Abstract
The human disease network (HDN) has become a powerful tool for revealing disease–disease associations. Some studies have shown that genes that share similar or same disease phenotypes tend to encode proteins that interact with each other. Therefore, protein–protein interactions (PPIs) may help us to further understand the relationships between diseases with overlapping clinical phenotypes. In this study, we constructed the expanded HDN (eHDN) by combining disease gene information with PPI information, and analyzed its topological features and functional properties. We found that the network is hierarchical and, most diseases are connected to only a few diseases, whereas a small part of diseases are linked to many different diseases. Diseases in a specific disease class tend to cluster together, and genes associated with the same disease are functionally related. Comparing the eHDN with the original HDN (oHDN, constructed using disease gene information) revealed high consistency over all topological and functional properties. This, to some extent, indicates that our eHDN is reliable. In the eHDN, we found some new associations among diseases resulting from the shared genes interacting with disease genes. The new eHDN will provide a valuable reference for clinicians and medical researchers.
Keywords: expanded human disease network (eHDN), protein–protein interactions (PPIs), disease–disease associations, diseasome, biological networks
Introduction
In past decades, biological questions were often approached by studying individual genes and their functions.1 Despite the enormous success of this reductionist approach, it has become increasingly clear that this method ignores the relationship between both genes and gene products.1 The recent availability of a wealth of functional genomic and proteomic (‘omic') information, and the development of high-throughput data-collection techniques, has resulted in a transition from individual gene-based traditional molecular biology to ‘network biology'.1, 2 In network biology, biological processes are considered as complex networks of interactions between the cell's numerous components rather than as independent interactions involving only a few molecules. Researchers have constructed various types of networks that include, protein–protein interaction network,3 metabolic network,4 transcription regulatory network5 and gene coexpression network.6 With the development of network biology, the nodes of biological networks are no longer limited to cell components alone. Instead, some researchers have introduced macroscopic concepts and biological networks, like the human disease network (HDN),7 the phenotype network8 and the drug-target network,9 have recently been constructed.
Goh, et al used the Online Mendelian Inheritance in Man (OMIM)10 knowledgebase to construct the HDN in which two diseases are connected to each other, if they share at least one gene. Initially, OMIM focused on high-quality data with high significance for Mendelian disorders. Although, in recent years, more complex traits have been included, this history still introduces some bias; most importantly, association studies of non-Mendelian, common complex diseases, often have low-significance values. The Genetic Association Database (GAD)11 collects, standardizes and archives almost all of the genetic association study data from published literature and is, therefore, more exhaustive than OMIM. In current biology research, the number of known disease genes is limited and so the identification of disease susceptibility genes remains an important issue. Some studies have indicated that genes that share similar or same disease phenotypes tend to encode proteins that interact with each other.12, 13 Indeed, the Hermansky–Pudlak syndrome14 and Fanconi anaemia15 are known to be caused by mutations affecting different interacting proteins.
Here, we describe the construction of the expanded human disease network (eHDN) by combining the available disease gene information in GAD with PPI data from the Human Protein Reference Database (HPRD),16 in which the PPIs are sourced from literature by manual curation. We analyzed the topological features and functional properties for the eHDN. Comparing the eHDN with oHDN revealed high consistency over all topological and functional properties. This is proven that our eHDN is credible. We hope that our study will provide a new approach for exploring the associations between diseases with overlapping clinical phenotypes.
Materials and methods
Disease-gene association and protein–protein interaction data
A compendium of human disease-gene associations with a total of 39 930 records corresponding to 5638 diseases/phenotypes and 2675 genes, was obtained from GAD on 6 June 2009. We selected 6350 records that described positive associations between genes and disorders. The PPI data set, obtained from HPRD on 14 July 2009, contains 37 107 interactions. After eliminating self-interactions and interactions that corresponded to the Entrez ID ‘None', 35 000 interactions between 9303 genes remained. For details of these datasets see Supplementary information.
Measuring the topological features of a network
The ‘degree' (k) is defined as the number of edges that point to a node. The ‘clustering coefficient' is defined as Ci=2n/ki (ki−1), where n is the number of links connecting the ki nearest neighbors of node i to each other, and ki (ki−1)/2 is the total number of triangles that can pass through node i. The clustering coefficient reflects the local clustering of a network, and the average of Ci over all nodes of a network characterizes the overall tendency of the nodes to form clusters or groups. The function C(k) is defined as the average clustering coefficient of all nodes with k links. For most networks, C(k) approximately follows C(k) ∼k,−1 indicating that the network has hierarchical features.1, 17 Dyadicity (D) and heterophilicity (H), which are two network properties of nodes, have been used to quantifying whether diseases in the same disease class tend to cluster together in a network.8, 18 Dyadicity D>1 (D<1) suggests that the diseases in a disease class tend to have more (fewer) links to each other than randomly expected. For heterophilicity H>1 (H<1), the diseases in a disease class have the tendency to connect more (less) loosely than expected randomly. For details of the computation of D and H see Supplementary information.
Randomization of disease-gene associations
To obtain random controls for the topological features of eHDN, we randomly shuffled the disease-gene associations while keeping unchanged both the number of genes that a disorder is associated with and the number of disorders that a gene is implicated in. From the randomized disease-gene associations, we created a randomized eHDN by projecting it onto disease space.7 We generated 104 independent randomized samples.
Measuring the functional properties of a network
The maximum fraction of genes annotated to the same disease that have the same GO (Gene Ontology)19 terms is defined as GO homogeneity. We introduced KEGG (Kyoto Encyclopedia of Genes and Genomes)20 homogeneity and defined it as the maximum fraction of genes assigned to the same disease that have the same KEGG terms. To reduce the bias, we removed the corresponding disease pathways from KEGG when the KEGG homogeneity is calculated for a certain disease. We also introduced subcellular location homogeneity, which we defined as the maximum fraction of genes assigned to the same disease that have the same subcellular location. Similarly, we used tissue homogeneity to measure the maximum fraction of genes implicated in the same disorders that are expressed in a specific tissue. The Pearson's correlation coefficient (PCC) and cosine correlation distance (CCD)21 for each gene pair associated with the same disease were calculated to measure the coexpression characteristic from different perspectives. The synchronized expression property of the genes in a specific disease was characterized by the average PCC and CCD. For details of the computation and randomization of functional properties, see Supplementary information. We performed 104 independent randomization runs over all the defined functional properties.
Gene expression microarray data
To calculate the tissue homogeneity, coexpression and synchronized expression, we used the microarray data that is available for normal human tissues. We selected the GSE7307 and GSE3526 (Affymetrix U133 plus 2.0 arrays) datasets from the Gene Expression Omnibus (GEO) repository.22 GSE7307 and GSE3526 contain 83 and 63 distinct tissue types referring to 20 080 and 17 906 human genes, respectively. To determine the tissue-selective genes, we used the Significance Analysis of Microarrays (SAM) algorithm23 as described previously.24 For details of the microarray data, see Supplementary information.
Results
Construction of the eHDN
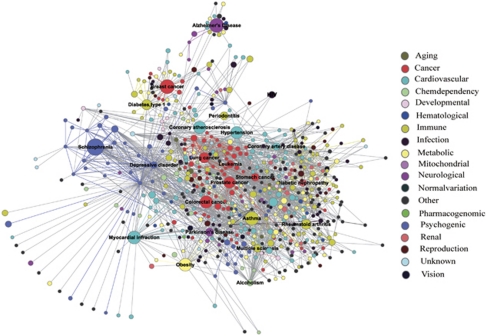
We obtained an association list, which contains 1336 diseases/phenotypes and 1639 disease genes from GAD. All the diseases were classified into 19 categories according to the Disease Class field of GAD. We constructed a bipartite graph to represent the associations between diseases and disease genes, and defined it as the GAD diseasome. Using the GAD diseasome, we generated the oHDN projection of the bipartite graph. In the oHDN, nodes represent diseases and two diseases are connected to each other if they share at least one disease gene. We then constructed the extended diseasome by combining the disease gene information with the protein–protein interaction information. We added, to the GAD diseasome, 332 genes that give proteins that interact with at least two proteins encoded by genes associated with a disorder, to generate the GAD-HPRD2 diseasome (Supplementary Table S1). We used the newly obtained GAD-HPRD2 diseasome to generate the eHDN projection (Figure 1). For details of the construction of eHDN see Supplementary information.
Figure 1.
The expanded HDN (eHDN). In the eHDN, each node corresponds to a distinct disease and is colored based on the disease class to which it belongs. The names of the 19 disease classes are shown on the right. Links between diseases in the same disease class are correspondingly colored and links connecting different disease classes are gray. The size of each node is proportional to the number of genes associated with the corresponding disease, and the thickness of the link is proportional to the number of genes shared by the diseases it connects. Diseases with >10 associated genes are named.
Using the PPI information, we obtained 1852 new disease-gene links. For example, CDK5, which was linked to Alzheimer's disease in the eHDN, is an important regulator of brain development, neuronal maturation and synaptic transmission, and participates in the Alzheimer's disease pathway. DLG4 was linked to Huntington's disease because of its interaction with the disease susceptibility genes, HTT, GRIK1 and GRIK2. DLG4 is annotated to the GO biological process terms, signal transduction, synaptic transmission and nervous system development, and, in KEGG, to the Huntington's disease pathway. Thus, our eHDN will help researchers investigate whether the genes that interact with the previously identified (real) disease genes and that are involved in the same cell functions or pathways also influence the occurrence of the disease.
A total of 1102 new connections were established between the diseases in the eHDN (Supplementary Table S2). As an example, type 2 diabetes mellitus (T2DM) is connected to kidney failure, nasopharyngeal carcinoma and skin carcinoma. T2DM is a metabolic disease, which is characterized by high blood glucose and related insulin deficiency. Some researchers have indicated that end-stage renal disease is one of the complications of T2DM.25 Chan, et al26 have reported that cancer is emerging as an important cause of morbidity and mortality for Asian patients with diabetes and at high risk of cardiorenal complications. The new links among diseases offers meaningful information for clinicians that will allow them to adopt early strategies to tackle a number of complications in the treatment of diseases.
Analysis of the topological features
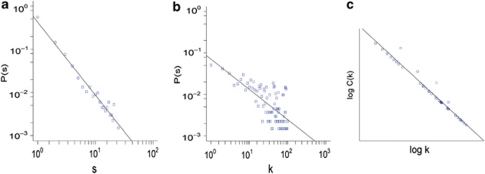
The eHDN displays many links between both individual diseases and disease classes (Figure 1). Of the 1336 diseases in eHDN, 1226 have at least one edge with other diseases, suggesting that most diseases have some common genetic origins with other diseases. A common biological basis for some complex diseases has been reported earlier.27 By calculating the distribution of the number of genes associated with a disorder, s, we found that most diseases have a few disease genes (Figure 2a, the average of s was 4.75). However, the top 10 diseases related to cancer, and metabolic, neurological, psychogenic, immune and cardiovascular diseases, have dozens of disease genes.
Figure 2.
Analysis of the topological features in the eHDN. (a) The distribution of the number of genes associated with a disorder, (s). (b) Distribution of the degree (k). (c) The distribution of the clustering coefficient follows C(k) ∼k−1, a straight line of slope-1 on a log-log plot.
Analysis of the degree and clustering coefficient
The degree (k) distribution of the eHDN approximates a power law (Figure 2b), showing that eHDN is scale-free.1 This result also indicates that most diseases are connected to only a few other diseases, whereas a small number of diseases, including breast cancer (k=584), atherosclerosis (k=519) and rheumatoid arthritis (k=433), are linked to many different diseases. Cancers are densely connected to each other because multiple types of cancer have common genes, like TP53, TGFB1, APC and PTEN, associated with them.28, 29, 30, 31 Most of the common genes have the GO biological process terms, cell growth, cell aging and apoptosis, cell cycle, DNA damage repair and embryonic development, associated with them. For the eHDN, C(k) approximately follows C(k) ∼k−1, indicating that it is a hierarchical network (Figure 2c). The distribution of k and C(k) are both significantly different from the random controls (P-value <2.2e−16). For details of the analysis of the degree and clustering coefficient see Supplementary information.
The modular structure of the eHDN
The dyadicity and heterophilicity values for the eHDN obtained using 19 disease classes illustrate that diseases in a specific-disease class are clustered into densely connected groups (Supplementary Table S3). There are, however, some differences for each disease class. The immune diseases class, for example, has a high-dyadicity value, suggesting a clearly modular structure. The high connectivity may be attributed to the common susceptibility genes associated with immune diseases, such as the major histocompatibility complex (MHC), CTLA4 and PTPN22, that encode molecules involved in the immune response.32, 33 Some disease classes, for example, the developmental diseases, are heterophilic indicating a tendency to connect to different categories of diseases. One possible explanation is that most developmental diseases influence multiple tissues or physiological systems.8
Analysis of the functional properties
Several studies have shown that genes related to the same disease tend to display functional relatedness.34, 35 These functionally related genes usually belong to common function modules, such as the coexpression modules, cellular pathways, or molecular complexes.8, 17 We analyzed the functional relationships of genes associated with the same diseases, by examining their functional annotation, tissue expression, coexpression and synchronized expression data.
GO and KEGG homogeneity analysis
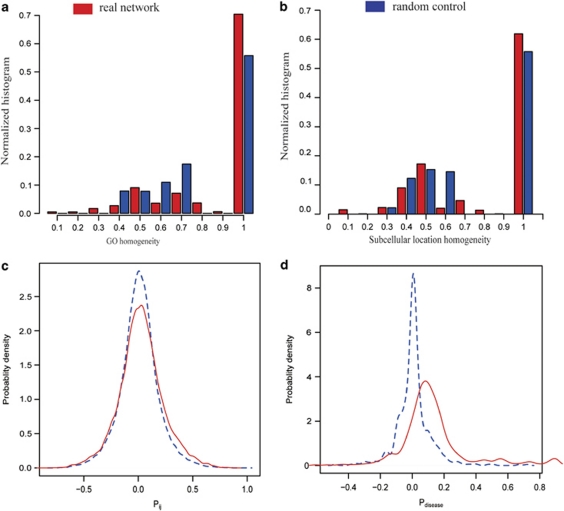
Of the genes in the eHDN, 1923 were annotated in GO, and 1189 of them had corresponding KEGG annotations. We measured the functional relatedness of genes within the same disease by analyzing GO and KEGG homogeneity. For the GO homogeneity, we not only considered the GO annotations as a whole, but we also calculated homogeneity separately for each branch of GO, biological process, molecular function and cellular component. We found a significant elevation of GO homogeneity in eHDN compared with the homogeneity in the random control. For the GO annotations, as a whole, the P-value was <2.2e−16 (Figure 3a) and the P-values were smaller than 2.2e−16 for all three branches. Similarly, we calculated KEGG homogeneity, and found that 54% of the diseases show almost perfect homogeneity compared with only 29% in the random control (P-value <1.0e−5). Thus, we concluded that, based on the GO and KEGG annotations, genes that belong to the same disease have similar cellular and functional characteristics. For details of the GO and KEGG homogeneity analysis see Supplementary information.
Figure 3.
Analysis of the functional properties in the eHDN. (a) Distribution of the GO homogeneity of a disease. A random control with the same number of genes chosen randomly is shown for comparison. (b) Distribution of the subcellular location homogeneity of a disease. A random control with the same number of genes chosen randomly is shown for comparison. (c) As an example of GSE7307, the distribution of Pij values for the expression profiles of each disease gene pair that belongs to the same disorder (solid line) and the control (dashed line), representing the PCC distribution between all gene pairs, is shown. (d) For the GSE7307, the distribution of the average PCC (Pdisease) between the expression profiles of all the genes associated with the same disorder (solid line) and the random control (dashed line) with the same number of genes chosen randomly is shown.
Subcellular location homogeneity analysis
The function of a protein and its role in a cell are closely correlated with the subcellular location or environment of the protein.36 For example, drug target proteins and non-drug target proteins have different subcellular locations.37 We introduced the subcellular location homogeneity to measure the tendency of the protein products of genes in a common disease to cluster in the same subcellular location. We calculated the subcellular location homogeneity for the eHDN and found that it differs significantly from the random control (P-value <7e−07, Figure 3b), indicating that genes in the same disease share similar subcellular location. For details of the subcellular location homogeneity analysis, see Supplementary information.
Tissue homogeneity analysis
We selected two microarray datasets of healthy human tissue to analyze tissue homogeneity. In all, 1925 and 1781 of the genes in the eHDN are expressed in GSE7307 and GSE3526, respectively. We found that the tissue homogeneity coefficients are significantly higher for the eHDN compared with the random controls in both GSE7307 (P-value <1.0e−5) and GSE3526 (P-value <4.9e−5). These results show that genes associated with the same disease are generally expressed in a specific tissue. For details of the tissue homogeneity analysis, see Supplementary information.
Coexpression and synchronized expression analysis
We used the Pearson's correlation coefficient (PCC) and cosine correlation distance (CCD) to measure the coexpression and synchronized expression features of genes assigned to the same disease. For the GSE7307 data, the distribution of PCC and average PCC in the eHDN are higher than in the random controls (P-value <1.0e−5, Figures 3c and d). When CCD was used as an evaluation indicator, the expression correlation of the disease genes was more significant than was indicated by the PCC. In the eHDN, 45 diseases with the average PCC>0.5 and CCD<0.1 display high synchronized expression characteristic, for example, pulmonary hypertension (PCC=0.902, CCD=0.007) and Gilbert syndrome (PCC=0.823, CCD=0.027). We carried out the same analysis for the GSE3526 data and obtained similar results. These results indicated that disease genes that are implicated in the same disease, display high expression profile correlation.
In summary, by analyzing the properties of the eHDN we conclude that: (1) there is a common genetic origin for most diseases, especially for complex diseases; (2) most diseases have only a few disease genes, whereas a small number of diseases are related to dozens of genes; (3) a power law degree distribution shows that the eHDN is scale-free; (4) the majority of diseases have links to only a few diseases, whereas a handful of diseases are connected to many different diseases; (5) the eHDN is hierarchical; (6) diseases in a specific disease class have a tendency to cluster into densely connected groups; and (7) genes associated with the same disease tend to: (i) share GO and KEGG terms, (ii) have similar subcellular location, (iii) be expressed in a specific tissue, (iv) exhibit high-coexpression levels, and (v) display synchronized expression characteristic.
To illustrate the credibility of our eHDN, we carried out a comparative analysis of eHDN and oHDN. We found that the eHDN is highly consistent with the oHDN over all topological and functional characteristics. To further demonstrate the reliability of eHDN, we eliminated the real disease genes from the GAD-HPRD2 diseasome and obtained the HPRD2 diseasome and its HDN projection. We then analyzed the topological and functional properties of the HDN projection and found them to be consistent with the oHDN (Supplementary information). This result, to some extent, confirms that the properties of the HDN projection obtained by eliminating the true disease genes from GAD-HPRD2 diseasome are similar to the oHDN. Therefore, we feel confident that our eHDN constructed by combining disease gene information with protein–protein interaction information is reliable.
Discussion
The integration analysis of various ‘omic' data has become increasingly widespread because each approach has intrinsic caveats.2 For instance, important information may be missing because of false negatives or misleading because of false positives. Therefore, the data emerging from any single omic approach should be cautiously interpreted because it only provides crude indications of gene or protein function.38, 39 Some studies have indicated that these limitations can be mitigated by integrating two or more omic datasets.2, 39 To explore disease–disease relationships from a functional perspective, we constructed the eHDN by integrating phenome and interactome information.
Here, the reasons for adding genes that give proteins interacting with at least two proteins encoded by genes associated with a disorder, to expand the oHDN are discussed. We first obtained GAD-HPRD1 diseasome by added 5175 genes that encoded proteins interacting with at least one proteins encoded by genes associated with a particular disease, to GAD diseasome. We analyzed the topological and functional properties of the HDN projection generated from the GAD-HPRD1 diseasome and found that it was inconsistent with the oHDN. To test whether the inconsistency arose from the genes that we added to expand GAD diseasome, we separately removed the real disease genes from GAD-HPRD1 diseasome and GAD-HPRD2 diseasome to obtain HPRD1 diseasome and HPRD2 diseasome, respectively. We then analyzed the topological and functional properties of the HDN projections generated from the HPRD1 diseasome and HPRD2 diseasome, and found that while the latter is consistent with the oHDN, the former is not. Thus, we concluded that genes that were added to obtain GAD-HPRD1 diseasome may affect its overall properties. By comparing the two expanded networks, we concluded that GAD-HPRD2 diseasome is more reasonable.
Generally speaking, the biological processes of living cells are attributable to complex interactions between multiple gene products.1, 2 Evidence from many resources has shown that diseases with overlapping clinical phenotypes are caused by mutations in functionally related genes34 and that protein–protein interactions are the strongest manifestation of a functional relationship between disease genes.13 Applying a network model to represent associations between diseases has proven to be an effective approach for revealing the relationship among diseases on a large scale.7, 8 Our study considered the interactions among gene products, and measured the topological and functional properties of eHDN from a network-based perspective. We discovered new links among diseases by comparing the eHDN with oHDN. The new links among diseases will provide some meaningful information for clinicians and medical researchers that may help them to understand the relation between diseases. Although 35 000 interactions between 9303 proteins were used in this study, the actual number of interactions between these proteins will be much greater. In addition, the disease phenotypic data are limited at present. With increasing quantity and quality of interaction and phenotypic data, the reliability and utility of eHDN will be further improved.
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China (Grant No. 30871394), the National High Tech Development Project of China, the 863 Program (Grant No. 2007AA02Z329), the National Basic Research Program of China, Natural Science Foundation of Heilongjiang Province (Grant No. F2008-02), and the Innovation Fund of Harbin Medical University (No. HCXS2010010).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Ge H, Walhout AJ, Vidal M. Integrating ‘omic' information: a bridge between genomics and systems biology. Trends Genet. 2003;19:551–560. doi: 10.1016/j.tig.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Yook SH, Oltvai ZN, Barabasi AL. Functional and topological characterization of protein interaction networks. Proteomics. 2004;4:928–942. doi: 10.1002/pmic.200300636. [DOI] [PubMed] [Google Scholar]
- Wagner A, Fell DA. The small world inside large metabolic networks. Proc Biol Sci. 2001;268:1803–1810. doi: 10.1098/rspb.2001.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc Natl Acad Sci USA. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Liu B, Jiang J, et al. Modularity in the genetic disease-phenotype network. FEBS Lett. 2008;582:2549–2554. doi: 10.1016/j.febslet.2008.06.023. [DOI] [PubMed] [Google Scholar]
- Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug-target network. Nat Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nat Genet. 2004;36:431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- Lage K, Karlberg EO, Storling ZM, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- Oti M, Snel B, Huynen MA, Brunner HG. Predicting disease genes using protein-protein interactions. J Med Genet. 2006;43:691–698. doi: 10.1136/jmg.2006.041376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro SM, Dell'Angelica EC. The cell biology of Hermansky-Pudlak syndrome: recent advances. Traffic. 2005;6:525–533. doi: 10.1111/j.1600-0854.2005.00299.x. [DOI] [PubMed] [Google Scholar]
- Mace G, Bogliolo M, Guervilly JH, Dugas du Villard JA, Rosselli F. 3R coordination by Fanconi anemia proteins. Biochimie. 2005;87:647–658. doi: 10.1016/j.biochi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Peri S, Navarro JD, Kristiansen TZ, et al. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 2004;32:D497–D501. doi: 10.1093/nar/gkh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasz E, Barabasi AL. Hierarchical organization in complex networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2003;67:026112. doi: 10.1103/PhysRevE.67.026112. [DOI] [PubMed] [Google Scholar]
- Park J, Barabasi AL. Distribution of node characteristics in complex networks. Proc Natl Acad Sci USA. 2007;104:17916–17920. doi: 10.1073/pnas.0705081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmeren P, van Berkum NL, Vilo J, et al. Protein interaction verification and functional annotation by integrated analysis of genome-scale data. Mol Cell. 2002;9:1133–1143. doi: 10.1016/s1097-2765(02)00531-2. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao LL, Dangond F, Yoshida T, et al. A compendium of gene expression in normal human tissues. Physiol Genomics. 2001;7:97–104. doi: 10.1152/physiolgenomics.00040.2001. [DOI] [PubMed] [Google Scholar]
- Chiasson JL. Early insulin use in type 2 diabetes: what are the cons. Diabetes Care. 2009;32 (Suppl 2:S270–S274. doi: 10.2337/dc09-S321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JC, Malik V, Jia W, et al. Diabetes in Asia: epidemiology, risk factors, and pathophysiology. Jama. 2009;301:2129–2140. doi: 10.1001/jama.2009.726. [DOI] [PubMed] [Google Scholar]
- Xie C, Wang ZC, Liu XF, Yang MS. The common biological basis for common complex diseases: evidence from lipoprotein lipase gene. Eur J Hum Genet. 2009. [DOI] [PMC free article] [PubMed]
- Liu Y, Bodmer WF. Analysis of P53 mutations and their expression in 56 colorectal cancer cell lines. Proc Natl Acad Sci USA. 2006;103:976–981. doi: 10.1073/pnas.0510146103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang DC, Luo YH, Yang SM, Li XA, Ling XL, Fang L. Mutation analysis of APC gene in gastric cancer with microsatellite instability. World J Gastroenterol. 2002;8:787–791. doi: 10.3748/wjg.v8.i5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- Saal LH, Gruvberger-Saal SK, Persson C, et al. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat Genet. 2008;40:102–107. doi: 10.1038/ng.2007.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plenge RM, Padyukov L, Remmers EF, et al. Replication of putative candidate-gene associations with rheumatoid arthritis in >4000 samples from North America and Sweden: association of susceptibility with PTPN22, CTLA4, and PADI4. Am J Hum Genet. 2005;77:1044–1060. doi: 10.1086/498651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumanovics A, Takada T, Lindahl KF. Genomic organization of the mammalian MHC. Annu Rev Immunol. 2003;21:629–657. doi: 10.1146/annurev.immunol.21.090501.080116. [DOI] [PubMed] [Google Scholar]
- Brunner HG, van Driel MA. From syndrome families to functional genomics. Nat Rev Genet. 2004;5:545–551. doi: 10.1038/nrg1383. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HB, Chou KC. Hum-mPLoc: an ensemble classifier for large-scale human protein subcellular location prediction by incorporating samples with multiple sites. Biochem Biophys Res Commun. 2007;355:1006–1011. doi: 10.1016/j.bbrc.2007.02.071. [DOI] [PubMed] [Google Scholar]
- Bakheet TM, Doig AJ. Properties and identification of human protein drug targets. Bioinformatics. 2009;25:451–457. doi: 10.1093/bioinformatics/btp002. [DOI] [PubMed] [Google Scholar]
- Ge H, Liu Z, Church GM, Vidal M. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat Genet. 2001;29:482–486. doi: 10.1038/ng776. [DOI] [PubMed] [Google Scholar]
- Vidal M. A biological atlas of functional maps. Cell. 2001;104:333–339. doi: 10.1016/s0092-8674(01)00221-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.