Abstract
Helicobacter pylori plays an essential role in the development of various gastroduodenal diseases; however, only a small proportion of people infected with H. pylori develop these diseases. Some populations that have a high prevalence of H. pylori infection also have a high incidence of gastric cancer (for example, in East Asia), whereas others do not (for example, in Africa and South Asia). Even within East Asia, the incidence of gastric cancer varies (decreasing in the south). H. pylori is a highly heterogeneous bacterium and its virulence varies geographically. Geographic differences in the incidence of gastric cancer can be explained, at least in part, by the presence of different types of H. pylori virulence factor, especially CagA, VacA and OipA. However, it is still unclear why the pathogenicity of H. pylori increased as it migrated from Africa to East Asia during the course of evolution. H. pylori infection is also thought to be involved in the development of duodenal ulcer, which is at the opposite end of the disease spectrum to gastric cancer. This discrepancy can be explained in part by the presence of H. pylori virulence factor DupA. Despite advances in our understanding of the development of H. pylori-related diseases, further work is required to clarify the roles of H. pylori virulence factors.
Introduction
Helicobacter pylori is a Gram-negative spiral bacterium whose ecological niche is the human stomach. H. pylori gastritis is etiologically associated with peptic ulcer, primary gastric B-cell lymphoma and gastric carcinoma. Despite a general decline in the incidence of gastric cancer, it remains the fourth most common cancer and second leading cause of cancer-related deaths worldwide.1 In 2010, it is estimated that there will be 21,000 new gastric cancer cases and 10,570 deaths from gastric cancer in the USA.2 In Africa and South Asia, the incidence of gastric cancer is much lower than in East Asian countries, although the prevalence of H. pylori infection is high in each of these regions (Table 1).3 Even within East Asia, the incidence of gastric cancer varies, decreasing at increasingly southerly latitudes. What is the cause of these geographic differences in disease pattern? In addition, H. pylori infection is thought to be involved in the development of both gastric cancer and duodenal ulcer, which are at opposite ends of the disease spectrum. What makes it possible for H. pylori infection to be involved in both of these gastroduodenal diseases?
Table 1.
Frequency of CagA phenotype and incidence of gastric cancer in 2008
| Geographic region/country | Number of strains studied | Number of sequences by CagA type
|
Age-standardized incidence rates per 100,000 population for gastric cancer
|
|||
|---|---|---|---|---|---|---|
| Western | East Asian | Overall incidence | Male | Female | ||
| World total | NA | NA | NA | 14.1 | 19.8* | 9.1 |
|
| ||||||
| Asia | NA | NA | NA | 18.6* | 25.9* | 11.7 |
|
| ||||||
| East Asia | NA | NA | NA | 30.0* | 42.4* | 18.3* |
|
| ||||||
| South Korea | 175 | 1 | 174 | 41.4* | 62.2* | 24.6* |
|
| ||||||
| Japan | 419 | 21 | 398 | 31.1* | 46.8* | 18.2* |
|
| ||||||
| China | 65 | 4 | 61 | 29.9* | 41.3* | 18.5* |
|
| ||||||
| Taiwan | 34 | 0 | 34 | 10.4 | 13.5 | 7.6 |
|
| ||||||
| West Asia | NA | NA | NA | 9.4 | 12.6 | 6.7 |
|
| ||||||
| Southeast Asia | NA | NA | NA | 8.6 | 10.9 | 6.7 |
|
| ||||||
| Vietnam | 122 | 4 | 118 | 18.9* | 24.4* | 14.6 |
|
| ||||||
| Malaysia | 3 | 2 | 1 | 8.4 | 10.7 | 6.4 |
|
| ||||||
| Thailand | 106 | 54 | 52 | 3.5 | 4.2 | 3.0 |
|
| ||||||
| South-Central Asia | NA | NA | NA | 5.3 | 6.7 | 3.9 |
|
| ||||||
| Kazakhstan | 4 | 4 | 0 | 20.6* | 31.7* | 13.7 |
|
| ||||||
| Iran | 91 | 91 | 0 | 15.6* | 21.9* | 9.0 |
|
| ||||||
| Turkey | 51 | 51 | 0 | 13.5 | 18.9* | 8.8 |
|
| ||||||
| Pakistan | 26 | 26 | 0 | 6.3 | 8.0 | 4.5 |
|
| ||||||
| India | 4 | 4 | 0 | 3.8 | 4.7 | 2.9 |
|
| ||||||
| Latin America and Caribbean | NA | NA | NA | 11.7 | 15.7* | 8.4 |
|
| ||||||
| South America | NA | NA | NA | 12.4 | 17.3* | 8.4 |
|
| ||||||
| Colombia | 147 | 147 | 0 | 17.4* | 23.4* | 12.5 |
|
| ||||||
| Chile | 1 | 1 | 0 | 17.9* | 27.3* | 10.3 |
|
| ||||||
| Brazil | 10 | 10 | 0 | 10.9 | 16.2* | 6.6 |
|
| ||||||
| Central America | NA | NA | NA | 10.9 | 12.7 | 9.3 |
|
| ||||||
| Costa Rica | 33 | 33 | 0 | 21.8* | 28.5* | 15.6* |
|
| ||||||
| Caribbean | NA | NA | NA | 8.5 | 11.2 | 6.1 |
|
| ||||||
| Europe | NA | NA | NA | 10.3 | 14.7 | 7.0 |
|
| ||||||
| Central-East Europe | NA | NA | NA | 14.7 | 22.2* | 9.7 |
|
| ||||||
| West Europe | NA | NA | NA | 6.5 | 9.0 | 4.4 |
|
| ||||||
| Italy | 57 | 57 | 0 | 10.9 | 14.8 | 7.7 |
|
| ||||||
| Germany | 1 | 1 | 0 | 7.7 | 10.3 | 5.5 |
|
| ||||||
| Ireland | 3 | 3 | 0 | 7.3 | 10.1 | 4.8 |
|
| ||||||
| Greece | 100 | 100 | 0 | 5.7 | 7.9 | 3.9 |
|
| ||||||
| France | 63 | 63 | 0 | 4.9 | 7.5 | 2.8 |
|
| ||||||
| Sweden | 5 | 5 | 0 | 4.3 | 5.7 | 3.1 |
|
| ||||||
| North Europe | NA | NA | NA | 6.2 | 8.6 | 4.2 |
|
| ||||||
| Oceania | NA | NA | NA | 5.5 | 7.5 | 3.7 |
|
| ||||||
| Australia | 1 | 1 | 0 | 5.2 | 7.3 | 3.2 |
|
| ||||||
| North America | NA | NA | NA | 4.2 | 5.8 | 2.8 |
|
| ||||||
| Canada | 20 | 20 | 0 | 4.8 | 6.7 | 3.1 |
|
| ||||||
| USA | 152 | 152 | 0 | 4.1 | 5.7 | 2.8 |
|
| ||||||
| Africa | NA | NA | NA | 4.0 | 4.7 | 3.3 |
|
| ||||||
| South Africa | 12 | 12 | 0 | 3.2 | 4.5 | 2.3 |
CagA sequencing data were initially obtained from the NCBI (April 16, 2007).26 Data from previous studies on genotyping of the cagA repeat region were also included for South Korea, Japan, China, Taiwan, Kazakhstan, Pakistan, Italy, France, South Africa, Canada, USA, Colombia and Brazil,20 in addition to Vietnam,20,24 Thailand,20,25 Iran23 and Turkey.22 Data on age-standardized incidence rates (ASRs) per 100,000 population obtained from the GLOBOCAN database, which provides access to the most recent (2008) estimates of the incidence of, and mortality from, 27 major cancers worldwide; organized by the International Agency for Research on Cancer (IARC).1
Regions and countries with an ASR ≥15.0. Abbreviation: NA, not available.
The questions above may be answered by considering various host-related factors, the duration of infection and/or environmental factors. Host genetics play an important role in interactions between the host and H. pylori. For example, polymorphisms in those genes that control the host’s inflammatory response can either accentuate or attenuate the inflammatory response and thus the incidence of adverse outcomes associated with an infection.4 The duration of infection and the age at acquisition of an H. pylori infection are also important, as is poor nutrition in childhood (or childhood infections), which lead to reduced acid secretion and increased susceptibility to the development of H. pylori-induced atrophic gastritis and gastric cancer.5 Environmental factors known to protect against gastric cancer include storage of food in a refrigerator (that is, a food preservation surrogate and an indication of reduced consumption of a seasonal diet) and the increased consumption of fresh fruit and vegetables; correlates with gastric cancer include a high salt intake and nitrate consumption.6 In addition to host-related factors, duration of infection and/or environmental factors, considering the bacterial factors present should provide one of the most important clues needed to answer the questions.
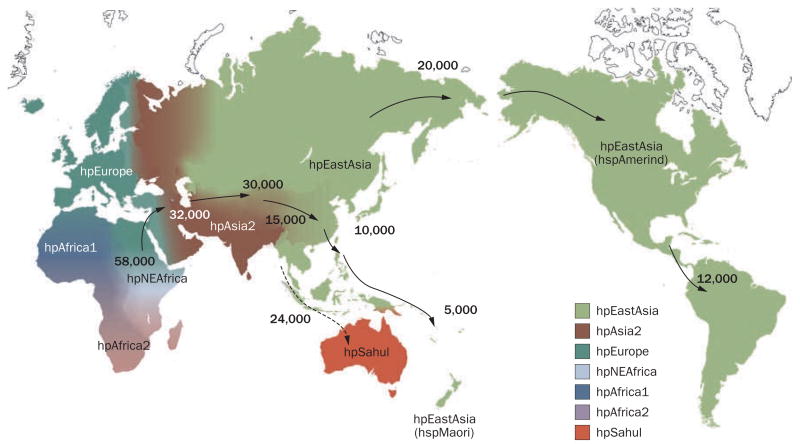
H. pylori has co-evolved with humans at least since they migrated out of Africa approximately 58,000 years ago and probably throughout their evolution (Figure 1); H. pylori is, therefore, a highly heterogeneous bacterium,7–10 the virulence of which has also changed geographically. In this Review, current knowledge of H. pylori virulence factors and how they contribute to the differing geographic gastric cancer disease patterns and to the development of both gastric cancer and duodenal ulcer is discussed, with a focus on cytotoxin-associated gene A product (CagA), vacuolating cytotoxin (VacA), outer inflammatory protein (OipA) and duodenal ulcer promoting (DupA).
Figure 1.
Distribution of Helicobater pylori genotypes before Columbus found the New World and human migration to America and Oceania began. There are seven modern H. pylori population types—hpEurope, hpEastAsia, hpAfrica1, hpAfrica2, hpAsia2, hpNEAfrica and hpSahul.7–9 hpEurope includes almost all H. pylori strains isolated from ethnic Europeans, including people from countries colonized by Europeans. Most H. pylori isolates from East Asia belong to hpEastAsia, which includes hspMaori (Polynesians, Melanesians, and native Taiwanese), hspAmerind (American Indians) and hspEAsia (East Asia) subpopulations. hpAsia2 strains are isolated in South, Southeast and Central Asia; hpAfrica1 in West Africa, South Africa and African Americans. hpNEAfrica is predominantly made up of isolates from Northeast Africa. hpAfrica2 is very distinct from any other type and has currently only been isolated in South Africa. hpSahul is a novel group specific to H. pylori strains isolated from Australian Aborigines and Highlanders of New Guinea. H. pylori is predicted to have spread from East Africa over the same time period as anatomically modern humans (~58,000 years ago), and has remained intimately associated with their human hosts ever since. Estimated global patterns of H. pylori migration are indicated by arrows and the numbers show the estimated time since they migrated (years ago). The broken arrow indicates an unconfirmed migration pattern. Detailed analyses of migration patterns have been explained previously.8,9
CagA
CagA is the most extensively studied H. pylori virulence factor. There are two types of clinical H. pylori isolate: CagA-producing (cagA positive) strains and CagA-nonproducing (cagA negative) strains. Studies of Mongolian gerbils showed that gastric cancer developed in those animals infected with wild-type H. pylori, but not in animals infected with isogenic cagA mutants.11,12 Since then, another study reported that gastric cancer and other malignant neoplasms occurred in some transgenic mice that had CagA protein artificially introduced.13 These data have led to increasing knowledge of CagA as a bacteria-derived carcinogen. In Western countries, it has been reported that individuals infected with cagA-positive strains of H. pylori are at a higher risk of peptic ulcer or gastric cancer than those infected with cagA-negative strains.14,15 In East Asia, however, most strains of H. pylori have the cagA gene irrespective of the disease; the pathogenic difference in East Asia is, therefore, difficult to explain simply in terms of the presence or absence of the cagA gene alone.16
Caga type: Western versus East Asian
cagA is a polymorphic gene. In particular, there are different numbers of repeat sequences located in the 3′ region of the cagA gene of different H. pylori strains.17–19 The repeat regions were initially classified into two types—the first repeat and the second repeat—and the sequence of the second repeat region was found to differ considerably between East Asian strains and Western strains of H. pylori.17–20
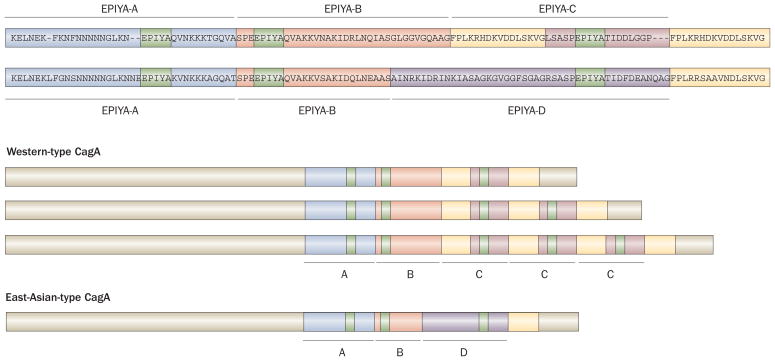
Each repeat region of the CagA protein contains Glu-Pro-Ile-Tyr-Ala (EPIYA) motifs, including a tyrosine phosphorylation site. It has now become more common to name the first repeat region as EPIYA-A and EPIYA-B segments and to name the second repeat region as EPIYA-C or EPIYA-D segments for Western and East Asian strains, respectively.21 As such, each CagA is assigned a sequence type consisting of the names of the EPIYA segments in its sequence (that is, ABC, ABCC or ABCCC for Western-type CagA and ABD for East-Asian-type CagA) (Figure 2). Analyses of the repeat regions of CagA has demonstrated that, although rare, some East Asian strains have a Western-type CagA sequence, especially strains isolated from Okinawa, the south islands of Japan (Table 1).20,22–26 By contrast, none of the Western strains studied have an East-Asian-type CagA sequence.20,26 A study published in 2009 reported that the Western-type CagA detected in strains from Okinawa formed a different cluster compared with the ordinary Western-type CagA and it was named as the J-Western-type CagA subtype.27 As H. pylori, together with humans, is thought to have migrated out of Africa approximately 58,000 years ago, and reached parts of Europe and finally East Asia via Central Asia (Figure 1), the original H. pylori strain is thought to have a Western-type CagA sequence that possibly evolved to the J-Western-type CagA and then to the East-Asian-type CagA. Further studies are needed to clarify the role of the different Western-type CagA sequences in the pathogenesis of disease.
Figure 2.
Structural polymorphism in CagA. Western-type CagA contain EPIYA-A, EPIYA-B, and EPIYA-C segments. By contrast, East-Asian-type CagA contain the EPIYA-A, EPIYA-B and EPIYA-D segments, but not the EPIYA-C segment. The EPIYA motif in each segment (shown in green) represents the tyrosine phosphorylation sites of CagA. The sequence flanking the tyrosine phosphorylation site of the EPIYA-D segment (EPIYATIDF), but not the EPIYA-C segment (EPIYATIDD), matches perfectly the consensus high-affinity binding sequence for the SH2 domains of SHP2. In Western countries, the incidence of gastric cancer is significantly higher in patients infected with strains containing multiple EPIYA-C segments than in patients infected with strains containing a single EPIYA-C segment (that is, ABCCC versus ABC). By contrast, almost all East Asian strains contain a single EPIYA-D segment. CagA forms dimers in cells in a phosphorylation-independent manner, and the CagA multimerization (CM) sequence (also named the conserved repeat responsible for phosphorylation-independent activity [CRPIA] or MARK2/PAR1b kinase inhibitor [MKI]) in yellow was identified as the site responsible for dimerization, for inhibition of MARK2/PAR1b kinase and for the interaction of CagA with activated c-Met.
Pathogenicity of multiple EPIYA segments
The first reports to suggest that the number of EPIYA segments in the second repeat region is associated with gastric cancer were published in the late 1990s.17,18 Subsequent studies confirmed that, in Western countries, the incidence of gastric cancer is notably higher in patients infected with strains containing multiple EPIYA-C segments than in patients infected with strains containing a single EPIYA-C segment (that is, ABCCC versus ABC).17,18,28,29 However, as almost all East Asian strains contain a single EPIYA-D segment,26 it is difficult to differentiate between simple gastritis and gastric cancer simply by considering the number of repeated sequences in East Asia.
With regard to the function of the repeat regions, initial demonstrations suggest that H. pylori strains that have a larger number of EPIYA segments in their repeat regions are less resistant to gastric acid.18 This finding seems to indicate that H. pylori strains containing many EPIYA segments can survive only in the presence of advanced atrophic gastritis (such as gastric cancer), in which gastric acid secretion is low. If this is the case, multiple EPIYA segments may have developed after the onset of atrophy and are not the cause, but rather the effect of atrophy.
Tyrosine phosphorylation and pathogenicity
There have been many reports that the site of tyrosine phosphorylation in the EPIYA motif of CagA plays a direct role in the pathogenicity of H. pylori. Following injection of CagA into epithelial cells, the EPIYA motifs are tyrosine phosphorylated by Src and Abl family kinases,30–34 which results in the impairment of a variety of intracellular signaling systems (Figure 3). Backert et al. reported that there are at least 20 known cellular binding partners of CagA, which is the highest number currently known for any virulence-associated effector protein in the microbial world.35 Of these binding partners, 10 host cell proteins bind CagA in a phosphorylation-dependent manner.35,36 The number of EPIYA motifs present is related to the level of CagA phosphorylation that occurs in epithelial cells infected by either East Asian37 or Western strains.28
Figure 3.
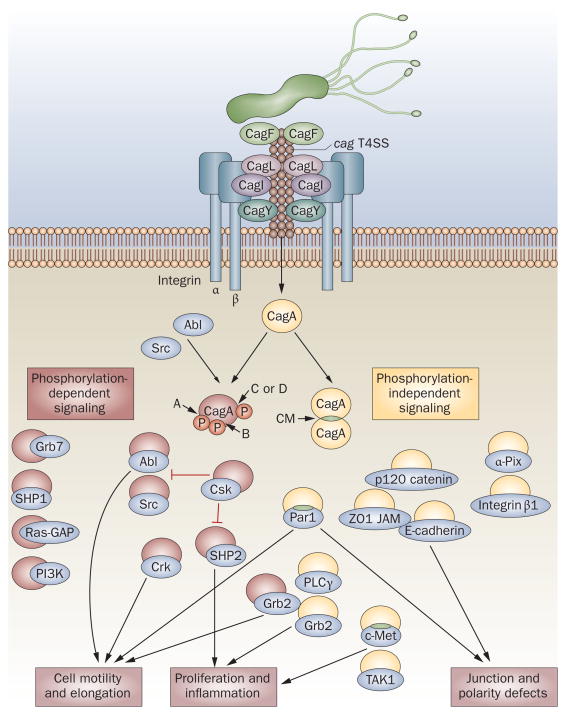
The pathogenesis of CagA-related signaling. In the early steps of CagA recognition, CagF binds CagA.117,118 CagL, CagY and probably CagI utilize host integrin β1 as a cell-surface receptor, which triggers delivery of CagA into target cells.41,119 Injected CagA can then undergo tyrosine phosphorylation by Src and Abl family kinases at the EPIYA motifs located near the C-terminal end (in EPIYA-A [A], EPIYA-B [B], EPIYA-C [C] or EPIYA-D [D] segments). CagA binds to and activates or inactivates multiple signaling proteins in both a phosphorylation-dependent and phosphorylation-independent manner. There are currently 20 known cellular binding partners of CagA, 10 of which form phosphorylation-dependent interactions with CagA.35 CagA itself forms dimers in a phosphorylation-independent manner, via the CagA multimerization (CM) sequence42 (also known as MKI or CRPIA). CM sequence is also essential for formation of the CagA–Par142 and CagA–c-Met complexes.46 The proposed functions of complexes formed by CagA with Grb7, SHP1, Ras-GAP, PI3K, α-Pix and integrin β1 are unknown. Experimental data obtained via natural infection versus transfection of CagA are conflicting—PI3K, Grb2 and Crk did not interact with CagA in transfection experiments.34
The best studied of the host signaling factors that interact with phosphorylated CagA is the Src homology-2 domain-containing phosphatase 2 (SHP2).38 SHP2 is able to bind EPIYA-B, EPIYA-C and EPIYA-D segments. Importantly, however, CagA with an EPIYA-D segment has a higher binding affinity for SHP2 than CagA with an EPIYA-C segment.21 The sequence flanking the tyrosine phosphorylation site of the EPIYA-D segment (EPIYATIDF), but not the EPIYA-C segment (EPIYATIDD), matches perfectly the consensus high-affinity binding sequence for the SH2 domains of SHP2 (Figure 2).
A study that used stable isotope labeling with amino acids in cell culture combined with mass spectrometry of bound protein (SILAC/MS) demonstrated that C-terminal Src tyrosine kinase (Csk) prefers to bind EPIYA-A and EPIYA-B segments, whereas Ras-GAP and Grb7 prefer to bind EPIYA-C segments.36 Malignant neoplasms have been shown to occur with greater frequency in transgenic mice in which East-Asian-type CagA had been introduced compared with transgenic mice in which Western-type CagA had been introduced.39 The discovery that, in vivo, East-Asian-type CagA is more carcinogenic than Western-type CagA is important; however, in this transgenic mouse model, the gastric cancer developed without inflammation and malignancy also occurred in nongastric organs; which raises questions about the applicability of this model to human disease.
Many previous in vitro studies of CagA involved transfection of a CagA expression vector instead of natural injection of CagA via the cag pathogenicity island (PAI) type IV secretion system (T4SS). However, transfection experiments have potentially confounded our understanding of the role of CagA. For example, Grb2 reportedly interacts with phosphorylated CagA during H. pylori infection,36 but such an interaction was not found in studies using transfected CagA.34,40 Importantly, transfection of CagA does not closely mimic natural injection of CagA by the T4SS. T4SS-injected CagA primarily localizes to focal adhesions,41 whereas vector-expressed CagA localizes to the entire host membrane.38 Most importantly, transfection prevents assessment of critical bacterial–host cell interactions, such as the activation of cellular signaling associated with VacA and OipA. Therefore, information gained from transfection experiments should be interpreted with caution and the findings confirmed by experiments using natural injection.
Phosphorylation-independent signaling
Nonphosphorylated CagA also impairs intracellular signaling systems. Currently, there are at least 10 known phosphorylation-independent CagA host interaction partners (Figure 3).35 In addition, CagA itself forms dimers in cells in a phosphorylation-independent manner, and the CagA multimerization (CM) sequence (FPLxRxxxVxDLSKVG) was identified by Ren et al. as the site responsible for dimerization.42 The CM sequence is located within the EPIYA-C segment, but is just downstream of the EPIYA-D segment (Figure 2).
The CM sequence is also essential for the formation of the CagA–PAR1 (MARK) complex—bound nonphosphorylated CagA inhibits the kinase activity of PAR1 to promote the loss of cell polarity42,43 and a loss of the elongation phenotype of host cells.44 The CM sequence has also been named by Nesic et al. as the MARK2/PAR1b kinase inhibitor (MKI).45 In addition, the same region was reported to be responsible for the interaction of CagA with activated c-Met and named as the conserved repeat responsible for phosphorylation-independent activity (CRPIA) by Suzuki and collegues.46 The interaction of CRPIA with c-Met subsequently led to the upregulation of β-catenin and nuclear factor κB (NFκB) transcriptional activities, which promoted proliferation and inflammation, respectively. Therefore, there are currently three acronyms that essentially correspond to the same sequence in CagA. Functional differences between the CM/MKI/CRPI sequence in Western-type CagA (FPLKRHDKVDDLSKVG) and East-Asian-type CagA (FPLRRSAAVNDLSKVG) have not been detected42,43 and have yet to be identified.45
Interleukin 8 production
Infection of the gastric mucosa by H. pylori is characterized by the production of proinflammatory cytokines, especially interleukin (IL)-8, a potent neutrophil-activating chemokine.47–50 The cag PAI, in which the cagA gene is localized at one end, is involved in the induction of gastric IL-8 production; whereas most reports have demonstrated that the CagA protein is not involved in IL-8 induction.51–53 However, one study has demonstrated that CagA is involved in IL-8 induction in a strain-dependent and time-dependent manner.54 IL-8 induction has typically been assessed 12–24 h after infection in vitro using gastric epithelial cells; however, the potentiating effects of CagA only become evident 36–48 h after infection.54 Interestingly, findings from transfection experiments suggest that the IL-8 levels induced by East-Asian-type CagA are higher than those induced by Western-type CagA.55
Clinical importance of CagA phenotype
As described above, there has been an increasing tendency in the past 10 years to explain the higher incidence of gastric cancer in East Asia using the concept of East-Asian-type CagA and Western-type CagA. The incidence of gastric cancer is clearly higher in East Asian countries than in any other countries when age-standardized rates (ASRs) are considered (Table 1). However, it should be noted that the incidence of gastric cancer is also high in some regions where Western-type CagA strains are reported to account for the majority of H. pylori strains (for example, in Peru and Columbia [ASR per 100,000 population 21.2 and 17.4, respectively]).20,56 In addition, in Africa the rate of H. pylori infection is high (for example, 70–97% of patients with dyspepsia are infected with H. pylori, as are 80% of asymptomatic volunteers57) but gastric cancer is generally uncommon; this seemingly contradictory situation is known as the “African Enigma”.57 The incidence of gastric cancer is, however, extremely high in Mali, West Africa (ASR per 100,000 population 20.3) and the frequency of gastric cancer among women in this country is higher than it is among women in Japan (ASR per 100,000 population 19.3 versus 18.2) (Table 1). These facts cannot be explained by the presence of East-Asian-type CagA versus Western-type CagA alone.
In an attempt to explain the geographic difference in the incidence of gastric cancer, my group compared the cagA gene repeat sequences found in Columbia (ASR per 100,000 population 17.4) with those found in the USA (ASR per 100,000 population 4.1). 100 H. pylori isolates from patients with simple gastritis (30 from Columbia and 70 from the USA) were analyzed—57% of the isolates from Columbia had two EPIYA-C segments, whereas only 4% of the isolates from the USA had two EPIYA-C segments (Y. Yamaoka, unpublished data). Overall, the number of EPIYA-C segments may explain, to some extent, the geographic difference in the incidence of gastric cancer in Western countries.
In Thailand, the incidence of gastric cancer is extremely low compared with that in the surrounding countries (ASR per 100,000 population 3.5) (Table 1). A previous study demonstrated that gastric cancer was common among ethnic Chinese and rare among the Thai indigenous population.25 All ethnic Chinese patients who had gastric cancer or peptic ulcers were infected with H. pylori with East-Asian-type CagA; whereas East-Asian-type CagA was found in only 40% of ethnic Chinese patients who had gastritis. These data suggest that East-Asian-type CagA may have increased virulence compared with Western-type CagA. However, interpretation of the relationship between the CagA type and outcome in Thailand is complicated by the fact that the majority of gastric cancers (82%) occurred among ethnic Chinese or Thai–Chinese individuals.25 Taken together, although there is increasing evidence from in vitro studies and animal models that East-Asian-type CagA is more virulent than Western-type CagA, the clinical concept that East-Asian-type CagA versus Western-type CagA explains the geographic difference in the incidence of gastric cancer is still not accepted.
In East Asia, the incidence of gastric cancer decreases at increasingly southerly latitudes. Indeed, the incidence of gastric cancer in Vietnam is half that in South Korea, although most Vietnamese strains of H. pylori (97%) have been reported to contain East-Asian-type CagA (Table 1). In addition, most of the East Asian strains of H. pylori have only one EPIYA-D segment.26 Therefore, in areas where infection with East-Asian-type CagA H. pylori strains is predominant, it is not possible to explain the difference in the incidence of gastric cancer by CagA alone. The structure of the East-Asian-type cagA gene in Vietnamese strains of H. pylori is, however, slightly different to that of strains from other East Asian countries.24 An 18 bp deletion unique to Vietnamese strains was identified in the region located slightly upstream of the EPIYA-A segment, whereas a 39 bp deletion was identified in strains common in East Asian countries such as Japan, and no deletion was identified in Western strains.24 Further research is required to determine whether these subtypes are involved in the pathogenesis of gastric cancer.
VacA
VacA is the second most extensively studied H. pylori virulence factor. In addition to inducing vacuolation, VacA can induce multiple cellular activities, including membrane-channel formation, cytochrome c release from mitochondria leading to apoptosis, and binding to cell-membrane receptors followed by initiation of a proinflammatory response.58–60 VacA can also specifically inhibit T-cell activation and proliferation.61–63 Studies indicate that VacA and CagA can even inhibit at least some of each other’s signaling pathways.64–67 For example, CagA has been shown to promote the expression of the apoptotic suppressor Mcl1, and inhibit epithelial cell apoptosis caused by VacA.64,65 These data again emphasize the importance of in vitro infection experiments in which interaction among H. pylori virulence factors can be taken into account.
Clinical importance of vacA genotype
Virtually all H. pylori strains have a functional vacA gene. There is variation in the vacuolating activity of different H. pylori strains,68,69 primarily due to differences in the vacA gene structure at the signal (s) region (s1 and s2) and the middle (m) region (m1 and m2).70 In vitro experiments demonstrated that s1/m1 strains are the most cytotoxic, followed by s1/m2 strains, whereas s2/m2 strains have no cytotoxic activity and s2/m1 strains are rare.70,71 In agreement with in vitro data, in Western countries, including Latin America, the Middle East and Africa, there have been many reports that individuals infected with s1 or m1 H. pylori strains have an increased risk of peptic ulcer or gastric cancer compared with individuals infected with s2 or m2 strains.14,70,72,73 In East Asia, however, most H. pylori strains have an s1-type s region; therefore the pathogenic difference cannot be explained by the type of s region present.16,20 Moreover, it is interesting to note that almost all cagA-positive strains are classified as an s1 strain, whereas almost all cagA-negative strains are classified as an s2/m2 strain.70
With respect to the m region, however, there is variation within East Asia. Although m1 strains are common in parts of north East Asia, such as Japan and South Korea, m2 strains are predominant in parts of south East Asia, such as Taiwan and Vietnam (Table 2).20,24 As the incidence of gastric cancer is higher in the north than in the south of East Asia, the m region may play a role in the regional difference in disease pattern. Even within Vietnam, the incidence of gastric cancer is approximately 1.5 times higher in Hanoi in the north than in Ho Chi Minh in the south of the country (Table 2). Research has shown that almost all H. pylori strains are classified as cagA-positive and vacA s1 strains in these two cities, but that the frequency of the vacA m1 strain is considerably higher in Hanoi.24
Table 2.
vacAm region genotype and incidence of gastric cancer in 2008
| Country/city | Number of strains studied |
vacA genotypes (%)
|
Age-standardized incidence rates per 100,000 population for gastric cancer
|
|||
|---|---|---|---|---|---|---|
| m1 | m2 | Overall incidence | Male | Female | ||
| South Korea | 87 | 83 (95.4) | 4 (4.6) | 41.4 | 62.2 | 24.6 |
|
| ||||||
| Japan | 83 | 79 (95.2) | 4 (4.8) | 31.1 | 46.8 | 18.2 |
|
| ||||||
| China | 27 | 18 (66.7) | 9 (33.3) | 29.9 | 41.3 | 18.5 |
|
| ||||||
| Vietnam* | 78 | 29 (37.1) | 45 (57.9) | 18.9 | 24.4 | 14.6 |
|
| ||||||
| Hanoi*‡ | 38 | 19 (50.0) | 17 (44.7) | NA | 27 | 13.2 |
|
| ||||||
| Ho Chi Minh*‡ | 40 | 10 (25.0) | 28 (70.0) | NA | 18.7 | 8.1 |
|
| ||||||
| Taiwan | 28 | 9 (32.1) | 19 (67.9) | 10.4 | 13.5 | 7.6 |
Data for vacA genotypes were obtained from previous studies by Yamaoka et al. (for South Korea, Japan, China and Taiwan)20 and by Uchida et al. (for Vietnam).24 To exclude the possible bias among countries, only simple gastritis cases were extracted from the published data. Data for gastric cancer incidence by country were obtained from the GLOBOCAN database, which provides access to the most recent estimates (for 2008) of the incidence of, and mortality from, 27 major cancers worldwide; organized by the International Agency for Research on Cancer (IARC).1
For four strains from Vietnam (two from Hanoi and two from Ho Chi Min) neither m1 nor m2 could be detected; therefore, the sum of m1 and m2 does not match the number of strains studied.
Data for gastric cancer incidence in two cities in Vietnam were obtained from the CI5 (Cancer Incidence in Five Continents) database, which provides access to detailed information on the incidence of cancer recorded by cancer registries (regional or national) worldwide; organized by the IARC (http://ci5.iarc.fr/). Abbreviations: m, middle; NA, not analyzed.
Novel genotypes
In 2007, a third disease-related region of vacA was identified between the s region and the m region; it was named the intermediate (i) region.74 According to the authors of the study, all s1/m1 strains were classified as type i1, and all s2/m2 strains were classified as type i2, but s1/m2 strains were classified as either type i1 or i2, and i1 strains were shown to be more pathogenic. In the same study, typing of the i region was also reported to be more effective for determining the risk of gastric cancer in Iranian patients than typing of the s region or m region.74 An additional study conducted by the same group showed that the i region plays a role in the development of peptic ulcer in people in Iraq and Italy.75,76 However, in a study of patients from East and Southeast Asia, there was no association between the i region and disease.77 More recently, a fourth disease-related region—the deletion (d) region—was identified between the i region and the m region.78 The d region is divided into d1 (no deletion) and d2 (a 69–81 bp deletion). The study of Western strains demonstrated that d1 was a risk factor for gastric mucosal atrophy; however, almost all East Asian strains are classified as s1/i1/d1.78 At present, therefore, typing of the m region may be the best overall marker for gastric cancer, especially in East Asia.
OipA
Approximately 4% of the H. pylori genome is predicted to encode outer membrane proteins (OMPs), some of which may function as adhesins.79–85 One such OMP is OipA, which was identified in 2000.86 The functional status of OipA is regulated by slipped strand mispairing that is determined by the number of CT dinucleotide repeats in the 5′ region of the gene (switch ‘on’ and OipA is functional; switch ‘off’ and OipA is nonfunctional).86
Interleukin 8 production
OipA was initially identified as a proinflammatory-response-inducing protein based, in part, on the fact that oipA isogenic mutants reduced the production of IL-8 from gastric epithelial cell lines.86 In vivo experiments using human gastric biopsy specimens also confirmed that a functional oipA was independently and significantly associated with high mucosal IL-8 levels.15 Some subsequent conflicting reports regarding IL-8 induction in vitro87–90 might be at least partially explained by the fact that multiple in vitro passages can allow factors other than OipA to enhance IL-8 induction.91 Binding sites for the transcription factors NFκB, activator protein 1 (AP1), and interferon-stimulated responsive element (ISRE)-like element within the IL-8 promoter are involved in regulating IL-8 gene transcription in H. pylori-infected gastric epithelial cells in both an OipA-dependent and cag PAI-dependent manner.92–94 However, data from my group demonstrated that the activation of NFκB is strain dependent and this might also be a reason for the conflicting reports (Figure 4).83
Figure 4.
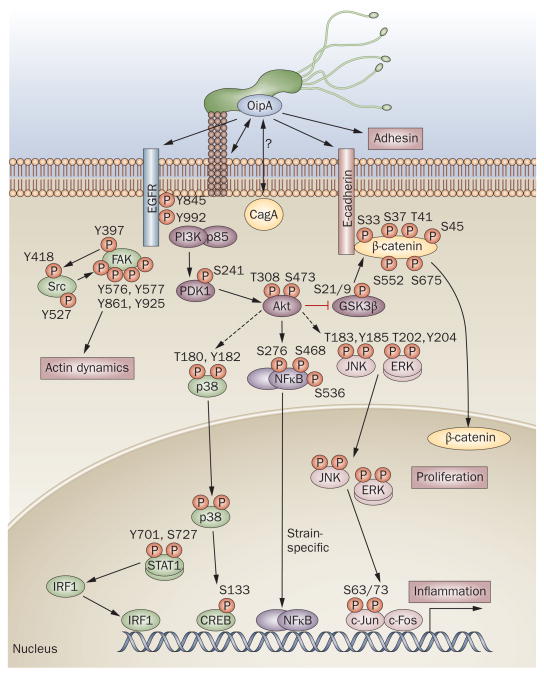
The pathogenesis of OipA-related signaling. Phosphorylation by OipA reportedly regulates various epithelial cell signaling pathways, most of which are also important for cag PAI-related signaling. p38/STAT1 pathways are regulated independently of the cag PAI. OipA and the cag PAI are both necessary for full activation of the IL-8 promoter, but act via different pathways that diverge upstream of IRF1: only OipA is involved in the STAT1/IRF1/ISRE pathway.92 OipA is believed to function as an adhesin; but its target receptors have not been identified. OipA phosphorylation sites are shown for all molecules. EGFR contains more than 10 phosphorylation sites but only the two investigated as part of OipA-related signaling pathways98,99 are shown. OipA is reportedly involved in phosphorylation of FAK at tyrosine (Y) 397, Y576, Y577, Y861 and Y925, but not Y407; whereas the cag PAI is involved in phosphorylation of FAK at Y407 only.98 However, another study reported that FAK phosphorylation at Y397 was cag PAI dependent.41 Broken arrows indicate unconfirmed interactions. Abbreviations: S, serine; T, threonine.
Gradually it has become clear that one of the functions of OipA is to induce inflammation and actin dynamics via phosphorylation of multiple signaling pathways (Figure 4).92,95–99 However, most of these signaling pathways, with the exception of p38-related pathways, are also involved in cag PAI-related pathways. Therefore, there might have been some interaction between OipA and cag PAI and/or CagA; however, evidence of such an interaction has not been reported and further studies will be necessary to clarify the specific roles of OipA on these cag PAI-related pathways. One study has demonstrated that OipA in addition to CagA is involved in β-catenin signaling, leading to the opening of cell–cell junctions and proliferation.12
Gastric colonization
OipA functions as an adhesin and is reported to be involved in the attachment of H. pylori to gastric epithelial cells in vitro.89,92,100 In addition, animal studies have revealed that OipA plays a role in the bacterial colonization of the gastric mucosa. In one study of mice infected with either wild-type H. pylori strain CPY2052 or its isogenic oipA mutant, the bacterial density was considerably decreased in the mice infected with the mutant strain compared with the mice infected with the wild-type strain.100 In a study of Mongolian gerbils, the isogenic oipA mutant of the TN2 wild-type H. pylori strain did not cause infection.90 By contrast, the isogenic oipA mutant of wild-type H. pylori strain 7.13 has been shown to infect animals; however, no animals infected with the oipA mutant developed gastric cancer, whereas 27% of those infected with the wild-type strain developed gastric cancer.12 These animal studies demonstrate that OipA alone plays a role in the development of gastric cancer.
Clinical importance of OipA
Interestingly, the oipA ‘on’ status and cagA positivity are closely linked with each other (correlation coefficient = 0.82).15 As described above, the cagA status is also linked to the vacA s region type and it is further closely linked to the presence of the babA gene; the BabA protein is another virulence factor that is also an OMP.80 The links among these factors should have a certain biological significance and they may somehow interact with each other. It might be more relevant to hypothesize that these factors interact synergistically with each other and induce serious diseases, rather than to discuss which factor is the most virulent. According to one investigation so far, it is interesting to note that all East Asian strains are classified as oipA status ‘on’ and as functional OipA-protein-producing strains.86 Half-collapsed CT repeat sequences in the s region of the oipA gene (for example, Japanese-derived strain JK51 contains a CTGCCTTTCT repeat sequence and its status is ‘on’) suggest that this sequence may result from an intentional change in the status in the course of the evolution of the bacteria to prevent the switch from being turned ‘off’ easily.
DupA
The production of CagA, VacA and OipA is linked and the majority of H. pylori strains produce either all of these proteins or none of them. Almost all East Asian strains of H. pylori are classified as CagA-producing, VacA-producing (vacA s1), and OipA-producing (oipA status ‘on’) strains and are highly pathogenic. In addition, CagA, VacA and OipA are all thought to be involved in the development of both gastric cancer and duodenal ulcer, which are at the opposite ends of the disease spectrum. In 2005, the first disease-specific H. pylori virulence factor that induced duodenal ulcer and had a suppressive action on gastric cancer was identified, and was named duodenal ulcer promoting gene A (DupA).101
Cytokine induction
Initial in vitro experiments using dupA-deleted and dupA-complemented mutants showed that the presence of dupA was associated with increased IL-8 production from both the antral gastric mucosa in vivo and gastric epithelial cells.101 The relationship between dupA and IL-8 induction from gastric epithelial cells in vitro is still controversial;102,103 however, one study supports the original data that the presence of the dupA gene is associated with increased IL-8 production from gastric mucosa in vivo.102 Importantly, in vitro experiments using dupA mutants showed that dupA substantially increased H. pylori-induced IL-12p40 and IL-12p70 production by CD14+ mononuclear cells.102 The production of other T-helper-1-associated cytokines and IL-8 were also modestly induced by the cells. These data suggest that virulent H. pylori strains cause inflammation by stimulating epithelial cells via CagA, OipA and/or VacA-related proteins and mononuclear inflammatory cells through DupA.
Clinical outcomes
In an initial study of a total of 500 H. pylori isolates, including 160 from Japan, 175 from South Korea and 165 from Colombia, the positive rate for the dupA gene was high in patients with duodenal ulcer and low in patients with gastric cancer, regardless of a patients’ nationality (42% versus 9% on average).101 Since this report was published, studies have been conducted all over the world.104 Reports from India,105 China106 and Iraq75 support the initial data, whereas reports from Iran,75,107 Brazil,108,109 Singapore,103 Malaysia103 and Japan110 failed to demonstrate a correlation between the presence of the dupA gene and disease.
A pooled analysis of data from three Western countries (the USA, Belgium and South Africa) showed that the presence of the dupA gene was correlated with peptic ulcer as well as with gastric cancer.111 One report from Iran, which was not supportive of the hypothesis that dupA contributes to the development of duodenal ulcer but not gastric cancer, showed that the incidence of dysplasia—a premalignant condition—was notably lower in dupA-positive patients,107 partially supporting the hypothesis. One report from Brazil108 was also unsupportive of the hypothesis, but showed that the rate of infection with dupA-positive strains in pediatric patients was 100% and higher than that in adult patients. This finding indicates that the number of dupA-positive strains decreases as gastric mucosal atrophy gradually progresses with aging, which also partially supports the hypothesis.108 In a review of previous reports on dupA that included a total of 2,358 cases, the positive rate for dupA was 48% and confirmed that dupA was a risk factor for duodenal ulcer.112 However, a large regional difference was also reported: dupA might, therefore, be a risk factor for both duodenal ulcer and gastric cancer in some regions.
Of note, an academic report on Brazilian strains that was presented at the Annual Meeting of the American Gastroenterological Association in 2008, showed that a dupA gene mutation (deletion or insertion) was found in 50% of patients with gastric cancer, whereas it was found in only approximately 20% of patients with duodenal ulcer.113 As a result, the positive rate for the functional dupA gene was considerably higher in patients with duodenal ulcer than in patients with gastric cancer, and this finding supported the original hypothesis. That such mutations occur at an unexpectedly high frequency should not be ignored. However, it is impossible to detect dupA mutations by using a simple PCR method; therefore, detection of functional dupA by measuring intact DupA protein using immunoblotting techniques, which has not been reported, should be attempted.
DupA as part of a novel T4SS
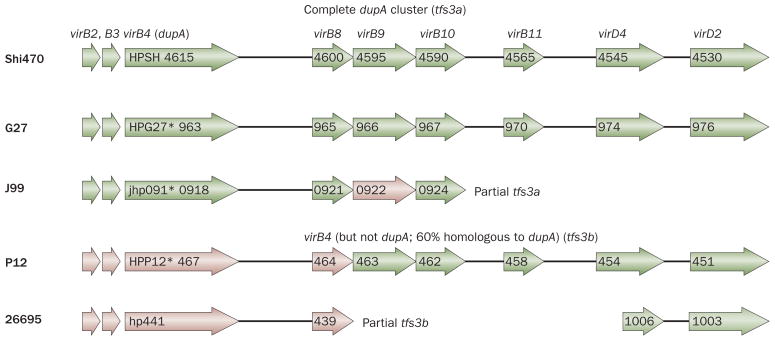
DupA is found in an area called the plasticity zone, in which many genes thought to be associated with pathogenicity can be found (Box 1).101,104 DupA has a high homology with the VirB4 ATPase, but there are vir genes before and after the region of the dupA locus. In the strain Shi470, for example, virB2, virB3, virB4 (dupA), virB8, virB9, virB10, virB11, virD4 and virD2 can all be found (Figure 5). These vir genes before and after the region of the dupA locus are structurally similar to the T4SS called cag PAI and ComB, and are thought to be the third T4SS. Kersulyte et al. named the third T4SS in the plasticity zone tfs3 (type IV secretion 3).114 The T4SS containing dupA has since been named as tfs3a, and T4SS containing the virB4 sequence but not dupA was named as tfs3b.115 The strain G27 that contains tfs3a is unlikely to be functional because dupA has a frame-shift mutation. In addition, the strain J99 has a mutation in dupA and contains only a part of tfs3a. These observations suggest that only strains with functional dupA and complete tfs3a may be considered as pathogenic strains that have the same action as T4SS, such as cag PAI and ComB. The study of the plasticity zone is only at its beginning and may be the most attractive area for future investigation.
Box 1. Helicobacter pylori plasticity zones.
There are several regions of the H. pylori genome in which the G+C content is lower than in the rest of the genome (33–34% vs 39%)
One region of such low G+C content is the cag PAI—the others have been named collectively as plasticity zones on the basis of the variability in gene content among different H. pylori isolates
Nearly half of the strain-specific genes of H. pylori are located in plasticity zones
Figure 5.
Type IV secretion system in the Helicobacter pylori plasticity zone. Gene arrangement in the plasticity region of H. pylori strain Shi470, which has a complete dupA cluster (named tfs3a [type IV secretion 3a]) and functional dupA gene sequence, in comparison with corresponding regions of genome sequences of other H. pylori strains (G27, J99, P12 and 26695). Genes encoding type IV secretion system (T4SS) components are represented by arrows, with frameshift mutations indicated by asterisks. Genes encoding proteins that have 90–95% sequence similarity to the Shi470 proteins are shown in green and genes encoding proteins that have 50–75% sequence similarity are shown in pink. It is possible that only those H. pylori strains that have both a functional dupA gene sequence and a complete tfs3a are pathogenic and have the same action as the cag PAI and ComB T4SS. The numbers shown represent the gene number of each strain deposited in GenBank (GenBank accession numbers for Shi470, G27, J99, P12 and 26695 are CP001072, CP001173, AE001439, CP001217 and AE000511, respectively).
Conclusions
So, can H. pylori factors explain both the geographic differences in gastric cancer disease pattern and the role of H. pylori infection in gastric cancer and duodenal ulcer? CagA, VacA and probably OipA (that is, different CT repeat sequence patterns) are key factors needed to answer the first question and DupA is a key factor needed to answer the second question. However, although these four virulence factors are obviously important they can provide only partial answers to the questions. First, since H. pylori consist of approximately 1,600 genes, there remains the possibility that additional important pathogenic genes will be identified. Second, it is also true that gastric cancer incidence has changed remarkably with environmental factors such as diet (for example, salt intake) or with migration. Host factors (for example, IL-1 polymorphisms) and duration of infection (for example, early infection with duodenal ulcer and late infection with gastric cancer) should also be taken into account. These various factors are thought to interact with each other in a complex manner in the actual development of disease.
In the past 10 years, tremendous progress has been made in understanding the mechanisms by which H. pylori virulence factors induce gastroduodenal damage. However, controversial data are also increasing as research moves forward. Transfection of CagA versus natural infection via T4SS gives us different results. The relationship between induction of IL-8 and CagA or OipA is also controversial, and ‘time-dependent’ and ‘strain-dependent’ phenomena have proved confusing. The terminology used in the field is also confusing, with CM, MKI and CRPIA all being used to describe the same sequence. In addition, tfs3a and tfs3b in strain P12 were both named tfs4.116 We should, therefore, take the time to understand and interpret each report carefully, with methodology and terminology used taken into account.
There also remains a considerable knowledge gap between the detailed dissection of events observed in vitro or in animal models and the clinical and/or epidemiological confirmation of their role in the pathogenesis of human disease. One reason for this discrepancy is that H. pylori is surprisingly heterogenic and we use only limited H. pylori strains in vitro or in animal models. By detailed typing of H. pylori in addition to searching for interactions between bacterial factors and other factors such as environmental factors and host factors, we will gradually understand the mechanisms by which H. pylori induces gastric injury, leading to gastric cancer.
Key points.
Experimental evidence suggests that East-Asian-type CagA is more virulent than Western-type CagA, but the concept that this explains geographic differences in the incidence of gastric cancer is still controversial
The number of CagA Glu-Pro-Ile-Tyr-Ala (EPIYA)-C segments (with multiple repeats increasing the virulence) may explain, to some extent, geographic differences in the incidence of gastric cancer in Western countries
The genotype of the vacA middle region (m1 versus m2; m1 being more virulent) may, in part, explain geographic differences in the incidence of gastric cancer in East Asian countries
CagA, OipA and VacA are thought to interact synergistically with each other to induce serious disease
The presence of functional DupA with an intact, full size dupA cluster (tfs3a) is associated with duodenal ulcers
Bacterial factors, environmental factors and host factors are thought to form complex interactions with each other in the actual development of disease
Review criteria.
The references cited in this review were identified by searching the MEDLINE database. No date restrictions were applied to the search, which was limited to articles published in the English language. The search terms used were “Helicobacter pylori”, “CagA”, “cag pathogenicity island”, “VacA”, “OipA”, “DupA”, and “plasticity”. The reference lists of the articles identified were also searched to identify additional relevant references.
Acknowledgments
This report is based on work supported in part by grants from the NIH (DK62813), and grants-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (22390085 and 22659087).
Footnotes
Competing interests
The author declares no competing interests.
References
- 1.International Agency for Research on Cancer. The Globocan Project. 2010 [online], http://globocan.iarc.fr/
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer Statistics. CA Cancer J Clin. 2010 doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Malaty HM. Epidemiology of Helicobacter pylori infection. Best Pract Res Clin Gastroenterol. 2007;21:205–214. doi: 10.1016/j.bpg.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 4.El-Omar EM. Role of host genes in sporadic gastric cancer. Best Pract Res Clin Gastroenterol. 2006;20:675–686. doi: 10.1016/j.bpg.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Graham DY. Helicobacter pylori infection in the pathogenesis of duodenal ulcer and gastric cancer: a model. Gastroenterology. 1997;113:1983–1991. doi: 10.1016/s0016-5085(97)70019-2. [DOI] [PubMed] [Google Scholar]
- 6.Graham DY, Lu H, Yamaoka Y. African, Asian or Indian enigma, the East Asian Helicobacter pylori: facts or medical myths. J Dig Dis. 2009;10:77–84. doi: 10.1111/j.1751-2980.2009.00368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falush D, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299:1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- 8.Linz B, et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445:915–918. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moodley Y, et al. The peopling of the Pacific from a bacterial perspective. Science. 2009;323:527–530. doi: 10.1126/science.1166083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaoka Y. Helicobacter pylori typing as a tool for tracking human migration. Clin Microbiol Infect. 2009;15:829–834. doi: 10.1111/j.1469-0691.2009.02967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franco AT, et al. Activation of beta-catenin by carcinogenic Helicobacter pylori. Proc Natl Acad Sci USA. 2005;102:10646–10651. doi: 10.1073/pnas.0504927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franco AT, et al. Regulation of gastric carcinogenesis by Helicobacter pylori virulence factors. Cancer Res. 2008;68:379–387. doi: 10.1158/0008-5472.CAN-07-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohnishi N, et al. Transgenic expression of Helicobacter pylori CagA induces gastrointestinal and hematopoietic neoplasms in mouse. Proc Natl Acad Sci USA. 2008;105:1003–1008. doi: 10.1073/pnas.0711183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Doorn LJ, et al. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology. 1998;115:58–66. doi: 10.1016/s0016-5085(98)70365-8. [DOI] [PubMed] [Google Scholar]
- 15.Yamaoka Y, et al. Importance of Helicobacter pylori oipA in clinical presentation, gastric inflammation, and mucosal interleukin 8 production. Gastroenterology. 2002;123:414–424. doi: 10.1053/gast.2002.34781. [DOI] [PubMed] [Google Scholar]
- 16.Yamaoka Y, et al. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome: studies in four different countries. J Clin Microbiol. 1999;37:2274–2279. doi: 10.1128/jcm.37.7.2274-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamaoka Y, Kodama T, Kashima K, Graham DY, Sepulveda AR. Variants of the 3′ region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases. J Clin Microbiol. 1998;36:2258–2263. doi: 10.1128/jcm.36.8.2258-2263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamaoka Y, et al. Relationship between the cagA 3′ repeat region of Helicobacter pylori, gastric histology, and susceptibility to low pH. Gastroenterology. 1999;117:342–349. doi: 10.1053/gast.1999.0029900342. [DOI] [PubMed] [Google Scholar]
- 19.Yamaoka Y, et al. Molecular epidemiology of Helicobacter pylori: separation of H. pylori from East Asian and non-Asian countries. Epidemiol Infect. 2000;124:91–96. doi: 10.1017/s0950268899003209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamaoka Y, et al. Helicobacter pylori in North and South America before Columbus. FEBS Lett. 2002;517:180–184. doi: 10.1016/s0014-5793(02)02617-0. [DOI] [PubMed] [Google Scholar]
- 21.Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer. 2004;4:688–694. doi: 10.1038/nrc1433. [DOI] [PubMed] [Google Scholar]
- 22.Saribasak H, Salih BA, Yamaoka Y, Sander E. Analysis of Helicobacter pylori genotypes and correlation with clinical outcome in Turkey. J Clin Microbiol. 2004;42:1648–1651. doi: 10.1128/JCM.42.4.1648-1651.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shokrzadeh L, et al. Analysis of 3′-end variable region of the cagA gene in Helicobacter pylori isolated from Iranian population. J Gastroenterol Hepatol. 2010;25:172–177. doi: 10.1111/j.1440-1746.2009.05979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uchida T, et al. Analysis of virulence factors of Helicobacter pylori isolated from a Vietnamese population. BMC Microbiol. 2009;9:175. doi: 10.1186/1471-2180-9-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vilaichone RK, et al. Molecular epidemiology and outcome of Helicobacter pylori infection in Thailand: a cultural cross roads. Helicobacter. 2004;9:453–459. doi: 10.1111/j.1083-4389.2004.00260.x. [DOI] [PubMed] [Google Scholar]
- 26.Xia Y, Yamaoka Y, Zhu Q, Matha I, Gao X. A comprehensive sequence and disease correlation analyses for the C-terminal region of CagA protein of Helicobacter pylori. PLoS ONE. 2009;4:e7736. doi: 10.1371/journal.pone.0007736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Truong BX, et al. Diverse characteristics of the CagA gene of Helicobacter pylori strains collected from patients from southern Vietnam with gastric cancer and peptic ulcer. J Clin Microbiol. 2009;47:4021–4028. doi: 10.1128/JCM.00504-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Argent RH, et al. Determinants and consequences of different levels of CagA phosphorylation for clinical isolates of Helicobacter pylori. Gastroenterology. 2004;127:514–523. doi: 10.1053/j.gastro.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 29.Azuma T, et al. Correlation between variation of the 3′ region of the cagA gene in Helicobacter pylori and disease outcome in Japan. J Infect Dis. 2002;186:1621–1630. doi: 10.1086/345374. [DOI] [PubMed] [Google Scholar]
- 30.Poppe M, Feller SM, Romer G, Wessler S. Phosphorylation of Helicobacter pylori CagA by c-Abl leads to cell motility. Oncogene. 2007;26:3462–3472. doi: 10.1038/sj.onc.1210139. [DOI] [PubMed] [Google Scholar]
- 31.Selbach M, Moese S, Hauck CR, Meyer TF, Backert S. Src is the kinase of the Helicobacter pylori CagA protein in vitro and in vivo. J Biol Chem. 2002;277:6775–6778. doi: 10.1074/jbc.C100754200. [DOI] [PubMed] [Google Scholar]
- 32.Stein M, et al. c-Src/Lyn kinases activate Helicobacter pylori CagA through tyrosine phosphorylation of the EPIYA motifs. Mol Microbiol. 2002;43:971–980. doi: 10.1046/j.1365-2958.2002.02781.x. [DOI] [PubMed] [Google Scholar]
- 33.Tammer I, Brandt S, Hartig R, Konig W, Backert S. Activation of Abl by Helicobacter pylori: a novel kinase for CagA and crucial mediator of host cell scattering. Gastroenterology. 2007;132:1309–1319. doi: 10.1053/j.gastro.2007.01.050. [DOI] [PubMed] [Google Scholar]
- 34.Tsutsumi R, Higashi H, Higuchi M, Okada M, Hatakeyama M. Attenuation of Helicobacter pylori CagA x SHP-2 signaling by interaction between CagA and C-terminal Src kinase. J Biol Chem. 2003;278:3664–3670. doi: 10.1074/jbc.M208155200. [DOI] [PubMed] [Google Scholar]
- 35.Backert S, Tegtmeyer N, Selbach M. The versatility of Helicobacter pylori CagA effector protein functions: The master key hypothesis. Helicobacter. 2010;15:163–176. doi: 10.1111/j.1523-5378.2010.00759.x. [DOI] [PubMed] [Google Scholar]
- 36.Selbach M, et al. Host cell interactome of tyrosine-phosphorylated bacterial proteins. Cell Host Microbe. 2009;5:397–403. doi: 10.1016/j.chom.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 37.Hirata Y, et al. Functional variability of cagA gene in Japanese isolates of Helicobacter pylori. Gene. 2004;343:165–172. doi: 10.1016/j.gene.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 38.Higashi H, et al. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science. 2002;295:683–686. doi: 10.1126/science.1067147. [DOI] [PubMed] [Google Scholar]
- 39.Miura M, Ohnishi N, Tanaka S, Yanagiya K, Hatakeyama M. Differential oncogenic potential of geographically distinct Helicobacter pylori CagA isoforms in mice. Int J Cancer. 2009;125:2497–2504. doi: 10.1002/ijc.24740. [DOI] [PubMed] [Google Scholar]
- 40.Churin Y, et al. Helicobacter pylori CagA protein targets the c-Met receptor and enhances the motogenic response. J Cell Biol. 2003;161:249–255. doi: 10.1083/jcb.200208039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwok T, et al. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature. 2007;449:862–866. doi: 10.1038/nature06187. [DOI] [PubMed] [Google Scholar]
- 42.Ren S, Higashi H, Lu H, Azuma T, Hatakeyama M. Structural basis and functional consequence of Helicobacter pylori CagA multimerization in cells. J Biol Chem. 2006;281:32344–32352. doi: 10.1074/jbc.M606172200. [DOI] [PubMed] [Google Scholar]
- 43.Saadat I, et al. Helicobacter pylori CagA targets PAR1/MARK kinase to disrupt epithelial cell polarity. Nature. 2007;447:330–333. doi: 10.1038/nature05765. [DOI] [PubMed] [Google Scholar]
- 44.Umeda M, et al. Helicobacter pylori CagA causes mitotic impairment and induces chromosomal instability. J Biol Chem. 2009;284:22166–22172. doi: 10.1074/jbc.M109.035766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nesic D, et al. Helicobacter pylori CagA inhibits PAR1-MARK family kinases by mimicking host substrates. Nat Struct Mol Biol. 2010;17:130–132. doi: 10.1038/nsmb.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki M, et al. Helicobacter pylori CagA phosphorylation-independent function in epithelial proliferation and inflammation. Cell Host Microbe. 2009;5:23–34. doi: 10.1016/j.chom.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 47.Yamaoka Y, Kita M, Kodama T, Sawai N, Imanishi J. Helicobacter pylori cagA gene and expression of cytokine messenger RNA in gastric mucosa. Gastroenterology. 1996;110:1744–1752. doi: 10.1053/gast.1996.v110.pm8964399. [DOI] [PubMed] [Google Scholar]
- 48.Yamaoka Y, et al. Chemokines in the gastric mucosa in Helicobacter pylori infection. Gut. 1998;42:609–617. doi: 10.1136/gut.42.5.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamaoka Y, et al. Relation between clinical presentation, Helicobacter pylori density, interleukin 1β and 8 production, and cagA status. Gut. 1999;45:804–811. doi: 10.1136/gut.45.6.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamaoka Y, et al. Relation between cytokines and Helicobacter pylori in gastric cancer. Helicobacter. 2001;6:116–124. doi: 10.1046/j.1523-5378.2001.00017.x. [DOI] [PubMed] [Google Scholar]
- 51.Censini S, et al. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fischer W, et al. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol Microbiol. 2001;42:1337–1348. doi: 10.1046/j.1365-2958.2001.02714.x. [DOI] [PubMed] [Google Scholar]
- 53.Al-Ghoul L, et al. Analysis of the type IV secretion system-dependent cell motility of Helicobacter pylori-infected epithelial cells. Biochem Biophys Res Commun. 2004;322:860–866. doi: 10.1016/j.bbrc.2004.07.199. [DOI] [PubMed] [Google Scholar]
- 54.Brandt S, Kwok T, Hartig R, Konig W, Backert S. NF-kappaB activation and potentiation of proinflammatory responses by the Helicobacter pylori CagA protein. Proc Natl Acad Sci USA. 2005;102:9300–9305. doi: 10.1073/pnas.0409873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim SY, Lee YC, Kim HK, Blaser MJ. Helicobacter pylori CagA transfection of gastric epithelial cells induces interleukin-8. Cell Microbiol. 2006;8:97–106. doi: 10.1111/j.1462-5822.2005.00603.x. [DOI] [PubMed] [Google Scholar]
- 56.Kersulyte D, et al. Differences in genotypes of Helicobacter pylori from different human populations. J Bacteriol. 2000;182:3210–3218. doi: 10.1128/jb.182.11.3210-3218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holcombe C. Helicobacter pylori: the African enigma. Gut. 1992;33:429–431. doi: 10.1136/gut.33.4.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Atherton JC. The pathogenesis of Helicobacter pylori-induced gastro-duodenal diseases. Annu Rev Pathol Mech Dis. 2006;1:63–96. doi: 10.1146/annurev.pathol.1.110304.100125. [DOI] [PubMed] [Google Scholar]
- 59.Cover TL, Blanke SR. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat Rev Microbiol. 2005;3:320–332. doi: 10.1038/nrmicro1095. [DOI] [PubMed] [Google Scholar]
- 60.Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19:449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boncristiano M, et al. The Helicobacter pylori vacuolating toxin inhibits T cell activation by two independent mechanisms. J Exp Med. 2003;198:1887–1897. doi: 10.1084/jem.20030621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gebert B, Fischer W, Weiss E, Hoffmann R, Haas R. Helicobacter pylori vacuolating cytotoxin inhibits T lymphocyte activation. Science. 2003;301:1099–1102. doi: 10.1126/science.1086871. [DOI] [PubMed] [Google Scholar]
- 63.Sundrud MS, Torres VJ, Unutmaz D, Cover TL. Inhibition of primary human T cell proliferation by Helicobacter pylori vacuolating toxin (VacA) is independent of VacA effects on IL-2 secretion. Proc Natl Acad Sci USA. 2004;101:7727–7732. doi: 10.1073/pnas.0401528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mimuro H, et al. Helicobacter pylori dampens gut epithelial self-renewal by inhibiting apoptosis, a bacterial strategy to enhance colonization of the stomach. Cell Host Microbe. 2007;2:250–263. doi: 10.1016/j.chom.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 65.Oldani A, et al. Helicobacter pylori counteracts the apoptotic action of its VacA toxin by injecting the CagA protein into gastric epithelial cells. PLoS Pathog. 2009;5:e1000603. doi: 10.1371/journal.ppat.1000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tegtmeyer N, et al. Importance of EGF receptor, HER2/Neu and Erk1/2 kinase signaling for host cell elongation and scattering induced by the Helicobacter pylori CagA protein: antagonistic effects of the vacuolating cytotoxin VacA. Cell Microbiol. 2009;11:488–505. doi: 10.1111/j.1462-5822.2008.01269.x. [DOI] [PubMed] [Google Scholar]
- 67.Yokoyama K, et al. Functional antagonism between Helicobacter pylori CagA and vacuolating toxin VacA in control of the NFAT signaling pathway in gastric epithelial cells. Proc Natl Acad Sci USA. 2005;102:9661–9666. doi: 10.1073/pnas.0502529102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cover TL, Blaser MJ. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J Biol Chem. 1992;267:10570–10575. [PubMed] [Google Scholar]
- 69.Leunk RD. Production of a cytotoxin by Helicobacter pylori. Rev Infect Dis. 1991;13 (Suppl 8):S686–S689. doi: 10.1093/clinids/13.supplement_8.s686. [DOI] [PubMed] [Google Scholar]
- 70.Atherton JC, et al. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pyloriAssociation of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 71.Letley DP, Lastovica A, Louw JA, Hawkey CJ, Atherton JC. Allelic diversity of the Helicobacter pylori vacuolating cytotoxin gene in South Africa: rarity of the vacA s1a genotype and natural occurrence of an s2/m1 allele. J Clin Microbiol. 1999;37:1203–1205. doi: 10.1128/jcm.37.4.1203-1205.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sugimoto M, Zali MR, Yamaoka Y. The association of vacA genotypes and Helicobacter pylori-related gastroduodenal diseases in the Middle East. Eur J Clin Microbiol Infect Dis. 2009;28:1227–1236. doi: 10.1007/s10096-009-0772-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sugimoto M, Yamaoka Y. The association of vacA genotype and Helicobacter pylori-related disease in Latin American and African populations. Clin Microbiol Infect. 2009;15:835–842. doi: 10.1111/j.1469-0691.2009.02769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rhead JL, et al. A new Helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology. 2007;133:926–936. doi: 10.1053/j.gastro.2007.06.056. [DOI] [PubMed] [Google Scholar]
- 75.Hussein NR, et al. Differences in virulence markers between Helicobacter pylori strains from Iraq and those from Iran: potential importance of regional differences in H. pylori-associated disease. J Clin Microbiol. 2008;46:1774–1779. doi: 10.1128/JCM.01737-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Basso D, et al. Clinical relevance of Helicobacter pylori cagA and vacA gene polymorphisms. Gastroenterology. 2008;135:91–99. doi: 10.1053/j.gastro.2008.03.041. [DOI] [PubMed] [Google Scholar]
- 77.Ogiwara H, Graham DY, Yamaoka Y. vacA i-region subtyping. Gastroenterology. 2008;134:1267. doi: 10.1053/j.gastro.2007.11.062. [DOI] [PubMed] [Google Scholar]
- 78.Ogiwara H, et al. Role of deletion located between the intermediate and middle regions of the Helicobacter pylori vacA gene in cases of gastroduodenal diseases. J Clin Microbiol. 2009;47:3493–3500. doi: 10.1128/JCM.00887-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Figueiredo C, Machado JC, Yamaoka Y. Pathogenesis of Helicobacter pylori Infection. Helicobacter. 2005;10 (Suppl 1):14–20. doi: 10.1111/j.1523-5378.2005.00339.x. [DOI] [PubMed] [Google Scholar]
- 80.Fujimoto S, et al. Helicobacter pylori BabA expression, gastric mucosal injury, and clinical outcome. Clin Gastroenterol Hepatol. 2007;5:49–58. doi: 10.1016/j.cgh.2006.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu H, Yamaoka Y, Graham DY. Helicobacter pylori virulence factors: facts and fantasies. Curr Opin Gastroenterol. 2005;21:653–659. doi: 10.1097/01.mog.0000181711.04529.d5. [DOI] [PubMed] [Google Scholar]
- 82.Yamaoka Y, et al. Helicobacter pylori outer membrane proteins and gastroduodenal disease. Gut. 2006;55:775–781. doi: 10.1136/gut.2005.083014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yamaoka Y, Alm R. In: Helicobacter pylori: Molecular Genetics and Cellular Biology. Yamaoka Y, editor. Caister Academic Press; 2008. pp. 37–60. [Google Scholar]
- 84.Yamaoka Y. Increasing evidence of the roles of Helicobacter pylori SabA in the pathogenesis of gastroduodenal disease. J Infect Dev Ctries. 2008;2:174–181. doi: 10.3855/jidc.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yamaoka Y. Roles of H. pylori BabA in gastroduodenal pathogenesis. World J Gastroenterol. 2008;14:4265–4272. doi: 10.3748/wjg.14.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yamaoka Y, Kwon DH, Graham DY. A M(r) 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc Natl Acad Sci USA. 2000;97:7533–7538. doi: 10.1073/pnas.130079797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Odenbreit S, Kavermann H, Puls J, Haas R. CagA tyrosine phosphorylation and interleukin-8 induction by Helicobacter pylori are independent from alpAB, HopZ and bab group outer membrane proteins. Int J Med Microbiol. 2002;292:257–266. doi: 10.1078/1438-4221-00205. [DOI] [PubMed] [Google Scholar]
- 88.Ando T, et al. Polymorphisms of Helicobacter pylori HP0638 reflect geographic origin and correlate with cagA status. J Clin Microbiol. 2002;40:239–246. doi: 10.1128/JCM.40.1.239-246.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dossumbekova A, et al. Helicobacter pylori HopH (OipA) and bacterial pathogenicity: genetic and functional genomic analysis of hopH gene polymorphisms. J Infect Dis. 2006;194:1346–1355. doi: 10.1086/508426. [DOI] [PubMed] [Google Scholar]
- 90.Akanuma M, et al. The evaluation of putative virulence factors of Helicobacter pylori for gastroduodenal disease by use of a short-term Mongolian gerbil infection model. J Infect Dis. 2002;185:341–347. doi: 10.1086/338772. [DOI] [PubMed] [Google Scholar]
- 91.Yamaoka Y. Helicobacter pylori outer membrane proteins and gastric inflammation: author’s reply. Gut. 2006;55:1361. [PMC free article] [PubMed] [Google Scholar]
- 92.Yamaoka Y, et al. Role of interferon-stimulated responsive element-like element in interleukin-8 promoter in Helicobacter pylori infection. Gastroenterology. 2004;126:1030–1043. doi: 10.1053/j.gastro.2003.12.048. [DOI] [PubMed] [Google Scholar]
- 93.Choi IJ, Fujimoto S, Yamauchi K, Graham DY, Yamaoka Y. Helicobacter pylori environmental interactions: effect of acidic conditions on H pylori-induced gastric mucosal interleukin-8 production. Cell Microbiol. 2007;9:2457–2469. doi: 10.1111/j.1462-5822.2007.00973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lu H, et al. Functional and intracellular signaling differences associated with the Helicobacter pylori AlpAB adhesin from Western and East Asian strains. J Biol Chem. 2007;282:6242–6254. doi: 10.1074/jbc.M611178200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kudo T, et al. Pattern of transcription factor activation in Helicobacter pylori-infected Mongolian gerbils. Gastroenterology. 2007;132:1024–1038. doi: 10.1053/j.gastro.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu JY, et al. Balance between polyoma enhancing activator 3 and activator protein 1 regulates Helicobacter pylori-stimulated matrix metalloproteinase 1 expression. Cancer Res. 2006;66:5111–5120. doi: 10.1158/0008-5472.CAN-06-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lu H, et al. Regulation of interleukin-6 promoter activation in gastric epithelial cells infected with Helicobacter pylori. Mol Biol Cell. 2005;16:4954–4966. doi: 10.1091/mbc.E05-05-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tabassam FH, Graham DY, Yamaoka Y. OipA plays a role in Helicobacter pylori-induced focal adhesion kinase activation and cytoskeletal re-organization. Cell Microbiol. 2008;10:1008–1020. doi: 10.1111/j.1462-5822.2007.01104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tabassam FH, Graham DY, Yamaoka Y. Helicobacter pylori activate epidermal growth factor receptor- and phosphatidylinositol 3-OH kinase-dependent Akt and glycogen synthase kinase 3β phosphorylation. Cell Microbiol. 2009;11:70–82. doi: 10.1111/j.1462-5822.2008.01237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yamaoka Y, et al. Helicobacter pylori infection in mice: Role of outer membrane proteins in colonization and inflammation. Gastroenterology. 2002;123:1992–2004. doi: 10.1053/gast.2002.37074. [DOI] [PubMed] [Google Scholar]
- 101.Lu H, Hsu PI, Graham DY, Yamaoka Y. Duodenal ulcer promoting gene of Helicobacter pylori. Gastroenterology. 2005;128:833–848. doi: 10.1053/j.gastro.2005.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hussein NR, et al. Helicobacter pylori dupA is polymorphic, and its active form induces proinflammatory cytokine secretion by mononuclear cells. J Infect Dis. 2010;202:261–269. doi: 10.1086/653587. [DOI] [PubMed] [Google Scholar]
- 103.Schmidt HM, et al. The prevalence of the duodenal ulcer promoting gene (dupA) in Helicobacter pylori isolates varies by ethnic group and is not universally associated with disease development: a case-control study. Gut Pathog. 2009;1:5. doi: 10.1186/1757-4749-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yamaoka Y. Roles of the plasticity regions of Helicobacter pylori in gastroduodenal pathogenesis. J Med Microbiol. 2008;57:545–553. doi: 10.1099/jmm.0.2008/000570-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Arachchi HSJ, et al. Prevalence of duodenal ulcer-promoting gene (dupA) of Helicobacter pylori in patients with duodenal ulcer in North Indian population. Helicobacter. 2007;12:591–597. doi: 10.1111/j.1523-5378.2007.00557.x. [DOI] [PubMed] [Google Scholar]
- 106.Zhang Z, et al. The Helicobacter pylori duodenal ulcer promoting gene, dupA in China. BMC Gastroenterol. 2008;8:49. doi: 10.1186/1471-230X-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Douraghi M, et al. dupA as a risk determinant in Helicobacter pylori infection. J Med Microbiol. 2008;57:554–562. doi: 10.1099/jmm.0.47776-0. [DOI] [PubMed] [Google Scholar]
- 108.Gomes LI, et al. Lack of association between Helicobacter pylori infection with dupA-positive strains and gastroduodenal diseases in Brazilian patients. Int J Med Microbiol. 2007;298:223–230. doi: 10.1016/j.ijmm.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 109.Pacheco AR, et al. Involvement of the Helicobacter pylori plasticity region and cag pathogenicity island genes in the development of gastroduodenal diseases. Eur J Clin Microbiol Infect Dis. 2008;27:1053–1059. doi: 10.1007/s10096-008-0549-8. [DOI] [PubMed] [Google Scholar]
- 110.Nguyen LT, et al. Helicobacter pylori dupA gene is not associated with clinical outcomes in the Japanese population. Clin Microbiol Infect. 2010;16:1264–1269. doi: 10.1111/j.1469-0691.2009.03081.x. [DOI] [PubMed] [Google Scholar]
- 111.Argent RH, Burette A, Miendje Deyi VY, Atherton JC. The presence of dupA in Helicobacter pylori is not significantly associated with duodenal ulceration in Belgium, South Africa, China, or North America. Clin Infect Dis. 2007;45:1204–1206. doi: 10.1086/522177. [DOI] [PubMed] [Google Scholar]
- 112.Hussein NR. The association of dupA and Helicobacter pylori-related gastroduodenal diseases. Eur J Clin Microbiol Infect Dis. 2010;29:817–821. doi: 10.1007/s10096-010-0933-z. [DOI] [PubMed] [Google Scholar]
- 113.Queiroz DM, et al. dupA polymorphisms and risk of distal gastric cancer. Gastroenterology. 2008;134:A-85. [Google Scholar]
- 114.Kersulyte D, et al. Cluster of type IV secretion genes in Helicobacter pylori’s plasticity zone. J Bacteriol. 2003;185:3764–3772. doi: 10.1128/JB.185.13.3764-3772.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kersulyte D, et al. Helicobacter pylori’s plasticity zones are novel transposable elements. PLoS ONE. 2009;4:e6859. doi: 10.1371/journal.pone.0006859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fischer W, et al. Strain-specific genes of Helicobacter pylori: genome evolution driven by a novel type IV secretion system and genomic island transfer. Nucleic Acids Res. doi: 10.1093/nar/gkq378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Couturier MR, Tasca E, Montecucco C, Stein M. Interaction with CagF is required for translocation of CagA into the host via the Helicobacter pylori type IV secretion system. Infect Immun. 2006;74:273–281. doi: 10.1128/IAI.74.1.273-281.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pattis I, Weiss E, Laugks R, Haas R, Fischer W. The Helicobacter pylori CagF protein is a type IV secretion chaperone-like molecule that binds close to the C-terminal secretion signal of the CagA effector protein. Microbiology. 2007;153:2896–2909. doi: 10.1099/mic.0.2007/007385-0. [DOI] [PubMed] [Google Scholar]
- 119.Jimenez-Soto LF, et al. Helicobacter pylori type IV secretion apparatus exploits beta1 integrin in a novel RGD-independent manner. PLoS Pathog. 2009;5:e1000684. doi: 10.1371/journal.ppat.1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]