Abstract
Active DNA demethylation is involved in many vital developmental and physiological processes of plants and animals. Recent genetic and biochemical studies in Arabidopsis have demonstrated that a subfamily of DNA glycosylases function to promote DNA demethylation through a base excision-repair pathway. These specialized bifunctional DNA glycosylases remove the 5-methylcytosine base and then cleave the DNA backbone at the abasic site, resulting in a gap that is then filled with an unmethylated cytosine nucleotide by as yet unknown DNA polymerase and ligase enzymes. Evidence suggests that active DNA demethylation in mammalian cells is also mediated at least in part by a base excision repair pathway where the AID/Apobec family of deaminases convert 5-methylcytosine to thymine followed by G/T mismatch repair by the DNA glycosylase MBD4 or TDG. This review also discusses other possible mechanisms of active DNA demethylation, how genome DNA methylation status might be sensed to regulate the expression of demethylase genes, and the targeting of demethylases by small RNAs.
Keywords: epigenetics, DNA demethylase, base excision repair, small RNA
INTRODUCTION
The methylated nucleotide 5-methyl-deoxycytidine (5-meC) is sometimes called the fifth nucleotide in DNA, and was identified long before DNA was recognized as genetic material (41). Although DNA demethylation is the focus of this review, a brief discussion of DNA methylation will provide useful background. Approximately 2% to 8% of cytosines in mammals and up to 50% in higher plants are methylated, but 5-meC is undetectable in budding and fission yeasts, nematodes, or adult Drosophila melanogaster (20). In most bacterial species, cytosine methylation serves an immune function, which protects the bacteria from bacteriophage infection by selectively degrading unmethylated foreign DNA using type 2 restriction-modification systems (104a). DNA methylation in promoter elements represses gene transcription directly by interfering with the binding of transcriptional activators and indirectly by favoring the formation of repressive chromatin by methyl DNA-binding proteins (9). In higher eukaryotes, DNA methylation is critical for a wide range of cellular functions such as genome stability and defense, imprinting, X chromosome inactivation, paramutation, tissue-specific gene regulation, carcinogenesis, and aging (9, 5).
DNA methylation is a postreplicative process. The methyl group is transferred from S-adenosyl methionine to cytosines in DNA by DNA methyltransferase enzymes in a reaction that involves base flipping, whereby a cytosine base is swung completely out of the DNA helix into an extrahelical position so that the enzyme can access and methylate the cytosine (86). Both de novo and maintenance DNA methyltransferases from mammals and plants contain a conserved methyltransferase catalytic domain in their C-terminal regions (7, 14). Dnmt3 and DRM2 (Domain Rearranged Methyltransferase 2) are the de novo DNA methyltransferases in mammals and plants, respectively (7, 14). Maintenance of CpG methylation is catalyzed by Dnmt1 in mammals and the Dnmt1 ortholog MET1 in plants, which recognize a hemi-methylated meCG/GC and methylate the unmodified C. Plants also have non-CpG methylation; CpNpG (N is A, T, or C) methylation is maintained by CMT3 (chromodomain methyltransferase 3), a plant-specific enzyme, whilst asymmetric CpNpN methylation cannot be maintained (it must occur de novo) and is carried out by DRM2 and directed by 24-nt small interfering RNAs (siRNAs) (14, 64).
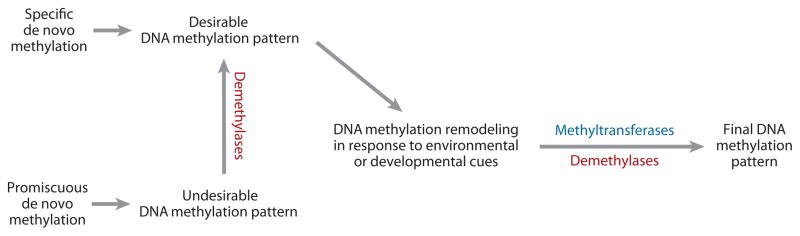
The level and pattern of 5-meC are determined by both DNA methylation and demethylation processes. For some genes, targeted or specific methylation by methyltransferases may be sufficient to create their methylation patterns, without the need for demethylases; for others, promiscuous methylation would need to be pruned by demethylases to generate the desired methylation pattern (Figure 1). In addition, demethylation may be needed to activate specific genes or to reset the epigenetic state of the genome during development or in response to environmental perturbations. Demethylation of DNA can be passive and/or active. Passive DNA demethylation occurs when maintenance methyltransferases are inactive during the cell cycle following DNA replication, which results in a retention of the unmethylated state of the newly synthesized strand. Active DNA demethylation involves one or more enzymes and can occur independently of DNA replication. The first enzyme in the active demethylation pathway has often been referred to as the demethylase. The identity of enzymes that promote active DNA demethylation has been elusive for many years. Recent work in plants demonstrated that a subfamily of DNA glycosylases can erase DNA methylation through a base excision repair pathway; this subfamily of DNA glycosylases thusly constitutes one type of DNA demethylase. Evidence suggests that active DNA demethylation in mammals is also achieved, at least in part, by a base excision repair pathway, although it appears that 5-meC is first converted to thymine through deamination before the DNA glycosylase acts.
Figure 1.
Role of active DNA demethylation in establishing DNA methylation patterns. DNA methylation patterns are established by the combined actions of DNA methyltransferases and demethylases. Demethylases are required for pruning unwanted DNA methylation generated by promiscuous methyltransferases, and DNA methylation reprogramming/remodeling during development and in response to environmental changes.
This review examines the expanding list of cellular activities and physiological processes that involve active DNA demethylation. The review also considers the genetic and biochemical evidence supporting a mechanism of active demethylation based on DNA glycosylases. In addition, the review discusses the regulation and targeting of demethylases and other possible mechanisms of active DNA demethylation.
THE MANY FUNCTIONS OF ACTIVE DNA DEMETHYLATION
Increasing evidence points to the importance of DNA demethylation in several cellular processes during development, defense, and disease. In plants, an important function of active DNA demethylation is to counteract the activities of the RNA-directed DNA methylation pathway to prevent the spreading of methylation from repetitive sequences to neighboring genes. The demethylases involved in active DNA demethylation in plants are known. In animals, however, the involvement of active DNA demethylation in most cases is supported only by the lack of a requirement for DNA replication or cell division for hypomethylation to occur. As we learn more about DNA demethylases and where to look for their effects on DNA methylation status, it is likely that active DNA demethylation will be found to contribute to most, if not all, processes where DNA methylation is important.
Mammals
There is extensive documentation of the roles of global and gene-specific active DNA demethylation in mammalian development, immune response, and diseases.
Early development
There is a very rapid demethylation of the male pronucleus in the zygote in mouse (65), human, pig, rat, and bovine preimplantation embryos (73). Because passive demethylation requires DNA replication and thusly takes time to occur, this rapid demethylation appears to be an active one. Active demethylation is presumably important for resetting the epigenetic state of the paternal genome to establish parent-specific developmental programs during early embryogenesis. Even during this so-called global demethylation, some regions of the genome, including certain heterochromatic sequences, retrotransposons, and paternally methylated imprinted genes, are not demethylated (73). PGC7/Stella, a DNA-binding protein, helps to protect the maternal genome and specific paternal genes against demethylation (75).
Reprogramming during gametogenesis and cloning
Genome-wide demethylation also occurs in primordial germ cells, where parental imprints are erased and totipotency is restored. Active demethylation is likely involved in this major reprogramming of the paternal and maternal genomes because demethylation occurs in the presence of the maintenance methylase Dnmt1 in the nucleus (73). It is unclear whether active demethylation in primordial germ cells occurs by mechanisms that are similar to those that function in the zygote.
During somatic cell nuclear transfer or cloning, a differentiated somatic cell nucleus must be reprogrammed in an enucleated oocyte to become totipotent. This epigenetic reprogramming involves active DNA demethylation (19). Active promoter demethylation precedes the reprogramming of the pluripotency regulator gene Oct4 after transfer of the somatic cell nucleus (91). The high rates of failure of cloning and frequent developmental abnormalities observed in cloned animals are presumed to reflect incomplete and aberrant reprogramming of somatic epigenetic marks (73, 108). In addition to embryonic stem cells and oocytes having the capacity to demethylate and reprogram other nuclei, differentiated mesodermal somatic cells have been shown to confer gene-specific active DNA demethylation in stable heterokaryons (110).
Memory formation and neurogenesis
A recent study found that dynamic regulation of DNA methylation by environmental cues is important for memory formation in rats (69). DNA methylation is required for the inactivation of the memory suppressor gene PP1, whereas active DNA demethylation is associated with the activation of the memory-promoting gene reelin (69).
Adult neurogenesis through continuous generation of new neurons in the mature brain represents one type of neural plasticity of the mammalian brain (70). The expression of BDNF (brain-derived neurotrophic factor) and FGF-1 (fibroblast growth factor-1) genes is critical for adult neurogenesis. 5-meC-immunoprecipitation-PCR and bisulfite sequencing analysis found demethylation in the promoter regions of BDNF and FGF-1 in response to electroconvulsive treatment that promotes adult neurogenesis (61). This active demethylation involves the induction of Gadd45b (DNA-damage-inducible protein 45 beta), a regulator of demethylation reactions (61).
Neurogenesis during the development of the zebrafish embryo also requires active DNA demethylation. Reducing the expression of Gadd45a or other proteins involved in demethylation causes the loss of neurons because of hypermethylation and consequent transcriptional silencing of genes required for neurogenesis (83).
Immune response
The cytokines IL-2 and IFN-γ are critical for the function of CD8 T cells. Upon re-encounter with antigens, the IL-2 and IFN-γ promoters are actively demethylated, which results in rapid cytokine production in memory CD8 T cells (12, 52, 76). Dynamic regulation of DNA methylation and demethylation appears to be an important part of epigenetic control in the immune response (85).
Tumorigenesis
In many cancers, bulk genome DNA methylation levels are reduced (22, 25, 104). Sequences with hypomethylation in cancers include repetitive sequences, imprinted genes, tissue-specific genes, oncogenes, and genes associated with invasion and metastases (54, 104). However, various loci, including many tumor suppressor genes, are hypermethylated and silenced during tumorigenesis (25, 42, 43). Although little is known about the mechanisms underlying the aberrant DNA methylation patterns, global DNA hypomethylation in cancers may involve both active and passive DNA demethylation, whereas inhibiting active DNA demethylation may contribute to the hypermethylation of tumor suppressor genes.
Plants
The elucidation of an active DNA demethylation mechanism in plants has helped to expose the roles of active DNA demethylation in genome regulation and plant development.
Prevention of transcriptional silencing of transgenes and endogenous genes
In plants, siRNAs of the 24-nt size class can trigger cytosine methylation and consequent transcriptional silencing of homologous DNA (4, 63, 64). These siRNAs are generated endogenously from transposons and other repetitive DNA sequences in a pathway involving the plant-specific RNA polymerase IV, RNA-dependent RNA polymerase 2 (RDR2), and Dicer-like 3 (DCL3). The siRNAs are loaded onto an effector complex that contains Pol V, Argonaute 4 (AGO4), or AGO6, and presumably the de novo DNA methyltransferase DRM2, which methylates cytosines in all sequence contexts, i.e., CpG, CpNpG, and CpNpN (14, 33, 63, 84). The KTF1 protein binds AGO4 and nascent transcripts generated by Pol V (103), thus serving as a bridge between the effector complex and siRNA target loci (33). When a trans-gene or endogenous gene promoter generates 24-nt siRNAs, the promoter is silenced by RNA-directed DNA methylation (63, 64). The ROS1 (repressor of silencing 1) gene, which encodes a 5-meC DNA glycosylase/demethylase, is required to maintain the expression of a trans-gene and its homologous endogenous gene (29). In the absence of ROS1 activity, the homologous genes are targets of RNA-directed DNA methylation and become heavily methylated and silenced transcriptionally. ROS1 is required to suppress the promoter methylation and silencing of a number of other endogenous genes (117). DML2 and DML3, two ROS1-like 5-meC DNA glycosylases, also prevent the hypermethylation of specific genomic loci in Arabidopsis vegetative tissues (80).
Using a genome tiling array, a study compared the DNA methylation profiles of wild-type plants with the demethylase triple mutant ros1 dml2 dml3 (80). The study found 179 loci with increased methylation in the triple mutant, indicating that these loci are normally targeted for demethylation. The majority of the identified loci are near genes or at the 5′ and 3′ ends of genes. Similarly, using a whole genome bisulfite sequencing approach, another study also found many hypermethylated genes in the ros1 dml2 dml3 mutant (60). However, there does not appear to be much overlap in the demethylation target genes identified from the two studies. This could be due to the different plant tissues used and possibly differences in growth conditions as well. The lack of extensive overlap also suggests that many more loci may be targeted by the demethylases during development and in response to environmental changes. Together, these studies provided evidence that active demethylation prevents the spreading of DNA methylation from repetitive sequences and thusly protects genes from deleterious methylation. The results suggest that many plant genes may be under the dynamic control of DNA methylation and active demethylation.
Regulation of imprinting
In Arabidopsis, active DNA demethylation is critical for activating the expression of the maternal allele of imprinted genes such as FWA (flowering wa-geningen) (53), the polycomb group genes MEA (MEDEA) (28) and FIS2 (fertilization independent seed 2) (49), and the C-terminal domain of poly(A)-binding protein MPC (maternally expressed PAB C-terminal ) (94). For these imprinted plant genes, the methylated inactive state is the default state, and demethylation and consequent expression take place only in the central cell of the female gametophyte and the endosperm where an active demethylase is expressed (36). The endosperm is derived from the fertilized central cell and supports embryo growth. It is a terminally differentiated tissue, so the methylation status of the hypomethylated maternal allele does not need to be reset. In the Arabidopsis DNA demethylase mutant dme, the imprinted MEA and FWA genes are not demethylated and the genes remain silent in the endosperm, which results in impaired seed development (36). In maize, the polycomb group gene FIE1 ( fertilization independent endosperm 1) is similarly imprinted (34). Only the maternal allele of FIE1 is expressed, and the expression is restricted to the endosperm, owing to active demethylation in this tissue.
Regulation of transposons
Active DNA demethylation is important for keeping transposons in a dynamic state that is not completely silenced. Most transposons and other repetitive DNA sequences in plants are considered to be silent because of heavy DNA methylation, particularly at CpG sites. However, low basal levels of expression are detected, suggesting that silencing of the transposons is incomplete (117). In the Arabidopsis ros1 mutants, some transposon or retrotransposon loci become more heavily methylated, especially at CpNpG and CpNpN sites (117). In association with this increased methylation, these loci show even lower levels of expression. Recent genome-wide methylation profiling in Arabidopsis identified transponsons among hundreds of loci that show hypermethylation and reduced expression in the demethylase triple mutant ros1 dml2 dml3 (60). These results suggest that active DNA demethylation maintains a basal level of expression of transposons. In rice, a ROS1-like 5-meC DNA glycosylase/DNA demethylase is important for maintaining the expression and promoting the transposition of the retrotransposon Tos17 (Guo-Liang Wang, personal communication). Transposons and other repetitive sequences make up the major parts of large plant genomes, and play important roles in shaping genome structure and in evolution by promoting genetic variability through transposition (6, 26). The dynamic control of transposons by both methylation and active demethylation may keep the plant epigenome plastic so that the plant can respond efficiently to environmental challenges during adaptation.
Decondensation of 5S rDNA chromatin
In Arabidopsis, 5S rDNA repeats within peri-centromeric heterochromatin are silenced by siRNA-directed DNA methylation and chromatin compaction (81). In early seedling development, there is a decondensation of 5S rDNA chromatin (21). The decondensation is caused by ROS1-mediated active DNA demethylation. Shortly after, the 5S rDNA chromatin is recondensed through the Pol IV-dependent RNA-directed DNA methylation pathway. The brief decondensation of 5S rDNA chromatin caused by active DNA demethylation may be important in unlocking a fraction of 5S rDNA units so that they can respond to environmental changes (21).
Plant responses to biotic and abiotic stresses
Global DNA methylation is substantially reduced in Arabidopsis plants infected with the bacterial pathogen Pseudomonas syringae (79). There is a marked decrease at the 180-bp centromeric repeat and other loci following pathogen attack. This change occurs in the absence of DNA replication, which suggests that it involves an active demethylation mechanism (79). In tobacco plants, DNA methylation is substantially and rapidly reduced in the coding region of a glycerophosphodiesterase-like gene one hour after treatment with aluminium, NaCl, cold, or oxidative stress (15). The reduced DNA methylation in the coding region correlates with stress induction of the glycerophosphodiesterase-like gene (15). Although the functional significance of gene-coding sequence methylation is unclear (119), the correlation suggests that active DNA demethylation is involved in permitting the induction of the glycerophosphodiesterase-like gene by stress.
POSSIBLE MECHANISMS OF ACTIVE DNA DEMETHYLATION
Base Excision Repair Initiated by 5-meC DNA Glycosylases
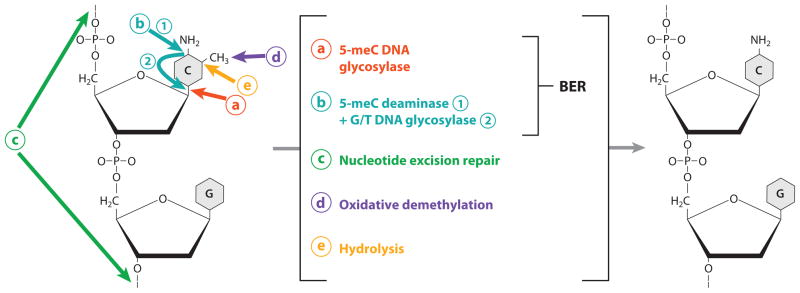
One proposed mechanism of active DNA demethylation involves base excision repair, which is initiated by DNA glycosylases (44, 47). The DNA glycosylases cleave the glycosidic bond between the 5-meC base and the deoxyribose, creating an abasic site or AP site; an AP endonuclease then removes the deoxyribose at the AP site; finally, the gap is filled by DNA polymerase and DNA ligase. The end result of this base excision repair pathway is the removal of methylated cytosine, and replacement by an unmethylated cytosine (Figure 2). An enzyme purified from chicken embryo nuclear extracts had weak 5-methylcytosine DNA glycosylase activity and strong G/T mismatch glycosylase activity (44, 47). Consistent with this observation, a partially purified enzyme preparation from HeLa nuclear extracts could also initiate DNA demethylation through a DNA glycosylase mechanism (100, 101). A thymine DNA glycosylase (TDG) cloned from chicken (116) had weak 5-methylcytosine DNA-glycosylase activity in vitro. Interestingly, the methylated DNA-binding protein (MBD4) from chicken and human (115) had a similar methylcytosine DNA glycosylase activity in vitro (115). Compared with their low methylcytosine DNA glycosylase activity, however, TDG and MBD4 have much stronger G/T mismatch repair activities in vitro. Furthermore, a defect in DNA methylation was not observed in mbd4 knockout mice (68, 105). Therefore, the significance of these DNA glycosylases in active DNA demethylation in vivo in mammals remains to be clarified. In contrast, considerable genetic and biochemical evidence indicates that a family of specialized DNA glycosylases are required for active DNA demethylation in plants, as discussed later.
Figure 2.
Diagram showing various possible levels and mechanisms of active DNA demethylation. (a) Base excision repair (BER) initiated by 5-methylcytosine (5-meC) DNA glycosylase. This is the predominant mechanism in plants but may also function in mammals. (b) Base excision repair initiated by coupled activities of 5-meC deaminase that converts 5-meC to T, and G/T mismatch DNA glycosylase that corrects the G/T mismatch. This appears to be the predominant mechanism in mammals but may also play a role in plants. (c) Nucleotide excision repair that removes methylated CpG dinucleotides. (d ) Oxidative removal of the methyl group. (e) Hydrolytic removal of the methyl group, releasing it as methanol.
Nucleotide Excision Repair and Hydrolysis
Another reported mechanism of active demethylation involves the excision of a methyl-CpG dinucleotide by an as yet unknown enzyme, and replacement of the methylated dinucleotide by an unmethylated CpG through DNA repair (102). This mechanism appears unlikely because the original report based on in vitro results has not been confirmed, and there is also no evidence for such a mechanism in vivo. A third reported mechanism of active DNA demethylation is the direct excision of the methyl group by hydrolysis, which results in the replacement of the methyl moiety by a hydrogen atom and the release of methanol. This energetically unfavorable reaction was reported to be carried out by MBD2, a methyl CpG-binding protein (8). However, this finding could not be replicated, and the conclusions have been contested (9).
5-meC Deamination Coupled with G/T Mismatch Repair
Active DNA demethylation could also occur through the enzymatic deamination of 5-meC to T, coupled with G/T mismatch repair by DNA glycosylases (72) (Figure 2). Recently, researchers proposed that in cultured human breast cancer cells the de novo DNA methyltransferases Dnmt3a and Dnmt3b can convert 5-meC to T through deamination; the resulting T is then removed by a G/T mismatch base excision repair pathway (66). The authors showed that this demethylation mechanism is important in the activation of the oestradiol-estrogen receptor target gene pS2 by E2. Remarkably, there is a rapid cycling of DNA methylation and demethylation at the promoters of PS2 and other oestradiol-estrogen receptor target genes, with a periodicity of approximately 100 min (50, 66). The cycling appears to correlate with the occupancy of the promoters by Dnmt3a and Dnmt3b (66). It is difficult to reconcile the findings with the known genetic functions of Dnmt3a and Dnmt3b (78), and thus these reports of rapid DNA methylation cycling and the DNMT3-based demethylation mechanism await confirmation by future studies. Dnmt3a (59) and Dnmt3b (10) are known to interact with the G/T mismatch repair DNA glycosylases. Such interactions may reflect the potential coupling between the 5-meC deamination activities of Dnmt3s and the G/T mismatch DNA glycosylase to achieve DNA demethylation (66). Alternatively, the interactions may simply allow the mismatch DNA glycosylase to be recruited to methylated DNA to prevent C to T mutations caused by spontaneous deamination of 5-meC (10, 59).
The AID/Apobec-1 (activation induced deaminase/apolipoprotein B RNA-editing catalytic component-1) family of RNA cytidine deaminases was also reported to have 5-meC deaminase activities (72), and if these deaminases are tightly and efficiently coupled to G/T mismatch repair systems, their activity could lead to DNA demethylation. Indeed, as discussed later, a recent study suggests that global active demethylation in zebrafish embryos can be achieved by the coupled action of AID and MBD4 (83).
Oxidative Demethylation
It is possible that active DNA demethylation may also be achieved by an oxidative demethylation mechanism (Figure 2). The Alkb family of enzymes can remove the methyl group from 1-methyladenine and 3-methylcytosine by oxidative demethylation, employing Fe(II) and alpha-ketoglutarate as cofactors, and release the methyl moiety as formaldehyde (24, 95). Oxidative demethylation of methylated histone H3 lysine 9 or lysine 36 by JmjC (Jumonji C) domain-containing proteins has been demonstrated (96, 107). The JmjC domain histone demethylases also use Fe(II) and alpha-ketoglutarate as cofactors, and the methyl moiety is released as formaldehyde during the reaction. In addition, demethylation of methylated histone H3 K4 by LSD1 involves an oxidative reaction, which uses FAD as a cofactor and releases formaldehyde as a by-product (90). Although the C-C bond in 5-meC is energetically much more difficult to break than the C-N bond in 1-meA, 3-meC, or methylated histones, it remains possible that a novel type of oxidase could demethylate 5-meC by an oxidative mechanism. In Arabidopsis, IBM1 (increase in bonsai methylation 1, a member of the JmjC domain histone demethylase-like protein family) is required to prevent DNA hypermethylation of the BNS (BONSAI ) locus (87). Although the authors suggested that the function of IBM1 is histone H3K9 demethylation, and its role in preventing DNA hypermethylation is indirect, the presumed histone demethylation activity of IBM1 has not been detected, and it remains possible that IBM1 has diverged from canonical histone demethylases and has acquired the ability to demethylate 5-meC DNA.
5-meC may also be converted to 5-hydroxymethylcytosine (hmC) through oxidation. hmC was recently found to be a nuclear DNA base in mammalian stem cells, Purkinje neurons and the brain (93a, 56a). TET1, an alpha-ketoglutarate and Fe(II)-dependent enzyme, can catalyze the conversion of 5-meC to hmC in cultured cells and in vitro (93a). The hmC then may be further converted to cytosine through a DNA glycosylase-based repair pathway or through as yet unknown mechanisms. The hmC may also facilitate passive DNA demethylation because it is not recognized well by the maintenance DNA methyltransferase DNMT1 (93a). At the present, it is not known whether hmC exists in plants.
In the possible mechanisms discussed above, hydrolysis and some oxidative demethylation mechanisms are direct, one-step reactions catalyzed by a single enzyme (Figure 2). The base and nucleotide excision repair pathways also can be considered as direct mechanisms but require multi-step reactions coordinated by several enzymes. In contrast, the mechanisms coupling deamination of 5-meC with base excision repair are indirect mechanisms and require multi-step reactions.
THE DISCOVERY OF DNA GLYCOSYLASES AS DNA DEMETHYLASES IN PLANTS
Two forward-genetic screens in Arabidopsis independently led to the discovery of DNA glycosylases that suppress DNA methylation and activate gene expression. Studies of the DNA glycosylase mutants provided strong genetic evidence that these enzymes are DNA demethylases.
ROS1
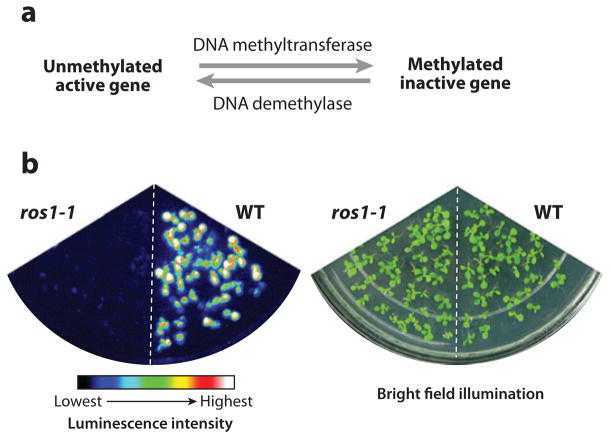
Our laboratory became interested in DNA methylation and demethylation because of our studies on plant responses to harsh environments. In these studies, we have been using the firefly luciferase (LUC) reporter gene driven by the salt-, drought-, cold-, or abscisic acid (ABA)-responsive RD29A promoter to study plant responses to harsh environments (118). The RD29A-LUC transgene in Arabidopsis (on chromosome III) behaves like the endogenous RD29A gene (on chromosome V), and plants containing this transgene emit bioluminescence in response to salt, drought, cold, or ABA treatment (Figure 3). These plants have facilitated genetic analysis of abiotic stress signal transduction, and many plant mutants with deregulated expression of the transgene and the homologous endogenous gene have been isolated and characterized (37). One group of mutants was intriguing because the RD29A-LUC trans-gene and the endogenous RD29A gene did not respond to any stress treatment, but all other stress-responsive genes examined were expressed normally. We found that the RD29A-LUC transgene and the endogenous RD29A were silenced transcriptionally in these mutants as a consequence of DNA hypermethylation (29).
Figure 3.
The Arabidopsis DNA demethylase ROS1 is required for preventing transcriptional silencing of genes that are under dynamic control by DNA methylation and demethylation. (a) Dynamic control of DNA methylation level and transcription activity by DNA methyltransferase and demethylase enzymes. (b) Silencing of the RD29A-LUC transgene in the ros1-1 mutant. Left, luminescence image of the ros1-1 mutant and WT plants; right, bright field illumination of all plants. The color scale under the luminescence image shows the luminescence intensity from black (lowest) to white (highest). Silencing in the ros1 mutant can be released by mutations in any component of the RNA-directed DNA methylation pathway, supporting dynamic control of the RD29A-LUC transgene by the opposing methylation and demethylation pathways. WT, wild type Arabidopsis seedlings.
Both the transgene RD29A promoter and the endogenous RD29A promoter from the mutants were heavily methylated in all sequence contexts, whereas only a low level of methylation was found in the promoters from wild-type plants. The transgene was inserted in the plant genome as a tandem repeat, and 24-nt siRNAs were generated from the RD29A promoter sequence in the transgene repeat (51). The silencing of the endogenous RD29A depends on the transgene. The silencing in the mutants is caused by the RD29A promoter hypermethylation, which in turn depends on the promoter siRNAs (32, 92, 113). Defects in any of the components of RNA-directed DNA methylation (RdDM) pathway can suppress the LUC silencing phenotype of the mutant (32). Therefore, although the promoter siRNAs are present in the wild type, they do not trigger sufficient promoter DNA methylation to cause transcriptional silencing. This indicates that antisilencing factors, later named ROS, exist in wild-type plants and that such factors are defective in the mutants (51).
Theoretically, antisilencing mechanisms could include factors that negatively regulate the production or action of 24-nt siRNAs so that RdDM does not occur, and could also include factors that reverse the effect of RdDM by erasing DNA methylation (e.g., DNA demethylase) or heterochromatic histone modification markers (e.g., histone H3 lysine 9 demethylase). The ROS1 gene isolated by map-based cloning encodes a large nuclear protein containing a C-terminal DNA glycosylase domain and an N-terminal histone H1-like basic region. The ros1-1 mutation creates a premature stop codon causing the truncation of much of the protein, including the DNA glycosylase domain, whereas the ros1-2 mutation causes a mis-sense mutation in a residue conserved in this subfamily of atypical DNA glycoslases (29). ROS1 is one of a small subfamily of four DNA glycosylases that also include Demeter (DME), DML2 (DME-like 2), and DML3. That DNA glycosylases are plausible DNA demethylases and the vital role of ROS1 in suppressing DNA methylation in vivo suggest that ROS1 is a DNA demethylase in Arabidopsis. Indeed, recombinant ROS1 protein was found to specifically cleave methylated but not unmethylated plasmid DNA in vitro (29).
Demeter
DME was identified because loss-of-function mutations in this gene resulted in impaired endosperm and embryo development, and consequently, in seed abortion (16). Compared with the widespread expression of ROS1 in all plant tissues examined (29), DME is preferentially expressed in the central cell and synergids of the female gametophyte (16). DME is required for the maternal allele-specific expression of MEDEA (MEA) in the central cell and endosperm. MEA, an imprinted gene, encodes a SET-domain polycomb group protein required for seed development (30, 55). DME was originally proposed to function by a mechanism not involving the demethylation of DNA, because no methylation was found in the region of the MEA locus that was bisulfite sequenced (16). More recent work, however, has demonstrated that the maternal allele of MEA in the seed is hypomethylated relative to the nonexpressed paternal allele, and that DME is required for this maternal allele-specific hypomethylation of MEA (28). The role of DME in preventing the methylation of the MEA locus or of an unknown positive regulator of MEA is also consistent with the finding that mutations in the maintenance DNA methyltransferase MET1 suppress the effect of dme mutations (106).
Another imprinted gene, FWA, also relies on DME for its maternal allele-specific expression in the endosperm (53). Unlike imprinting in mammals (57), imprinting of FWA and MEA in plants does not result from allele-specific de novo methylation but rather from maternal gametophyte-specific gene activation by DME-mediated DNA demethylation.
BIOCHEMICAL MECHANISM OF DNA GLYCOSYLASE-MEDIATED ACTIVE DNA DEMETHYLATION IN PLANTS
DNA glycosylases, proteins that excise damaged or mismatched bases, can be classified as monofunctional or bifunctional. The bi-functional glycosylases catalyze not only the hydrolysis of a glycosylic bond between the base and deoxyribose but also possess apurinic/apyrimidinic lyase activity that nicks the DNA backbone at the abasic site (67). Because of its DNA glycosylase domain sequences, ROS1 is predicted to be a bifunctional DNA glycosylase/lyase (29). DME was initially suggested to be a monofunctional DNA glycosylase (16). However, DME as well as DML2 and DML3 are in fact bifunctional DNA glycosylases like ROS1 (28, 71, 80).
Recombinant ROS1 protein, purified from Escherichia coli, had incision activity against methylated but not unmethylated plasmid DNA in vitro (1, 29). Purified recombinant ROS1, DME, DML2, and DML3 proteins also had incision activity against methylated oligonucleotide substrates (1, 28, 71, 80). Borohydride-dependent trapping assays confirmed the formation of a Schiff base intermediate between ROS1 or DME and a ring-opened sugar, which demonstrates that the reaction proceeds through a bifunctional DNA glycosylase/lyase mechanism (1, 28, 71). When the conserved glutamic acid-1303 was changed to lysine, the resulting mutant version of the ROS1 protein lacked DNA incision activity. Replacing a conserved aspartic acid with alanine in the active site of the glycosylase domain of ROS1 (D971A) also abolished the activities of the protein (71). Complementation assays using the ros1 mutant showed that the conserved glutamic acid-1303 is essential for ROS1 function in vivo (1). Similarly, changing the invariant lysine (position 1286) to glutamine or aspartic acid (position 1304 or 1562 depending on the splicing variant) to asparagine or alanine blocked the in vitro activities of DME (28, 71). The D1304N mutation disrupted the in vivo function of DME (17).
The use of oligonucleotide substrates with 5-meC in different sequence contexts allowed the characterization of substrate preferences of ROS1 and DME in vitro. One study found that ROS1 and DME are equally active on both fully and hemimethylated substrates (71). The results of Gehring et al. (28) and Penterman et al. (80) are consistent with this observation. Agius et al. (1), however, used different oligonucleotide substrates and found that ROS1 prefers fully methylated over hemimethylated sequences in vitro. This preference was also observed for a partially purified 5-methylcytosine DNA glycosylase from HeLa nuclear extracts (99, 101). The discrepancies between the different studies may be explained by the different sequences of the substrates used or different assay conditions. The different sequences of the substrates also likely explain discrepancies regarding the preference for 5-meC in CpG, CpNpG, or CpNpN sequence contexts. ROS1, DME, DML2, and DML3 can excise 5-meC from all sequence contexts (80), but Agius et al. (1) found a preference of ROS1 for CpNpG over CpG in vitro, whereas Morales-Ruiz et al. (71) observed opposite preference for ROS1 and DME. ROS1 has been shown to erase 5-meC in vivo from all sequence contexts for some target genes such as the RD29A promoter, but it preferentially removes 5-meC from non-CpG sites in most other target loci (117). In vitro, methylated RD29A promoter serves as a better substrate than the oligonucleotides for the detection of the 5-methylcytosine glycosylase activity of ROS1 (1). This is probably because the used promoter DNA was much longer than the oligonucleotide substrates.
ROS1 and DME can also remove mismatched thymine from DNA (1, 28, 71). However, unlike the mammalian MBD4, which has a strong preference for G/T mismatch substrates over 5-meC, ROS1 (1) and DME (28) prefer methylated substrates. ROS1 and DME are not active on U/G mismatch substrates (71) or substrates with damaged bases such as 8-oxo-7,8-dihydroguanine (1).
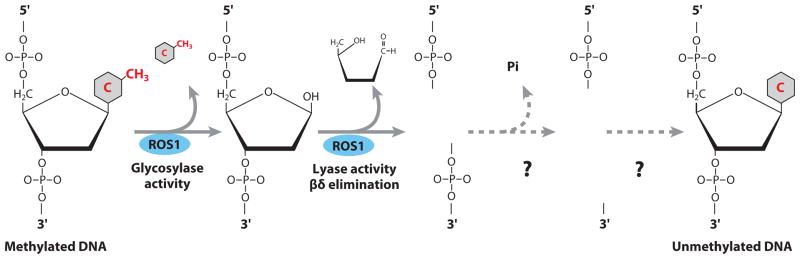
ROS1 predominantly generates β, δ elimination products (1) (Figure 4). In contrast, DME appears to generate a mixture of β and β, δ elimination products (28, 71). In the β elimination reaction, the lyase activity of the bifunctional DNA glycosylase causes one cleavage of the DNA backbone, whereas β, δ elimination results in two cleavages to release the abasic residue (1, 67). So, ROS1 and DME not only excise the 5-meC base through their DNA glycosylase activity but can also, via successive β, δ elimination, twice cleave the phosphodiester backbone at the abasic site through their lyase activity (Figure 4) (51). The final product is a single nucleoside gap, which must be further processed to generate a 3′ OH group, after which the gap is filled by an as yet unknown polymerase and ligase.
Figure 4.
Diagram of ROS1-mediated DNA demethylation by a base excision repair pathway. Question marks indicate as yet unidentified enzymes in the pathway. ROS1 is a bifunctional DNA glycosylase/lyase that removes the 5-methylcytosine base and then cleaves the DNA backbone at the abasic site, resulting in a gap that is then filled with an unmethylated cytosine nucleotide by as yet unknown DNA polymerase and ligase enzymes.
As discussed above, ROS1 and DME do have G/T mismatch repair DNA glycosylase activities, although the activities are weak (1, 28, 71). It is possible that these enzymes may occasionally or under certain in vivo conditions work together with a 5-meC deaminase to achieve demethylation. In such cases, we may expect increased C to T mutations in the demethylase mutant plants, unless there are other types of G/T mismatch repair DNA glycosylases that are redundant with the demethylases or unless the unknown 5-meC deaminase is active only in the presence of a demethylase. These predictions can be tested experimentally.
ACTIVE DNA DEMETHYLATION IN MAMMALS BY DEAMINATION COUPLED WITH G/T MISMATCH REPAIR
With the realization of the critical role of DNA methylation in epigenetic regulation and the increasing documentation of the involvement of active DNA demethylation in development, diseases, and many other important cellular processes, there is a pressing need to identify DNA demethylases in mammals. These demethylases and other proteins in the demethylation pathway could be excellent targets of drugs for cancers and other epigenetic-related diseases. As described above, biochemical support exists for active DNA demethylation through a DNA glycosylase pathway in mammalian cells (115, 116). In mammals, however, a major concern about the function of MBD4 and TDG DNA glycosylases as demethylases is that their 5-methylcytosine DNA glycosylase activity is very weak compared with their strong activities toward G/T mismatch DNA substrates. These enzymes may have stronger 5-methylcytosine DNA glycosylase activities in vivo, which may require other cofactors or interacting proteins. Alternatively, the G/T mismatch repair DNA glycosylase activity of MBD4 and TDG may be an essential part of demethylation if this activity is coordinated with the activity of a 5-meC deaminase.
Recently, additional evidence for DNA-repair mechanisms of demethylation in mammalian cells has been reported. Glucocorticoid treatment of cultured embryonic day-15 fetal hepatocytes or of a rat hepatoma cell line triggers active demethylation of the Tat enhancer (56). Data suggest that the DNA backbone is cleaved 3′ to the 5-meC, resulting in detection of a 5′ phosphate-containing 3′ cleavage fragment (56). The results are consistent with a base or nucleotide excision repair mechanism of demethylation. Furthermore, growth arrest and DNA-damage-inducible protein 45 alpha (Gadd45a), a small acidic nuclear protein induced by stress, was found to promote active DNA demethylation (3). Transfection of Gadd45a in cultured cells led to the demethylation and activation of methylated reporter and endogenous genes. Interestingly, a global reduction in DNA methylation was also observed in the transfected cells, which suggests a role for Gadd45a in genome-wide demethylation. Knockdown of Gadd45a and Gadd45b with siRNAs led to hypermethylation and gene inactivation as well as impaired UV light-induced DNA hypomethylation. Gadd45a was shown to interact with the nucleotide excision repair endonuclease XPG, which is also required for DNA demethylation (3). Thus, active demethylation in mammalian cells may be mediated by a pathway involving nucleotide excision, long-patch base excision, or mismatch repair; Gadd45a is a critical component of this repair pathway. In support of this hypothesis, Schmitz et al. (88) found that Gadd45a and its interacting nucleotide excision repair machinery are recruited to the rDNA promoter by TAF12, a TBP-associated factor in Pol I-and Pol II-specific TBP-TAF complexes. This leads to the demethylation of the rDNA promoter, keeping the rDNA in an active state. An independent study, however, failed to substantiate a functional role of Gadd45a in DNA demethylation (39), and global or locus-specific DNA hypermethylation was not observed in Gadd45a-deficient mice (23). A careful examination of mice deficient in all three Gadd45 genes (Gadd45a, Gadd45b, and Gadd45g) for DNA methylation profiles of genes known to be controlled by active demethylation (31) will help to resolve the controversy and definitively establish whether Gadd45 proteins function in locus-specific and global demethylation.
In zebrafish embryos, Gadd45a overexpression elicited global genome demethylation as well as demethylation of a methylated DNA fragment that was injected into the embryos (83). The study found evidence that locus-specific and global demethylation in zebrafish embryos is mediated by the AID/Apobec family of deaminases and MBD4. Overexpression of AID and MBD4 together in zebrafish embryos causes demethylation of the bulk genome and injected methylated DNA fragments. Overexpression of either protein alone does not elicit DNA demethylation. The results suggest that in zebrafish embryos AID acts as a 5-meC deaminase to convert 5-meC to T, and that the G/T mismatch DNA glycosylase activity of MBD4 then initiates a base excision repair reaction, with the final output of changing 5-meC to C. Zebrafish embryos overexpressing AID and a catalytically inactive MBD4 cannot demethylate injected methylated DNA fragments but instead accumulate C to T mutations in the injected DNA. The results strongly support the idea that AID and MBD4 are functionally coupled and that their overexpression causes DNA demethylation via a G:T intermediate. Although a DNA methylation defect was not detected in mbd4 deficient mice, the mutant mice did have a higher frequency of mutations at CpG sites (68, 105). It is possible that the increased mutations at CpG sites in mbd4 knockout mice result from 5-meC deamination without coupling to efficient G/T mismatch repair by MBD4.
In addition, morpholino injection to knockdown AID, MBD4, or Gadd45a in zebrafish embryos causes the loss of neurons at 24 hours post fertilization (83). The transcription factors neurod2, sox1a, sox-2, and other genes important for neurogenesis are affected by the knockdown. The knockdown causes a pronounced increase in CpG methylation at the promoter of neurod2. Gadd45a promotes the interaction between MBD4 and the deaminase Apobec2b. Co-immunoprecipitation analysis suggested that Gadd45a may form a complex with Apobec2a, Apobec2b, AID, and MBD4, and help bridge the deaminase and glycosylase enzymes (83).
Clearly, evidence is accumulating for pathways of active DNA demethylation based on DNA repair in mammals. Evidence is also increasingly indicating that these demethylation pathways require Gadd45a, the AID/Apobec family of deaminases, and MBD4. The model of demethylation by coupled action of AID and MBD4 is very attractive, and there is considerable experimental support for it. However, there are many unanswered questions regarding this model (40). For example, Apobecs are known to efficiently deaminate cytosines in only single-stranded RNA (Apobec1) or single-stranded DNA (AID), but the injected demethylation substrate DNA fragment in the zebrafish embryos is double stranded and is not expected to be made transiently single stranded because it is not replicated or transcribed. Because the research on active DNA demethylation in mammals has often been controversial and includes a number of reports that cannot be substantiated (78), current models of repair-based demethylation in mammals must be further tested by independent studies. Thus far, studies that support or rebut the repair-based demethylation pathways all have relied solely on transient overexpression or knockdown experiments for genetic evidence. Firm genetic evidence in the form of DNA methylation profiling in stable transgenic and knockout animals for candidate demethylases is of critical importance.
REGULATION OF DEMETHYLASE GENE EXPRESSION
The level of genome DNA methylation appears to be strictly controlled, and therefore, the levels and activities of DNA demethylases as well as methyltransferases must be tightly regulated. As discussed above, global DNA demethylation in mammals only occurs at certain developmental stages, and locus-specific demethylation is subjected to developmental and environmental control. In plants, the demethylase DME is expressed primarily in certain reproductive tissues (16). In contrast, the ROS1 transcript is widespread in plants (29). Although the ROS1 protein accumulation pattern has not been examined, it may vary in different tissues and in response to environmental perturbations.
Interestingly, the ROS1 transcript level appears to correlate with plant genome DNA methylation status. In the maintenance DNA methyltransferase mutant met1, genome DNA methylation is drastically decreased (60). ROS1 mRNA is virtually undetectable in met1 mutant plants (35, 62). Similarly, in the RdDM mutants nrpd1a, rdr2, dcl3, and drm2, ROS1 mRNA level is also very low (35, 62). In addition, the RdDM mutants drd1, nrpd2a, nrpd1b, and ago6 also have reduced ROS1 transcript levels (35, 112). In these RdDM mutants, locus-specific DNA methylation is blocked, but the total level of genome DNA methylation is not severely affected. These results suggest that ROS1 expression responds to the methylation levels of certain loci in the genome. It is likely that the methylation level of these sensor loci is sensed to regulate the expression of ROS1. Such sensing could be accomplished by a methyl DNA-binding protein (109) that has a fixed level and is normally occupied by methyl DNA at the sensor loci. Presumably, DML2 and DML3 expression levels are also sensitive to DNA methylation, although this is more difficult to determine because their expression levels are generally very low even in wild-type plants (62). The methylation-sensitive expression of ROS1 helps to explain a mysterious phenotypic reversion of the met1 mutant (62). Although newly generated homozygous met1 mutant plants have a very low level of genome DNA methylation, by the third or fourth generation no methylation defect can be found in the progeny of met1 plants (62). This gradual and unexpected recovery of DNA methylation is catalyzed by the residual DNA methyltransferase activity from DRM2 and also results from the lack of ROS1 expression in the mutant (62). Future experiments may determine whether forced ectopic expression of ROS1 blocks or slows down the recovery of DNA methylation in met1 progenies.
In addition to ROS1 expression, ROS3 expression is also regulated by the level of genome DNA methylation (112). ROS3 is an RNA-binding protein required for active DNA demethylation at some ROS1 target loci. In the ago6 (argonaute 6 ) mutant, the mRNA level of ROS3, like that of ROS1, is drastically reduced. It appears that the expression of the entire demethylation machinery, not just the demethylase, is responsive to DNA methylation. Interestingly, ROS1 expression is increased in ros3 mutant plants, and ROS3 expression is enhanced in the ros1 mutant plants (112). Because a number of loci have increased DNA methylation in the ros1 and ros3 mutants (112), this observation suggests that the expression of demethylation pathway components also is enhanced by increased methylation levels of certain sensor loci.
The expression of demethylases in mammals may also be affected by the level of DNA methylation. Injection of large amounts of methylated DNA into zebrafish embryos triggers not only the demethylation of the injected DNA but also of the endogenous genomic DNA of the embryo (83). Injection of unmethylated DNA is less effective in triggering DNA demethylation. The induced demethylation coincides with the up-regulation of AID/Apobec deaminase genes. Overexpression of Gadd45a also elicits DNA demethylation, and this is correlated with the enhanced expression of AID and Apobec2b (83).
TARGETING OF DNA DEMETHYLASES
The identification of DNA demethylases has generated many new questions regarding the mechanism of targeting demethylation to specific loci and the interplay between DNA demethylation and other epigenetic modifications (such as histone modifications, histone variants, and chromatin remodeling). In contrast to the well-documented global demethylation in mammals, there is no strong evidence for genome-wide demethylation in plants. It was reported that during tobacco pollen development there is a drastic reduction in DNA methylation in the nucleus of the generative cell, the progenitor of male gametes (77). The conclusion, however, was based solely on immunostaining using antibodies against 5-meC, a method prone to artifacts, and there has been no further report to support this finding. The known demethylases in Arabidopsis do not seem to function in global demethylation because ros1, dme, or ros1 dml2 dml3 mutations affect the methylation status of only a relatively small number of loci (up to several hundred) and do not substantially change the methylation level of the bulk genome DNA (28, 60, 80, 117). The locus-specific effects of the demethylases suggest that there are mechanisms for targeting the demethylases.
One possible mechanism is targeting by small RNAs. MicroRNAs and small interfering RNAs (siRNAs) are sequence-specific guides for gene silencing (4, 11, 13, 63, 93). De novo DNA methylation in plants is guided by 24-nt siRNAs (63, 64, 81). Sequence-specific active DNA demethylation may also be guided by certain small RNAs. ROS3, a regulatory factor for DNA demethylation in Arabidopsis (112), was identified from the same genetic screen that led to the discovery of ROS1. A loss-of-function mutation in ROS3 causes DNA hypermethylation and transcriptional gene silencing at a number of loci, some of which overlap with ROS1 targets. ROS3 encodes an RRM (RNA Recognition Motif) protein that binds to single-stranded small RNAs of specific sequences. Although the sequence features of ROS3-binding small RNAs have yet to be fully defined, they appear to be highly rich in G. ROS3 colocalizes with ROS1 in discrete nucleoplasmic foci and in the nucleolus, suggesting that the two proteins may function in a demethylation complex. If ROS3-binding small RNAs indeed guide sequence-specific demethylation, these small RNAs may be referred to as saRNAs (small activating RNAs) rather than siRNAs. It would be interesting to determine whether the biogenesis of ROS3-binding small RNAs differs from that of the heterochromatic siRNAs or any other known small RNAs.
Most of the endogenous small RNAs in Arabidopsis are 24-nt heterochromatic siRNAs generated by a pathway dependent on the plant-specific DNA-dependent RNA polymerase Pol V and/or Pol IV (63, 64, 74, 81, 84). In Pol IV and Pol V mutants, these siRNAs are not produced and DNA methylation of the corresponding loci is reduced. However, a study has found some 24-nt small RNAs corresponding to loci where the DNA methylation level is increased in the Pol IV and Pol V mutants (74). The results suggest that this type of small RNAs could function in guiding DNA demethylation and that Pol IV and Pol V may have a role in such demethylation (74).
In addition to its ability to bind specific small RNAs, ROS3 may also be capable of binding long RNAs, and the long RNAs could be targeting ROS3 and the demethylase complex to complementary loci. Targeting may be achieved by the pairing of ROS3-binding small RNAs and/or long RNAs with complementary DNA or complementary, nascent RNA transcript from the target DNA.
Some promoter-directed small RNAs appear to be able to cause gene activation in human cells, a phenomenon termed RNAa (38, 58). Even though RNAa in human cells does not appear to involve DNA methylation changes, it still could be mechanistically related to small RNA-directed DNA demethylation. Demethylation of DNA by purified chick embryo 5-methylcytosine-DNA glycosylase was found to require both protein and RNA (27, 45). 5-meC-DNA glycosylase activity from chicken embryo (27) or G8 myoblasts (48) was lost following RNase treatment but was restored by the addition of synthetic RNA complementary to the methylated strand of the substrate DNA. In addition, a DEAD-box RNA helicase is associated with the demethylase complex purified from chicken embryos (46, 89). Although the RNA involvement in these in vitro studies could be caused by artifacts, the results are consistent with the notion that the mammalian 5-meC DNA glycosylase may be targeted by RNAs. Interestingly, the RNAs targeting the demethylase are rich in CpGs (45, 89). It is not known whether the CpG-rich RNAs may give rise to small RNAs.
DNA demethylases may also be guided directly or indirectly by sequence-specific DNA-binding proteins. Human thymine DNA glycosylase (TDG) associates physically with the retinoid receptor (97, 114). Overexpression of the TDG causes demethylation of a retinoic acid-responsive promoter linked to a beta-galactosidase reporter gene, thusly activating the reporter (114). The results suggest that the hormone receptor may target the putative demethylase to the reporter gene promoter through physical interaction.
Certain structural domains in DNA demethylases may help target the enzymes to their DNA substrates. The ROS1 family of plant DNA demethylases have an N-terminal domain with sequence similarity to histone H1 (51). This domain may help target the demethylases to substrates by binding to DNA. The putative mammalian DNA demethylase MBD4 contains a methyl DNA-binding domain (2). This methyl DNA-binding domain may help target MBD4 to methylated DNA substrates. Recognition of a specific methylated DNA substrate would depend on other more precise targeting mechanisms.
The targeting of demethylases may be influenced by the chromatin environment. The function of DNA methyltransferases depends on histone modification patterns, such as histone H3 lysine 9 methylation, histone deacetylation, and deubiquitination (92, 98). Similarly, certain histone modifications may be required for DNA demethylase function. For example, specific histone modifications may be needed for binding or access of the demethylases to target sequences. Although nothing is currently known about the requirements of histone modifications in active DNA demethylation, the interplay between histone modifications and DNA demethylation is expected to be an important part of epigenetic regulation and thus a focal point of future research.
It appears that DNA demethylases and their regulators are not uniformly distributed in the nucleus. Immunostaining showed that ROS1 and ROS3 are colocalized in discrete foci in the nucleoplasm (112). These foci do not correspond to chromocenters where methylated DNA is most concentrated. A fraction of the ROS1 and ROS3 proteins is found in the nucleolus (112), suggesting that active DNA demethylation may be involved in the epigenetic regulation of rRNA genes and nucleolar dominance (82). Although it is possible that these sites are storage forms of the ROS proteins, it is also conceivable that DNA demethylases and their regulators are organized into active demethylation factories where specific methylated sequences are gathered for efficient demethylation.
CONCLUDING REMARKS
Genetic and biochemical evidence has established that the ROS1 subfamily of DNA glycosylases are locus-specific DNA demethylases in plants. It appears that locus-specific demethylation in mammals employs a different but related mechanism in which a deaminase first converts 5-meC to T before the G/T mismatch repair DNA glycosylases MBD4 and TDG can act. However, MBD4 and TDG also have 5-meC DNA glycosylase activities (albeit weak) in vitro (44, 47, 116). DNA demethylation in vivo in mammalian cells may in part be mediated directly by MBD4 or TDG without the need for AID/Apobec. This would be more similar to the plant demethylation pathway. It is possible that both deamination-dependent and -independent demethylation via MBD4 occur in mammals in vivo, and that the two modes of demethylation are complementary. Because the plant demethylases have some weak G/T mismatch DNA glycosylase activities (1, 28, 71), they may also be capable of functioning together with AID/Apobec-like deaminases to carry out DNA demethylation. Perhaps the two related mechanisms can both take place in plants and mammals, but plants rely mainly on the direct, deamination-independent pathway whereas mammals rely on the indirect, deamination-dependent pathway.
Although it remains unclear whether global demethylation takes place in plants during gametogenesis or early embryo development, the occurrence of rapid genome-wide active demethylation in developing mammals is well documented (65). Despite recent reports of DNA repair-based global active demethylation (3, 83), it is still possible that global demethylation does not involve DNA repair because repair-based reactions may generate numerous strand breaks that are potentially detrimental to genome integrity. On the other hand, with efficient functioning of the repair-pathway enzymes and tight coupling between the various components, strand breaks may be kept very transient and may not be exposed or accumulate. Whatever the mechanisms and candidate demethylases in mammals, critically examining the DNA methylation status of mice deficient in the candidate genes is important.
In plant mutants lacking ROS1 or related demethylases, the overall 5-meC levels are not affected, although specific loci are hypermethylated (29, 60, 80, 117). Therefore, knowing where to look for methylation changes is important. Genome-wide methylation profiling in knockout mice for candidate demethylases at various developmental stages and in various tissues may be necessary. Such profiling can be done by genome bisulfite sequencing (18, 60) or 5-meC immunoprecipitation coupled with tiling array analysis (111). In addition, researchers should remember that the phenotypic analysis may be complicated by the existence of multiple mechanisms and multiple demethylases that may be functionally redundant. Multiple mutants with several related genes knocked out simultaneously may be needed to overcome gene redundancy problems.
In addition to the DNA glycosylases and deaminases, the DNA repair-based demethylation pathways require other components such as DNA polymerase and ligase. Unlike ROS1 and other plant demethylases, MBD4 and TDG are not bifunctional DNA glycosylases and do not possess lyase activities, so a lyase remains to be identified in the mammalian demethylation pathway. In addition, tight regulation of active demethylation during development and environmental responses necessitates the existence of demethylation regulators. The RNA-binding protein ROS3 in plants and Gadd45a in mammals are examples of such regulators, but more need to be identified. The elucidation of active DNA demethylation pathways and the mechanisms of demethylase targeting and regulation will contribute greatly to our understanding of epigenetics and its role in the development, environmental responses, and evolution of plants and animals.
NOTE ADDED IN PROOF
Two recent studies found DME-dependent genome-wide demethylation in the Arabidopsis endosperm, supporting that DNA repair-based active DNA demethylation can contribute to global DNA demethylation.
Gehring M, Bubb KL, Henikoff S. 2009. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324:1447–51
Hsieh TF, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D. 2009. Genome-wide demethylation of Arabidopsis endosperm. Science 324:1451–54
SUMMARY POINTS.
Active DNA demethylation plays critical roles in development, diseases, and environmental responses. It counteracts the activities of the RNA-directed DNA methylation pathway to prevent the spreading of methylation from repetitive sequences in plants.
Several mechanisms are possible for active DNA demethylation, but the ones employed by cells have been difficult to pinpoint.
Forward genetic screens in Arabidopsis identified two bifunctional DNA glycosylase enzymes (ROS1 and DME) critical for active DNA demethylation in vivo.
Biochemical studies showed that ROS1, DME, and two related proteins (DML2 and DML3) are 5-methylcytosine DNA glycosylases that initiate a base excision pathway for active DNA demethylation.
Accumulating evidence supports a DNA repair-mediated pathway for locus-specific and possibly global active DNA demethylation in mammals. It is likely that active DNA demethylation in mammals is largely achieved by the coupled action of 5-methylcytosine deaminases (converting 5-meC to T) and DNA glycosylases (correcting G/T mismatches).
DNA demethylases may be guided to specific loci by factors such as small RNAs, transcription factors, and chromatin status.
FUTURE ISSUES.
Genetic evidence in the form of DNA methylation phenotypes in stable transgenic and knockout animals for candidate demethylases is needed to firmly establish the active DNA demethylation mechanisms in mammals.
Is global DNA demethylation in mammals mediated by a DNA repair mechanism?
Several components in the base excision repair pathway of active DNA demethylation are still missing (e.g., DNA polymerase and ligase) and need to be identified in plants and mammals.
What is the role of hydroxymethylcytosine? Is it an intermediate in DNA demethylation?
The mechanisms of targeting and regulation of demethylases need to be better understood.
Glossary
- 5-methylcytosine (5-meC)
a major form of DNA modification and a stable epigenetic mark that represses gene transcription
- siRNA
small interfering RNA
- Passive DNA demethylation
in the absence of maintenance DNA methylation, methylation marks are lost in newly synthesized DNA during DNA replication
- Active DNA demethylation
enzymatic conversion of 5-methylcytosine to cytosine
- Base excision repair
a multi-step reaction that is initiated when a DNA glycosylase removes a modified or damaged base in DNA
- RNA-directed DNA methylation
sequence-specific DNA methylation by the de novo DNA methyltransferase DRM2 in plants is directed by 24-nt siRNAs
- Bisulfite sequencing
a method for determining methylation status of individual cytosines. Sodium bisulfite converts cytosine to uracil but leaves 5-methylcytosine unchanged
- Gadd45a (DNA-damage-inducible protein 45 alpha)
a small acidic nuclear protein induced by stress; a putative regulator of active DNA demethylation in mammals
- repressor of silencing 1 (ROS1)
a 5-meC DNA glycosylase/DNA demethylase that prevents DNA hypermethylation and transcriptional silencing in Arabidopsis
- 5-meC DNA glycosylase
DNA glycosylase that removes 5-methylcytosine bases from DNA, also referred to as DNA demethylase
- Thymine DNA glycosylase (TDG)
a G/T mismatch DNA glycosylase in mammals; has a weak 5-meC DNA glycosylase activity in vitro
- Methyl DNA-binding protein 4 (MBD4)
a G/T mismatch DNA glycosylase in mammals; has a weak 5-meC DNA glycosylase activity in vitro
- Activation induced deaminase (AID)
a member of the family of RNA cytidine deaminases; can deaminate 5-meC to convert it to thymine
- Apolipoprotein B RNA-editing catalytic component (Apobec)
member of the family of RNA cytidine deaminases; can deaminate 5-meC to convert it to thymine
- 5-meC deaminase
the AID/Apobec family enzyme or other enzyme that can deaminate 5-methylcytosine to thymine
- RdDM
RNA-directed DNA methylation
- Demeter (DME)
a 5-meC DNA glycosylase/DNA demethylase important for gene imprinting in Arabidopsis
- Demeter-like proteins (DML)
including DML1/ ROS1, DML2 and DML3 in Arabidopsis
- Bifunctional DNA glycosylase
an enzyme with both DNA glycosylase and endonuclease/lyase activities
Footnotes
DISCLOSURE STATEMENT
The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc Natl Acad Sci USA. 2006;103:11796–801. doi: 10.1073/pnas.0603563103. Provided evidence that the Arabidopsis ROS1 protein can specifically recognize and process 5-methylcytosine DNA through a DNA glycosylase mechanism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballestar E, Wolffe AP. Methyl-CpG-binding proteins. Targeting specific gene repression. Eur J Biochem. 2001;268:1–6. doi: 10.1046/j.1432-1327.2001.01869.x. [DOI] [PubMed] [Google Scholar]
- 3.Barreto G, Schäfer A, Marhold J, Stach D, Swaminathan SK, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–75. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 4.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–63. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 5.Bender J. DNA methylation and epigenetics. Annu Rev Plant Biol. 2004;55:41–68. doi: 10.1146/annurev.arplant.55.031903.141641. [DOI] [PubMed] [Google Scholar]
- 6.Bennetzen JL. Transposable elements, gene creation and genome rearrangement in flowering plants. Curr Opin Genet Dev. 2005;15:621–27. doi: 10.1016/j.gde.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 8.Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–83. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 9.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 10.Boland MJ, Christman JK. Characterization of Dnmt3b:thymine-DNA glycosylase interaction and stimulation of thymine glycosylase-mediated repair by DNA methyltransferase(s) and RNA. J Mol Biol. 2008;379:492–504. doi: 10.1016/j.jmb.2008.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–91. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4:235–40. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- 13.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–39. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 14.Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–60. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- 15.Choi CS, Sano H. Abiotic-stress induces demethylation and transcriptional activation of a gene encoding a glycerophosphodiesterase-like protein in tobacco plants. Mol Genet Genomics. 2007;277:589–600. doi: 10.1007/s00438-007-0209-1. [DOI] [PubMed] [Google Scholar]
- 16.Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 17.Choi Y, Harada JJ, Goldberg RB, Fischer RL. An invariant aspartic acid in the DNA glycosylase domain of DEMETER is necessary for transcriptional activation of the imprinted MEDEA gene. Proc Natl Acad Sci USA. 2004;101:7481–86. doi: 10.1073/pnas.0402328101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–19. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dean W, Santos F, Stojkovic M, Zakhartchenko V, Walter J, et al. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc Natl Acad Sci USA. 2001;98:13734–38. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doerfler W. DNA methylation and gene activity. Annu Rev Biochem. 1983;52:93–124. doi: 10.1146/annurev.bi.52.070183.000521. [DOI] [PubMed] [Google Scholar]
- 21.Douet J, Blanchard B, Cuvillier C, Tourmente S. Interplay of RNA Pol IV and ROS1 during post-embryonic 5S rDNA chromatin remodeling. Plant Cell Physiol. 2008;49:1783–91. doi: 10.1093/pcp/pcn152. [DOI] [PubMed] [Google Scholar]
- 22.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–13. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 23.Engel N, Tront JS, Erinle T, Nguyen N, Latham KE, et al. Conserved DNA methylation in Gadd45a(−/−) mice. Epigenetics. 2009;4:98–99. doi: 10.4161/epi.4.2.7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falnes PØ, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–82. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 25.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 26.Feschotte C, Jiang N, Wessler SR. Plant transposable elements: where genetics meets genomics. Nat Rev Genet. 2002;3:329–41. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- 27.Frémont M, Siegmann M, Gaulis S, Matthies R, Hess D, Jost JP. Demethylation of DNA by purified chick embryo 5-methylcytosine-DNA glycosylase requires both protein and RNA. Nucleic Acids Res. 1997;25:2375–80. doi: 10.1093/nar/25.12.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, et al. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. Found evidence that DME has 5-meC DNA glycosylase activity and functions to demethylate and activate the MEDEA gene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong Z, Morales-Ruiz T, Ariza RR, Roldán-Arjona T, David L, Zhu JK. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–14. doi: 10.1016/s0092-8674(02)01133-9. The first genetic characterization of active DNA demethylation pathway and its role in antagonizing DNA methylation and gene silencing. [DOI] [PubMed] [Google Scholar]
- 30.Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB. Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science. 1998;280:446–50. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- 31.Gupta M, Gupta SK, Balliet AG, Hollander MC, Fornace AJ, et al. Hematopoietic cells from Gadd45a- and Gadd45b-deficient mice are sensitized to genotoxic-stress-induced apoptosis. Oncogene. 2005;24:7170–79. doi: 10.1038/sj.onc.1208847. [DOI] [PubMed] [Google Scholar]
- 32.He XJ, Hsu YF, Pontes O, Zhu J, Lu J, et al. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerase IV and V and is required for RNA-directed DNA methylation. Genes Dev. 2009;23:318–30. doi: 10.1101/gad.1765209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He XJ, Hsu YF, Zhu S, Wierzbicki A, Pontes O, et al. An effector of RNA-directed DNA methylation in Arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009;137:498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hermon P, Srilunchang KO, Zou J, Dresselhaus T, Danilevskaya ON. Activation of the imprinted Polycomb Group Fie1 gene in maize endosperm requires demethylation of the maternal allele. Plant Mol Biol. 2007;64:387–95. doi: 10.1007/s11103-007-9160-0. [DOI] [PubMed] [Google Scholar]
- 35.Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJM, Matzke M. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 2006;25:2828–36. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huh JH, Bauer MJ, Hsieh TF, Fischer RL. Cellular programming of plant gene imprinting. Cell. 2008;132:735–44. doi: 10.1016/j.cell.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 37.Ishitani M, Xiong L, Stevenson B, Zhu JK. Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell. 1997;9:1935–49. doi: 10.1105/tpc.9.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3:166–73. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 39.Jin SG, Guo C, Pfeifer GP. GADD45A does not promote DNA demethylation. PloS Genet. 2008;4:e1000013. doi: 10.1371/journal.pgen.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiricny J, Menigatti M. DNA cytosine demethylation: are we getting close? Cell. 2008;135:1167–69. doi: 10.1016/j.cell.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 41.Johnson TB, Coghill RD. Researches on pyrimidines. C111 The discovery of 5-methyl-cytosine in tuberculinic acid, the nucleic acid of the tubercle bacillus. J Am Chem Soc. 1925;47:2838–44. [Google Scholar]
- 42.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 43.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–7. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 44.Jost JP. Nuclear extracts of chicken embryos promote an active demethylation of DNA by excision repair of 5-methyldeoxycytidine. Proc Natl Acad Sci USA. 1993;90:4684–88. doi: 10.1073/pnas.90.10.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jost JP, Frémont M, Siegmann M, Hofsteenge J. The RNA moiety of chick embryo 5-methylcytosine-DNA glycosylase targets DNA demethylation. 1997;25:4545–50. doi: 10.1093/nar/25.22.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jost JP, Schwarz S, Hess D, Angliker H, Fuller-Pace FV, et al. A chicken embryo protein related to the mammalian DEAD box protein p68 is tightly associated with the highly purified protein-RNA complex of 5-MeC-DNA glycosylase. Nucleic Acids Res. 1999;27:3245–52. doi: 10.1093/nar/27.16.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jost JP, Siegmann M, Sun L, Leung R. Mechanisms of DNA demethylation in chicken embryos—purification and properties of a 5-methylcytosine-DNA glycosylase. J Biol Chem. 1995;270:9734–97. doi: 10.1074/jbc.270.17.9734. [DOI] [PubMed] [Google Scholar]
- 48.Jost JP, Siegmann M, Thiry S, Jost YC, Benjamin D, Schwarz S. A re-investigation of the ribonuclease sensitivity of a DNA demethylation reaction in chicken embryo and G8 mouse myoblasts. FEBS Lett. 1999;449:251–54. doi: 10.1016/s0014-5793(99)00454-8. [DOI] [PubMed] [Google Scholar]
- 49.Jullien PE, Kinoshita T, Ohad N, Berger F. Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell. 2006;18:1360–72. doi: 10.1105/tpc.106.041178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kangaspeska S, Stride B, Métivier R, Polycarpou-Schwarz M, Ibberson D, et al. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–15. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 51.Kapoor A, Agius F, Zhu JK. Preventing transcriptional gene silencing by active DNA demethylation. FEBS Lett. 2005;579:5889–98. doi: 10.1016/j.febslet.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 52.Kersh EN, Fitzpatrick DR, Murali-Krishna K, Shires J, Speck SH, et al. Rapid demethylation of the IFN-gamma gene occurs in memory but not I CD8 T cells. J Immunol. 2006;176:4083–93. doi: 10.4049/jimmunol.176.7.4083. [DOI] [PubMed] [Google Scholar]
- 53.Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, et al. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science. 2004;303:521–23. doi: 10.1126/science.1089835. [DOI] [PubMed] [Google Scholar]
- 54.Kisseljova NP, Kisseljov FL. DNA demethylation and carcinogenesis. Biochemistry (Mosc) 2005;70:743–52. doi: 10.1007/s10541-005-0179-z. [DOI] [PubMed] [Google Scholar]
- 55.Kiyosue T, Ohad N, Yadegari R, Hannon M, Dinneny J, et al. Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:4186–91. doi: 10.1073/pnas.96.7.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kress C, Thomassin H, Grange T. Active cytosine demethylation triggered by a nuclear receptor involves DNA strand breaks. Proc Natl Acad Sci USA. 2006;103:11112–17. doi: 10.1073/pnas.0601793103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56a.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–30. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–65. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 58.Li LC, Okino ST, Zhao H, Pookot D, Place RF, et al. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci USA. 2006;103:17337–42. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li YQ, Zhou PZ, Zheng XD, Walsh CP, Xu GL. Association of Dnmt3a and thymine DNA glycosylase links DNA methylation with base-excision repair. Nucleic Acids Res. 2007;35:390–400. doi: 10.1093/nar/gkl1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–36. doi: 10.1016/j.cell.2008.03.029. Whole genome bisulfite sequencing of the demethylase triple mutant ros1dml2dml3 identified hundreds of demethylation target loci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–77. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007;130:851–62. doi: 10.1016/j.cell.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 63.Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJ. Targets of RNA-directed DNA methylation. Curr Opin Plant Biol. 2007;10:512–19. doi: 10.1016/j.pbi.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 64.Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- 65.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–2. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 66.Métivier R, Gallais R, Tiffoche C, Le Péron C, Jurkowska RZ, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 67.McCullough AK, Dodson ML, Lloyd RS. Initiation of base excision repair: glycosylase mechanisms and structures. Annu Rev Biochem. 1999;68:255–85. doi: 10.1146/annurev.biochem.68.1.255. [DOI] [PubMed] [Google Scholar]
- 68.Millar CB, Guy J, Sansom OJ, Selfridge J, MacDougall E, et al. Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science. 2002;297:403–5. doi: 10.1126/science.1073354. [DOI] [PubMed] [Google Scholar]
- 69.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–69. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 70.Ming GL, Song H. Adult neurogenesis in the mammalian central nervous system. Annu Rev Neurosci. 2005;28:223–50. doi: 10.1146/annurev.neuro.28.051804.101459. [DOI] [PubMed] [Google Scholar]
- 71.Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marín MI, Martínez-Macías MI, Ariza RR, Roldán-Arjona T. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc Natl Acad Sci USA. 2006;103:6853–58. doi: 10.1073/pnas.0601109103. Nice biochemical characterization of the 5-meC DNA glycosylase activities of ROS1 and DME. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–60. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 73.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14(Spec 1):R47–58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 74.Mosher RA, Schwach F, Studholme D, Baulcombe DC. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci USA. 2008;105:3145–50. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- 76.Northrop JK, Thomas RM, Wells AD, Shen H. Epigenetic remodeling of the IL-2 and IFN-gamma loci in memory CD8 T cells is influenced by CD4 T cells. J Immunol. 2006;177:1062–69. doi: 10.4049/jimmunol.177.2.1062. [DOI] [PubMed] [Google Scholar]
- 77.Oakeley EJ, Podestà A, Jost JP. Developmental changes in DNA methylation of the two tobacco pollen nuclei during maturation. Proc Natl Acad Sci USA. 1997;94:11721–25. doi: 10.1073/pnas.94.21.11721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–48. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 79.Pavet V, Quintero C, Cecchini NM, Rosa AL, Alvarez ME. Arabidopsis displays centromeric DNA hypomethylation and cytological alterations of heterochromatin upon attack by Pseudomonas syringae. Mol Plant Microbe Interact. 2006;19:577–87. doi: 10.1094/MPMI-19-0577. [DOI] [PubMed] [Google Scholar]
- 80.Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci USA. 2007;104:6752–57. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pikaard CS. Cell biology of the Arabidopsis nuclear siRNA pathway for RNA-directed chromatin modification. Cold Spring Harb Symp Quant Biol. 2006;71:473–80. doi: 10.1101/sqb.2006.71.046. [DOI] [PubMed] [Google Scholar]
- 82.Preuss S, Pikaard CS. rRNA gene silencing and nucleolar dominance: insights into a chromosome-scale epigenetic on/off switch. Biochim Biophys Acta. 2007;1769:383–92. doi: 10.1016/j.bbaexp.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–12. doi: 10.1016/j.cell.2008.11.042. Elegant characterization of an active DNA demethylation activity in zebrafish embryos. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ream TS, Haag JR, Wierzbicki AT, Nicora CD, Norbeck AD, et al. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell. 2009;33:192–203. doi: 10.1016/j.molcel.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reiner SL. Epigenetic control in the immune response. Hum Mol Genet. 2005;14(Spec 1):R41–46. doi: 10.1093/hmg/ddi115. [DOI] [PubMed] [Google Scholar]
- 86.Roberts RJ, Cheng X. Base flipping. Annu Rev Biochem. 1998;67:181–98. doi: 10.1146/annurev.biochem.67.1.181. [DOI] [PubMed] [Google Scholar]
- 87.Saze H, Shiraishi A, Miura A, Kakutani T. Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science. 2008;319:462–65. doi: 10.1126/science.1150987. [DOI] [PubMed] [Google Scholar]
- 88.Schmitz KM, Schmitt N, Hoffmann-Rohrer U, Schäfer A, Grummt I, Mayer C. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol Cell. 2009;33:344–53. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 89.Schwarz S, Bourgeois C, Soussaline F, Homsy C, Podestà A, Jost JP. A CpG-rich RNA and an RNA helicase tightly associated with the DNA demethylation complex are present mainly in dividing chick embryo cells. Eur J Cell Biol. 2000;79:488–94. doi: 10.1078/0171-9335-00070. [DOI] [PubMed] [Google Scholar]
- 90.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 91.Simonsson S, Gurdon J. DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat Cell Biol. 2004;6:984–90. doi: 10.1038/ncb1176. [DOI] [PubMed] [Google Scholar]
- 92.Sridhar VV, Kapoor A, Zhang K, Zhu J, Zhou T, et al. Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature. 2007;447:735–38. doi: 10.1038/nature05864. [DOI] [PubMed] [Google Scholar]
- 93.Sunkar R, Chinnusamy V, Zhu J, Zhu JK. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007;12:301–9. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 93a.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tiwari S, Schulz R, Ikeda Y, Dytham L, Bravo J, et al. MATERNALLY EXPRESSED PAB C-TERMINAL, a novel imprinted gene in Arabidopsis, encodes the conserved C-terminal domain of polyadenylate binding proteins. Plant Cell. 2008;20:2387–98. doi: 10.1105/tpc.108.061929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–78. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 96.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–16. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 97.Um S, Harbers M, Benecke A, Pierrat B, Losson R, Chambon P. Retinoic acid receptors interact physically and functionally with the T:G mismatch-specific thymine-DNA glycosylase. J Biol Chem. 1998;273:20728–36. doi: 10.1074/jbc.273.33.20728. [DOI] [PubMed] [Google Scholar]
- 98.Vaillant I, Paszkowski J. Role of histone and DNA methylation in gene regulation. Curr Opin Plant Biol. 2007;10:528–33. doi: 10.1016/j.pbi.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 99.Vairapandi M. Characterization of DNA demethylation in normal and cancerous cell lines and the regulatory role of cell cycle proteins in human DNA demethylase activity. J Cell Biochem. 2004;91:572–83. doi: 10.1002/jcb.10749. [DOI] [PubMed] [Google Scholar]
- 100.Vairapandi M, Duker NJ. Enzymic removal of 5-methylcytosine from DNA by a human DNA glycosylase. Nucleic Acids Res. 1993;21:5323–27. doi: 10.1093/nar/21.23.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vairapandi M, Liebermann DA, Hoffman B, Duker NJ. Human DNA-demethylating activity: a glycosylase associated with RNA and PCNA. J Cell Biochem. 2000;79:249–60. doi: 10.1002/1097-4644(20001101)79:2<249::aid-jcb80>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 102.Weiss A, Keshet I, Razin A, Cedar H. DNA demethylation in vitro: Involvement of RNA. Cell. 1996;86:709–18. doi: 10.1016/s0092-8674(00)80146-4. [DOI] [PubMed] [Google Scholar]
- 103.Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–48. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775:138–62. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 104a.Wilson GG. Type II restriction–modification systems. Trends Genet. 1988;4:314–18. doi: 10.1016/0168-9525(88)90109-6. [DOI] [PubMed] [Google Scholar]
- 105.Wong E, Yang K, Kuraguchi M, Werling U, Avdievich E, et al. Mbd4 inactivation increases C → T transition mutations and promotes gastrointestinal tumor formation. Proc Natl Acad Sci USA. 2002;99:14937–42. doi: 10.1073/pnas.232579299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xiao W, Gehring M, Choi Y, Margossian L, Pu H, et al. Imprinting of the MEA Polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Dev Cell. 2003;5:891–901. doi: 10.1016/s1534-5807(03)00361-7. [DOI] [PubMed] [Google Scholar]
- 107.Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–95. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 108.Yang X, Smith SL, Tian XC, Lewin HA, Renard JP, Wakayama T. Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat Genet. 2007;39:295–302. doi: 10.1038/ng1973. [DOI] [PubMed] [Google Scholar]
- 109.Zemach A, Grafi G. Methyl-CpG-binding domain proteins in plants: interpreters of DNA methylation. Trends Plant Sci. 2007;12:80–85. doi: 10.1016/j.tplants.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 110.Zhang F, Pomerantz JH, Sen G, Palermo AT, Blau HM. Active tissue-specific DNA demethylation conferred by somatic cell nuclei in stable heterokaryons. Proc Natl Acad Sci USA. 2007;104:4395–400. doi: 10.1073/pnas.0700181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 112.Zheng X, Pontes O, Zhu J, Miki D, Zhang F, et al. ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature. 2008;455:1259–62. doi: 10.1038/nature07305. Identified ROS3, a small RNA-binding protein, as a regulator of demethylation, suggesting that small RNAs may guide active DNA demethylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zheng X, Zhu J, Kapoor A, Zhu JK. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 2007;26:1691–701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhu B, Benjamin D, Zheng Y, Angliker H, Thiry S, et al. Overexpression of 5-methylcytosine DNA glycosylase in human embryonic kidney cells EcR293 demethylates the promoter of a hormone-regulated reporter gene. Proc Natl Acad Sci USA. 2001;98:5031–36. doi: 10.1073/pnas.091097298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhu B, Zheng Y, Angliker H, Schwarz S, Thiry S, et al. 5-methylcytosine DNA glycosylase activity is also present in the human MBD4 (G/T mismatch glycosylase) and in a related avian sequence. Nucleic Acids Res. 2000;28:4157–65. doi: 10.1093/nar/28.21.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhu B, Zheng Y, Hess D, Angliker H, Schwarz S, et al. 5-methylcytosine-DNA glycosylase activity is present in a cloned G/T mismatch DNA glycosylase associated with the chicken embryo DNA demethylation complex. Proc Natl Acad Sci USA. 2000;97:5135–39. doi: 10.1073/pnas.100107597. The authors cloned the G/T mismatch DNA glycosylase TDG and showed that it has a weak 5-meC DNA glycosylase activity in vitro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr Biol. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 118.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–73. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhu JK. Epigenome sequencing comes of age. Cell. 2008;133:395–97. doi: 10.1016/j.cell.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]