Abstract
In the uropathogenic Escherichia coli strain F11, in silico genome analysis revealed the dicistronic iron uptake operon fetMP, which is under iron-regulated control mediated by the Fur regulator. The expression of fetMP in a mutant strain lacking known iron uptake systems improved growth under iron depletion and increased cellular iron accumulation. FetM is a member of the iron/lead transporter superfamily and is essential for iron uptake by the Fet system. FetP is a periplasmic protein that enhanced iron uptake by FetM. Recombinant FetP bound Cu(II) and the iron analog Mn(II) at distinct sites. The crystal structure of the FetP dimer reveals a copper site in each FetP subunit that adopts two conformations: CuA with a tetrahedral geometry composed of His44, Met90, His97, and His127, and CuB, a second degenerate octahedral geometry with the addition of Glu46. The copper ions of each site occupy distinct positions and are separated by ∼1.3 Å. Nearby, a putative additional Cu(I) binding site is proposed as an electron source that may function with CuA/CuB displacement to reduce Fe(III) for transport by FetM. Together, these data indicate that FetMP is an additional iron uptake system composed of a putative iron permease and an iron-scavenging and potentially iron-reducing periplasmic protein.
Keywords: Copper, Crystal Structure, Iron, Reductase, Transport Metals, F11, FetMP, ILT, Uropathogenic
Introduction
Iron plays a critical role in biological systems; however, the physicochemical properties of iron dictate cells to develop sophisticated methods for iron acquisition. This is especially true for pathogenic bacteria, which have to compete with their hosts for this important transition metal. Under oxic conditions and neutral pH values, iron is predominantly in its ferric Fe(III) form, which precipitates as a hydroxide complex. This limits free iron availability to ∼1 atomolar, whereas cells need about 1 μm of bioavailable iron for growth (1–3).
To overcome the low availability of iron, most bacteria produce and utilize siderophores, small organic compounds that chelate ferric iron. Typically, ferric siderophores are recognized and transported by receptors in the outer membrane and cognate ABC3 permeases (4). Uptake of free iron via non-ABC permeases of the cytoplasmic membrane is far less studied. However, the ferrous iron transporting systems FeoABC, MntH, and ZupT from Escherichia coli and other bacteria have been identified (2). Recently, an additional bacterial iron uptake permease, EfeU, was identified that belongs to the OfeT (oxidase-dependent ferrous iron transporter) protein family (Transporter Classification Database entry 9.A.10.1) within the iron/lead transporter (ILT) superfamily (Transporter Classification Database entry 9.A.10) (5). Members of this group are best characterized in lower eukaryotes, such as Saccharomyces cerevisiae, where the transport of ferrous iron by Ftr1p (an OfeT member) is coupled with the oxidation of the transported iron by a multicopper oxidase (Fet3p) (6).
Some E. coli strains and other bacterial species (7) harbor a second ILT-type iron permease that is overall much more narrowly distributed than EfeU (2). The predicted proteins belong to a subgroup of ILT proteins (Transporter Classification Database entry 9.A.10.2.3) different from OfeT proteins. They are more closely related to the PbrT protein (Transporter Classification Database entry 9.A.10.2.1) involved in Pb(II) resistance in the metal-resistant bacterium Cupriavidus metallidurans strain CH34 (8). The gene fetM (locus tag EcF11_1995, ZP_03035145) from the uropathogenic E. coli strain F11 (genomic sequence NZ_AAJU02000030) has a much longer open reading frame than efeU and is probably part of a dicistronic operon. The 276-residue EfeU protein contains seven predicted transmembrane helices (I–VII), as is typical for Ftr1-like ILT proteins, and includes two conserved “REGLE” consensus sequences on helices I and IV that may be involved in iron binding (9). Conversely, FetM is much longer, with a predicted size of 646 residues and eight transmembrane α-helices. The first putative transmembrane α-helix is followed by a 349-residue putative periplasmic domain that is followed by the remaining 268 residues, which can be aligned with the N terminus EfeU (29% sequence identity, E-value 10−10). Thus, FetM resembles EfeU fused to an N-terminal 39-kDa periplasmic protein with a potential membrane anchor or leader peptide.
FetP (locus tag EcF11_1994, ZP_03035154), located directly downstream of fetM, encodes a putative periplasmic protein. A general relationship between FetP homologues and iron metabolism has been reported previously. For example, ChpA from the marine magnetotactic Vibrio strain MV-1 is required for magnetosome production (10); P19 from pathogenic Campylobacter jejuni is required for growth under iron limitation (11); Tp34 (TP0971) from Treponema pallidum is a lactoferrin-binding protein (12). All three homologs can also form homodimers and bind either copper or zinc.
In this study, we demonstrate that FetM constitutes a functional iron transport system that is enhanced by FetP. Furthermore, we have characterized the biochemical and structural properties of FetP, giving mechanistic insight into the potential role of copper binding in iron uptake.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
Strains and plasmids used in this study are listed in Table 1, and primers are listed in supplemental Table S1. E. coli was grown in Luria-Bertani (LB) medium (13) or in Tris-buffered mineral salts medium (TMM, pH 7) (14) containing 2 ml of glycerol and 3 g of casamino acids per liter with or without additional iron. Antibiotics (15 μg/ml gentamicin, 20 μg/ml chloramphenicol, or 125 μg/ml ampicillin), metals, and chelators were added as indicated. Reagents used in FetP analysis experiments were the highest purity available (≥99%). All metal ions (as chloride or sulfate salts), MES, BisTris, and ACES were obtained from Sigma-Aldrich. Glassware was acid-treated to reduce contamination with iron.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strains and plasmids | Relevant genotype | Reference/Source |
|---|---|---|
| E. coli strains | ||
| F11 | Extraintestinal pathogenic E. coli (ExPEC), fetMP− | Ref. 59 |
| ECA150 | W3110 ΔlacZYA | This study |
| ECA458 | W3110 ΔentC ΔfecABCDE ΔfeoABC ΔmntH ΔzupT | This study |
| ECA611 | ECA458 glmS-Gm | This study |
| ECA612 | ECA458 glmS-fetMP-Gm | This study |
| ECA613 | ECA458 glmS-fetM-Gm | This study |
| ECA614 | ECA458 glmS-fetP-Gm | This study |
| ECA615 | W3110 ΔlacZYA glmS-fetMp-lacZ | This study |
| XL1-Blue | recA1, endA1, gyrA96, thi-1, hsdR17, relA1, supE44, lac (F¢, proAB, lacIqZ_M15, Tn10) | Stratagene (La Jolla, CA) |
| EC100DTMpir-116 | F−mcrA Δ(mrr-hsdRMS-mcrBC) Φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ−rpsL nupG pir-116(DHFR) | Epicentre Biotechnologies (Madison, WI) |
| BL21 | F−dcm, omp, hsdS, (rB−mB−), lon−, galλ, DE3 | Novagen (Darmstadt, Germany) |
| Vectors | ||
| pGEM®-T Easy | Promega (Mannheim, Germany) | |
| pASK-IBA3 | IBA (Göttingen, Germany) | |
| pET22b(+) | Novagen | |
| pTNS1 | Ref. 15 | |
| pUC18R6K-mini-Tn7T-Gm | Ref. 15 | |
| pUC18-mini-Tn7T-Gm-lacZ | Ref. 15 | |
| Plasmids | ||
| pECD967 | pGEM-fhuFp | Ref. 19 |
| pECD1097 | pGEM-fetMp | This study |
| pECD1098 | pASK-IBA3-fetM | This study |
| pECD1099 | pASK-IBA3-fetP | This study |
| pECD1100 | pASK-IBA3-fetMP | This study |
| pECD1101 | pET22b(+)-fetP | This study |
| pECD1102 | pUC18R6K-mini-Tn7T-Gm-fetMP | This study |
| pECD1103 | pUC18R6K-mini-Tn7T-Gm-fetM | This study |
| pECD1104 | pUC18R6K-mini-Tn7T-Gm-fetP | This study |
| pECD1105 | pUC18R6K-mini-Tn7T-Gm-fetMp | This study |
| pECD1106 | pUC18R6K-mini-Tn7T-Gm-fetMp-lacZ | This study |
Strain Constructions
To analyze the FetMP system from uropathogenic E. coli strain F11, single fetMP, fetM, fetP or fetMp-lacZ chromosomal integrations were generated at the glmS-linked attTn7 sites of E. coli strains ECA458 (ΔentC ΔfecABCDE ΔfeoABC ΔmntH ΔzupT), ECA150 (ΔlacZYA), or E. coli strain F11 as published (15). The complete fet operon or fetM, including its promoter region (fetMp), was amplified by PCR from genomic F11 DNA using primer pairs fetMPpromd/pGEM fetMPu (for fetMP) and fetMPpromd/pGEM fetMu (for fetM), and cloned into the pGEM®-T Easy vector. Primer pair pGEM Delta fetM NcoId/pGEM Delta fetM NcoIu and pGEM-fetMP as template were used to generate pGEM-fetP. The fetMP promoter region was amplified with primer fetMPpromd and fetMPpromu and cloned into pGEM vector. Each construct was subcloned into plasmid pUC18R6K-mini-Tn7T-Gm. The lacZ gene was amplified by PCR with pUC18-mini-Tn7T-Gm-lacZ as template using primer pUC18 lacZ KpnI d and pUC18 lacZ KpnI u for subcloning into pECD1105 (pUC18R6K-mini-Tn7T-Gm-fetMp) to generate plasmid pECD1106 (pUC18R6K-mini-Tn7T-Gm-fetMp-lacZ). In the presence of the helper plasmid pTNS1, genes were integrated into attTn7 sites of the chromosomes of E. coli strains ECA458 (ΔentC ΔfecABCDE ΔfeoABC ΔmntH ΔzupT), ECA150 (ΔlacZYA), and E. coli strain F11.
Growth Experiments
To produce iron-limited cell cultures of various E. coli strains, LB overnight cultures were diluted 1:400 in TMM without iron and cultivated overnight at 37 °C with shaking. Cultures were diluted a second time 1:400 into fresh TMM without iron. After 2 h of growth at 37 °C with shaking, cultures were diluted again 1:400 in TMM without iron and used for further experiments.
For dose-response experiments at different pH values, FeCl3, FeSO4 with 1 mm ascorbate, or cyclohexane 1,2-diaminetetraacetic acid (CDTA) was added to iron-limited cultures of strains ECA611 (glmS-Gm) or ECA612 (glmS-fetMP) in MES or TMM (pH 5–9). After 16 and 20 h of incubation at 37 °C with shaking, the turbidity was measured at 600 nm.
For anaerobic dose-response experiments, 0.2% glucose as the carbon source and FeSO4 or FeCl3 were added at various concentrations to iron-limited cultures of E. coli strains ECA611 (glmS-Gm), ECA613 (glmS-fetM), ECA614 (glmS-fetP), and ECA612 (glmS-fetMP). Cells were cultivated anaerobically for 17 h in Hungate tubes. Growth was measured as turbidity at 600 nm.
For time course growth experiments, a 1 μm concentration of the iron chelator CDTA was added to iron-limited cultures of the strains ECA611 (glmS-Gm), ECA613 (glmS-fetM), ECA614 (glmS-fetP), and ECA612 (glmS-fetMP). Growth was measured as Klett units over a time period of 20 h.
Periplasmic Fractionation
Periplasmic fractionation was performed as published (16). E. coli cells were cultivated until they reached the mid to late exponential phase, harvested by centrifugation, and suspended in 0.4 volume of Tris-EDTA-sucrose buffer (10 mm Tris-HCl, pH 8.0, 0.1 mm EDTA, 20% (w/v) sucrose) for 10 min at room temperature. Cells were sedimented by centrifugation and suspended in 0.02 volume of ice-cold 5 mm MgSO4 for 20 min at 4 °C. Cells were sedimented by centrifugation, and the resulting supernatant contained the periplasmic fraction.
Fur Titration Assays
Fur Titration Assays were performed as described (17). A high copy number pGEM®-T Easy vector derivative containing the promoter region of fetMP was transformed into E. coli strain H1717 (18) that harbored a chromosomal fhuFp-lacZ fusion. E. coli H1717 transformants were cultivated in LB medium and streaked on MacConkey lactose agar supplemented with 30 μm Fe(NH4)2(SO4)2. After incubation for 18 h at 37 °C, the Lac phenotype was recorded. For a positive control, cells harboring the fhuF promoter region were cloned in the same plasmid (19).
β-Galactosidase Assays
50 μm 2,2′-dipyridyl, 10 μm FeCl3, or 10 μm FeSO4 and 1 mm ascorbate were added to iron-limited cultures of E. coli W3110 cells (ECA615) containing a lacZYA deletion and a single fetMp-lacZ chromosomal integration. Incubation was continued with shaking for 2 h at 37 °C, and the β-galactosidase activity was determined (20). For E. coli strain F11 also harboring a single fetMp-lacZ chromosomal integration, the experiment was performed as described above, but the medium contained 0.2% glucose instead of glycerol to repress expression of the wild-type lacZYA operon of F11.
Sandwich Hybridization Assay
To quantify fetP mRNAs from E. coli strain ECA612 (glmS-fetMP), a sandwich hybridization assay was performed as described previously (21) using two unlabeled helper probes, a biotin-labeled capture probe and a detection probe (supplemental Table S1) that was labeled with a second generation DIG oligonucleotide 3′-end-labeling kit (Roche Applied Science). E. coli strain ECA611 (glmS-Gm) lacking fetMP was used as a negative control. Overnight cultures of both strains were cultivated in TMM without iron (pH 7), diluted 1:20 into fresh medium, and grown to an optical density of 0.7 at 600 nm. Cultures were treated consecutively with 100 μm FeCl3 and 200 μm CDTA, samples were taken (1 ml), cell densities were measured, and the fetP mRNA content was determined by sandwich hybridization. As a reference for this experimental series, the fetP gene was amplified by PCR using primers fetP T7 new d and fetP u, fetP mRNA was synthesized with the MAXIscript T7 kit (Ambion), and the resulting mRNA concentration was determined using the Quant-IT RiboGreen RNA assay kit (Invitrogen). Different amounts of this fetP mRNA were added to clarified cell extracts from negative control cells, a sandwich hybridization assay was performed, and the results were used as a calibration curve.
Expression of fetM, fetP, and fetMP
Strains ECA611 (glmS-Gm), ECA613 (glmS-fetM), ECA614 (glmS-fetP), and ECA612 (glmS-fetMP) were cultivated in TMM (14) containing 2 ml of glycerol and 3 g of casamino acids per liter without additional iron until the turbidity reached 100 Klett units. Total RNA was isolated as described (22). DNase treatment was performed, followed by purification with phenol/chloroform and precipitation with ethanol. RNA concentrations were photometrically determined, and RNA quality was checked on formamide gels (13). To exclude experimental artifacts resulting from DNA contaminations, only RNA was used that did not generate products in a PCR with chromosomal primers. For the RT reaction, 1 μg of total RNA and 0.1 μg of hexamer primers were incubated at 65 °C for 5 min and snap-cooled on ice. After the addition of 0.5 mm dNTP-mix, 10 mm DTT, and 100 units of reverse transcriptase (Superscript II) in reaction buffer (Invitrogen), reverse transcription proceeded for 10 min at room temperature, followed by 1 h at 50 °C. After finishing the RT reaction, the enzyme was inactivated at 70 °C for 15 min. The resulting cDNA was amplified by PCR (for primers, see supplemental Table S1) as published (22) and separated on an ethidium bromide-stained agarose gel. RT-PCR-based amplification of rpoZ-specific RNA served as loading and process control. No fetP-specific band was visible when the RNA came from a fetM-carrying strain or the negative control, and no fetM-specific band was visible when the RNA came from fetP-containing bacteria or the negative control. However, fetP-specific bands occurred when the RNA came from fetP- and fetMP-carrying strains, and fetM-specific bands could be seen when the RNA came from fetM- or fetMP-possessing bacteria. The intensities of the bands in the agarose gel were analyzed using ImageJ (National Institutes of Health, Bethesda, MD). Finally, a band of 1636 bp was visible when the primers 1149 fetM down (RT-PCR) and 2785 fetP up (RT-PCR) were used to PCR-amplify the cDNA. This band corresponded in size with a band coming from the positive control, which used DNA instead of cDNA as template of the PCR. The 1636 bp band did not appear when the RNA was isolated from the fetP strain, the fetM strain, or the negative (water) control (data not shown).
Iron Uptake Experiments
A filtration assay using E. coli strain ECA458 (ΔentC ΔfecABCDE ΔfeoABC ΔmntH ΔzupT) harboring plasmid pECD1098 (pASK-IBA3-fetM), pECD1099 (pASK-IBA3-fetP), or pECD1100 (pASK-IBA3-fetMP) or the empty vector pASK-IBA3 as control was performed as published (23). Briefly, cells were cultivated overnight in LB medium, diluted 1:400 into TMM, cultivated overnight, and diluted into fresh medium to a final turbidity of 30 Klett units. The cultures were incubated with shaking at 37 °C to a turbidity of 60 Klett units, 200 μg of anhydrotetracycline per liter was added to induce expression of the cloned genes, and incubation was continued for an additional 1 h. The cells were then harvested by centrifugation and washed twice with TMM without casamino acids, phosphate, trace elements, and iron. Metal uptake was started by the addition of a reaction mix, leading to final concentrations of 1 μCi of 55Fe, 5 μm FeSO4, and 1 mm ascorbate. The 55FeCl3 (PerkinElmer Life Sciences) had a specific activity of 89.81 Ci/g.
For iron uptake experiments at different pH values, E. coli strains ECA612 (glmS-fetMP), ECA613 (glmS-fetM), ECA614 (glmS-fetP), and ECA611 (glmS-Gm) were incubated in MES-Tris-buffered mineral salts medium (pH 5, 7, and 9) and grown until turbidity doubled. The cells were harvested and washed in fresh buffer before the uptake assay was started (see above).
Expression of fetP-Strep-tag
The fetP gene was amplified by PCR from strain F11 genomic DNA as template using the primer pair pASK3 fetP EcoRId and pASK3 fetP PstIu and cloned into vector pASK-IBA3. E. coli strain BL21 with pECD1099 (pASK-IBA3-fetP) was cultivated in LB medium with shaking at 37 °C until an optical density of 0.8 at 600 nm. Expression of fetP was induced with 200 μg of anhydrotetracycline per liter, and incubation was continued for 3 h at 30 °C. Cells were harvested by centrifugation, and the periplasmic fraction was isolated as described (16).
FetP protein was expressed as a Strep-tagged protein and purified using a Strep-tactin affinity chromatography column according to the manufacturer's protocol (IBA GmbH). Protein concentrations were determined with a NanoDrop ND-1000 spectrophotometer using the molecular mass of the mature periplasmic protein and an extinction coefficient of 29,910 m−1 cm−1 at 280 nm, which was calculated from the primary sequence with the ProtParam tool at ExPASy (available on the World Wide Web) (24). FetP was detected by Western blot analyses after SDS-PAGE with Strep-tactin-horseradish peroxidase conjugate (IBA), and the molecular mass was determined by MALDI-TOF (UltraflexII, Bruker Daltonik GmbH).
Expression of FetP without Strep-tag
The fetP gene without the coding region for the leader sequence was amplified by PCR using primer pair pET22b(+) fetP NcoId/pET22b(+) fetP XhoIu and cloned into the pET22b(+) vector (Novagen), leading to plasmid pECD1101. Recombinant FetP was then expressed with an N-terminal artificial leader sequence and a C-terminal His6 tag, including a thrombin cleavage site. E. coli strain BL21 (pECD1101) was cultivated with shaking in LB medium at 37 °C to an optical density of 0.8. Gene expression was induced with 1 mm isopropyl-β-d-thiogalactopyranoside for 3 h at 30 °C. Cells were harvested by centrifugation, and the periplasmic fraction was isolated as described (16). FetP-His6 was purified by nickel-NTA-agarose affinity chromatography (Qiagen). The tag was removed by treatment with biotinylated thrombin at room temperature for 16 h according to the manufacturer's directions (Novagen). Streptavidin-agarose chromatography and a second affinity chromatography step on nickel-NTA-agarose were used to eliminate thrombin and uncleaved FetP protein, respectively. Successful purification was determined by SDS-PAGE. The untagged protein was concentrated using Amicon ultracentrifuge filter devices (10,000 molecular weight cut-off; Millipore), incubated with 10 mm EDTA for 30 min on ice, and finally dialyzed against different buffers for subsequent experiments. Protein concentrations were determined by absorbance at 280 nm as described above.
For crystallographic studies, FetP was overproduced in E. coli strain BL21 (pECD1101) grown in 2× YT. Each 1-liter culture was inoculated with 2 ml of overnight culture and incubated at 37 °C with shaking to an optical density of 1.0. The culture temperature was reduced to 25 °C for 30 min, gene expression was induced with 0.25 mm isopropyl-β-d-thiogalactopyranoside. After overnight incubation, the cells were harvested by centrifugation, and the periplasmic fraction was isolated as described above. The periplasmic fraction was dialyzed into 30 mm phosphate buffer, pH 7.8, 300 mm NaCl, loaded onto a HisTrap nickel resin affinity column, and FetP was eluted with increasing imidazole. The buffer was exchanged with 30 mm phosphate, pH 7.8, or 20 mm BisTris, pH 6.5, by ultrafiltration, and the protein was digested with thrombin (Hematologic Technologies). Benzamidine beads were used to remove the thrombin. The resulting FetP protein (referred to as “as-isolated”) was concentrated to at least 25 mg/ml using Amicon filter devices (10,000 molecular weight cut-off; Millipore).
Copper-reconstituted FetP
As-isolated FetP was applied to a Source Q ion exchange column and eluted with a NaCl gradient in 30 mm phosphate, pH 7.8. Apo-FetP was prepared by treatment with a 20-fold molar excess of EDTA in 20 mm BisTris, pH 6.5, for 3 h on ice, followed by repeated buffer exchange by ultrafiltration. Copper-reconstituted FetP (Cu-FetP) was prepared by the direct addition of a 1.5-fold molar excess of CuCl2 to apo-FetP and incubation of the mixture on ice for 20 min, followed by ultrafiltration to at least 25 mg/ml protein.
Crystallization and Structure Determination
As-isolated FetP and Cu-FetP were crystallized by the sitting drop method. A single, large crystal of as-isolated FetP appeared within 2 weeks of incubation with a reservoir of 20% PEG 3350, 0.2 m ammonium citrate, pH 7. The crystals were soaked in mother liquor supplemented with 20% ethylene glycol as a cryoprotectant and flash frozen using liquid nitrogen, and data were collected at 100 K at the Stanford Synchrotron Radiation Lightsource (SSRL) on beamline 9-2. An x-ray fluorescence scan was used to detect transition metals in the crystal. A 1.6 Å resolution data set was collected using incident radiation with a wavelength of 0.97964 Å. Data were processed using iMosflm (25). MolRep (26) was used to determine initial phases of as-isolated FetP using one chain from the crystal structure of P19 (Protein Data Bank code 3LZO). Cycles of structure refinement and building were performed using Refmac 5 (27) from the CCP4 suite of programs (28) and the program Coot (29). The crystal structure of as-isolated FetP was determined in space group P32 with four subunits per asymmetric unit and twinning with four twin domains as defined by Refmac (calculated distribution of 41.40, 40.74, 9.00, and 8.85%). Sufficient electron density was observed to build residues 2–153 of chains A and B and residues 2–152 of chains C and D. A clear break in the electron density was observed at residue 34 of Chain B and is therefore not modeled. A Ramachandran plot, with 96.6% of the residues within the favored regions and only Phe3 of chains B and C flagged as outliers, shows that the structure has excellent stereochemistry (29).
Cu-FetP was crystallized in 0.1 m BisTris, pH 6.5, 25% pentaerythritol ethoxylate (15:4 EO/OH), 25 mm ammonium sulfate. Small (∼0.05 mm), thin crystals of Cu-FetP in the orthorhombic space group P212121 appeared within a week and were harvested 3 months later to improve thickness. The crystals were flash frozen using liquid nitrogen with no additional cryoprotectant, and data were collected at 100 K at the SSRL on beamline 7-1. An x-ray fluorescence scan was used to ensure no detectable contamination by metals other than copper in the sample and to determine the optimal wavelength for maximal copper anomalous signal. A 1.7 Å resolution data set at 0.97641-Å wavelength and a copper anomalous data set at 1.37725-Å wave length were collected on a single crystal. MolRep was used to determine initial phases using one chain from the crystal structure of as-isolated FetP. The crystal structure of Cu-FetP was determined with two subunits per asymmetric unit. Electron density was observed to build residues 4–153 of chain A and 4–152 of chain B. Other than the two residues at the C terminus of chain A, the average B-factors of all residues in both chains are under 35 Å2, and 98.9% of the residues in the Cu-FetP structure fall within the preferred regions, with a single outlier (Gly33) observed in the Ramachandran plot. All structural figures have been generated with the program PyMOL (Version 1.2r3pre, Schrödinger, LLC).
Ferric Reductase Assay
The iron reduction assay was performed as described earlier (30). E. coli strains ECA611 (glmS-Gm) and ECA614 (glmS-fetP) were grown in TMM without additional iron until the turbidity at 600 nm reached 0.5 units. Cells of 6 ml of each culture were harvested by centrifugation, washed twice, and resuspended in 1 ml of assay buffer (50 mm sodium citrate, pH 4.0, 0.5% glucose). For the assay, 132 μg, dry weight, of cells were incubated with 1 mm bathophenanthroline disulfonic acid and increasing concentrations of FeCl3 at 37 °C for 2 h. Absorbance at 520 nm was kinetically recorded. The amount of ferrous ions produced was estimated by means of a calibration curve using a solution of known ferrous ion concentrations. Ferric reductase activity was expressed in nmol of ferrous ions produced under the assay conditions.
Analytical Ultracentrifugation
FetP (0.3 mg/ml) in 25 mm Tris/HCl, pH 7.2, in ultrapure water supplemented with 100 or 20 mm NaCl or a 300 μm concentration of either ZnCl2, CuCl2, or FeCl3 or no additives was centrifuged at 25 °C in a Beckman Optima XL-A centrifuge equipped with an An50Ti rotor and double sector cells. Sedimentation equilibrium measurements were performed at 8,000 rpm, and sedimentation velocity was monitored at 40,000 rpm. The data were recorded at 280 nm and analyzed using the software provided by Beckman Instruments.
Atomic Absorption Spectroscopy (AAS)
FetP-Strep-tag protein (7 μm) was dialyzed against 10 mm EDTA on ice for 30 min and incubated with 35 μm ZnCl2, CuCl2, MnCl2, FeCl3, or FeSO4 or no additives in 25 mm Tris-HCl, pH 7.2, for 30 min on ice. In the case of FeSO4, 1 mm ascorbate was added to maintain Fe(II). Excess metal was removed by washing five times with ice-cold buffer using Amicon ultracentrifuge filter devices (10,000 molecular weight cut-off; Millipore). Protein concentrations were determined as above, and samples were mineralized with 0.5% HNO3 and 0.5% H2O2 for 5 min at 95 °C before AAS (ZEEnit®600/650, Analytik Jena AG).
Circular Dichroism Spectroscopy
Far-UV CD spectra were recorded on a Jasco J710 spectropolarimeter. Spectra of periplasmic FetP without a tag (0.9 mg/ml) were recorded in 25 mm Tris-HCl, pH 7.2, using ultrapure water at 25 °C in a 0.1-mm cuvette.
Isothermal Titration Calorimetry (ITC)
Metal ion binding affinities of tagless FetP were analyzed in 25 mm BisTris, pH 7.2, or 25 mm ACES, pH 7.2, by monitoring heat changes upon injection of metal ion solutions into a FetP solution in a VP-ITC microcalorimeter (MicroCal) at 25 °C. Concentrations of the stock metal solutions were initially determined by AAS and diluted into the respective dialysis buffers for the protein. Titration was carried out by 28 or 56 injections of 10 and 5 μl of metal ion solution, respectively, at concentrations of 300–800 μm into the stirred reaction cell of the calorimeter (1.4 ml) containing 35 μm FetP. Heats of titrant dilutions were measured by identical injections into buffer without protein and subtracted to yield net reaction heats. At least three titrations were carried out for each metal. Results were evaluated using the MicroCal Origin software.
Diethyl Pyrocarbonate Titrations
Diethyl pyrocarbonate titrations of the histidine residues of FetP were performed as published (31, 32).
RESULTS
The FetMP System Is Required for Growth under Iron Limitation
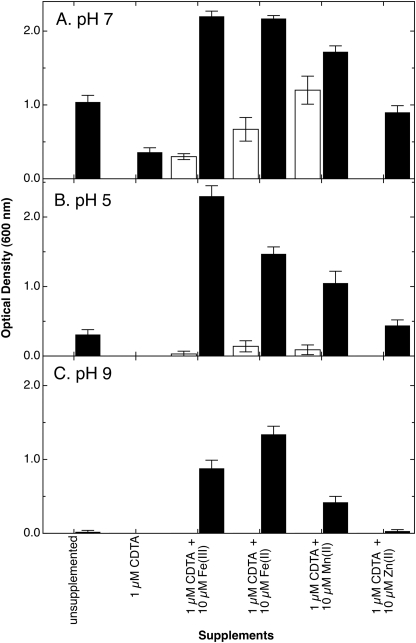
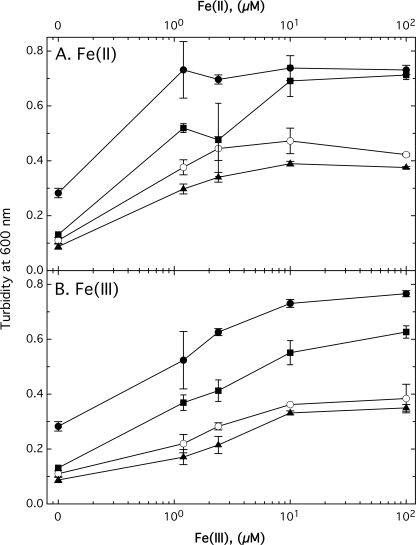
To investigate the contribution of FetMP to growth, a single copy of fetMP was inserted into the chromosome of E. coli strain ECA458, a derivative of wild type strain W3110 devoid of all characterized iron uptake systems. Growth of strain ECA612 (glmS-fetMP) was compared with that of the negative control strain ECA611 (glmS-Gm). Cells harboring fetMP grew in minimal medium without added iron at pH 7, whereas the control strain was unable to grow under the same iron-limited conditions (Fig. 1A). Growth of strain ECA612 was still observed in the presence of the metal chelator CDTA added to diminish the bioavailability of trace iron. Supplementing cultures with low concentrations of Fe(II), Fe(III), and to some extent Mn(II) in the presence of CDTA greatly improved growth of ECA612 in contrast to the control strain lacking fetMP (Fig. 1A). The growth difference of both strains with CDTA plus Mn(II) was less prominent compared with CDTA plus iron. Nevertheless, the residual growth of the control strain indicates the presence of at least one other unknown manganese/iron uptake system despite the ΔentC ΔfecABCDE ΔfeoABC ΔmntH ΔzupT deletions. Supplementation with Zn(II) did not result in the growth of ECA612 beyond the level observed in cultures without added CDTA (Fig. 1A). The slight increase in cell densities with Zn(II) may be due to displacement of trace iron bound by CDTA.
FIGURE 1.
FetMP stimulates growth of an E. coli strain devoid of iron and manganese uptake systems. The fetMP operon or an antibiotic cassette control was inserted into the chromosome of strain ECA458 (ΔentC ΔfecABCDE ΔfeoABC ΔmntH ΔzupT), resulting in strains ECA612 (glmS-fetMP) (black bars) and ECA611 (glmS-Gm) (white bars). Iron-limited cultures of both strains were diluted 400-fold into Tris-buffered (A and C) or MES-buffered (B) mineral salts medium at pH 7 (A), pH 5 (B), or pH 9 (C) without added iron or containing the chelator CDTA without further additives or containing CDTA plus the indicated metal salts (FeCl3, FeSO4 with 1 mm ascorbate, MnCl2, or ZnCl2). The cultures were cultivated at 37 °C with shaking. Turbidity was determined at 600 nm after 16 h (A) or 20 h (B and C). Averages of three independent experiments with S.D. values (error bars) are shown.
Under acidic (pH 5) or alkaline (pH 9) conditions (Fig. 1, B and C, respectively), the presence of fetMP also supported the growth of strain ECA612, whereas no growth was observed for the control. At either pH, the addition of CDTA abolished growth of strain ECA612. However, supplementation with Fe(II) or Fe(III) can rescue growth of the fetMP-containing strain but not that of the negative control. This effect is more pronounced than the addition of either Mn(II) or Zn(II) at both acidic and alkaline conditions. Growth with the addition of CDTA and Zn(II) was similar to that of unsupplemented cultures. Other divalent transition metal cations (Co(II), Cu(II), Cd(II), and Ni(II)) were also tested, but growth response of strain ECA612 was similar to that of CDTA plus Zn(II), even if these metals were supplied at concentrations as high as 50 μm (data not shown). The latter dosage implies that FetMP-mediated iron uptake is not inhibited by these divalent ions. Thus, FetMP probably serves as an iron import system and may also transport Mn(II).
Expression of fetMP Is under the Control of the Iron Regulator Fur
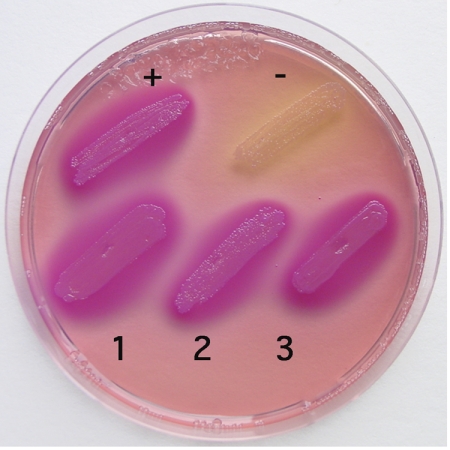
In silico analysis indicated the presence of a binding site (Fur box) for the global repressor of iron homeostasis, Fur, upstream of the putative fetMP operon in E. coli strain F11 (data not shown). To investigate regulation of fetMP expression by Fur in response to iron, a Fur titration assay was conducted (17). In this assay, derepression by iron results in cell colorization if the promoter of interest is Fur-regulated. This was the case when fetM promoter (fetMp)-harboring strains were assayed (Fig. 2), which demonstrated that a functional Fur box regulated the expression of fetMP in an iron-dependent manner.
FIGURE 2.
Iron-dependent regulation of fet in a Fur titration assay. E. coli cells containing a fhuF-lacZ fusion and the vector plasmid pGEM®-T-Easy with the fetM promoter (1, 2, and 3; three independent clones), the positive control fhuFp, or no insert (negative control) were cultivated overnight on MacConkey lactose agar plates supplemented with 30 μm Fe(NH4)2(SO4)2 at 37 °C. The presence of a Fur box on a high copy number plasmid titrates the Fur repressor away from fhuF-lacZ and leads to β-galactosidase activity. This results in lactose consumption and purple staining of the colonies.
To corroborate regulation of fetMP expression, a fetMp-lacZ reporter gene fusion was transferred as a single-copy insertion into the chromosome of strain ECA150, a ΔlacZYA derivative of E. coli wild type strain W3110, and the original strain F11. When both strains were cultivated at various pH values in the presence of the iron chelator 2,2′-dipyridyl, maximum up-regulation of reporter gene (4.5-fold) occurred at pH 6.5 (supplemental Fig. S1) (data not shown).
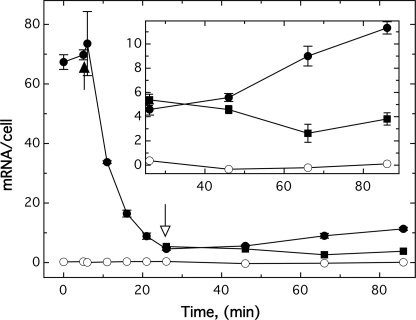
The total mRNA transcripts in E. coli strain ECA612 cultivated in TMM without added iron were also examined. The starting culture contained about 70 fetP-specific mRNAs per cell (Fig. 3). This number dropped to about 5 copies per cell within 20 min after the addition of 100 μm FeCl3, indicating an apparent half-life of 5 min (regression coefficient 99.1%) for fetP mRNA. The subsequent addition of 200 μm of the metal cation chelator CDTA led to a slow increase of fetP mRNAs per cell (Fig. 3, inset). CDTA treatment did not fully recover the fetP expression level probably due to iron storage during the initial growth at 100 μm iron. Taken together, fetMP expression is indeed controlled by the availability of iron through Fur, indicating that fet may be involved in iron uptake.
FIGURE 3.
Expression levels of fetMP depend on iron availability. E. coli strains ECA612 (glmS-fetMP) (●) and negative control ECA611 (glmS-Gm) (○) were cultivated in TMM (pH 7) without added iron at 37 °C with shaking. After 5 min of incubation, 100 μm FeCl3 was added (black arrow). At the 25 min time point, the cultures were divided, and 200 μm CDTA was added to one set of cultures (open arrow, ●), whereas no addition was made to the other set (■). The cells were harvested at various time points, and the fetP mRNA content was determined using the sandwich hybridization technique. The inset indicates the mRNA levels of fetMP-containing cells after the addition of 200 μm CDTA. Averages of three independent experiments with S.D. values (error bars) are shown.
Contribution of FetM and FetP to Growth under Iron Limitation and Iron Uptake
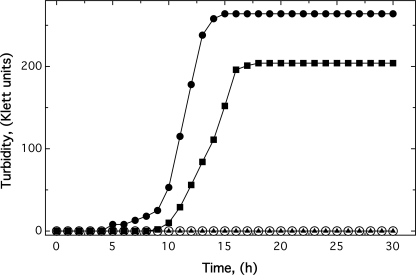
E. coli strains ECA612 (glmS-fetMP) and ECA611 (glmS-Gm) were cultivated in TMM with glycerol as the carbon source in the presence of 1 μm CDTA without added iron. The negative control strain ECA611 was unable to grow under these conditions (30-h incubation at 37 °C; Fig. 4). In contrast, E. coli strain ECA612 with glmS-fetMP exited lag phase after 5 h and reached a final turbidity of 264 Klett units (Fig. 4).
FIGURE 4.
The permease FetM is required for growth of E. coli strain ECA458 (ΔentC ΔfecABCDE ΔfeoABC ΔmntH ΔzupT) in the presence of the iron chelator CDTA. The fetMP operon, fetM, fetP, or empty vector control was inserted into the chromosome of strain ECA458, resulting in strains ECA612 (glmS-fetMP) (●), ECA613 (glmS-fetM) (■), ECA614 (glmS-fetP) (▴), and ECA611 (glmS-Gm) (○). Iron-limited cultures of these four E. coli strains were diluted 400-fold into TMM containing no iron but 1 μm CDTA. The cultures were cultivated at 37 °C with shaking, and the turbidity was measured as Klett units. Shown are averages of four independent experiments.
To investigate the individual contributions of FetM and FetP for growth, their genes were inserted as single copies into the chromosome of E. coli strain ECA458 (ΔentC ΔfecABCDE ΔfeoABC ΔmntH ΔzupT) to give strains ECA613 (glmS-fetM) and ECA614 (glmS-fetP). Expression of either gene was verified by RT-PCR experiments, which indicated that fetP or fetM mRNA levels in the respective single gene strains were 53–67% of those in the fetMP-containing strain. Moreover, using RNA from the fetMP-harboring strain, an RT-PCR product was obtained using a fetP-specific primer with a fetM-specific primer (data not shown) to show that fetMP was transcribed as an operon. The presence of fetP alone did not rescue the iron uptake-deficient strain ECA458, whereas fetM expression led to a retarded onset of growth that reached a lower final turbidity (204 Klett units) compared with strain ECA612 (glmS-fetMP) (Fig. 4). Thus, FetM alone was essential for growth in the presence of 1 μm CDTA, and FetP co-expression was required for maximal growth.
Under anaerobic conditions, the presence of FetMP also stimulated growth of E. coli strain ECA458 (ΔentC ΔfecABCDE ΔfeoABC ΔmntH ΔzupT) (Fig. 5). Relative to the control strain (ECA611), the presence of FetMP enhanced growth in the absence of added iron, which is in contrast to either FetM or FetP alone. Interestingly, the presence of FetP alone inhibited Fe(II) acquisition slightly (Fig. 5). At iron concentrations of 1 μm and above, FetM-containing cells generally reached cell densities between that of FetMP-containing and control cells. The presence of high concentrations (100 μm) of Fe(II)SO4 but not Fe(III)Cl3 stimulated the growth of ECA613 (glmS-fetM) and ECA612 (glmS-fetMP) similarly (Fig. 5). The presence of FetP in addition to FetM seemed to be required for maximal growth at high Fe(III) but not at high Fe(II) concentrations, with a 1.35 ± 0.02-fold difference between the two strains at 100 μm Fe(III) (t test, p > 0.999) (Fig. 5 and Table 2). This effect was inhibited in the presence of 1 μm Ag(I), which decreased the growth advantage to 1.06 ± 0.03-fold. Ag(I) is an analog of Cu(I) that can compete for the copper-binding sites of FetP (see below), preventing electron transfer. These results suggest involvement of FetP in reduction of Fe(III) as part of an iron uptake system.
FIGURE 5.
FetMP facilitate iron acquisition of E. coli strain ECA458 (ΔentC ΔfecABCDE ΔfeoABC ΔmntH ΔzupT) derivatives under anaerobic conditions. E. coli strains ECA612 (glmS-fetMP) (●), ECA613 (glmS-fetM) (■), ECA614 (glmS-fetP) (▴), and ECA611 (glmS-Gm) (○) were cultivated for 17 h at 37 °C anaerobically in TMM with 0.2% glucose as carbon source. Additionally, the growth medium contained the indicated concentrations of Fe(II)SO4 (A) or Fe(III)Cl3 (B). Turbidity at 600 nm was determined, and averages of four independent experiments with S.D. values (error bars) are shown.
TABLE 2.
Anaerobic growth in the presence of Fe(III) and Ag(I)
E. coli strains ECA613 (glmS-fetM) and ECA612 (glmS-fetMP) were anaerobically cultivated in the dark for 17 h at 37 °C in TMM with 100 μm Fe(III) and 0.2% glucose as a carbon source. In half of the samples, the growth medium was additionally supplemented with 1 μm Ag(I). Turbidity at 600 nm was determined, and averages of three independent experiments with S.D. values are shown. Statistics were done with Student's t test, 4 degrees of freedom. For comparison of ECA612/ECA613 without silver, F(t) = 41.6 (p > 0.999) and with silver F(t) = 5.6 (p > 0.995). For ECA613 with or without silver, F(t) = 6.1 (p > 0.995), and for ECA612 with or without silver, F(t) = 18.6 (p > 0.999).
| Bacterial strains | Turbidity at 600 nm |
|
|---|---|---|
| 0 μm Ag(I) | 1 μm Ag(I) | |
| ECA613 (glmS-fetM) | 0.647 ± 0.005 | 0.670 ± 0.008 |
| ECA612 (glmS-fetMP) | 0.873 ± 0.005 | 0.713 ± 0.012 |
Increased Iron Reduction by Cells Expressing fetP
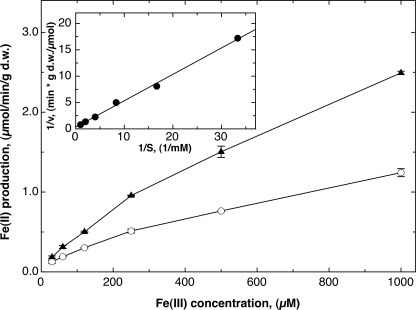
To test the hypothesis that FetP is an Fe(III) reductase, an iron reductase assay was performed (30). The formation of Fe(II) from Fe(III) was quantified with a colorimetric assay using bathophenanthroline disulfonic acid. The negative control strain ECA611 (glmS-Gm) was able to reduce Fe(III) to Fe(II), but the presence of FetP in strain ECA614 (glmS-fetP) doubled the reduction rate (Fig. 6). Analysis of the reduction rate difference by a Lineweaver-Burk plot yielded an apparent vmax of 3 μmol of Fe(II)/min/g dry weight and an apparent Km of 1.5 mm Fe(III) (regression coefficient 99.8%; Fig. 6, inset). Therefore, FetP-producing E. coli cells reduced Fe(III) twice as fast as control cells, supporting the hypothesis that FetP functions as a Fe(III) reductase.
FIGURE 6.
FetP functions as an iron reductase in vivo. An iron reductase assay was performed with E. coli strains ECA614 (glmS-fetP) (▴) and ECA611 (glmS-Gm) (○) in the presence of various concentrations of Fe(III) chloride. Formation of Fe(II) was recorded with 1 mm bathophenanthroline disulfonic acid (BPS) for 2 h at 37 °C at 520 nm using 0.132 mg dry weight (d.w.) of cells, obtaining a reduction rate of 1/min. An equilibration experiment yielded an absorption coefficient of 0.0695/nmol of Fe(II) under these conditions. This value was used to calculate the iron reduction rate in μmol/min/g dry weight, which was plotted against the Fe(III) concentration (three experiments, deviation bars shown). The inset shows a Lineweaver-Burk plot of the reciprocal difference between both curves (1/(vfetP − vcontrol)) against 1/S in 1/mm. A linear regression yielded an apparent vmax of 3 μmol of Fe(II)/min/g dry weight and an apparent Km of 1.5 mm Fe(III) (regression coefficient 99.8%).
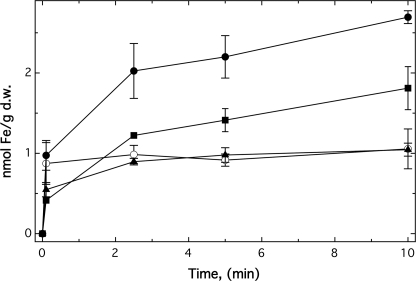
Expression of fetMP Leads to Increased Iron Uptake
No significant differences in iron accumulation were observed when 55Fe uptake of strain ECA612 (glmS-fetMP) was compared with the negative control strain ECA611 (glmS-Gm) in TMM at pH 7 (data not shown). Therefore, the fet genes were cloned into pASK-IBA3 to increase gene expression. In the resulting strains, expression of fetMP led to a 2.8-fold increase in iron accumulation after 10 min (Fig. 7). Whereas FetP alone did not enhance iron uptake by the cells, FetM alone increased this process 1.7-fold (Fig. 7). In contrast to pH 7, enhanced uptake of 55Fe was observed for strain ECA612 (glmS-fetMP) compared with ECA611 (glmS-Gm) at pH 9 and pH 5 (supplemental Fig. S2). As at pH 7, FetM was necessary for iron uptake, and FetP enhanced iron import. Increasing the pH value from 5 to 9 led to a 20-fold higher iron accumulation of the cells after 10 min.
FIGURE 7.
FetMP is an iron uptake system. Uptake of 55Fe by cells of strain ECA458 (ΔentC ΔfecABCDE ΔfeoABC ΔmntH ΔzupT) and its fet derivatives were compared using the filtration method. Strain ECA458 containing the vector pASK-IBA3 only (○), fetMP (●), fetM (■), or fetP (▴) was incubated with 200 μg of anhydrotetracycline per liter for 1 h to induce expression of the cloned genes. For the uptake experiments, 55Fe(II)Cl3 at a final 1 μCi, 1 mm ascorbate, 5 μm FeSO4 were added to the cells, and samples were removed at the indicated time points. Averages of three independent experiments with S.D. values (error bars) are shown. d.w., dry weight.
FetP Is a Dimeric, Periplasmic Metal-binding Protein
A unique feature of the FetMP system is the presence of FetP, a predicted 19,224-Da, 175-residue protein. SignalP analysis (see the SignalP 3.0 site on the World Wide Web) predicted a highly probable 24-residue N-terminal leader featuring the n-region MTMKKT, the h-region LIASAVMASIFI, and the c-region APAAFAF. To test this prediction, the fetP gene was cloned into vector pASK3 (producing plasmid pECD1099), which added a C-terminal Strep-tag epitope to FetP and changed the predicted sizes of the preprotein and the mature protein to 21,888 and 18,662 Da, respectively. Transformation of pECD1099 into E. coli strain BL21 allowed for FetP-Strep-tag protein purification from the periplasmic fraction of these bacteria (supplemental Fig. S3, inset), demonstrating that FetP was indeed a periplasmic protein.
MALDI-TOF analysis (supplemental Fig. S3) of purified FetP-Strep-tag protein yielded two major peaks corresponding to 18,676 and 18,739 Da. The 18,676 Da signal agreed with the predicted size of 18,662 Da of the periplasmic protein lacking its leader sequence within an error of 0.075%. The size difference of 63 Da was close to the atomic mass of copper of 63.43 Da. Therefore, it was likely that the mature periplasmic FetP-Strep-tag protein was purified from E. coli in two forms: with and without one tightly bound copper cation.
To analyze the metal-binding properties of the protein, purified FetP-Strep-tag was dialyzed against EDTA to remove any bound copper and other metal cations; incubated in the presence of a 5-fold excess of Fe(II), Fe(III), Mn(II), Cu(II), or Zn(II); and washed with buffer to remove excess unbound metal ions. Metal content was quantified by AAS. Molar metal/protein ratios of 0.6 iron, 0.7 manganese, 1.5 copper, or 1.0 zinc, respectively, were determined (data not shown). When FetP-Strep-tag was cross-linked using tris-2,2‘-bispyridylruthenium(II), a band corresponding to a FetP dimer appeared after 10 s of incubation time (data not shown).
Because the Strep-tag may interfere with the metal-binding process, the fetP gene was recloned into pET22b(+) to produce a His-tagged protein with an artificial periplasmic leader (pECD1101). FetP was then purified by nickel-NTA-agarose affinity chromatography from the periplasmic fraction of E. coli strain BL21, and the His tag was removed by thrombin digest. The success of removal of the His tag and of the biotinylated protease was controlled by SDS-PAGE (data not shown).
Because metal binding was required for dimerization of Tp34, a FetP homolog from T. pallidum (12), dimerization of FetP was studied using analytical ultracentrifugation (Table S2). The experimentally determined mass of FetP was 35,283 Da, which was independent of variations in sodium chloride concentration or the presence or absence of transition metals, indicating a stable dimer. The observed sedimentation coefficient of 2.7–2.8 S also fitted well to a dimeric species. Thus, FetP was a dimer even in the absence of transition metals.
CD spectroscopy was used to investigate if transition metals changed the structure of FetP (supplemental Fig. S4). Almost no changes in the spectrum were visible in the presence of a 2:1 ratio of Fe(II), 2:1 Mn(II), or 3:1 Zn(II), but the addition of Cu(II) in a ratio of copper/FetP of 2:1 altered the spectrum significantly (supplemental Fig. S4A). The difference spectrum (no addition minus Cu(II) addition) started with a strong minimum at 190 nm and displayed two maxima at 202 and 233 nm (supplemental Fig. S4B). Thus, binding of copper but not of other metal cations may lead to conformational changes of FetP.
To quantify the metal binding affinities of FetP, ITC (Table 3) was employed. These ITC metal-binding studies used 34.3 μm FetP, including negligible amounts of free metal. Therefore, the data obtained represent minimum (weakest) affinity values. A variety of buffers were tested, but most buffers caused either Cu(II) or Zn(II) to precipitate. Neither 25 mm ACES nor BisTris, both at pH 7.2, caused precipitation and were finally used. However, Zn(II) binding to FetP in Bis-Tris and Cu(II) binding in ACES buffer was not detected by ITC, probably due to buffer effects (Table 3).
TABLE 3.
Metal binding to FetP (34.3 μm) as determined by ITC
| Conditionsa | Metal titrated | n | Kd | ΔH | ΔS | ΔG |
|---|---|---|---|---|---|---|
| μm | kcal/mol | cal/mol/K | kcal/mol | |||
| BisTris | Cu2+ | 0.915 | 4.1 | −2.18 | +17.3 | −7.33 |
| BisTris, 27.4 μm Mn2+ | Cu2+ | (1.0)b | 69.4 | −1.52 | +13.9 | −5.66 |
| BisTris, 160 μm Zn2+ | Cu2+ | NBc | NB | NB | NB | NB |
| BisTris | Mn2+ | 0.404 | 0.6 | −3.82 | +15.5 | −8.44 |
| BisTris, 94 μm Cu2+ | Mn2+ | PEd | PE | PE | PE | PE |
| BisTris, 160 μm Zn2+ | Mn2+ | 0.372 | 10.9 | −3.49 | +11 | −6.77 |
| BisTris | Zn2+ | NB | NB | NB | NB | NB |
| ACES | Zn2+ | 1.4 | 15.2 | +2.27 | +29.6 | −6.55 |
| ACES, 34.3 μm Mn2+ | Zn2+ | (1.0)b | 348 | +8.89 | +45.6 | −4.70 |
| ACES, 34.3 μm Mn2+ | Zn2+ | (1.5)b | 303 | +5.48 | +34.5 | −4.80 |
| ACES, 94 μm Cu2+ | Zn2+ | NB | NB | NB | NB | NB |
| ACES | Mn2+ | 0.426 | 3.6 | −4.66 | +9.27 | −7.42 |
| ACES, 160 μm Zn2+ | Mn2+ | 0.548 | 14.4 | −9.37 | −9.3 | −6.60 |
| ACES, 94 μm Cu2+ | Mn2+ | 0.435 | 45 | −22.0 | −46.1 | −8.26 |
| ACES | Cu2+ | NB | NB | NB | NB | NB |
a 25 mm buffer, pH 7.2.
b n assumed in the modeling to allow calculation of the other values.
c NB, no binding was observed.
d PE, positive enthalpy. ΔH of about +1 kcal/mol, but the data could not be used in binding site models to calculate discrete values for Kd.
FetP bound ∼2 coppers/dimer in BisTris with a Kd of 4.1 μm in a process that was enthalpy- and entropy-driven. In contrast, FetP bound between 2 and 3 Zn(II)/dimer in ACES buffer with a Kd of 15.2 μm, but binding was exclusively driven by a gain in entropy (Table 3 and supplemental Fig. S5, a and b). Mn(II) was used as a substitute for Fe(II). FetP bound ∼1 Mn(II)/dimer in either buffer with Kd values of 0.6 μm (BisTris) or 3.6 μm (ACES). Similar to copper binding, manganese binding was enthalpy- and entropy-driven (Table 3 and supplemental Fig. S5, c and d). Interestingly, neither copper nor zinc prevented binding of manganese by FetP (Table 3). In the presence of Cu(II) or Zn(II), FetP bound ∼1 Mn(II) per dimer with Kd values between 14 and 45 μm (ACES). Likewise, the presence of Mn(II) did not prevent binding of copper or zinc by FetP. Thus, there is one manganese (or, by analogy, one iron) binding site per FetP dimer. This site is different from the binding sites for copper or zinc, which do not interfere with the binding of Mn(II). Although no binding of Zn(II) in BisTris and Cu(II) in ACES was measured, introducing Zn(II) or Cu(II) to FetP prevented binding of the other metal in these two buffer systems (Table 3). Therefore, copper and zinc appear to compete for the same binding sites or at least for the same ligands. The negative enthalpy and a low Kd value of the copper binding, the presence of Cu(II) in FetP periplasmic preparations, and the conformational effects of copper ions from the CD spectra support Cu(II), rather than Zn(II), as the native cation in FetP.
FetP was also titrated with diethyl pyrocarbonate prior to the ITC measurements to chemically modify surface-exposed histidine residues. The occlusion of accessible histidine residues did not prevent binding of Zn(II) or Cu(II) by FetP, although a stepwise increase in the absorbance at 243 nm during diethyl pyrocarbonate titration (32) was observed (data not shown). This indicates that if histidines are ligands of these cations, they are buried within FetP.
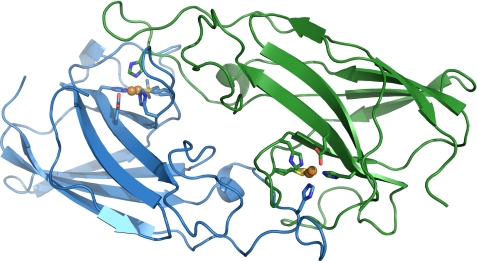
Overall Structure of FetP
The crystal structure of recombinant FetP expressed and isolated from the periplasm of E. coli was solved to 1.6 Å resolution (Fig. 8 and Table 4). The four molecules of FetP in the asymmetric unit are arranged in two dimers in which each subunit is related by non-crystallographic 2-fold rotational symmetry (root mean square deviation of 0.6 Å over all Cα). Each FetP dimer is a relatively flat, oval-shaped macromolecule, with dimensions of ∼20 × 35 × 75 Å. A gap in the electron density suggests proteolytic cleavage at residue 34 on chain B, which was left unmodeled. Inspection of the protein sample on an SDS-polyacrylamide gel confirmed the presence of two additional distinct bands correlating with the FetP cleavage products along with the band for full-length FetP (data not shown).
FIGURE 8.
General structure of the Cu-FetP dimer with bound copper. The two subunits are shown in blue and green, respectively. Copper atoms are shown in orange. Copper ligands are represented as stick models.
TABLE 4.
Data collection and refinement statistics for FetP
| As-isolated FetP | Cu-FetP | |
|---|---|---|
| Data collectiona | ||
| Resolution range (Å) | 50.00–1.60 | 38.41–1.70 |
| (1.69–1.60) | (1.79–1.70) | |
| Space group | P32 | P212121 |
| Cell dimensions (Å) | a = 134.85 | a = 38.41 |
| b = 134.85 | b = 51.97 | |
| c = 45.42 | c = 146.65 | |
| Wavelength (Å) | 0.97964 | 0.97641 |
| Unique reflections | 114166 | 31725 |
| Completeness (%) | 93.6 (88.1) | 96.7 (93.5) |
| Average I/σI | 8.3 (2.4) | 11.5 (2.9) |
| Redundancy | 4.1 (3.6) | 4.8 (4.2) |
| Rmerge | 0.080 (0.393) | 0.030 (0.143) |
| Refinement | ||
| Rwork/Rfree | 0.171/0.208 | 0.196/0.235 |
| No. of waters | 356 | 223 |
| Average B-factors (Å2) | ||
| All atoms | 21.1 | 32.8 |
| Protein | 20.8 | 32.2 |
| Copper | 22.7 | |
| Waters | 25.4 | 40.3 |
| Root mean square deviation, bond lengths (Å) | 0.012 | 0.012 |
| Ramachandran plot | ||
| Most favorable (%) | 96.6 | 98.9 |
| Allowed (%) | 3.1 | 0.7 |
| Protein Data Bank code | 3NRP | 3NRQ |
a Values for the highest resolution shell are shown in parentheses.
Examination of the metal content of as-isolated FetP crystals by an x-ray fluorescence scan revealed the presence of copper and nickel in approximately equal amounts. Fluorescence signal strength was weak (data not shown), consistent with low metal occupancy in the structure. Nickel contamination of the sample is probably due to purification with a Ni(II) affinity chromatography. Because no other exogenous transition metals were introduced during the protein purification process and in agreement with the MALDI-TOF result for purified FetP-Strep-tag (supplemental Fig. S3), copper was probably acquired from the medium during FetP expression in E. coli. Based on these results, FetP was treated with EDTA, and the crystal structure of copper-reconstituted FetP (Cu-FetP) was solved to 1.7 Å resolution. Structural comparisons between the as-isolated and copper-reconstituted FetP crystal structures reveal minimal overall fold differences (root mean square deviation of 0.4–0.6 Å over all Cα as calculated by TopMatch (33)). Unless specified, any subsequent descriptions of the structural features of FetP will be based on the copper-reconstituted structure.
A single dimer was observed in the Cu-FetP crystal structure, with each subunit composed primarily of two 4-stranded anti-parallel β-sheets stacked upon each other in an immunoglobulin-like fold (Fig. 8). Structural alignments of Cu-FetP reveal greater overall fold similarity with P19 (Protein Data Bank code 3LZO; root mean square deviation of 0.7 Å over all Cα) than Tp34 (Protein Data Bank code 2O6E; root mean square deviation of 1.5 Å).
The dimer interface was analyzed with the program PISA (34), which reveals an extensive interface area of ∼1600 Å2 (or ∼20% of the total solvent-accessible surface area of each subunit) and involves 45 residues from each subunit. An estimated solvation free energy gain of −45 kcal/mol upon interface formation indicates a predicted stable dimer in solution. Tight packing of the dimer interface is also supported by lower than average B-factors in this region (data not shown).
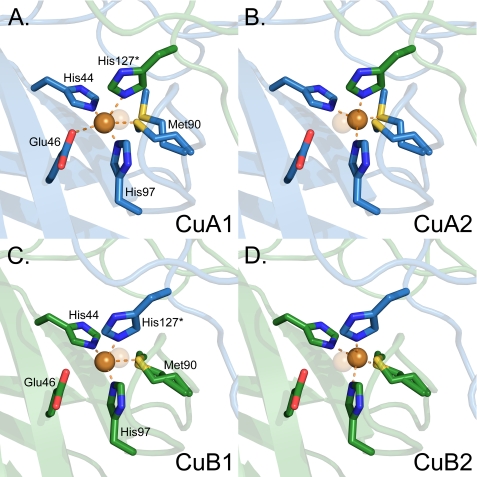
The FetP Copper-binding Site
Anomalous dispersion diffraction data collected at the copper anomalous edge indicate the locations of the copper ions in the structure. Two copper ions (CuA and CuB) were identified from ∼30 σ symmetry-related peaks in the anomalous dispersion maps. The copper ions are buried, separated by ∼29 Å and are related by the 2-fold symmetry of the FetP molecule. Each copper site (Fig. 9) is located at the dimer interface and is formed by three histidines (His44, His97, and His127* (an asterisk denotes a ligand originating from the symmetry-opposing subunit)), Met90, and Glu46. The buried nature of the copper sites agrees with the inaccessibility of the histidine ligands for modification by diethyl pyrocarbonate mentioned above. The sites are located at the surface of the core β-sandwich with two residues (His44 and Glu46) originating directly from a β-strand and His127* that is derived from an extended loop from the opposing subunit. In each site, the copper ion and Met90 were observed in two positions (supplemental Fig. S6) denoted 1 and 2 (Fig. 9). Each conformer is refined at 50% occupancy with similar B-factors. Analysis of the two copper binding modes indicates that in both conformers, the copper is four-coordinate, involving a sulfur atom of Met90 and the three imidazole ring nitrogens of His44, His97, and His127* (Table 5). In conformer 2, the copper sites adopt a tetrahedral geometry (supplemental Fig. S7, C and F). In conformer 1 of CuA, however, the oxygen atom of Glu46 may form a weak fifth ligand (supplemental Fig. S7A and Table 5) in a degenerated octahedral complex with a Cu-O distance of 2.7 Å, drawing the copper below the plane defined by the three His-N atoms. No suitable residue is present to serve as a potential sixth ligand. In contrast, the Cu-O distance is 3 Å in conformer 1 of CuB (Table 5 and Fig. 9B), leaving the copper atom in the plane defined by the three His-Nδ atoms (supplemental Fig. S7D). Met90 and Glu46 are located on opposing sides of a plane formed by the three His residues, with Met90 in multiple conformations to maintain a Met-S-Cu association during the copper displacement.
FIGURE 9.
The copper centers of FetP are in differing conformations. Copper sites A (A and B) and B (C and D) are shown as ball and stick representations. The two conformations of the copper cation and its ligands at each site are shown side by side to highlight the changes in ligand identity and geometry in each conformation. The copper cation is shown as an orange sphere and is always bound to His44, Met90, and His97 from the same subunit and His127* from the other subunit. Similar to the copper cation, Met90 is also observed in two conformations. In CuA1 (A), copper is additionally bound to Glu46 (CuA1-O distance 2.7 Å) in a degenerated octahedral arrangement with five copper ligands. The three His-N atoms all lie on the same plane, and the Cu-N bonds form nearly right angles (97–105°), whereas the Glu46-O-Cu-Met90-S atoms almost form a straight line. In CuA2, the copper atom is in the center of a tetrahedral complex (B), similar to that of CuB2 (D).
TABLE 5.
Ligand bond lengths (Å) in the primary site of the Cu2+-FetP crystal structure
Only ligand distances up to 3 Å are listed
| Atom | Distance to copper site |
|||
|---|---|---|---|---|
| CuA1 | CuA2 | CuB1 | CuB2 | |
| Å | ||||
| His44 | 2.1 | 2.1 | 1.9 | 2.1 |
| His97 | 1.8 | 2.3 | 1.8 | 2.4 |
| His127a | 2.3 | 2.2 | 2.1 | 2.2 |
| Met90-1b | 2.7 | 2.7 | ||
| Met90-2b | 2.3 | 2.2 | ||
| Glu46 | 2.7 | |||
a His127 originates from the symmetry-related subunit.
b Two conformations of Met90 are observed.
Adjacent to each of the copper sites, three residues from the same subunit (Met29, Met34, and Met88) and His125* from the other subunit form another putative metal binding site, “CuC” (supplemental Fig. S8). Met34 is situated on a flexible lid-like loop at the molecular surface of FetP that is observed in differing conformations in each subunit (supplemental Fig. S9). In the more open loop conformation, the distance between Met34-S and His125-Nϵ is 7.6 Å, whereas in the closed conformation, this distance is reduced to 3.5 Å. A metal ion modeled in site CuC within coordinating distance of these four putative ligands in the closed conformation would be ∼7 Å from the nearest copper ion (supplemental Fig. S8).
The Copper-binding Site Is Preformed
Examination of the 2Fo − Fc maps of the as-isolated FetP structure reveals no electron density for an ion or molecule bound in the copper-binding site of chain A and either a full occupancy water or a low occupancy copper ion in chains B, C, and D (data not shown). Because all of the interactions are within hydrogen-bonding distance, a water molecule has been modeled into these three sites. The longer hydrogen-bonding distances between the copper ligands and the modeled waters in these sites suggest that when little to no metal is bound, the loops in this region experience some flexibility but preform a binding site ready for incoming copper. The presence of copper, on the other hand, tightens the protein fold in the copper-binding region, as demonstrated in the Cu-FetP structure and by CD spectroscopy (supplemental Fig. S4).
DISCUSSION
The FetMP system from the uropathogenic E. coli strain F11 was able to supply iron (and potentially manganese) to E. coli cells, especially under acidic or alkaline conditions. The integral membrane protein FetM was central to this task, whereas the periplasmic protein FetP enhanced iron acquisition at low ferrous iron concentrations and in the presence of ferric iron in general. FetP is a dimeric protein that bound copper, zinc, iron, and manganese. In the copper-loaded FetP crystal structure, copper was coordinated by residues His44, Met90, His97, and His127* from the other respective subunit as well as forming a weaker interaction with Glu46. These five aminoacyl residues are conserved in the 100 closest homologs of FetP in a BLAST search (data not shown), including the three previously characterized homologs, ChpA from the marine magnetotactic Vibrio strain MV-1 (10), P19 from the pathogenic Campylobacter jejuni (11), and Tp34 (TP0971) from Treponema pallidum (12).
Recombinant Tp34 from T. pallidum, soaked in 10 mm Zn(II), bound a Zn(II) atom in a degenerated octahedral geometry that is coordinated by 3 His, 1 Glu, and 1 Met residue, corresponding to the copper-coordinating residues from FetP (12). The authors discuss that this geometry may represent a novel zinc-binding site or that zinc could have bound adventitiously to a site that would bind a different metal in vivo. In FetP, copper and zinc binding exclude each other (Table 3), and thus, both metals bind to the same site. The binding of different metals to sites with very similar ligand identities and geometries has been described before (35), such as in the case of the manganese-binding periplasmic protein MncA and the copper-binding CucA from the cyanobacterium Synechocystis strain PCC6801 (36). Similar to FetP, ChpA from the marine Vibrio contained copper after purification of the periplasmic protein fraction from its native host (10). P19 from C. jejuni was shown to preferentially bind copper over zinc (11). In this protein, copper is also bound to 3 His and 1 Met residues plus a nearby Glu residue, all corresponding to the respective residues in FetP, Together with the MALDI-TOF and CD data for FetP, all of these lines of evidence indicate that copper rather than zinc is the likely cofactor of FetP and its relatives.
In contrast to the other previously characterized FetP homologs, the copper ion in each binding site in the crystal structure of FetP exhibited two residence probability maxima (1 and 2) and had conformation differences between the two copper-binding sites in the FetP dimer (CuA and CuB). Copper plasticity has been also been shown recently for the copper-dependent tyrosinase from Bacillus megaterium (37). Histidine-rich copper-binding motifs as in CuA and CuB of FetP is Cu(II)-specific in the copper homeostasis protein CopC, whereas a more methionine-rich binding motif is Cu(I)-specific (38, 39). Interestingly, an additional copper-binding site CuC might exist in FetP adjacent to the other copper sites and is composed of Met29, Met34, Met88, and His125. Both Met29 and Met88 are conserved in the 100 closest relatives of FetP in a BLAST search (data not shown), including P19, ChpA, and Tp34 (10–12), whereas Met34 and His125 are not. CuC was in a tetrahedral “metal-ready” conformation when the adjacent site was in the CuB conformation, whereas the Met34 “lid” of CuC was in the open conformation when the adjacent site was in the CuA conformation. Although a stoichiometry of three bound copper ions was not observed by ITC analysis, the binding of a third copper to the FetP dimer was demonstrated by AAS. Possibly, CuC may preferentially bind Cu(I) and thus may not be detected in the ITC titrations.
In addition to copper, FetP also bound iron (as shown by AAS) and manganese (as demonstrated by AAS and ITC). Mn(II) has been used in other studies as a substitute for Fe(II) because the latter is highly reactive and can denature or damage proteins by the Fenton reaction (40–42). Binding of manganese is not hindered by the presence of either zinc or copper ions (Table 3), indicating that Mn(II) binds to a site distinct from the copper-binding sites. This is interesting because manganese is far down below in the Irving-Williams series (43), such that Zn(II) or Cu(II) should easily be able to displace this cation if they compete for the same site (44). Besides its role as an iron proxy in biochemical experiments, however, Mn(II) was also a substrate for FetMP, as indicated by growth experiments (Fig. 1).
A manganese- or iron-binding site was identified in P19 from C. jejuni, composed of 2 Glu and 1 Asp residue that correspond to residues Glu3, Glu46, and Asp94 of FetP (11). These residues are all conserved in FetP relatives, including ChpA and Tp34 (data not shown). Interestingly, Glu46 seems to be involved in the binding of manganese/iron and/or copper in the CuA1 conformation. Binding of copper in this conformation might thus prevent binding of manganese and iron to this site but would allow binding if the copper site is in the CuB conformation. This may explain why FetP bound only one manganese per dimer in the AAS and ITC experiments and one iron in the AAS determination although two iron/manganese binding sites should exist in a homodimer. Alternatively, the copper ion in the CuA1 conformation may displace the carboxylic group of Glu46 in such a way that one oxygen interacts with copper and the other allows binding of iron or manganese. Regardless, occupation of CuA1 may interfere with iron or manganese binding.
The membrane-bound FetM protein was essential for iron supply by FetMP. In agreement with the greater peptide sequence similarity of FetM with the Pb(II) transporter PbrT (8) than to the iron transporter EfeU (9) and its ability to import Mn(II), FetM may import Fe(II) rather than Fe(III). Periplasmic FetP was important for FetMP-mediated iron uptake at low iron concentrations and at higher Fe(III) concentrations but not at higher Fe(II) concentrations (Fig. 5). Fe(III)-reducing activity of E. coli cells was also doubled when FetP was present (Fig. 6). Thus, FetP may have a dual function as a periplasmic Fe(II) chaperone and a Fe(III) reductase, possibly using a temporarily bound Cu(I) in CuC as an electron donor with copper ion displacement between sites CuA and CuB as a mechanism for electron transfer. Moreover, inhibition by Ag(I) of the specific growth-stimulating effect of FetP in the presence of 100 μm Fe(III) may indicate that the silver ion can compete with Cu(I) for the CuC site or may remove copper from CuA or CuB, thereby preventing reduction of Fe(III) to Fe(II). Nevertheless, the in vivo copper reduction assays demonstrate that E. coli can reduce Fe(III) in the absence of FetP, which may explain why FetP is not essential for iron uptake by FetM. In this model, the function of FetP is analogous to that of the S. cerevisiae ferrireductases Fre1p and Fre2p, which solubilize iron from ferric hydroxide complexes for its uptake (45).
To our knowledge, neither Fre1p nor Fre2p has been purified, and their Km values for iron have not yet been determined, so that the Km values of FetP and a Fre protein cannot be compared. Moreover, the apparent Km value of FetP was determined with whole cells as a difference between FetP-expressing and control cells, which might explain the high value of 1.5 mm. Nevertheless, the vmax for iron reduction of FetP-expressing cells would equal a reduction rate of about 1.8 million irons/cell/min, which would be sufficient even at micromolar iron concentrations to provide the 300,000 iron atoms that E. coli cells contain.4
Prior to high affinity Fe(III) uptake by Ftr1p, ferrous iron is oxidized again by the multicopper oxidase Fet3p, a glycosylated ferroxidase at the yeast plasma membrane and a homolog of the human plasma protein ceruloplasmin (46). Fet3p contains a mononuclear copper site that reacts with Fe(II) (or free Cu(I)) and a trinuclear copper cluster that transfers the metal-derived electrons to reduce molecular oxygen to water (47, 48). FetP does not contain a copper cluster similar to that of Fet3p and is unlikely to have ferroxidase activity.
The other E. coli gene that is a member of the ILT family encodes the EfeU protein and is part of a tricistronic operon, which is under dual control by the Fur repressor in response to the iron status (9) and by CpxAR repression at high pH (49). When the efe operon is expressed in a strain lacking all known iron uptake systems, the cells gained a major growth advantage (9, 49). The other two Efe proteins are both periplasmic proteins. EfeB is a hemoprotein proposed to promote iron extraction from heme (50), whereas EfeO is a novel cupredoxin-like protein (51, 52). In contrast to Efe, the Fet system is not involved in heme-iron acquisition (data not shown). Reminiscent of the yeast model, the current working hypothesis for the Efe system is that EfeB mediates iron extraction from heme, EfeO oxidizes Fe(II) to Fe(III), and the Fe(III) is delivered to EfeU for uptake with the electron given to an unknown electron acceptor via EfeB (51).
E. coli strain F11 is able to colonize the bladder and is therefore exposed to urine, a liquid with changing pH values, oxygen tension, and concentration of organic matter (53). These parameters strongly influence the solubility of Fe(III) (54). At moderately acidic or alkaline conditions, more soluble Fe(III)(OH)2+ or Fe(III)(OH)4− complexes, respectively, exist instead of the insoluble Fe(III)(OH)3 complex, facilitating the use by strain F11 of Fe(III) in the bladder. We have shown that FetMP is useful for growth under these conditions (Fig. 1, B and C). If FetP uses Cu(I) as electron donor for Fe(III) mobilization, it would link iron and copper homeostasis as shown in yeast. FetP would also change the more toxic Cu(I) into the less toxic Cu(II), reminiscent of the function of the copper resistance protein CueO (55–58). In summary, E. coli F11 can be expected to have the ability to use both oxidation states of iron efficiently under conditions of ever changing Fe(II)/Fe(III) ratios, probably aiding in colonization of the urinary tract.
Supplementary Material
Acknowledgments
We thank Dr. Wolfgang Rabsch of the Robert Koch-Institut for allowing us to perform β-galactosidase assays with E. coli strain F11 under S2 biosafety conditions in Wernigerode, Cecile Wandersman (Insitut Pasteur, Paris) for assaying heme-iron acquisition via Fet, and Dr. Angelika Schierhorn for the MALDI-TOF and the MALDI-In-Source-Decay-TOF-MS analysis. We thank Dr. Christian Lange for discussions and suggestions concerning ITC experiments. We thank Karola Otto for skillful technical assistance. Strain ECA458 was constructed by Nadine Taudte. We thank Herbert P. Schweizer (Colorado State University) for Tn7-bearing plasmids. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a national user facility operated by Stanford University on behalf of the United States Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the NCRR, National Institutes of Health (NIH), Biomedical Technology Program, and NIGMS, NIH.
This work was supported by Deutsche Forschungsgemeinschaft Grants Ni262/8-2 (to D. H. N. and G. G.) and GR2061/1-3 (to G. G.) and a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (to M. E. P. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S9.
The atomic coordinates and structure factors (codes 3NRP and 3NRQ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
D. H. Nies, unpublished results.
- ABC
- ATP-binding cassette
- ILT
- iron/lead transporter
- LB
- Luria-Bertani
- TMM
- Tris-buffered mineral salts medium
- BisTris
- bis(2-hydroxyethyl)aminotris(hydroxymethyl)methane
- ACES
- N-(2-acetamido)-2-aminoethanesulfonic acid
- CDTA
- cyclohexane-1,2-diaminetetraacetic acid
- AAS
- atomic absorption spectroscopy
- ITC
- isothermal titration calorimetry
- Cu-FetP
- copper-reconstituted FetP
- SSRL
- Stanford Synchrotron Radiation Lightsource.
REFERENCES
- 1. Neilands J. B., Peterson T., Leong S. A. (1980) ACS Symp. Ser. 140, 264–278 [Google Scholar]
- 2. Nies D. H., Grass G. (2009) in EcoSal: Escherichia coli and Salmonella, Cellular and Molecular Biology (Böck A., Curtiss R., 3rd, Kaper J. B., Neidhardt F. C., Nyström T., Rudd K. E., Squires C. L. eds) American Society for Microbiology Press, Washington, D. C [Google Scholar]
- 3. Andrews S. C., Robinson A. K., Rodríguez-Quiñones F. (2003) FEMS Microbiol. Rev. 27, 215–237 [DOI] [PubMed] [Google Scholar]
- 4. Braun V., Braun M. (2002) Curr. Opin. Microbiol. 5, 194–201 [DOI] [PubMed] [Google Scholar]
- 5. Debut A. J., Dumay Q. C., Barabote R. D., Saier M. H., Jr. (2006) J. Mol. Microbiol. Biotechnol. 11, 1–9 [DOI] [PubMed] [Google Scholar]
- 6. Stearman R., Yuan D. S., Yamaguchi-Iwai Y., Klausner R. D., Dancis A. (1996) Science 271, 1552–1557 [DOI] [PubMed] [Google Scholar]
- 7. Forman S., Paulley J. T., Fetherston J. D., Cheng Y. Q., Perry R. D. (2010) Biometals 23, 275–294 [DOI] [PubMed] [Google Scholar]
- 8. Borremans B., Hobman J. L., Provoost A., Brown N. L., van Der Lelie D. (2001) J. Bacteriol. 183, 5651–5658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grosse C., Scherer J., Koch D., Otto M., Taudte N., Grass G. (2006) Mol. Microbiol. 62, 120–131 [DOI] [PubMed] [Google Scholar]
- 10. Dubbels B. L., DiSpirito A. A., Morton J. D., Semrau J. D., Neto J. N., Bazylinski D. A. (2004) Microbiology 150, 2931–2945 [DOI] [PubMed] [Google Scholar]
- 11. Chan A. C., Doukov T. I., Scofield M., Tom-Yew S. A., Ramin A. B., Mackichan J. K., Gaynor E. C., Murphy M. E. (2010) J. Mol. Biol. 401, 590–604 [DOI] [PubMed] [Google Scholar]
- 12. Deka R. K., Brautigam C. A., Tomson F. L., Lumpkins S. B., Tomchick D. R., Machius M., Norgard M. V. (2007) J. Biol. Chem. 282, 5944–5958 [DOI] [PubMed] [Google Scholar]
- 13. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 14. Mergeay M., Nies D., Schlegel H. G., Gerits J., Charles P., Van Gijsegem F. (1985) J. Bacteriol. 162, 328–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Choi K. H., Schweizer H. P. (2006) Nat. Protoc. 1, 153–161 [DOI] [PubMed] [Google Scholar]
- 16. Brüser T., Deutzmann R., Dahl C. (1998) FEMS Microbiol. Lett. 164, 329–336 [DOI] [PubMed] [Google Scholar]
- 17. Stojiljkovic I., Bäumler A. J., Hantke K. (1994) J. Mol. Biol. 236, 531–545 [DOI] [PubMed] [Google Scholar]
- 18. Hantke K. (1987) Mol. Gen. Genet. 210, 135–139 [DOI] [PubMed] [Google Scholar]
- 19. Koch D., Nies D. H., Grass G. (2007) Biometals 20, 759–771 [DOI] [PubMed] [Google Scholar]
- 20. Miller J. H. (1972) Experiments in Molecular Genetics, Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- 21. Thieme D., Neubauer P., Nies D. H., Grass G. (2008) Appl. Environ. Microbiol. 74, 7463–7470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grosse C., Grass G., Anton A., Franke S., Santos A. N., Lawley B., Brown N. L., Nies D. H. (1999) J. Bacteriol. 181, 2385–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grass G., Otto M., Fricke B., Haney C. J., Rensing C., Nies D. H., Munkelt D. (2005) Arch. Microbiol. 183, 9–18 [DOI] [PubMed] [Google Scholar]
- 24. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., Bairoch A. (2005) in The Proteomics Protocols Handbook (Walker J. M. ed) pp. 571–607, Humana Press Inc., Totowa, NJ [Google Scholar]
- 25. Leslie A. G. W. (1992) Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography 26, 27–33 [Google Scholar]
- 26. Vagin A. A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 27. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 28. Collaborative Computation Project 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 29. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30. Georgatsou E., Alexandraki D. (1994) Mol. Cell. Biol. 14, 3065–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Anton A., Weltrowski A., Haney C. J., Franke S., Grass G., Rensing C., Nies D. H. (2004) J. Bacteriol. 186, 7499–7507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schiavo G., Rossetto O., Santucci A., DasGupta B. R., Montecucco C. (1992) J. Biol. Chem. 267, 23479–23483 [PubMed] [Google Scholar]
- 33. Sippl M. J., Wiederstein M. (2008) Bioinformatics 24, 426–427 [DOI] [PubMed] [Google Scholar]
- 34. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 35. Chakrabarti P. (1989) Biochemistry 28, 6081–6085 [DOI] [PubMed] [Google Scholar]
- 36. Tottey S., Waldron K. J., Firbank S. J., Reale B., Bessant C., Sato K., Cheek T. R., Gray J., Banfield M. J., Dennison C., Robinson N. J. (2008) Nature 455, 1138–1142 [DOI] [PubMed] [Google Scholar]
- 37. Sendovski M., Kanteev M., Ben-Yosef V. S., Adir N., Fishman A. (2011) J. Mol. Biol. 405, 227–237 [DOI] [PubMed] [Google Scholar]
- 38. Arnesano F., Banci L., Bertini I., Mangani S., Thompsett A. R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3814–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang L., Koay M., Maher M. J., Xiao Z., Wedd A. G. (2006) J. Am. Chem. Soc. 128, 5834–5850 [DOI] [PubMed] [Google Scholar]
- 40. Haber F., Weiss J. (1932) Naturwissenschaften 20, 948–950 [Google Scholar]
- 41. Langlois d'Estaintot B., Santambrogio P., Granier T., Gallois B., Chevalier J. M., Précigoux G., Levi S., Arosio P. (2004) J. Mol. Biol. 340, 277–293 [DOI] [PubMed] [Google Scholar]
- 42. Frolow F., Kalb A. J., Yariv J. (1994) Nat. Struct. Biol. 1, 453–460 [DOI] [PubMed] [Google Scholar]
- 43. Irving H., Williams R. J. (1948) Nature 162, 746–747 [Google Scholar]
- 44. Nies D. H. (2007) in Molecular Microbiology of Heavy Metals (Nies D. H., Silver S. eds) pp. 118–142, Springer-Verlag, Berlin [Google Scholar]
- 45. Kosman D. J. (2003) Mol. Microbiol. 47, 1185–1197 [DOI] [PubMed] [Google Scholar]
- 46. de Silva D., Davis-Kaplan S., Fergestad J., Kaplan J. (1997) J. Biol. Chem. 272, 14208–14213 [DOI] [PubMed] [Google Scholar]
- 47. Taylor A. B., Stoj C. S., Ziegler L., Kosman D. J., Hart P. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15459–15464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sedlák E., Ziegler L., Kosman D. J., Wittung-Stafshede P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19258–19263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cao J., Woodhall M. R., Alvarez J., Cartron M. L., Andrews S. C. (2007) Mol. Microbiol. 65, 857–875 [DOI] [PubMed] [Google Scholar]
- 50. Létoffé S., Heuck G., Delepelaire P., Lange N., Wandersman C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 11719–11724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rajasekaran M. B., Nilapwar S., Andrews S. C., Watson K. A. (2010) Biometals 23, 1–17 [DOI] [PubMed] [Google Scholar]
- 52. Sturm A., Schierhorn A., Lindenstrauss U., Lilie H., Brüser T. (2006) J. Biol. Chem. 281, 13972–13978 [DOI] [PubMed] [Google Scholar]
- 53. Brooks T., Keevil C. W. (1997) Lett. Appl. Microbiol. 24, 203–206 [DOI] [PubMed] [Google Scholar]
- 54. Millero F. J. (1998) Earth Planet Sci. Lett. 154, 323–329 [Google Scholar]
- 55. Grass G., Rensing C. (2001) Biochem. Biophys. Res. Commun. 286, 902–908 [DOI] [PubMed] [Google Scholar]
- 56. Outten F. W., Huffman D. L., Hale J. A., O'Halloran T. V. (2001) J. Biol. Chem. 276, 30670–30677 [DOI] [PubMed] [Google Scholar]
- 57. Roberts S. A., Weichsel A., Grass G., Thakali K., Hazzard J. T., Tollin G., Rensing C., Montfort W. R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2766–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Singh S. K., Grass G., Rensing C., Montfort W. R. (2004) J. Bacteriol. 186, 7815–7817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stapleton A., Moseley S., Stamm W. E. (1991) J. Infect. Dis. 163, 773–779 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.