonium bicarbonate was added. For reduction the gel slices were incubated with 45 mm DTT at 50 °C for 30 min. After cooling to room temperature the cysteines were then alkylated by incubation with 100 mm iodoacetamide for 30 min at room temperature. The gel slices were washed twice with water, which was then removed and a 50:50 (50 mm ammonium bicarbonate: acetonitrile) mixture was placed in each tube, and samples were incubated at room temperature for 1 h. The solution was then removed and 200 μl of acetonitrile was added to each tube at, which point the gels slices turned opaque white. The acetonitrile was removed, and gel slices were further dried in a SpeedVac. The gel slices were then rehydrated in a 2 ng/μl solution of trypsin (Sigma) in 0.01% ProteaseMAX Surfactant (Promega): 50 mm ammonium bicarbonate for 21 h at 37 °C. The supernatant of each sample was then removed and placed in a separate 0.5 ml Eppendorf tube. Gel slices were further dehydrated with 60 ml of 80:20 (acetonitrile: 1% formic acid). The extract was combined with the previous supernatants of each sample and further purified using a micro Zip Tip (Millipore). The tryptic fragments were analyzed using matrix-assisted-laser desorption/ionization Time-of-Flight (MALDI-TOF) and subsequent MS/MS using a Shimadzu Biotech Axima TOF2 (Shimadzu Instruments) mass spectrometer to determine the maturity of the chromophore. The observed mass loss in GFP is due to oxidation of the Tyr-66 (loss of two daltons) and dehydration (loss of eighteen daltons) during the chromophore maturation process.
Circular Dichroism
Repeating structures such as α-helices and β-sheets rotate circularly polarized light differently such that the degree of protein secondary structure can be estimated from a circular dichroism spectra. Purified CA-GFP and GFP were prepared at a 10 μm concentration as described for absorbance measurements. CD spectra were measured on a J-715 circular dichroism spectrometer (Jasco) at 25 °C.
Size Exclusion Chromatography
The size of CA-GFP, GFP, and cleaved CA-GFP was determined by SEC using a Superdex 200 10/300 GL column (GE Healthcare). The sample was run in a buffer of 100 mm NaCl, 20 mm Tris pH 8.0 and compared with molecular weight standards albumin, carbonic anhydrase, ovalbumin, and ribonuclease A with blue dextran 2000 used to determine the void volume of the column (Sigma product No. MW-GF-1000).
RESULTS
Within the goal of developing a new dark-to-bright version of GFP, we sought an application area that would provide stringent activity requirements and a useful and interesting framework for characterization of the quenching and dark-to-bright transition both in vitro and intracellularly. We selected the proteolytically controlled pathway of apoptotic cell death as a first application for the dark-to-bright version of GFP.
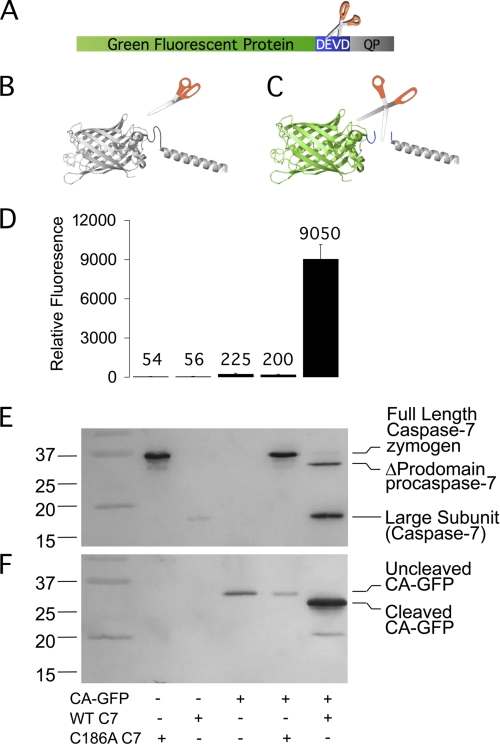
Fusion of a variety of proteins has been observed to silence GFP fluorescence (22) which led us to design a fluorescent reporter based on this quenching principle (Fig. 1A). We discovered that fluorescence of GFP (S65T) is quenched completely (0.9–1.5% of GFP fluorescence remained) when our first candidate, a 27-amino acid peptide derived from the tetrameric proton channel domain of influenza M2 protein, was fused to the GFP carboxy terminus (Fig. 1B). This full quenching proved to be an important component in maximizing sensitivity. To make a caspase activatable GFP (CA-GFP) the caspase recognition sequence (DEVD) was inserted between GFP and the quenching peptide (Fig. 1C).
FIGURE 1.
CA-GFP becomes fluorescent only upon cleavage by active caspase. A, CA-GFP fusion protein consists of green fluorescent protein (GFP, green), the four amino acid caspase-7 cleavage site Asp-Glu-Val-Asp (DEVD, blue) and the quenching peptide (QP, gray). B, model of uncleaved CA-GFP (non-fluorescent). C, model of cleaved CA-GFP (fluorescent). D–F, cleavage and fluorescence of bacterially co-expressed CA-GFP with active (WT) or inactive (C186A) caspase-7. Full-length caspase-7 undergoes auto-zymogen processing to generate the active, two-chain form of caspase-7. Mutation of the catalytic cysteine in the C186A mutant yields the inactive single-chain procaspase-7 zymogen. D, fluorescence of CA-GFP in lysates (n = 3 (S.E.)). E, immunoblot probed with anti-caspase-7 large subunit antibody. F, immunoblot probed with anti-GFP antibody.
CA-GFP Can Be Activated and Observed in Cells
To assess the response of CA-GFP to active caspase, CA-GFP was expressed in bacterial cells in the presence or absence of full-length active caspase-7 or the catalytically inactive C186A mutant. Only co-expression with active caspase-7 resulted in a 45-fold increase in CA-GFP fluorescence (Fig. 1D), suggesting that caspase activity is responsible for the fluorescence increase. Fluorescence in cells and lysates was observed within 1 h and was bright after 18 h. In bacterial cells, zymogen activation of full-length procaspase-7 to mature (active, two-chain) caspase-7 occurs with similar kinetics (23), further supporting the conclusion that the increase in fluorescence is due to caspase proteolytic activity. The gain of GFP fluorescence in the presence of wild-type caspase-7 (Fig. 1E) correlated with cleavage of CA-GFP by caspase (Fig. 1F). Cleaved CA-GFP was only observed in the presence of active caspase-7.
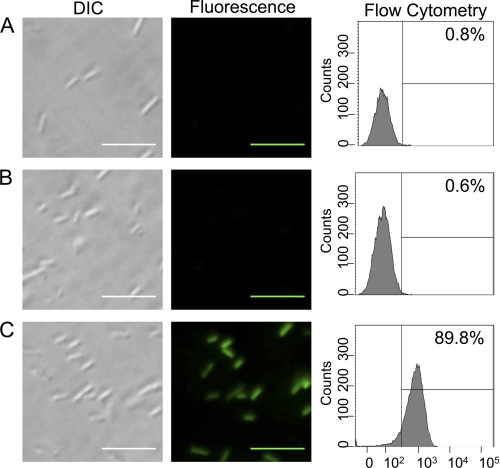
E. coli expressing CA-GFP alone or with caspase-7 (wild-type or C186A) were visualized by fluorescence microscopy. Cells expressing CA-GFP alone or with inactive caspase-7 showed no green fluorescence (Fig. 2, A and B), whereas CA-GFP expressed with wild-type caspase-7 yielded cells that were visibly green (Fig. 2C). These populations are also distinguishable by flow cytometry. 90% of cells expressing CA-GFP with active caspase-7 are fluorescent, whereas fewer than 1% are green in the absence of caspase activity.
FIGURE 2.
CA-GFP fluorescence is robust enough for fluorescence microscopy and flow cytometry. BL21 (DE3) E. coli cells expressing (A) CA-GFP alone or (B) CA-GFP with inactive caspase-7 C186A or (C) CA-GFP with active wild-type caspase-7. DIC images (left). Fluorescent microscopic images (center) of live cells. Intact cell flow cytometric analysis of GFP fluorescence (right) listing the fraction of cells with GFP fluorescence. Scale bars represent 10 μm.
CA-GFP Is an Apoptosis Reporter in Mammalian Cells
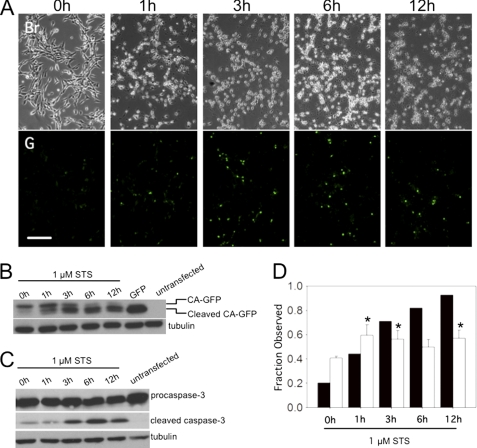
While CA-GFP responds robustly to the activity of human caspase expressed heterologously in bacteria, perhaps the most significant application for CA-GFP is as a reporter of apoptosis in mammalian cells, where caspase activity governs apoptotic cell death. Treatment of CA-GFP-transfected NIH 3T3 cells with staurosporine (STS), a documented apoptosis inducer (24–26), resulted in a time-dependent change in cell morphology, which is consistently observed during apoptosis (Fig. 3A). A concomitant increase in cellular CA-GFP fluorescence was also observed as expected (Fig. 3A, lower panels). Cleavage of CA-GFP was monitored by Western blot for GFP at various times after induction of apoptosis. A smaller fragment of cleaved CA-GFP appeared as a function of time after treatment with staurosporine (Fig. 3B). Cleavage of CA-GFP was consistent with the appearance of cleaved (active) caspase-3 (Fig. 3C). The observation that both GFP fluorescence and CA-GFP cleavage increase during apoptosis (Fig. 3D) indicates that CA-GFP functions as a mammalian apoptosis reporter on a population level. In control cells, increases in fluorescence, CA-GFP cleavage and the appearance of active (cleaved) caspase-3 were all correlated (supplemental Fig. S1), suggesting that CA-GFP is sensitive enough to report on even low levels of active caspases.
FIGURE 3.
CA-GFP is fluorescently activated in mammalian cells undergoing apoptosis. A, fluorescent response of CA-GFP-transfected NIH 3T3 cells at indicated times after induction of apoptosis with staurosporine (STS). Scale bar represents 50 μm. Br; Brightfield microscopy. G, GFP fluorescence. B, CA-GFP cleavage after induction of apoptosis is observed as a function of time by immunoblotting with an anti-GFP antibody. C, appearance of cleaved caspase-3 was observed after induction of apoptosis by immunoblotting with an anti-caspase-3 antibody. Tubulin was probed as a loading control. D, fraction of transfected cells that are fluorescent (white bars) and fraction of cleaved CA-GFP (black bars) in cells induced to undergo apoptosis. *, significance level; p < 0.05 relative to zero time point.
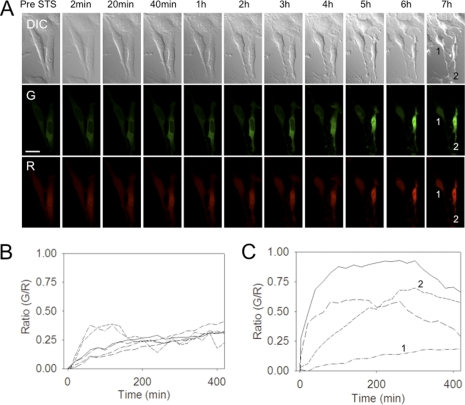
To uncover the details of CA-GFP expression and activation in mammalian cells, we performed time-lapse imaging of single cells expressing CA-GFP. To afford an internal, optically distinct fluorescent control, we generated the expression construct CA-GFP-IRES-mLumin that constitutively expresses both CA-GFP and the red fluorescent protein mLumin, which is a brighter, more photostable derivative of mKate (20). After addition of staurosporine to induce apoptosis, GFP fluorescence increased markedly (Fig. 4A). CA-GFP begins to be activated immediately and appears to remain localized in the cytoplasm for ∼3 h (supplemental movie S1) until the nuclear membrane is permeablized. This is consistent with previously reported executioner caspase activity, which is cytoplasmic in the early stages of apoptosis (27), however this is the first observation of the direct relationship of caspase activity to the loss of the nuclear membrane integrity. Notably, the red fluorescence of mLumin is present throughout the cell, including in the nucleus, prior to apoptosis. In untreated control cells, green and red fluorescence increased minimally and to approximately the same degree over the time course of the observation (supplemental Fig. S2 and Movie S2). Red fluorescence also increases during apoptosis, due to shrinking of the volume of the cell, which is a hallmark of apoptosis (28). Quantification of the green and red signals showed that only CA-GFP responds to apoptotic stimuli.
FIGURE 4.
The response of CA-GFP can be measured in single cells undergoing apoptosis. A, time-lapse confocal images of NIH 3T3 cells co-expressing CA-GFP and mLumin were recorded at the indicated times following treatment with STS to induce apoptosis. DIC; differential interference contrast images showing changes in cell morphology. G, green channel monitoring CA-GFP fluorescence. R, red channel, monitoring mLumin (control) fluorescence. Scale bar represents 25 μm. B and C, ratio of green (GFP)/red (mLumin) fluorescence for (B) untreated control or (C) staurosporine-treated cells. Ratios for six (B) or four (C) independent cells were measured. The G/R ratio of cells 1 and 2 shown in panel (A) are indicated as 1 or 2 in plot (C).
The ratios of green/red fluorescence for the untreated control cells (Fig. 4B) and for the staurosporine-treated cells induced to undergo apoptosis (Fig. 4C) were quantified. The appearance of GFP fluorescence in apoptosing cells appeared with related kinetics in all cells, however cell-to-cell variations were observed. The variation in the kinetics of GFP fluorescence occurred in a manner that is consistent with cell-to-cell differences in morphological changes and cell death kinetics. One of the apoptosing cells observed had very low fluorescence, which may be due to lower efficiency of transfection in that cell (the lowest green/red ratio Fig. 4C). The ratio of green/red fluorescence in the apoptotic cells was ∼3-fold higher than in control untreated cells, indicating that CA-GFP responds to apoptotic signals with a 3-fold increase in signal over background.
CA-GFP Requires No Cofactors and Can Be Activated in Vitro
The response of CA-GFP in mammalian systems is robust enough to make CA-GFP a reliable reporter of apoptosis, however the sensitivity of CA-GFP in bacterial systems is markedly greater. To better understand the mechanism by which CA-GFP is activated, and to determine whether activation of CA-GFP was dependent on some intracellular components, cleavage of CA-GFP in vitro was also tested. Purified active caspase-7 and purified CA-GFP were incubated together in a minimal buffer and cleavage of CA-GFP by wild-type caspase-7 was observed as a function of time (Fig. 5A). During the cleavage assay, a gain in fluorescence was also observed for CA-GFP (Fig. 5B). This suggests that the transition from the dark state to the brightly fluorescent state is dependent only on cleavage and not on any intracellular condition such as co-translational cleavage or the activity of chaperones to refold GFP. Cleavage of CA-GFP preceded fluorescence by ∼60 min (Fig. 5C). The appearance of fluorescence upon cleavage of purified CA-GFP seems to be slower than in mammalian cells where CA-GFP fluorescence appears within 20 min after activation of caspases (Fig. 4C). This lag in acquisition of fluorescence for purified CA-GFP is similar to the in vitro chromophore maturation time for purified GFP S65T, which occurs on the timescale of 27 to 122 min depending on the starting state of the protein (29, 30).
FIGURE 5.
CA-GFP can be activated in vitro. A, cleavage of CA-GFP by wild-type caspase-7 from 0 min to 18 h. (overnight). B, gain of fluorescence during in vitro cleavage of CA-GFP by caspase-7 (black line) with error bars (gray). C, CA-GFP cleavage precedes appearance of fluorescence. Percentage of total fluorescence (white) and cleaved CA-GFP (black) at each time point (n = 3 (S.D.)).
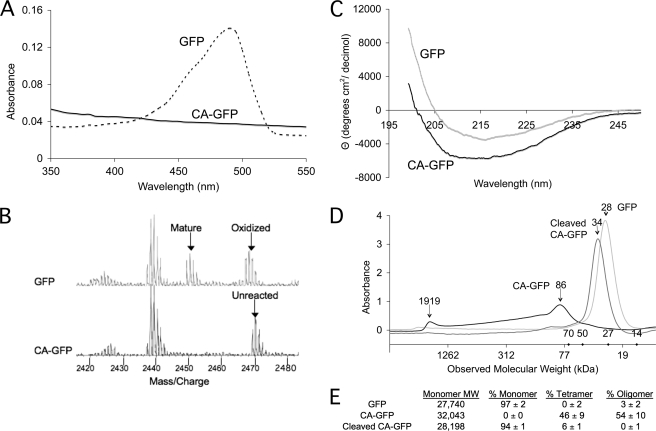
Dark CA-GFP Is Folded with an Immature Chromophore
CA-GFP cleavage and fluorescence kinetics suggest that the GFP chromophore is not mature in the dark state. The absorbance spectra of CA-GFP does not have the characteristic 488 nm peak of GFP (Fig. 6A), also suggesting that the CA-GFP chromophore has not undergone chromophore maturation. In GFP the chromophore is formed by a cyclization reaction in which the amide nitrogen of Gly-67 initiates nucleophilic attack on the carbonyl carbon of Thr-65, forming an imidazolone ring. Next the ring is oxidized (loss of 2 Da) to a cyclic imine and the carbonyl oxygen of Thr65 is dehydrated (loss of 18 Da), fully conjugating the system (Fig. 7) (29, 31). Thus the maturation state of the chromophore can be observed as a change in the mass of the protein (loss of 20 Da). We compared the mass spectra of tryptic peptides from dark CA-GFP and mature, green GFP (Fig. 6B). The mature chromophore-containing fragment (2449.2 Da) was observed for GFP as expected. The partially reacted intermediate in which only ring oxidation has occurred was also observed (2467.2 Da). This direct observation of the oxidized intermediate is the first observation for an S65T intermediate and supports other mechanistic studies on maturation of other versions of GFP (32) in which the oxidation step occurs prior to dehydration. In dark CA-GFP only the unreacted, immature peptide (2469.3 Da) was observed, indicating that the CA-GFP quenching peptide prevents chromophore maturation.
FIGURE 6.
CA-GFP silencing relies on tetramerization and prevention of chromophore maturation. A, absorbance spectra of GFP and CA-GFP suggest that CA-GFP chromophore is non-functional. B, mass spectra of GFP and CA-GFP tryptic fragments indicate that the CA-GFP chromophore has not undergone the maturation reaction. C, circular dichroism spectra of GFP and CA-GFP indicate that both proteins are folded into a predominantly β-sheet structure. D, size exclusion chromatogram for GFP, CA-GFP, and cleaved CA-GFP. The observed molecular weights demonstrate that GFP and cleaved CA-GFP are monomers, while uncleaved CA-GFP is predominantly tetramer. The molecular weights for the standards are marked as diamonds on the x-axis. E, expected and observed molecular weights by size exclusion chromatography for GFP, CA-GFP, and cleaved CA-GFP.
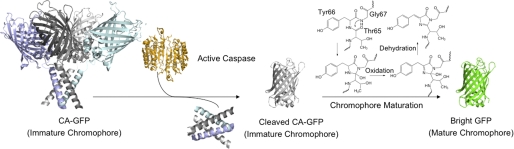
FIGURE 7.
CA-GFP is activated by cleavage and maturation of the chromophore. This scheme depicts the model of how fluorescence of tetrameric CA-GFP is activated. Dark CA-GFP contains an immature chromophore. Cleavage by active caspase releases GFP from the tetramer. Free GFP is then able to undergo the required conformational changes allowing chromophore maturation by the reaction scheme shown and production of fluorescent, bright GFP.
Like native GFP, CA-GFP is soluble when over-expressed in bacterial cells (30), suggesting that the GFP β-barrel domain of CA-GFP is globally folded. To assess the fold of CA-GFP in the dark state, purified CA-GFP and GFP were analyzed by circular dichroism spectroscopy. While the spectra of non-fluorescent CA-GFP and fluorescent wild-type GFP are not super-imposable (Fig. 6C), the similarity in the spectral features were coincident with published spectra (33) and suggest that CA-GFP forms the β-barrel observed in all GFPs. The gain in the intensity of the circular dichroism signal is consistent with an increase in the multimeric state of CA-GFP. The oligomeric state of CA-GFP, GFP, and cleaved CA-GFP were assessed by size exclusion chromatography (Fig. 6D). The majority of the GFP and cleaved CA-GFP in solution are monomeric (Fig. 6E). Uncleaved CA-GFP exists predominantly as a soluble tetramer, which is consistent with the oligomeric state of the parent quenching peptide. The tetramers may have exposed hydrophobic patches, as some portion of the CA-GFP is also found in higher order oligomers, presumably clusters of tetramers. Thus tetramerization of CA-GFP appears to be sufficient for quenching GFP fluorescence.
DISCUSSION
One of the key features of CA-GFP is the mechanism by which the quenching peptide silences fluorescence. The M2 protein, from which the CA-GFP quenching peptide is derived, forms active tetramers. To our knowledge, this is the first report of an appended peptide that can quench fluorescence of GFP without a change in the solubility. The dark state of CA-GFP appears to be folded by circular dichroism and is soluble, forming tetramers under identical conditions in which GFP is fully fluorescent. The tetramerization of CA-GFP prevents formation of the mature chromophore and thus fluorescence (Fig. 7). Cleavage of CA-GFP releases GFP, which can then undergo the necessary conformational rearrangements enabling chromophore maturation. Thus, it appears that tetramerization is sufficient to induce a slightly strained, but folded conformation of GFP that disallows chromophore formation. It is not unprecedented that subtle changes to GFP, similar to tetramerization, prevent chromophore maturation. Although GFP is a stable and independently folding protein, subtle changes in the environment around the chromophore can dramatically affect fluorescence and the rate of chromophore maturation. For example, maturation of the chromophore in the S65T version of GFP, which adds but a single methyl group to the chromophore cavity is 4.4-fold faster than wild-type GFP. In the R96M variant, the lack of a positive charge drastically changes maturation from hours to months (34). When GFP is compressed with an atomic force microscope tip, the hydrogen-bond network inside the GFP barrel is broken, resulting in a non-fluorescent chromophore (35). Even a subtle change in pH (to < pH 6.5) is sufficient to quench fluorescence (36, 37). When GFP is split to remove the last of the 11 β-strands in the GFP barrel, fluorescence is silenced. Association of GFP1–10 (amino acids 1–214) with GFP11 containing only the 11th strand (amino acids 215–230) results in recovered fluorescence (38). In CA-GFP interactions enforced by tetramerization are likewise sufficient to silence fluorescence. This mechanism of fluorescence quenching is likely to be applicable in a variety of GFP-based reporters.
CA-GFP is directly activated by caspase activity and no other cellular components are essential. On the other hand, one possible reason for the more effective fluorescent conversion of CA-GFP in bacterial cells may be the presence of additional cellular factors, such as bacterial chaperone proteins. In mammalian cells CA-GFP conversion to the bright state can be seen in 20–40 min, which is consistent with the timing of caspase activation and the irreversible initiation of apoptosis (39), suggesting that some cellular factors (potentially macromolecules or metabolites) accelerate the maturation process over in vitro conditions where maturation of purified protein is slower (60 min). Therefore although some maturation time is required, CA-GFP still is able to accurately report caspase activity during apoptosis. Similarly in split GFP, association of the two halves of the protein is not the rate-limiting step but rather subsequent maturation of the chromophore dictates the kinetics of the appearance of fluorescence (38). The maturation of CA-GFP in vitro is similar to that of split GFP (38), which has also proven to be an incredibly useful technology.
CA-GFP is unique among fluorescent protease reporters in its transition from a quenched dark state to a bright state upon cleavage. This type of reporter was inspired by small molecule fluorogenic substrates that transition from dark to bright fluorescence upon protease activity. The utility of a dark-to-bright transition stems from its low fluorescent background and use of a single fluorophore. The caspase-mediated increase in CA-GFP fluorescence is 45-fold in bacteria compared with the next most sensitive reporter, CyPet-YPet, in which a 3.3-fold increase in FRET fluorescence is observed (40). This gain in fluorescence makes CA-GFP readily applicable to protease profiling in bacteria and for engineering protease specificity by directed evolution methods using sorting by flow cytometry. In mammalian cells, the increase in CA-GFP fluorescence is also higher than other apoptosis reporters. CA-GFP fluorescence increases 3-fold upon induction of apoptosis and activation of caspases, whereas the next two strongest reporters give 0.6-fold (19) and 2-fold (16) increases in fluorescence upon caspase cleavage.
The response of CA-GFP to caspases is not reliant on any other biological processes, and is therefore suitable for monitoring caspase activity under any cellular conditions. Other classes of caspase reporters rely on nuclear localization of fluorescent proteins (17, 19) or on proteosome-based degradation and thus are dependent on processes such as nuclear translocation, the integrity of the nuclear pores and membrane or proper functioning of the proteosome to observe apoptosis. During apoptotic cell death all biological processes ultimately shut down, potentially impacting the utility of reporters that rely on other biological pathways. The fact that localization of CA-GFP is not essential for monitoring caspase activity also allows CA-GFP to report on the integrity of the organellar membranes, as we observed for the first time using our genetically-encoded reporter and time-lapse imaging. In mammalian cells CA-GFP has proven to be an effective tool for time-resolved observation of subcellular events in apoptosis. The fact that CA-GFP shows no fluorescence in the nucleus in early apoptosis suggests that it is working accurately. The executioner caspases have been shown to localize in the cytoplasm in the early stages of apoptosis, only entering the nucleus in later stages after the nuclear membrane has been compromised (27). Caspases have been shown to target two of the twenty nuclear pore components, Nup93 and Nup96 (41). Once the nuclear pore is compromised larger proteins such as caspases and CA-GFP tetramers are able to pass into the nucleus. CA-GFP requires neither translocation nor additional cofactors and thus is useful for a wider array of applications than other reporters. These include monitoring apoptosis in intact living organisms without the need for sectioning or high-resolution microscopy and flow-cytometric sorting of apoptosing cells from healthy cells.
The fact that several caspase-reporting technologies have been developed prior to CA-GFP reflects the importance of apoptosis in modern biomedical research and the demand for new, better apoptosis reporting methods. CA-GFP combines dark-to-bright fluorescence, the most positive property of the synthetic peptide-based fluorophores, with the important advantage of being genetically encoded, to produce a new class of apoptosis reporter. We recognize that the CA-GFP platform is also amenable to substitution of both the protease recognition site and the fluorescent protein leading to reporters for many proteases in a range of colors. A panel of CA-GFP derivatives of various colors and with various protease sensitivities would enable simultaneous imaging of a number of proteolytic cascades. It is also possible to envision engineering reporters based on the silenced state of GFP that respond to the action of other enzyme families.
Supplementary Material
Acknowledgments
We thank Jessica Bauer, Mara Isel Guerrero, and Nagedra Yadava for preliminary work on the project and Dr. John Leszyk for MS analysis.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM080532 and a grant from the Center of Excellence in Apoptosis Research. The work was also supported by NSF Grant IGERT DGE-0654128 (to S. B. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and movies S1 and S2.
- FRET
- fluorescence resonance energy transfer
- CA-GFP
- caspase activatable-GFP
- STS
- staurosporine.
REFERENCES
- 1. Shaner N. C., Steinbach P. A., Tsien R. Y. (2005) Nat. Methods 2, 905–909 [DOI] [PubMed] [Google Scholar]
- 2. Lukyanov K. A., Chudakov D. M., Lukyanov S., Verkhusha V. V. (2005) Nat. Reviews 6, 885–891 [DOI] [PubMed] [Google Scholar]
- 3. Reed J. C., Tomaselli K. J. (2000) Curr Opin Biotechnol 11, 586–592 [DOI] [PubMed] [Google Scholar]
- 4. Mahrus S., Trinidad J. C., Barkan D. T., Sali A., Burlingame A. L., Wells J. A. (2008) Cell 134, 866–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garcia-Calvo M., Peterson E. P., Rasper D. M., Vaillancourt J. P., Zamboni R., Nicholson D. W., Thornberry N. A. (1999) Cell Death Differ. 6, 362–369 [DOI] [PubMed] [Google Scholar]
- 6. Thornberry N. A., Rano T. A., Peterson E. P., Rasper D. M., Timkey T., Garcia-Calvo M., Houtzager V. M., Nordstrom P. A., Roy S., Vaillancourt J. P., Chapman K. T., Nicholson D. W. (1997) J. Biol. Chem. 272, 17907–17911 [DOI] [PubMed] [Google Scholar]
- 7. Timmer J. C., Zhu W., Pop C., Regan T., Snipas S. J., Eroshkin A. M., Riedl S. J., Salvesen G. S. (2009) Nat. Struct. Mol. Biol. 16, 1101–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boulware K. T., Daugherty P. S. (2006) Proc. Natl. Acad. Sci. U.S.A 103, 7583–7588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goode D. R., Sharma A. K., Hergenrother P. J. (2005) Org Lett 7, 3529–3532 [DOI] [PubMed] [Google Scholar]
- 10. Lee A. Y., Park B. C., Jang M., Cho S., Lee D. H., Lee S. C., Myung P. K., Park S. G. (2004) Proteomics 4, 3429–3436 [DOI] [PubMed] [Google Scholar]
- 11. Albeck J. G., Burke J. M., Aldridge B. B., Zhang M., Lauffenburger D. A., Sorger P. K. (2008) Mol. Cell 30, 11–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Berger A. B., Witte M. D., Denault J. B., Sadaghiani A. M., Sexton K. M., Salvesen G. S., Bogyo M. (2006) Mol. Cell 23, 509–521 [DOI] [PubMed] [Google Scholar]
- 13. Chu J., Wang L., Luo Q. M., Zhang Z. H. (2008) P. Soc. Photo-Opt. Ins. 6857, R8570–R8570 [Google Scholar]
- 14. Kawai H., Suzuki T., Kobayashi T., Sakurai H., Ohata H., Honda K., Momose K., Namekata I., Tanaka H., Shigenobu K., Nakamura R., Hayakawa T., Kawanishi T. (2005) J. Pharmacol. Sci. 97, 361–368 [DOI] [PubMed] [Google Scholar]
- 15. Wu X., Simone J., Hewgill D., Siegel R., Lipsky P. E., He L. (2006) Cytometry A 69, 477–486 [DOI] [PubMed] [Google Scholar]
- 16. Ray P., De A., Patel M., Gambhir S. S. (2008) Clin. Cancer Res. 14, 5801–5809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bardet P. L., Kolahgar G., Mynett A., Miguel-Aliaga I., Briscoe J., Meier P., Vincent J. P. (2008) Proc. Natl. Acad. Sci. U.S.A 105, 13901–13905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Clontech (2009) ApoAlert Annexin V User Manual, Clontech, Mountain View, CA [Google Scholar]
- 19. Lee P., Beem E., Segal M. S. (2002) BioTechniques 33, 1284–1287, 1289–1291 [DOI] [PubMed] [Google Scholar]
- 20. Chu J., Zhang Z., Zheng Y., Yang J., Qin L., Lu J., Huang Z. L., Zeng S., Luo Q. (2009) Biosens. Bioelectron 25, 234–239 [DOI] [PubMed] [Google Scholar]
- 21. Riedl S. J., Renatus M., Schwarzenbacher R., Zhou Q., Sun C., Fesik S. W., Liddington R. C., Salvesen G. S. (2001) Cell 104, 791–800 [DOI] [PubMed] [Google Scholar]
- 22. Waldo G. S., Standish B. M., Berendzen J., Terwilliger T. C. (1999) Nature Biotechnology 17, 691–695 [DOI] [PubMed] [Google Scholar]
- 23. Stennicke H. R., Salvesen G. S. (1999) Methods 17, 313–319 [DOI] [PubMed] [Google Scholar]
- 24. Belmokhtar C. A., Hillion J., Ségal-Bendirdjian E. (2001) Oncogene 20, 3354–3362 [DOI] [PubMed] [Google Scholar]
- 25. Bertrand R., Solary E., O'Connor P., Kohn K. W., Pommier Y. (1994) Exp. Cell Res. 211, 314–321 [DOI] [PubMed] [Google Scholar]
- 26. Stepczynska A., Lauber K., Engels I. H., Janssen O., Kabelitz D., Wesselborg S., Schulze-Osthoff K. (2001) Oncogene 20, 1193–1202 [DOI] [PubMed] [Google Scholar]
- 27. MacFarlane M., Merrison W., Dinsdale D., Cohen G. M. (2000) J. Cell Biol. 148, 1239–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maeno E., Ishizaki Y., Kanaseki T., Hazama A., Okada Y. (2000) Proc. Natl. Acad. Sci. U.S.A 97, 9487–9492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heim R., Cubitt A. B., Tsien R. Y. (1995) Nature 373, 663–664 [DOI] [PubMed] [Google Scholar]
- 30. Reid B. G., Flynn G. C. (1997) Biochemistry 36, 6786–6791 [DOI] [PubMed] [Google Scholar]
- 31. Ormö M., Cubitt A. B., Kallio K., Gross L. A., Tsien R. Y., Remington S. J. (1996) Science 273, 1392–1395 [DOI] [PubMed] [Google Scholar]
- 32. Zhang L., Patel H. N., Lappe J. W., Wachter R. M. (2006) J. Am. Chem. Soc. 128, 4766–4772 [DOI] [PubMed] [Google Scholar]
- 33. Visser N. V., Hink M. A., Borst J. W., van der Krogt G. N., Visser A. J. (2002) FEBS Lett. 521, 31–35 [DOI] [PubMed] [Google Scholar]
- 34. Wood T. I., Barondeau D. P., Hitomi C., Kassmann C. J., Tainer J. A., Getzoff E. D. (2005) Biochemistry 44, 16211–16220 [DOI] [PubMed] [Google Scholar]
- 35. Gao Q., Tagami K., Fujihira M., Tsukada M. (2006) Jap. J. Appl. Physics 45, L929–L931 [Google Scholar]
- 36. Alkaabi K. M., Yafea A., Ashraf S. S. (2005) Appl Biochem. Biotechnol. 126, 149–156 [DOI] [PubMed] [Google Scholar]
- 37. Enoki S., Saeki K., Maki K., Kuwajima K. (2004) Biochemistry 43, 14238–14248 [DOI] [PubMed] [Google Scholar]
- 38. Cabantous S., Terwilliger T. C., Waldo G. S. (2005) Nat. Biotechnol. 23, 102–107 [DOI] [PubMed] [Google Scholar]
- 39. Li Y., Chopp M., Jiang N., Zhang Z. G., Zaloga C. (1995) Stroke 26, 1252–1257; discussion 1257–1258 [DOI] [PubMed] [Google Scholar]
- 40. Nguyen A. W., Daugherty P. S. (2005) Nat. Biotechnol. 23, 355–360 [DOI] [PubMed] [Google Scholar]
- 41. Patre M., Tabbert A., Hermann D., Walczak H., Rackwitz H. R., Cordes V. C., Ferrando-May E. (2006) J. Biol. Chem. 281, 1296–1304 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.