Abstract
Species–area relationships (SARs) have mostly been treated from an ecological perspective, focusing on immigration, local extinction and resource-based limits to species coexistence. However, a full understanding across large regions is impossible without also considering speciation and global extinction. Rates of both speciation and extinction are known to be strongly affected by area and thus should contribute to spatial patterns of diversity. Here, we explore how variation in diversification rates and ecologically mediated diversity limits among regions of different sizes can result in the formation of SARs. We explain how this area-related variation in diversification can be caused by either the direct effects of area or the effects of factors that are highly correlated with area, such as habitat diversity and population size. We also review environmental, clade-specific and historical factors that affect diversification and diversity limits but are not highly correlated with region area, and thus are likely to cause scatter in observed SARs. We present new analyses using data on the distributions, ages and traits of mammalian species to illustrate these mechanisms; in doing so we provide an integrated perspective on the evolutionary processes shaping SARs.
Keywords: mammals, macroevolution, speciation, extinction, ecological limits, biogeography
1. Introduction
The species–area relationship (SAR), which describes an increase in the number of species as region size increases, is a nearly ubiquitous pattern of biodiversity. SARs exist at a wide range of spatial scales, from local to global, and in a wide range of taxa, including mammals [1]. In the ecological literature, SARs have been explained by considering the factors that limit species from immigrating into, establishing, and persisting in a region [2–4]. However, at large geographical scales, in situ diversification contributes significantly to generating diversity, and so a full understanding of the generation of SARs at such scales is impossible without also considering the macroevolutionary processes of speciation and extinction [5–7].
Here, we explore the evolutionary underpinnings of large-scale SARs, outlining the roles of area itself, environmental variation, clade traits and historical contingency. We adopt a model of clade diversity in which clade diversification within regions is diversity-dependent and SARs are created by the scaling of both diversity limits and diversification rates with area. We support this discussion with new analyses using mammals as they are a well-known, diverse, and globally distributed group with a wide variety of life histories, occupying a wide range of habitats and with robust data for many key traits [8].
SARs have traditionally been treated as the outcome of differences between regions in the balance between immigration and local extinction [4] and in the number of species that can coexist [2,3]. However, it was later recognized that SARs may not be controlled by the same processes at all spatial scales [6,9]. At the smallest scales, SARs result from more complete sampling of the local biota as the area sampled increases, and as such they are sampling rather than biological phenomena. At larger scales (sampling all of the local biota), classical ecological explanations apply, with SARs emerging as a result of more species being able to immigrate into and persist in larger areas. Finally, at the largest scales, differences between regions in rates of speciation and extinction should be the main factor generating SARs [5–7]. Here, we focus on SARs at the largest geographical scale. For mammals, this large-scale phase is likely to occur only when considering quite large regions: in Kisel & Barraclough's [10] study of the spatial scale of speciation, both the mammal groups represented (bats and carnivores) required a region larger than 400 000–500 000 km2 for any in situ speciation to occur.
We use a framework of diversity-dependent cladogenesis (§3) to explore how the area (§4) and environment (§5) of regions affect diversification and diversity limits in the generation of SARs. We also examine the role of clade traits (§6) and temporal patterns of diversification (§7) in modulating the shape of SARs. See table 1 for a summary of the factors addressed.
Table 1.
Summary of factors affecting diversification rates and diversity limits. Up arrows, increases/ing; down arrows, decreases/ing.
| type of factor | factor | effects on speciation rate | effects on extinction rate | effects on diversity limits |
|---|---|---|---|---|
| area | ↑ potential for geographical isolation of separated populations | ↓ by ↑ survival of refuge populations | ||
| environmental factors strongly correlated with area | population size | ↑ rate of appearance of beneficial mutations, standing genetic variation, persistence of incipient species | ↓ by buffering populations from demographic stochasticity, environmental disasters, habitat loss | ↑ number of species with viable populations supported |
| habitat diversity | ↑ population divergence through local adaptation | ↑ niche space available | ||
| fragmentation /topographic diversity | ↑ isolation of populations; however past a certain point, will ↓ speciation by ↓ population size | if fragmentation results in too small patches of area or habitat, will ↑ extinction rate | ↑ by allowing more ecologically equivalent species to be supported | |
| environmental factors not strongly correlated with area | energy availability | ↑ rate of molecular evolution, rate of coevolutionary dynamics, size of populations supported | ↑ by facilitating specialization to narrow niches | |
| clade traits | life-history traits | faster life cycle ↑ speciation by increasing mutation rate | faster life cycle ↓ extinction by ↑ resilience to disturbance | |
| range size | larger range sizes ↑ speciation by ↑ potential for isolation of populations | larger range sizes ↓ extinction by ↑ survival of refuge populations | smaller range sizes ↑ diversity limit by allowing more species to pack into same area | |
| niche breadth | narrower niche breadths associated with ↑ speciation | narrower niche breadths associated with ↑ extinction | narrower niche breadths ↑ diversity limit by allowing finer subdivision of niche space | |
| dispersal | ↓ by reducing potential isolation of populations, but can also ↑ speciation rate by ↑ rate at which species colonize new regions | ↓ by ↑ resilience to disturbance | ↓ if high dispersal ability is associated with large, non-overlapping species ranges |
2. Methods
We used the geographical distributions of 4650 terrestrial mammal species within PanTHERIA [8] to explore the scaling of species richness with area. The choice of appropriate regions at a global scale is not obvious, so we have taken two approaches to identify provinces. First, we used botanical sampling regions based on geopolitical units (Taxonomic Database Working Group (TDWG) [11]) to subdivide continental landmasses, although we further separated disjunct sub-regions, such as islands. Second, we identified species presence in equal-area grid cells at a resolution (96.5 km) comparable to a 1° grid. We then used complete linkage hierarchical clustering on the Jaccard distance ([12]; but see [13]) between grid cells to identify approximate mammalian biotic regions. Both methods are hierarchically nested between levels but regions within the same level are not nested. The fineness of subdivision can also be varied: the TDWG standard defines four levels, ranging roughly from different biomes at the coarsest scale (level 1) to subdivisions within countries at the finest scale (level 4); the hierarchical clustering can be cut at different ‘heights’ to give different numbers of regions and we have used 50, 100, 150 and 200 regions (mapped in the electronic supplementary material, figure S1). The two region types differ in ways that are likely to affect the outcome: for example, political boundaries are likely to more finely partition large biotically homogeneous regions in the temperate zone and agglomerate smaller biotically heterogeneous tropical regions. We used both methods and the variety of scales to assess the robustness of our conclusions to the details of sampling. Separating discontinuous parts of detailed polygons of TDWG regions, in combination with the imprecision in global species distribution maps, led to a large number of tiny islands and boundary regions with implausible biotas. We, therefore, removed all regions at the coarsest TDWG scale that did not contain at least one species endemic to that region, reducing 3974 candidate regions to 117. All nested subdivisions of these 117 regions at the finer TDWG scales were retained.
The areas of both geopolitical and clustered regions were calculated using an equal-area projection of the land within each region (electronic supplementary material, figure S2). We recorded both the total and endemic mammalian species richness for each region and fitted SARs at each scale of subdivision using linear models on log–log axes to estimate the slope. We modelled species richness (S) as a power of area (A) as S = cAz ([2,6]): although there has been considerable debate about the shape of SARs [14,15], our results should be general to alternative functions. For all further analyses, we used the most finely divided regions and compared results using TDWG level 4 and 200 biotic regions. We also explored the differences between slopes of SARs arising from species endemic to a region versus those occurring in more than one region, and the variation among mammalian orders in slopes of SARs.
To investigate the additional explanatory power of habitat diversity and environmental variables, we used two variables to capture different elements of habitat diversity: the diversity of land cover classes (GLCC v. 2.0, http://edc2.usgs.gov/glcc/glcc.php) calculated as the inverse of Simpson's diversity index (1-D) on the relative areas of the classes within each region; and the log range in elevation (GTOPO30, http://eros.usgs.gov/) within each region. We considered two environmental variables within regions: the mean annual temperature (www.worldclim.org) and the mean normalized difference vegetation index (NDVI; [16]). We fitted multiple regressions with log area and each of these four variables in turn as predictors of log species richness. For each variable, we tested whether it showed a significant interaction with area as well as its significance as a main effect. All covariates were mean centred and standardized to facilitate the interpretation and comparison of these models [17].
An approximate measure of habitat breadth for mammalian species was found by counting the number of Global Land Cover Characteristics (GLCC) habitat cover classes across all the 96.5 km cells intersecting each species' range. This number correlates strongly with the species' geographical range (Kendall's tau = 0.61) and we therefore also estimated a number of major habitats by counting only those habitats with a proportional contribution of at least 0.142. This cutoff was selected because it minimizes the observed correlation between the resulting number of major habitats and the species' range size (Kendall's tau = −0.0002). We then calculated the Kendall's correlation between family species richness and both the number of habitats and number of major habitats.
In order to explore the effects of area on the temporal patterns of recent diversification within mammals, we identified two sets of monophyletic clades from the mammal supertree [18,19], excluding monotypic clades. One set had crown ages younger than 20 Myr (421 clades), the other had crown ages younger than 10 Myr (616 clades) and was nested inside the older set. We recorded each clade's species richness, stem-group age and present-day area (either the total area of all TDWG level 4 provinces or of all biotic regions (finest scale) in which the component species occurred). We then fitted a suite of six models of diversification rate across each set [20,21]. The most complex model is an extension of those outlined in Rabosky [20] and Phillimore [21] and fits an exponential decline in diversification with rate z over clade age (t) from an initial diversification rate (λ), but where log present-day area (A) contributes to both initial λ (scaling by c) and the rate of decline (scaling by p); the overall diversification rate is always scaled by the relative extinction rate (ɛ):
We also fitted five simplifications of this model by fixing sets of parameters at zero: a constant diversification rate across clades (c, z and p fixed), a constant diversification rate scaled by individual clade area (z and p fixed), an exponential decline in rate within clades (c and p fixed), an exponential decline from an initial λ scaled by area (p fixed) and an exponential decline at a rate z scaled by area (c fixed). We optimized parameter estimates for the free variables in each model by maximizing the sum of log likelihoods of the observed species richness (n) across clades given clade age and the model estimates (following [21–23]). The models were not nested and we therefore used Akaike Information Criterion (AIC) to assess relative model support. As the two methods to define regions gave qualitatively similar results, we report only the TDWG analysis here (see the electronic supplementary material for biotic region results).
3. A verbal model for clade diversification in space
Diversity-dependent models of diversification have two main features: a growth phase, where the clade in question diversifies until it reaches an external limit; and an equilibrium phase, where species identity turns over but clade size fluctuates about that limit [24,25]. The precise shape of diversity-dependent diversification has been debated [26,27], but the exact shape of the diversification trajectory should not change the broad-scale implications of the existence of diversity-dependent diversification. There is taxonomic, phylogenetic and palaeontological evidence to support the existence of diversity-dependent diversification in many cases, described variously as ‘ecological limits on diversity,’ ‘diversification slowdowns’ and ‘diversity equilibria’ [25–32].
A variety of processes could generate diversity-dependent diversification. Perhaps the most commonly referenced is a model of ecological limits wherein, as available niches are filled, speciation declines and new species are only added to a region following extinctions and release of sufficient niche space [27,33]. Such a mechanism would provide a link between the ecological processes typically associated with SARs and the evolutionary processes being proposed here. Alternatively, reduction of both population and range sizes as diversity increases could lead to decreased rates of speciation and increased rates of extinction, and thus a diversification slowdown conceivably divorced from any niche-based mechanism [34,35].
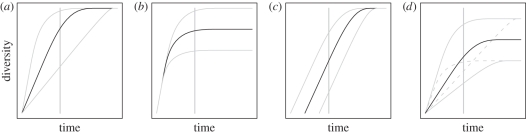
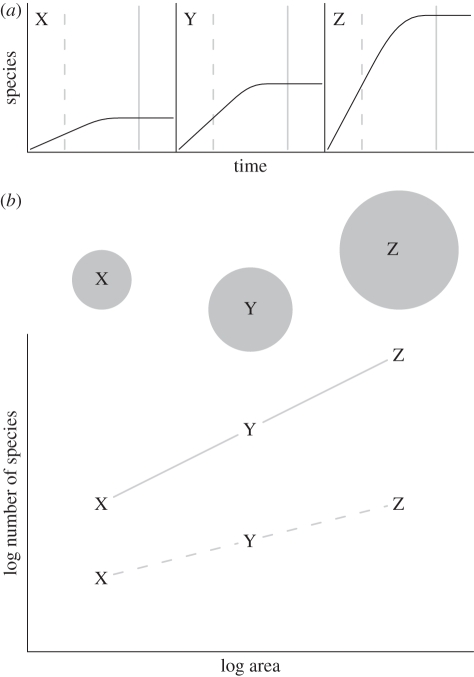
Within our diversity-dependent framework, there are only three features of a clade's diversification curve that can vary: the speed at which a region initially accumulates species (figure 1a), the diversity limit (or equilibrium species richness, figure 1b) and the age at which diversification begins (figure 1c; see also [27]). Before equilibrium is reached, the richness of clades depends only on their age and their rate of diversification. In contrast, clade sizes at equilibrium depend on their diversity limits, which are controlled by the interaction of external factors with clade traits ([36] and see below). SARs will emerge from this model whenever diversification rates and/or diversity limits are higher in larger regions (figure 2). When a clade inhabits multiple separate regions of different areas, the species richness of that clade will be higher in the larger regions, creating a SAR.
Figure 1.
Variation in patterns of clade diversification arising from variation in (a) initial rate of diversification, (b) equilibrium diversity, (c) clade age and (d) reinforcing (solid grey) and opposing (dashed grey) combinations of rate and equilibrium diversity. Sampling clade diversity at the time specified by the vertical line demonstrates the variation possible.
Figure 2.
(a) The development of a species–area relationship (SAR) across three regions (X, Y and Z), in which both initial rate of diversification and equilibrium diversity increase with area. (b) The resulting SAR across regions exhibits power-law scaling both before (dashed line) and after (solid line) the regions have reached equilibrium diversity. It is important to discriminate between (a) the clade diversification curves and (b) SARs; each region will follow a particular diversification trajectory but contributes a single point to the SAR.
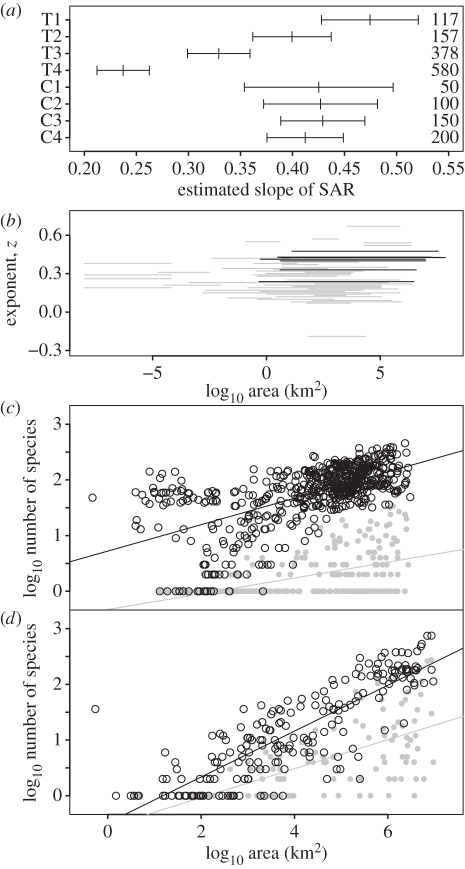
Globally, mammalian species richness shows strong scaling with area between non-nested provinces for both TDWG and clustered regions at all four scales (figure 3). These are well described by power laws but there are differences between the two region types (figure 3a): clustered regions show consistent slopes across changing scales (0.41–0.43), whereas TDWG regions show a decline in slope from 0.47 to 0.24 with increasing subdivision. These slopes lie within the range of 65 previously reported slopes from mammal power law SARs (figure 3b; [37]), but the higher values fall towards the top of the reported range (92% quantile). The decreases in slope between TDWG scales are accompanied by higher intercepts (electronic supplementary material, table S1, figures S3 and S4) and are primarily driven by small political units, such as the Vatican City and Likoma, within species-rich areas (figure 3c); these outliers are not found in small regions based on mammalian biotas (figure 3d). In all cases, endemic species also show significant scaling with area but with reduced slopes compared with total and non-endemic species richness (figure 3b,c and electronic supplementary material, table S1, figures S3 and S4).
Figure 3.
(a) Slopes and their standard errors of SARs for 4560 terrestrial mammals at four different scales across geopolitical regions (T1–4) and biotic regions (C1–4). (b) Distribution of power-law exponents from mammalian SARs showing the range of non-nested region sizes considered (grey lines—data from [37]; black lines—values from panel above). Scatterplots show the distribution and least-squares fit of SARs for (c) T4 and (d) C4 for total (black) and endemic species richness (grey). See also electronic supplementary material, table S1, figures S3 and S4.
We also tested how well area explains variation in diversification rate across sets of mammalian clades. For both sets of clades (crown group age less than 20 Myr and less than 10 Myr, table 2 and electronic supplementary material, table S2), an exponential decline in diversification rate is best supported, demonstrating apparent limits to diversity. For clades younger than 20 Myr, the most complex model was best supported, with clades occupying larger areas having increased initial diversification rates and decreased rate of decline. For clades younger than 10 Myr, a simpler model, with area affecting only the rate of decline, could not be rejected. These results suggest that for mammals the decline in diversification rate as a region fills is more strongly affected by available area than the initial rate. Nevertheless, support for an effect of available area on initial rate was still found for both clade sets and the similar likelihoods for the younger clades may simply reflect individual clade differences within the set tested (see also [38,39]).
Table 2.
Summary of diversification models fitted to mammalian clades. Models were fit using TDWG level 4 data to calculate the present clade distribution area, for clades with crown ages younger than (a) 20 or (b) 10 Myr before present. Six models of diversification were fitted representing: 1, constant rate; 2, constant rate scaled by region area; 3, exponential decline and exponential decline, with region area scaling; 4, initial rate; 5, rate of decline or 6, both. In each case, the maximum-likelihood estimate of the model is reported for each free parameter within the bounds shown. Dashed parameter estimates were fixed at zero. The overall best-fit model for each set of clades is shown in bold. Results using biotic regions to calculate the present clade distributions are presented in electronic supplementary material, table S2.
| lambda | c | z | p | epsilon | |||
|---|---|---|---|---|---|---|---|
| [−1,1] | [−0.2,0.2] | [−0.2,0.2] | [−0.2,0.2] | [0.5,0.999] | ΔAICc | likelihood | |
| (a) 20 Myr | |||||||
| 1 | 0.340 | — | — | — | 0.990 | 222.4 | −1410.0 |
| 2 | −0.300 | 0.040 | — | — | 0.990 | 136.8 | −1366.2 |
| 3 | 0.790 | — | −0.030 | — | 0.990 | 187.2 | −1391.4 |
| 4 | −0.300 | 0.040 | −0.030 | — | 0.610 | 53.1 | −1323.3 |
| 5 | 0.474 | — | −0.138 | 0.007 | 0.814 | 19.1 | −1306.3 |
| 6 | − 0.260 | 0.040 | − 0.100 | 0.004 | 0.610 | 0.0 | − 1295.8 |
| (b) 10 Myr | |||||||
| 1 | 0.265 | — | — | — | 0.999 | 164.2 | −1578.3 |
| 2 | −0.223 | 0.030 | — | — | 0.990 | 93.4 | −1541.9 |
| 3 | 0.530 | — | −0.043 | — | 0.999 | 110.6 | −1550.5 |
| 4 | −0.193 | 0.031 | −0.043 | — | 0.520 | 30.0 | −1509.1 |
| 5 | 0.377 | — | − 0.232 | 0.012 | 0.711 | 0.0 | − 1494.1 |
| 6 | − 0.064 | 0.023 | − 0.120 | 0.005 | 0.500 | 1.16 | − 1493.7 |
4. generating species–area relationships in an evolutionary framework
In explanations of SARs, area is frequently viewed as a proxy or summary variable [40] acting only indirectly via other variables, such as population size and habitat diversity, that are highly correlated with area [4]. The individual effects of area and such correlated factors are difficult to separate in practice [41,42], and their relative importance is likely to vary depending on the taxon concerned [6,43]. However, we believe that area could conceivably have some direct effects, and we discuss these first.
(a). Direct effects of area
We can see only two ways that area could control diversity directly (i.e. without invoking increased population sizes or habitat variety). Firstly, extinction rates should be lower in larger regions, in which refuge populations are more likely to survive after any catastrophic disturbance affecting only part of the region [44]. Secondly, if populations are patchily distributed, speciation rates should be higher in larger areas [7], where distances between populations can be larger and barriers that can cause vicariant speciation are likely to be larger and more numerous [6]. It could be argued that the effect of barriers is really an indirect effect of area via fragmentation, and we discuss this point further below. Greater geographical isolation between populations will lead to higher speciation rates if: (i) there is sufficient selection pressure and/or genetic drift to drive population divergence through to reproductive isolation (although there is no evidence for speciation via genetic drift on its own [45]); (ii) gene flow is the main force preventing population divergence and speciation [46]; and (iii) the regions considered are large enough for populations to be sufficiently isolated to permit speciation. The definition of ‘large enough’ will depend on the dispersal ability of the organism and the strength of selection relative to gene flow, as poorer dispersers will attain sufficient isolation in smaller regions [10], as will species whose populations experience stronger divergent selection [47,48].
(b). Effects of area via population size
Because larger regions are able to support greater total numbers of individuals [49], and thus are also likely to have species with larger population sizes, the effects of population size on diversification can contribute to the generation of SARs. In fact, many of the effects of population size that we describe below have previously been described as direct effects of area itself [4,43]. It is well established that larger populations are less likely to go extinct, as they are more buffered from the effects of demographic stochasticity, environmental disasters and habitat loss [6,50]. Additionally, there are three ways that larger population size may drive higher speciation rates. First, new beneficial mutations will arise faster in larger populations [51], allowing faster divergence between separated populations if mutation limits speciation [52]. Second, larger populations hold more standing genetic variation [53,54] for selection to work on [55,56]. Third, newly isolated populations resulting from the break-up of larger populations will also be larger, and therefore more likely to survive long enough to diverge into new species [57]. In addition to effects on rates of diversification, the total abundance of individuals supported by a region places a hard limit on the number of species that the region can hold. If we assume that all species are ecologically identical and so have the same minimum viable population size [40,58], then larger regions will be able to support more species at sustainable equilibrium population sizes.
(c). Effects of area via habitat diversity and fragmentation
Some authors have suggested that SARs are only a proxy for the scaling of species richness with habitat diversity [4,41,59,60], and indeed habitat diversity and area are typically very highly correlated. Along steep environmental gradients, and in heterogeneous habitats, populations can more easily become specialized to different habitats, making ecological speciation more likely and perhaps more rapid [52]. Regions with high habitat diversity also have a higher number of possible distinct niches or niche combinations [61], thus increasing the number of species that can coexist at equilibrium.
High levels of regional fragmentation can also elevate diversification rate and diversity limits, by providing a textured landscape with subunits that are physically isolated from one another but environmentally equivalent. Barrier formation can occur through many processes, including river formation, mountain building, sea-level fluctuation, volcanic uplift and habitat fragmentation, and is more likely in larger regions. Barriers elevate diversification rate by separating previously interacting populations, which are then more likely to evolve reproductive isolation [6]. In addition, fragmentation can boost equilibrium diversity, as ecologically equivalent species can be maintained in separated sub-regions [62,63]. For example, Esselstyn et al. [64] suggest that tree shrew diversity in the Philippines has arisen predominantly via speciation in allopatry on newly formed islands, with limited apparent morphological or ecological differentiation. One particularly important measure of regional fragmentation is topographic complexity, as environmental turnover along altitude gradients is a barrier to many species' ranges [65]. The richness of uniquely adapted, restricted-range endemics found along altitudinal transects in tropical mountains is perhaps the classic example of such fine-scale spatial partitioning [66,67].
The effects of fragmentation on species richness will show a complex relationship with the total summed area of the subunits. While greater fragmentation of a region may permit more species to exist within the same total area, it may also push the area of the component fragments below a size which can maintain viable populations [58,68] or generate endemics [7,10]. Thus, plots of species richness against total area occupied may not yield significant relationships unless the degree of fragmentation is also considered and the total area is scaled appropriately (see [63]). In addition, the dispersal ability of a clade in combination with the geographical structure of the fragments will influence the number of fragments that can be occupied. Finally, the effect of barriers will depend on the average range sizes of species in a region: if the average range size is small, barriers need not be large or bisect an entire region to cause speciation [35].
Attesting to the importance of environmental features in the generation of SARs, increased elevational range is associated with higher diversity in both geopolitical and biotic regions; habitat diversity also drives higher diversity, but only in geopolitical regions (table 3 and electronic supplementary material, figure S4). This arises from differences between the clustering methods: areas with similar habitat are likely to be biotically homogeneous and therefore form a single biotic region, whereas political boundaries are more likely to cut across such regions. As a result, Simpson's index (1-D) of habitat diversity is low in biotic clusters and scales extremely weakly with region area (intercept: 0.227, s.e. = 0.042, t = 3.83; slope: 0.018, s.e. = 0.014, t = 1.27; d.f. = 148) whereas in TDWG regions it is higher and scales strongly with area (intercept: 0.356, s.e. = 0.025, t = 14.39; slope: 0.055, s.e. = 0.005, t = 10.27; d.f. = 578). In all these models, the high relative magnitude of the standardized parameter estimate for area also implies that it is not simply acting as a proxy for either variable.
Table 3.
Multi-variate SARs, with mammal species richness regressed against area and other environmental variables. In addition to area, models included (a) habitat diversity, (b) log range in elevation, (c) mean annual temperature and (d) mean NDVI. The models were fitted to log 10 species richness within both geopolitical and biotic regions and the explanatory covariates in all models were centred and standardized to facilitate model comparison. The statistical significance of each parameter is given (**p < 0.01, ***p < 0.001), as well as the number of regions with available data (n) for each model.
| (a) geopolitical regions |
(b) biotic regions |
||||||
|---|---|---|---|---|---|---|---|
| estimate | s.e. | p | estimate | s.e. | p | ||
| (a) | n | 580 | 150 | ||||
| intercept | 1.7419 | 0.0186 | *** | 1.2670 | 0.0342 | *** | |
| habitat diversity | 0.0234 | 0.0194 | -0.0649 | 0.0340 | |||
| log area | 0.3407 | 0.0203 | *** | 0.7608 | 0.0352 | *** | |
| interaction | 0.0410 | 0.0152 | ** | -0.0340 | 0.0325 | ||
| (b) | n | 578 | 200 | ||||
| intercept | 1.6810 | 0.0202 | *** | 1.0211 | 0.0391 | *** | |
| log elevation range | 0.0533 | 0.0297 | 0.2383 | 0.0573 | *** | ||
| log area | 0.3956 | 0.0237 | *** | 0.5869 | 0.0478 | *** | |
| interaction | 0.1114 | 0.0162 | *** | 0.1923 | 0.0343 | *** | |
| (c) | n | 477 | 130 | ||||
| intercept | 1.8201 | 0.0160 | *** | 1.0802 | 0.0496 | *** | |
| NDVI | 0.0142 | 0.0174 | 0.1947 | 0.0521 | *** | ||
| log area | 0.3075 | 0.0218 | *** | 0.8997 | 0.0536 | *** | |
| interaction | 0.1007 | 0.0218 | *** | −0.0442 | 0.0550 | ||
| (d) | n | 525 | 196 | ||||
| intercept | 1.7463 | 0.0158 | *** | 1.1339 | 0.0343 | *** | |
| temperature | 0.0577 | 0.0168 | *** | 0.2750 | 0.0407 | *** | |
| log area | 0.4110 | 0.0201 | *** | 0.8411 | 0.0341 | *** | |
| interaction | 0.0979 | 0.0179 | *** | −0.0545 | 0.0432 | ||
5. Abiotic factors modulating the species–area relationship
Some abiotic factors, such as energy availability, do not correlate closely with area but may still affect diversification rates or diversity limits of different regions, leading to departures from SARs that depend on a region's prevailing environmental conditions.
Energy availability is one of the key variables thought to contribute to large-scale spatial patterns of diversity, and has mainly been discussed for its part in generating latitudinal differences in diversity (reviewed in [69,70] and see [71]). On average, energy availability (either ambient, e.g. temperature, or productive, e.g. plant biomass) explains 60 per cent of the variation in broad-scale richness across a range of plant and animal groups ([72] and see [71]). This variation should lead to consistent differences between SARs of high- and low-energy regions.
As expected, increases in both mean annual temperature and mean NDVI act to significantly elevate both overall mammal diversity and slopes of mammalian SARs (table 3 and electronic supplementary material, figure S5). Again though, as in analyses including habitat and topographical diversity, the relative magnitudes of standardized regression coefficients show that area is the main driver of diversity within regions.
We expect energy to affect SARs through both diversification rates and diversity limits. First, it could affect speciation rates through faster rates of molecular evolution, with increased metabolic rates in higher energy regions leading to both shorter generation times and higher mutation rates [73,74]. There has been mixed evidence for this molecular rate hypothesis, with particularly weak support in endotherms [38] and no support in angiosperms (although a direct effect of energy on species richness is supported [75]). However, Gillman et al. [76] recently presented evidence for higher rates of microevolution in tropical mammals and explained this as an indirect consequence of more rapid coevolution with other tropical ectotherms (see also [77,78]). Energy is also expected to increase diversification rates through effects on population dynamics, as aseasonal and elevated productive energy can support larger populations, resulting in increased speciation and reduced extinction, as described above (and see [71]). Such an aseasonal and high-energy environment will also increase the equilibrium diversity limit by increasing resource availability, facilitating specialization to very narrow niches, and thus increasing the number of distinct niches available [66]. Conversely, seasonal habitats in temperate regions may select for more motile, generalist species. These traits should decrease both speciation rate and the number of species that can be supported in a region [79,80]. Although not attempted here, incorporating ecological covariates into our diversification models could lend insight into the effects of, for example, energy availability on the diversification trajectory of clades in different regions [31].
6. Clade traits modulating the species–area relationship
So far our framework has considered species richness within a region as an outcome of solely environmental and geographical influences, taking a neutral view of the organisms themselves [4]. However, there is abundant research (reviewed in [45]) indicating that species traits affect clade diversity. Any clade traits that affect diversity will give rise to clade-specific SARs, and create scatter around SARs that aggregate species richness across multiple clades. The effects of clade traits on SARs are reflected in the clear differences between mammalian orders in the scaling of species richness with area: order-specific slopes vary between −1.71 and 0.59 with medians of 0.16 for clustered regions and 0.11 for geopolitical regions (electronic supplementary material, table S3; because regions are not nested, negative slopes arise simply where orders have high diversity in small regions).
According to our general model, clade traits can modulate SARs by modifying the net rate of diversification (figure 1a) and/or the diversity limit (figure 1b). It is not straightforward to assign traits to one of these mechanisms. Firstly, data are lacking: studies analysing differences between clades in diversification (reviewed in [81,82]) have not discriminated between effects on diversification rate and effects on diversity limits (but see [31]), and studies of diversification slowdowns in phylogenies (e.g. [83]) have not investigated the influence of species' traits. Secondly, individual traits are unlikely to act solely through modification of either diversification rates or diversity limits [36]. Finally, many clade traits are strongly correlated (for example, geographical range size, dispersal distance and body size [8,81]) and so any traits acting through one mechanism are likely to be associated with traits acting through the other. Below, we discuss traits expected to influence SARs, with particular emphasis on those that affect species' use of space.
While most traits are likely to influence both diversification rates and diversity limits, life-history traits are perhaps the only class of traits expected to influence only diversification rate. Typically, r-selected species exhibit higher net rates of diversification than K-selected species, and several mechanisms have been proposed to explain this [74,84,85]. Short generation times are associated with high rates of population increase and the ability to rapidly exploit favourable conditions [84], conferring resilience to disturbance and leading to lower rates of extinction. They are also associated with increased rates of evolution owing to shorter nucleotide generation times [70,74,86], higher metabolic rates [74,86], larger population sizes and increased fecundity [74], in all cases leading to higher rates of speciation. In addition, the larger population sizes associated with r selection should directly increase speciation rates and decrease extinction rates, as discussed in §2.
Clade traits that determine how space is occupied within a region also affect both the generation and maintenance of SARs. Larger species ranges are associated with lower clade diversity limits as well as reduced rates of extinction (e.g. [87,88]) and increased rates of speciation ([89], but see [90]). Regarding diversity limits, there is evidence from both mammals [63] and birds [91] that increasing species' range overlap is a stronger predictor of increased species richness than decreased median range size.
Similarly to species' range size, several aspects of narrow niche breadth, such as ecological specialization, high host specificity and narrow environmental tolerances, have been associated with increased diversity limits as well as increased rates of extinction and speciation [71,81]. Increased clade diversity is also associated with greater niche overlap rather than decreased niche breadth (see also [92]). Ricklefs [23] has shown that South American bird families of varying species richness do not differ in the average number of habitats occupied by species, suggesting that niche overlap between species increases as family size increases. We find the same in mammals, using simple measures of the number of habitats used by species. There is no significant correlation between the richness of mammalian families and either the average total number of habitats occupied (tau = −0.076, p = 0.21) or the average number of major habitats occupied (tau = −0.043, p = 0.49) nor is there a decrease in mean species range size with increasing family richness (tau = −0.001, p = 0.99).
Finally, increased dispersal ability has been found to reduce speciation and extinction rates in some cases [93], while in others it has been shown to increase diversification rate [89,94]. With respect to diversity limits, high dispersal ability may lead to low equilibrium diversity within a region if it leads to clades consisting of few species with large ranges [71]. At the other extreme, strong philopatry, where individuals retain or return to natal locations, might both increase rates of diversification by accelerating rates of genetic differentiation [95] and increase equilibrium diversity by impeding range expansion and boosting the number of equivalent species that can persist in a region [62,96]. Alternatively, high dispersal ability can increase the rate at which new regions are occupied, increasing clade richness through occupation of multiple regions. Such long-distance dispersal may significantly distort SARs if newly colonized regions harbour clades with higher diversity owing to competitive release [28].
7. Historical and temporal effects on the species–area relationship
In general, diversification in any region is influenced by the climatic, geological and biogeographic history of the region [28,64,97], and as a result, SARs should be affected by history as well. SARs will be clearest when clades have reached equilibrium throughout their ranges, but this requires that they have had enough time to diversify to their limit in each region that they occupy. Thus, in parts of the world where the current habitat has only recently become available, current diversity is likely to be lower than expected (e.g. a recently formed island, [64], or a recently deglaciated region [71,98]) and may be biased towards large-ranged generalists [71,79,92]. In contrast, a comparison of mammalian sister taxon pairs with disjunct distributions across two realms indicated that sisters remaining in the realm unambiguously reconstructed as ancestral (using DIVA [99]) are significantly less species-rich (12 out of 41, binomial p = 0.004; electronic supplementary material, table S4) than sisters that dispersed. This suggests a diversification burst in newly colonized regions, driven by competitive release (more in [28]). Finally, if a region is subject to frequent extrinsic perturbations (such as an archipelago subject to repeated sea-level changes), fluctuating extinction rates make it unlikely that equilibrium diversity will ever be reached or maintained [64,100]. Indeed, explanations for high tropical diversity, such as the time-for-speciation effect [101] and reduced extinction owing to long-term climatic stability [77], are compatible with tropical regions being able to more closely approach diversity limits (more in [71]).
Diversity may also transiently over- or under-shoot the diversity limit of a region if speciation or extinction occurs very rapidly, or if perturbations occur that suddenly alter clade diversity limits [102]. Alternatively, non-ecological modes of speciation (e.g. via sexual selection or polyploidy) may produce transient species that are unable to persist in the long term given the niche space available, and thus are committed to eventual extinction [6,103,104]. This may also apply to ecologically equivalent species formed in allopatry, if the barriers separating them are themselves transient. Transient dynamics are now thought to be crucial in predicting biodiversity responses to current global changes (recently reviewed in [105]); though the changes will probably not be as immediately apparent as for ecological processes such as community assembly, evolutionary clade dynamics will certainly be affected as well [106].
8. Conclusions
We have presented a framework, based on a diversity-dependent model of clade diversification, for understanding how evolutionary processes contribute to the creation of large-scale SARs. This framework is supported by analyses on mammals using data from the PanTHERIA database [8]. SARs themselves result from direct and positive effects of area on diversification rates and diversity limits, as well as indirect effects of area through population size, habitat diversity and habitat fragmentation. We found that these effects are apparent in the histories of mammal diversification—clades occupying larger areas had higher initial diversification rates and lower rates of decline in diversification. We also confirmed that habitat and topographical diversity are significant predictors of regional diversity in mammals, but found that neither is a proxy for area—the most predictive models of diversity always include area as well. Environmental factors and clade traits that are not tightly correlated with area also cause systematic differences in SARs between clades or regions, and cause scatter around any general SAR generated without accounting for them. We tested the influence of energy availability on mammal diversity and showed that high energy availability significantly increases the slopes and intercepts of SARs. In addition, mammal orders vary greatly in the slopes of their SARs. Finally, we provide evidence that historical contingencies impact SARs, demonstrating that mammal clades able to colonize new, competitor-free regions are more diverse than their stay-at-home sisters.
Schoener [107] referred to the SAR as the phenomenon closest to attaining rule status in ecology, and SARs are indeed one of the most general diversity patterns, existing for a wide range of organisms across a range of spatial scales. However, we argue here that in addition to the processes most discussed in the ecological literature—immigration, local extinction and species coexistence—SARs are also influenced by macroevolutionary processes, in particular speciation and global extinction. None of these processes operates in isolation, and every SAR is the result of interplay between both ecological and evolutionary processes. Diversity limits, for instance, must ultimately result from ecological limits on the number of species that can coexist in a region, though the speed at which they are reached may depend on evolutionary processes. We suggest that a full understanding of SARs will require integrating both ecological and evolutionary perspectives on the processes that generate and constrain diversity.
Acknowledgements
We thank Kate Jones and Kamran Safi for the invitation to contribute to this special issue and Tim Barraclough, Natalie Cooper, Susanne Fritz, Alex Pigot, James Rosindell and two anonymous reviewers for comments on previous versions of the manuscript. Special thanks are extended to Ally Phillimore for insightful discussion and comments, and R code for implementing the diversification models. Y.K. was supported by a U.S. National Science Foundation Graduate Research Fellowship and a Deputy Rector's Award from Imperial College London, L.M. by a Grantham Institute studentship and C.D.L.O. by an RCUK fellowship and a Natural Environment Research Council grant (NE/B503492/1).
References
- 1.Pagel M. D., May R. M., Collie A. R. 1991. Ecological aspects of the geographical distribution and diversity of mammalian species. Am. Nat. 137, 791–815 10.1086/285194 (doi:10.1086/285194) [DOI] [Google Scholar]
- 2.Arrhenius O. 1921. Species and area. J. Ecol. 9, 95–99 10.2307/2255763 (doi:10.2307/2255763) [DOI] [Google Scholar]
- 3.Preston F. W. 1960. Time and space and the variation of species. Ecology 41, 612–627 10.2307/1931793 (doi:10.2307/1931793) [DOI] [Google Scholar]
- 4.MacArthur R. H., Wilson E. O. 1967. The theory of island biogeography. Princeton, NJ: Princeton University Press [Google Scholar]
- 5.Rosenzweig M. L. 1998. Preston's ergodic conjecture: the accumulation of species in space and time. In Biodiversity dynamics: turnover of populations, taxa and communities (eds McKinney M. L., Drake J. A.), pp. 311–348 New York, NY: Columbia University Press [Google Scholar]
- 6.Rosenzweig M. L. 1995. Species diversity in space and time. Cambridge, UK: Cambridge University Press [Google Scholar]
- 7.Losos J. B., Schluter D. 2000. Analysis of an evolutionary species–area relationship. Nature 408, 847–850 10.1038/35048558 (doi:10.1038/35048558) [DOI] [PubMed] [Google Scholar]
- 8.Jones K. E., et al. 2009. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology 90, 2648–2648 10.1890/08-1494.1 (doi:10.1890/08-1494.1) [DOI] [Google Scholar]
- 9.Palmer M. W., White P. S. 1994. Scale dependence and the species–area relationship. Am. Nat. 144, 717–740 10.1086/285704 (doi:10.1086/285704) [DOI] [Google Scholar]
- 10.Kisel Y., Barraclough T. G. 2010. Speciation has a spatial scale that depends on levels of gene flow. Am. Nat. 175, 316–334 10.1086/650369 (doi:10.1086/650369) [DOI] [PubMed] [Google Scholar]
- 11.Brummit R. K. 2001. World geographical scheme for recording plant distributions, 2nd edn. Pittsburgh, PA: Hunt Institute for Botanical Documentation, Carnegie-Mellon University (for the International Working Group on Taxonomic Databases for Plant Sciences) [Google Scholar]
- 12.Linder H. P., Lovett J. C., Mutke J., Barthlott W., Jürgens N., Rebelo T., Küper W. 2005. A numerical re-evaluation of the sub-Saharan phytochoria of mainland Africa. Biol. Skrifter 55, 229–252 [Google Scholar]
- 13.Kreft H., Jetz W. 2010. A framework for delineating biogeographical regions based on species distributions. J. Biogeogr. 37, 2029–2053 10.1111/j.1365-2699.2010.02375.x (doi:10.1111/j.1365-2699.2010.02375.x) [DOI] [Google Scholar]
- 14.Lomolino M. V. 2000. Ecology's most general, yet protean pattern: the species–area relationship. J. Biogeogr. 27, 17–26 10.1046/j.1365-2699.2000.00377.x (doi:10.1046/j.1365-2699.2000.00377.x) [DOI] [Google Scholar]
- 15.Scheiner S. M. 2003. Six types of species-area curves. Glob. Ecol. Biogeogr. 12, 441–447 10.1046/j.1466-822X.2003.00061.x (doi:10.1046/j.1466-822X.2003.00061.x) [DOI] [Google Scholar]
- 16.Los S. O., et al. 2000. A global 9-year biophysical land-surface data set from NOAA AVHRR data. J. Hydrometerol. 1, 183–199 (doi:10.1175/1525-7541(2000)001<0183:AGYBLS>2.0.CO;2) [DOI] [Google Scholar]
- 17.Schielzeth H. 2010. Simple means to improve the interpretability of regression coefficients. Methods Ecol. Evol. 1, 103–113 10.1111/j.2041-210X.2010.00012.x (doi:10.1111/j.2041-210X.2010.00012.x) [DOI] [Google Scholar]
- 18.Bininda-Emonds O. R. P., et al. 2007. The delayed rise of present-day mammals. Correction. Nature 456, 274. 10.1038/nature05634 (doi:10.1038/nature05634) [DOI] [PubMed] [Google Scholar]
- 19.Bininda-Emonds O. R. P., et al. 2007. The delayed rise of present-day mammals. Nature 446, 507–512 10.1038/nature07347 (doi:10.1038/nature07347) [DOI] [PubMed] [Google Scholar]
- 20.Rabosky D. L. 2009. Ecological limits on clade diversification in higher taxa. Am. Nat. 173, 662–674 10.1086/597378 (doi:10.1086/597378) [DOI] [PubMed] [Google Scholar]
- 21.Phillimore A. B. 2010. Subspecies origination and extinction in birds. Ornithol. Monogr. 67, 42–53 10.1525/om.2010.67.1.42 (doi:10.1525/om.2010.67.1.42) [DOI] [Google Scholar]
- 22.Bokma F. 2003. Testing for equal rates of cladogenesis in diverse taxa. Evolution 57, 2469–2474 [DOI] [PubMed] [Google Scholar]
- 23.Ricklefs R. E. 2009. Speciation, extinction and diversity. In Speciation and patterns of diversity (eds Butlin R., Bridle J. R., Schluter D.), pp. 257–278 Cambridge, UK: Cambridge University Press [Google Scholar]
- 24.Sepkoski J. J. 1978. Kinetic model of Phanerozoic taxonomic diversity: I. Analysis of marine orders. Paleobiology 4, 223–251 [Google Scholar]
- 25.Alroy J. 1998. Equilibrial diversity dynamics in North American mammals. In Biodiversity dynamics: turnover of populations, taxa and communities (eds McKinney M. L., Drake J. A.), pp. 232–288 New York, NY: Columbia University Press [Google Scholar]
- 26.Nee S., Mooers A., Harvey P. 1992. Tempo and mode of evolution revealed from molecular phylogenies. Proc. Natl Acad. Sci. USA 89, 8322–8326 10.1073/pnas.89.17.8322 (doi:10.1073/pnas.89.17.8322) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabosky D. L. 2009. Ecological limits and diversification rate: alternative paradigms to explain the variation in species richness among clades and regions. Ecol. Lett. 12, 735–743 10.1111/j.1461-0248.2009.01333.x (doi:10.1111/j.1461-0248.2009.01333.x) [DOI] [PubMed] [Google Scholar]
- 28.Purvis A., Fritz S. A., Rodrígues J., Harvey P. H., Grenyer R. 2011. The shape of mammalian phylogeny: patterns, processes and scales. Phil. Trans. R. Soc. B 366, 2462–2477 10.1098/rstb.2011.0025 (doi:10.1098/rstb.2011.0025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sepkoski J. J., Jr 1976. Species diversity in the Phanerozoic: species-area effects. Paleobiology 2, 298–303 [Google Scholar]
- 30.Rabosky D. L., Lovette I. J. 2008. Density-dependent diversification in North American wood warblers. Proc. R. Soc. B 275, 2363–2371 10.1098/rspb.2008.0630 (doi:10.1098/rspb.2008.0630) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vamosi J. C., Vamosi S. M. 2010. Key innovations within a geographical context in flowering plants: towards resolving Darwin's abominable mystery. Ecol. Lett. 13, 1270–1279 10.1111/j.1461-0248.2010.01521.x (doi:10.1111/j.1461-0248.2010.01521.x) [DOI] [PubMed] [Google Scholar]
- 32.Cardillo M. 2011. Phylogenetic structure of mammal assemblages at large geographic scales: linking phylogenetic community ecology with macroecology. Phil. Trans. R. Soc. B 366, 2545–2553 10.1098/rstb.2011.0021 (doi:10.1098/rstb.2011.0021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKinney M. L. 1998. Biodiversity dynamics: niche preemption and saturation in diversity equilibria. In Biodiversity dynamics: turnover of populations, taxa and communities (eds McKinney M. L., Drake J. A.), pp. 1–18 New York, NY: Columbia University Press [Google Scholar]
- 34.Pigot A. L., Phillimore A. B., Owens I. P. F., Orme C. D. L. 2010. The shape and temporal dynamics of phylogenetic trees arising from geographic speciation. Syst. Biol. 59, 615–618 10.1093/sysbio/syq058 (doi:10.1093/sysbio/syq058) [DOI] [PubMed] [Google Scholar]
- 35.Rosenzweig M. L. 1975. On continental steady states of species diversity. In The ecology and evolution of communities (eds Cody M., Diamond J.), pp. 121–140 Cambridge, MA: Harvard University Press [Google Scholar]
- 36.Mallet J. Submitted The ‘struggle for existence:' why the mismatch of theory in ecology and evolution. Am. Nat. [Google Scholar]
- 37.Drakare S., Lennon J. J., Hillebrand H. 2006. The imprint of the geographical, evolutionary and ecological context on species–area relationships. Ecol. Lett. 9, 215–227 10.1111/j.1461-0248.2005.00848.x (doi:10.1111/j.1461-0248.2005.00848.x) [DOI] [PubMed] [Google Scholar]
- 38.Cardillo M., Orme C. D. L., Owens I. P. F. 2005. Testing for latitudinal bias in diversification rates: an example using New World birds. Ecology 86, 2278–2287 10.1890/05-0112 (doi:10.1890/05-0112) [DOI] [Google Scholar]
- 39.Linder H. P. 2008. Plant species radiations: where, when, why? Phil. Trans. R. Soc. B 363, 3097–3105 10.1098/rstb.2008.0075 (doi:10.1098/rstb.2008.0075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hubbell S. P. 2001. The unified neutral theory of biodiversity and biogeography. Princeton, NJ: Princeton University Press [Google Scholar]
- 41.Triantis K. A., Mylonas M., Lika K., Vardinoyannis K. 2003. A model for the species-area-habitat relationship. J. Biogeogr. 30, 19–27 10.1046/j.1365-2699.2003.00805.x (doi:10.1046/j.1365-2699.2003.00805.x) [DOI] [Google Scholar]
- 42.Kallimanis A. S., Mazaris A. D., Tzanopoulos J., Halley J. M., Pantis J. D., Sgardelis S. P. 2008. How does habitat diversity affect the species–area relationship? Glob. Ecol. Biogeogr. 17, 532–538 10.1111/j.1466-8238.2008.00393.x (doi:10.1111/j.1466-8238.2008.00393.x) [DOI] [Google Scholar]
- 43.Ricklefs R. E., Lovette I. J. 1999. The roles of island area per se and habitat diversity in the species–area relationships of four Lesser Antillean faunal groups. J. Anim. Ecol. 68, 1142–1160 10.1046/j.1365-2656.1999.00358.x (doi:10.1046/j.1365-2656.1999.00358.x) [DOI] [Google Scholar]
- 44.Wiley J. W., Wunderle J. M. 1994. The effects of hurricanes on birds, with special reference to Caribbean islands. Bird Conserv. Int. 3, 319–349 10.1017/S0959270900002598 (doi:10.1017/S0959270900002598) [DOI] [Google Scholar]
- 45.Coyne J. A., Orr H. A. 2004. Speciation. Sunderland. MA: Sinauer Associates, Inc [Google Scholar]
- 46.Slatkin M. 1987. Gene flow and the geographic structure of natural populations. Science 236, 787–792 10.1126/science.3576198 (doi:10.1126/science.3576198) [DOI] [PubMed] [Google Scholar]
- 47.Slatkin M. 1973. Gene flow and selection in a cline. Genetics 75, 733–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slatkin M. 1985. Gene flow in natural populations. Annu. Rev. Ecol. Syst. 16, 393–430 10.1146/annurev.ecolsys.16.1.393 (doi:10.1146/annurev.ecolsys.16.1.393) [DOI] [Google Scholar]
- 49.Brown J. H. 1995. Macroecology. Chicago, IL: University of Chicago Press [Google Scholar]
- 50.Lande R. 1993. Risks of population extinction from demographic and environmental stochasticity and random catastrophes. Am. Nat. 142, 911–927 10.1086/285580 (doi:10.1086/285580) [DOI] [PubMed] [Google Scholar]
- 51.Willi Y., Van Buskirk J., Hoffmann A. A. 2006. Limits to the adaptive potential of small populations. Annu. Rev. Ecol. Evol. Syst. 37, 433–458 10.1146/annurev.ecolsys.37.091305.110145 (doi:10.1146/annurev.ecolsys.37.091305.110145) [DOI] [Google Scholar]
- 52.Schluter D. 2009. Evidence for ecological speciation and its alternative. Science 323, 737–741 10.1126/science.1160006 (doi:10.1126/science.1160006) [DOI] [PubMed] [Google Scholar]
- 53.Frankham R. 1996. Relationship of genetic variation to population size in wildlife. Conserv. Biol. 10, 1500–1508 10.1046/j.1523-1739.1996.10061500.x (doi:10.1046/j.1523-1739.1996.10061500.x) [DOI] [Google Scholar]
- 54.Leimu R., Mutikainen P., Koricheva J., Fischer M. 2006. How general are positive relationships between plant population size, fitness and genetic variation? J. Ecol. 94, 942–952 10.1111/j.1365-2745.2006.01150.x (doi:10.1111/j.1365-2745.2006.01150.x) [DOI] [Google Scholar]
- 55.Weber K. E. 1990. Increased selection response in larger populations. I. Selection for wing-tip height in Drosophila melanogaster at 3 population sizes. Genetics 125, 579–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schluter D., Conte G. L. 2009. Genetics and ecological speciation. Proc. Natl Acad. Sci. USA 106, 9955–9962 10.1073/pnas.0901264106 (doi:10.1073/pnas.0901264106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chown S. L., Gaston K. J. 2000. Areas, cradles and museums: the latitudinal gradient in species richness. Trends Ecol. Evol. 15, 311–315 10.1016/S0169-5347(00)01910-8 (doi:10.1016/S0169-5347(00)01910-8) [DOI] [PubMed] [Google Scholar]
- 58.Gilpin M. E., Soule M. E. 1986. Minimum viable populations: processes of species extinction. In Conservation biology: the science of scarcity and diversity (ed. Soule M. E.), pp. 19–34 Sunderland, MA: Sinauer Associate, Inc [Google Scholar]
- 59.Baldi A. 2008. Habitat heterogeneity overrides the species–area relationship. J. Biogeogr. 35, 675–681 10.1111/j.1365-2699.2007.01825.x (doi:10.1111/j.1365-2699.2007.01825.x) [DOI] [Google Scholar]
- 60.Losos J. B., Parent C. E. 2010. The speciation–area relationship. In The theory of island biogeography revisited (eds Losos J. B., Ricklefs R. E.), pp. 415–438 Oxford, UK: Princeton University Press [Google Scholar]
- 61.Hutchinson G. E., MacArthur R. H. 1959. A theoretical ecological model of size distributions among species of animals. Am. Nat. 93, 117. 10.1086/282063 (doi:10.1086/282063) [DOI] [Google Scholar]
- 62.Shmida A., Wilson M. V. 1985. Biological determinants of species diversity. J. Biogeogr. 12, 1–20 10.2307/2845026 (doi:10.2307/2845026) [DOI] [Google Scholar]
- 63.Orme C. D. L., et al. In preparation. The phylogenetic species-area relationship. [Google Scholar]
- 64.Esselstyn J. A., Timm R. M., Brown R. M. 2009. Do geological or climatic processes drive speciation in dynamic archipelagos? The tempo and mode of diversification in southeast Asian shrews. Evolution 63, 2595–2610 10.1111/j.1558-5646.2009.00743.x (doi:10.1111/j.1558-5646.2009.00743.x) [DOI] [PubMed] [Google Scholar]
- 65.McInnes L., Purvis A., Orme C. D. L. 2009. Where do species' geographic ranges stop and why? Landscape impermeability and the Afrotropical avifauna. Proc. R. Soc. B 276, 3063–3070 10.1098/rspb.2009.0656 (doi:10.1098/rspb.2009.0656) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Janzen D. 1967. Why mountain passes are higher in the tropics. Am. Nat. 101, 233–249 10.1086/282487 (doi:10.1086/282487) [DOI] [Google Scholar]
- 67.Rahbek C., Graves G. R. 2001. Multiscale assessment of patterns of avian species richness. Proc. Natl Acad. Sci. USA 98, 4534–4539 10.1073/pnas.071034898 (doi:10.1073/pnas.071034898) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maurer B. A., Nott M. P. 1998. Geographic range fragmentation and the evolution of biological diversity. In Biodiversity dynamics: turnover of populations, taxa and communities (eds McKinney M. L., Drake J. A.), pp. 31–50 New York, NY: Columbia University Press [Google Scholar]
- 69.Willig M., Kaufmann D., Stevens R. 2003. Latitudinal gradients of biodiversity: pattern, process, scale, and synthesis. Annu. Rev. Ecol. Syst. 34, 273–309 10.1146/annurev.ecolsys.34.012103.144032 (doi:10.1146/annurev.ecolsys.34.012103.144032) [DOI] [Google Scholar]
- 70.Mittelbach G. G., et al. 2007. Evolution and the latitudinal diversity gradient: speciation, extinction and biogeography. Ecol. Lett. 10, 315–331 10.1111/j.1461-0248.2007.01020.x (doi:10.1111/j.1461-0248.2007.01020.x) [DOI] [PubMed] [Google Scholar]
- 71.Davies T. J., Buckley L. B., Grenyer R., Gittleman J. L. 2011. The influence of past and present climate on the biogeography of modern mammal diversity. Phil. Trans. R. Soc. B 366, 2526–2535 10.1098/rstb.2011.0018 (doi:10.1098/rstb.2011.0018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hawkins B. A., et al. 2003. Energy, water, and broad-scale geographic patterns of species richness. Ecology 84, 3105–3117 10.1890/03-8006 (doi:10.1890/03-8006) [DOI] [Google Scholar]
- 73.Rohde K. 1992. Latitudinal gradients in species diversity—the search for the primary cause. Oikos 65, 514–527 10.2307/3545569 (doi:10.2307/3545569) [DOI] [Google Scholar]
- 74.Bromham L. 2011. The genome as a life history character: why rate of molecular evolution varies between mammal species. Phil. Trans. R. Soc. B 366, 2503–2513 10.1098/rstb.2011.0014 (doi:10.1098/rstb.2011.0014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Davies T. J., Savolainen V., Chase M. W., Moat J., Barraclough T. G. 2004. Environmental energy and evolutionary rates in flowering plants. Proc. R. Soc. B 271, 2195–2200 10.1098/rspb.2004.2849 (doi:10.1098/rspb.2004.2849) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gillman L. N., Keeling D. J., Ross H. A., Wright S. D. 2009. Latitude, elevation and the tempo of molecular evolution in mammals. Proc. R. Soc. B 276, 3353–3359 10.1098/rspb.2009.0674 (doi:10.1098/rspb.2009.0674) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fischer A. G. 1960. Latitudinal variations in organic diversity. Evolution 14, 64–81 10.2307/2405923 (doi:10.2307/2405923) [DOI] [Google Scholar]
- 78.Schemske D. W. 2002. Tropical diversity: patterns and processes. In Ecological and evolutionary perspectives on the origins of tropical diversity: key papers and commentaries (eds Chazdon R., Whitmore T.), pp. 163–173 Chicago, IL: University of Chicago Press [Google Scholar]
- 79.Dynesius M., Jansson R. 2000. Evolutionary consequences of changes in species' geographical distributions driven by Milankovitch climate oscillations. Proc. Natl Acad. Sci. USA 97, 9115–9120 10.1073/pnas.97.16.9115 (doi:10.1073/pnas.97.16.9115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sheldon P. R. 1996. Plus ca change—a model for stasis and evolution in different environments. Palaeogeogr. Palaeoclimatol. Palaeoecol. 127, 209–227 10.1016/S0031-0182(96)00096-X (doi:10.1016/S0031-0182(96)00096-X) [DOI] [Google Scholar]
- 81.Jablonski D. 2008. Species selection: theory and data. Annu. Rev. Ecol. Evol. Syst. 39, 501–524 10.1146/annurev.ecolsys.39.110707.173510 (doi:10.1146/annurev.ecolsys.39.110707.173510) [DOI] [Google Scholar]
- 82.Rabosky D. L., McCune A. R. 2010. Reinventing species selection with molecular phylogenies. Trends Ecol. Evol. 25, 68–74 10.1016/j.tree.2009.07.002 (doi:10.1016/j.tree.2009.07.002) [DOI] [PubMed] [Google Scholar]
- 83.Phillimore A. B., Price T. D. 2008. Density-dependent cladogenesis in birds. PLoS Biol. 6, 483–489 10.1371/journal.pbio.0060071 (doi:10.1371/journal.pbio.0060071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mayhew P. J. 2007. Why are there so many insect species? Perspectives from fossils and phylogenies. Biol. Rev. 82, 425–454 10.1111/j.1469-185X.2007.00018.x (doi:10.1111/j.1469-185X.2007.00018.x) [DOI] [PubMed] [Google Scholar]
- 85.Marzluff J. M., Dial K. P. 1991. Life history correlates of taxonomic diversity. Ecology 72, 428–439 10.2307/2937185 (doi:10.2307/2937185) [DOI] [Google Scholar]
- 86.Martin A. P., Palumbi S. R. 1993. Body size, metabolic rate, generation time and the molecular clock. Proc. Natl Acad. Sci. USA 90, 4087–4091 10.1073/pnas.90.9.4087 (doi:10.1073/pnas.90.9.4087) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Payne J. L., Finnegan S. 2007. The effect of geographic range on extinction risk during background and mass extinction. Proc. Natl Acad. Sci. USA 104, 10 506–10 511 10.1073/pnas.0701257104 (doi:10.1073/pnas.0701257104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jablonski D. 2008. Extinction and the spatial dynamics of biodiversity. Proc. Natl Acad. Sci. USA 105, 11 528–11 535 10.1073/pnas.0801919105 (doi:10.1073/pnas.0801919105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Phillimore A. B., Freckleton R. P., Orme C. D. L., Owens I. P. F. 2006. Ecology predicts large-scale patterns of phylogenetic diversification in birds. Am. Nat. 168, 220–229 10.1086/505763 (doi:10.1086/505763) [DOI] [PubMed] [Google Scholar]
- 90.Jablonski D., Roy K. 2003. Geographical range and speciation in fossil and living molluscs. Proc. R. Soc. B 270, 401–406 10.1098/rspb.2002.2243 (doi:10.1098/rspb.2002.2243) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Phillimore A. B., Orme C. D. L., Thomas G. H., Blackburn T. M., Bennett P. M., Gaston K. J., Owens I. P. F. 2008. Sympatric speciation in birds is rare: insights from range data and simulations. Am. Nat. 171, 646–657 10.1086/587074 (doi:10.1086/587074) [DOI] [PubMed] [Google Scholar]
- 92.Safi K., Cianciaruso M. V., Loyola R. D., Brito D., Armour-Marshall K., Diniz-Filho J. A. F. 2011. Understanding global patterns of mammalian functional and phylogenetic diversity. Phil. Trans. R. Soc. B 366, 2536–2544 10.1098/rstb.2011.0024 (doi:10.1098/rstb.2011.0024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xiang Q. Y. J., Zhang W. H., Ricklefs R. E., Qian H., Chen Z. D., Wen J., Li J. H. 2004. Regional differences in rates of plant speciation and molecular evolution: a comparison between eastern Asia and eastern North America. Evolution 58, 2175–2184 [DOI] [PubMed] [Google Scholar]
- 94.Phillimore A. B., Price T. D. 2009. Ecological influences on the temporal pattern of speciation. In Speciation and patterns of diversity (eds Butlin R., Bridle J. R., Schluter D.), pp. 240–256 Cambridge, UK: Cambridge University Press [Google Scholar]
- 95.Peterson A. T. 1992. Philopatry and genetic differentiation in the Aphelocoma jays (Corvidae). Biol. J. Linn. Soc. 47, 249–260 10.1111/j.1095-8312.1992.tb00669.x (doi:10.1111/j.1095-8312.1992.tb00669.x) [DOI] [Google Scholar]
- 96.Seehausen O. 2006. African cichlid fish: a model system in adaptive radiation research. Proc. R. Soc. B 273, 1987–1998 10.1098/rspb.2006.3539 (doi:10.1098/rspb.2006.3539) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Springer M. S., Meredith R. W., Janecka J. E., Murphy W. J. 2011. The historical biogeography of Mammalia. Phil. Trans. R. Soc. B 366, 2478–2502 10.1098/rstb.2011.0023 (doi:10.1098/rstb.2011.0023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pielou E. C. 1979. Biogeography. New York, NY: Wiley [Google Scholar]
- 99.Ronquist F. 1997. Dispersal-vicariance analysis: a new approach to the quantification of historical biogeography. Syst. Biol. 46, 195–203 10.1093/sysbio/46.1.195 (doi:10.1093/sysbio/46.1.195) [DOI] [Google Scholar]
- 100.Whittaker R. J., Triantis K. A., Ladle R. J. 2008. A general dynamic theory of oceanic island biogeography. J. Biogeogr. 35, 977–994 10.1111/j.1365-2699.2008.01892.x (doi:10.1111/j.1365-2699.2008.01892.x) [DOI] [Google Scholar]
- 101.Stephens P. R., Wiens J. J. 2003. Explaining species richness from continents to communities: the time-for-speciation effect in emydid turtles. Am. Nat. 161, 112–128 10.1086/345091 (doi:10.1086/345091) [DOI] [PubMed] [Google Scholar]
- 102.Gavrilets S., Vose A. 2005. Dynamic patterns of adaptive radiation. Proc. Natl Acad. Sci. USA 102, 18 040–18 045 10.1073/pnas.0506330102 (doi:10.1073/pnas.0506330102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McPeek M. A. 2008. The ecological dynamics of clade diversification and community assembly. Am. Nat. 172, E270–E284 10.1086/593137 (doi:10.1086/593137) [DOI] [PubMed] [Google Scholar]
- 104.Chesson P. 2000. Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Syst. 31, 343–366 10.1146/annurev.ecolsys.31.1.343 (doi:10.1146/annurev.ecolsys.31.1.343) [DOI] [Google Scholar]
- 105.Jackson S. T., Sax D. F. 2010. Balancing biodiversity in a changing environment: extinction debt, immigration credit and species turnover. Trends Ecol. Evol. 25, 153–160 10.1016/j.tree.2009.10.001 (doi:10.1016/j.tree.2009.10.001) [DOI] [PubMed] [Google Scholar]
- 106.Rosenzweig M. L. 2001. Loss of speciation rate will impoverish future diversity. Proc. Natl Acad. Sci. USA 98, 5404–5410 10.1073/pnas.101092798 (doi:10.1073/pnas.101092798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schoener T. W. 1976. The species–area relationship within archipelagoes: models and evidence from island birds.In Proc. of XVI Int. Ornithological Congress, vol. 6 (eds Frith H. J., Calaby J. H.), pp. 629–642 Canberra, Australia: Australian Academy of Science [Google Scholar]