Abstract
The alcohol-tolerant AT and -non-tolerant ANT rat lines have been selectively bred for innate sensitivity to ethanol-induced motor impairment. The cerebellar GABAA receptor α6 subunit alleles α6-100R and α6-100Q are segregated in the AT and ANT rats, respectively. This α6 polymorphism might explain various differences in pharmacological properties and density of GABAA receptors between the rat lines. In the present study we have used non-selected outbred Sprague-Dawley rats homozygous for the α6-100RR (RR) and α6-100QQ (QQ) genotypes to show that these RR and QQ rats display similar differences between genotypes as AT and ANT rat lines. The genotypes differed in their affinity for [3H]Ro 15–513 and classical benzodiazepines to cerebellar “diazepam-insensitive” binding sites, in density of cerebellar [3H]muscimol binding and in the antagonizing effect of furosemide on GABA-induced inhibition of [3H]EBOB binding. The results suggest the involvement of α6-R100Q polymorphism in these line differences and in the differences previously found between AT and ANT rats. In addition, the α6-R100Q polymorphism induces striking differences in [3H]Ro 15-4513 binding kinetics to recombinant α6β3γ2s receptors as well as cerebellar diazepam-insensitive sites. Association of [3H]Ro 15-4513 binding was ~10 fold faster and dissociation was ~3–4 fold faster in diazepam-insensitive α6βγ2 receptors containing the α6-100Q allele, with a resulting change of ~2.5-fold in equilibrium dissociation constant (KD). The results indicate that in addition to the central role of the homologous α6-100R/Q (α1-101H) residue in benzodiazepine binding and efficacy, this critical benzodiazepine binding site residue has a major impact on benzodiazepine binding kinetics.
Keywords: Cerebellar granule cell, Benzodiazepine, GABAA receptors, Ethanol sensitivity, Ro 15-4513, Selected rat lines
Introduction
Selective breeding of rodent lines has been used as a tool to identify genes that mediate behavioral differences related to the selection criteria used. At the Biomedical Research Center, Alko Ltd, two rat lines, the AT and ANT rats, differing in their innate sensitivity to motor-impairing effects of ethanol have been produced (Eriksson and Rusi, 1981). The ethanol-sensitive ANT rats are also more sensitive than the ethanol-insensitive AT rats to GABAA receptor- (GABAAR) positive modulators lorazepam, diazepam and barbital (Hellevuo et al., 1989; Wong et al., 1996b) suggesting the involvement of GABAAergic mechanisms mediating the sensitivity differences between the rat lines.
Various differences have been found between the AT and ANT lines in characteristics of cerebellar GABAARs. The density of high affinity GABAA agonist ([3H]muscimol) binding sites is lower in ANT than in AT rats (Malminen and Korpi, 1988; Uusi-Oukari and Korpi, 1989). The affinity of “classical” benzodiazepine (BZ) agonists to α6 subunit-containing, “diazepam-insensitive” (DZ-IS) binding sites in α6β 2 receptors is about 100-fold higher in ANT than in AT rats (Uusi-Oukari and Korpi, 1990, 1991). This line difference is dependent on a single point mutation in the α6 codon 100 (CGA→CAA) in ANT rats leading to an amino acid change from arginine (R) to glutamine (Q) (Korpi et al., 1993). The homologous position in α1,2,3,5 receptors is a histidine residue which has been shown to be a critical residue for classical benzodiazepine binding at the αγ2 subunit interface (Wieland et al., 1992). The α6-R100Q mutation increases the affinity of [3H]Ro 15-4513 to DZ-IS binding sites (Mäkelä et al., 1995; Uusi-Oukari and Korpi, 1990). The allosteric interaction between GABA and classical BZs in DZ-IS sites was found to be present only in ANT rats (Korpi et al., 1993; Uusi-Oukari and Korpi, 1992). Furthermore, furosemide, a GABAAR antagonist selective for α6 subunit-containing receptors (Korpi et al., 1995) is less efficient in enhancing basal binding of the GABAAR-selective channel blocker [35S]TBPS and less efficient in antagonizing GABA-induced inhibition of [35S]TBPS binding in ANT rats than in AT rats (Mäkelä et al., 1996, 1999). The same α6-R100Q mutation was later found to be enriched in the Sardinian non-alcohol-preferring (sNP) rat line (Saba et al., 2001), and also in rats selected for alcohol (non)preference (Carr et al., 2003), and it was shown to be a rather frequent naturally occurring α6 polymorphism in some Sprague-Dawley laboratory rat colonies (Hanchar et al., 2005).
The higher affinity and functional sensitivity of cerebellar α6β 2 receptors in ANT than in AT rats to classic BZs suggest that the α6-100Q mutation determines the increased BZ-induced motor impairment in ANT rats (Korpi et al., 1993). However, the α6-R100Q mutation expressed in α6β2/3γ2 combination did not affect GABA or ethanol sensitivity (Hanchar et al., 2005; Korpi et al., 1993). In contrast, it was shown that α6-100Q mutation in the ethanol-sensitive receptor subtype α6β3δ (Wallner et al., 2003), while not affecting GABA sensitivity, dramatically increases ethanol sensitivity in the ethanol concentration range of 3–30 mM in recombinantly expressed receptors and in cerebellar granule cells in slices (Hanchar et al., 2005) suggesting the involvement of α6β3δ receptors in alcohol sensitivity differences between AT and ANT rats. The increased ethanol sensitivity seen with the R100Q mutation is consistent with earlier work using receptors reconstituted (by injection of cerebellar vesicles from α6-100RR and α6-100QQ rats) in oocytes (Sanna et al, 2003). Higher functional sensitivity to ethanol and BZs was also demonstrated in ANT when compared to AT cerebellar synaptoneurosomes (Schmid et al., 1999). However, using patch-clamp electrophysiological techniques in cerebellar slices of outbred Spraque-Dawley rats homozygous for the α6-100R and −100Q alleles, Botta et al., 2007 were not able to detect genotype dependent ethanol sensitivity differences. In addition, crossbreeding studies using AT and ANT animals did not support the conclusion that the α6R100Q polymorphism is important for differential ethanol sensitivity (Radcliffe et al., 2004). The controversy surrounding the ethanol sensitivity of α4/6βδ-GABAARs and whether this is a solution for the ethanol/GABAAR dilemma is discussed in detailed in a special issue of the Journal Alcohol, where the issue is introduced by an editorial by Lovinger and Homanics (2007).
In the present study we have compared cerebellar and hippocampal GABAARs of Sprague-Dawley rats homozygous for the α6-100R and −100Q alleles. We focused on receptor differences found between AT and ANT rats to reveal if these differences are related to α6-R100Q polymorphism. In addition, we characterized the effects of the polymorphism on cerebellar [3H]Ro 15-4513 binding kinetics to cerebellar DZ-IS binding sites and recombinant α6β3γ2 receptors.
Materials and methods
Animals
Sprague-Dawley rats obtained from Charles River (Hollister, CA) were genotyped as described in Hanchar et al. (2005) to identify animals homozygous for the α6-100R and α6-100Q polymorphisms. A total of 14 RR and 17 QQ adult rats were used for the studies. The rats used in experiments of [3H]Ro 15-4513 binding kinetics were 6–12 month old males, while rats used in other experiments were 3–4 month old males. The animals were killed by decapitation, their brains were removed, and the cerebelli and hippocampi frozen on dry ice and stored at −70 oC. All procedures were in accordance with protocols approved by the University of California at Los Angeles (UCLA) Chancellor’s Animal Research Committee and by the Institutional Animal Care and Use Committee of the University of Turku.
Reagents
The radioligands [propyl-2,3-3H]EBOB (specific activity 48 Ci/mmol), [methylene-3H]muscimol (18 Ci/mmol), and [7,9-3H]Ro 15-4513 (28 Ci/mmol) were purchased from Perkin Elmer Life and Analytical Sciences (Boston, MA, USA). Flumazenil (Ro 15–1788) was a gift from F. Hoffmann-La Roche Ltd (Basel, Switzerland). Diazepam, GABA and picrotoxin were from Sigma Chemical Co. (St. Louis, MO, USA).
Recombinant GABAA receptor expression in HEK 293 cells
Human embryonal kidney (HEK) 293 cells were transfected with rat cDNAs under the control of CMV promoter (α1, α6-100R or α6-100Q:β3:γ2S, 1:1:2) as described in Meera et al. (1997) and the cells were harvested 48 h after transfection. The cells were washed with PBS, homogenized in PBS using an Ultra-Turrax and stored frozen at −70 oC. Before binding assays the suspensions were thawed, washed once with ice-cold assay buffer by resuspension and centrifugation, and homogenized in ice-cold assay buffer with an Ultra-Turrax.
[3H]Ro 15-4513 binding assay
Cerebellar and hippocampal membranes were prepared and [3H]Ro 15-4513 binding assays performed essentially as described in Uusi-Oukari and Korpi (1990). Tris-HCl (50 mM, pH 7.4) containing 120 mM NaCl was used as the incubation buffer. Non-specific binding was determined in the presence of 10 μM flumazenil. Triplicate samples were incubated in an ice-water bath in the dark with shaking for 1 h in a total volume of 300 μl. Diazepam-insensitive (DZ-IS) [3H]Ro 15-4513 binding was determined in the presence of 100 μM diazepam to differentiate between RR and QQ binding sites. Receptors containing the α6-100Q allele display 1 μM diazepam affinity (Mäkelä et al., 1995; Uusi-Oukari and Korpi, 1992). Diazepam at 100 μM displaces essentially all binding in QQ rats while hardly affecting α6-100R receptors (Mäkelä et al., 1995; Uusi-Oukari and Korpi, 1992). The incubation was terminated by filtration of the samples with a Brandel Cell Harvester (model 48R, Gaithesburg, Maryland, USA) onto Whatman GF/B filters (Whatman International Ltd., Maidstone, UK). The samples were rinsed twice with 4–5 ml of ice-cold incubation buffer. Air-dried filters were immersed in 4 ml of Optiphase HiSafe 2 scintillation fluid (Wallac, Turku, Finland) and radioactivity determined in a Wallac model 1410 liquid scintillation counter (Wallac, Turku, Finland). Non-specific binding was subtracted from total binding and from binding in the presence of diazepam to get total specific binding and DZ-IS binding, respectively. DZ-IS binding was subtracted from total specific binding to get diazepam-sensitive (DZ-S) binding.
Measurement of [3H]Ro 15-4513 binding kinetics
To measure association of [3H]Ro 15-4513 binding, cerebellar membranes of RR and QQ rats were incubated in incubation buffer with 5 nM [3H]Ro 15-4513 in the absence and presence of 1 μM diazepam (DZ-IS binding) or 10 μM flumazenil (non-specific binding) in conditions described above. Diazepam at 1 μM displaces essentially all DZ-S binding, but only 50% of DZ-IS binding in QQ membranes thus allowing measurement of DZ-IS binding selectively also in QQ cerebellum. The incubations were terminated at varíous time points. To measure dissociation of [3H]Ro 15-4513 binding triplicate samples of membranes were first preincubated in a total volume of 300 μl for 1 h with 5 nM [3H]Ro 15-4513 in the absence and presence of 1 μM diazepam or 10 μM flumazenil. The dissociation was started by adding 100 μl of 40 μM flumazenil to the incubation mixtures to reach a final 10 μM flumazenil concentration in all tubes. The tubes were mixed and incubations terminated at varíous time points as described above.
[3H]Muscimol binding assay
Cerebellar membranes were prepared and saturation analysis of [3H]muscimol binding performed essentially as described in Uusi-Oukari and Korpi (1989). The radioligand was used at a concentration range of 0.5–20 nM. Non-specific binding was determined in the presence of 1 mM GABA. Triplicate reaction mixtures in 50 mM Tris-citrate buffer, pH 7.4, were incubated in ice-water bath in the dark with shaking for 30 min. The final incubation volume was 300 μl. The incubation was terminated as descibed for [3H]Ro 15-4513 binding.
[3H]EBOB binding assay
Cerebellar membranes for binding of [3H]EBOB, a ligand binding to the same site as the more commonly used [35S]TBPS, were prepared and [3H]EBOB binding assay performed essentially as described in Uusi-Oukari and Maksay (2006). Triplicate samples of cerebellar membranes were incubated at room temperature with shaking for 2 h in 50 mM Tris-HCl, pH 7.4, containing 120 mM NaCl with 1 nM [3H]EBOB in a total volume of 400 μl in the absence and presence of 5 μM GABA with or without 300 μM furosemide. Non-specific binding was determined in the presence of 100 μM picrotoxin. The incubation was terminated as descibed for [3H]Ro 15-4513 binding.
Protein measurement
In all ligand binding studies, protein concentrations of membranes were determined with the Bio-Rad Coomassie blue dye-based protein assay kit (Hercules, CA, USA) according to manufacturer’s instructions.
Data analysis
Saturation isotherms for estimation of KD and Bmax values, and association and dissociation curves for estimation of association and dissociation rate constants were analysed with Prism 5 software (Graph Pad, San Diego, CA, USA). Two-tailed unpaired t-test or one-way ANOVA followed by Tukey’s post hoc test was used to assess statistical significances of the differences between the groups. P-values of less than 0.05 were considered significant.
Results
[3H]Ro 15-4513 binding to brain membranes and to recombinant receptors
Cerebellar DZ-IS [3H]Ro 15-4513 binding was determined from all animals used in the study. The portion of 5 nM [3H]Ro 15-4513 binding in the presence of 100 μM diazepam was 21.9 ± 0.9 % and 1.0 ± 0.1 % of total binding in membranes of RR and QQ rats, respectively. The portion of DZ-IS binding was 20–25% in all RR samples, while QQ samples under these conditions displayed only small amounts of DZ-IS [3H]Ro 15-4513 binding. The values are consistent with previous studies on AT and ANT rat lines and confirm the GABAAR α6-100RR and α6-100QQ rat genotypes.
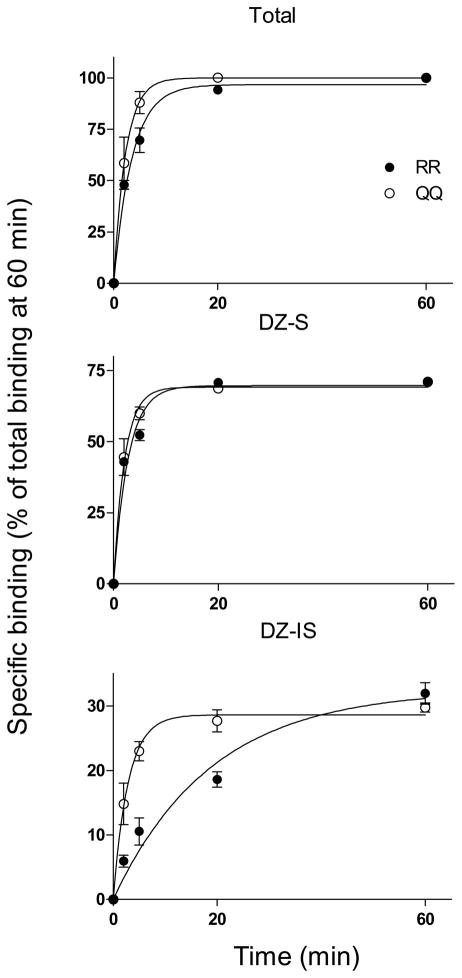
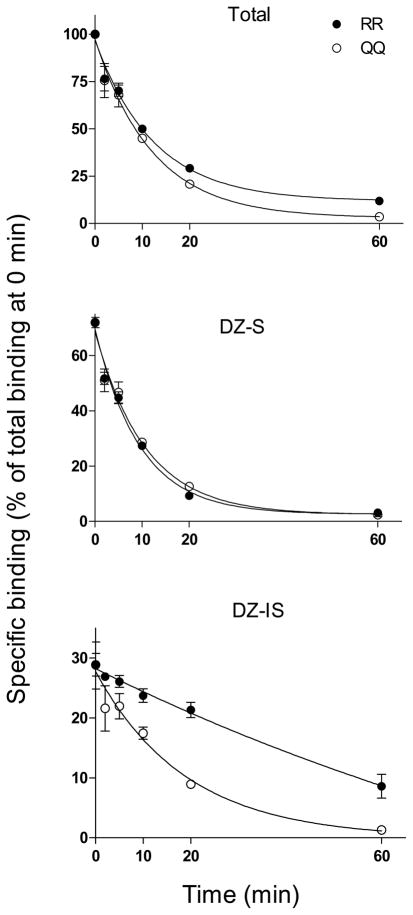
The association and dissociation rates of total, DZ-S and DZ-IS [3H]Ro 15-4513 binding were determined using cerebellar membranes from RR and QQ rats (Fig. 1 and 2, Table 1). Association and dissociation rates of total and DZ-S binding were very similar in RR and QQ membranes (Fig. 1, Table 1). In contrast, the Kon value of DZ-IS binding was 7.4-fold lower in RR than in QQ membranes indicating slower association of [3H]Ro 15-4513 to DZ-IS receptors in RR membranes. The same applies to dissociation of DZ-IS [3H]Ro 15-4513 binding. The Koff value was 2.9-fold lower in RR than in QQ membranes indicating slower [3H]Ro 15-4513 dissociation in RR membranes (Fig. 2, Table 1). The calculation of KD values (Koff/Kon) for [3H]Ro 15-4513 binding were 1.6 nM and 1.4 nM (DZ-S) and 2.3 nM and 0.9 nM (DZ-IS) for RR and QQ rats, respectively (Table 1).
Fig. 1.
Association of total, DZ-S and DZ-IS [3H]Ro 15-4513 binding to cerebellar membranes from RR and QQ rats (mean ± SEM of three independent experiments made in triplicate). DZ-IS binding was measured in the presence of 1 μM diazepam.
Fig. 2.
Dissociation of total, DZ-S and DZ-IS [3H]Ro 15-4513 binding from cerebellar membranes from RR and QQ rats (mean ± SEM of three independent experiments made in triplicate). DZ-IS binding was measured in the presence of 1 μM diazepam.
Table 1.
Association and dissociation rate constants of [3H]Ro 15-4513 binding in cerebellar membranes of RR and QQ rats and in recombinant receptors expressed in HEK cells
| Kon (M−1 × min−1] | Koff (min−1) | KD (Koff/Kon) (nM) | |
|---|---|---|---|
| Total binding | |||
| RR | 4.6 ± 0.4 × 107 | 0.063 ± 0.002 | 1.4 ± 0.1 |
| 7.6 ± 1.1 × 107 | 0.080 ± 0.006 | 1.1 ± 0.3 | |
| DZ-S binding | |||
| RR | 6.0 ± 0.2 × 107 | 0.097 ± 0.004 | 1.6 ± 0.1 |
| 6.9 ± 0.9 × 107 | 0.090 ± 0.008 | 1.4 ± 0.3 | |
| DZ-IS binding | |||
| RR | 8.2 ± 0.3 × 106 | 0.019 ± 0.001 | 2.3 ± 0.1 |
| 6.1 ± 0.8 × 107## | 0.055 ± 0.001### | 0.9 ± 0.2### | |
| Recombinant recept. | |||
| α1β3γ2 | 3.4 ± 0.2 × 107*** | 0.098 ± 0.010** | 2.9 ± 0.3* |
| α6-100R-β3γ2 | 8.0 ± 0.8 × 106*** | 0.018 ± 0.002 | 2.4 ± 0.4* |
| α6-100Q-β3γ2 | 8.7 ± 0.2 × 107*** | 0.083 ± 0.013** | 1.0 ± 0.1 |
DZ-S = diazepam-sensitive, DZ-IS = diazepam-insensitive, KD = dissociation constant, Kon = association rate constant, Koff = dissociation rate constant. Total, DZ-S and DZ-IS binding to cerebellar membranes, mean ± SEM values (n=3, measured in triplicate).
p<0.001,
p<0.01, significantly different from the corresponding RR value (unpaired t-test); Recombinant receptors, mean ± SEM (n=3, measured in triplicate).
p<0.001, significantly different from two other groups;
p<0.01, significantly different from α6-100R value;
p<0.05, significantly different from α6-100Q value; one-way ANOVA followed by Tukey’s post hoc test.
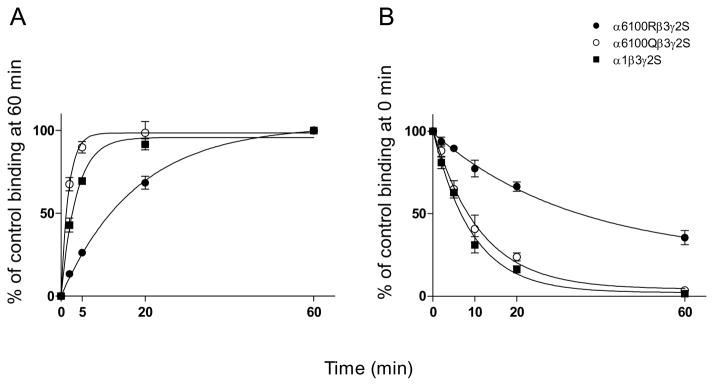
Association rate of [3H]Ro 15-4513 binding to DZ-S α1β3γ2 recombinant receptors was comparable to that of [3H]Ro 15-4513 binding to DZ-S receptors in RR and QQ membranes (Table 1, Fig. 3A). Similarly, dissociation rate of the binding to α1β3γ2 receptors was indistinguishable from dissociation of DZ-S binding in cerebellar membranes (Table 1, Fig. 3B). Association and dissociation rates of [3H]Ro 15-4513 binding to α6-100R-β3γ2 recombinant receptors and DZ-IS binding to cerebellar membranes of RR rats were almost identical, while the association rate constant was lower in DZ-IS binding to QQ rat cerebellar membranes than to α6-100Q-β3γ2 recombinant receptors (Table 1, Fig. 3A, 3B). This difference between QQ recombinant receptors and membranes is likely due to 1 μM diazepam present with cerebellar membranes. The Ki of diazepam to DZ-IS sites in QQ animals is 1 μM (Uusi-Oukari and Korpi, 1992) indicating that diazepam competes with [3H]Ro 15-4513 in binding to these sites. The association rate constant of [3H]Ro 15-4513 binding was 11 fold lower in α6-100R-β3γ2 than in α6-100Q-β3γ2 receptors (Table 1, tFig. 3A) (p<0.001, two-tailed unpaired -test). The dissociation rate constant in α6-100Q-β3γ2 receptors was 4.6-fold larger than that in α6-100R-β3γ2 receptors (Table 1, Fig. 3B) (p<0.05, two-tailed unpaired t-test). The calculated KD values (Koff/Kon) of [3H]Ro 15-4513 binding to recombinant receptors were 2.9 nM (α1β3γ2), 2.3 nM (α6-100R-β3γ2) and 1.0 nM (α6-100Q-β3γ2) (Table 1). The association and dissociation kinetics of [3H] Ro15-4513 binding were in a similar manner slower also in α6-100R-β2γ2 recombinant receptors as compared to α6-100Q-β2γ2 receptors (L.-S. Kontturi and M. Uusi-Oukari, unpublished results) indicating that the difference in binding kinetics is independent on β variant present in the receptor.
Fig. 3.
A. Association kinetics of 5 nM [3H]Ro 15-4513 to α1β3γ2, α6-100R-β3γ2 and α6-100Q-β3γ2 recombinant receptors expressed in HEK cells. 3B. Dissociation of [3H]Ro 15-4513 binding from α1β3γ2, α6-100R-β3γ2 and α6-100Q-β3γ2 recombinant receptors expressed in HEK cells (mean ± SEM, n=3 independent experiments made in triplicate).
Since association of [3H]Ro 15-4513 binding to RR cerebellar membranes in the presence of diazepam (DZ-IS binding) and to α6-100R-β3γ2 recombinant receptors was very slow, we tested longer incubation periods. A 2 h incubation with 5 nM [3H] Ro15-4513 yielded 23% higher binding both to RR cerebellar membranes in the presence of diazepam and to α6-100R-β3γ2 recombinant receptors (Fig. 4).
Fig. 4.
Association of [3H]Ro 15-4513 to α6-100R-β3γ2 recombinant receptors and to cerebellar membranes from RR rats using longer incubation times (mean values from two independent experiments made in triplicate). DZ-IS binding was measured in the presence of 1 μM diazepam.
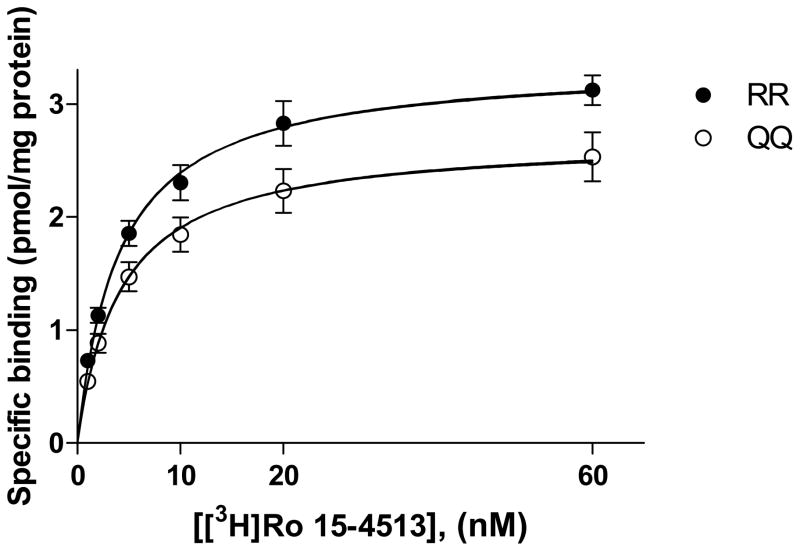
Saturation analysis of [3H]Ro 15-4513 binding to hippocampal membranes revealed 23% higher Bmax value in RR than in QQ rats (Fig. 5). This difference, however, did not reach statistical significance (p=0.063).
Fig. 5.
Saturation analysis of [3H]Ro 15-4513 binding to hippocampal membranes of RR and QQ rats (mean ± SEM; RR, n= 4; QQ, n=5), Difference in Bmax between the rat lines: p=0.062, two-tailed unpaired t-test.
[3H]Muscimol binding to cerebellar membranes
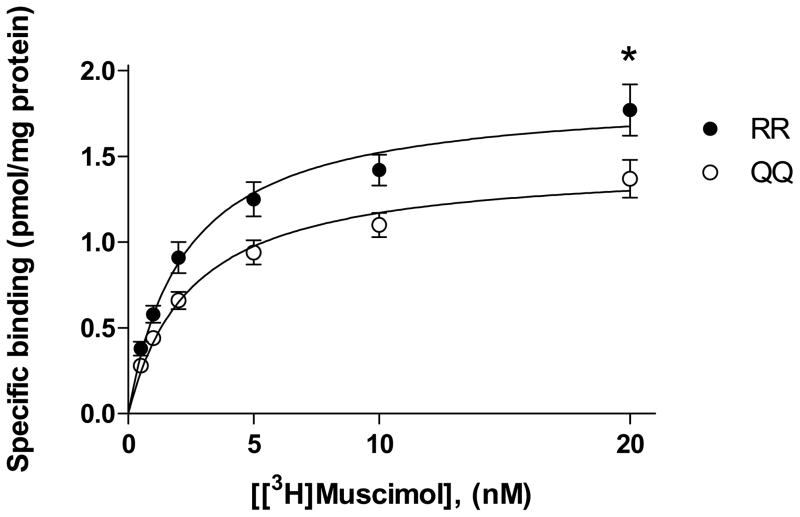
The Bmax value of [3H]muscimol binding to cerebellar membranes was lower in QQ (1.46 ± 0.10 pmol/mg protein, mean ± SEM) than in RR (1.84 ± 0.14 pmol/mg protein) animals (p<0.05, two-tailed unpaired t-test; RR, n=7; QQ, n=8) (Fig. 6). The KD values of cerebellar [3H]muscimol binding did not differ between the rat lines (2.21 ± 0.13 and 2.52 ± 0.22 nM for RR and QQ rats, respectively).
Fig. 6.
Saturation analysis of [3H]muscimol binding to cerebellar membranes of RR and QQ rats (mean ± SEM; RR, n=7; QQ, n=8). *p<0.05, significance of the difference in Bmax between the rat lines, two-tailed unpaired t-test.
[3H]EBOB binding to cerebellar membranes
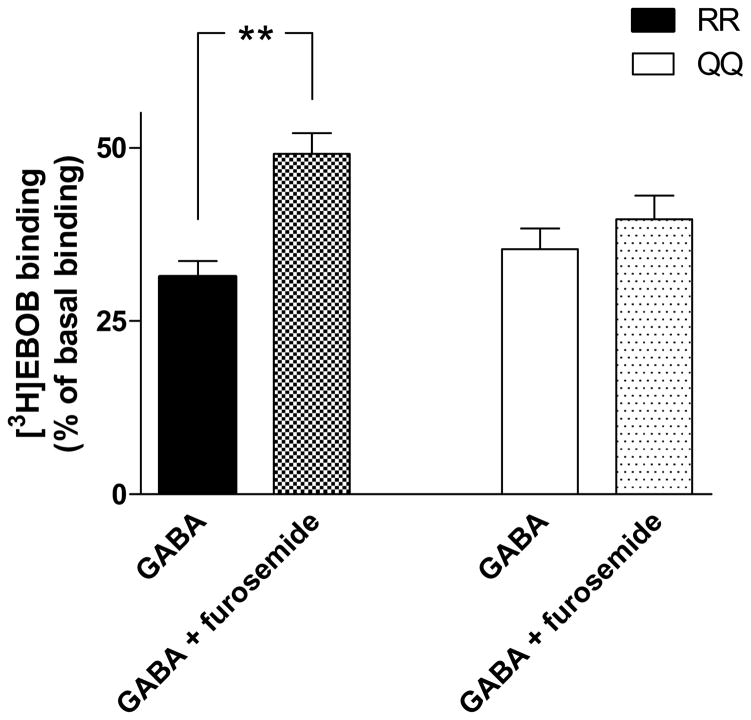
The GABA-antagonizing effect of furosemide on [3H]EBOB binding was studied in cerebellar membranes of RR and QQ rats. GABA (5 μM) inhibited basal binding of 1 nM [3H]EBOB by 65–70% (Fig. 7). The effect of GABA on the binding did not significantly differ between RR and QQ rats. However, furosemide (300 μM) significantly antagonized the effect of GABA on [3H]EBOB binding only in RR rats (p<0.05, one-way ANOVA followed by Tukey’s post hoc test).
Fig. 7.
Antagonizing effect of furosemide on GABA-induced displacement of [3H]EBOB in cerebellar membranes from RR and QQ rats (mean ± SEM; RR, n=11; QQ, n=13). **p<0.01, significance of the difference between GABA-induced displacement and antagonism of this displacement in RR rats, one-way ANOVA followed by Tukey’s post hoc test.
Discussion
Using two groups of Sprague-Dawley rats homozygous for GABAAR alleles α6-100R and α6-100Q we found several differences between the groups in their cerebellar GABAARs previously found between the AT and ANT rat lines: the affinity of cerebellar DZ-IS [3H]Ro 15-4513 binding sites is higher to (1) [3H]Ro 15-4513 and to (2) classical BZs in QQ than in RR animals; (3) the number of cerebellar high-affinity [3H]muscimol binding sites is lower in QQ than in RR animals; and (4) the GABA-antagonizing effect of furosemide on GABA-induced inhibition of [3H]EBOB binding is lower in QQ than in RR animals. The affinity differences (1) and (2) have been previously published between alcohol-nonpreferring sNP rats homozygous for QQ and RR alleles (Sanna et al., 2003), between Spraque-Dawley rats with α6-100RR and α6-100QQ genotypes (Hanchar et al., 2005) and using α6-100R/Q-β2/3γ2 recombinant receptors (Hanchar et al., 2005; Korpi et al., 1993). The results clearly show that the differences (1, 2) are caused by the R to Q substitution in α6 residue 100.
As a surprise, this amino acid change affects association and dissociation kinetics of [3H]Ro 15-4513 binding. Residue 100 in α6 (and the homologous His residue in α1) reside in loop A of the BZ binding pocket (Tan et al., 2007). Association and dissociation rates of [3H]Ro 15-4513 binding appear to be very similar in α1-101H-β3γ2 and α6-100Q-β3γ2 receptors. It has been previously shown, that the α1-H101Q mutation in α1β2γ2 recombinant receptors increases [3H]Ro 15-4513 binding affinity (Davies et al., 1998). In fact, α1-H101 substitutions F, K, R, E, Q and C increase [3H]Ro 15-4513 affinity while drastically decreasing flunitrazepam affinity (Davies et al., 1998, Wieland et al., 1992). These substitutions also affect efficacy of BZs, including Ro 15-4513, in α1β2γ2 receptors indicating the central role of the residue in BZ binding and function (Dunn et al., 1999). We performed [3H]Ro 15-4513 association and dissociation measurements at 0 oC degrees. The association rates in vivo at 37 oC would be much faster and although obviously slower in α6-100R receptors, equilibrium would be reached within minutes. Our data suggest that in vitro binding studies using [3H]Ro 15-4513 as a ligand, 1 h incubation times at concentrations ≤ 5 nM at 0 oC have obviously led to an underestimation of the number of cerebellar [3H]Ro 15-4513 DZ-IS binding sites.
The other two AT/ANT line differences, (3) and (4), were present in RR/QQ rats, but the molecular basis of these differences is less clear. Cerebellar high affinity [3H]muscimol binding is mainly localized in cerebellar granule layer and associated with α6β γ2 and α6βδ receptor subtypes (Jones et al., 1997; Korpi et al., 2002; Mihalek et al., 1999). The higher Bmax of high affinity [3H]muscimol binding therefore suggests a higher amount of α6β γ2 and/or α6βδ receptors in AT/RR than in ANT/QQ cerebellar granule cells. The basal level α6 mRNA expression has been shown to be higher in sNP rats with RR than with QQ genotype (Saba et al., 2005). This difference was suggested to be due to differences in α6 promoter sequences between RR and QQ genotypes (Saba et al., 2005). However, no difference has been found between AT and ANT rats in the number of cerebellar [3H]Ro 15-4513 binding sites (Uusi-Oukari and Korpi, 1990) or in cerebral granule cell tonic GABAergic currents (Valenzuela et al., 2005), and only a tendency (~10%) to higher level of α6 mRNA expression in AT than in ANT rats (Uusi-Oukari et al., 2000). However, the slow association of [3H]Ro 15-4513 to DZ-IS binding sites in RR rats suggests that the Bmax value of AT/RR rats might have been underestimated because the binding does not reach equilibrium within 1 h of incubation. Due to high BZ sensitivity it is not possible to determine Bmax of DZ-IS [3H]Ro 15-4513 binding in QQ animals. It might be also difficult to see a 30% difference corresponding to difference in [3H]muscimol binding in DZ-IS [3H]Ro 15-4513 binding by determining total [3H]Ro 15-4513 binding, because a 30% difference in 20–25% DZ-IS portion of total binding would make a 7% difference in Bmax of total binding. In addition to a difference in α6 mRNA transcription there may be differences in the efficiency between α6-100R and α6-100Q proteins to combine with other subunits to form α6β γ2 and/or α6βδ receptors.
The blunted furosemide action in antagonising GABA-induced inhibition of [3H]EBOB binding was also seen in QQ rats in the present study. The result suggests the involvement of residue 100Q on the mechanism(s) of blunted furosemide response in ANT/QQ rats. However, the inability to reproduce this line difference using α6-100Q-β3γ2 recombinant receptors (Mäkelä et al., 1996) suggests that in addition to α6-R100Q amino acid change there are other unidentified alteration(s) in ANT/QQ receptors necessary for the insensitivity to furosemide. There may also be GABAAR-associated component(s) present in cerebellar granule cells needed for the ANT/QQ furosemide insensitivity not present in HEK cells used for recombinant receptor expression. Granule cell-specific posttranslational modification is also possible. Anyway, the data shown here suggest that the furosemide-insensitive property seems to be linked to α6-100Q allele.
There was a tendency for lower [3H]Ro 15-4513 binding in hippocampal membranes of QQ rats as compared to RR rats. The result is in accordance with the hippocampal line difference between AT and ANT rats (Uusi-Oukari and Korpi, 1990). GABAAR subunits are clustered in genome as clusters of 3–4 subunits (Darlison et al., 2005). The α6 gene is clustered with genes for α1, β2, and γ2 subunits, the latter three subunits forming the major GABAAR subtype α1β2γ2 in the brain (Garrett et al., 1997; McKernan and Whiting, 1996). The α6-100Q genotype is strictly associated with various silent polymorphisms/single nucleotide polymorphisms (SNPs) in the other three GABAAR subunit genes of the cluster (Congeddu et al., 2003). Therefore, it remains possible that SNPs in α1, β2 and γ2 subunit genes (that co-segregate with the α6-100Q genotype) might contribute to lower number of hippocampal [3H]Ro 15-4513 binding sites in ANT/QQ than in AT/RR rats.
While the results suggest that most of the differences in cerebellar GABAARs are dependent on α6-R100Q polymorphism, it is possible that other polymorphisms found in the genes of the β2-α6-α1-γ2 cluster and linked to α6-R100Q polymorphism might play a role in some of the differences between RR and QQ rats. The R100Q mutation is enriched in two alcohol-non-preferring rats lines (sNP and Alko Non-Alcohol, ANA) suggesting that in addition to ethanol and BZ sensitivity to motor impairment the polymorphism and/or the associated SNPs of the cluster might be associated in alcohol preference and avoidance (Carr et al., 2003; Saba et al., 2001; Wong et al., 1996a). Genes in β2-α6-α1-γ2 cluster have also been associated to ethanol-related behaviors in humans. Several genetic association studies with human alcohol-dependent subjects suggest a role for genes in β2-α6-α1-γ2 cluster in the development of alcohol dependence (Chang et al., 2002; Loh et al., 1999, 2002; Radel et al., 2005; Sander et al., 1999).
Acknowledgments
This study was supported by grants from the Finnish Society of Sciences and Letters (MU-O), Oskar Öflund Foundation (MU-O) and the Turku University Foundation (MU-O) and by NIH grant AA017891 (MW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Botta P, Mameli M, Floyd KL, Radcliffe RA, Valenzuela CF. Ethanol sensitivity of GABAergic currents in cerebellar granule neurons is not increased by a single amino acid change (R100Q) in the α6 GABAA receptor subunit. J Pharmacol Exp Ther. 2007;323:684–691. doi: 10.1124/jpet.107.127894. [DOI] [PubMed] [Google Scholar]
- Carr LG, Spence JP, Eriksson CJ, Lumeng L, Li TK. AA and ANA rats exhibit the R100Q mutation in the GABAA receptor α6 subunit. Alcohol. 2003;31:93–97. doi: 10.1016/j.alcohol.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Chang YT, Sun HS, Fann CS, Chang CJ, Liao ZH, Huang JL, Loh EW, Yu WY, Cheng AT. Association of the γ-aminobutyric acid A receptor gene cluster with alcohol dependence in Taiwanese Han. Mol Psychiatry. 2002;7:828–829. doi: 10.1038/sj.mp.4001110. [DOI] [PubMed] [Google Scholar]
- Congeddu E, Saba L, Porcella A, Sanna A, Marchese G, Lobina C, Gessa GL, Pani L. Molecular characterization of new polymorphisms at the β2, α1, γ2 GABAA receptor subunit genes associated to a rat nonpreferring ethanol phenotype. Brain Res, Mol Brain Res. 2003;110:289–297. doi: 10.1016/s0169-328x(02)00660-5. [DOI] [PubMed] [Google Scholar]
- Darlison MG, Pahal I, Thode C. Consequences of the evolution of the GABAA receptor gene family. Cell Mol Neurobiol. 2005;25:607–624. doi: 10.1007/s10571-005-4004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies M, Bateson AN, Dunn SM. Structural requirements for ligand interactions at the benzodiazepine recognition site of the GABAA receptor. J Neurochem. 1998;70:2188–2194. doi: 10.1046/j.1471-4159.1998.70052188.x. [DOI] [PubMed] [Google Scholar]
- Dunn SM, Davies M, Muntoni AL, Lambert JJ. Mutagenesis of the rat α1 subunit of the γ-aminobutyric acidA receptor reveals the importance of residue 101 in determining the allosteric effects of benzodiazepine site ligands. Mol Pharmacol. 1999;56:768–774. [PubMed] [Google Scholar]
- Eriksson K, Rusi M. Finnish selection studies on alcohol-related behaviours: General outline. In: McClearn GE, Deitrich RA, Erwin G, editors. Development of Animal Models as Pharmacogenetic tools, NIAAA Research Monograph No 6. U.S. Government Printing Office; Washington, DC: 1981. pp. 87–117. [Google Scholar]
- Garrett KM, Haque D, Berry D, Niekrasz I, Gan J, Rotter A, Seale TW. The GABAA receptor α6 subunit gene (Gabra6) is tightly linked to the α1-γ2 subunit cluster on mouse chromosome 11. Brain Res, Mol Brain Res. 1987;45:133–137. doi: 10.1016/s0169-328x(96)00290-2. [DOI] [PubMed] [Google Scholar]
- Hanchar HJ, Dodson PD, Olsen RW, Otis TS, Wallner M. Alcohol-induced motor impairment caused by increased extrasynaptic GABAA receptor activity. Nat Neurosci. 2005;8:339–345. doi: 10.1038/nn1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellevuo K, Kiianmaa K, Korpi ER. Effect of GABAergic drugs on motor impairment from ethanol, barbital and lorazepam in rat lines selected for differential sensitivity to ethanol. Pharmacol Biochem Behav. 1989;34:399–404. doi: 10.1016/0091-3057(89)90333-x. [DOI] [PubMed] [Google Scholar]
- Jones A, Korpi ER, McKernan RM, Pelz R, Nusser Z, Mäkelä R, Mellor JR, Pollard S, Bahn S, Stephenson FA, et al. Ligand-gated ion channel subunit partnerships: GABAA receptor α6 subunit gene inactivation inhibits delta subunit expression. J Neurosci. 1997;17:1350–1362. doi: 10.1523/JNEUROSCI.17-04-01350.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpi ER, Kleingoor C, Kettenmann H, Seeburg PH. Benzodiazepine-induced motor impairment linked to point mutation in cerebellar GABAA receptor. Nature. 1993;361:356–359. doi: 10.1038/361356a0. [DOI] [PubMed] [Google Scholar]
- Korpi ER, Kuner T, Seeburg PH, Lüddens H. Selective antagonist for the cerebellar granule cell-specific γ-aminobutyric acid type A receptor. Mol Pharmacol. 1995;47:283–289. [PubMed] [Google Scholar]
- Korpi ER, Mihalek RM, Sinkkonen ST, Hauer B, Hevers W, Homanics GE, Sieghart W, Lüddens H. Altered receptor subtypes in the forebrain of GABAA receptor delta subunit-deficient mice: recruitment of γ2 subunits. Neuroscience. 2002;109:733–743. doi: 10.1016/s0306-4522(01)00527-9. [DOI] [PubMed] [Google Scholar]
- Loh EW, Ball D. Role of the GABAA β2, GABAA α6, GABAA α1 and GABAA γ2 receptor subunit genes cluster in drug responses and the development of alcohol dependence. Neurochem Int. 2000;37:413–423. doi: 10.1016/s0197-0186(00)00054-1. [DOI] [PubMed] [Google Scholar]
- Loh EW, Smith I, Murray R, McLaughlin M, McNulty S, Ball D. Association between variants at the GABAA β2, GABAA α6 and GABAA γ2 gene cluster and alcohol dependence in a Scottish population. Mol Psychiatry. 1999;4:539–544. doi: 10.1038/sj.mp.4000554. [DOI] [PubMed] [Google Scholar]
- Lovinger DM, Homanics GE. Tonic for what ails us? High-affinity GABAA receptors and alcohol. Alcohol. 2007;41:139–143. doi: 10.1016/j.alcohol.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malminen O, Korpi ER. GABA/benzodiazepine receptor/chloride ionophore complex in brains of rat lines selectively bred for differences in ethanol-induced motor impairment. Alcohol. 1988;5:239–249. doi: 10.1016/0741-8329(88)90059-6. [DOI] [PubMed] [Google Scholar]
- Mäkelä R, Lehtonen M, Wisden W, Lüddens H, Korpi ER. Blunted furosemide action on cerebellar GABAA receptors in ANT rats selectively bred for high alcohol sensitivity. Neuropharmacology. 1996;35:1493–1502. doi: 10.1016/s0028-3908(96)00073-1. [DOI] [PubMed] [Google Scholar]
- Mäkelä R, Uusi-Oukari M, Oja SS, Alho H, Anghelescu I, Klawe C, Lüddens H, Korpi ER. Furosemide action on cerebellar GABAA receptors in alcohol-sensitive ANT rats. Alcohol. 1999;19:197–205. doi: 10.1016/s0741-8329(99)00040-3. [DOI] [PubMed] [Google Scholar]
- Mäkelä R, Wong G, Lüddens H, Korpi ER. Phenotypic and genotypic analysis of rats with cerebellar GABAA receptors composed from mutant and wild-type α6 subunits. J Neurochem. 1995;65:2401–2408. doi: 10.1046/j.1471-4159.1995.65062401.x. [DOI] [PubMed] [Google Scholar]
- McKernan RM, Whiting PJ. Which GABAA-receptor subtypes really occur in the brain? Trends Neurosci. 1996;19:139–143. doi: 10.1016/s0166-2236(96)80023-3. [DOI] [PubMed] [Google Scholar]
- Meera P, Wallner M, Song M, Toro L. Large conductance voltage- and calcium-dependent K+ channel, a distinct member of voltage-dependent ion channels with seven N-terminal transmembrane segments (S0–S6), an extracellular N terminus, and an intracellular (S9–S10) C terminus. Proc Natl Acad Sci USA. 1997;94:14066–14071. doi: 10.1073/pnas.94.25.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalek RM, Banerjee PK, Korpi ER, Quinlan JJ, Firestone LL, Mi ZP, Lagenaur C, Tretter V, Sieghart W, Anagnostaras SG, et al. Attenuated sensitivity to neuroactive steroids in γ-aminobutyrate type A receptor δ subunit knockout mice. Proc Natl Acad Sci USA. 1999;96:12905–12910. doi: 10.1073/pnas.96.22.12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radel M, Vallejo RL, Iwata N, Aragon R, Long JC, Virkkunen M, Goldman D. Haplotype-based localization of an alcohol dependence gene to the 5q34 γ-aminobutyric acid type A gene cluster. Arch Gen Psychiatry. 2005;62:47–55. doi: 10.1001/archpsyc.62.1.47. [DOI] [PubMed] [Google Scholar]
- Saba L, Porcella A, Congeddu E, Colombo G, Peis M, Pistis M, Gessa GL, Pani L. The R100Q mutation of the GABAA α6 receptor subunit may contribute to voluntary aversion to ethanol in the sNP rat line. Brain Res, Mol Brain Res. 2001;87:263–270. doi: 10.1016/s0169-328x(01)00003-1. [DOI] [PubMed] [Google Scholar]
- Saba L, Porcella A, Sanna A, Congeddu E, Marziliano N, Mongeau R, Grayson D, Pani L. Five mutations in the GABAA α6 gene 5′ flanking region are associated with a reduced basal and ethanol-induced α6 upregulation in mutated Sardinian alcohol non-preferring rats. Brain Res, Mol Brain Res. 2005;137:252–257. doi: 10.1016/j.molbrainres.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Sander T, Ball D, Murray R, Patel J, Samochowiec J, Winterer G, Rommelspacher H, Schmidt LG, Loh EW. Association analysis of sequence variants of GABAA α6, β2, and γ2 gene cluster and alcohol dependence. Alcohol Clin Exp Res. 1999;23:427–431. [PubMed] [Google Scholar]
- Sanna A, Congeddu E, Porcella A, Saba L, Pistis M, Peis M, Marchese G, Ruiu S, Lobina C, Grayson DR, et al. Characterization of wild-type (R100R) and mutated (Q100Q) GABAA α6 subunit in Sardinian alcohol non- preferring rats (sNP) Brain Res. 2003;967:98–105. doi: 10.1016/s0006-8993(02)04230-0. [DOI] [PubMed] [Google Scholar]
- Schmid G, Bonanno G, Raiteri L, Sarviharju M, Korpi ER, Raiteri M. Enhanced benzodiazepine and ethanol actions on cerebellar GABAA receptors mediating glutamate release in an alcohol-sensitive rat line. Neuropharmacology. 1999;38:1273–1279. doi: 10.1016/s0028-3908(99)00025-8. [DOI] [PubMed] [Google Scholar]
- Tan KR, Baur R, Gonthier A, Goeldner M, Sigel E. Two neighboring residues of loop A of the α1 subunit point towards the benzodiazepine binding site of GABAA receptors. FEBS Lett. 2007;581:4718–4722. doi: 10.1016/j.febslet.2007.08.068. [DOI] [PubMed] [Google Scholar]
- Uusi-Oukari M, Heikkilä J, Sinkkonen ST, Mäkelä R, Hauer B, Homanics GE, Sieghart W, Wisden W, Korpi ER. Long-range interactions in neuronal gene expression: evidence from gene targeting in the GABAA receptor β2-α6-α1-γ2 subunit gene cluster. Mol Cell Neurosci. 2000;16:34–41. doi: 10.1006/mcne.2000.0856. [DOI] [PubMed] [Google Scholar]
- Uusi-Oukari M, Kleinz R, Mäkelä R, Lüddens H, Korpi ER. Quantification of GABAA receptor subunit mRNAs by non-radioisotopic competitive RT-PCR utilizing plate-based EIA methodology. J Neurosci Methods. 2000;95:65–73. doi: 10.1016/s0165-0270(99)00158-2. [DOI] [PubMed] [Google Scholar]
- Uusi-Oukari M, Korpi ER. Cerebellar GABAA receptor binding and function in vitro in two rat lines developed for high and low alcohol sensitivity. Neurochem Res. 1989;14:733–739. doi: 10.1007/BF00964950. [DOI] [PubMed] [Google Scholar]
- Uusi-Oukari M, Korpi ER. Diazepam sensitivity of the binding of an imidazobenzodiazepine, [3H]Ro 15-4513, in cerebellar membranes from two rat lines developed for high and low alcohol sensitivity. J Neurochem. 1990;54:1980–1987. doi: 10.1111/j.1471-4159.1990.tb04901.x. [DOI] [PubMed] [Google Scholar]
- Uusi-Oukari M, Korpi ER. Specific alterations in the cerebellar GABAA receptors of an alcohol-sensitive ANT rat line. Alcohol Clin Exp Res. 1991;15:241–248. doi: 10.1111/j.1530-0277.1991.tb01864.x. [DOI] [PubMed] [Google Scholar]
- Uusi-Oukari M, Korpi ER. Functional properties of GABAA receptors in two rat lines selected for high and low alcohol sensitivity. Alcohol. 1992;9:261–269. doi: 10.1016/0741-8329(92)90063-g. [DOI] [PubMed] [Google Scholar]
- Uusi-Oukari M, Maksay G. Allosteric modulation of [3H]EBOB binding to GABAA receptors by diflunisal analogues. Neurochem Int. 2006;49:676–682. doi: 10.1016/j.neuint.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Valenzuela CF, Mameli M, Carta M. Letter to the Editor. Alcohol, Clin Exp Res. 2005;29:1356–1357. doi: 10.1097/01.alc.0000171926.66397.94. [DOI] [PubMed] [Google Scholar]
- Wallner M, Hanchar HJ, Olsen RW. Ethanol enhances α4β3δ and α6β3δ γ-aminobutyric acid type A receptors at low concentrations known to affect humans. Proc Natl Acad Sci USA. 2003;100:15218–15223. doi: 10.1073/pnas.2435171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland HA, Lüddens H, Seeburg PH. A single histidine in GABAA receptors is essential for benzodiazepine agonist binding. J Biol Chem. 1992;267:1426–1429. [PubMed] [Google Scholar]
- Wong G, Ovaska T, Korpi ER. Brain regional pharmacology of GABAA receptors in alcohol-preferring AA and alcohol-avoiding ANA rats. Addict Biol. 1996a;1:263–272. doi: 10.1080/1355621961000124876. [DOI] [PubMed] [Google Scholar]
- Wong G, Sarviharju M, Toropainen M, Matecka D, Korpi ER. Pharmacologic actions of subtype-selective and novel GABAergic ligands in rat lines with differential sensitivity to ethanol. Pharmacol Biochem Behav. 1996b;53:723–730. doi: 10.1016/0091-3057(95)02076-4. [DOI] [PubMed] [Google Scholar]