Abstract
Fragile X syndrome (FXS), the most common form of hereditary mental retardation, is caused by a loss-of-function mutation of the Fmr1 gene, which encodes fragile X mental retardation protein (FMRP). FMRP affects dendritic protein synthesis, thereby causing synaptic abnormalities. Here, we used a quantitative proteomics approach in an FXS mouse model to reveal changes in levels of hippocampal synapse proteins. Sixteen independent pools of Fmr1 knock-out mice and wild type mice were analyzed using two sets of 8-plex iTRAQ experiments. Of 205 proteins quantified with at least three distinct peptides in both iTRAQ series, the abundance of 23 proteins differed between Fmr1 knock-out and wild type synapses with a false discovery rate (q-value) <5%. Significant differences were confirmed by quantitative immunoblotting. A group of proteins that are known to be involved in cell differentiation and neurite outgrowth was regulated; they included Basp1 and Gap43, known PKC substrates, and Cend1. Basp1 and Gap43 are predominantly expressed in growth cones and presynaptic terminals. In line with this, ultrastructural analysis in developing hippocampal FXS synapses revealed smaller active zones with corresponding postsynaptic densities and smaller pools of clustered vesicles, indicative of immature presynaptic maturation. A second group of proteins involved in synaptic vesicle release was up-regulated in the FXS mouse model. In accordance, paired-pulse and short-term facilitation were significantly affected in these hippocampal synapses. Together, the altered regulation of presynaptically expressed proteins, immature synaptic ultrastructure, and compromised short-term plasticity points to presynaptic changes underlying glutamatergic transmission in FXS at this stage of development.
Keywords: Mass Spectrometry (MS), Neurobiology, Neuroscience, Proteomics, Synapses
Introduction
Fragile X syndrome (FXS)4 is the most common form of hereditary mental retardation disease, affecting one in 4000 males and one in 8000 females. Patients suffer from multiple behavioral problems including cognitive impairments, impaired visuo-spatial processing, hyperactivity, anxiety, and in a number of cases, autism and/or epilepsy (1). FXS is caused by mutations in the Fmr1 gene resulting in the absence, significant down-regulation, or loss-of-function of the corresponding protein product, fragile X mental retardation protein (FMRP) (2). In particular, aberrant synapse function caused by reduced FMRP expression is considered to be the major factor underlying FXS.
FMRP is expressed abundantly in neurons and travels into dendritic spines in an activity-dependent manner (3), where it affects local synaptic protein synthesis and plasticity. FMRP is an mRNA-binding protein, associated to many types of mRNAs, encoding both presynaptic and postsynaptic proteins (4). By controlling several aspects of mRNA biology, e.g. transport, translation, and metabolism, FMRP may regulate synaptic structure and function (5, 6). Indeed, both FXS patients and Fmr1 knock-out (KO) mice display an abnormal appearance of generally long and thin dendritic spines that resemble an immature postsynaptic phenotype (7, 8). Presynaptically, lack of FMRP significantly affects the motility and dynamics of axonal growth cone development in hippocampal neurons, and alterations in terminal branching of hippocampal mossy fibers are found in the Fmr1 KO mouse (9, 10). Thus, the effects of FMRP upon both presynaptic and postsynaptic structure are likely to alter functional aspects of glutamatergic signaling during synaptic development in Fmr1 KO mice.
Recent studies of molecular mechanisms underlying reduced cognitive function in FXS have focused on glutamatergic synapse function in several brain regions including hippocampus, cortex, and cerebellum. In these brain areas, long-term synaptic plasticity is strongly affected. The relationship between impaired FMRP expression and synaptic plasticity is complex (11, 12). In the cortex, the absence of FMRP results in reduced long-term potentiation (13, 14). In the hippocampus and cerebellum, enhanced metabotropic glutamate receptor-dependent long-term depression via an increased internalization of membrane-localized AMPA receptors is observed in Fmr1 KO mice (15, 16). Activity-dependent changes of synaptic efficacy are encoded by sequential molecular events at the synapse, including FMRP-mediated de novo protein synthesis during the late phase of long-term potentiation and long-term depression (17). Interestingly, in addition, Gq-coupled M1 muscarinic acetylcholine receptor-dependent long-term depression requires protein synthesis and is enhanced in Fmr1 KO mice (18, 19). However, an overview of synaptic proteins that are affected by the lack of FMRP and to what extent their expression levels are altered is largely unknown. Many forms of short-term plasticity, such as synaptic facilitation, are affected by rapid changes occurring at the presynaptic terminal. FMRP is expressed at presynaptic sites (20), where it may regulate establishment and/or maintenance of synaptic connections (21, 22). A direct relation between the absence of FMRP and its regulated proteins upon synaptic ultrastructure and fast presynaptic-mediated forms of synapse function in Fmr1 KO mice has not been fully investigated in mammalian neurons.
To address these issues, we have taken a quantitative proteomics approach to reveal changes in the hippocampal synaptic membrane proteome in Fmr1 KO mice as compared with the wild type (WT) mice during an early developmental period of synapse formation and refinement. We observed changes in a group consisting of known regulators of cell differentiation, filopodia protrusion, and neurite outgrowth by remodeling the actin cytoskeleton. Increase in their expression may retain synapses in an immature state, as we confirmed for Fmr1 KO synapses at the ultrastructural level. In addition, a second group of presynaptic vesicle-associated proteins was dysregulated, in line with our observed functional changes in presynaptic plasticity. Our findings show that changes in the expression levels of synaptic proteins in the Fmr1 KO mice correlate to changes in both excitatory synapse structure and excitatory synapse function, demonstrating a presynaptic locus for mechanisms underlying FXS.
EXPERIMENTAL PROCEDURES
Animals
Fmr1 KO1 mice on a background of C57Bl/6J from Harlan Netherlands (23), a kind gift of Dr. B. Oostra and Dr. R. Willemsen, were bred in our facility. Pregnant WT mice C57Bl/6JOlaHsd were obtained from Harlan Netherlands, and pups were born in our facility. For the proteomics experiment, 16 Fmr1 KO mice and 16 WT mice were used.
Synaptic Membrane Preparation
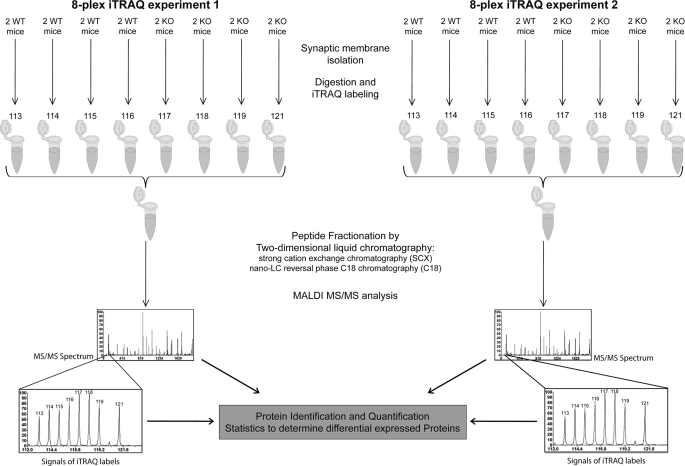
Synaptic membranes from predominately glutamatergic synapses were isolated from PN14 Fmr1 KO and WT mice as described previously (24, 25). A short overview of the experimental setup is represented in Fig. 1. In brief, for each sample, hippocampi from two mice were pooled and homogenized in a glass Potter-Elvehjem homogenizer containing 5 ml of ice-cold homogenization buffer (320 mm sucrose in 5 mm Hepes, pH 7.4) at 900 rpm with 12 up and down strokes of the piston. The lysate was centrifuged at 1000 × g, 4 °C, for 10 min. Supernatant was loaded on top of a sucrose step gradient consisting of 0.85 and 1.2 m sucrose. After ultracentrifugation at 100,000 × g, 4 °C for 2 h, the synaptosome fraction at the interface of 0.85/1.2 m sucrose was collected, diluted six times with 5 mm HEPES buffer (pH 7.4), and centrifuged at 80,000 × g, 4 °C for 40 min. The osmotic shock of synaptosomes was done by resuspending the pellet in HEPES buffer and stirring slowly over ice for 15 min. The resulting synaptic membrane fraction was recovered by ultracentrifugation using the sucrose step gradient as outlined above. Protein concentration was determined by Bradford measurement. The obtained synaptic membranes were subjected to trypsin digestion and iTRAQ reagent tagging or quantitative immunoblotting.
FIGURE 1.
Setup of the iTRAQ experiments. The hippocampi from two mice were pooled together to obtain enough material as a single sample for proteomics analysis. Together, synaptic membrane fractions were isolated from 32 animals representing 16 independent pools of samples of Fmr1 KO mice and WT mice, respectively. Tryptic digests of synaptic membranes from these 16 samples were tagged with two sets of 8-plex iTRAQ reagents. Peptides from each set of iTRAQ experiments were pooled together, fractionated by two-dimensional liquid chromatography, and subjected to tandem mass spectrometric analysis. Protein identification and quantification were performed as detailed under “Results”.
Protein Digestion and iTRAQ Labeling
In two independent 8-plex iTRAQ experiments, we have compared eight WT samples with eight Fmr1 KO samples. The relatively high number of biological replicas (n = 8) facilitates subsequent statistical analysis. The digestion and iTRAQ labeling of the synaptic membranes have been described in previous studies (24, 25). In short, for each sample, 75 μg of membrane proteins was solubilized in 28 μl of 0.85% RapiGest (Waters Corp.) reconstituted with dissolution buffer (iTRAQ reagent kit, AB Sciex). A 2-μl cleavage reagent (iTRAQ reagent kit, AB Sciex) was added and incubated at 55 °C for 1 h, after which 1 μl of Cys blocking reagent (iTRAQ reagent kit, AB Sciex) was added and samples were vortexed for 10 min. Subsequently, 5 μg of trypsin (sequencing grade, Promega) was added and incubated overnight at 37 °C. The trypsinized peptides were then tagged with iTRAQ reagents dissolved in 85 μl of ethanol. After incubation for 3 h, the samples were pooled and acidified with 10% trifluoroacetic acid to pH 2.5–3. After 1 h, the sample was centrifuged, and the supernatant was dried in a SpeedVac. We performed two 8-plex iTRAQ experiments to cover the 16 samples. In each iTRAQ experiment, four WT samples were labeled respectively with iTRAQ reagents 113–116 and four Fmr1 KO mice samples were labeled respectively with iTRAQ reagents 117–119 and 121 (Fig. 1).
Two-dimensional Liquid Chromatography
The dried iTRAQ labeled sample was dissolved in 250 μl of loading buffer (20% acetonitrile, 10 mm KH2PO4, pH 2.9), whereas 200 μl was injected into a strong cation exchange column (2.1 × 150-mm PolySULFOETHYL A column, PolyLC Inc.). Peptides were eluted with a linear gradient of 0–500 mm KCl in 20% acetonitrile, 10 mm KH2PO4, pH 2.9, over 25 min at a flow rate of 200 μl/min. Fractions were collected at 1-min intervals and dried in a SpeedVac. The peptides were redissolved in 20 μl of 0.1% TFA, delivered with a FAMOS autosampler at 30 μl/min to a reverse phase C18 trap column (1 mm × 300-μm inner diameter column), and separated on an analytical capillary reverse phase C18 column (150 mm × 100 μm-inner diameter column) at 400 nl/min using the LC-Packing Ultimate system. The peptides were separated using a linear increase in concentration of acetonitrile from 6 to 45% in 50 min and to 90% in 1 min. The eluent was mixed with matrix (7 mg of recrystallized α-cyano-hydroxycinnamic acid in 1 ml of 50% acetonitrile, 0.1% TFA, 10 mm ammonium dicitrate) delivered at a flow rate of 1.5 μl/min and deposited off-line to an Applied Biosystems metal target every 15 s using an automatic robot (Probot, Dionex).
MALDI-MS/MS
The sample was analyzed on an ABI 4800 proteomics analyzer (AB Sciex). Peptide collision induced dissociation was performed at 2 kV; the collision gas was air. MS/MS spectra were each collected from 2500 laser shots. Peptides with signal-to-noise ratio above 50 at the MS mode were selected for an MS/MS experiment; a maximum of 25 MS/MS was allowed per spot. The precursor mass window was 200 relative to resolution (full width at half maximum).
Protein Identification and Quantitation
To annotate spectra, Mascot (MatrixScience) searches were performed against the SwissProt database (release 12.6) and the larger but more redundant National Center for Biotechnology Information (NCBI) database (release 081003) using the GPS Explorer (AB Sciex version 3.6). MS/MS spectra were searched against mouse databases with trypsin specificity and fixed iTRAQ modifications at lysine residues and N termini of the peptides. Mass tolerance was 150 ppm for precursor ions and 0.5 Da for fragment ions; one missed cleavage was allowed. The false discovery rate (percentage) for peptide identification was calculated using a randomized database. Protein redundancy in the result files was removed by clustering the precursor protein sequences at a threshold of 90% sequence similarity over 85% of the sequence length (blastclust, version 20041205). Subsequently, all peptides were matched against the protein clusters; those that were matched to more than one protein cluster were not considered for protein identification and quantification, leaving only “unique” peptides in the analysis. Only proteins identified with at least two peptides with a confidence interval ≥95% (AB Sciex CI, percentage) were considered identified, and of these proteins, only those with three or more quantifiable peptides in both iTRAQ experiments were included in subsequent quantitative analyses. Peak areas for each iTRAQ signature peak (m/z 113.1, 114.1, 115.1, 116.1, 117.1, 118.1, 119.1, 121.1) were obtained and corrected according to the manufacturers' instructions to account for the differences in isotopic overlap.
To compensate for the possible variations in the starting amounts of the samples, the individual peak areas of each iTRAQ signature peak were log 2-transformed and normalized to the total peak area of the signature peak. Peptides with iTRAQ signature peaks of less than 2000 were not considered for quantification. Within each experiment, for each peptide, the peak area in each sample was mean-centered. Protein averages were calculated from these mean-centered peak areas of multiple peptides. Finally, the eight mutant and eight wild type protein means of both experiments were used to calculate the average difference between WT and Fmr1 KO mice. To assess whether this difference had occurred by chance due to the multiple testing problem or could be deemed significant, we calculated the permutation-derived false discovery rate (q-value) using the Excel plug-in of the Significance Analysis of Microarrays (SAM) program (26, 27). The settings for the SAM program were: two class unpaired, log 2-scaled, t-statistic, 1000 permutations, automatic estimation of s0 factor, 10 k-nearest neighbors.
Immunoblotting
Samples were mixed with SDS sample buffer and heated to 90 °C for 5 min. Proteins were separated on an SDS-polyacrylamide gel in a Mini-PROTEAN electrophoresis system (Bio-Rad) and electroblotted onto the PVDF membrane overnight at 40 V. Membranes were incubated with primary antibodies followed by an alkaline phosphatase-conjugated secondary antibody (Dako; 1:1000). The signal was developed with ECF substrate (GE Healthcare); the image was scanned with an FLA 5000 instrument (Fujifilm) and analyzed with Quantity One software (Bio-Rad). The following antibodies were used: mouse anti-calcium/calmodulin-dependent protein kinase type II subunit β (Zymed Laboratories Inc.; 1:1000), mouse anti-PSD-95 (Abcam; 1:1000), mouse anti-glutamate receptor (NMDAR1) (BD Biosciences; 1:1000), rabbit anti-syntaxin 1B (gift from M. Verhage; 1:1000), rabbit anti-synaptophysin (gift from M. Verhage; 1:2000), rabbit anti-synapsin I and II (gift from M. Verhage; 1:1000), rabbit anti-NAP-22 (Basp1) (Chemicon; 1:1000), rabbit anti-BM88 (Cend1; gift from R. Matsas; 1:2000), and mouse anti-β-catenin (BD Biosciences; 1:1000).
Ultrastructure Analysis by Electron Microscopy
Mice (n = 3/genotype) were subjected to transcardiac perfusion with a mixture of glutaraldehyde (2.5%) and paraformaldehyde (4%) in 0.1 m cacodylate buffer, pH 7.4. From the fixed brain, 50-μm-thick coronal slices containing the stratum radiatum were post-fixed (1% OsO4) and stained with 1% uranyl acetate. After embedding in Epon, ultrathin sections (∼90 nm) were collected on 400-mesh copper grids and stained with uranyl acetate and lead citrate. For each condition, docked vesicles, total vesicle number, postsynaptic density (PSD), active zone (AZ) length, and vesicle cluster surface (nm2) were measured on digital images taken at 100,000× magnification using software in individual electron micrographs, in which a single synapse was counted from each of the three perfusion-fixed mice per genotype using a Jeol (Peabody, MA) 1010 electron microscope. The observer was blind to genotype during the procedure.
Electrophysiological Analysis of Miniature Excitatory Postsynaptic Currents (mEPSCs)
Horizontal hippocampal brain slices (300 μm thick) were prepared from 13–15-day-old WT and Fmr1 KO mice. Brains were rapidly removed and dissected in ice-cold artificial cerebrospinal fluid containing (in mm): 125 NaCl; 3 KCl; 1.25 NaH2PO4; 3 MgSO4; 1 CaCl2; 26 NaHCO3; 10 glucose (∼300 mosm). Slices were cut on a vibrating microtome and placed in artificial cerebrospinal fluid in a submerged-style holding chamber, bubbled with carbogen (95% O2, 5% CO2) containing the following (in mm): 125 NaCl; 3 KCl; 1.25 NaH2PO4; 1 MgSO4; 2 CaCl2; 26 NaHCO3; 10 glucose.
Slices were left for 1 h to recover before recording began. Tetrodotoxin (1 μm) was added to the recording artificial cerebrospinal fluid (same as holding chamber), and slices were perfused in a submerged recording chamber at 28–32 °C. SR-95531 (gabazine, 10 μm, Tocris) was added to the bath to block GABAA receptor-mediated synaptic currents. Whole-cell patch clamp recordings were made from pyramidal cells in the CA1 region under visual guidance by differential interference contrast microscopy. Patch pipettes (3–5 megaohms) were pulled from standard-wall borosilicate tubing and were filled with intracellular solution containing the following (in mm): 140 potassium gluconate; 9 KCl; 10 HEPES; 4 K2-phosphocreatine; 4 ATP (magnesium salt); 0.4 GTP (pH 7.2–7.3, pH adjusted with KOH; 290–300 mosm). For recording mEPSCs, cells were held in a voltage clamp and recorded for on average 8 min at −70 mV. Events were analyzed using Mini Analysis (Synaptosoft, NJ) with event detection levels for synaptic currents set at 8 pA. Rise and decay times were calculated from average event fits. Statistical comparisons were made using analyses of variance with post hoc t tests and Levene's analysis of variance as appropriate in SPSS software with p values < 0.05 being considered statistically significant. Synaptic current parameters are reported as mean ± S.E.
Electrophysiological Analysis of Short-term Plasticity
Horizontal hippocampal brain slices (300 μm thick) were prepared from 14–18-day-old WT and Fmr1 KO mice, and CA1 pyramidal cells were selected and recorded as described previously for mEPSCs but without the addition of tetrodotoxin to artificial cerebrospinal fluid. Patch pipettes were filled with intracellular solution containing the following (in mm): 129 cesium gluconate; 1 CsCl; 10 HEPES; 4 K2-phosphocreatine; 10 tetraethylammonium; 4 ATP (magnesium salt); and 0.4 GTP (pH 7.2–7.3, pH adjusted to 7.3 with CsOH; 290–300 mosm). After whole-cell configuration, the membrane potential was held at −70 mV, and the internal solution was allowed to diffuse for 5 min into the cell prior to the onset of recording. The low intracellular chloride concentration in the pipette allowed identification and exclusion of experiments in which inhibitory events occurred close to the EPSCs measured.
Schaffer collateral fibers were stimulated using an extracellular electrode positioned in the stratum radiatum on the CA3 side. Moderate stimulation was used to measure synaptic facilitation or depression in response to presynaptic trains (of up to 10 pulses) over a range of frequencies (5–50 Hz). For each frequency, the stimulus train was repeated 20 times, with a 15-s delay between each sweep. Sweeps at each frequency were equally divided into two groups, one at the start and one at the end of the experiment, allowing time-dependent changes in the responses to be identified. Synaptic currents sampled at intervals of 100 μs were digitized and analyzed off-line as described previously (28). To assess significance, analysis of variance with repeated measures was used (for within subjects effect test, Huynh-Feldt was used after Mauchly's test for sphericity) as well as post hoc unpaired Student's t test. Data are given as mean ± S.E., with p < 0.05 indicating significance.
RESULTS
MS/MS Analysis, Protein Identification, and Quantification
To reveal the molecular basis of synapse dysfunction that underlies the FXS phenotype, we compared the proteomes of the synaptic membrane fractions of hippocampus from Fmr1 KO and WT mice. In previous studies, we showed the robustness of the synaptic proteome to resist perturbation, which argues for the use of a large sample size to detect subtle changes (24, 25). Therefore, we analyzed eight independent pools of both Fmr1 KO and WT mice. Altogether ∼50,000 MS/MS analyses were performed, and 347 proteins were identified with at least two distinct peptides with a confidence of ≥95% present in both iTRAQ experiments. The false discovery rate (percentage) for peptide identification using randomized database from NCBI and SwissProt was about 2% (the false discovery rate from the first iTRAQ experiment is 1.6% for NCBI and 2.1% for SwissProt; for the second iTRAQ experiment, it is 1.9 and 2.1%, respectively). As iTRAQ-based quantitation relies on the iTRAQ signature peak intensities generated from MS/MS analysis of a single selected parent ion, the presence of a co-eluting but distinct peptide of similar mass would contribute to the noise depending on the intensity and mass difference of the interfering peptide. For the sake of increased reliability of quantification of protein abundance, we focused on proteins with ≥3 distinct peptides in both iTRAQ experiments (n = 205, supplemental Table 1). The observed -fold changes were low with the largest difference of 1.24-fold (Table 1; log 2 = 0.31), but these were similar as observed previously for relevant biological changes (29). Using the Significance Analysis of Microarray program (SAM, q-value) (27) with false discovery rate <5%, 23 proteins were considered to be significantly different between KO and WT mice. Among them, several presynaptic proteins were present, including syntaxin, synapsin, synaptophysin, piccolo, Rab3A, and vacuolar proton pump subunit E, as well as proteins involved in cell differentiation/neurite outgrowth including Cend1, Basp1, and Gap43.
TABLE 1.
Proteins with significant changes in the hippocampal synaptic membrane fraction of Fmr1 KO mice
The proteins with at least three distinct peptides in both iTRAQ experiments with the q-values <5% are shown. The accession number for each protein is indicated. Furthermore, the total number of peptides identified, the coverage of the protein, the -fold difference, and the corresponding standard deviation between Fmr1 KO mice and WT mice are shown. CI, confidence interval; SAM, Significance Analysis of Microarrays program.
| UniProt accession number | Protein name | % of coverage of the protein | No. of peptides CI ≥ 95% | Difference KO-WT (log 2 scale) | Standard deviation KO-WT (log 2 scale) | q-value (%) using SAM |
|---|---|---|---|---|---|---|
| Cell differentiation/neurite outgrowth | ||||||
| Q91XV3 | Brain acid-soluble protein 1 | 58.1 | 11 | 0.31 | 0.20 | 0.00 |
| Q9JKC6 | Cell cycle exit and neuronal differentiation protein 1 | 61.3 | 7 | 0.27 | 0.18 | 0.00 |
| Q02248 | Catenin β-1 | 31.1 | 19 | −0.16 | 0.12 | 0.00 |
| P18760 | Cofilin-1 | 55.7 | 7 | 0.13 | 0.11 | 0.00 |
| P06837 | Gap43 | 86.8 | 14 | 0.23 | 0.23 | 0.00 |
| Presynaptic transmission | ||||||
| P61264 | Syntaxin-1B2 | 52.6 | 11 | 0.16 | 0.14 | 0.00 |
| P63011 | Ras-related protein Rab-3A | 30.3 | 5 | 0.15 | 0.13 | 0.00 |
| Q9QYX7 | Protein piccolo | 8.1 | 17 | 0.13 | 0.12 | 1.80 |
| O88935 | Synapsin-1 | 46.1 | 22 | 0.11 | 0.12 | 3.50 |
| Q62277 | Synaptophysin | 21.0 | 5 | 0.12 | 0.14 | 3.50 |
| Q9DBG3 | AP-2 complex subunit β-1 | 8.6 | 5 | 0.15 | 0.19 | 3.50 |
| P50518 | Vacuolar ATP synthase subunit E | 47.6 | 9 | 0.18 | 0.14 | 0.00 |
| Mitochondrial/metabolic proteins | ||||||
| Q9CR68 | Ubiquinol-cytochrome c reductase iron-sulfur subunit | 30.2 | 7 | 0.15 | 0.12 | 0.00 |
| O35143 | ATPase inhibitor, mitochondrial precursor | 28.0 | 3 | 0.19 | 0.15 | 0.00 |
| Q9WUA3 | 6-Phosphofructokinase type C | 16.9 | 8 | 0.13 | 0.10 | 0.00 |
| Q9CQ69 | Ubiquinone-binding protein QP-C | 54.2 | 5 | 0.17 | 0.17 | 0.00 |
| P17182 | α-Enolase | 29.2 | 10 | 0.14 | 0.15 | 1.80 |
| P05064 | Fructose-bisphosphate aldolase A | 74.2 | 19 | 0.13 | 0.14 | 1.80 |
| P16858 | Glyceraldehyde-3-phosphate dehydrogenase | 43.4 | 11 | 0.12 | 0.11 | 1.80 |
| Q9CPQ8 | ATP synthase subunit g, mitochondrial | 59.6 | 4 | 0.14 | 0.13 | 1.80 |
| Other | ||||||
| P17742 | Peptidyl-prolyl cis-trans isomerase A | 81.2 | 11 | 0.17 | 0.14 | 0.00 |
| P05132 | cAMP-dependent protein kinase, α-catalytic subunit | 15.3 | 3 | 0.18 | 0.18 | 0.00 |
| P58252 | Elongation factor 2 | 15.0 | 5 | 0.18 | 0.15 | 0.00 |
Confirmation of iTRAQ Data by Immunoblotting
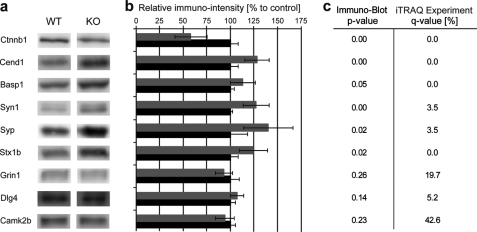
Quantitative immunoblotting analysis was performed to verify the quantitative proteomics data obtained by iTRAQ. Fig. 2 shows the good agreement between the two methods, i.e. syntaxin, synapsin, synaptophysin, Basp1, Cend1, and Ctnnb1 (q <5%) showed significant difference by immunoblotting (p ≤ 0.05), whereas calcium/calmodulin dependent protein kinase type II subunit β (Camk2β), N-methyl-d-aspartate receptor subunit NR1 (Grin1), and PSD-95 (Dlg4) (q >5%) failed to show changes by immunoblotting.
FIGURE 2.
Immunoblot analysis of selected synaptic proteins from Fmr1 KO and WT mice. a, the gene names of the proteins and the corresponding immunoreactive bands on the Western blots made from the WT and KO mice synaptic membrane samples are shown. b, the average intensity of the immunoreactive bands for each protein measured in the Fmr1 KO (gray bars) is normalized to that of the WT group (black bar; set to 100%, control). c, the Student's t test p value of the immunoblot analysis (n = 5) and corresponding q-values measured by iTRAQ-based quantitative proteomics are shown. p values below 0.005 are indicated as 0.00.
Ultrastructural Analysis of Synapses from the Hippocampal CA1 Region
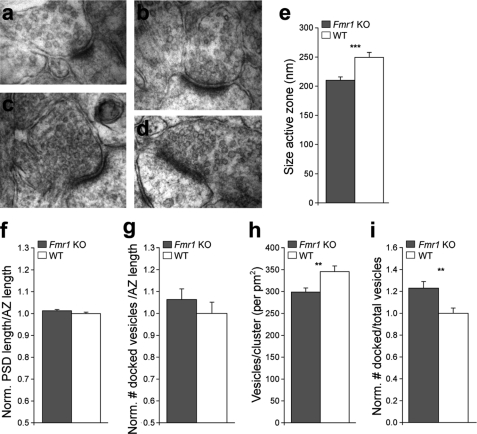
The proteomics data implicate a dysregulation of proteins localized to the presynaptic compartment in Fmr1 KO mice (Table 1). The up-regulation of presynaptic proteins could be due to a mismatch between AZ and PSD size, a higher number of docked vesicles, or a change of protein composition of synaptic vesicles. To obtain insight into the subcellular changes of synapses correlated with these dysregulated proteins, we performed an ultrastructural analysis using electron microscopy (Fig. 3, a–d). We found that the AZ length of the presynaptic elements in Fmr1 KO mice was shorter by 19% (p = 0.000141) as compared with WT mice (Fig. 3e). A similar decrease was observed for the length of the PSD of 16% (p = 0.00046) (data not shown). When the PSD was normalized to the AZ length, there was no difference between Fmr1 KO and WT animals with both lengths shortened accordingly, resulting in a smaller synaptic structure in the Fmr1 KO mice (Fig. 3f). In addition, this result excludes the possibility that a mismatched ratio between active zone and PSD length leads to up-regulation of presynaptic proteins.
FIGURE 3.
Ultrastructural analysis of CA3 to CA1 synapses in the stratum radiatum. a–e, Fmr1 KO (a and b) as compared with WT (c and d) animals revealed a smaller length of the AZ (e). f and g, no significant differences were observed for the ratio of PSD to AZ length (f; p = 0.154), and the number of docked vesicles per AZ length (g; p = 0.375). h and i, Fmr1 KO mice showed fewer vesicles per cluster surface (h) and an increase of the ratio of docked to total number of vesicles (i) indicative for their immature phenotype. Data are mean ± S.E. The analysis consisted of n = 86–87 Fmr1 KO and n = 68–71 WT synapses from three animals per genotype. **, p < 0.01; ***, p < 0.001 using a Newman-Keuls test. Norm. PSD, normalized PSD.
Furthermore, by counting synaptic vesicles, we observed that there was a trend for a higher number of docked vesicles per AZ length in the Fmr1 KO mice (Fig. 3g), the value of 10% being similar to the observed changes in protein regulation ∼10%. The number of vesicles per cluster surface in Fmr1 KO mice was significantly decreased (p = 0.00413) (Fig. 3h). As a result, the fraction of docked vesicles per total number of vesicles in the presynapse was increased in Fmr1 KO mice (Fig. 3i). Taken together these data suggest that synapses of Fmr1 KO mice are less developed as compared with WT mice. More importantly, these results provide clear ultrastructural evidence for a presynaptic phenotype in the CA1 region of hippocampus.
mEPSCs to Determine Basic Parameters of Vesicle Release
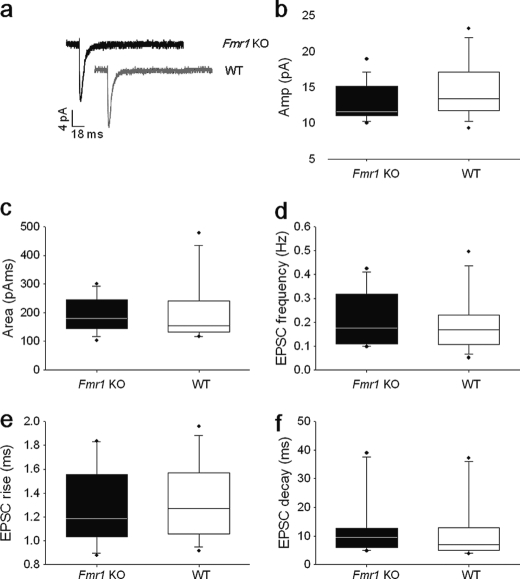
The changes in presynaptic morphology and global changes in presynaptic protein levels are suggestive for changes in presynaptic function and plasticity and prompted us to test whether these structural alterations are reflected in changes in the basic quantal units of synaptic transmission. We measured mEPSCs from hippocampal slices of 2-week-old WT and Fmr1 KO mice (Fig. 4a). We observed no differences in baseline parameters of amplitude (WT = 14.7 ± 1.0 pA; Fmr1 KO = 12.9 ± 0.7 pA), charge transfer (WT = 204 ± 30 pA ms; Fmr1 KO = 194 ± 16 pA ms), or frequency (Fmr1 KO = 0.2 ± 0.03 Hz; WT = 0.19 ± 0.03 Hz) of responses between the two groups (WT, n = 15; Fmr1 KO, n = 14; Fig. 4, b–d). Furthermore, we found no difference in rise (WT = 1.5 ± 0.2 ms; Fmr1 KO = 1.6 ± 0.2 ms) or decay times (WT = 9.8 ± 2.5 ms; Fmr1 KO = 12.7 ± 2.9 ms) of the mEPSCs between the two groups. The absence of effect upon baseline parameters of synaptic amplitude and frequency indicates that no detectable changes in synapse strength, glutamate release probability, or vesicle release occur in Fmr1 KO pyramidal neurons under baseline conditions at this developmental stage.
FIGURE 4.
No significant differences in mEPSCs between CA1 pyramidal neurons of 2-week-old WT and Fmr1 KO mice. a, averaged traces of mEPSCs held at −70 mV from Fmr1 KO (black) and WT neurons (gray). b–f, no significant differences were observed between mean amplitude (Amp), area, frequency, rise time, or decay time of events recorded for WT (n = 15) and Fmr1 KO (n = 14) mice. Mean ± S.E. is shown with individual data points to the right.
Short-term Plasticity
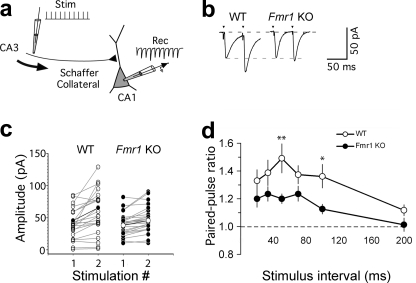
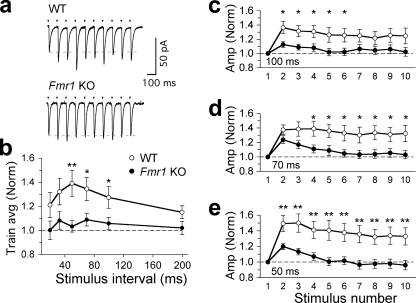
Because we did not observe any effect on basic quantal units of synaptic transmission, we investigated whether the observed changes in presynaptic proteins and the increased fraction of docked vesicles per total number of vesicles in Fmr1 KO mice might affect the rapid dynamics of synaptic plasticity. For this, we performed paired-pulse and short-term facilitation on Schaffer collateral inputs onto CA1 hippocampal pyramidal cells (Fig. 5a). Paired-pulse facilitation was significantly lower in Fmr1 KO mice than in WT mice in response to low frequency synaptic input (50-ms interpulse intervals; PPR WT = 1.38 ± 0.07, n = 19; PPR Fmr1 KO = 1.21 ± 0.04, n = 23; p = 0.016 (Fig. 5, b–d), or 100-ms interpulse intervals; PPR WT = 1.37 ± 0.1, n = 19; PPR Fmr1 KO = 1.13 ± 0.04, n = 23; p = 0.034 (Fig. 5d)). This decreased facilitation at low input frequencies of 10–20 Hz was also observed when the amplitudes of all the EPSC responses in a train (Fig. 6a) were analyzed (50 ms; WT = 1.28 ± 0.1, Fmr1 KO = 1.05 ± 0.05, p = 0.03; 70 ms; WT = 1.31 ± 0.11, Fmr1 KO = 1.08 ± 0.06, p = 0.048; 100 ms; WT = 1.29 ± 0.1, Fmr1 KO = 1.05 ± 0.06, p = 0.037; WT, n = 19, Fmr1 KO, n = 23; Fig. 6, b–e). Similar significant differences were also observed with shorter trains of five pulses between 20- and 200-Hz stimulation frequency input (supplemental Fig. 1). Kinetic parameters of first and second pulses during paired-pulse facilitation at 50-ms time intervals between WT (n = 11) and Fmr1 KO (n = 12) mice showed no significant differences between rise (analysis of variance p = 0.59) and decay times (analysis of variance p = 0.68) of first (P1) and second (P2) pulses for WT and KO recordings (supplemental Fig. 2). Meanwhile, at higher stimulus frequencies, the degree of paired-pulse facilitation and the degree of facilitation during the EPSC train became more similar in WT and Fmr1 KO mice (Fig. 6b). Thus, although basal transmission remains unchanged, the short-term dynamics of the Schaffer collateral input are significantly altered over a specific 10–20-Hz range of synaptic input frequencies to CA1 pyramidal cells.
FIGURE 5.
Paired-pulse ratio is affected in Fmr1 KO mice. a, schematic showing the short-term plasticity protocol. Schaffer collateral fibers were stimulated (Stim) using a 10-pulse variable frequency, and the stimulation train and EPSCs were recorded (Rec) in CA1 pyramidal cells. b, example of single experiments (50-ms interpulse interval) showing paired-pulse facilitation in response to the first two stimuli in the train (arrowheads). EPSCs are averaged responses from 20 sweeps (stimulus artifacts truncated). c, paired-pulse facilitation in WT and Fmr1 KO pyramidal cells (interpulse interval; 50 ms). Each pair of points (connected by gray lines) shows the average first and second EPSCs from individual experiments (mean of 20 stimulus trains). Points connected by a thick black line show group averages. d, effect of interstimulus interval on paired-pulse facilitation. PPR is plotted against the interpulse interval (each point shows the mean ± S.E., *, p < 0.05, **, p < 0.01).
FIGURE 6.
Hippocampal short-term synaptic plasticity is affected in Fmr1 KO mice. a, example single experiments (100-ms interpulse interval) showing short-term synaptic plasticity in response to a 10-pulse stimulus train (arrowheads). EPSC trains are the averaged responses from 20 sweeps (stimulus artifacts truncated). b, averaged synaptic facilitation during the stimulus train. Each point represents the averaged EPSC amplitude from all 10 pulses, normalized to the amplitude of the first EPSC in the train (bars show average (Train avg) S.E.). c–e, facilitation is reduced in Fmr1 KO mice, with low frequency synaptic input. Interpulse intervals are as follows: 100 (c), 70 (d), and 50 (e) ms. Each point shows the EPSC amplitudes, averaged from 20 sweeps, normalized to the amplitude (Amp (Norm)) of the first EPSC (*, p < 0.05; **, p < 0.01; bars show S.E.).
DISCUSSION
In this study, we investigated levels of synaptic proteins, ultrastructure, and the physiology of the synapses in the hippocampus of Fmr1 KO mice, a model for FXS. Although the mRNA targets of FMRP have been well characterized in neuronal synapses in brain tissue, it is not known how this is reflected in a dysregulated synaptic proteome. We first took a quantitative approach using 8-plex iTRAQ reagents in conjunction with LC-LC MALDI MS/MS analysis to compare the proteomes of hippocampal synaptic membrane fractions from WT and Fmr1 KO mice. The high number of independent biological samples (n = 8) was instrumental in revealing subtle, statistically significant differences in protein levels of the synaptic proteome. Indeed, small differences are observed that have been reported to have significant physiological and/or morphological implications for FXS (7, 16, 30–32). Interestingly, only one of the differentially expressed proteins, Basp1, has been reported as an mRNA target of FMRP. This protein is implicated in regulation of cell morphology via the modulation of actin dynamics, together with e.g. Gap43, and both show increased levels in Fmr1 KO mice. These proteins are cytoskeleton-associated and regulate actin dynamics through a similar mechanism (33, 34). Their regulatory roles in actin dynamics may contribute to molecular mechanisms underlying neuronal development of Fmr1 KO mice by influencing synapse morphology. The expression of Basp1 and Gap43 is developmentally regulated and is high in axons during outgrowth and regeneration. Overexpression of Gap43 and Basp1 has been shown to promote axon sprouting (33). Taken together Basp1 and Gap43 are likely to be important regulators of synapse morphology via remodeling of the actin cytoskeleton. In agreement with this, a downstream effector of actin dynamics, cofilin-1, was also dysregulated in KO synapses. Indeed, dysregulated actin dynamics have been proposed previously as an important factor in FXS (35, 36). In addition, Cend1, a protein important for cell differentiation, is significantly up-regulated. Cend1 is expressed throughout all brain regions with a high level in the hippocampus. It is involved in neuronal differentiation in the developing embryonic brain and was shown to enhance morphological differentiation of mouse neuroblastoma cells in vitro (37, 38). Thus, Cend1 up-regulation may represent an upstream effector underlying the altered hippocampal synapse morphology in Fmr1 KO mice. Previous studies in the Drosophila model of FXS are in agreement with our findings; the Drosophila fragile X gene dFXR was shown to negatively regulate neuronal elaboration and synaptic differentiation, whereas overexpression had the opposite effect (39).
The down-regulation of β-catenin we observed in the Fmr1 KO synapses may have both structural and functional implications for hippocampal neurons. β-Catenin is involved in activity-dependent synaptic remodeling and gene transcription (40) and is present in both the presynaptic active zone and the postsynaptic density. Deletion of β-catenin in the presynaptic terminal has been shown to reduce the reserve pool of synaptic vesicles, resulting in synaptic depression during repeated stimulation (41). In facilitating synapses between CA3-CA1 hippocampal neurons, we observed a significantly reduced facilitation in Fmr1 KO mice. In addition, ablating β-catenin in the postsynaptic neuron results in thin and elongated immature spines (42), an observation in line with the immature ultrastructural findings we reported, and has been observed in many brain regions in Fmr1 KO mice (7).
In the present study, we showed that specific synaptic vesicle proteins were up-regulated in the synaptic membrane preparation of Fmr1 KO mice. Previous studies examined long-term plasticity with an underlying postsynaptic locus in Fmr1 KO synapses and revealed significant impairments in long-term potentiation (28). We wished to test whether glutamate release and its short-term dynamics were altered by regulation of proteins in the axon terminal of KO neurons. Recording mEPSCs samples synaptic currents from all regions of the dendrite, including basal and proximal branches of a neuron. We found no basal changes in mEPSCs at these synapses, in line with observations at other hippocampal and cortical synapses (43–45). However, the short-term dynamics of this Schaffer collateral input pathway were significantly modified. In Fmr1 KO mice, the response to a second stimulus in a paired-pulse ratio was significantly lower than in WT mice. Paired-pulse facilitation is generally acknowledged to have a presynaptic locus; thus, the lower ratio in Fmr1 KO mice points toward a presynaptic defect in function. In line with these findings, the smaller AZ and PSD observed in Fmr1 KO synapses together with the overall decrease in numbers of synaptic vesicles and down-regulation of β-catenin correlate with the reduced ability of the synapse to respond to rapid, repeated stimulation. Furthermore, these synapses were also impaired in their facilitating response to a train of synaptic stimuli in Fmr1 KO, at interstimulus intervals of between 50 and 100 ms. This physiological range of stimulus frequencies could reflect an altered processing of the hippocampal network as a whole during β-frequency (10–20 Hz) activity, as observed during hippocampal oscillations (46).
In our study, all proteomics analysis, electrophysiology, and electron microscopy were carried out at 2 postnatal weeks of development. Postnatal age has a significant effect upon the alteration in spine morphology reported in the Fmr1 KO mouse; in sensory cortex, abnormalities are observed in spine length at both 1 and 2 postnatal weeks, but not at 4 weeks old (7). The dendritic spine abnormality returns, however, later than 4 weeks of age in the cortex (47). Synaptic function in the somatosensory barrel cortex is also age-dependent, with significantly altered synaptic AMPA and NMDA ratios being observed during the 1st postnatal week but normalizing by 2–3 weeks of postnatal age (32). Thus, the alterations we observed at this key period for spine formation and pruning could be restricted to specific developmental time points. Indeed, no changes have been observed in PPR ratios in adolescent (4–6 weeks) Fmr1 KO mice as compared with WT mice (48), thus showing evidence that there may be critical developmental time windows for the specific effects of FMRP upon hippocampal synapse structure and resulting function.
Although our data from three differing techniques reveal clear presynaptic changes, it does not rule out additional postsynaptic factors from affecting the synapse at this developmental age. Both NMDA and AMPA receptor subunits showed slight small down-regulation in our Fmr1 KO samples, but the changes were not statistically significant. Previous studies reported no overall changes in NMDA and AMPA receptors in the hippocampus of Fmr1 KO1 mice (14, 49), but only signaling via an mGluR5-mediated activity-dependent increase in AMPA receptors internalization in Fmr1 KO mice (15, 16). Alteration of other signal transduction pathways, especially the decrease in production of cAMP, has been reported in several model systems of FXS (50). We detected an increase in PKA levels in Fmr1 KO synapses, which may in part compensate for a decreased production of cAMP.
In our experiments, we used the Fmr1 KO1 mouse model for FXS, which is in widespread usage and has a total absence of FMRP but traces of Fmr1 mRNA. One recent study using an “Fmr1 KO2” mouse model that is deficient in both Fmr1 RNA and FMRP showed opposing changes in synaptic function as compared with other current research in the field (16). Also, in contrast to our study using Fmr1 KO1 mice, Pilpel et al. (16) observed a normal paired-pulse ratio. These observations might point to the importance of the remaining levels of mRNA of Fmr1 in the conventional Fmr1 KO1 model in the regulation of synapse function.
In conclusion, our quantitative proteomics data set reports the global changes in FMRP-regulated expression at this critical point in hippocampal synapse development. Regulation of specific proteins with presynaptic vesicle and morphological growth function correlate with the ultrastructural changes we observed in synaptic vesicles and active zone size at the EM level in Fmr1 KO synapses. These changes can be attributed to the absence of FMRP during early development of the hippocampus. Alteration in protein expression and immature morphology at presynaptic loci correlate with impairments in rapid functional dynamics of short-term plasticity. In combination, we propose that these abnormalities show clear alterations of presynaptic structure and function in the Fmr1 KO mouse and contribute to the underlying molecular basis of hippocampal deficits in FXS.
Supplementary Material
Acknowledgments
The Fmr1 KO mice were a kind gift of Prof. B. Oostra and Dr R. Willemsen (Erasmus MC, Rotterdam, The Netherlands). Prof. M. Verhage (VU University, Amsterdam, The Netherlands) and Prof. R. Matsas (Hellenic Pasteur Institute, Athens, Greece) provided some of the antibodies for immunoblotting. We thank Dr. E. W. van Zwert (Leiden University Medical Center, Leiden, The Netherlands) for comments on the statistical analysis.
This work was supported by the NeuroBsik grant to the Dutch Mouse Phenomics consortium and support of the Center for Medical Systems Biology (CMSB) Netherlands Organization for Scientific Research (NWO) (Grant 91710372) (to A. B. S., K. W. L., and R. C. v. d. S.), funding from Hersenstichting Nederland (to R. M. M.), and funding from Neuromics Marie Curie Early stage Training Grant MEST-CT-2005-020919 (to P. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and Table 1.
- FXS
- fragile X syndrome
- FMRP
- fragile X mental retardation protein
- AZ
- active zone
- PSD
- postsynaptic density
- EPSC
- excitatory postsynaptic current
- mEPSC
- miniature EPSC
- PPR
- paired-pulse ratio
- WT
- wild type
- KO
- knock-out.
REFERENCES
- 1. Garber K. B., Visootsak J., Warren S. T. (2008) Eur. J. Hum. Genet. 16, 666–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koukoui S. D., Chaudhuri A. (2007) Brain Res. Rev. 53, 27–38 [DOI] [PubMed] [Google Scholar]
- 3. Dictenberg J. B., Swanger S. A., Antar L. N., Singer R. H., Bassell G. J. (2008) Dev. Cell 14, 926–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown V., Jin P., Ceman S., Darnell J. C., O'Donnell W. T., Tenenbaum S. A., Jin X., Feng Y., Wilkinson K. D., Keene J. D., Darnell R. B., Warren S. T. (2001) Cell 107, 477–487 [DOI] [PubMed] [Google Scholar]
- 5. Bagni C., Greenough W. T. (2005) Nat. Rev. Neurosci. 6, 376–387 [DOI] [PubMed] [Google Scholar]
- 6. Bassell G. J., Warren S. T. (2008) Neuron 60, 201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nimchinsky E. A., Oberlander A. M., Svoboda K. (2001) J. Neurosci. 21, 5139–5146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Irwin S. A., Galvez R., Greenough W. T. (2000) Cereb. Cortex 10, 1038–1044 [DOI] [PubMed] [Google Scholar]
- 9. Antar L. N., Li C., Zhang H., Carroll R. C., Bassell G. J. (2006) Mol. Cell. Neurosci. 32, 37–48 [DOI] [PubMed] [Google Scholar]
- 10. Ivanco T. L., Greenough W. T. (2002) Hippocampus 12, 47–54 [DOI] [PubMed] [Google Scholar]
- 11. Meredith R. M., de Jong R., Mansvelder H. D. (2011) Neurobiol. Dis. 41, 104–110 [DOI] [PubMed] [Google Scholar]
- 12. Meredith R. M., Mansvelder H. D. (2010) Front. Synaptic Neurosci. 10, 2–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilson B. M., Cox C. L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2454–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li J., Pelletier M. R., Perez Velazquez J. L., Carlen P. L. (2002) Mol. Cell. Neurosci. 19, 138–151 [DOI] [PubMed] [Google Scholar]
- 15. Nosyreva E. D., Huber K. M. (2006) J. Neurophysiol. 95, 3291–3295 [DOI] [PubMed] [Google Scholar]
- 16. Pilpel Y., Kolleker A., Berberich S., Ginger M., Frick A., Mientjes E., Oostra B. A., Seeburg P. H. (2009) J. Physiol. 587, 787–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Costa-Mattioli M., Sossin W. S., Klann E., Sonenberg N. (2009) Neuron 61, 10–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Massey P. V., Bhabra G., Cho K., Brown M. W., Bashir Z. I. (2001) Eur. J. Neurosci. 14, 145–152 [DOI] [PubMed] [Google Scholar]
- 19. Volk L. J., Pfeiffer B. E., Gibson J. R., Huber K. M. (2007) J. Neurosci. 27, 11624–11634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Christie S. B., Akins M. R., Schwob J. E., Fallon J. R. (2009) J. Neurosci. 29, 1514–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bureau I., Shepherd G. M., Svoboda K. (2008) J. Neurosci. 28, 5178–5188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gibson J. R., Bartley A. F., Hays S. A., Huber K. M. (2008) J. Neurophysiol. 100, 2615–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dutch-Belgian Fragile X Consortium (1994) Cell 78, 23–33 [PubMed] [Google Scholar]
- 24. Li K. W., Miller S., Klychnikov O., Loos M., Stahl-Zeng J., Spijker S., Mayford M., Smit A. B. (2007) J. Proteome Res. 6, 3127–3133 [DOI] [PubMed] [Google Scholar]
- 25. Klychnikov O. I., Li K. W., Sidorov I. A., Loos M., Spijker S., Broos L. A., Frants R. R., Ferrari M. D., Mayboroda O. A., Deelder A. M., Smit A. B., van den Maagdenberg A. M. (2010) Proteomics 10, 2531–2535 [DOI] [PubMed] [Google Scholar]
- 26. Tusher V. G., Tibshirani R., Chu G. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roxas B. A., Li Q. (2008) BMC Bioinformatics 9, 187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meredith R. M., Holmgren C. D., Weidum M., Burnashev N., Mansvelder H. D. (2007) Neuron 54, 627–638 [DOI] [PubMed] [Google Scholar]
- 29. Van den Oever M. C., Goriounova N. A., Li K. W., Van der Schors R. C., Binnekade R., Schoffelmeer A. N., Mansvelder H. D., Smit A. B., Spijker S., De Vries T. J. (2008) Nat. Neurosci. 11, 1053–1058 [DOI] [PubMed] [Google Scholar]
- 30. Irwin S. A., Patel B., Idupulapati M., Harris J. B., Crisostomo R. A., Larsen B. P., Kooy F., Willems P. J., Cras P., Kozlowski P. B., Swain R. A., Weiler I. J., Greenough W. T. (2001) Am. J. Med. Genet. 98, 161–167 [DOI] [PubMed] [Google Scholar]
- 31. Comery T. A., Harris J. B., Willems P. J., Oostra B. A., Irwin S. A., Weiler I. J., Greenough W. T. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5401–5404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harlow E. G., Till S. M., Russell T. A., Wijetunge L. S., Kind P., Contractor A. (2010) Neuron 65, 385–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Frey D., Laux T., Xu L., Schneider C., Caroni P. (2000) J. Cell Biol. 149, 1443–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Laux T., Fukami K., Thelen M., Golub T., Frey D., Caroni P. (2000) J. Cell Biol. 149, 1455–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Castets M., Schaeffer C., Bechara E., Schenck A., Khandjian E. W., Luche S., Moine H., Rabilloud T., Mandel J. L., Bardoni B. (2005) Hum. Mol. Genet. 14, 835–844 [DOI] [PubMed] [Google Scholar]
- 36. Reeve S. P., Bassetto L., Genova G. K., Kleyner Y., Leyssen M., Jackson F. R., Hassan B. A. (2005) Curr. Biol. 15, 1156–1163 [DOI] [PubMed] [Google Scholar]
- 37. Georgopoulou N., Hurel C., Politis P. K., Gaitanou M., Matsas R., Thomaidou D. (2006) J. Biol. Chem. 281, 33606–33620 [DOI] [PubMed] [Google Scholar]
- 38. Mamalaki A., Boutou E., Hurel C., Patsavoudi E., Tzartos S., Matsas R. (1995) J. Biol. Chem. 270, 14201–14208 [DOI] [PubMed] [Google Scholar]
- 39. Tessier C. R., Broadie K. (2008) Development 135, 1547–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Arikkath J. (2009) Mol. Neurobiol. 40, 46–54 [DOI] [PubMed] [Google Scholar]
- 41. Bamji S. X., Shimazu K., Kimes N., Huelsken J., Birchmeier W., Lu B., Reichardt L. F. (2003) Neuron 40, 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Okuda T., Yu L. M., Cingolani L. A., Kemler R., Goda Y. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13479–13484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Braun K., Segal M. (2000) Cereb Cortex 10, 1045–1052 [DOI] [PubMed] [Google Scholar]
- 44. Desai N. S., Casimiro T. M., Gruber S. M., Vanderklish P. W. (2006) J. Neurophysiol. 96, 1734–1745 [DOI] [PubMed] [Google Scholar]
- 45. Pfeiffer B. E., Huber K. M. (2007) J. Neurosci. 27, 3120–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mann E. O., Radcliffe C. A., Paulsen O. (2005) J. Physiol. 562, 55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Galvez R., Greenough W. T. (2005) Am. J. Med. Genet. A 135, 155–160 [DOI] [PubMed] [Google Scholar]
- 48. Zhang J., Hou L., Klann E., Nelson D. L. (2009) J. Neurophysiol. 101, 2572–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Giuffrida R., Musumeci S., D'Antoni S., Bonaccorso C. M., Giuffrida-Stella A. M., Oostra B. A., Catania M. V. (2005) J. Neurosci. 25, 8908–8916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kelley D. J., Davidson R. J., Elliott J. L., Lahvis G. P., Yin J. C., Bhattacharyya A. (2007) PLoS One 2, e931 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.