Abstract
We recently discovered that in rat spermatids, kinesin light chain KLC3 can associate with outer dense fibers, major sperm tail components, and accumulates in the sperm midpiece. Here we show that mitochondria isolated from rat elongating spermatids have bound KLC3. Immuno electron microscopy indicates that the association of KLC3 with mitochondria coincides with the stage in spermatogenesis when mitochondria move from the plasma membrane to the developing midpiece. KLC3 is able to bind in vitro to mitochondria from spermatids as well as somatic cells employing a conserved kinesin light chain motif, the tetratrico-peptide repeats. Expression of KLC3 in fibroblasts results in formation of large KLC3 clusters close to the nucleus, which also contain mitochondria: no other organelles were present in these clusters. Mitochondria are not present in KLC3 clusters after deletion of KLC3’s tetratrico-peptide repeats. Our results indicate that the rat spermatid kinesin light chain KLC3 can associate with mitochondria.
Keywords: Spermatogenesis, spermatids, midpiece, mitochondria, kinesin, kinesin light chain, motor molecule, transfection
INTRODUCTION
Spermatogenesis is characterized by continuous germ cell proliferation and differentiation and takes place in seminiferous tubules, where developing germ cells undergo a series of characteristic differentiation steps in close association with somatic Sertoli cells. After an initial proliferative phase, mitotically active spermatogonia enter a pre-meiotic phase and become spermatocytes that undergo meiotic division. Upon completion of meiosis, spermatids emerge that differentiate into spermatozoa through a process known as spermiogenesis. Dramatic morphological changes occur in this phase including the formation of the sperm tail, which contains two unique structures: outer dense fibers (ODF) and fibrous sheath (FS). Several ODF components and associated proteins were recently cloned (van der Hoorn et al., 1990; Burfeind and Hoyer-Fender, 1991; Morales et al., 1994; Shao et al., 1997; Turner et al., 1997; Schalles et al., 1998; Hoyer-Fender et al., 1998; Shao et al., 1999; Shao et al., 2001; Petersen et al., 2002) and we demonstrated that they interact specifically via leucine zipper motifs (Shao et al., 1997; Kierszenbaum, 2001). A distinct region of the tail is the midpiece, which contains mitochondria surrounding the ODF and axoneme, but lacks the FS. The mitochondria form a helical mitochondrial sheath. The basis for the formation of the midpiece is not understood: in step 15 rat spermatids a constriction point called the annulus (Irons and Clermont, 1982a; Irons and Clermont, 1982b) that separates the bulk of spermatid cytoplasm from the growing tail descends. In its wake mitochondria, which until that time are located at the plasma membrane, migrate to ODF that are now accessible in the future midpiece region. In mature spermatozoa mitochondria appear connected to ODF by a proteinaceous structure, the submitochondrial reticulum (SMR) (Olson and Winfrey, 1990; Olson and Winfrey, 1992). Its composition and involvement in aligning mitochondria is not clear.
We recently reported the isolation of the novel kinesin light chain KLC3 (KLC3 mRNA GenBank accession nr. AF166267) that is expressed in rat testis as well as in some other tissues (Junco et al., 2001). Together with kinesin heavy chains (KHC) KLC form kinesin motor molecules. Kinesins and kinesin related proteins (KRPs) transport cargo along microtubules (reviewed in (Hackney, 1996; Howard, 1996; Moore and Endow, 1996; Scholey, 1996; Block, 1998; Hirokawa, 1998; Goldstein, 2001a; Goldstein, 2001b; Higuchi and Endow, 2002; Kamal and Goldstein, 2002; Endow, 2003; Schnapp, 2003). The role of KLC may be regulation of motor activity (Verhey et al., 1998) and/or binding of cargo, which can include proteins, mitochondria (Khodjakov et al., 1998), and various vesicles (Johnson et al., 1996; Gindhart, Jr. et al., 1998). Kinesins and KRPs have been detected in mammalian testis including six KRPs (Sperry and Zhao, 1996) and a conventional kinesin associated with the manchette, a transient microtubule structure that may function as an organizational scaffold for proteins that are required in the sperm tail or head (Hall et al., 1992; Kierszenbaum, 2001). KRP3A and KRP3B localize to spermatid nuclei (Zou et al., 2002), KIFC5 kinesin associates with all microtubule-containing spermatid structures (Navolanic and Sperry, 2000) and Kinesin II (Miller et al., 1999), kinesin associated protein 3 KAP3 (Yamazaki et al., 1996) and kinesin KIFC1 are found in close association with the developing acrosome (Yang and Sperry, 2003). These results indicate a role for kinesins in sperm tail development, including organelle movement and transport in the tail. No kinesin or kinesin components have been described thus far in association with mitochondria in elongating spermatids.
The germ cell expression pattern of KLC3 is interesting: mRNA is present before and after meiosis, in contrast to the neuronal KLC1 and ubiquitous KLC2 (Rahman et al., 1998) that are only expressed before meiosis (Junco et al., 2001). KLC3 has domains characteristic of KLC including the conserved heptad repeat (HR) that binds to KHC (Gauger and Goldstein, 1993; Diefenbach et al., 1998) and tandem tetratrico-peptide repeats (TPR) that in other proteins mediate protein interactions (Gindhart, Jr. and Goldstein, 1996). Our recent results suggested that KLC3 might be involved in sperm tail midpiece formation: KLC3 accumulates in the sperm tail midpiece, which contains the axoneme, ODF and the mitochondrial sheath (Junco et al., 2001). We recently described that KLC3 can bind to ODF (Bhullar et al., 2003).
Here we studied the developmental expression of KLC3 and discovered its interaction with rat spermatid mitochondria both in vitro and in vivo. We asked which region is responsible for this interaction and the in vitro assays demonstrate that KLC3 can bind to mitochondria mediated by specific repeats in the TPR domain. Expression of KLC3 in fibroblasts produces KLC3 protein clusters which contain mitochondria but not other organelles.
MATERIALS AND METHODS
Immuno-electron microscopy
Processing of tissues for Lowicryl K4M embedding followed a standard protocol used in our laboratory (Oko et al., 1996). Lowicryl-embedded ultra thin sections of 5 independently processed testes and epididymides preparations were mounted on 200-mesh, Formavar-coated nickel grids, transferred and floated tissue side down on 10 – 20 μl drops of the following solutions: 10 % goat serum (gs) in 20 mM Tris-HCl buffered saline (TBS), pH 7.4, 15 min; anti-KLC3 MAb diluted 1:10 in TBS, 1 hr; TBS containing 0.1 % Tween-20, 5 × 5 min; 10 % gs in TBS, 15 min; colloidal gold (10 nm)-conjugated goat anti-mouse IgG diluted 1:20 in TBS, 45 min; TBS-Tween-20, 3 × 5 min; and dH2O), 2 × 5 min. Sections were counterstained with uranyl acetate and lead citrate and 35 and 20 representative sections of step 15/16 elongating spermatids and step 19 elongated spermatids, respectively, were examined by electron microscopy. Controls consisted of replacing the primary antibody (anti-KLC3) step with TBS and pre-immune mouse sera diluted 1:10 in TBS.
Constructs used in this study
KLC3 mutants were made by PCR using pBS(ATG)KLC3 (Junco et al., 2001) as template. To produce KLC3-ΔC the following primers were used: Cf1478 (5′ CGCTAAGTGGACTGGCTGCAG 3′) and Cr 1174 (5′ GCTGAGGATCTCCTTGTATAGCTCC 3′). To make KLC3ΔTPR we used as primers: Tf2012 (5′ GATGTGGGCCAAGCAGCTCAAC 3′) and Tr2012 (5′ GTCGACCATGGTGGGTACGTC 3′). The KLC3-ΔHR mutant has been described before (Junco et al., 2001). Mutant forms of KLC3 were cloned into the pCI-HA vector for in vitro translation and binding experiments and into the pGFP vector for cell transfection studies.
Isolation of liver and spermatid mitochondria
Round and elongating spermatids were isolated from rat testes by centrifugal elutriation as described previously (Higgy et al., 1995). Mitochondria were isolated from rat liver and epididymal spermatozoa as follows. Cells were resuspended in ice cold RSB (10mM NaCl, 1.5mM MgCl2, 10mM Tris-HCL, pH 7.5) and transferred to a 15ml Dounce homogenizer. Cells were disrupted with several strokes of the B pestle, 2.5xMS buffer was added immediately to a final concentration of 1xMS (210 mM mammitol, 70mM sucrose, 5mM Tris-HCl, pH7.5, 1mM EDTA). The homogenate was transferred to a centrifuge tube and spun at 1300g for 5 minutes to remove nuclei, unbroken cells and large membrane fragments. This was repeated twice. The supernatant was transferred to a clean centrifuge tube and mitochondria were pelleted at 17,000g for 15 minutes. The crude mitochondrial pellet was purified on a sucrose step density gradient (1.0M sucrose and 1.5M sucrose in 10mM Tris-HCl, pH 7.5, 1mM EDTA) at 60,000g for 20 minutes and resuspended in test buffer (1M KCl, 0.1M KH2PO4, 50mM succinate, 1M Hepes, pH 7.4). All preparations were examined under the microscope and were found not to contain major contaminating cellular structures. Also, they do not contain appreciable amounts of tubulins as determined by western blotting assays. Total protein extracts were also prepared from elongating spermatids.
Mitochondrial binding assay
KLC3, p21ras, which is not known to bind mitochondria, and mutant KLC3 proteins, which allow determination of the region(s) involved in mitochondrial binding, were translated in vitro using the TNT reticulocyte transcription and translation system (Promega, Madison, WI) in the presence of [35S]-methionine. Radiolabeled proteins were incubated with purified mitochondria at 30°C for 30 min. The mitochondria were pelleted at 17,000g for 15min at 22°C through a sucrose cushion. Supernatants were saved for SDS-PAGE analysis, and two subsequent pelleting steps were performed. Aliquots of both supernatants (called s1 and s2) and the final pellet (indicated as p) were boiled in SDS sample buffer and analyzed by electrophoresis on 10% w/v SDS-PAGE gels. Gels were dried and exposed to Kodak BIOMAX film. Radiolabeled proteins present in the mitochondrial pellets were identified by autoradiography, and the intensity of bands was quantitated. In control experiments the above procedures were followed except that no mitochondria were added.
Western blot analysis of mitochondrial KLC3
Mitochondria were isolated from elutriated elongating spermatids, spermatozoa and liver, resuspended in SDS loading buffer, boiled for 5 min and analyzed by SDS-PAGE. Gel-separated proteins were transferred to nitrocellulose membranes. Membranes were blocked in 5% w/v non-fat dry milk in TBSTN (25mM Tris-HCl, pH7.5, 150 mM NaCl, 0.1% Tween-20, 0.1%NP-40) and incubated with 1–5 μg/ml primary polyclonal anti-KLC3 antibody in blocking solution for 1 hour at room temperature, followed by goat anti-rabbit IgG at 1:12,000 dilutions. Finally, chemiluminescent reagents (KPL) were used for detection.
Cell culture and transfection
NIH 3T3 cells were cultured in DMEM supplemented with 10% fetal bovine serum and 1% sodium pyruvate on 6-well plates containing glass coverslips. Cultures were transfected with the indicated GFP-KLC3, GFP-KLC1 and GFP-Odf2 fusion construct DNAs using Lipofectamine Plus reagent as recommended by the manufacturer. GFP-c2 DNA (1 μg) was used as control. Cells were analyzed 48 hr after transfection.
Organelle labeling and microscopy
Mitochondria were stained as follows: transfected and control NIH 3T3 cells grown on glass coverslips were incubated for 30min at 37°C in DMEM medium containing 10 nM MitoTracker Red CMXRos and washed three times with dye-free medium. Next, cells were fixed in 3.7 % formaldehyde. Transfected cells were observed with a Nikon fluorescence microscope or with a Zeiss confocal microscope.
Golgi vesicles and lysosomes were visualized by indirect immunofluorescence microcopy of transfected cells using anti-Golgi and anti-lysosomal primary antibodies (gift of Dr. M. Fritzler) and Cy-3 labeled secondary antibodies. Images obtained by confocal microscopy were analyzed by deconvolution processing (Microscopy and Imaging Unit, University of Calgary). Images were merged using Adobe Photoshop.
RESULTS
KLC3 is associated with mitochondria from epididymal spermatozoa
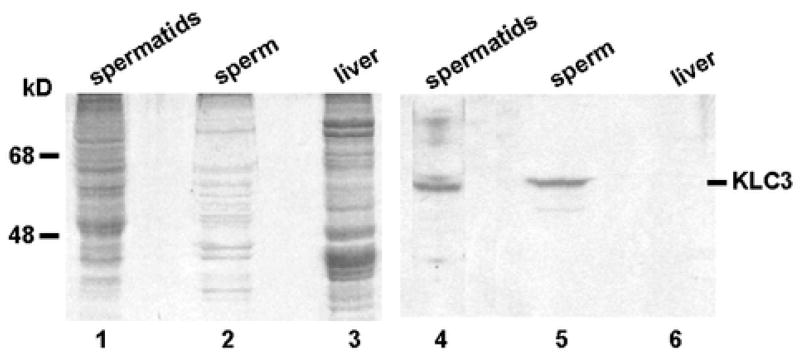
The KLC3-staining pattern observed in previous confocal analysis of epididymal spermatozoa resembled that of mitochondria in the sperm midpiece (Junco et al., 2001). Since virtually nothing is known about molecules that play a role in sperm midpiece formation we investigated if mitochondria isolated from elongating spermatids and epididymal sperm contain associated KLC3. As control we used mitochondria from liver that expresses little if any KLC3 (Junco et al., 2001). Mitochondria were isolated from spermatozoa and liver and protein extracts were prepared and analyzed by western blotting. Total elongating spermatid proteins acted as positive control. Figure 1 shows a Coomassie-stained gel of the indicated separated proteins (lanes 1–3), and the corresponding western blot result is shown in lanes 4–6. As expected, we observe KLC3 in elongating spermatids (lane 4), but not in liver mitochondria (lane 6). The result in lane 5 shows that KLC3 is indeed associated with mitochondria from epididymal spermatozoa.
Figure 1. KLC3 is associated with sperm mitochondria.
Protein extracts prepared from total elongating spermatids (lanes 1 and 4), from mitochondria isolated from spermatozoa (lanes 2 and 5), and from mitochondria isolated from liver, which does not express detectable levels of KLC3(lanes 3 and 6), were analyzed by western blotting using anti-KLC3 MAb. Lanes 1 – 3, Coomassie stained gel; lanes 4 – 6, western blotting results of indicated protein extracts.
Ultrastructural developmental analysis of KLC3 expression
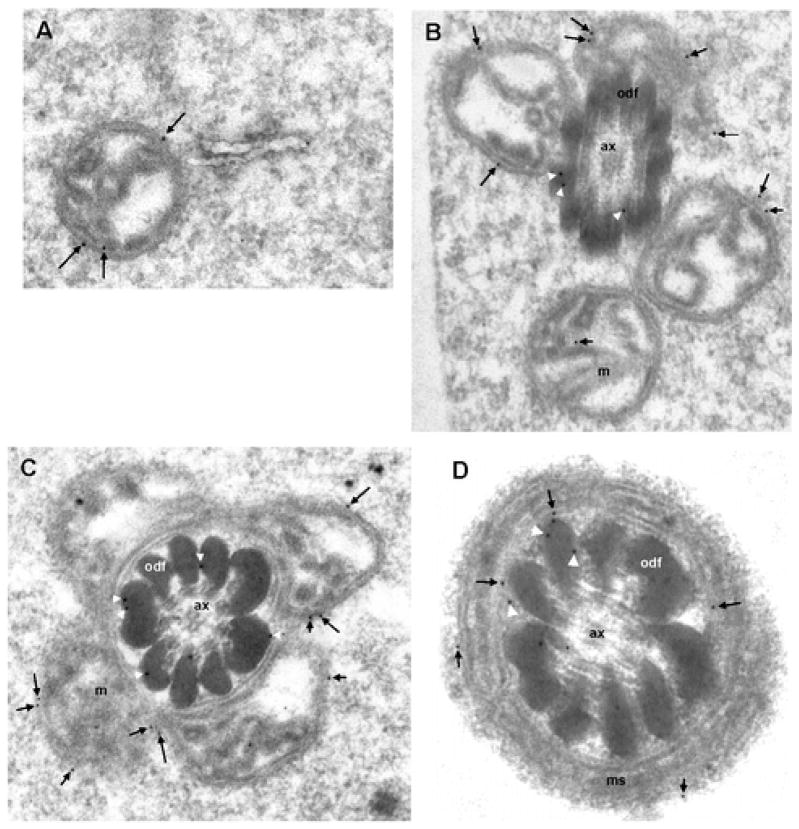
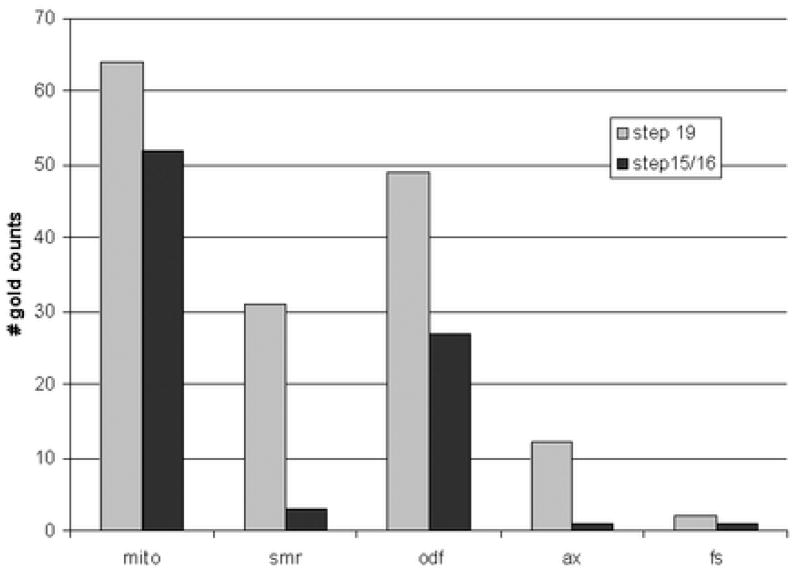
Previous immunocytochemical analysis using monoclonal antibodies had shown that KLC3 expression can first be detected in step 12 spermatids reaching its highest levels in step 15 – 17 elongating spermatids (Bhullar et al., 2003). Peak production thus coincides with the period in spermiogenesis when mitochondria move away from the cell periphery to the developing spermatid midpiece where they encircle the growing ODF. The association of KLC3 with sperm mitochondria suggested that KLC3 might also be found in association with mitochondria in step 15 – 17 elongating spermatids. To examine this we carried out immuno electron microscopy of elongating spermatids of different developmental stages using anti-KLC3 MAb that specifically recognizes the 58 kD KLC3 protein. The results are shown in Figure 2. Since the intensity of gold label was low we quantitated label in a large number of sections at different stages of spermatid development (step 15/16 and step 19). The results of quantitation are shown in Figure 3. Figure 2, panel A shows that KLC3 label can be detected in association with a step 15 spermatid mitochondrion that is not yet adjacent to ODF. Panel B shows KLC3 label on membranes of step 15 spermatid mitochondria that have moved in close proximity to ODF (arrows). At this stage, KLC3 label can also be observed on ODF (panel B, arrowheads), extending our previous observations made in mature sperm (Bhullar et al., 2003). In step17 spermatids (panel C) KLC3 localizes to the surface of mitochondria (arrows) and between ODF and mitochondria or adjacent ODF (arrowheads). Note that in step 17 spermatids mitochondria are beginning to reshape and become more intimately associated with ODF (compare mitochondria in panels B and C). Panel D shows a cross section through the midpiece of a step 19 mature elongated spermatid about to be released in the lumen of the seminiferous epithelium: KLC3 label can be observed associated with mitochondria (arrows), as well as with ODF and the SMR (arrowheads). Little labeling was seen over the axoneme (Figure 2B – 2D) or the fibrous sheath (not shown) in agreement with earlier observations (Bhullar et al., 2003). Quantitation of these results thus demonstrated that KLC3 not only associates with ODF in elongating (step 15/16) and elongated (step 19) spermatids but also with mitochondria. We also observed significant label in the SMR in step 19 spermatids.
Figure 2. KLC3 is associated with mitochondria in elongating spermatids.
Anti-KLC3 MAbs were used in immuno electron microscopy of rat testis sections to investigate the KLC3 expression pattern and ultrastructural localization during spermiogenesis. Panels A and B show electron micrographs of step 15 spermatids: KLC3 label (arrows) is over the periphery of individual mitochondria (panel A) and mitochondria that are congregating at the ODF (panel B, arrows). Arrowheads indicate label associated with ODF. Panel C shows KLC3 label on mitochondrial membranes in step 17 elongating spermatids (arrows) and ODF (arrowheads). Note the more advanced condensation of mitochondria in step 17 spermatids. In mature step 19 elongated spermatids, KLC3 gold label can be observed on mitochondrial membranes (arrows) as well as in association with ODF (arrowheads). Ax, axoneme; m, mitochondria; odf, outer dense fibers. Magnification: panels A – C, 50,000×; panel D, 75,000×.
Figure 3. Quantitation of gold label observed in immuno electron microscopy.
To quantitate gold label observed over different subcellular structures in step 15/16 elongating spermatids and step 19 elongated spermatids, label was counted in 35 and 20 representative sections of these cells, respectively. The Y-axis shows the total number of gold label associate with indicated structures within the indicated cells: “mito”, mitochondria; “smr”, submitochondrial reticulum; “odf”, outer dense fibers; “ax”, axoneme; “fs”, fibrous sheath.
KLC3 forms clusters that contain mitochondria in transfected fibroblasts
Organelles can be cargo of kinesin motor molecules: however, the only light chain shown to bind mitochondria (Khodjakov et al., 1998) is not expressed in spermatids (Junco et al., 2001). The timing of KLC3 expression in elongating spermatids and the association of KLC3 with mitochondria suggested that KLC3 might play a role similar to that of rat KLC1 in somatic cells (Khodjakov et al., 1998). However, an effect of KLC3 on mitochondria cannot be investigated in isolated elongating spermatids, which are short lived and refractory to transfection. Therefore we analyzed possible effects of KLC3 on cellular organelles in transfected fibroblasts that do not express endogenous KLC3 protein (not shown). To visualize KLC3 in transfected cells we produced GFP-KLC3 fusion protein expression constructs.
Fibroblasts were transfected with GFP or GFP-KLC3 fusion protein. After transfection cells were stained with MitoTracker to visualize mitochondria, fixed and analyzed by fluorescence microscopy. In some experiments transfected cells were fixed, incubated with antibodies specific for Golgi or lysosomes, and analyzed by fluorescence microscopy. Figure 4 shows the results. As expected, GFP is present throughout the transfected cells (panel A). In GFP-positive cells mitochondria have a normal distribution (panel B). GFP does not co-localize with mitochondria (panel C, merged images). In contrast to GFP, we observed that expression of GFP-KLC3 caused formation of large KLC3-positive cytoplasmic clusters (panels D, G, J). These clusters appear close to the cell nucleus. Interestingly, we observed that these KLC3-positive clusters contain mitochondria (MitoTracker stain, panel E; merged images, panel F). Examination of over 200 KLC3-cluster positive cells indicated that they all demonstrated mitochondrial clustering as well. However, clusters do not contain lysosomes (panel H; merged images, panel I) or Golgi apparatus (panel K; merged images, panel L). These experiments indicate that in transfected cells KLC3 and mitochondria can accumulate in artificial structures (clusters).
Figure 4. Association of mitochondria with KLC3 clusters.
To investigate the specificity of binding of KLC3 to mitochondria 3T3 fibroblasts (KLC3 negative) were transfected with GFP (panels A – C), GFP-KLC3 (panels D – L), GFP-KLC1 (panel M) and GFP-Odf2 (panel N). GFP signal (green) was observed by fluorescent microscopy. Mitochondria were labeled with MitoTracker (red stain: panels B, C, E, F, M, and N), and lysosomal vesicles (red stain: panels H and I) and Golgi apparatus (red stain: panels K and L) were visualized with antibodies in indirect immunofluorescence microscopy. Panels C, F, I, L, M and N show merged images (overlays) of green and red signals. In panels M and N, nuclear DNA was stained with DAPI (blue). Note that GFP-KLC3, GFP-KLC1 and GFP-Odf2 all cause cluster formation, but that only mitochondria co-localize with only KLC3 clusters as demonstrated by the yellow signal in the merged image (panel F). Magnification in panels A – I, 40×. Magnification in panels J – N, 100×.
We next investigated if the observed effect is specific for KLC3 or could also be seen using other proteins that we had determined previously can form clusters. Cells were transfected with GFP-KLC1 or with GFP-Odf2 fusion proteins. The results show that both GFP-KLC1 and GFP-Odf2 form cytoplasmic clusters (green signal: panels M and N, respectively). However, in contrast to GFP-KLC3 neither GFP-KLC1 nor GFP-Odf2 containing clusters contained mitochondria (red signal: panels M and N, respectively). Together these results indicate that mitochondria are only captured by KLC3-containing clusters.
Binding of KLC3 to mitochondria is mediated by TPR sequences
To delineate sequences involved in KLC3-mitochondrial associations observed in spermatids we designed an in vitro binding assay where mitochondria isolated from sperm or from liver are incubated with in vitro translated KLC3 or KLC3 deletion mutants. Figure 5, panel A, shows the results of binding experiments using liver mitochondria and KLC3 or p21ras, which protein does not bind mitochondria. Identical results were obtained using sperm mitochondria (not shown). KLC3 (lane 2) was mixed with mitochondria, which were pelleted through a cushion resulting in supernatant “s1” (lane 4), washed, re-pelleted resulting in supernatant “s2” (lane 6) and the final pellet “p” (lane 8). KLC3 can be observed in the pellet (lane 8), but not in the corresponding supernatant “s2” (lane 6). Thus KLC3 can bind mitochondria from liver (and sperm) in vitro. These results do not differentiate if binding is direct or indirect. As expected, p21ras (lane 1) does not associate detectably with mitochondria as evidenced by its absence from the pellet fraction “p” (lane 7). To demonstrate that in vitro translated KLC3 is not insoluble, which property would allow it to partition to the pellet fractions, we carried out the same binding procedure, wash and pellet steps in the absence of added mitochondria: the results in panel B demonstrate that in this case no KLC3 is observed in the pellet fractions (lane p). Thus, in vitro translated KLC3 is not insoluble.
Figure 5. In vitro binding analysis of KLC3-mitochondrial interactions.
A) To analyze if KLC3 can bind to purified mitochondria an in vitro binding assay was done: liver mitochondria were incubated with in vitro translated, radiolabeled KLC3 or p21ras (negative control), pelleted, resuspended, washed and pelleted once more before analysis. Protein extracts from the input in vitro translation products (TNT), from the first supernatant (“s1”) and from the final supernatant (“s2”) and corresponding mitochondrial pellet (“p”) were analyzed by SDS PAGE and autoradiography. Lanes 1, 3, 5 and 7; p21ras and lanes 2, 4, 6 and 8; KLC3. Note that KLC3, but not p21ras, binds mitochondria.
B) To ensure that the presence of KLC3 in the mitochondrial pellets did not result from the KLC3 protein being insoluble, the binding experiments as described under section A were done with in vitro translated KLC3 in the absence of mitochondria. Lane TNT shows in vitro translated KLC3, lane “s1” shows protein in the first supernatant, and lanes “s2” and “p” show the corresponding final supernatant and pellet fractions.
C) Schematic representation of KLC3 deletion mutants that were tested for mitochondrial binding in comparison to wild type KLC3 (WT): ΔHR lacks the heptade repeat, ΔTPR lacks the tetratrico-peptide repeat sequences, ΔC lacks the variable C-terminus. The approximate extent of the deletions is shown.
D) The in vitro binding assay was next used to analyze KLC3 sequences involved in binding to mitochondria. In vitro translated proteins (“TNT”, lanes 1 – 4) were incubated with liver mitochondria and examined as described in section A. Radiolabeled proteins bound to mitochondria (“pellet”: lanes 5 – 8) are shown. Lanes 1 and 5: KLC3-ΔHR (“ΔHR”) lacks sequences that bind to KHC. Lanes 2 and 6: wild type KLC3 (“wt”). Lanes 3 and 7: KLC3-ΔC (“ΔC”) lacks the variable C-terminus. Lanes 4 and 8: KLC3-ΔTPR (“ΔTPR”) lacks the TPR. Note the significant decrease in binding upon deletion of TPR sequences.
We next employed this assay to delineate the mitochondrial-binding domain of KLC3 using the KLC3 deletion mutants that are schematically represented in Figure 5, panel C. We focused on three important kinesin light chain regions and constructed the following three mutants: a 52 kD KLC3-ΔHR mutant, which lacks the heptade repeat sequence that binds to the kinesin heavy chain, the 28 kD KLC3-ΔTPR mutant, which lacks TPR repeats, and the 43 kD KLC3-ΔC mutant, which has a deletion in the C-terminal variable region, which in somatic rat KLC1 was shown to bind mitochondria (Khodjakov et al., 1998). These mutant proteins and wild type KLC3 were translated in vitro and incubated with liver mitochondria. Binding was analyzed as described for panel A and results are shown in Figure 5, panel D. Lanes 1 – 4 show the translation products for KLC3-ΔHR, wild type KLC3, KLC3-ΔC and KLC3-ΔTPR, respectively, and lanes 5 – 8 show the KLC3 proteins present in the final mitochondrial pellets. We observe that mitochondrial binding of the KLC3-ΔHR mutant (lane 5) was similar to that of wild type KLC3 (lane 6) suggesting that HR sequences do not mediate mitochondrial binding. The binding efficiency of the KLC3-ΔC mutant (lane 7) appeared slightly decreased compared to wild type KLC3. However, deletion of TPR sequences significantly reduced binding (lane 8). Signals observed for bound KLC3 variants were quantitated and normalized for input: compared to KLC3 (set at 100 %) the binding efficiency was approximately 100 %, 75 % and 9 % for KLC3-ΔHR, KLC3-ΔC and KLC3-ΔTPR, respectively. These data indicate that KLC3 TPR sequences mediate the in vitro association with mitochondria –be it direct or indirect-, a result which differs from that of rat KLC1.
To analyze if TPR sequences also play a role in the observed inclusion of mitochondria in KLC3-positive clusters in transfected cells the following experiments were performed. Fibroblasts were transfected with three mutant GFP-fusion constructs, which contain GFP linked to the deletion mutants analyzed for binding as described in Figure 5B, and mitochondria were detected in transfected cells using MitoTracker. The results are presented in Figure 6. First, the results show that all three deletion constructs cause formation of KLC3-positive clusters in transfected cells (panels A, D and G) similar to wild type KLC3. Second, clusters formed in cells expressing either the HR domain deletion mutant (KLC3-ΔHR) or the C-terminus deletion mutant (KLC3-ΔC) contain mitochondria similar to clusters formed in wild type KLC3 expressing cells (note the overlap of the GFP-KLC3 deletion mutant signals and MitoTracker signals in merged images in panels C and F, respectively). In contrast to these results, KLC3-ΔTPR-positive clusters fail to capture mitochondria (merged images, panel I). Together with the binding data this suggests that the KLC3 TPR region is involved in mitochondrial binding, but not clustering.
Figure 6. KLC3 TPR domain is required for mitochondrial clustering.

To analyze the region in KLC3 responsible for capturing mitochondria in clusters, 3T3 fibroblasts were transfected with GFP-KLC3-ΔHR (panels A – C), GFP-KLC3-ΔC (panels D – F) and GFP-KLC3-ΔTPR (panels G - I) and analyzed by fluorescent microscopy. Mitochondria were stained using MitoTracker. GFP signal (green), MitoTracker signal (red) and merged images (panels C, F and I) are indicated. Note that colocalization is evident from the yellow color in merged images.
DISCUSSION
Kinesins and KRPs are expressed during spermiogenesis. Their presence makes sense: from morphogenetic studies it appears that a flagellar transport mechanism must exist, because during post-meiotic differentiation of spermatids a structure called the annulus separates the narrow and long membrane bound periaxonemal compartment from the bulk of the cytoplasm (Cesario and Bartles, 1994): proteins that are needed for the development of the FS and ODF are transported past the annulus. The axoneme, which represents the first tail structure to be formed during spermiogenesis, likely provides the molecular tracks for microtubule-based transport by kinesins and perhaps other motor proteins. There has however not been any evidence for involvement of kinesins or other motor molecules in processes involving spermatid mitochondria.
Kinesins are involved in mitochondrial transport in somatic cells
Several results gathered from neurons and in vitro systems support a role in the transport of mitochondria by specific kinesins and KRPs (Leopold et al., 1992; Jellali et al., 1994): KIF1B transports mitochondria along microtubules in vitro (Nangaku et al., 1994; Conforti et al., 1999); KIF3A functions in fast axonal transport of mitochondria (Kondo et al., 1994; Elluru et al., 1995); activated PKA negatively regulates kinesin-mediated mitochondrial transport (Okada et al., 1995); Drosophila KHC mutants display disrupted fast axonal transport and organelle jams involving mitochondria (Hurd and Saxton, 1996); KIF5B−/− knockout mice are embryonic lethal and –among other defects- show perinuclear aggregations of mitochondria (Tanaka et al., 1998); and overexpression of tau slows down fast axonal transport of mitochondria (Ebneth et al., 1998; Trinczek et al., 1999). Recent evidence suggests that the mutation in Charcot-Marie-Tooth disease type 2A, which involves a human peripheral neuropathy, is in kinesin KIF1B, one of the main neural transporters of mitochondria (Zhao et al., 2001). Direct evidence for the involvement of kinesin light chains in binding to somatic mitochondria is limited to one report describing binding of the C-terminus of rat KLC1 to mitochondria (Khodjakov et al., 1998). Circumstantial evidence for KLC involvement comes from a study showing that tumor necrosis factor causes hyperphosphorylation of KLC, impairs kinesin motor activity and results in perinuclear aggregation of mitochondria (De Vos et al., 2000). Recently we demonstrated that of the three known mammalian kinesin light chains only KLC3 is expressed in spermatids (Junco et al., 2001) and it was suggested that KLC3 might thus play roles in many transport processes in those cells. Here we show that KLC3 associates with mitochondria in elongating spermatids and spermatozoa in vivo and in vitro, and KLC3-positive clusters in transfected fibroblasts contain mitochondria.
KLC3 TPR domain mediates mitochondrial binding
The developmental study carried out at the ultrastructural level indicates that in rat spermatids KLC3 associates with mitochondria at stages during spermiogenesis when mitochondria relocate and attach to ODF in the future sperm tail midpiece. This region, containing the axoneme surrounded by ODF, becomes accessible in step 15 elongating spermatids after descent of the annulus. Mature spermatozoa also show KLC3-specific label surrounding ODF and in the protein-rich SMR located between mitochondria and ODF (Olson and Winfrey, 1990). Isolated sperm mitochondria contain associated KLC3. Our binding studies further indicate that mitochondria from somatic cells that do not express endogenous KLC3 (liver) are able to bind KLC3 directly or indirectly. This indicates that the mitochondrial KLC3 binding site is ubiquitous, rather than spermatid-specific. We further discovered that KLC3 TPR repeats mediate mitochondrial binding, in contrast to results obtained for rat KLC1 (Khodjakov et al., 1998). TPR repeats have previously been shown to act in binding membrane vesicles in axonal transport (Stenoien and Brady, 1997; Tsai et al., 2000; Bowman et al., 2000) and our findings now expand that repertoire to include mitochondria. Our transfection data show that KLC3 clusters contain somatic cell mitochondria. The specificity of the observed association of mitochondria with the KLC3 clusters is demonstrated by the absence of Golgi and lysosomal vesicles in such clusters. In addition, we show that clusters formed by ODF2 do not contain mitochondria. A similar observation was made for KLC1: it is not clear at present why KLC1 clusters do not contain detectable amounts of mitochondria. Possibilities are that regions involved in KLC1 clustering and mitochondrial binding are mutually exclusive, or that the KLC1 C-terminus which binds mitochondria is not accessible in KLC1 clusters. Importantly, the in vitro binding experiments and transfection experiments gave the same results indicating that the TPR region is involved in mitochondrial binding.
We do not at present know if the KLC3 clusters containing mitochondria that we observe are similar to the aggregates reported in KIF5B−/− knockout cells (Tanaka et al., 1998) or to aggregates observed after inactivation of KLC and kinesin motor activity (De Vos et al., 2000). These studies suggest that the action of KIF1B, KIF5B or other mitochondrial kinesins are required for maintenance of cytoplasmic mitochondrial localization and axonal transport in somatic cells (Nangaku et al., 1994; Tanaka et al., 1998). As reported by several groups (Tanaka et al., 1998; De Vos et al., 2000) intervention results in abnormal clustering of mitochondria which usually has adverse effects on the cell: indeed we have thus far not been able to obtain fibroblast cell lines that stably express KLC3 (not shown).
A similarity in underlying mechanism would imply that mitochondrial aggregation in KLC3-positive clusters might result from an inactivation of a kinesin or kinesin component that normally maintains the subcellular mitochondrial distribution. The role of KLC3 in mitochondrial localization and/or movement in spermatids is unclear at present: in step 15 spermatids microtubules are present in the axoneme. However, they are surrounded by ODF and not accessible to KLC3-labeled mitochondria. Possible models for KLC3 function include an unconventional role for KLC3 in bringing together mitochondria and ODF that does not involve classical kinesin-microtubule mediated transport (see below), movement along tracks other than microtubules by association with other motor molecules (spermatids contain actin filaments that support myosin –related transport), and a role as a “glue” bringing together ODF fragments or components and mitochondria. At present, based on our results, it can be suggested that a role of KLC3 in spermatid midpiece formation might include mitochondrial processes. Recently, a 14 kD spermatid protein called Spergen1 was shown to bind mitochondria (Doiguchi et al., 2002). Spergen1 shares no homology with KLC3. Its relationship, if any, with KLC3 and spermatid mitochondrial associations in elongating spermatids remains to be determined.
The observations presented here may help address an aspect of the phenotype of certain mouse mutants that fail to assemble an axoneme during spermiogenesis: homozygous male sterility-inducing mutation hpy mice (Bryan, 1981) and infertile transgenic OVE 129 mice (Russell et al., 1994) lack normal axonemes and as a result ODF assembly is abnormal. In these mice, abnormal ODF fragments are found grouped with mitochondria. It had been concluded that ODF assembly and organization requires an axoneme, and we recently provided evidence in support of this notion by showing that SPAG4 may be a crucial factor in this process (Shao et al., 1999). The molecular basis of the association of ODF fragments with mitochondria is not known. Our results together with previous observations show that KLC3 can bind to ODF (Bhullar et al., 2003), employing the HR region, and to mitochondria, using the TPR region. Thus, KLC3 may play multiple independent roles in spermatid development, or these observations may be related suggesting the possibility that KLC3 might bring together ODF fragments and mitochondria.
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (to R.O. and F.A.v.d.H.) and from the Natural Sciences and Engineering Council of Canada (to R.O.).
Reference List
- Bhullar B, Zhang Y, Junco A, Oko R, van der Hoorn FA. Association of Kinesin light chain with Outer dense fibers in a Microtubule-independent fashion. J Biol Chem. 2003;278:16159–16168. doi: 10.1074/jbc.M213126200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block SM. Kinesin: what gives? Cell. 1998;93:5–8. doi: 10.1016/s0092-8674(00)81138-1. [DOI] [PubMed] [Google Scholar]
- Bowman AB, Kamal A, Ritchings BW, Philp AV, McGrail M, Gindhart JG, Goldstein LS. Kinesin-dependent axonal transport is mediated by the sunday driver (SYD) protein. Cell. 2000;103:583–594. doi: 10.1016/s0092-8674(00)00162-8. [DOI] [PubMed] [Google Scholar]
- Bryan JH. Spermatogenesis revisited. V Spermiogenesis in mice homozygous for two different male-sterile mutations (ps and hpy) Cell Tiss Res. 1981;221:169–180. doi: 10.1007/BF00216579. [DOI] [PubMed] [Google Scholar]
- Burfeind P, Hoyer-Fender S. Sequence and developmental expression of a mRNA encoding a putative protein of rat sperm outer dense fibers. Dev Biol. 1991;148:195–204. doi: 10.1016/0012-1606(91)90329-2. [DOI] [PubMed] [Google Scholar]
- Cesario MM, Bartles JR. Compartmentalization, processing and redistribution of the plasma membrane protein CE9 on rodent spermatozoa. Relationship of the annulus to domain boundaries in the plasma membrane of the tail. J Cell Sci. 1994;107:561–570. [PubMed] [Google Scholar]
- Conforti L, Buckmaster EA, Tarlton A, Brown MC, Lyon MF, Perry VH, Coleman MP. The major brain isoform of kif1b lacks the putative mitochondria-binding domain. Mamm Genome. 1999;10:617–622. doi: 10.1007/s003359901056. [DOI] [PubMed] [Google Scholar]
- De Vos K, Severin F, Van Herreweghe F, Vancompernolle K, Goossens V, Hyman A, Grooten J. Tumor necrosis factor induces hyperphosphorylation of kinesin light chain and inhibits kinesin-mediated transport of mitochondria. J Cell Biol. 2000;149:1207–1214. doi: 10.1083/jcb.149.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbach RJ, Mackay JP, Armati PJ, Cunningham AL. The C-terminal region of the stalk domain of ubiquitous human kinesin heavy chain contains the binding site for kinesin light chain. Biochemistry. 1998;37:16663–16670. doi: 10.1021/bi981163r. [DOI] [PubMed] [Google Scholar]
- Doiguchi M, Mori T, Toshimori K, Shibata Y, Iida H. Spergen-1 might be an adhesive molecule associated with mitochondria in the middle piece of spermatozoa. Dev Biol. 2002;252:127–137. doi: 10.1006/dbio.2002.0833. [DOI] [PubMed] [Google Scholar]
- Ebneth A, Godemann R, Stamer K, Illenberger S, Trinczek B, Mandelkow E. Overexpression of tau protein inhibits kinesin-dependent trafficking of vesicles, mitochondria, and endoplasmic reticulum: implications for Alzheimer’s disease. J Cell Biol. 1998;143:777–794. doi: 10.1083/jcb.143.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elluru RG, Bloom GS, Brady ST. Fast axonal transport of kinesin in the rat visual system: functionality of kinesin heavy chain isoforms. Mol Biol Cell. 1995;6:21–40. doi: 10.1091/mbc.6.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow SA. Kinesin motors as molecular machines. Bioessays. 2003;25:1212–1219. doi: 10.1002/bies.10358. [DOI] [PubMed] [Google Scholar]
- Gauger AK, Goldstein LS. The Drosophila kinesin light chain. Primary structure and interaction with kinesin heavy chain. J Biol Chem. 1993;268:13657–66. [PubMed] [Google Scholar]
- Gindhart JG, Jr, Desai CJ, Beushausen S, Zinn K, Goldstein LS. Kinesin light chains are essential for axonal transport in Drosophila. J Cell Biol. 1998;141:443–454. doi: 10.1083/jcb.141.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindhart JG, Jr, Goldstein LS. Tetratrico peptide repeats are present in the kinesin light chain. Trends Biochem Sci. 1996;21:52–53. [PubMed] [Google Scholar]
- Goldstein LS. Kinesin molecular motors: transport pathways, receptors, and human disease. Proc Natl Acad Sci U S A. 2001a;98:6999–7003. doi: 10.1073/pnas.111145298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein LS. Molecular motors: from one motor many tails to one motor many tales. Trends Cell Biol. 2001b;11:477–482. doi: 10.1016/s0962-8924(01)02143-2. [DOI] [PubMed] [Google Scholar]
- Hackney DD. The kinetic cycles of myosin, kinesin, and dynein. Annu Rev Physiol. 1996;58:731–750. doi: 10.1146/annurev.ph.58.030196.003503. [DOI] [PubMed] [Google Scholar]
- Hall ES, Eveleth J, Jiang C, Redenbach DM, Boekelheide K. Distribution of the microtubule-dependent motors cytoplasmic dynein and kinesin in rat testis. Biol Reprod. 1992;46:817–828. doi: 10.1095/biolreprod46.5.817. [DOI] [PubMed] [Google Scholar]
- Higgy NA, Zackson SL, van der Hoorn FA. Cell interactions in testis development: overexpression of c-mos in spermatocytes leads to increased germ cell proliferation. Dev Genet. 1995;16:190–200. doi: 10.1002/dvg.1020160211. [DOI] [PubMed] [Google Scholar]
- Higuchi H, Endow SA. Directionality and processivity of molecular motors. Curr Opin Cell Biol. 2002;14:50–57. doi: 10.1016/s0955-0674(01)00293-9. [DOI] [PubMed] [Google Scholar]
- Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- Howard J. The movement of kinesin along microtubules. Annu Rev Physiol. 1996;58:703–729. doi: 10.1146/annurev.ph.58.030196.003415. [DOI] [PubMed] [Google Scholar]
- Hoyer-Fender S, Petersen C, Brohmann H, Rhee K, Wolgemuth DJ. Mouse Odf2 cDNAs consist of evolutionary conserved as well as highly variable sequences and encode outer dense fiber proteins of the sperm tail. Mol Reprod Dev. 1998;51:167–175. doi: 10.1002/(SICI)1098-2795(199810)51:2<167::AID-MRD6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Hurd DD, Saxton WM. Kinesin mutations cause motor neuron disease phenotypes by disrupting fast axonal transport in Drosophila. Genetics. 1996;144:1075–1085. doi: 10.1093/genetics/144.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irons MJ, Clermont Y. Formation of the outer dense fibers during spermiogenesis in the rat. Anat Rec. 1982a;202:463–471. doi: 10.1002/ar.1092020405. [DOI] [PubMed] [Google Scholar]
- Irons MJ, Clermont Y. Kinetics of fibrous sheath formation in the rat spermatid. Am J Anat. 1982b;165:121–130. doi: 10.1002/aja.1001650204. [DOI] [PubMed] [Google Scholar]
- Jellali A, Metz-Boutigue MH, Surgucheva I, Jancsik V, Schwartz C, Filliol D, Gelfand VI, Rendon A. Structural and biochemical properties of kinesin heavy chain associated with rat brain mitochondria. Cell Motil Cytoskeleton. 1994;28:79–93. doi: 10.1002/cm.970280108. [DOI] [PubMed] [Google Scholar]
- Johnson KJ, Hall ES, Boekelheide K. Kinesin localizes to the trans-Golgi network regardless of microtubule organization. Eur J Cell Biol. 1996;69:276–287. [PubMed] [Google Scholar]
- Junco A, Bhullar B, Tarnasky HA, van der Hoorn FA. Kinesin light-chain KLC3 expression in testis is restricted to spermatids. Biol Reprod. 2001;64:1320–1330. doi: 10.1095/biolreprod64.5.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal A, Goldstein LS. Principles of cargo attachment to cytoplasmic motor proteins. Curr Opin Cell Biol. 2002;14:63–68. doi: 10.1016/s0955-0674(01)00295-2. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Lizunova EM, Minin AA, Koonce MP, Gyoeva FK. A specific light chain of kinesin associates with mitochondria in cultured cells. Mol Biol Cell. 1998;9:333–343. doi: 10.1091/mbc.9.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszenbaum AL. Spermatid manchette: plugging proteins to zero into the sperm tail. Mol Reprod Dev. 2001;59:347–349. doi: 10.1002/mrd.1040. [DOI] [PubMed] [Google Scholar]
- Kondo S, Sato-Yoshitake R, Noda Y, Aizawa H, Nakata T, Matsuura Y, Hirokawa N. KIF3A is a new microtubule-based anterograde motor in the nerve axon. J Cell Biol. 1994;125:1095–1107. doi: 10.1083/jcb.125.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopold PL, McDowall AW, Pfister KK, Bloom GS, Brady ST. Association of kinesin with characterized membrane-bounded organelles. Cell Motil Cytoskeleton. 1992;23:19–33. doi: 10.1002/cm.970230104. [DOI] [PubMed] [Google Scholar]
- Miller MG, Mulholland DJ, Vogl AW. Rat testis motor proteins associated with spermatid translocation (dynein) and spermatid flagella (kinesin-II) Biol Reprod. 1999;60:1047–1056. doi: 10.1095/biolreprod60.4.1047. [DOI] [PubMed] [Google Scholar]
- Moore JD, Endow SA. Kinesin proteins: a phylum of motors for microtubule-based motility. Bioessays. 1996;18:207–219. doi: 10.1002/bies.950180308. [DOI] [PubMed] [Google Scholar]
- Morales CR, Oko R, Clermont Y. Molecular cloning and developmental expression of an mRNA encoding the 27 kDa outer dense fiber protein of rat spermatozoa. Mol Reprod Dev. 1994;37:229–40. doi: 10.1002/mrd.1080370215. [DOI] [PubMed] [Google Scholar]
- Nangaku M, Sato-Yoshitake R, Okada Y, Noda Y, Takemura R, Yamazaki H, Hirokawa N. KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell. 1994;79:1209–1220. doi: 10.1016/0092-8674(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Navolanic PM, Sperry AO. Identification of Isoforms of a Mitotic Motor in Mammalian Spermatogenesis. Biol Reprod. 2000;62:1360–1369. doi: 10.1095/biolreprod62.5.1360. [DOI] [PubMed] [Google Scholar]
- Okada Y, Sato-Yoshitake R, Hirokawa N. The activation of protein kinase A pathway selectively inhibits anterograde axonal transport of vesicles but not mitochondria transport or retrograde transport in vivo. J Neurosci. 1995;15:3053–3064. doi: 10.1523/JNEUROSCI.15-04-03053.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oko RJ, Jando V, Wagner CL, Kistler WS, Hermo LS. Chromatin reorganization in rat spermatids during the disappearance of testis-specific histone, H1t, and the appearance of transition proteins TP1 and TP2. Biol Reprod. 1996;54:1141–1157. doi: 10.1095/biolreprod54.5.1141. [DOI] [PubMed] [Google Scholar]
- Olson GE, Winfrey VP. Mitochondria-cytoskeleton interactions in the sperm midpiece. J Struct Biol. 1990;103:13–22. doi: 10.1016/1047-8477(90)90081-m. [DOI] [PubMed] [Google Scholar]
- Olson GE, Winfrey VP. Structural organization of surface domains of sperm mitochondria. Mol Reprod Dev. 1992;33:89–98. doi: 10.1002/mrd.1080330113. [DOI] [PubMed] [Google Scholar]
- Petersen C, Aumuller G, Bahrami M, Hoyer-Fender S. Molecular cloning of Odf3 encoding a novel coiled-coil protein of sperm tail outer dense fibers. Mol Reprod Dev. 2002;61:102–112. doi: 10.1002/mrd.1136. [DOI] [PubMed] [Google Scholar]
- Rahman A, Friedman DS, Goldstein LS. Two kinesin light chain genes in mice. Identification and characterization of the encoded proteins. J Biol Chem. 1998;273:15395–15403. doi: 10.1074/jbc.273.25.15395. [DOI] [PubMed] [Google Scholar]
- Russell LD, Ying L, Overbeek PA. Insertional mutation that causes acrosomal hypo-development: its relationship to sperm head shaping. Anat Rec. 1994;238:437–453. doi: 10.1002/ar.1092380403. [DOI] [PubMed] [Google Scholar]
- Schalles U, Shao X, van der Hoorn FA, Oko R. Developmental expression of the 84-kDa ODF sperm protein: localization to both the cortex and medulla of outer dense fibers and to the connecting piece. Dev Biol. 1998;199:250–260. doi: 10.1006/dbio.1998.8931. [DOI] [PubMed] [Google Scholar]
- Schnapp BJ. Trafficking of signaling modules by kinesin motors. J Cell Sci. 2003;116:2125–2135. doi: 10.1242/jcs.00488. [DOI] [PubMed] [Google Scholar]
- Scholey JM. Kinesin-II, a membrane traffic motor in axons, axonemes, and spindles. J Cell Biol. 1996;133:1–4. doi: 10.1083/jcb.133.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X, Tarnasky HA, Lee JP, Oko R, van der Hoorn FA. Spag4, a novel sperm protein, binds outer dense-fiber protein Odf1 and localizes to microtubules of manchette and axoneme. Dev Biol. 1999;211:109–123. doi: 10.1006/dbio.1999.9297. [DOI] [PubMed] [Google Scholar]
- Shao X, Tarnasky HA, Schalles U, Oko R, van der Hoorn FA. Interactional cloning of the 84-kDa major outer dense fiber protein Odf84. Leucine zippers mediate associations of Odf84 and Odf27. J Biol Chem. 1997;272:6105–6113. doi: 10.1074/jbc.272.10.6105. [DOI] [PubMed] [Google Scholar]
- Shao X, Xue J, van der Hoorn FA. Testicular protein Spag5 has similarity to mitotic spindle protein Deepest and binds outer dense fiber protein Odf1. Mol Reprod Dev. 2001;59:410–416. doi: 10.1002/mrd.1047. [DOI] [PubMed] [Google Scholar]
- Sperry AO, Zhao LP. Kinesin-related proteins in the mammalian testes: candidate motors for meiosis and morphogenesis. Mol Biol Cell. 1996;7:289–305. doi: 10.1091/mbc.7.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenoien DL, Brady ST. Immunochemical analysis of kinesin light chain function. Mol Biol Cell. 1997;8:675–89. doi: 10.1091/mbc.8.4.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Kanai Y, Okada Y, Nonaka S, Takeda S, Harada A, Hirokawa N. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell. 1998;93:1147–1158. doi: 10.1016/s0092-8674(00)81459-2. [DOI] [PubMed] [Google Scholar]
- Trinczek B, Ebneth A, Mandelkow EM, Mandelkow E. Tau regulates the attachment/detachment but not the speed of motors in microtubule-dependent transport of single vesicles and organelles. J Cell Sci. 1999;112:2355–2367. doi: 10.1242/jcs.112.14.2355. [DOI] [PubMed] [Google Scholar]
- Tsai MY, Morfini G, Szebenyi G, Brady ST. Release of kinesin from vesicles by hsc70 and regulation of fast axonal transport. Mol Biol Cell. 2000;11:2161–2173. doi: 10.1091/mbc.11.6.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner KJ, Sharpe RM, Gaughan J, Millar MR, Foster PM, Saunders PT. Expression cloning of a rat testicular transcript abundant in germ cells, which contains two leucine zipper motifs. Biol Reprod. 1997;57:1223–1232. doi: 10.1095/biolreprod57.5.1223. [DOI] [PubMed] [Google Scholar]
- van der Hoorn FA, Tarnasky HA, Nordeen SK. A new rat gene RT7 is specifically expressed during spermatogenesis. Dev Biol. 1990;142:147–154. doi: 10.1016/0012-1606(90)90158-f. [DOI] [PubMed] [Google Scholar]
- Verhey KJ, Lizotte DL, Abramson T, Barenboim L, Schnapp BJ, Rapoport TA. Light chain-dependent regulation of Kinesin’s interaction with microtubules. J Cell Biol. 1998;143:1053–1066. doi: 10.1083/jcb.143.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Nakata T, Okada Y, Hirokawa N. Cloning and characterization of KAP3: a novel kinesin superfamily-associated protein of KIF3A/3B. Proc Natl Acad Sci U S A. 1996;93:8443–8448. doi: 10.1073/pnas.93.16.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WX, Sperry AO. C-terminal kinesin motor KIFC1 participates in acrosome biogenesis and vesicle transport. Biol Reprod. 2003;69:1719–1729. doi: 10.1095/biolreprod.102.014878. [DOI] [PubMed] [Google Scholar]
- Zhao C, Takita J, Tanaka Y, Setou M, Nakagawa T, Takeda S, Yang HW, Terada S, Nakata T, Takei Y, Saito M, Tsuji S, Hayashi Y, Hirokawa N. Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bbeta. Cell. 2001;105:587–597. doi: 10.1016/s0092-8674(01)00363-4. [DOI] [PubMed] [Google Scholar]
- Zou Y, Millette CF, Sperry AO. KRP3A and KRP3B: Candidate Motors in Spermatid Maturation in the Seminiferous Epithelium. Biol Reprod. 2002;66:843–855. doi: 10.1095/biolreprod66.3.843. [DOI] [PubMed] [Google Scholar]