Abstract
In response to severe stress, apoptotic cell death is engaged. Apoptosis is a well orchestrated process that involves the activation and implication of many factors. In this study, we identified a role for the nuclear trafficking factor TRN2 (transportin 2) in cell death. TRN2 is normally responsible for the nuclear import of the RNA-binding protein HuR. During apoptosis, however, HuR accumulates in the cytoplasm. This is due to the caspase-mediated cleavage of the cytoplasmic fraction of HuR. One of the cleavage fragments generated by this processing of HuR interacts with TRN2 and thus blocks the re-import of HuR into the nucleus. This concentrates HuR in the cytoplasm, advancing apoptosis. Therefore, increasing or decreasing the levels of TRN2 has an inverse consequential effect on cell death, demonstrating for the first time the role of a nucleocytoplasmic transport factor in apoptosis.
Keywords: Cell Death, Cellular Regulation, mRNA, RNA-binding Protein, Trafficking, HuR, Stress Response, Transportin 2
Introduction
When confronted with stress, eukaryotic cells undergo different responses depending on the severity and duration of the stress (1). Although adaptation and survival are at one end of these responses, cell death can also be the outcome. Apoptotic cell death is a complex pathway involving the activation and cleavage of several proteins, some of the most important of which are cysteine-aspartic proteases (caspases). This family of proteases then cleaves specific substrates, allowing the progression of cell death (2–5). In addition to causing the organized destruction of a cell, caspase-mediated cleavage can also serve to activate target proteins that have important roles in advancing apoptosis themselves. Recently, the RNA-binding protein HuR was identified as one such factor (6). Under normal conditions, HuR is an important regulator of gene expression, controlling the localization, stabilization, and translation of many different mRNAs (7, 8). The targets of HuR are involved in many cellular processes, including cell growth, differentiation, and survival and apoptotic cell death. Likewise, the importance of HuR in regulating these processes has been shown, given that its absence or overexpression in different systems can have potent effects on cell physiology (6, 9–12).
The role of HuR as a promoter of cell death was identified when it was found that caspase-3 and caspase-7 cleave HuR in response to lethal stress (6), such as with the use of the apoptotic stimulator drug staurosporine (STS).4 The cleavage of HuR, occurring at Asp-226 of this protein, yields two cleavage products: HuR-CP1 (26 kDa) and HuR-CP2 (8 kDa). Mutation of this cleavage site prevents apoptosis, whereas providing these two cleavage products can stimulate cell death under nonlethal conditions (13). It was shown that HuR-CP2 helps promote apoptosis by binding to putative HLA-associated protein-I (PHAPI) (6), an activator of apoptosis and a known ligand of full-length HuR. However, the implication of other protein partners has not yet been explored.
Intriguingly, prior to its cleavage, HuR, which typically shuttles between the nucleus and the cytoplasm, must have an increased cytoplasmic localization. We previously observed that this occurs as soon as 1 h following STS treatment, whereas HuR cleavage only appears on a Western blot after 1.5–2 h (6). Furthermore, although only ∼10% of total HuR is cleaved during stress (6), this is ∼50% of the total percentage of HuR found in the cytoplasm (22). Similar results were also found during muscle differentiation. HuR and its cleavage are crucial for the fusion of muscle cells (myoblasts) to form fibers (myotubes) (9, 10, 14). In a recent study (14), we showed that this cleavage, which also depends on caspases and occurs at Asp-226, is also related to the cytoplasmic accumulation of HuR. Upon being cleaved, HuR-CP1 competes with full-length HuR for binding to its import factor TRN2 (transportin 2) (14, 15). TRN2 was previously shown to promote the nuclear import of HuR (16, 17), and in our muscle differentiation study, we found that, by binding TRN2, HuR-CP1 prevents the nuclear reuptake of HuR, thus causing it to accumulate in the cytoplasm (14). The consequence of this accumulation the promotion of myogenesis and HuR cleavage.
These data have raised the possibility that a similar mechanism may be taking place during the stress response. The region of HuR previously shown to associate with TRN2, the HuR nucleocytoplasmic shuttling (HNS) domain (16–18), is in fact the location of the Asp-226 caspase cleavage site of HuR (6, 14). This further supports that the cleavage of HuR may interfere with the normal localization and nuclear import of HuR during apoptosis. Therefore, in this study, we investigated the role of TRN2 in regulating HuR cleavage and also if TRN2 itself contributes to the apoptotic response.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfections, and Treatments
HeLa CCL-2TM cells (American Type Culture Collection) were grown and maintained in l-glutamine-containing DMEM (Sigma) supplemented with fetal bovine serum (10%) and penicillin/streptomycin (Sigma). Plasmids (described below) and siRNA duplexes (control and HuR siRNAs (9) and TRN2 siRNA (15)) were transfected when cells were at ∼70% confluency (for plasmid transfections) or 50% confluency (for siRNA transfections) as described previously. When transfecting cells with TRN2 siRNA, this transfection was repeated 24 h following the first transfection. In all cases, samples were harvested 48 h following transfections. Lipofectamine and PLUS reagents (Invitrogen) were used according to the manufacturer's recommendations. STS (Sigma) was used at 1 μm for 3 h to induce cell death as described previously (6).
Immunofluorescence and Quantification of Nuclear Fragmentation
HeLa cells plated on coverslips were treated as described in the figure legends. Cells were fixed, and immunofluorescence was performed as described (19). Anti-HuR monoclonal antibody (3A2) (20), anti-G3BP (Ras GTPase-activating protein SH3 domain-binding protein) polyclonal antibody (23), and Alexa Fluor 488-conjugated secondary antibody (Molecular Probes) were used along with DAPI stain. Images were taken at room temperature with a ×63 oil objective using an Axiovert 200M microscope (Carl Zeiss, Inc.) as described previously (6). To determine nuclear fragmentation, pictures of 10 fields/condition were taken, and the sum of all fragmented nuclei in these fields was divided by the sum of all nuclei (whole and fragmented) in the same fields.
Cellular Fractionation, Cell Extract Preparation, and Western Blotting
Cytoplasmic and nuclear fractions, along with total cell extracts, were prepared as described previously (13, 21). Western blotting was performed as described (22) using the following antibodies: anti-HuR (3A2) (20); anti-heterogeneous nuclear ribonucleoprotein A1 and anti-cleaved caspase-3 and caspase-7 (Cell Signaling); anti-FLAG (Sigma); anti-α-tubulin (Developmental Studies Hybridoma Bank); anti-G3BP (23); and anti-GFP (which also recognizes cyan fluorescent protein (CFP); Clontech). Quantification of bands on Western or slot blots was done using ImageQuant software as described in the figure legends. Statistical analysis for significance was performed using on-line GraphPad software with a two-tailed unpaired t test.
RNA Isolation and Slot Blotting
Cells were collected in TRIzol (Invitrogen), and mRNA was isolated as described according to the manufacturer's protocol. RNA was then loaded onto a slot blot and analyzed as described (19) using radiolabeled probes complementary to TRN2 and 18 S RNAs as described previously (15, 24).
Immunoprecipitation
HeLa cells were treated as detailed in the figure legend and collected. Immunoprecipitation was performed as described previously (14) using protein A beads (GE Healthcare) with anti-TRN2 (15) or IgG control (Jackson ImmunoResearch Laboratories) antibody.
Plasmid Construction
GFP, GFP-HuR, and GFP-HuR-CP1 plasmids were described previously (6, 13). The CFP-TRN2 plasmid was generated as described previously (14) except that CFP-containing pcDNA3 (generously donated by J. Pelletier) was used in place of the YFP-containing plasmid. The primers used were the same as for YFP-TRN2 (14). FLAG-HuR-CP1 and FLAG-HuR-CP2 plasmids were generated by PCR amplification with the GFP-HuR plasmid as a template using the following primers (the forward primers used also contained a sequence to produce a FLAG tag at the 5′-end of the proteins): FLAG-CP1-F, 5′-ccg gaa ttc atg gac tac aaa gac gac gac gac aaa tct aat ggt tat gaa gac cac-3′; FLAG-CP1-R, 5′-taa agc ggc cgc tta atc gac gcc cat ggg gga-3′; FLAG-CP2-F, 5′-ccg gaa ttc atg gac tac aaa gac gac gac gac aaa cac atg agc ggg ctc tct-3′; and FLAG-CP2-R, 5′-gaa agc ggc cgc tta ttt gtg gga ctt gtt ggt ttt-3′. For both HuR-CP1 and HuR-CP2 amplification, this created an EcoRI/NotI PCR product, which was restriction-digested and ligated into the pcDNA3 plasmid (Invitrogen).
RESULTS
TRN2 Regulates HuR Localization and Cleavage
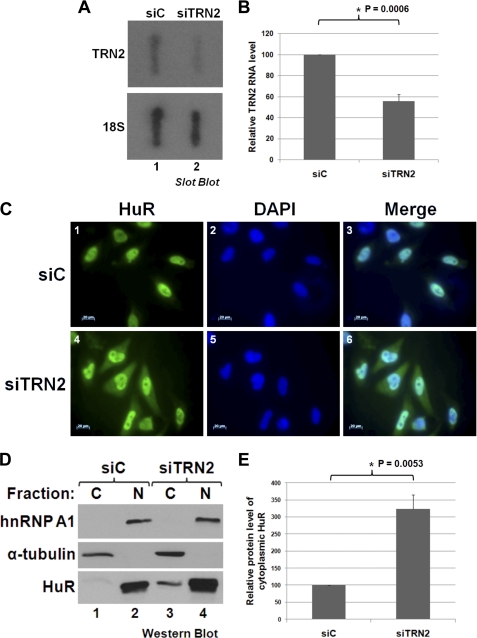
To investigate if there is a link between TRN2 and the cytoplasmic accumulation of HuR during apoptosis, we first depleted TRN2 expression and followed HuR localization. We observed that the knockdown of TRN2 (TRN2 siRNA), compared with control siRNA (Fig. 1, A and B), resulted in an increase in the portion of HuR that could be found in the cytoplasm (Fig. 1, C–E). This was seen visually both by performing immunofluorescence staining against HuR (Fig. 1C) and by isolating the nuclear and cytoplasmic fractions of transfected cells and examining the amount of HuR present in each fraction (Fig. 1, D and E). These data suggest that, in HeLa cells, TRN2 regulates the nuclear localization of HuR seen under normal conditions. This does not evaluate, however, the implication of TRN2 in the stress response.
FIGURE 1.
TRN2 regulates the nuclear localization of HuR. A and B, TRN2 siRNA reduces the level of TRN2 RNA. HeLa cells were transfected with either an siRNA that targets TRN2 (siTRN2) or control siRNA (siC). Their total RNA content was isolated and analyzed by slot blotting, probing for TRN2 and 18 S (loading control) RNAs. Quantification was performed using ImageQuant software, and average values of TRN2 RNA levels, relative to 18 S RNA levels, are graphed in B, with error bars representing S.E. of four independent experiments. The asterisk indicates that the observed difference is significant, with p = 0.0006. C–E, knockdown of TRN2 causes HuR to accumulate in the cytoplasm. Cells were treated as described for A. They were fixed, and immunofluorescence was performed by staining with anti-HuR antibody and DAPI (C). Scale bars = 20 μm. Alternatively, the total content of lysed cells was separated into nuclear (N) and cytoplasmic (C) fractions and analyzed by Western blotting (D) with antibodies against HuR, heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1; nuclear marker), and α-tubulin (cytoplasmic marker). Quantification of HuR and α-tubulin band intensities was determined using ImageQuant software, and average values of cytoplasmic HuR levels relative to α-tubulin are graphed in E, with error bars representing S.E. of three independent experiments. The asterisk indicates that the observed difference is significant, with p = 0.0053. All images shown in C and the blot in D are representative of three independent experiments.
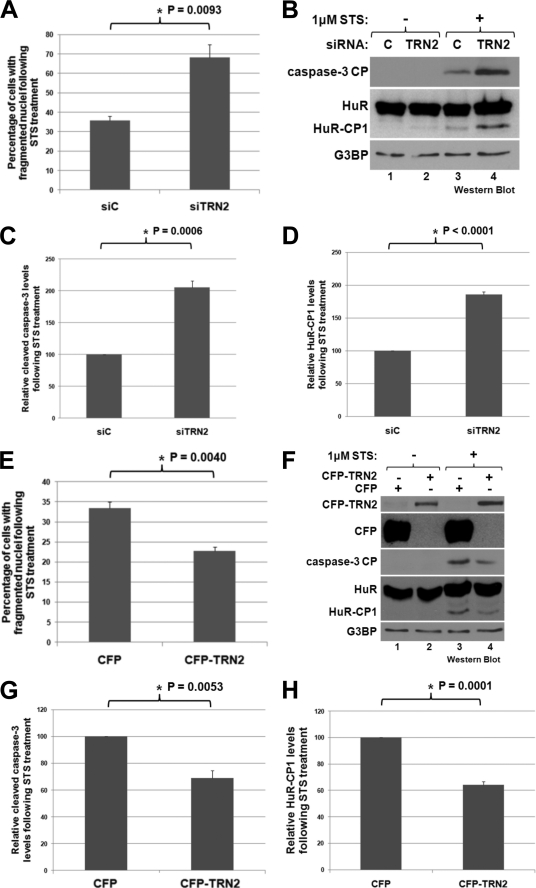
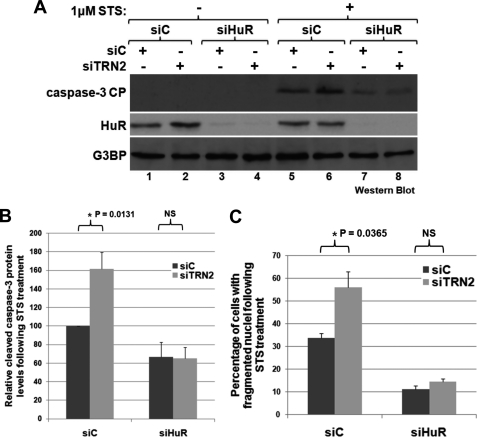
To determine whether TRN2 is involved in apoptosis, knockdown of its expression was repeated, and cells were then treated or not with STS to induce death. The reduction of TRN2 under these conditions resulted in a significant increase in the fragmentation of cellular nuclei as seen by performing DAPI staining on fixed cells (Fig. 2A and supplemental Fig. 1). During apoptosis, nuclei condense to become smaller and then fragment as the cells die, so quantification of the percentage of cells with fragmented nuclei is a reflection of the apoptotic state of the population (6). Additionally, the cleavage of caspase-3, an important caspase and a known marker of apoptosis (5), was assessed and quantified (Fig. 2, B and C). Both of these measures of apoptosis indicated that a reduction of TRN2 expression increased the apoptotic response to STS. When TRN2 was overexpressed by transfecting a CFP-tagged TRN2 plasmid into HeLa cells, followed by treatment with STS, the opposite response was observed (Fig. 2, E–G). In this situation, nuclear fragmentation and caspase-3 cleavage significantly increased. Intriguingly, upon both the reduction and overexpression of TRN2, HuR cleavage also changed (Fig. 2, B, D, F, and H), similar to caspase-3 cleavage. This raised the possibility that the effect on apoptosis of modifying TRN2 expression may depend on HuR cleavage. Conversely, because HuR was identified as a target of caspase-3, the changes observed in HuR cleavage may simply be secondary. To determine whether this was the case or not, TRN2 and HuR were knocked down simultaneously to assess whether TRN2 knockdown would still promote apoptosis in the absence of HuR (Fig. 3). As expected (6), cell death was reduced in the absence of HuR (determined by caspase-3 cleavage (Fig. 3, A and B) and the percentage of fragmented nuclei (Fig. 3C)). Importantly, the significant increase in caspase-3 cleavage and nuclear fragmentation that occurs during knockdown of TRN2 was lost in cells depleted of endogenous HuR (Fig. 3, A, compare lanes 5 and 6 with lanes 7 and 8; and C). This supports that it is through HuR cleavage that TRN2 regulates apoptosis and that the observed changes in the level of caspase-3 cleavage and nuclear fragmentation following knockdown and overexpression of TRN2 are a consequence of this regulation.
FIGURE 2.
TRN2 affects apoptosis and HuR cleavage. A, STS treatment causes an increase in nuclear fragmentation following TRN2 knockdown. HeLa cells were transfected with control siRNA (siC) or TRN2 siRNA (siTRN2) as described in the legend to Fig. 1, and 48 h after transfection, they were treated for 3 h with 1 μm STS. Cells were then fixed and stained with DAPI, a nuclear fluorescent marker, and images were taken. 10 fields/condition were used to determine the percentage of cells in which nuclei appeared fragmented. Average percentages are graphed, with error bars representing S.E. of three independent experiments. The asterisk indicates that the observed difference is significant, with p = 0.0093. B–D, knockdown of TRN2 results in an increase in the cleavage of caspase-3 and HuR. Cells treated as described for A were collected after STS treatment, and their lysates were analyzed by Western blotting (B) using antibodies against HuR, the caspase-3 cleavage product (CP; a known marker of apoptosis), and G3BP (loading control). Cleavage products of caspase-3 and HuR were quantified as described in the legend to Fig. 1, and average values relative to the loading control are graphed in C and D, with error bars representing S.E. of three independent experiments. The asterisks indicate that the observed differences between control siRNA (C)- and TRN2 siRNA-treated cells are significant, with p = 0.0006 for caspase-3 cleavage and p < 0.0001 for HuR cleavage. E, STS treatment causes a decrease in nuclear fragmentation following TRN2 overexpression. HeLa cells were transfected with plasmids encoding CFP or CFP-tagged TRN2 and were treated with STS as described for A. Cells were then fixed and stained, and quantification of 10 fields/condition for three independent experiments was also done as described for A. The average percentages of cells with fragmented nuclei are graphed, with error bars representing S.E. of three independent experiments. The asterisk indicates that the observed difference is significant, with p = 0.0040. F–H, overexpression of TRN2 causes a decrease in the cleavage of caspase-3 and HuR. Total cell lysates were prepared from samples treated as described for E and were analyzed by Western blotting using antibodies against CFP, cleaved caspase-3, HuR, and G3BP (loading control). Quantification of cleavage products was performed as described above and is shown in G and H, with error bars representing S.E. of three independent experiments. The asterisks indicate that the observed differences are significant, with p = 0.0053 for caspase-3 cleavage and p = 0.0001 for HuR cleavage.
FIGURE 3.
TRN2 influences apoptosis in an HuR-dependent manner. HeLa cells were transfected with either control siRNA (siC) or an siRNA targeting HuR (siHuR) in combination with an additional amount of control siRNA or TRN2 siRNA (siTRN2). 48 h post-transfection, cells were treated with STS as described in the legend to Fig. 2A and analyzed by Western blotting (A and B) using antibodies against HuR, cleaved caspase-3, and G3BP (loading control) or by microscopy with DAPI stain (C). Quantification of caspase-3 cleavage was performed as described in the legend to Fig. 2 and is graphed in B. Images were taken of DAPI-stained samples, and 10 fields were used for each condition so that the percentage of cells with fragmented nuclei could be assessed. The average percentages of three independent experiments are graphed in C. Error bars in B and C indicate S.E. of three independent experiments, and the asterisks indicate that the observed differences are significant, with p = 0.0131 (B) and p = 0.0365 (C). NS, not statistically significant.
HuR-CP1 Binds TRN2 to Promote Apoptosis
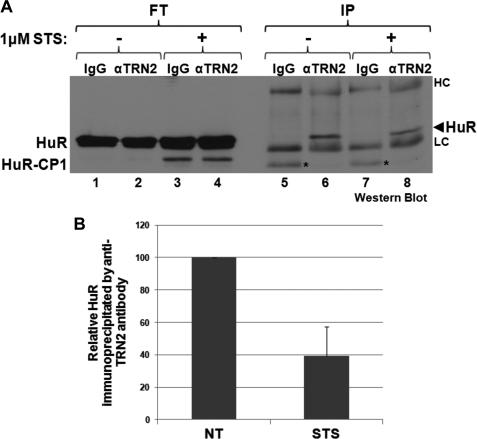
The observations described above identified a role for TRN2 in regulating apoptosis. A role for TRN2 in the stress response was previously proposed when we showed that the interaction between HuR and TRN2 is lost following heat shock (18), but the mechanism behind this effect was not determined. To better understand how TRN2 (through HuR cleavage) is implicated in the stress response and cell death, we wished to determine whether the HuR/TRN2 interaction in HeLa cells would also change following STS treatment. HeLa cells were treated or not with STS, and immunoprecipitation was performed using anti-TRN2 antibody to evaluate the association of HuR with this transport factor (Fig. 4). As expected, HuR interacted with TRN2, but in response to STS, this interaction was reduced (Fig. 4, compare lanes 6 and 8). A similar change in HuR/TRN2 association was reported to occur during muscle differentiation (15). It was later shown that this dissociation is due to an interaction between TRN2 and the larger HuR cleavage product, HuR-CP1 (14). HuR-CP1 was actually shown in vitro to outcompete HuR for binding to TRN2, whereas HuR-CP2 has no effect (see Fig. 5d of Ref. 14). These data raise the possibility that, in our apoptotic system, HuR-CP1 may play a similar role.
FIGURE 4.
HuR loses association with TRN2 following STS treatment. A, HeLa cells were treated with 1 μm STS for 3 h or not, and their lysates were used for immunoprecipitation with either anti-TRN2 antibody or IgG control antibody. Flow-through fractions (FT) and immunoprecipitates (IP) were analyzed by Western blotting and probed with anti-HuR antibody. The heavy (HC) and light (LC) chains of the antibodies appeared on the Western blot. Additionally, a nonspecific band appeared in lanes 5 and 7, indicated by asterisks. Shown is a representative blot of two independent experiments. B, the band intensities of HuR in lanes 6 and 8 were quantified, and the averages of the relative band intensities are graphed, with error bars representing S.E. of two independent experiments. NT, no treatment.
FIGURE 5.
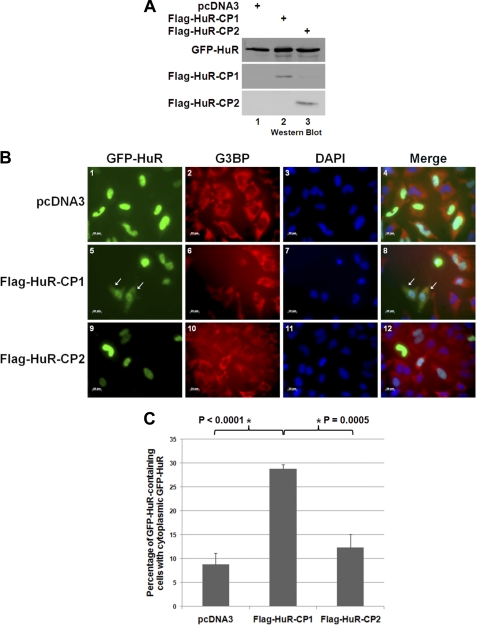
HuR-CP1 is responsible for the cytoplasmic accumulation of HuR. A–C, HeLa cells were cotransfected with GFP-HuR and either the pcDNA3 empty vector or the pcDNA3 vector containing FLAG-tagged HuR-CP1 or HuR-CP2. Lysates from these cells were prepared and analyzed by Western blotting 48 h after transfection using anti-FLAG and anti-GFP antibodies (A), or cells were fixed and immunofluorescence was performed using anti-G3BP antibody (a cytoplasmic marker) along with DAPI stain (B). Scale bars = 20 μm. The arrows indicate cells in which GFP-HuR was seen in the cytoplasm. The number of cells that contained GFP-HuR in 12 fields/condition was counted, and the number of cells in which GFP-HuR was found in the cytoplasm was determined. Average percentages from five independent experiments are graphed in C, with error bars representing S.E. of these five independent experiments, a representative field of which is shown in B. The asterisks indicate that the observed differences are significant, with p < 0.0001 when comparing pcDNA3 and FLAG-HuR-CP1 and p = 0.0005 when comparing FLAG-HuR-CP1 and HuR-CP2.
If HuR-CP1 indeed binds to TRN2 and thus causes HuR to accumulate in the cytoplasm in HeLa cells undergoing apoptosis, expressing this cleavage product should cause accumulation of full-length HuR in the cytoplasm. To determine whether this is the case, we wished to see if HuR-CP1 could affect the localization of HuR. Because our anti-HuR monoclonal antibody (3A2) recognizes the N terminus of the HuR protein, it is incapable of distinguishing between full-length HuR and HuR-CP1 in an immunofluorescence staining experiment. Therefore, GFP-tagged HuR was cotransfected into HeLa cells with either the pcDNA3 empty vector or the FLAG-tagged HuR-CP1 or HuR-CP2 plasmid (Fig. 5A). The localization of full-length HuR was monitored in these cells by fluorescence microscopy (Fig. 5B), and we observed that FLAG-HuR-CP1 caused significantly more cells to have HuR in the cytoplasmic compartment (co-staining with G3BP, a cytoplasmic marker) than either HuR-CP2 or the empty plasmid (Fig. 5, B and C). Because both TRN2 knockdown and HuR-CP1 overexpression (Figs. 1 and 5) caused an increase in the portion of HuR that was found in the cytoplasm, similar to lethal stress treatment (6), this supports our hypothesis that TRN2 and HuR-CP1 work together to coordinate this effect.
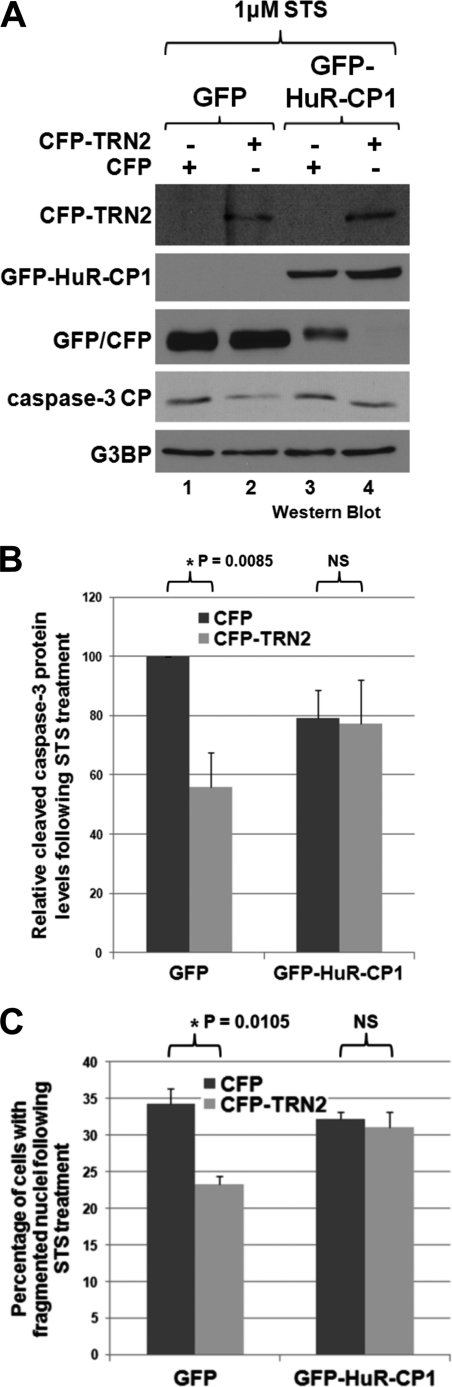
If TRN2 actually affects apoptosis by regulating the localization and cleavage of HuR, then it should be possible to negate the inhibitory effect of overexpressing TRN2 on apoptosis by providing HuR-CP1. CFP-TRN2 and GFP-HuR-CP1 (or CFP and GFP as respective controls) were cotransfected into HeLa cells. 48 h following these transfections, cells were treated with STS, and caspase-3 cleavage was evaluated by Western blotting as an indicator of cell death (Fig. 6, A and B). Nuclear fragmentation was also determined as described above (Fig. 6C). As expected, overexpression of TRN2 caused a decrease in both the cleavage of caspase-3 and the fragmentation of nuclei (Fig. 6, A, compare lanes 1 and 2; and C), but in the presence of HuR-CP1, these decreases were lost (Fig. 6A, compare lanes 3 and 4). Together, these results clearly show that the role of TRN2 in cell death is linked to its regulation of HuR localization and cleavage.
FIGURE 6.
TRN2 does not inhibit apoptosis in the presence of HuR-CP1. A–C, HeLa cells were cotransfected with either a plasmid containing GFP or GFP-tagged HuR-CP1 and either CFP or CFP-TRN2. 48 h following this transfection, cells were treated with 1 μm STS for 3 h and collected or fixed. Lysates were analyzed by Western blotting using antibodies against GFP/CFP (both are recognized), cleaved caspase-3, and G3BP (loading control), a representative blot of which is shown in A. The relative levels of cleaved caspase-3 were determined and normalized to loading controls as described above, and average values are graphed in B, with error bars representing S.E. of at least three independent experiments. Alternatively, fixed cells were stained with DAPI, and 10 images were taken per condition, which allowed the percentage of cells in which nuclei were fragmented to be determined. The average percentages of cells with fragmented nuclei of three independent experiments are graphed in C, with error bars representing S.E. of three independent experiments. The asterisks indicate significant differences, with p = 0.0085 (B) and p = 0.0105 (C). NS, not statistically significant.
DISCUSSION
TRN2 was previously implicated in regulating HuR localization in both HeLa and C2C12 muscle cells (15–17). It was later found that TRN2 plays an important role during the process of muscle differentiation, where the cytoplasmic accumulation and caspase-mediated cleavage of HuR are necessary (14). This accumulation is achieved by the binding of HuR-CP1 to TRN2, which prevents the nuclear import of full-length HuR. Thus, HuR moves out of the nucleus but fails to return, concentrating in the cytoplasmic compartment. In this study, we have found that a similar mechanism takes place following response to lethal stress. HuR is cleaved by caspases in the cytoplasm, and the HuR-CP1 fragment that is generated interferes with the TRN2-mediated import of HuR back into the nucleus.
The data presented here (Fig. 2) demonstrate that TRN2 is also involved in apoptosis. When TRN2 levels are modified during the stress response, caspase-3 levels and the fragmentation state of nuclei are altered, suggesting that TRN2 is an anti-apoptotic factor. By showing that this effect is dependent on HuR and can be negated by the presence of HuR-CP1, however (Figs. 3 and 6), we have demonstrated that the role of TRN2 in cell death is actually a consequence of the pro-apoptotic cleavage of HuR. Recently, protein kinase RNA (PKR) was also linked to regulation of HuR cleavage during the stress response (13). However, PKR has been strongly implicated in cell death, whereas this is the first time that a transport factor has been linked to regulating this process. Together, the previous PKR study and the results reported here reveal an added complexity to the apoptotic cascades that occur in response to lethal stimuli. Unlike PKR, which is involved in many cellular pathways (25), there have been only limited functions and partners identified for TRN2. Our data thus suggest that TRN2 levels could be modulated to influence cell fate. The fact that cell death can be affected by modifying the cytoplasmic accumulation of HuR as shown here suggests that clinical induction of apoptosis could be achieved or facilitated by blocking the HuR/TRN2 interaction.
The smaller cleavage fragment of HuR, HuR-CP2, is capable of interacting with the pro-apoptotic protein partner of HuR, PHAPI (6). PHAPI itself is also linked with HuR localization, as PHAPI contains a nuclear export signal and is essential for the cytoplasmic accumulation of HuR during the stress response (6, 18, 26). We previously showed that the association between HuR and PHAPI is mediated through the HNS domain and RNA recognition motif 3 of HuR, whereas the HNS domain was also shown to be necessary for the HuR/TRN2 interaction (18, 22). Intriguingly, although PHAPI associates with RNA recognition motif 3-containing HuR-CP2, only a portion of the HNS domain is part of this second HuR cleavage product. However, the bulk of the HNS domain remains within HuR-CP1, which we showed can interact with TRN2 to even a greater extent than full-length HuR (14). On the other hand, HuR-CP2 neither interacts with TRN2 (14) nor affects the localization of HuR (Fig. 5, B and C). Collectively, these data support the idea that interactions between full-length HuR and protein partners can be altered by the presence of caspase-generated HuR cleavage products. This also raises the possibility that HuR cleavage products can interact with protein partners that do not associate with full-length HuR. This may explain how the presence of these two cleavage products can trigger cell death under nonlethal conditions (13).
Intriguingly, the cleavage of HuR is detectable by Western blotting only after HuR begins to accumulate in the cytoplasm. In addition, we previously showed that HuR cleavage activates the apoptosome complex, although cleavage occurs downstream of this part of the apoptotic caspase cascade (6). One explanation for this is that some HuR cleavage occurs early following lethal stress. Although not detectable by Western blotting, the low amount of HuR cleavage products that are generated may cause cytoplasmic accumulation of HuR (HuR-CP1) and activation of the apoptosome (HuR-CP2), which is then capable of cleaving more HuR. This feedback loop mechanism was originally proposed upon the discovery of HuR cleavage (6) and is supported by the observations in this work. Intriguingly, the inhibition of caspases by benzyloxycarbonyl-VAD in our previous study did not appear to prevent the cytoplasmic localization of HuR after 3 h of STS treatment. Because some cleavage of HuR could be seen at that late time point, however, it is possible that this effect is due to the generation of some HuR-CP1, capable of binding TRN2. Of course, there is also the possibility that another unknown mechanism (such as a post-translational modification or a competing factor) may influence the association between HuR and TRN2, prior to HuR-CP1 production. In this case, HuR-CP1 could enhance the cytoplasmic accumulation of HuR that began prior to its appearance. Further studies on TRN2 ligands and modifications under both untreated and stress response conditions will help explore this possibility.
HuR is the best studied stabilizer of mRNA and has important roles in regulating the localization, translation, and stability of its targets, thus having a substantial ability to impact gene expression. HuR functions can be modulated by alterations such as the cleavage or localization of HuR itself, however, and upon the discoveries of such mechanisms, reconsideration of HuR function is required. Here, we have shown that the TRN2-mediated localization of HuR during the stress response is important for its pro-apoptotic role. This also implicates TRN2 in cell death, demonstrating that the ubiquitously expressed RNA-binding protein HuR can have extensive control over not only gene expression but also protein function.
Supplementary Material
Acknowledgments
We thank Dr. Sergio Di Marco for critical review of this manuscript and insightful discussions and Vincent Nadeau, Justin Chen, and Yu Yao for assisting with the production of the CFP- and FLAG-tagged constructs.
This work was supported in part by National Cancer Institute of Canada Operating Grant 016247 and Canadian Institutes of Health Research Operating Grant MOP-89798 (to I.-E. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- STS
- staurosporine
- PHAPI
- putative HLA-associated protein-I
- HNS
- HuR nucleocytoplasmic shuttling
- CFP
- cyan fluorescent protein
- PKR
- protein kinase RNA.
REFERENCES
- 1. Beere H. M. (2004) J. Cell Sci. 117, 2641–2651 [DOI] [PubMed] [Google Scholar]
- 2. Fadeel B., Ottosson A., Pervaiz S. (2008) Cell Death Differ. 15, 443–452 [DOI] [PubMed] [Google Scholar]
- 3. Johnson C. R., Jarvis W. D. (2004) Apoptosis 9, 423–427 [DOI] [PubMed] [Google Scholar]
- 4. Breckenridge D. G., Stojanovic M., Marcellus R. C., Shore G. C. (2003) J. Cell Biol. 160, 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Green D. R. (2005) Cell 121, 671–674 [DOI] [PubMed] [Google Scholar]
- 6. Mazroui R., Di Marco S., Clair E., von Roretz C., Tenenbaum S. A., Keene J. D., Saleh M., Gallouzi I. E. (2008) J. Cell Biol. 180, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fan X. C., Steitz J. A. (1998) EMBO J. 17, 3448–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abdelmohsen K., Lal A., Kim H. H., Gorospe M. (2007) Cell Cycle 6, 1288–1292 [DOI] [PubMed] [Google Scholar]
- 9. van der Giessen K., Di-Marco S., Clair E., Gallouzi I. E. (2003) J. Biol. Chem. 278, 47119–47128 [DOI] [PubMed] [Google Scholar]
- 10. Figueroa A., Cuadrado A., Fan J., Atasoy U., Muscat G. E., Muñoz-Canoves P., Gorospe M., Muñoz A. (2003) Mol. Cell. Biol. 23, 4991–5004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Katsanou V., Milatos S., Yiakouvaki A., Sgantzis N., Kotsoni A., Alexiou M., Harokopos V., Aidinis V., Hemberger M., Kontoyiannis D. L. (2009) Mol. Cell. Biol. 29, 2762–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Papadaki O., Milatos S., Grammenoudi S., Mukherjee N., Keene J. D., Kontoyiannis D. L. (2009) J. Immunol. 182, 6779–6788 [DOI] [PubMed] [Google Scholar]
- 13. von Roretz C., Gallouzi I. E. (2010) J. Biol. Chem. 285, 16806–16813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beauchamp P., Nassif C., Hillock S., van der Giessen K., von Roretz C., Jasmin B. J., Gallouzi I. E. (2010) Cell Death Differ. 17, 1588–1599 [DOI] [PubMed] [Google Scholar]
- 15. van der Giessen K., Gallouzi I. E. (2007) Mol. Biol. Cell 18, 2619–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rebane A., Aab A., Steitz J. A. (2004) RNA 10, 590–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Güttinger S., Mühlhäusser P., Koller-Eichhorn R., Brennecke J., Kutay U. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2918–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gallouzi I. E., Steitz J. A. (2001) Science 294, 1895–1901 [DOI] [PubMed] [Google Scholar]
- 19. Lian X. J., Gallouzi I. E. (2009) J. Biol. Chem. 284, 8877–8887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gallouzi I. E., Brennan C. M., Stenberg M. G., Swanson M. S., Eversole A., Maizels N., Steitz J. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 3073–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Di Marco S., Hel Z., Lachance C., Furneaux H., Radzioch D. (2001) Nucleic Acids Res. 29, 863–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brennan C. M., Gallouzi I. E., Steitz J. A. (2000) J. Cell Biol. 151, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mazroui R., Sukarieh R., Bordeleau M. E., Kaufman R. J., Northcote P., Tanaka J., Gallouzi I., Pelletier J. (2006) Mol. Biol. Cell 17, 4212–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Di Marco S., Mazroui R., Dallaire P., Chittur S., Tenenbaum S. A., Radzioch D., Marette A., Gallouzi I. E. (2005) Mol. Cell. Biol. 25, 6533–6545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. García M. A., Gil J., Ventoso I., Guerra S., Domingo E., Rivas C., Esteban M. (2006) Microbiol. Mol. Biol. Rev. 70, 1032–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pan W., da Graca L. S., Shao Y., Yin Q., Wu H., Jiang X. (2009) J. Biol. Chem. 284, 6946–6954 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.