Abstract
The extracellular effect of fibroblast growth factor-12 (FGF12) remains unknown because FGF12 cannot activate any fibroblast growth factor receptors (FGFRs), and FGF12 is not currently thought to be released from cells. We reported previously that FGF12 plays an intracellular role in the inhibition of radiation-induced apoptosis. In this study, we demonstrated that recombinant FGF12 was able to be internalized into the cytoplasm of a rat intestinal epithelial cell line, IEC6, and this process was dependent on two novel cell-penetrating peptide (CPP) domains (CPP-M and CPP-C). In particular, CPP-C, composed of ∼10 amino acids, was identified as a specific domain of FGF12 and its subfamily in the C-terminal region (residues 140–149), although CPP-M was a common domain in the internal region of the FGF family. The absence of CPP-C from FGF12 or a mutation (E142L) in the CPP-C domain drastically reduced the internalization of FGF12 into cells. Therefore, CPP-C played an essential role in the internalization of FGF12. In addition, CPP-C was able to deliver other polypeptides into cells as a CPP because an FGF1/CPP-C chimeric protein was internalized into IEC6 cells more efficiently than wild-type FGF1. Finally, intraperitoneally added FGF12 inhibited radiation-induced apoptosis in the intestinal epithelial cells of BALB/c mice, and deletion of the CPP-C domain decreased the inhibition of the apoptosis. These findings suggest that exogenous FGF12 can play a role in tissues by translocating into cells through the plasma membrane, and the availability of this novel CPP provides a new tool for the intracellular delivery of bioactive molecules.
Keywords: Apoptosis, Growth Factors, Intestine, Peptides, Protein Domains, FGF, Cell-penetrating Peptide, Cellular Internalization, Radiation-induced Apoptosis
Introduction
Fibroblast growth factors (FGFs)2 play important roles in embryogenesis, angiogenesis, and wound repair, and the FGF family is currently composed of 22 members in humans (1). FGF12, initially designated as FGF homologous factor 1 (FHF1), was identified by its sequence homology to known FGFs, and it has a high degree of homology with FGF11 (FHF2), FGF12 (FHF3), and FGF14 (FHF4), with a 58–71% amino acid sequence identity with these FGF11 subfamily members (2). However, FGF12 shows less than 30% amino acid identity with other FGFs (2). In addition, FGF12 and other FGF11 subfamily members do not activate any fibroblast growth factor receptors (FGFRs), although they can bind to heparin with high affinity like other FGFs (3). FGF12 has structural similarity with FGF1 and FGF2, in that it lacks a classical signal sequence and contains a nuclear localization signal (NLS), resulting in the accumulation of FGF12 in the nucleus without any release from cells (2). Therefore, it remains unknown whether FGF12 is able to act extracellularly like other FGFs. FGF12 has two forms, a long form (FGF12A) and a short form (FGF12B). FGF12B lacks the NLS because the N-terminal 66 amino acid residues of FGF12A are substituted by 4 amino acids by means of alternative splicing.
Several FGFs (FGF1, FGF2, FGF4, FGF7, FGF10, and FGF20) are able to inhibit radiation-induced tissue damage and are expected to have clinical uses (4–6). Therefore, a lot of effort has been made to improve FGFs for greater biological activity and therapeutic effects (6–8). Generally, FGFs can function through the activation of surface FGFRs, while receptor-bound FGF1 can be endocytosed to reach the nucleus via the presence of a NLS, leading to DNA synthesis and cell proliferation (9, 10). Its translocation depends on binding with FGFR1 and FGFR4 (11) and requires phosphatidylinositol 3-kinase (PI3K) activity (12) and Hsp90 (13). Moreover, FGF1 can interact with intracellular proteins such as FIBP (14), p34 (15), casein kinase 2 (CK2) (16), and mortalin (17), suggesting that endocytosed FGF1 plays multiple roles inside cells. Interestingly, overexpression of the FGF12 gene down-regulated radiation-induced apoptosis in the human leukemic mast cell line HMC-1, indicating that FGF12 is able to inhibit radiation-induced tissue damage like other FGFs (18). Moreover, it was reported that FGF12 could bind to the C terminus of the cardiac voltage-gated sodium channel to modulate its properties (19). Thus, we speculated that externally added FGF12 might translocate into the cytoplasm through the plasma membrane to exert its effects on various cells and tissues.
Cell-penetrating peptides (CPPs) or protein transduction domains are short peptide sequences identified from cellular and viral proteins that mediate cellular internalization through the cell membranes. They can be efficiently taken up by cells, and CPPs are expected to be able to deliver a variety of bioactive molecules into cells. For example, the transactivator protein of human HIV-1 (Tat) (20, 21), fragments of antennapedia of Drosophila (penetratin) (22), and transportan (TP-10) (23, 24) are the principal CPPs that have been suggested as intracellular delivery vectors. Among the FGF family, a hydrophobic peptide in FGF4 was identified as a CPP (25). The TAT peptide is one of the most common CPPs and is characterized as rich in arginine residues. Therefore, artificial arginine-rich peptides were developed as more effective transporters (26, 27). Internalization of CPPs was initially thought to be independent of endocytosis because it was reported that cellular uptake was not affected by incubation at low temperature (27–29) and CPPs were internalized by a receptor-independent mechanism (30). However, it was recognized later that cell fixation caused the artifactual uptake of CPPs and further study using live cells showed that endocytic pathways were involved in the cellular uptake of TAT and arginine-rich peptides (31).
This study describes that (i) FGF12 can be internalized into cells depending on a novel CPP domain (CPP-C); (ii) CPP-C can play a role in delivering other polypeptides into cells as a CPP; and (iii) intraperitoneally added FGF12 inhibited radiation-induced apoptosis in mice and deletion of the CPP-C domain decreased the inhibition of apoptosis. This provides the first evidence that exogenous FGF12 can play a role in vivo.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
The rat intestinal epithelial cell line IEC6 was provided by RIKEN BioSource Center (Tsukuba, Japan). IEC6 cells were maintained in medium consisting of Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Grand Island, NY) supplemented with 5% fetal calf serum (FCS) and 4 μg/ml insulin. Anti-cleaved caspase-3 antibody (Asp-175) was purchased from Cell Signaling (Danvers, MA). Recombinant human FGF12B without any tags was purchased from R&D Systems (Minneapolis, MN). Recombinant human FGF12A linked to a 3xFLAG-His6 tag was produced using the same procedure as described previously (32). Recombinant human FGF1 without any tags was produced as described previously (9).
Mice
Male BALB/c mice weighing 23–28 g were obtained from Japan Clea (Tokyo, Japan). Mice were housed in an air-conditioned room in a specific-pathogen-free facility, and they were irradiated at 8 weeks old with γ-rays from a 137Cs source (Gammacell 40, Atomic Energy of Canada, Ottawa, Canada). All protocols complied with the Guidelines on Animal Experiments from National Institute of Radiological Sciences (NIRS) and were approved by the Laboratory Animal Safety and Ethics Council of NIRS.
Construction of FGF12B Mutants with C-terminal Deletions
The coding sequence of FGF12A was obtained from Invitrogen. Each gene was amplified using a gene-specific primer set. Forward primer: 5′-ggg gac aag ttt gta caa aaa agc agg ctt cac cat gta ccc ata cga tgt tcc aga tta cgc tga gag caa aga acc cca gct caa agg gat tgt gac aag gt-3′ containing an HA tag encoding sequence. Reverse primers were as follows: FGF12B, 5′-ggg gac cac ttt gta caa gaa agc tgg gtc cta cta tgt tga atc ttg att cac aac ttt gcc-3′; Δ170–181, 5′-ggg gac cac ttt gta caa gaa agc tgg gtc cta ggt tgg tgt tcc aga act ttt cct tga acg-3′; Δ160–181, 5′-ggg gac cac ttt gta caa gaa agc tgg gtc cta ccc ttg ttt ttc tcc aat ttc atg tag cga-3′; Δ150–181, 5′-ggg gac cac ttt gta caa gaa agc tgg gtc cta tgg ttc tct gta cat aca cac ttc aat agg-3′, and Δ140–181, 5′-ggg gac cac ttt gta caa gaa agc tgg gtc cta ttt cgg tac aaa atg tga tga ggg ctt ggt-3′. Each PCR fragment was introduced into a pDONR221 vector for cloning of the gene in accordance with the instructions for GATEWAY Cloning Technology (Invitrogen) and confirmed by sequencing. Then, each gene was transferred by recombination from its entry clone into a pDEST17 vector, which is an N-terminal fusion vector and containing an ATG initiation codon upstream a sequence encoding a His6 tag.
Construction of FGF12B Mutants with Internal Deletions
Each of the amino acid residues 30–59, 50–79, 70–99, and 80–109 of FGF12B were deleted from the FGF12B protein to create FGF12B mutants (Δ30–59, Δ50–79, Δ70–99, and Δ80–109, respectively). Two HindIII sites were introduced into the appropriate sites of pDONR221-HA-FGF12B using a QuikChange site-directed mutagenesis kit in accordance with the manufacturer's protocol (Stratagene, La Jolla, CA). These sites were digested with HindIII and self-ligated to delete a 90-base fragment from the FGF12B gene. Then each gene was transferred by recombination from the entry clone into a pET57 (Novagen) expression vector, which is a N-terminal fusion vector containing an ATG initiation codon upstream sequences encoding a His6 tag and a Nus tag, in accordance with the instructions for GATEWAY Cloning Technology (Invitrogen).
Construction of an FGF12B Mutant with a Single Substitution
Single substitutions of E142L were created in the CPP-C domains of FGF12B and protein Δ80–109 (E142L and Δ80–109/E142L). A mutation was introduced into each gene in the pDONR221 vector using a QuikChange site-directed mutagenesis kit. The primer set was 5′-gta ccg aaa cct att cta gtg tgt atg tac-3′ and 5′-gta cat aca cac tag aat agg ttt cgg tac-3′. The E142L gene was transferred by recombination from its entry clone into a pDEST17 vector, whereas the Δ80–109/E142L gene was cloned into a pET57 vector and fused to a Nus Tag.
Construction of an FGF1/CPP-C Fusion Protein
The coding sequence of FGF1 was obtained from Invitrogen. EcoRI-SalI sites were created in the C-terminal end of the FGF1 gene by PCR using a gene-specific primer set; 5′-ggg gac aag ttt gta caa aaa agc agg ctt cac cat ggc tga agg gga aat cac cac ctt cac a-3′ and 5′-ggg gac cac ttt gta caa gaa agc tgg gtc tta gtc gac gaa ttc cag ggg gag aaa caa gat tgc ttt ctg gcc-3′. The PCR fragment was introduced into a pDONR221 vector and confirmed by sequencing. The DNA fragment for FGF12B residues 140–149 (CPP-C) was synthesized and cloned into the FGF1 construct pre-digested with EcoRI and SalI. Finally, the insert in the pDONR221 vector was transferred into a pDEST17 expression vector by recombination in accordance with the instructions for GATEWAY Cloning Technology (Invitrogen).
Expression and Purification of Recombinant Proteins
BL21(DE3)pLysS E. coli cells were transformed with pDEST17 or pET57 expression constructs. Protein expression was induced using an Overnight Express Autoinduction System 1 in accordance with the manufacturer's instructions (Novagen). Briefly, a non-inducing, log phase starter culture was diluted 1:100 in Overnight Express System medium supplemented with ampicillin and chloramphenicol, and incubated for 18 h at 37 °C with shaking. The cell pellet was suspended in BugBuster Master Mix (Novagen) containing 1 mm phenylmethylsulfonyl fluoride (PMSF), 1 mm benzamidine and EDTA-free mixture (Complete) (Roche Applied Science) and incubated with slow shaking for 20 min at room temparture. The insoluble cell debris was removed by centrifugation, and then the soluble extracts were purified using His SpinTrap columns (GE Healthcare).
Fluorescent Labeling of Recombinant Proteins
Recombinant proteins were labeled with Alexa Fluor 568 dye using an Alexa Fluor 568 Protein Labeling kit (Invitrogen). After 25 μg of each protein was diluted to a final volume of 0.1 ml with phosphate-buffered saline (PBS), sodium bicarbonate was added into the protein solution at a final concentration of 0.1 m. The protein solution was transferred to the vial of Alexa Fluor 568 reactive dye. This reaction mixture was stirred for 1 h at room temperature using a magnetic stir bar. The labeled proteins were purified using BioSpin 6 columns in accordance with the manufacturer's instructions (Bio-Rad).
Flow Cytometry
IEC6 cells were subjected to flow cytometry to quantify cellular uptake of fluorescently labeled FGFs and peptides. The cells were harvested in trypsin-EDTA and washed twice with PBS containing 0.2% bovine serum albumin (BSA). A suspension of cells was subjected to FACS Calibur flow cytometry (BD Biosciences, CA) without any cell fixation to measure the intensity of fluorescence.
TUNEL Assay
BALB/c mice were given intraperitoneally each FGF diluted in saline containing 5% mouse serum 24 h before total body irradiation (TBI) at 12 Gy, and the jejunum was removed 24 h after irradiation. Apoptosis was evaluated with paraffin-embedded sections of the jejunum by analysis of terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick end labeling (TUNEL) using an ApopTag Plus Peroxidase In Situ Apoptosis Detection kit in accordance with the manufacturer's protocol (Chemicon, Temecula, CA) (33). The number of TUNEL+ cells was determined in 10 sequential cells of each crypt.
RESULTS
Externally Added FGF12 Inhibits Radiation-induced Apoptosis in the Intestinal Crypt Cells
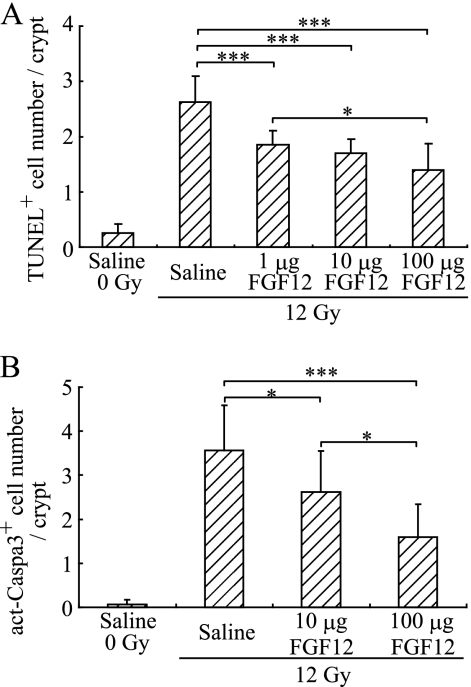
TUNEL assay was performed on paraffin-embedded sections of the jejunum from BALB/c mice to evaluate apoptosis 24 h after TBI at 12 Gy as described previously (4, 6). Recombinant FGF12B without heparin was administered intraperitoneally to the mice at a dose of 1, 10, or 100 μg of 24 h before irradiation. As a result, intraperitoneally administered FGF12B significantly inhibited radiation-induced apoptosis almost in a dose-dependent manner (Fig. 1A), and after treatment with 1–100 μg of FGF12B, the level of apoptosis declined to the range from 50–70% compared with untreated crypts. In addition, radiation induced the activation of caspase-3 in crypt cells 8 h after irradiation because the cells underwent apoptosis. Indeed, FGF12B prominently decreased the appearance of activated caspase-3-positive cells in a dose-dependent manner (Fig. 1B). These findings indicate that extracellular FGF12 is able to decrease radiation-induced apoptosis.
FIGURE 1.
Effects of FGF12 on radiation-induced apoptosis in intestinal crypt cells. BALB/c mice were given TBI at 12 Gy to induce apoptosis in the crypts of the jejunum. Recombinant FGF12B (R&D) was administered intraperitoneally without heparin to BALB/c mice at the indicated doses 24 h before TBI. A TUNEL assay and staining with anti-cleaved caspase 3 antibody were performed on paraffin-embedded sections to evaluate apoptosis in the crypts of the jejunum, as described previously (4). A, jejunum was removed from treated mice 24 h after irradiation. The number of TUNEL+ cells was determined in each crypt by screening more than 10 crypts. B, number of cleaved caspase 3-positive cells was determined in each crypt 8 h after irradiation. All values are means ± S.D. (n = 6). Similar findings were observed in two independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Extracellular FGF12 Can Be Internalized into the Cytoplasm
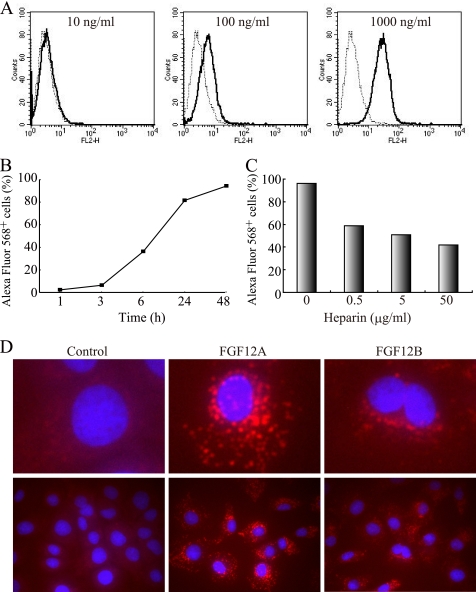
The fluorescence level of a rat intestinal epithelial cell line, IEC6, was evaluated after 24 h culture with Alexa Fluor 568-labeled recombinant FGF12 to examine internalization of FGF12 into cells. IEC6 cells were cultured for 24 h in complete medium with 10, 100, or 1000 ng/ml of Alexa Fluor 568-labeled FGF12B and then subjected to flow cytometric analysis (Fig. 2A). The level of fluorescence of cells increased in a dose-dependent manner after treatment with FGF12. Representative data of FACS histograms showed that the proportion of fluorescence-positive cells was 96% in cells incubated with 1000 ng/ml FGF12. The level of Alexa Fluor 568-positive cells was determined up to 48 h after the addition of FGF12 (Fig. 2B). The kinetics of cellular uptake of extracellular FGF12 was examined by flow cytometry. An increase in cell fluorescence was detected 1 h after administration of FGF12 to the cell culture, and it rose gradually up to 48 h. The proportion of fluorescence-positive cells at 1, 3, and 6 h were 2.4, 6.3, and 36.1%, respectively and reached more than 80% after 24 h. In addition, FGFs have heparin-binding domains, so that FGF12 can also bind to heparin with high affinity (3). Therefore, the presence of heparin in the culture medium affected the proportions of Alexa Fluor 568-positive cells (Fig. 2C). IEC6 cells were cultured for 24 h in medium with 1 μg/ml of Alexa Fluor 568-labeled FGF12B in the presence of 0.5, 5, or 50 μg/ml of heparin, and the proportions of fluorescence-positive cells decreased drastically in a dose-dependent manner with the increasing concentration of heparin. The rates of Alexa Fluor 568-positive cells ranged from 40% to 60% in the cultured cells with heparin; however, heparin could not completely prevent FGF12 entry even at 500 μg/ml (data not shown). Finally, the distribution of incorporated FGF12 in cells was examined by confocal microscopy (Fig. 2D). The confocal microscopy images indicated that FGF12 was localized mainly in the cytoplasm of IEC6 cells after 24 h culture and not on the cell surface. In addition, FGF12A was also detected in the nuclei of the cells, although FGF12B was not detected in the nuclei. These findings indicate that recombinant FGF12 protein is able to specifically enter IEC6 cells with relatively slow kinetics and FGF12 internalization can be inhibited strongly by heparin.
FIGURE 2.
Internalization of FGF12 into IEC6 cells. Human recombinant FGF12 was labeled with Alexa Fluor 568. FGF12B (R&D) had no tags, whereas FGF12A was linked to a 3xFLAG-His6 tag as described previously (32). A rat intestinal epithelial cell line, IEC6, was cultured in complete medium with Alexa Fluor 568-labeled FGF12 A, IEC6 cells were incubated with FGF12B at a dose of 10, 100, or 1000 ng/ml for 24 h. They were harvested in trypsin-EDTA, washed twice with phosphate-buffered saline (PBS) containing 0.2% bovine serum albumin (BSA), and subjected to FACS Calibur flow cytometry to estimate fluorescence intensity. A dotted line shows the control cells and a solid line shows the FGF12-treated cells. B, kinetics of cellular uptake of extracellular FGF12 was examined over 48 h by flow cytometry. IEC6 cells were incubated in culture medium with 1 μg/ml of FGF12B and subjected to flow cytometry to determine the percentage of Alexa Fluor 568-positive cells at the indicated time points. C, IEC6 cells were cultured for 24 h in medium with 1 μg/ml of FGF12B and 0.5, 5, or 50 μg/ml of heparin. They were subjected to flow cytometry. D, IEC6 cells were cultured for 24 h in medium with 1 μg/ml of FGF12A or FGF12B. The cells were fixed in 1% glutaraldehyde, and the nuclei were visualized by staining with 20 μg/ml of Hoechst 33342 (blue) and the fluorescence confocal images were acquired using a IX81 fluorescence microscope with a disk scanning unit (Olympus, Tokyo, Japan).
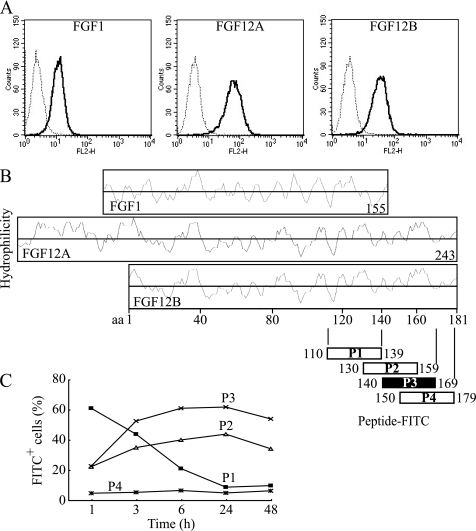
The C-terminal Amino Acid Sequence of FGF12 Is Involved in Cellular Internalization
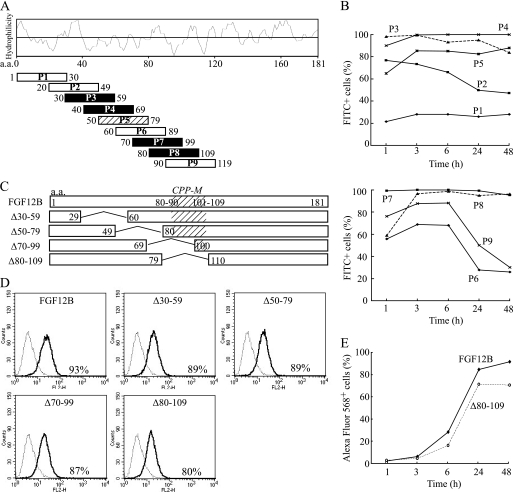
FGF12 has structural similarity with FGF1; however, FGF1 was internalized much less than FGF12 (Fig. 3A). A significant structural difference between these FGFs is the C-terminal sequence because FGF12 possesses a unique C-terminal polypeptide next to a highly homologous C-terminal region of both FGFs (Fig. 3B). Therefore, this C-terminal region was suspected to play a role in cellular internalization. Four peptides containing 30 amino acids each were synthesized on the basis of the FGF12B C-terminal polypeptide sequence and conjugated with fluorescein isothiocyanate (FITC). Each peptide was added to the culture of IEC6 cells at a concentration of 10 μg/ml, and the kinetics of peptide internalization was analyzed by FACS. Peptide P3, which corresponded to FGF12B residues 140–169, was internalized most readily after 24 h culture (supplemental Fig. S1A). Peptide P3 was rapidly internalized, reaching a maximum at 24 h after addition to the medium (Fig. 3C). Peptide P2 (130–159) showed a similar kinetics to peptide P3 (140–149). In contrast, peptide P4 (150–179) was not internalized at all. The kinetics of peptide P1 (110–139) internalization were completely different from that of FGF12, although peptide P1 was internalized most readily at 1 h. Therefore, the region of FGF12 corresponding to peptides P1 and P4 was not likely to be involved in FGF12 internalization. Confocal microscopy confirmed that peptide P3 could be internalized because of its distribution in the cytoplasm (supplemental Fig. S1B). As a result, FGF12B residues 140–149 were suggested to be the most likely as the cell-penetrating domain of FGF12.
FIGURE 3.
C-terminal amino acid sequence of FGF12 involved in cellular internalization. A, human recombinant FGF1, FGF12A, and FGF12B (R&D) were labeled with Alexa Fluor 568. FGF1 and FGF12B had no tag, whereas FGF12A had a 3xFLAG-His6 tag. IEC6 cells were incubated with 1 μg/ml of each Alexa Fluor 568-labeled FGF for 24 h. The cells were harvested in trypsin-EDTA, washed twice with PBS containing 0.2% BSA, and subjected to FACS Calibur flow cytometry to estimate fluorescence intensity. A dotted line shows the control cells, and a solid line shows the FGF-treated cells. B, each Hopp-Woods scale, hydrophilicity plot, for FGF1, FGF12A, and FGF12B was compared with identify the possible domains of FGFs. Four peptides containing 30 amino acid residues conjugated with FITC were synthesized on the basis of the C-terminal sequence of FGF12, and then purified to more than 95% purity using HPLC as specified by the manufacturer (Invitrogen). C, kinetics of cellular uptake of FITC-labeled peptides were examined over 48 h by FACS. IEC6 cells were cultured with 10 μg/ml of each FITC-labeled peptide and subjected to flow cytometry to assess the level of cellular uptake of each peptide.
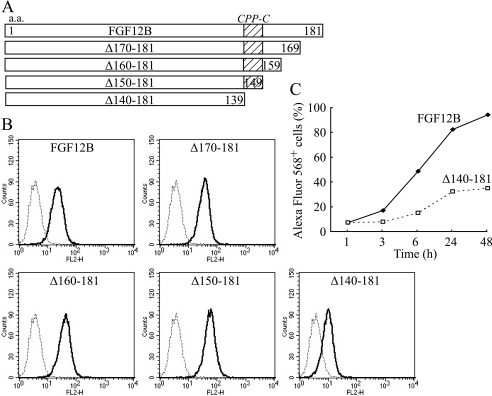
C-terminal Deletion of FGF12 Reduces Cellular Internalization
FGF12 mutant proteins were created on the basis of peptide studies to identify CPP domains. Amino acid residues 170–181, 160–181, 150–181, and 140–181 of FGF12B were deleted from the FGF12B protein (Δ170–181, Δ160–181, Δ150–181, and Δ140–181, respectively) (Fig. 4A) and labeled with Alexa Fluor 568. IEC6 cells were cultured for 24 h in complete medium with 1000 ng/ml mutant proteins and then subjected to flow cytometric analysis (Fig. 4B). Δ140–181 drastically reduced cellular internalization to 32% after 24 h culture. Cellular internalization increased gradually up to 48 h after administration of Δ140–181 to the cell culture but reached only 35% at 48 h (Fig. 4C). In contrast, Δ170–181, Δ160–181, and Δ150–181 did not reduce it, but rather slightly increased cellular uptake of the mutant proteins. Therefore, residues 140–149 of FGF12B may play an essential role in FGF12 internalization, and this region was named CPP-C.
FIGURE 4.
Decrease in FGF12 internalization through C-terminal deletion. A, four FGF12B mutants were created by C-terminal deletion of FGF12B. Amino acid residues 170–181, 160–181, 150–181, and 140–181 of FGF12B were deleted from the FGF12B protein (Δ170–181, Δ160–181, Δ150–181, and Δ140–181, respectively). B, IEC6 cells were incubated in complete medium with 1 μg/ml Alexa Fluor 568-labeled FGF12 mutant protein for 24 h. They were analyzed by flow cytometry to estimate fluorescence intensity. C, kinetics of cellular uptake of Alexa Fluor 568-labeled Δ140–181 protein were examined over 48 h by FACS.
An Internal Region of FGF12 May Be Involved in Cellular Internalization
Nine peptides containing 30 amino acid residues each were synthesized on the basis of the FGF12B polypeptide sequence to examine the involvement in cellular internalization except C-terminal peptides (Fig. 5A). Unexpectedly, several peptides were internalized very quickly and efficiently. In particular, peptides P3, P4, P7, and P8 could translocate into almost all the cells after 3 h culture and remained as intracellular peptides up to 48 h (Fig. 5B). Confocal microscopy images indicated that these peptides were actually localized in the cytoplasm of IEC6 cells (data not shown). However, the kinetics of internalization of these peptides were different from that of FGF12. Therefore, FGF12 deletion mutants were created to examine the involvement of these peptide sequences in cellular internalization of FGF12 protein. Amino acid residues 30–59, 50–79, 70–99, and 80–109 of the FGF12B were deleted from the FGF12B protein (Δ30–59, Δ50–79, Δ70–99, and Δ80–109, respectively) (Fig. 5C). The mutant proteins were fused to a Nus tag to enhance their solubility using pET57 expression vector because all the mutant proteins were accumulated in insoluble inclusion bodies of E. coli without the Nus tag. As a result, internalization of these mutant proteins decreased slightly (Fig. 5D). In particular, Δ80–109 reduced cellular internalization more than the other mutant proteins, although the level of internalization of Δ80–109 was still high (Fig. 5, D and E). Thus, amino acid residues 80–109 of FGF12B include another cell-penetrating peptide domain (CPP-M), which may be distributed from amino acid position 80–90 to 101–109 (Fig. 5C).
FIGURE 5.
Possible involvement of an internal region of FGF12 in cellular internalization. A, each 30 amino acid peptide was synthesized on the basis of the N-terminal and internal FGF12B sequence and conjugated with FITC. A Hopp-Woods scale, hydrophilicity plot for FGF12B, is shown to compare the hydrophilicity of all peptides. Black squares indicated the most likely CPPs on the basis of FACS analysis and a slash square is the next most likely. B, kinetics of cellular uptake of FITC-labeled peptides were examined over 48 h by flow cytometry. IEC6 cells were cultured with 10 μg/ml of each FITC-labeled peptide and subjected to flow cytometry to assess the level of cellular uptake of each peptide. C, each of the amino acid residues 30–59, 50–79, 70–99, and 80–109 of FGF12B were deleted from the FGF12B protein to create FGF12B mutants (Δ30–59, Δ50–79, Δ70–99, and Δ80–109, respectively). As a result, FGF12B and mutant proteins were fused to a Nus tag. D, IEC6 cells were incubated in complete medium with 1 μg/ml Alexa Fluor 568-labeled FGF12 mutant proteins for 24 h. They were analyzed by flow cytometry to estimate fluorescence intensity. E, kinetics of cellular uptake of Alexa Fluor 568-labeled wild-type FGF12B and Δ80–109 were examined over 48 h by FACS.
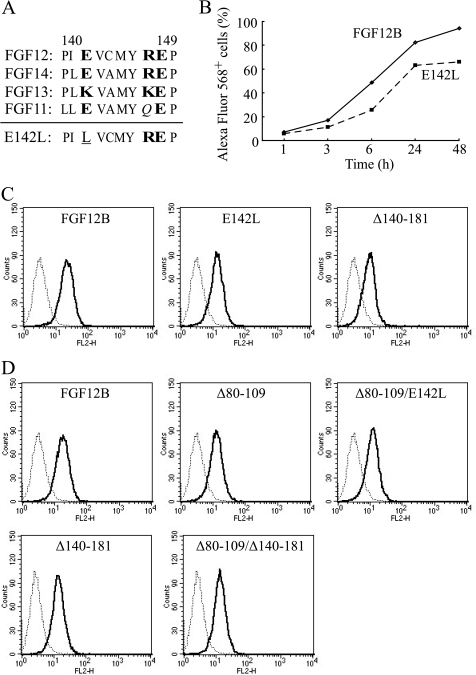
A Single Mutation of CPP-C Decreases Cellular Internalization
Studies of synthesized peptides and deletion mutant proteins suggested that amino acid residues 140–149 (CPP-C) were an essential a CPP domain for FGF12 internalization. To further characterize the CPP-C domain for cellular internalization, a single mutation was introduced into the CPP-C sequence in FGF12B. FGF12 belongs to an FGF11 subfamily consisting of FGF11-FGF14, which share high homology at the CPP-C domain (Fig. 6A). Seven hydrophobic and three hydrophilic amino acids constituted the CPP-C. Therefore, we chose residue 142 for mutation because it was a single hydrophilic residue among a hydrophobic sequence and was conserved well in this subfamily. Hydrophilic glutamic acid (E) at position 142 in FGF12B was substituted with hydrophilic lysine (L) (E142L). This mutation reduced cellular internalization significantly and was shown to be an essential region for cellular uptake (Fig. 6, B and C), although E142L substitution reduced uptake less than the deletion mutation of residues 140–181 (Fig. 6C). Taken together, residues 140–149 (CPP-C) of FGF12B functioned as a CPP domain and played an important role in cellular internalization of FGF12.
FIGURE 6.
Decrease in FGF12 internalization through a single mutation in the C-terminal region of FGF12. A, amino acid sequences of the FGF11 subfamily corresponding to amino acids 140–149 (CPP-C) of FGF12B are shown. Plain letters indicate hydrophobic amino acids. Hydrophilic amino acid residues are printed in bold letters and neutral amino acid residues are in italics. A single substitution, E142L, was introduced into the CPP-C domain, shown underlined, to examine the contribution of CPP-C to cellular uptake. B, kinetics of cellular uptake of Alexa Fluor 568-labeled wild-type FGF12B and FGF12B mutant E142L were examined over 48 h by FACS. C, representative FACS histograms are shown in red-orange fluorescence Alexa 568-positive cell populations after 24 h culture with 1 μg/ml of FGF12B, E142L, and Δ140–181. D, single substitution E142L and C-terminal deletion of residues 140–181 were made in the Δ80–109 proteins (Δ80–109/E142L and Δ80–109/Δ140–181, respectively) to examine any additive effect of CPP-M and CPP-C on cellular internalization. Each recombinant protein was fused to a Nus tag. IEC6 cells were incubated in complete medium with 1 μg/ml Alexa Fluor 568-labeled FGFs for 24 h. The cells were analyzed by flow cytometry to estimate fluorescence intensity.
Both CPP-C and CPP-M Are Necessary for Internalization of FGF12
Two CPP domains, CPP-M and CPP-C, were found in FGF12. The level of internalization of Δ140–181 with a Nus tag (Fig. 6D) was higher than that of Δ140–181 without a Nus tag (Fig. 6C). Therefore, Δ80–109 and Δ140–181 were fused to a His6-Nus tag to compare the extent of cellular internalization (Fig. 6D). Under these experimental conditions, Δ80–109 and Δ140–181 were internalized almost equally. In addition, a single substitution E142L and C-terminal deletion of residues 140–181 were made in protein Δ80–109 (Δ80–109/E142L and Δ80–109/Δ140–181) to examine any mutual effects of CPP-M and CPP-C on cellular internalization. If the effects of CPP-M or CPP-C are additive, Δ80–109/E142L and Δ80–109/Δ140–181 would be internalized much less than Δ80–109. However, Δ80–109, Δ80–109/E142L, and Δ80–109/Δ140–181 were internalized almost equally, suggesting that CPP-M and CPP-C did not exert an additive independent effect on internalization. Therefore, both of them were necessary for internalization.
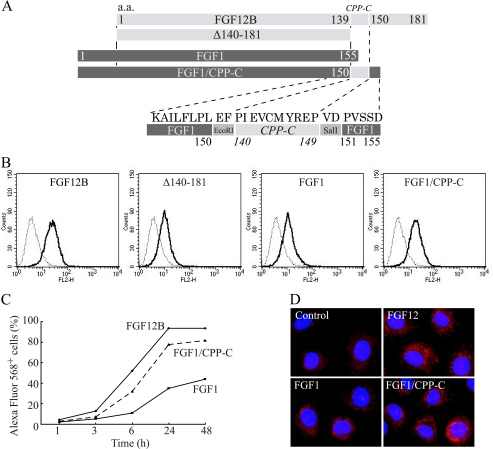
FGF1 Internalization Is Enhanced by the Addition of CPP-C to the C terminus
To examine whether the CPP-C peptide sequence could deliver other polypeptide into cells, CPP-C was fused to the C-terminal end of human FGF1 (Fig. 7A). FGF12 and FGF1 have structural similarity and share a similar alignment of their amino acid sequences. However, FGF1 lacks the corresponding CPP-C domain (Fig. 3B). Therefore, a FGF1/CPP-C fusion protein was created following the alignment of the FGF12 C-terminal region. FGF1 and Δ140–181 were internalized very weakly because they had no CPP-C domains. On the other hand, FGF1/CPP-C was internalized into IEC6 cells after culture for 24 h much more than wild-type FGF1 (Fig. 7B). In addition, the kinetics of cellular uptake of FGF1/CPP-C were almost consistent with that of FGF12 (Fig. 7C). Confocal microscopy images indicated that FGF1/CPP-C translocated into cells and localized mainly in the cytoplasm (Fig. 7D). These findings suggested that the CPP-C peptide could deliver other polypeptides into cells and serve as an intracellular delivery vector.
FIGURE 7.
Enhancement of FGF1 internalization by CPP-C. A, amino acid residues 140–149 (CPP-C) of FGF12B were fused to the C-terminal end of the human FGF1 sequence to create an FGF1/CPP-C fusion protein (FGF1/CPP-C) to examine the capability of CPP-C to deliver FGF1 into cells. B, IEC6 cells were incubated in complete medium with 1 μg/ml Alexa Fluor 568-labeled FGF12B, Δ140–181, FGF1, or FGF1/CPP-C for 24 h. The cells were analyzed by flow cytometry to estimate fluorescence intensity. C, kinetics of cellular uptake of FGF12B, FGF1, and FGF1/CPP-C were examined over 48 h by FACS. D, IEC6 cells were cultured for 18 h in medium with 1 μg/ml Alexa Fluor 568-labeled FGF12, FGF1, or FGF1/CPP-C. The nuclei were visualized by staining with Hoechst 33342 (blue) and fluorescence confocal images were acquired using an IX81 fluorescence microscope with disk-scanning unit (DSU) (Olympus, Tokyo, Japan).
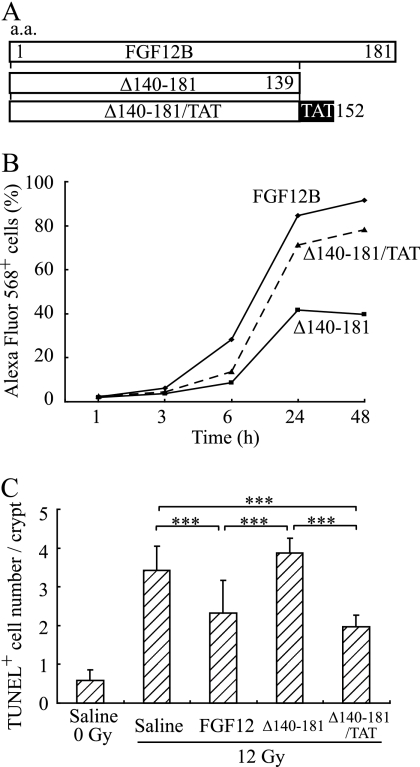
FGF12 Internalization by CPP-C Causes an Anti-apoptotic Effect of FGF12 in the Intestine
Intracellular expression of the FGF12 gene decreased radiation-induced apoptosis as described previously (18). Therefore, it was examined whether cellular uptake of FGF12 was involved in radiation-induced apoptosis, because FGF12 was able to translocate into the cytoplasm using the CPP-C domain. Interestingly, Δ140–181 did not inhibit radiation-induced apoptosis in the crypts because it could not enter cells without the CPP-C domain (Fig. 8, B and C). However, Δ140–181 may have lost not only the domain for internalization but also that for the anti-apoptotic function owing to the C-terminal deletion. Therefore, the TAT peptide, one of the most well-studied CPPs, was fused to the C-terminal end of Δ140–181 to evaluate the anti-apoptotic effect of this deletion mutant protein Δ140–181 (Fig. 8A), and this protein recovered the ability to achieve cellular internalization. Δ140–181/TAT significantly inhibited radiation-induced apoptosis in the crypts, indicating that Δ140–181 still possessed the anti-apoptotic domain of FGF12. Accordingly, a lack of uptake of the protein reduced the anti-apoptotic effect in the intestine, suggesting that CPP-C is involved in internalization of FGF12 to affect apoptosis of cells in vivo.
FIGURE 8.
FGF12 internalization is responsible for its anti-apoptotic effects. A, TAT peptide coding sequence was fused to the coding sequence for the C-terminal end of Δ140–181 (Δ140–181/TAT). FGF12B and protein Δ140–181 were produced without fusion to a Nus Tag, whereas protein Δ140–181/TAT was fused to a Nus tag. B, IEC6 cells were incubated in complete medium with 1 μg/ml of Alexa Fluor 568-labeled FGF12B, Δ140–181, or Δ140–181/TAT. They were analyzed in a time course up to 48 h by flow cytometry to estimate fluorescence intensity. C, 10 μg of FGF12B, Δ140–181, or Δ140–181/TAT was administered intraperitoneally without heparin to BALB/c mice 24 h before TBI at 12 Gy, and apoptosis in the crypts of the jejunum was evaluated by TUNEL assay 24 h after TBI. All values are means ± S.D. (n = 6). Similar findings were observed in two independents experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
DISCUSSION
In this study, we demonstrated for the first time that exogenous FGF12 can be internalized into cells to play a role in physiological events. FGF12 was able to translocate into many cell lines, including IEC6 cells (data not shown). In addition, a novel CPP (CPP-C) was identified from the C-terminal region of the FGF12 protein, which was specific to the FGF11 subfamily, and other FGFs had no corresponding peptide sequence. CPP-C was not only a protein transduction domain for FGF12 but also a cell-penetrating peptide that was able to deliver other polypeptides into cells. Peptide 140–169, including CPP-C, was internalized moderately (Fig. 3) because amino acid residues 150–169 may have an inhibitory effect on cellular internalization. Actually, peptide 150–179 did not translocate into cells at all, and in addition, the deletion mutant Δ150–181 increased cellular internalization compared with wild-type FGF12 (Fig. 4B). Accordingly, a ten-amino acid sequence, PIEVCMYREP, was indentified as CPP-C. Although CPP-C has not been identified precisely, it is an appropriate region to deliver FGF1 efficiently into cells. CPP-C consists of seven hydrophobic and three hydrophilic residues, including one cationic and two anionic residues. CPP-C and corresponding peptides share highly homologous sequences and structures in the FGF11 subfamily (Fig. 6A). Therefore, other members might also be internalized efficiently into cells. Indeed, E142L decreased cellular internalization because this mutation destroyed the homologous structure of this region (Fig. 6C).
In contrast, CPP-M and corresponding peptides also share highly homologous sequences in not only the FGF11 subfamily but also other FGFs and may be constitutive components of FGF structures (supplemental Fig. S3). Therefore, CPP-M might be generally involved in the internalization of FGF family members. FGF1 has structural similarity with FGF12; in particular FGF1 and Δ140–181 share CPP-M or corresponding domains without CPP-C, so that FGF1 can be internalized into cells as slight as Δ140–181 (Fig. 7B). Interestingly, reconstitution of FGF1 with the addition of CPP-C to the C terminus prominently increased cellular internalization (Fig. 7), indicating that FGF1/CPP-C protein has great potential for intracellular function like DNA synthesis and cell proliferation (9). On the other hand, C-terminal deletion of FGF1 caused its post-translational degradation of FGF1 by proteasomes (34). Hence, C-terminal deletion of residues 140–181 from FGF12B protein may also enhance its degradation in cells, resulting in the decline in Alexa Fluor 568-positive cell numbers after culturing with Δ140–181. Even if so, CPP-C must play a role in the cellular internalization of FGF12 because a single mutation in the CPP-C domain decreased FGF12 internalization (Fig. 6), and CPP-C was capable of delivering FGF1 into the cells efficiently. In addition, the involvement of the structural stability of FGF1 in cellular uptake was examined using mutants of FGF1. As a result, the increased structural stability of FGF1 did not lead to marked cellular internalization (data not shown). Therefore, the CPP-C domain is significantly effective for the cellular internalization of FGF12.
FGF12 can bind heparin with high affinity, as for other FGFs, and a heparin-binding site of FGF12 seems to be similar to that of other FGFs (3). However, FGF12 does not activate any of the seven principal FGFRs because two unique residues of FGF12 make an extracellular FGF12-FGFR interaction impossible (3). Interestingly, FGF12 cellular entry was inhibited by free heparin (Fig. 2C), whereas HSPG on the cell surface was not involved in FGF12 internalization because FGF12 was able to translocate into HSPG-repressed cells created by siRNA transfection (35) or by heparitinase treatment (supplemental Fig. S2). The affinity of FGF12 for heparin is significantly lower than that of FGF1 (3). Therefore, inhibition of FGF12 entry by heparin may not depend on specific structural requirements of heparin but simply by its ionic charge. In contrast, the transactivator protein of human HIV-1 (Tat) can be also internalized into a variety of cells (20, 21); however, cell membrane HSPG acts as a receptor for extracellular Tat uptake (36). The binding of Tat to HSPG followed by low-density lipoprotein receptor-related protein (LPR)-mediated endocytosis is a pathway for the entry of Tat into neurons (37). Islet-brain 2 (IB2) is the only known molecule to which FGF12 can bind intracellularly, and it is expressed in the brain, pancreas, and specific cell lines (38, 39). However, IB2 may not be a receptor for internalization because repression of IB2 did not affect the cellular uptake of FGF12 in a human mast cell line HMC-1 (data not shown). Futhermore, the binding affinity of Alexa Fluor 568-labeled FGF12 and FITC-labeled peptides to IEC6 cells was examined by flow cytometry 1 h after incubation at 4 °C. Alexa Fluor 568-labeled FGF12 protein had no binding affinity after incubation of the harvested cells at 4 °C (data not shown). In addition, cell surface binding of FGF12 was not detected, even without trypsinization, by confocal microscopy (data not shown). The peptide including CPP-C also showed no affinity for IEC6 cells and the peptide including CPP-M showed the lowest affinity (supplemental Fig. S4). Therefore, CPP-C and CPP-M play a role in penetrating through plasma membranes, but not in initial binding.
The mechanism of internalization of CPPs had been under discussion; however, it was found that cell fixation caused an artifactual redistribution of CPPs, leading to the misunderstanding of cellular uptake (31). In this study, FACS was used to evaluate the extent of cellular internalization of FGF12 without any cell fixation. IEC6 cells were harvested in trypsin-EDTA and washed twice. The artifact was thought to be related to the extremely cationic nature of CPPs; however, CPP-C is not a cationic peptide. Therefore, adsorbed FGF12 does not remain on the cell surface to allow reliable FACS analysis. Under the current experimental conditions, FGF12 was internalized at 37 °C but not at 4 °C (data not shown). Therefore, internalization of FGF12 needs at least an active process involving endocytosis depending on energy.
FGF12 is expressed in the developing and adult nervous systems, suggesting that it is related to nervous system development and function (2). In addition, we reported previously that FGF12 is expressed in mast cells (18). FGF2, FGF7, and FGF10 are also expressed in mast cells, which seem to contribute to wound healing and tissue homeostasis directly or indirectly via the stimulation of dermal fibroblasts (40, 41). FGF12 is not currently thought to be released from the cells because it lacks a classical signal sequence and it is not secreted from FGF12 transfectants (2). However, without any classical signal sequences, FGF1 and FGF2 may be released from cells by an alternative pathway such as cell injury (42–45). Therefore, it is likely that FGF12 may also be secreted from mast cells via other unknown pathways. If so, FGF12 could exert various effects in the tissue via its cellular internalization.
Surprisingly, exogenous FGF12 showed an anti-apoptotic effect on IEC6 cells in vitro (supplemental Fig. S5). However, this effect did not significantly depend on CPP-C in vitro (data not shown). Although CPPs-deficient mutant proteins decreased cellular internalization, they still possessed their anti-apoptotic capability (Fig. 6). Because of the strong anti-apoptotic effect of FGF12, even small amounts of cellular uptake of Δ140–181 may be effective enough to inhibit apoptosis in vitro. However, exogenous heparin can block internalization of FGF12 to inhibit the anti-apoptotic effect in IEC6 cells (supplemental Fig. S5). Therefore, internalization of FGF12 is important for the initiation of the anti-apoptotic effect of FGF12. In contrast, intraperitoneally added FGF12B inhibited radiation-induced apoptosis in intestinal crypt cells depending on the presence of CPP-C. A number of cells are related with the absorption, circulation and metabolism of FGF12 in tissues, so that the effect of CPP-C may be amplified enough to show the physiological effects significantly.
It is possible that endocytosed FGF12B could translocate from endosomes into the cytosol to function because such translocation was observed with FGF1. Endocytosed FGF1 can also translocate from endosomes into the cytosol (46) to reach the nucleus through utilization of the NLS (9, 47). In addition, the nuclear location of FGF1 may be important for DNA synthesis and cell proliferation (9, 10). Therefore, exogenous NLS peptide of FGF1 can induce DNA synthesis if it is taken up by cells (48), while loss of the NLS increased the anti-apoptotic effect of FGF1 in the intestine (49). This is consistent with the findings in this study where FGF12B exerted a significant anti-apoptotic effect in the intestine because FGF12B did not translocate into the nucleus. Externally added FGF12A was detected in the nuclei of cells, whereas FGF12B was mainly detected in the cytoplasm (Fig. 2D). Therefore, a comparative study of FGF12A and FGF12B would be useful to clarify the difference between the cytosolic and nuclear functions of FGF. The mitogenic effect of FGF12 has not been reported because only intracellular FGF12B has been studied well. These findings suggest that signals for FGF functions come from not only from activated FGFRs but also internalized FGF. In particular, FGF12 seems to transmit the signal mainly through internalization, and internalization may be important in the signaling pathways for other FGFs.
Accordingly, we obtained the striking result that exogenous FGF12 was able to react with cells as well as other FGFs. In addition, our finding suggests that cellular internalization of FGFs is important for their function as well as FGFR signaling. Therefore, investigation of FGF12 may lead to further clarification of FGF functions related to wound healing and the regeneration of tissues. We concluded that FGF12 was a potential radioprotector that could exert therapeutic effects on radiation damage and the CPP-C might be useful to create a bioactive molecule that could function via its cellular internalization.
Supplementary Material
Acknowledgment
We thank Dr. Takehito Uruno for technical assistance.
This work was supported as a part of the Research Project for High-Dose Radiation Injuries at NIRS and by KAKENHI (22510065).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- FGF
- fibroblast growth factor
- CPP
- cell-penetrating peptide
- FGFR
- fibroblast growth factor receptor
- HSPG
- heparan sulfate proteoglycan
- Tat
- transactivator of transcription
- TBI
- total body irradiation
- TUNEL
- terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick end-labeling
- FITC
- fluorescein isothiocyanate
- FACS
- fluorescent-activated cell sorting.
REFERENCES
- 1. Itoch N., Ornitz D. M. (2011) J. Biochem. 149, 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smallwood P. M., Munoz-Sanjuan I., Tong P., Macke J. P., Hendry S. H., Gilbert D. J., Copeland N. G., Jenkins N. A., Nathans J. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 9850–9857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Olsen S. K., Garbi M., Zampieri N., Eliseenkova A. V., Ornitz D. M., Goldfarb M., Mohammadi M. (2003) J. Biol. Chem. 278, 34226–34236 [DOI] [PubMed] [Google Scholar]
- 4. Hagiwara A., Nakayama F., Motomura K., Asada M., Suzuki M., Imamura T., Akashi M. (2009) Radiat. Res. 172, 58–65 [DOI] [PubMed] [Google Scholar]
- 5. Nakayama F., Hagiwara A., Kimura M., Akashi M., Imamura T. (2009) Exp. Dermatol. 18, 889–892 [DOI] [PubMed] [Google Scholar]
- 6. Nakayama F., Hagiwara A., Umeda S., Asada M., Goto M., Oki J., Suzuki M., Imamura T., Akashi M. (2010) Int. J. Radiat. Oncol. Biol. Phys. 78, 860–867 [DOI] [PubMed] [Google Scholar]
- 7. Imamura T., Friedman S. A., Gamble S., Tokita Y., Opalenik S. R., Thompson J. A., Maciag T. (1995) Biochim. Biophys. Acta 1266, 124–130 [DOI] [PubMed] [Google Scholar]
- 8. Zakrzewska M., Krowarsch D., Wiedlocha A., Olsnes S., Otlewski J. (2005) J. Mol. Biol. 352, 860–875 [DOI] [PubMed] [Google Scholar]
- 9. Imamura T., Engleka K., Zhan X., Tokita Y., Forough R., Roeder D., Jackson A., Maier J. A., Hla T., Maciag T. (1990) Science 249, 1567–1570 [DOI] [PubMed] [Google Scholar]
- 10. Wesche J., Malecki J., Wiedlocha A., Ehsani M., Marcinkowska E., Nilsen T., Olsnes S. (2005) Biochemistry 44, 6071–6080 [DOI] [PubMed] [Google Scholar]
- 11. Sørensen V., Wiedlocha A., Haugsten E. M., Khnykin D., Wesche J., Olsnes S. (2006) J. Cell Sci. 119, 4332–4341 [DOI] [PubMed] [Google Scholar]
- 12. Klingenberg O., Wiedlocha A., Citores L., Olsnes S. (2000) J. Biol. Chem. 275, 11972–11980 [DOI] [PubMed] [Google Scholar]
- 13. Wesche J., Malecki J., Wiedlocha A., Skjerpen C. S., Claus P., Olsnes S. (2006) J. Biol. Chem. 281, 11405–11412 [DOI] [PubMed] [Google Scholar]
- 14. Kolpakova E., Wiedlocha A., Stenmark H., Klingenberg O., Falnes P. O., Olsnes S. (1998) Biochem. J. 336, 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Skjerpen C. S., Wesche J., Olsnes S. (2002) J. Biol. Chem. 277, 23864–23871 [DOI] [PubMed] [Google Scholar]
- 16. Skjerpen C. S., Nilsen T., Wesche J., Olsnes S. (2002) EMBO J. 21, 4058–4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mizukoshi E., Suzuki M., Loupatov A., Uruno T., Hayashi H., Misono T., Kaul S. C., Wadhwa R., Imamura T. (1999) Biochem. J. 343, 461–466 [PMC free article] [PubMed] [Google Scholar]
- 18. Nakayama F., Müller K., Hagiwara A., Ridi R., Akashi M., Meineke V. (2008) J. Radiat. Res. 49, 491–501 [DOI] [PubMed] [Google Scholar]
- 19. Liu C. J., Dib-Hajj S. D., Renganathan M., Cummins T. R., Waxman S. G. (2003) J. Biol. Chem. 278, 1029–1036 [DOI] [PubMed] [Google Scholar]
- 20. Frankel A. D., Pabo C. O. (1988) Cell 55, 1189–1193 [DOI] [PubMed] [Google Scholar]
- 21. Green M., Loewenstein P. M. (1988) Cell 55, 1179–1188 [DOI] [PubMed] [Google Scholar]
- 22. Perez F., Joliot A., Bloch-Gallego E., Zahraoui A., Triller A., Prochiantz A. (1992) J. Cell Sci. 102, 717–722 [DOI] [PubMed] [Google Scholar]
- 23. Pooga M., Hällbrink M., Zorko M., Langel U. (1998) FASEB J. 12, 67–77 [DOI] [PubMed] [Google Scholar]
- 24. Soomets U., Lindgren M., Gallet X., Hällbrink M., Elmquist A., Balaspiri L., Zorko M., Pooga M., Brasseur R., Langel U. (2000) Biochim. Biophys. Acta 1467, 165–176 [DOI] [PubMed] [Google Scholar]
- 25. Lin Y. Z., Yao S. Y., Veach R. A., Torgerson T. R., Hawiger J. (1995) J. Biol. Chem. 270, 14255–14258 [DOI] [PubMed] [Google Scholar]
- 26. Wender P. A., Mitchell D. J., Pattabiraman K., Pelkey E. T., Steinman L., Rothbard J. B. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13003–13008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Futaki S., Suzuki T., Ohashi W., Yagami T., Tanaka S., Ueda K., Sugiura Y. (2001) J. Biol. Chem. 276, 5836–5840 [DOI] [PubMed] [Google Scholar]
- 28. Derossi D., Joliot A. H., Chassaing G., Prochiantz A. (1994) J. Biol. Chem. 269, 10444–10450 [PubMed] [Google Scholar]
- 29. Vivès E., Brodin P., Lebleu B. (1997) J. Biol. Chem. 272, 16010–16017 [DOI] [PubMed] [Google Scholar]
- 30. Derossi D., Calvet S., Trembleau A., Brunissen A., Chassaing G., Prochiantz A. (1996) J. Biol. Chem. 271, 18188–18193 [DOI] [PubMed] [Google Scholar]
- 31. Richard J. P., Melikov K., Vives E., Ramos C., Verbeure B., Gait M. J., Chernomordik L. V., Lebleu B. (2003) J. Biol. Chem. 278, 585–590 [DOI] [PubMed] [Google Scholar]
- 32. Asada M., Shinomiya M., Suzuki M., Honda E., Sugimoto R., Ikekita M., Imamura T. (2009) Biochim. Biophys. Acta 1790, 40–48 [DOI] [PubMed] [Google Scholar]
- 33. Gavrieli Y., Sherman Y., Ben-Sasson S. A. (1992) J. Cell Biol. 119, 493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miyakawa K., Ozawa K., Uruno T., Imamura T. (1999) Growth Factors 16, 191–200 [DOI] [PubMed] [Google Scholar]
- 35. Sasaki N., Okishio K., Ui-Tei K., Saigo K., Kinoshita-Toyoda A., Toyoda H., Nishimura T., Suda Y., Hayasaka M., Hanaoka K., Hitoshi S., Ikenaka K., Nishihara S. (2008) J. Biol. Chem. 283, 3594–3606 [DOI] [PubMed] [Google Scholar]
- 36. Tyagi M., Rusnati M., Presta M., Giacca M. (2001) J. Biol. Chem. 276, 3254–3261 [DOI] [PubMed] [Google Scholar]
- 37. Liu Y., Jones M., Hingtgen C. M., Bu G., Laribee N., Tanzi R. E., Moir R. D., Nath A., He J. J. (2000) Nat. Med. 6, 1380–1387 [DOI] [PubMed] [Google Scholar]
- 38. Schoorlemmer J., Goldfarb M. (2001) Curr. Biol. 11, 793–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schoorlemmer J., Goldfarb M. (2002) J. Biol. Chem. 277, 49111–49119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reed J. A., Albino A. P., McNutt N. S. (1995) Lab. Invest. 72, 215–222 [PubMed] [Google Scholar]
- 41. Artuc M., Steckelings U. M., Henz B. M. (2002) J. Invest. Dermatol. 118, 391–395 [DOI] [PubMed] [Google Scholar]
- 42. Mignatti P., Morimoto T., Rifkin D. B. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 11007–11011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jackson A., Friedman S., Zhan X., Engleka K. A., Forough R., Maciag T. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 10691–10695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Florkiewicz R. Z., Majack R. A., Buechler R. D., Florkiewicz E. (1995) J. Cell Physiol. 162, 388–399 [DOI] [PubMed] [Google Scholar]
- 45. Florkiewicz R. Z., Anchin J., Baird A. (1998) J. Biol. Chem. 273, 544–551 [DOI] [PubMed] [Google Scholar]
- 46. Olsnes S., Klingenberg O., Wiedlocha A. (2003) Physiol. Rev. 83, 163–182 [DOI] [PubMed] [Google Scholar]
- 47. Imamura T., Oka S., Tanahashi T., Okita Y. (1994) Exp. Cell Res. 215, 363–372 [DOI] [PubMed] [Google Scholar]
- 48. Komi A., Suzuki M., Imamura T. (1998) Exp. Cell Res. 243, 408–414 [DOI] [PubMed] [Google Scholar]
- 49. Fu X. B., Li X. K., Wang T., Cheng B., Sheng Z. Y. (2004) World J. Gastroenterol. 10, 3590–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.