Abstract
We have shown that Alox5 is a critical regulator of leukemia stem cells (LSCs) in a BCR-ABL–induced chronic myeloid leukemia (CML) mouse model, and we hypothesize that the Alox5 pathway represents a major molecular network that regulates LSC function. Therefore, we sought to dissect this pathway by comparing the gene expression profiles of wild type and Alox5−/− LSCs. DNA microarray analysis revealed a small group of candidate genes that exhibited changes in the levels of transcription in the absence of Alox5 expression. In particular, we noted that the expression of the Msr1 gene was upregulated in Alox5−/− LSCs, suggesting that Msr1 suppresses the proliferation of LSCs. Using CML mouse model, we show that Msr1 is downregulated by BCR-ABL and this down-regulation is partially restored by Alox5 deletion, and that Msr1 deletion causes acceleration of CML development. Moreover, Msr1 deletion markedly increases LSC function through its effects on cell cycle progression and apoptosis. We also show that Msr1 affects CML development by regulating the PI3K-AKT pathway and β-Catenin. Together, these results demonstrate that Msr1 suppresses LSCs and CML development. The enhancement of the tumor suppressor function of Msr1 may be of significance in the development of novel therapeutic strategies for CML.
Introduction
Cancer stem cells (CSCs) are believed to be promising targets with the potential to offer curative therapies for some types of cancer, especially leukemias. Several genes and their pathways, including Wnt/β-Catenin,1,2 Hedgehog,3 Notch,4 Bim-1,5 p53,4 p166 and p19,7 Pten,8 PML,9 PP2A,10 Alox5,11 TGF-β/FOXO,12 and Musashi,13 have been found to promote or inhibit CSC proliferation. Some of these genes and pathways also play similar roles in regulating the function of normal stem cells.1,6,14 There is a need to identify CSC-specific genes for developing anti–stem cell strategies. Chronic myeloid leukemia (CML) is a stem cell–derived hematologic malignancy and serves as a good disease model for the study of CSC behavior and function. We have previously identified BCR-ABL–expressing Lin−Sca-1+c-Kit+ cells as leukemia stem cells (LSCs) in CML mice.15 We have demonstrated the critical role of the Alox5 gene in functional regulation of LSCs.11 We believe that the Alox5 pathway represents a major pathway that regulates the function of LSCs, and it will be important to identify other genes that interact with Alox5.
In theory, genes regulating LSC function can be grouped in 2 categories: those that enhance or promote LSC function or those that suppress and/or negatively regulate LSC function. Genetic deletions and/or mutations in the latter group are likely to favor leukemogenesis and a few genes in this category have been identified in LSCs. For example, Pten is down-regulated by BCR-ABL in LSCs. Pten deletion accelerates CML development and delays the disease on overexpression.16 p53 is also down-regulated by BCR-ABL and loss of p53 accelerates CML development.17,18 In this study, we identified that macrophage scavenger receptor (Msr1) suppresses BCR-ABL-expressing LSCs and CML development. Msr1 is a member of scavenger receptor family and is mostly expressed in hematopoietic cells.19 Msr1 is important for mediating host-cell interactions, macrophage adhesion and phagocytosis of apoptotic cells.20 Using Msr1−/− mice, we show that Msr1 delays CML development and regulates the self-renewal and differentiation capacity of LSCs through PI3K-AKT-GSK-3β and β-Catenin pathways.
Methods
Mice
C57BL/6, B6.SJL-Ptprca Pepcb/BoyJ, homozygous Alox5 knockout (Alox5−/−) mice and homozygous Msr1 knockout (Msr1−/−) mice in a C57BL/6 background were obtained from The Jackson Laboratory. Mice were maintained in a temperature- and humidity-controlled environment and given unrestricted access to 6% chow diet and acidified water.
DNA microarray
Bone marrow cells from C57BL/6 (B6) or Alox5−/− mice pretreated with 5-fluorouracil (5-FU) were transduced with BCR-ABL, followed by transplantation into B6 recipient mice (30 recipient mice for each group). Fourteen days later, bone marrow cells were isolated and subsequently sorted by FACS for LSCs (GFP+Lin−c-Kit+Sca-1+). Total RNA was isolated from these BCR-ABL–expressing LSCs or from the GFP+Lin−c-Kit+Sca-1+ cells that only expressed GFP, and DNA microarray analysis was carried out using the Affymetrix chips. All microarray data related to this Blood article have been deposited into the GEO public database, with accession numbers GSE10912 and GSE29347.
DNA constructs
Human Msr1 cDNA are kindly provided by Dr Sulahian (Harvard School of Public Health). To make the MSCV-BCR-ABL-Msr1 construct, Msr1 cDNA was amplified by Msr1-EcoRI 5′GAATTCATGGAGCAGTGGGATCACTTTC3′ and Msr1-ClaI 5′ATCGATTTATAAAGTGCAAGTGAC3′. The cDNA was sequenced from both ends to confirm the sequence. The Msr1 cDNA was cloned into the pMSCV-BCR-ABL-GFP vector between MfeI and ClaI sites. To make the pMSCV-Msr1-GFP construct, Msr1 cDNA was amplified by Msr1-EcoRI 5′GAATTCATGGAGCAGTGGGATCACTTTC3′ and Msr1-EcoRI 5′GAATTCTTATAAAGTGCAAGTGAC3′. The cDNA was sequenced from both ends to confirm the sequence. The Msr1 cDNA was cloned into the pMSCV-GFP vector at the EcoRI site.
Cell lines
Human K562 myeloid leukemia cell line was grown in RPMI 1640 medium containing 10% FBS. To generate the Msr1-expressing K562 cell lines, K562 cells were transduced with the pMSCV-Msr1-GFP retrovirus, and the Msr1-expressing cells were sorted for GFP by FACS. K562 cells transduced with the pMSCV-GFP retrovirus were used as a control.
Flow cytometry
Hematopoietic cells were collected from the bone marrow and peripheral blood of the normal and diseased mice. Erythrocytes were lysed in NH4Cl red blood cell lysis buffer (pH 7.4). The cells were washed with PBS and stained with B220-PE for B cells, Gr-1–APC for neutrophils, Mac-1–PE for macrophages, CD3E-APC for T cells and Sca-1/c-Kit/CD34/CD135 (Flt3)/CD204 for HSCs. All of these antibodies were purchased from Ebioscience Inc and Serotec Inc. After staining, the cells were washed once with PBS and subjected to FACS analysis.
Bone marrow transduction/transplantation
The retroviral vector MSCV-IRES-EGFP carrying the p210 BCR-ABL cDNA was used to make high-titer, helper-free, replication-defective ecotropic virus stock by transient transfection of 293T cells using the kat system as previously described.21,22 6- to 10-week-old WT C57BL/6 mice (The Jackson Laboratory) were used for leukemogenesis experiments. Induction of CML and B-acute lymphoblastic leukemia (ALL) was as previously described.21,22 Briefly, to model CML, bone marrow from 5-FU–treated (200 mg/kg) donor mice was transduced twice with BCR-ABL retrovirus by cosedementation in the presence of IL-3, IL-6, and SCF. To model B-ALL, bone marrow from non–5-FU–treated donors was transduced without cytokines. Wild-type recipient mice were prepared by 1100 cGy γ irradiation (given by 2 split 550 cGy doses) and 0.5 × 106 (CML) or 1.0 × 106 (B-ALL) cells were transplanted into recipient mice via tail vein injection. To make the retrovirus infecting human K562 cells, the MSCV-ecopac was replaced by pkat2ampac.23
Western blot analysis and antibodies
Antibodies against c-ABL, β-actin, β-Catenin and MSR1 were purchased from Santa Cruz Biotechnology and antibodies against p-Pten, Pten, p-PI3K, PI3K, p-AKT, AKT, p-GSK-3β and GSK-3β from Cell Signaling Technology. Protein lysates were prepared by lysing cells in RIPA buffer and Western blotting was carried out as described previously.24
In vitro culture of LSCs and drug treatment
Bone marrow cells isolated from CML mice were cultured in vitro in the presence of Stemspan SFEM (StemCell Technologies), stem cell factor, insulin-like growth factor-2, thrombopoietin, heparin, and α-fibroblast growth factor as described previously.25 PMA (Sigma-Aldrich) were dissolved in DMSO to make stock solution at 10mM, and were then diluted in culture media to different concentrations for use.
Histology
The lungs from the placebo- or drug-treated mice were fixed in Bouin fixative (Fisher Scientific) for 24 hours at room temperature, followed by an overnight rinse in water. 10-μm sections were stained with hematoxylin and eosin (H&E) and observed by a model DMRE compound microscope (Leica). All sections were imaged with a 2.5× PH1 objective (NPLan, NA 0.25) and 10× PH1 objective (NPLan, NA 0.40). All images were imported into MetaMorph Basic software (Molecular Devices) as a series of tagged image files. All images were then constructed in Adobe Photoshop 7.0 (Adobe).
Statistics
Statistical analyses were performed using Student t test (*P < .05, **P < .01; GraphPad Prism v5.01 software for Windows).
Results
Loss of Msr1 accelerates CML development
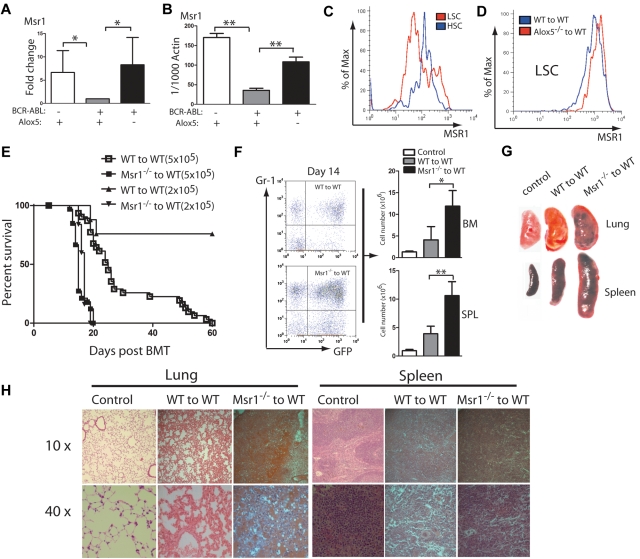
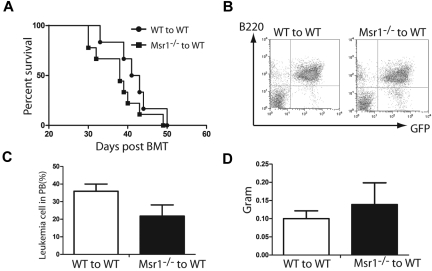
We previously demonstrated that Alox5 deficiency impairs the function of LSCs and prevents the initiation of BCR-ABL–induced CML. To identify the pathways in which Alox5 gene regulates function of LSCs, we performed a comparative DNA microarray analysis using total RNA isolated from non–BCR-ABL–expressing wild-type (WT) Lin−Sca-1+c-Kit+ cells, BCR-ABL–expressing WT LSCs and BCR-ABL–expressing Alox5−/− LSCs. Our data identified Msr1 as a candidate gene important for LSC function, as it was downregulated by BCR-ABL and restored by loss of Alox5 in LSCs (Figure 1A). This result was validated by quantitative real-time PCR (Figure 1B). The BCR-ABL downregulation of Msr1 in LSCs could not be restored by the treatment with the BCR-ABL kinase inhibitor imatinib (supplemental Figure 1, available on Blood Web site; see the Supplemental Materials link at the top of the online article). To further demonstrate the regulation of Msr1 by BCR-ABL, we compared the cell surface expression levels of MSR1 between LSCs and HSCs by FACS and found that MSR1 was downregulated by BCR-ABL (Figure 1C). This downregulation was rescued by the loss of Alox5 (Figure 1D). These results suggest that Msr1 suppresses LSCs and CML development. To test whether Msr1 affects CML development, we transduced bone marrow cells from 5-FU–treated WT or Msr1−/− mice with BCR-ABL-GFP retrovirus and transplanted these cells into lethally irradiated recipient mice. Recipients of BCR-ABL–transduced bone marrow cells from Msr1−/− donor mice developed CML significantly faster than those receiving BCR-ABL–transduced bone marrow cells from WT donor mice (Figure 1E). The accelerated death of CML mice in the absence of Msr1 also correlated with an elevated percentage of GFP+Gr1+ myeloid leukemia cells and an elevated number of total leukemia cells in the bone marrow and spleens of recipient mice (Figure 1F). Accelerated CML development in the absence of Msr1 correlated with more severe infiltration of leukemia cells in the lung and spleen (Figure 1G-H). These results show a potent tumor suppressor role for Msr1 in BCR-ABL–induced CML.
Figure 1.
Msr1 deletion accelerates CML development. (A) Expression of the Msr1 gene was compared among non–BCR-ABL–expressing Lin−Sca-1+c-Kit+ cells, BCR-ABL-expressing WT LSCs and BCR-ABL-expressing Alox5−/− LSCs. Msr1 expression was down-regulated by BCR-ABL and restored on Alox5 deletion in LSCs. (B) Quantitative real-time PCR analysis of non–BCR-ABL–expressing Lin−Sca-1+c-Kit+ cells, BCR-ABL–expressing WT LSCs and BCR-ABL–expressing Alox5−/− LSCs. Expression of the Msr1 gene was significantly down-regulated by BCR-ABL and partially rescued after Alox5 deletion in LSCs compared with WT LSCs by RT-PCR. Mean value (± SD) for each group is shown (**P < .01). (C) FACS analysis showed that MSR1 protein was down-regulated by BCR-ABL in LSCs compared with GFP−Lin−c-Kit+Sca-1+ cells that did not express BCR-ABL. (D) FACS analysis showed that MSR1 protein was up-regulated by Alox5 deletion in LSCs compared with WT LSCs. (E) Kaplan-Meier survival curves for recipients of BCR-ABL–transduced bone marrow cells from WT or Msr1−/− donor mice. All recipients of BCR-ABL–transduced bone marrow cells from Msr1−/− donor mice developed CML and died within 3 weeks post bone marrow transplantation, whereas recipients of BCR-ABL–transduced bone marrow cells from WT donor mice survived longer. (F) FACS analysis showed a higher percentage of GFP+Gr-1+ cells in the bone marrow and spleens of recipients of BCR-ABL–transduced bone marrow cells from Msr1−/− than WT donor mice (**P < .01). (G) Gross pathology and (H) H&E staining of the lungs and spleens showed severe lung hemorrhages and splenomegaly in recipients of BCR-ABL–transduced bone marrow cells from Msr1−/− donor mice. Recipient of pMSCV-GFP–transduced bone marrow cells was shown as a control.
Msr1 overexpression delays CML development
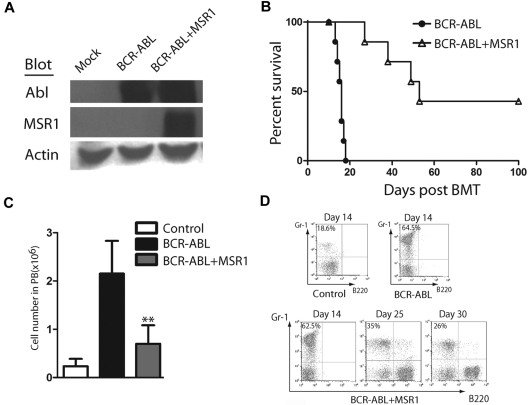
To further test whether the Msr1 can inhibit CML development, we coexpressed BCR-ABL and Msr1 in WT bone marrow cells by retroviral transduction and transplanted the transduced cells into recipient mice to induce CML. The BCR-ABL-IRES-GFP-pMSCV–expressing retrovirus was used as a control. The BCR-ABL-IRES-Msr1-pMSCV construct expressed BCR-ABL and MSR1 in 293T cells (Figure 2A). We next transduced wild-type donor bone marrow cells with BCR-ABL-IRES-Msr1-pMSCV or BCR-ABL-IRES-GFP-pMSCV retrovirus and transplanted these cells into recipient mice. CML development was significantly slower in mice receiving bone marrow cells transduced with BCR-ABL-IRES-Msr1-pMSCV than in those receiving bone marrow cells transduced with BCR-ABL-IRES-GFP-pMSCV. Some recipient mice did not even develop the CML (Figure 2B). The delayed CML development correlated with fewer leukemia cells in peripheral blood compared with recipients of pMSCV-GFP–transduced bone marrow cells (Figure 2C). In addition, FACS analysis of peripheral blood of recipients of BCR-ABL-IRES-Msr1-pMSCV–transduced bone marrow cells showed that the percentage of Gr-1+ myeloid leukemia cells declined with time (Figure 2D). These data further support the tumor suppressor function of Msr1 in CML development.
Figure 2.
Msr1 over-expression causes a delay of CML development. (A) BCR-ABL and MSR1 were detected by protein blotting using antibodies against ABL and MSR1. MSR1 protein was detected in cells transfected with BCR-ABL-IRES-Msr1-pMSCV. (B) Kaplan-Meier survival curves for recipients of BCR-ABL-IRES-pMSCV–transduced bone marrow cells (n = 7) or BCR-ABL-IRES-Msr1-pMSCV–transduced bone marrow cells (n = 7) donor mice. FACS analysis showed that the total number (C) and percentage (D) of Gr-1+ cells in peripheral blood of recipients of BCR-ABL-IRES-pMSCV–transduced bone marrow cells or BCR-ABL-IRES-Msr1-pMSCV–transduced bone marrow cells (*P < .05, **P < .01). Recipient of pMSCV-GFP–transduced bone marrow cells was shown as a control.
Msr1 deletion causes an increase in LSC function
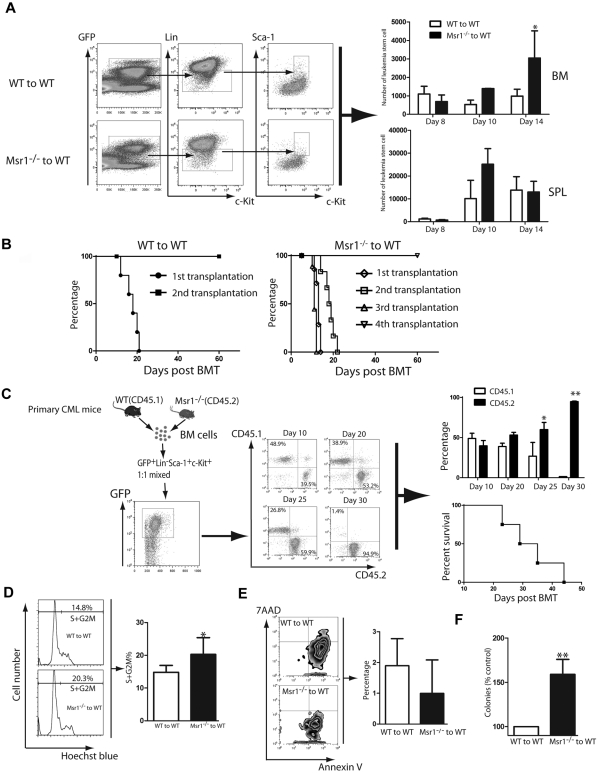
CML is a stem cell–derived disease and the accelerated development of CML in absence of Msr1 (Figure 1) prompted us to examine whether Msr1 regulates the function of LSCs. The accelerated CML development in the absence of Msr1 could be caused by an increase in the number of LSCs. To test this hypothesis, we quantified LSCs in the bone marrow and spleens of CML mice at 8, 10 and 14 days after CML induction. At day 8, Msr1 deficiency did not cause an increase of LSCs in either bone marrow or spleen compared with CML mice receiving BCR-ABL–tranduced wild-type bone marrow cells. At day 14, a significant increase in Msr1-deficient bone marrow LSCs was observed (Figure 3A), but Msr1 deficiency did not change the number of Msr1−/− LSCs in the spleen of CML mice (Figure 3A).
Figure 3.
Msr1 deletion affects the function of LSCs. (A) Loss of Msr1 caused a significant increase in LSCs in bone marrow (*P < .05). Mean value (± SD) for each group is shown (*P < .05). (B) Kaplan-Meier survival curves for a serial transplantation of recipients of 1 × 106 bone marrow cells from mice receiving BCR-ABL–transduced WT (n = 5) or Msr1−/− (n = 5) donor bone marrow cells. (C) Bone marrow cells derived from CML mice induced by transplanting BCR-ABL–transduced WT (CD45.1) or Msr1−/− (CD45.2) donor bone marrow cells were isolated. 5 × 103 WT and Msr1−/− LSCs were sorted by FACS, mixed by 1:1 ratio, and transplanted into lethally irradiated secondary recipient mice. At days 10, 20, 25 and 30 after BMT, FACS analysis showed that the percentages of CD45.2+ cells were much higher than those of CD45.1+ cells. All these mice died of CML, presumably because of the development of CML from CD45.1+ cells. (D) At 14 days post transplantation, bone marrow cells were isolated from recipients of BCR-ABL–transduced bone marrow cells from WT or Msr1−/− donor mice. The cells were stained with Hoechst Blue, and the DNA content, represented by the percentages of LSC populations in the S+G2M phase of the cell cycle, was examined by FACS. Mean percentage for each cell population (n = 4) is shown (*P < .05). (E) At 14 days post transplantation, bone marrow cells were isolated from recipients of BCR-ABL–transduced bone marrow cells from WT or Msr1−/− donor mice. The cells were stained with 7AAD and annexin V, and the percentages of LSCs positive for 7AAD and annexin V, representing apoptotic cells, were determined by FACS. (F) LTC-IC assay was performed to compare the function of LSCs. GFP+ bone marrow cells were sorted from recipients of BCR-ABL–transduced from WT or Msr1−/− donor bone marrow cells. The cells were cultured for 5 weeks in the presence of stromal cells. Colony formation was assessed 2 weeks later.
The ability to transfer leukemia to next generation recipient mice can be used to examine the biologic function of LSCs. We transduced WT or Msr1−/− donor bone marrow cells with a lower titer BCR-ABL retrovirus and transferred the transduced cells to recipient mice to induce primary CML. At day 13 after CML induction, bone marrow cells from primary CML mice were transplanted into secondary recipient mice. Subsequently, leukemic bone marrow cells were serially transferred to recipient mice for 2 more generations. We found that BCR-ABL–expressing Msr1−/− bone marrow cells could transfer CML up to the fourth generation of recipient mice, while BCR-ABL–expressing WT bone marrow cells only transferred CML once in recipient mice (Figure 3B). These results suggest that Msr1 deficiency causes an increase in LSC function. To further evaluate the effect of Msr1 on LSC function, we sorted WT (CD45.1) and Msr1−/− (CD45.2) LSCs from bone marrow of CML mice by FACS. The sorted WT and Msr1−/− LSCs were mixed in a 1:1 ratio (5000 LSCs for each group), and were transplanted into recipient mice. We observed that at day 10, the percentages of GFP+Gr-1+ WT (CD45.1) and Msr1−/− (CD45.2) cells in peripheral blood of the mice were similar, and then gradually increased and eventually became a dominant leukemic cell population (Figure 3C). We also observed that Msr1 deficiency did not cause an increase in the homing ability of bone marrow cells (supplemental Figure 2), which could result in accelerated CML development. To explain how the loss of Msr1 caused an increase in LSC function, we performed a DNA content analysis to examine the effect of Msr1 deficiency on cell cycle progression of LSCs. We found that the percentage of LSCs in the S+G2M phase was significantly higher in the Msr1−/− group than in the WT group (Figure 3D), indicating that the loss of Msr1 causes more LSCs to enter the cell cycle, thereby enhancing the proliferation of LSCs. Furthermore, we examined whether Msr1 deletion reduces apoptosis of LSCs by staining the cells with 7AAD and annexin V. We observed that apoptosis of Msr1−/− LSCs was reduced compared with WT LSCs (Figure 3E). Thus, Msr1 regulates cell cycle progression and apoptosis of LSCs. To further demonstrate the role of Msr1 in suppressing LSC function, we performed the long-term culture-initiating cell assay (LTC-IC). GFP+ bone marrow cells were collected from recipients of BCR-ABL–transduced WT or Msr1−/− donor bone marrow cells, and then the cells were cultured for 5 weeks in the presence of stromal cells. Colony formation was then analyzed 2 weeks later. Compared to WT CML cells, the ability to form colonies was significantly increased in Msr1−/− CML cells (Figure 3F).
Induction of Msr1 expression by PMA causes inhibition of human CML cells and LSCs
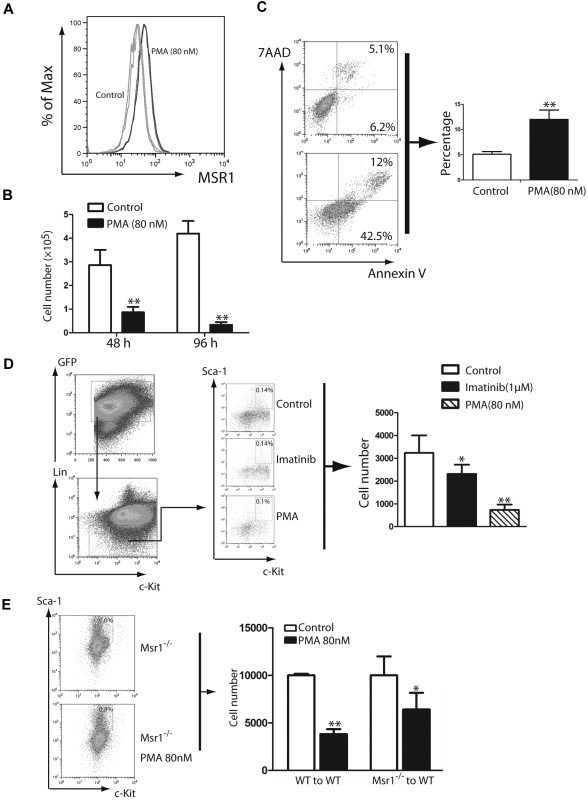
Next, we tested whether Msr1 inhibits human CML cells. Phorbol 12-myristate 13-acetate (PMA) induces differentiation of THP-1 cells, leading to a marked increase in the expression of MSR1 on the cell surface.26 We found that the level of MSR1 was significantly increased in K562 cells treated with PMA for 48 hours (Figure 4A). We also noted that PMA significantly inhibited the proliferation of K562 cells (Figure 4B). The decreased proliferation of K562 cells was consistent with an increase in apoptosis (Figure 4C). We also tested whether PMA inhibits LSCs from CML in vitro under stem cell culture conditions. We cultured bone marrow cells from CML mice in the presence of imatinib (1μM) or PMA (80nM) for 3 days, and calculated the total number and percentage of LSCs that remained at the end of the culture based on FACS analysis and total cell counts. We showed that PMA significantly inhibited LSCs in vitro (Figure 4D). To further confirm that PMA inhibits LSCs through inducing Msr1, we treated bone marrow cells from recipients of BCR-ABL–transduced WT or Msr1−/− donor bone marrow cells with PMA (80 nM) for 3 days and calculated the total numbers of LSCs at the end of the culture based on FACS analysis and total cell counts. We found that the number of Msr1−/− LSCs was reduced in a much lesser degree than the reduction of WT LSCs by PMA (Figure 4E), indicating that the induction of Msr1 contributes to the inhibition of LSCs by PMA.
Figure 4.
PMA inhibits proliferation and induces apoptosis of human CML cells. (A) FACS analysis showed that MSR1 protein was up-regulated in human leukemic cells treated with PMA for 48h compared with untreated cells. (B) PMA inhibits proliferation of human leukemic cells. K562 cells were treated with DMSO or PMA (80nM) for 48 and 96 hours, and live cells were counted (**P < .01). (C) PMA induces apoptosis of K562 cells. K562 cells were treated with DMSO or PMA (80nM) for 48 hours. Apoptotic cells (annexin V+/7AAD+) cells were analyzed by FACS (**P < .01). (D) PMA inhibits LSCs from CML mice in vitro. Bone marrow cells isolated from mice with CML induced by BCR-ABL-GFP were cultured (2 × 106 cells/6 cm plate) under stem cell conditions in the presence of DMSO, imatinib (1μM) or PMA (80nM) for 3 days, followed by FACS analysis of LSCs (*P < .05, **P < .01). (E) Bone marrow cells from recipients of BCR-ABL–transduced bone marrow cells from Msr1−/− donor mice were treated with PMA (80nM) for 3 days. The total number LSCs at the end of the culture were calculated based on FACS analysis and total cell counts.
Msr1 deletion does not affect the function of normal HSCs
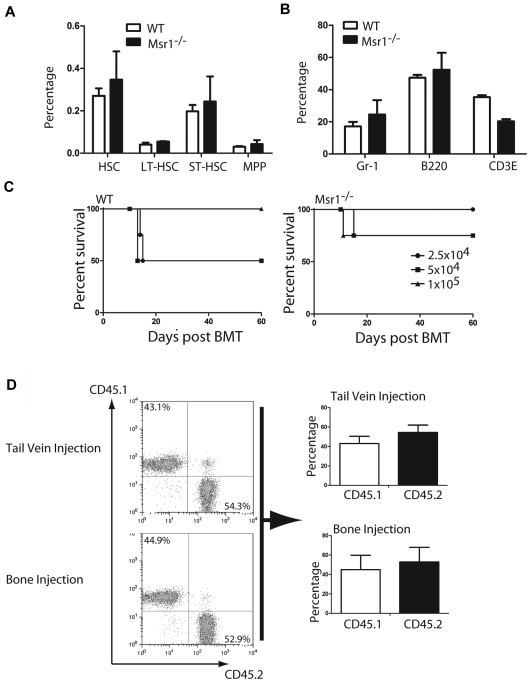
We further tested whether the loss of Msr1 affects the function of normal HSCs. We first compared hematopoietic cell lineages in bone marrow and peripheral blood of Msr1−/− mice with those of WT mice. The percentages of HSCs (Lin−c-Kit+Sca-1+), LT-HSCs (Lin−c-Kit+Sca-1+CD34−), ST-HSCs (Lin−c-Kit+Sca-1+CD34+Flt3−) and MPPs (Lin−c-Kit+Sca-1+CD34+Flt3+) in bone marrow of Msr1−/− mice were only slightly higher than those of WT mice (Figure 5A), and the percentages of Gr-1+, Mac-1+, B220+ or CD3E+ cell populations in bone marrow were similar between WT and Msr1−/− mice (Figure 5B). To compare the function of HSCs between Msr1−/− and WT mice, we conducted an irradiation rescue assay, in which several doses of Msr1−/− or WT bone marrow cells were transplanted into lethally irradiated WT mice. Survival of mice receiving WT or Msr1−/− bone marrow cells was similar (Figure 5C). To more stringently compare the stem cell function, we carried out a competitive reconstitution analysis, in which WT (CD45.1) and Msr1−/− (CD45.2) bone marrow cells were mixed in a 1:1 ratio and then transferred into recipient mice by tail vein or bone injection. Twelve weeks after the transplantation, the percentages of WT (CD45.1) and Msr1−/− (CD45.2) were similar in peripheral blood of the mice (Figure 5D), indicating that Msr1 did not suppress the function of normal HSCs.
Figure 5.
Msr1 deletion does not affect the function of normal HSCs. (A) Bone marrow cells from WT and Msr1−/− mice were analyzed by FACS for the percentages of total HSCs (Lin−c-Kit+Sca-1+), LT-HSCs (Lin−c-Kit+Sca-1+CD34−), ST-HSCs (Lin−c-Kit+Sca-1+CD34+Flt3−) and MPPs (Lin−c-Kit+Sca-1+CD34+Flt3+). (B) Cells from bone marrow and peripheral blood of WT and Msr1−/− mice were analyzed by FACS for the percentages of Gr-1+, B220+ and CD3E+cells. (C) 3 doses (1 × 105, 5 × 104 and 2.5 × 104) of WT or Msr1−/− bone marrow cells were injected into lethally irradiated recipient mice. Survival curves showed that there was only a minor engraftment defect of bone marrow cells in Msr1−/− mice. (D) Msr1−/− (CD45.2) and WT (CD45.1) bone marrow cells were 1:1 mixed and then transferred into recipient mice (n = 5). 12 weeks after transplantation, FACS analysis showed the percentages of WT and Msr1−/− cells in peripheral blood of recipient mice.
Msr1 affects CML development by regulating the PI3K-AKT-GSK-3β and β-Catenin pathways
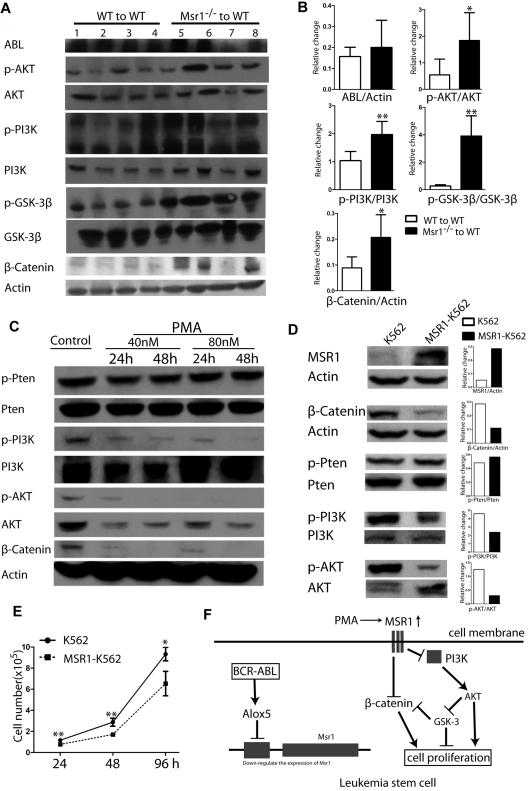
To determine the signaling pathways involved in Msr1 function, we analyzed protein lysates isolated from the spleens of CML mice receiving BCR-ABL–transduced WT or Msr1−/− donor bone marrow cells. Loss of Msr1 resulted in a marked activation of the PI3K-AKT-GSK3β pathway and increased expression of β-Catenin (Figure 6A). Conversely, we tested whether the upregulation of Msr1 expression inhibits these signaling molecules by treating K562 cells with PMA. PMA markedly inhibited the activation of the PI3K-AKT and reduced expression of β-Catenin (Figure 6B). To confirm the inhibitory effects of Msr1 on these signal pathways in leukemia cells, we overexpressed Msr1 gene in K562 cells by retroviral infection of the cells with pMSCV-Msr1-GFP or pMSCV-GFP (as a control; supplemental Figure 3A). The pMSCV-Msr1-GFP construct expressed MSR1 and GFP and the pMSCV-GFP construct expressed GFP in 293T cells (supplemental Figure 3B). K562 cells transduced with pMSCV-Msr1-GFP or pMSCV-GFP were sorted by FACS for GFP+ cells and overexpression of MSR1 was confirmed by FACS (supplemental Figure 3C) and Western blotting (Figure 6D). We found that overexpression of Msr1 inhibited the activation of the PI3K and AKT, reduced expression of β-Catenin (Figure 6D), and suppressed proliferation of leukemia cells (Figure 6E). Together, these results suggest that Msr1 is a negative regulator between BCR-ABL–activated Alox5 and the PI3K-AKT-GSK-3β and β-Catenin pathways (Figure 6F).
Figure 6.
Msr1 affects CML development by regulating PI3K-AKT-GSK-3β pathway and β-Catenin. (A) Msr1 regulates signaling pathways involved in the regulation of LSC functions. Spleens of recipients of BCR-ABL–transduced WT or Msr1−/− donor bone marrow cells (n = 4) at 14 days after transplantation were collected and protein lysates were prepared for expression of a group of signaling molecules. Loss of Msr1 markedly activated the PI3K-AKT pathway and increased expression of β-Catenin. (B) Quantification of relative protein expression levels in spleens of recipients of BCR-ABL-transduced WT or Msr1−/− donor bone marrow cells. Mean value (± SD) for each group (n = 4) is shown (P < .05, **P < .01). (C) PMA inhibits PI3K-AKT signaling pathways and β-Catenin expression in human CML cells. K562 cells were treated with PMA, and protein lysates were collected for expression of a group of signaling molecules. PMA markedly inhibits activity of PI3K-AKT and reduced expression of β-Catenin. (D) Over-expression of Msr1 inhibits the PI3K-AKT signaling pathways and reduces β-Catenin expression in human CML cells. Protein lysates from K562 transduced with pMSCV-Msr1-GFP or pMSCV-GFP retrovirus were analyzed by Western blotting for expression of a group of signaling molecules indicated. (E) Msr1 suppresses the proliferation of human leukemic cells. The same number of K562 cells transduced with pMSCV-GFP or pMSCV-Msr1-GFP was cultured in 24-well plates. Live cells were counted at 24h, 48h and 96h (*P < .05, **P < .01). (F) A summary of Msr1 pathway in LSCs of CML.
Msr1 does not affect BCR-ABL–induced lymphoid leukemia
We also tested whether Msr1 affects BCR-ABL–induced acute lymphoblastic leukemia (ALL), which originates from committed lymphoid progenitors. WT or Msr1−/− donor bone marrow cells were transduced by BCR-ABL-GFP retrovirus, followed by transplantation cells into lethally irradiated WT recipient mice. Both groups of mice developed and died of ALL with similar disease latency and survival time (Figure 7A). FACS analysis of lymphoid leukemia cells showed that both groups of ALL mice had similar percentages of B220+ leukemia cells in peripheral blood of these mice (Figure 7B-C) although the mass of spleen in Msr1−/− donor group was slightly more elevated than WT group (Figure 7D). Together, these results showed that Msr1 did not affect the development of BCR-ABL–induced ALL, and suggest that Msr1 is not required by ALL stem cells.
Figure 7.
Msr1 does not affect BCR-ABL–induced lymphoid leukemia. (A) Kaplan-Meier survival curves for recipients of BCR-ABL–transduced bone marrow cells from WT (n = 10) or Msr1−/− (n = 8) donor mice. Both groups of mice developed and died of ALL. (B-C) FACS analysis showed similar numbers of GFP+B220+ cells in peripheral blood of recipients of BCR-ABL–transduced bone marrow cells from WT (n = 5) or Msr1−/− (n = 5) donor mice. (D) Spleen masses of recipients of BCR-ABL–transduced bone marrow cells from Msr1−/− (n = 9) donor mice is similar to WT (n = 4) donor mice.
Discussion
In this study, we show that the reduction of LSCs in the absence of Alox5 is partially because of upregulated expression of Msr1 because Msr1 inhibits the proliferation of LSCs. Our finding suggest that both Msr1 and Alox5 are in the same pathway and that Msr1 functions downstream of Alox5 and suppresses BCR-ABL–induced CML. Compared with well-studied tumor suppressor genes like Pten and p53, Msr1 may represent a new group of genes with tumor suppressive activities, because deletion of this gene in mice does not result in tumor development (data not shown). Genetic changes to Msr1 have been shown to be related to prostate cancer risk, although the underlying mechanism is unclear.27 Our data demonstrate that the expression of Msr1 is downregulated by BCR-ABL. Loss of Msr1 function leads to accelerated development of CML in mice, and this disease phenotype can be rescued by ectopic Msr1 expression. Together, the results in this study reveal the functional relationship between Msr1 and Alox5, that is, BCR-ABL activates Alox5 in LSCs, leading to reduced expression of Msr1. Thus, restoration of Msr1 expression provides a strategy for shutting down the Alox5 pathway required for LSC function.11 Thus, the impetus for the identification of genes critical to LSC function in CML is the opportunity to find pathways that may be targeted for eradicating CML LSCs. Remarkably, enhanced Msr1 expression may provide a means for eliminating LSCs and preventing CML. Our findings that the pharmacologic induction of Msr1 impairs BCR-ABL expressing LSCs, as well as human CML cells, lend credence to the notion that Msr1 agonists may be used as a novel therapeutic strategy for eradicating LSCs in CML. Targeting Alox5 and Msr1 in combination with the BCR-ABL kinase inhibitor imatinib or a BCR-ABL protein inhibitor, such as an HSP90 inhibitor, may provide a novel approach that leads to better clinical control of CML and the depletion of LSCs.
Our results also provide insight into the cellular mechanism for CML development in the absence of Msr1. The CML serial transplantation assay and LSC competition assay suggest that the accelerated CML development is caused by the significantly enhanced ability of LSCs to self-renew and induce leukemia in the absence of Msr1. The enhanced function of LSCs in the absence of Msr1 is because of an increase in the number of LSCs in S+G2M phase and a decrease in LSC apoptosis. However, Msr1 deficiency does not appear to significantly affect normal HSCs, which is consistent with our observation that Alox5 deletion only affects LSCs instead of normal HSCs.11,28
The molecular mechanism for the functional regulation of LSCs by Msr1 remains largely unknown. We show that the loss of Msr1 function leads to activation of the PI3K-AKT pathway and upregulated expression of β-Catenin. By contrast, overexpression of Msr1 in human K562 leukemia cells or up-regulation of Msr1 by PMA inhibits the activity of the PI3K-AKT pathway and reduces the expression of β-Catenin. We and others have recently shown that β-Catenin plays a critical role in CML LSCs,1,2 and that Akt affects the progression of BCR-ABL–induced leukemia in a mouse model.16 Thus, the regulation of these LSC-related genes by Msr1 indicates its potential role in the development of BCR-ABL–induced CML. Because Alox5 regulates expression of β-Catenin in LSCs11 and loss of Alox5 upregulates the expression of Msr1, we propose that Alox5 and Msr1 are functionally linked and critical for LSC function. More studies are needed to explain how Msr1 suppresses LSCs in CML.
Supplementary Material
Acknowledgments
We thank Dr Sulahian (Harvard School of Public Health) for providing the Msr1 cDNA.
This work was supported by the grants from the Leukemia & Lymphoma Society and the National Institutes of Health (R01-CA122142, R01-CA114199 to S.L.). S.L. is a Scholar of the Leukemia & Lymphoma Society.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: Y.C. designed and performed experiments and analyzed the data; C.S. performed experiments and analyzed the data; C.P. and Y.H. performed experiments; Y.S. performed experiments and analyzed the data; D.L. helped with the experiments; and S.L. designed and performed experiments, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Shaoguang Li, University of Massachusetts Medical School, Dept of Medicine, 364 Plantation St, Worcester, MA 01605; e-mail: shaoguang.li@umassmed.edu.
References
- 1.Zhao C, Blum J, Chen A, et al. Loss of beta-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer Cell. 2007;12(6):528–541. doi: 10.1016/j.ccr.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu Y, Chen Y, Douglas L, Li S. beta-Catenin is essential for survival of leukemic stem cells insensitive to kinase inhibition in mice with BCR-ABL-induced chronic myeloid leukemia. Leukemia. Jan;23(1):109–116. doi: 10.1038/leu.2008.262. [DOI] [PubMed] [Google Scholar]
- 3.Zhao C, Chen A, Jamieson CH, et al. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature. 2009;458(7239):776–779. doi: 10.1038/nature07737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Molofsky AV, Pardal R, Morrison SJ. Diverse mechanisms regulate stem cell self-renewal. Curr Opin Cell Biol. 2004;16(6):700–707. doi: 10.1016/j.ceb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 5.Park IK, Qian D, Kiel M, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423(6937):302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 6.Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19(12):1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr Opin Genet Dev. 2003;13(1):77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 8.Yilmaz OH, Valdez R, Theisen BK, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441(7092):475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 9.Ito K, Bernardi R, Morotti A, et al. PML targeting eradicates quiescent leukaemia-initiating cells. Nature. 2008;453(7198):1072–1078. doi: 10.1038/nature07016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neviani P, Santhanam R, Trotta R, et al. The tumor suppressor PP2A is functionally inactivated in blast crisis CML through the inhibitory activity of the BCR/ABL-regulated SET protein. Cancer Cell. 2005;8(5):355–368. doi: 10.1016/j.ccr.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y, Hu Y, Zhang H, Peng C, Li S. Loss of the Alox5 gene impairs leukemia stem cells and prevents chronic myeloid leukemia. Nat Genet. 2009;41(7):783–792. doi: 10.1038/ng.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naka K, Hoshii T, Muraguchi T, et al. TGF-beta-FOXO signalling maintains leukaemia-initiating cells in chronic myeloid leukaemia. Nature. 2010;463(7281):676–680. doi: 10.1038/nature08734. [DOI] [PubMed] [Google Scholar]
- 13.Ito T, Kwon HY, Zimdahl B, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466(7307):765–768. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tothova Z, Kollipara R, Huntly BJ, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128(2):325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Hu Y, Swerdlow S, Duffy TM, Weinmann R, Lee FY, Li S. Targeting multiple kinase pathways in leukemic progenitors and stem cells is essential for improved treatment of Ph+ leukemia in mice. Proc Natl Acad Sci U S A. 2006;103(45):16870–16875. doi: 10.1073/pnas.0606509103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng C, Chen Y, Yang Z, et al. PTEN is a tumor suppressor in CML stem cells and BCR-ABL-induced leukemias in mice. Blood. 2010;115(3):626–635. doi: 10.1182/blood-2009-06-228130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wendel HG, de Stanchina E, Cepero E, et al. Loss of p53 impedes the antileukemic response to BCR-ABL inhibition. Proc Natl Acad Sci U S A. 2006;103(19):7444–7449. doi: 10.1073/pnas.0602402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 19.Rohrer L, Freeman M, Kodama T, Penman M, Krieger M. Coiled-coil fibrous domains mediate ligand binding by macrophage scavenger receptor type II. Nature. 1990;343(6258):570–572. doi: 10.1038/343570a0. [DOI] [PubMed] [Google Scholar]
- 20.Platt N, Gordon S. Is the class A macrophage scavenger receptor (SR-A) multifunctional? - The mouse's tale. J Clin Invest. 2001;108(5):649–654. doi: 10.1172/JCI13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu Y, Liu Y, Pelletier S, et al. Requirement of Src kinases Lyn, Hck and Fgr for BCR-ABL1-induced B-lymphoblastic leukemia but not chronic myeloid leukemia. Nat Genet. 2004;36(5):453–461. doi: 10.1038/ng1343. [DOI] [PubMed] [Google Scholar]
- 22.Li S, Ilaria RL, Jr, Million RP, Daley GQ, Van Etten RA. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J Exp Med. 1999;189(9):1399–1412. doi: 10.1084/jem.189.9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finer MH, Dull TJ, Qin L, Farson D, Roberts MR. kat: a high-efficiency retroviral transduction system for primary human T lymphocytes. Blood. 1994;83(1):43–50. [PubMed] [Google Scholar]
- 24.Li S, Couvillon AD, Brasher BB, Van Etten RA. Tyrosine phosphorylation of Grb2 by Bcr/Abl and epidermal growth factor receptor: a novel regulatory mechanism for tyrosine kinase signaling. EMBO J. 2001;20(23):6793–6804. doi: 10.1093/emboj/20.23.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng C, Brain J, Hu Y, et al. Inhibition of heat shock protein 90 prolongs survival of mice with BCR-ABL-T315I-induced leukemia and suppresses leukemic stem cells. Blood. 2007;110(2):678–685. doi: 10.1182/blood-2006-10-054098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao HS, Matsumoto A, Itakura H, et al. Transcriptional inhibition by interleukin-6 of the class A macrophage scavenger receptor in macrophages derived from human peripheral monocytes and the THP-1 monocytic cell line. Arterioscler Thromb Vasc Biol. 1999;19(8):1872–1880. doi: 10.1161/01.atv.19.8.1872. [DOI] [PubMed] [Google Scholar]
- 27.Xu J, Zheng SL, Komiya A, et al. Germline mutations and sequence variants of the macrophage scavenger receptor 1 gene are associated with prostate cancer risk. Nat Genet. 2002;32(2):321–325. doi: 10.1038/ng994. [DOI] [PubMed] [Google Scholar]
- 28.Chen XS, Sheller JR, Johnson EN, Funk CD. Role of leukotrienes revealed by targeted disruption of the 5-lipoxygenase gene. Nature. 1994;372(6502):179–182. doi: 10.1038/372179a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.