Abstract
Little is known about endosomal pathway proteins involved in arthropod-borne virus (arbovirus) assembly and cell-to-cell spread in vector mosquitoes. UNC93A and Synaptic vesicle-2 (SV2) proteins are involved in intracellular transport in mammals. They show amino acid sequence conservation from mosquitoes to humans, and their transcripts are highly-enriched in Aedes aegypti during arbovirus infection. Transient gene silencing of SV2 or UNC93A in mosquitoes infected with the recombinant alphavirus Sindbis MRE16-eGFP (SINV; family Togaviridae) resulted in the accumulation of viral positive- and negative-strand RNA, congregation of virus envelope antigen in intracellular networks, and reduced virus dissemination outside of the midgut. Further, UNC93A silencing, but not SV2 silencing, resulted in a 10-fold reduction in viral titers at 4 days post-infection. Together, these data support a role for UNC93A and SV2 in virus assembly or budding. Cis-regulatory elements (CREs) were identified at the 5′-ends of genes from the original dataset in which SV2 and UNC93A were identified. Common CREs at the 5′-end genomic regions of a subset of enriched transcripts support the hypothesis that UNC93A transcription may be co-regulated with that of other ion transport and endosomal trafficking proteins.
Keywords: mosquito, arbovirus, vector biology, SV2, UNC93A
Introduction
Enveloped arboviruses, such as alphaviruses, exploit multiple biological pathways in vector arthropod or mammalian host cells during successful infection. In mammals, alphaviruses globally control transcription, translation, and endosomal pathways to quickly establish an acute, cytopathic infection (Frolov and Schlesinger 1994; Gorchakov et al. 2005). In contrast, these viruses establish persistent infections in susceptible mosquitoes (Bowers et al. 1995). Some intracellular components, such as endosomal pathway Rab protein family members, are conserved among mosquitoes and humans (Mudiganti et al.; Colpitts et al. 2007). However, the features of persistent arbovirus infection of vector mosquitoes are not understood. Identification of common features of infection among mammals and vector mosquitoes may enhance discovery of new anti-viral therapeutics.
We use the alphavirus, Sindbis virus (SINV, Togaviridae), as a model to characterize the involvement of conserved endosomal pathway proteins during infection of Aedes aegypti. Synaptic vesicle-2 (SV2) and UNC93A are involved in intracellular transport mechanisms in mammals (Chang and Sudhof 2009). These transcripts are enriched more than 40-fold at 4 days post-infection (dpi) during SINV or Dengue virus (DENV) (Flaviviridae) infection of Ae. aegypti (Sanders et al. 2005; Gill, unpublished). This remarkable transcript enrichment, coupled with the inferred function from orthologous systems, supports the hypothesis that the corresponding proteins may be recruited to facilitate virus replication and dissemination. Transcripts from the Sanders et al. (2005) dataset, commonly enriched with SV2 or UNC93A, could be co-regulated by a viral transcriptional control mechanism. To test this, we analyzed 5′-end genomic regions to find common cis-regulatory elements (CREs), as has been done previously for mosquitoes (Campbell et al. 2008; Sieglaff et al. 2009). The presence of such elements would be consistent with a model in which virus-controlled gene expression co-regulates genes that aid viral propagation.
Both SV2 and UNC93A (Vectorbase accession numbers, AAEL005849 and AAEL004048, respectively) are members of the Major Facilitator Superfamily. Members of this large protein superfamily participate in solute transport. SV2 is a synaptic vesicle protein that acts during calcium-dependent exocytosis of synaptic vesicles (Chang and Sudhof 2009). Although there is no direct evidence of transporter activity by SV2, these studies demonstrate its role in transport.
Transient transcript silencing was used to explore the function of SV2 and UNC93A during alphavirus infection, using the same virus as the Sanders et al 2005 study. Detection of negative-strand RNA, positive-strand RNA, eGFP fluorescence, and viral antigen immunofluorescence were used as indicators of viral replication efficiency, viral transcription and protein translation, respectively, and successful viral assembly was assessed by plaque titration. The data provided here support a role for both SV2 and UNC93A in viral infection in adult mosquitoes.
Results
SV2 and UNC93A are conserved among mammals and insects
Aedine SV2 has 20% amino acid identity to human SV2 (Supplemental Fig. 1A). UNC93A is a component of a two-pore potassium transporter and is part of a multi-gene family that is conserved in humans, mosquitoes, and worms (de la Cruz et al. 2003). A mouse paralog, UNC93B, localizes to the endoplasmic reticulum, and mutation of this gene increases susceptibility to virus infection by preventing proper endosomal trafficking of Toll-like receptors (Tabeta et al. 2006). The aedine UNC93A ortholog has 17% amino acid identity to mouse UNC93B and 36% identity to UNC93A (Supplemental Fig. 1B).
Promoter Analysis
The 5′-end regions of genes for which transcripts were enriched at least 2-fold at 4 dpi post SINV infection were chosen using published microarray data (Sanders et al. 2005). 5′-end genomic regions 800, 1600 or 2000 nucleotides (nt) in length were analyzed to identify putative CREs using SCOPE with a significance cut-off of 5.0, where the significance value represents the negative log2 of the expectation value (Chakravarty et al. 2007). SCOPE uses three algorithms to identify bi-partite, degenerate and non-degenerate motifs (Carlson et al. 2006; Chakravarty et al. 2007). Some transcription factors work as dimers, requiring that contact with the DNA surface occurs at two half-sites of only 3 or 4 nt with intervening sequences of varied length (Landschulz et al. 1988).
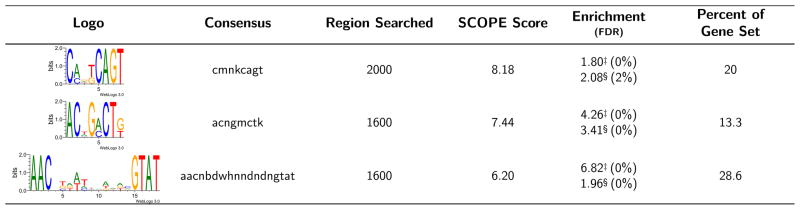
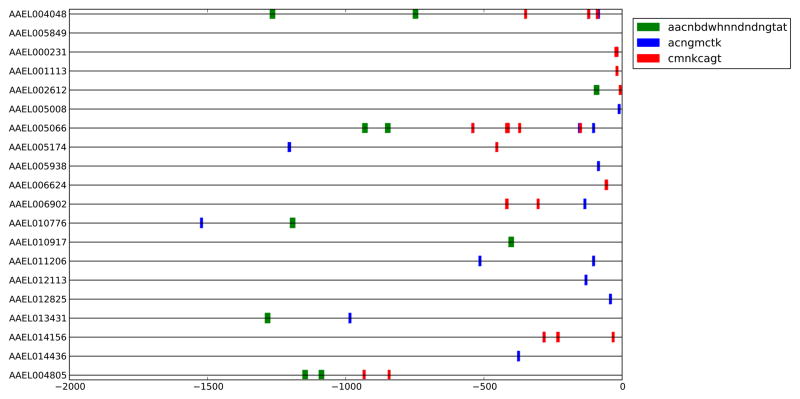
SCOPE analyses returned CREs common to genes involved in ion transport, endosomal function or nucleic acid metabolism. Of 31 promoters analyzed, 19 contained motifs in common with UNC93A, however none were shared with SV-2 (Fig. 1; Fig. 2; Supplemental Fig. 2). One or two copies of the bipartite motif “AACNBDWHNNDNDNGTAT” are present in 5′-end regions of UNC93A, potassium-dependent Na+/Ca2+ exchanger (NCKX, AAEL004805), lysosomal trafficking regulator (AAEL005066), and an organic ion transporter (AAEL010917) (Fig. 2), as well as other genes. One to five copies of “ACNGMCTK” are present at the 5′-ends of UNC93A, NCKX, Aquaporin 2 (AAEL005008), lysosomal trafficking regulator, vacuolar ATPase proteolipid subunit (AAEL012113), zinc transporter (AAEL014156), and a choline-kinase like protein (AAEL014436). The motif “CMNKCAGT” is present in 1–5 copies at the 5′-ends of Picot (AAEL001113), a chitin-binding protein (AAEL002612), UNC93A, NCKX, lysosomal trafficking regulator, and zinc transporter.
Figure 1.
Sequence Logo indicates predominant nucleotides in the consensus for each motif identified (http://weblogo.threeplusone.com/create.cgi). ‘Percent of Gene Set’ indicates the percent of all 5′ genomic regions bearing the motif of interest. ‘SCOPE Score’ indicates the negative log of the expectation as calculated by SCOPE. “Enrichment” presents the hypergeometric enrichment values (−log10(hypergeometric p-value)) for position weight matrix thresholds of 75% (‡) and 90% (§) of the maximum score with FDRs listed in parentheses. “Region Searched” indicates the 5′ genomic region of interest that yielded the given motif.
Figure 2.
Schematic shows the location of motifs along 5′ genomic regions of genes of interest. AAEL005849 (SV2) and AAEL004048 (UNC93A) are at the top of the figure. Only those motifs common to UNC93A were included.
Virus Feeding and Gene Silencing
A recombinant SINV, MRE16-eGFP, expressing enhanced green fluorescent protein (eGFP) was used to infect mosquitoes. MRE16-eGFP allows easy assessment of viral protein translation and dissemination to peripheral tissues. Laboratory infection of mosquitoes with arboviruses typically requires high-titer blood meals. Such high-titer infections could mask subtle differences in virus infection kinetics. Therefore, we used a blood meal virus titer of 5.4 log plaque-forming units (pfu) per ml, which is two orders of magnitude lower than that normally required to establish a productive disseminated infection (Foy et al. 2004).
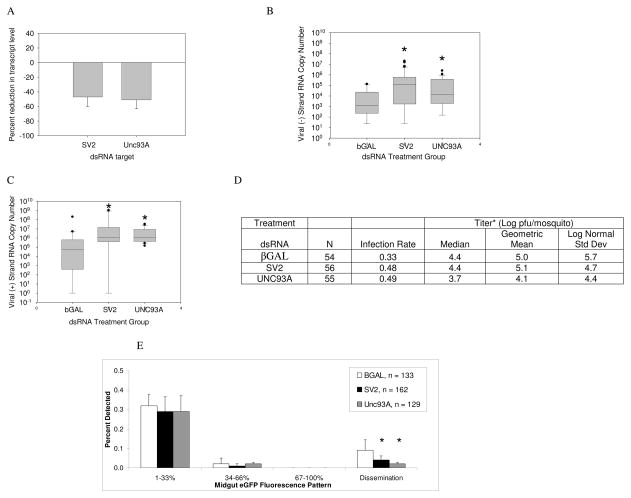
SV2 and UNC93A were silenced following injection of long double-stranded RNA (dsRNA) of approximately 500 nts in length. Because previous studies indicated that long dsRNA-induced gene silencing could be transient in SINV-infected mosquitoes, mosquitoes were injected with dsRNA within three hours of the virus meal (Campbell et al. 2008). Animals were harvested for all analyses at 4 dpi to match the period of reported UNC93A and SV2 transcript enrichment. Both SV2 and UNC93A transcripts were depleted ~50% at 4dpi (Fig. 3A).
Figure 3.
Viral RNA accumulates in UNC93A and SV2-silenced mosquitoes over βGAL-injected controls, but dissemination is reduced. (A) Evidence of gene silencing. Mosquitoes were injected with dsRNA immediately following a virus blood meal, and at 4 dpi, transcripts were detected using qRT-PCR. Relative transcript levels were normalized to an RPS7 reference standard and those of βGAL injected controls to calculate percent reduction. Error bars indicate standard error of the mean of three independent experiments. (B) Negative strand viral RNA copy number per mosquito. Levels are statistically significant difference from control per Mann-Whitney U Test (SV2, p=0.002; UNC93A, p=0.02). (C) Positive strand viral RNA copy number per mosquito. Mosquitoes were harvested at 4 dpi and subjected to two step qRT-PCR, (SV2, p=0.02; UNC93A, p=0.02). The data are representative of at least three biological replicates. Box outline displays 75th and 25th percentiles, while the whiskers show the 90th and 10th percentiles. (D) MRE16-eGFP virus production. *, Titer median and geometric means were calculated from infected mosquitoes. (E) Percentage of cells expressing eGFP is significantly reduced in both UNC93A- and SV2-silenced mosquitoes. Mosquitoes were harvested at 4 dpi and subjected to fluorescence microscopy at a 488nm excitation wavelength. Midgut infections were classified by the percentage of expressing eGFP. “Dissemination” refers to the percentage of mosquitoes displaying fluorescence in peripheral tissues. The data are representative of at least three biological replicates. Error bars indicate standard error of the mean. Mann-Whitney test (p<0.05).
Viral Replication and Dissemination
Negative-strand RNA is transcribed from the positive-sense genome following viral entry, uncoating and translation of non-structural proteins. Nascent viral genomes are transcribed from the negative-sense replicative strand. Both SV2- and UNC93A-silenced mosquitoes showed enriched negative-strand RNA compared to β-galactosidase (βGAL) - injected controls. SV2-silenced mosquitoes accumulated 180-fold higher median copy number negative-strand RNA than controls (Fig. 3B). UNC93A-silenced mosquitoes showed a 15-fold enrichment of median viral negative strand over controls. SV2 and UNC93A-silenced mosquitoes showed 14- and 20-fold enrichment of median viral positive strand RNA, respectively, over that of controls (Fig. 3C).
In contrast to viral RNA abundance, MRE16-eGFP titers in dsRNA-treated animals showed a much different pattern. While there was no difference in titers between controls and SV2-silenced mosquitoes, UNC93A silencing resulted in a 10-fold decrease in viral titers (Mann-Whitney U test of positive samples, p <0.03) (Fig. 3D; Supplemental Fig. 3). Furthermore, we used eGFP detection as an indication of viral protein translation; however, translation would not necessarily be indicative of successful virus assembly and maturation. Although there was no significant difference in detection of eGFP in midguts among SV2 dsRNA, UNC93A dsRNA and control dsRNA treatment groups, there were significantly fewer disseminated infections in both SV2- and UNC93A-silenced mosquitoes (Fig. 3E). Thus, translation of viral genomes was not affected, but dissemination was reduced. Figure 3D shows an increase in infection rate in SV2 and UNC93A-silenced mosquitoes over βGAL controls. This result was unexpected, because the increased infection rates are not coupled with similar trends in either viral titers or dissemination.
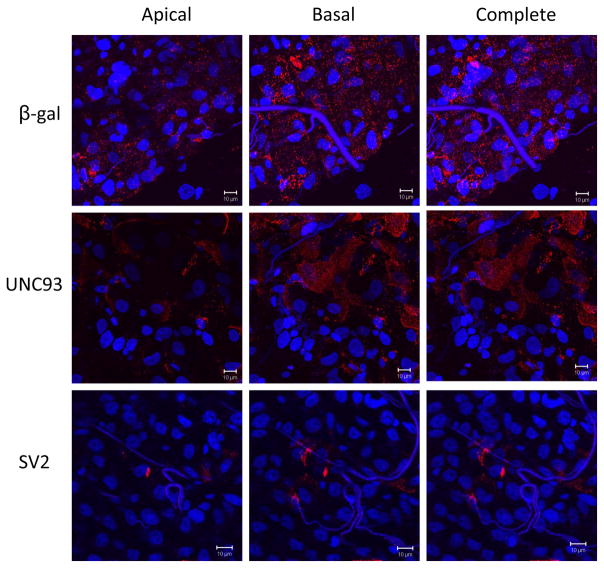
Confocal microscopy of infected mosquitoes provides visual corroboration of the indicated phenotypes (Fig. 4 and Supplemental Fig 4). βgal dsRNA-treated midguts show an even distribution of punctate SINV E1 antigen staining across the epithelia, especially along the basolateral surface; this even distribution is not seen in either SV2- or UNC93A-silenced midguts. Instead, the SV2-silenced and UNC93A-silenced midguts show variations of planar clustering of the E1 antigen, with heavy patches of E1 in some areas and no antigen in others.
Figure 4.
Spatial location of SINV E1 antigen in infected midguts. Rows indicate dsRNA treatment group (β-gal, UNC93 or SV2). Columns indicate which z-planes were involved in the compression. The basal column includes z-plane images on the basal half of the image set. The apical column shows z-plane images on the apical (luminal) half of the image set. ‘Complete’ images represent compressed versions of the entire Z stack. Virus envelope antigen is stained red and cell nuclei are stained blue. The images constituting the basal and apical columns were chosen by dividing all images in the full z-stack into two sets. 630× magnification.
Discussion
SINV buds preferentially from the apical surface of some epithelial cells and from the basolateral surface of others (Zurzolo et al. 1992). Viral proteins, nucleic acid and host lipid membranes must come together in specific orientation within an infected cell to produce a viable particle. As with other viruses, alphaviruses enter cells through a low pH fusion event. Nascent viral RNA replication occurs in modified endosomal vesicles, as well as along plasma membranes (Frolova et al. 2010). The accumulation of viral RNA and its apparent protection from nuclease digestion following UNC93A and SV2 silencing is consistent with the interpretation that the viral RNA is protected in endosomal compartments.
Although, the experimental system is benefitted by use of an in vivo model wherein virus disseminates in situ through polarized cells, its use limits characterization of a detailed mechanism. Corresponding to viral RNA accumulation in midgut epithelial cells, SV2 and UNC93A-silencing resulted in higher midgut infection rates, and significantly less virus dissemination from the midgut. Simultaneously, viral antigen aggregated within infected midgut epithelial cells in both treatments, but only UNC93A-silencing resulted in significantly lower viral titers per mosquito. Viral RNA accumulation might be directly associated with increased midgut infection rates, or it could be indicative of a delay in virus assembly, maturation or egress from midgut epithelial cells. The reason for the observed differences in virus titers between UNC93A-silenced mosquitoes and SV2-silenced mosquitoes is unknown, but they may reflect differences in the viral maturation process. UNC93A may act during virus assembly prior to maturation. In contrast, SV2 silencing causes accumulation of viral antigen in intracellular networks, but there is no reduction in overall viral titers, suggesting that SV2 may act after virus assembly but prior to budding from midgut cells, thus allowing virus maturation but preventing egress. The observation of reduced virus dissemination in the same treatments supports the possibility that viable virus particles were retained in the midgut. Interestingly, SV2 acts in the nervous system in the maturation of synaptic vesicles (Chang and Sudhof 2009); in the mosquito midgut, SV2 may also be involved in maturation of virus-containing vesicles.
Finally, epithelial cells maintain apical-basal polarity in both the asymmetrical distribution and secretion of intracellular components. Polarity is maintained by an ion gradient across one or more membrane surfaces. Sustainable ion gradients require active solute transport systems to maintain ionic charge differentials and protein or component transport across membranes. Identification of common CREs at the 5′-ends of genes encoding ion transporters and the putative endosomal protein UNC93A supports the hypothesis that endosomal and ion transport processes are coupled at the transcriptional level. Together these data provide supportive evidence for conservation of endosomal pathway components as a common feature of the arbovirus infection cycle among mammalian hosts and invertebrate vector mosquitoes. Characterization of conserved pathway components among humans and mosquitoes could lead to the development of novel anti-viral strategies.
Experimental Procedures
CRE Analysis
SCOPE (http://genie.dartmouth.edu/scope/) was used to search 5′-ends of 31 genes that were >2-fold enriched during SINV infection (Sanders et al. 2005; Carlson et al. 2006; Chakravarty et al. 2007). Promoter lengths of 800, 1600 and 2000 nts were used. A significance threshold of 5.0 was used for all output and represents the negative log of the expectation with a Bonferroni correction (Chakravarty et al. 2007). An additional level of stringency was imposed that was designed to reveal motifs that may be most associated with the 31 genes up-regulated at 4 dpi from the Sander’s (2005) dataset compared to transporter-related genes in general. SCOPE motifs were assigned hypergeometric enrichment scores (−log10(p-value)) and then compared to those of all Ae. aegypti genes bearing a “transporter activity” Gene Ontology function parent term (~730 genes) AegyXcel (http://exon.niaid.nih.gov/transcriptome.html#aegyxcel). The hypergeometric enrichment score measures the probability that the observed number or more of 5′-flanking regions of genes with at least one occurrence of a motif in the gene-set of interest might have occurred by chance given the total number of 5′-flanking regions in the genome that contain at least one instance of the motif. Motif threshold scores of both 75% and 90% of the maximum position weight matrix scores were used to determine presence/absence of motifs. Motifs were retained if the motif’s enrichment scored was higher in the 4 dpi gene set using both motif-threshold levels. False discovery rates (FDR) were estimated for these enrichment scores by repeating the enrichment analysis using 100 sets of 31 randomly chosen genes.
Mosquito/Virus Stocks
Colonized Ae. aegypti, Higgs White-eye strain, were reared at 28°C, 80% humidity and a 14:10 photo period (light:dark). Females, 4 to 7 days post adult emergence, were fed a blood meal containing recombinant SINV strain MRE16-eGFP at ~5.4 logs plaque-forming units per ml. A recombinant SINV, 5′ dsMRE16-eGFP, which expresses eGFP from the 5′ end of the subgenomic promoter, was used in this study (Foy et al. 2004). Third passage recombinant SINV stocks and virus/blood meals were prepared in mammalian Vero maru cells and titered according to methods reported previously (Campbell et al. 2008).
Transcript Silencing
dsRNA primers and the relative nucleotide position are shown in Supplemental File 2. Long dsRNA, of ~500nts in length, was transcribed from cDNA PCR products and the protocol described with the Megascript dsRNA kit (Ambion, Austin, TX). Nonspecific control dsRNA was generated from a portion of the β-galactosidase gene from E. coli. dsRNA was purified with phenol/chloroform extraction and diluted to a concentration of 7.248 μg/μl in phosphate-buffered saline (PBS). Immediately following the virus meal, females were anesthetized with cold, held on ice and intrathoracically injected with 500 ng dsRNA in a volume of 0.069 μl using a Nanoject II microinjector (Drummond). Mosquitoes were harvested four days later for analysis. Three replicate pools of three whole mosquitoes were harvested for RNA extraction and qRT-PCR to confirm gene silencing.
Visualization of MRE16-eGFP infection of Ae. aegypti mosquitoes
Evidence of viral protein translation and dissemination was assessed at 4 dpi by dissecting SINV-infected mosquitoes in PBS and observing eGFP fluorescence of midguts and peripheral tissues by epifluorescence microscopy at 488nm (Olympus, Center Valley, PA).
Confocal microscopy
Aedes aegypti mosquitoes were fed SINV MRE16-eGFP at ~5.4 logs plaque-forming units per ml and injected with 500 ng of β-gal, UNC93A, and SV2 dsRNA within three hours. Mosquito midguts were dissected three days later, briefly fixed in cold 1% paraformaldehyde, and fluorescently labeled with MAB 8742 mouse anti-Western Equine Encephalitis Virus (Chemicon, Billerica, MA) and goat anti-mouse tagged Alexafluor® 555 (Invitrogen, Carlsbad, CA). The midguts were then mounted onto slides with Vectashield® DAPI (Vector Laboratories, Burlingame, CA) and visualized with a Zeiss LSM 510 Meta® (Carl Zeiss International, Oberkochen, Germany) confocal microscope using Argon/2, HeNe543, and Diode 405-30 lasers.
Quantitative Real-time PCR
Total RNA was extracted from individual whole mosquitoes at 4dpi. RNAs were diluted to 10 ng/μl for detection of viral negative- and positive- strand RNAs, as well as confirmation of SV2 and UNC93A transcript silencing. qRT-PCR was performed on an iCycler real-time PCR thermocycler (BioRad, Hercules, CA) in triplicate using SYBR Green One step qRT-PCR reagents (Invitrogen, Carlsbad, CA) in 20 μl reactions. Supplemental File 2 shows the qPCR primers used. Cycling parameters followed the manufacturer’s protocol. Viral RNA copy number was calculated by use of a standard curve generated from serial diluted pCR 4.0 plasmid (Invitrogen) carrying a portion of the nsP 3 gene. Transcript silencing was confirmed using the comparative Ct analytical method with a ribosomal protein subunit 7 (RPS7) reference standard and the βGal dsRNA injected control as calibrator. Percent transcript reduction was calculated by normalizing relative transcript levels to that of the βGal dsRNA-injected control.
Plaque titrations
Whole mosquitoes were triturated, filtered using a 0.2 micron filter, and serially diluted onto Vero maru cells. Standard methods were used to calculate plaque-forming units per mosquito (Foy et al. 2004). The data are representative of at least three biological replicates.
Supplementary Material
Supplemental Figure 2. Excel file shows promoter motif data.
Supplemental Figure 1. 1A. Aedine SV2 aligned with human SV2. 1B. Aedine Unc93A aligned to Human Unc93A and mouse Unc93A. Identical amino acids are shown in reverse contrast. Gray highlighted amino acids are conserved among two of the three proteins.
Supplemental Figure 3. Distribution of viral titers among dsRNA treatment groups. Mosquitoes were fed a SINV meal and injected with dsRNA as indicated. Data represent at least three biological replicates.
Supplemental Figure 4. Differential spatial location of SINV E1 antigen in infection foci of UNC93A- and SV2-silenced midguts as determined by confocal microscopy. Files are 3 dimensional .avi files of rotating z-stack images. They correspond directly to the panel images in Figure 4. Website: http://www.cvmbs.colostate.edu/mip/aidl/imb-manuscript. Username: cvmbsdom\csufoyweb. Password: Pass#9752*.
Acknowledgments
We thank the Arthropod-borne Infectious Diseases Lab Core Support facility for help with viral titrations. CLC and BDF were supported by grants AI060960 AI079528, and by contract N01-AI-25489 from the U.S. National Institutes of Allergy and Infectious Diseases. WAD was supported in part by a predoctoral biomedical informatics training fellowship through the NIH grant T15LM07443, WAD and AAJ by the NIH grant R01AI29746, and SSG in part by NIAID grants 32572 and 48049. CJL was supported by NIH grant AI046435.
References
- Bowers DF, Abell BA, Brown DT. Replication and tissue tropism of the alphavirus Sindbis in the mosquito Aedes albopictus. Virology. 1995;212(1):1–12. doi: 10.1006/viro.1995.1447. [DOI] [PubMed] [Google Scholar]
- Campbell CL, Black WCt, Hess AM, Foy BD. Comparative genomics of small RNA regulatory pathway components in vector mosquitoes. BMC Genomics. 2008;9(1):425. doi: 10.1186/1471-2164-9-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CL, Keene KM, Brackney DE, Olson KE, Blair CD, Wilusz J, Foy BD. Aedes aegypti uses RNA interference in defense against Sindbis virus infection. BMC Microbiol. 2008;8:47. doi: 10.1186/1471-2180-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson JM, Chakravarty A, Gross RH. BEAM: a beam search algorithm for the identification of cis-regulatory elements in groups of genes. J Comput Biol. 2006;13(3):686–701. doi: 10.1089/cmb.2006.13.686. [DOI] [PubMed] [Google Scholar]
- Carlson JM, Chakravarty A, Khetani RS, Gross RH. Bounded search for de novo identification of degenerate cis-regulatory elements. BMC Bioinformatics. 2006;7:254. doi: 10.1186/1471-2105-7-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty A, Carlson JM, Khetani RS, DeZiel CE, Gross RH. SPACER: identification of cis-regulatory elements with non-contiguous critical residues. Bioinformatics. 2007;23(8):1029–31. doi: 10.1093/bioinformatics/btm041. [DOI] [PubMed] [Google Scholar]
- Chang WP, Sudhof TC. SV2 renders primed synaptic vesicles competent for Ca2+-induced exocytosis. J Neurosci. 2009;29(4):883–97. doi: 10.1523/JNEUROSCI.4521-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colpitts TM, Moore AC, Kolokoltsov AA, Davey RA. Venezuelan equine encephalitis virus infection of mosquito cells requires acidification as well as mosquito homologs of the endocytic proteins Rab5 and Rab7. Virology. 2007;369(1):78–91. doi: 10.1016/j.virol.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz IP, Levin JZ, Cummins C, Anderson P, Horvitz HR. sup-9, sup-10, and unc-93 may encode components of a two-pore K+ channel that coordinates muscle contraction in Caenorhabditis elegans. J Neurosci. 2003;23(27):9133–45. doi: 10.1523/JNEUROSCI.23-27-09133.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foy BD, Myles KM, Pierro DJ, Sanchez-Vargas I, Uhlirova M, Jindra M, Beaty BJ, Olson KE. Development of a new Sindbis virus transducing system and its characterization in three Culicine mosquitoes and two Lepidopteran species. Insect Mol Biol. 2004;13(1):89–100. doi: 10.1111/j.1365-2583.2004.00464.x. [DOI] [PubMed] [Google Scholar]
- Frolov I, Schlesinger S. Comparison of the effects of Sindbis virus and Sindbis virus replicons on host cell protein synthesis and cytopathogenicity in BHK cells. J Virol. 1994;68(3):1721–7. doi: 10.1128/jvi.68.3.1721-1727.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova EI, Gorchakov R, Pereboeva L, Atasheva S, Frolov I. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J Virol. 2010 doi: 10.1128/JVI.01441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov R, Frolova E, Frolov I. Inhibition of transcription and translation in Sindbis virus-infected cells. J Virol. 2005;79(15):9397–409. doi: 10.1128/JVI.79.15.9397-9409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240(4860):1759–64. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Mudiganti U, Hernandez R, Brown DT. Insect Response To Alphavirus Infection-Establishment Of Alphavirus Persistence In Insect Cells Involves Inhibition Of Viral Polyprotein Cleavage. Virus Res. doi: 10.1016/j.virusres.2010.02.016. [DOI] [PubMed] [Google Scholar]
- Sanders HR, Foy BD, Evans AM, Ross LS, Beaty BJ, Olson KE, Gill SS. Sindbis virus induces transport processes and alters expression of innate immunity pathway genes in the midgut of the disease vector, Aedes aegypti. Insect Biochem Mol Biol. 2005;35(11):1293–307. doi: 10.1016/j.ibmb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Sieglaff DH, Dunn WA, Xie XS, Megy K, Marinotti O, James AA. Comparative genomics allows the discovery of cis-regulatory elements in mosquitoes. Proc Natl Acad Sci U S A. 2009;106(9):3053–8. doi: 10.1073/pnas.0813264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, Crozat K, Mudd S, Mann N, Sovath S, Goode J, Shamel L, Herskovits AA, Portnoy DA, Cooke M, Tarantino LM, Wiltshire T, Steinberg BE, Grinstein S, Beutler B. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol. 2006;7(2):156–64. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- Zurzolo C, Polistina C, Saini M, Gentile R, Aloj L, Migliaccio G, Bonatti S, Nitsch L. Opposite polarity of virus budding and of viral envelope glycoprotein distribution in epithelial cells derived from different tissues. J Cell Biol. 1992;117(3):551–64. doi: 10.1083/jcb.117.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 2. Excel file shows promoter motif data.
Supplemental Figure 1. 1A. Aedine SV2 aligned with human SV2. 1B. Aedine Unc93A aligned to Human Unc93A and mouse Unc93A. Identical amino acids are shown in reverse contrast. Gray highlighted amino acids are conserved among two of the three proteins.
Supplemental Figure 3. Distribution of viral titers among dsRNA treatment groups. Mosquitoes were fed a SINV meal and injected with dsRNA as indicated. Data represent at least three biological replicates.
Supplemental Figure 4. Differential spatial location of SINV E1 antigen in infection foci of UNC93A- and SV2-silenced midguts as determined by confocal microscopy. Files are 3 dimensional .avi files of rotating z-stack images. They correspond directly to the panel images in Figure 4. Website: http://www.cvmbs.colostate.edu/mip/aidl/imb-manuscript. Username: cvmbsdom\csufoyweb. Password: Pass#9752*.