Abstract
Radiology images are generally disconnected from the metadata describing their contents, such as imaging observations (“semantic” metadata), which are usually described in text reports that are not directly linked to the images. We developed a system, the Biomedical Image Metadata Manager (BIMM) to (1) address the problem of managing biomedical image metadata and (2) facilitate the retrieval of similar images using semantic feature metadata. Our approach allows radiologists, researchers, and students to take advantage of the vast and growing repositories of medical image data by explicitly linking images to their associated metadata in a relational database that is globally accessible through a Web application. BIMM receives input in the form of standard-based metadata files using Web service and parses and stores the metadata in a relational database allowing efficient data query and maintenance capabilities. Upon querying BIMM for images, 2D regions of interest (ROIs) stored as metadata are automatically rendered onto preview images included in search results. The system’s “match observations” function retrieves images with similar ROIs based on specific semantic features describing imaging observation characteristics (IOCs). We demonstrate that the system, using IOCs alone, can accurately retrieve images with diagnoses matching the query images, and we evaluate its performance on a set of annotated liver lesion images. BIMM has several potential applications, e.g., computer-aided detection and diagnosis, content-based image retrieval, automating medical analysis protocols, and gathering population statistics like disease prevalences. The system provides a framework for decision support systems, potentially improving their diagnostic accuracy and selection of appropriate therapies.
Key words: Imaging informatics, data mining, databases, decision support, body imaging, cancer detection, computed tomography, computer-aided diagnosis (CAD), image retrieval, PACS, ROC curve, ROC-based analysis, web technology, digital imaging and communications in medicine (DICOM), algorithms
Introduction
Purpose
Explosive growth in the number of biomedical images obtained in radiology requires new techniques to manage the information collected by radiologists.1 “Image metadata” refers to information about an image, e.g., the modality from which it was derived, the acquisition parameters, the timing of imaging, regions of interest (ROI) it contains, anatomy, radiology observations, and annotations created by radiologists. These metadata provide crucial descriptive information that potentially could be used to search for images. Picture archiving and communication systems (PACS) can address the image data management problem,2 but PACS generally lack any facilities for managing or querying images based on image metadata that is recorded during interpretation. Our work addresses both (1) the needs of managing image metadata and (2) using image metadata in applications. We are developing a system that interfaces with a PACS and uses emerging standards for managing image metadata and enables their use by radiologists, students, and researchers. This tool could improve radiologist interpretations by enabling online decision support and might facilitate biomedical researchers to access image information.
The annotations that radiologists create on images are particularly important image metadata since they are used to identify and delineate lesion borders, to quantify features of ROI in lesions, and to note visual observations of those lesions. However, these annotations are not directly accessible for future analyses because they are typically captured as image graphics. Imaging observation characteristics made by radiologists may also be recorded in radiology reports, but there is usually no reference to the specific image pixels that correspond to the observation, and problems with synonymy, negation, and completeness hinder machine processing and search.3 As a result, it is currently not possible to retrieve images based on their features in a clinical environment, e.g., one cannot query the PACS to retrieve all “hypervascular” lesions. In order to achieve such functionality, methods are needed to make medical image metadata accessible for search, which, in turn, could enable newly annotated images to be queried against the database of existing images and the results ordered by decreasing similarity to support diagnostic decisions. In fact, previous work has shown that querying a database of medical image metadata and retrieving similar images can improve diagnosis.4–9 However, these systems were specifically designed to answer research questions about decision support and are not easily integrated into a PACS-based environment.
The primary goal of this study was to develop and evaluate methods to manage semantic annotation metadata and to create an application enabling users to search for and retrieve images according to particular image metadata. A secondary goal was to develop a demonstration application that uses image metadata for decision support in liver CT by retrieving similar images based on the image metadata related to radiologist observations. We implemented a Web-based application that enables radiologists and researchers to search for images in flexible ways, by anatomy, findings, or other semantic metadata. Our application also allows one to use an annotated image as a query to find similar images.
Materials and Methods
Our system, the Biomedical Image Metadata Manager (BIMM), is driven by a Web application (http://bimm.stanford.edu) that makes image metadata searchable and linked to the images, enabling query of image databases and retrieval of similar images based on image metadata. Examples of image metadata are the quantitative and qualitative information in images and the content normally stored in free text reports. We followed a spiral model of system design, iteratively gathering requirements, identifying tasks, building prototypes, and acquiring feedback. In the next subsection, we describe the system’s design and architecture. Then, we cover four main use cases covered in the following subsections: sending metadata to BIMM, viewing summary tables, text image search, and similar image retrieval.
System Design and Architecture
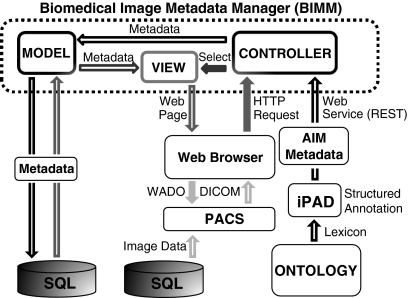
Figure 1 depicts the system architecture and its interactions with client applications. Components of the Web application are enclosed in the dotted box. The Web application was developed using Ruby on Rails [http://rubyonrails.org] and MySQL and hosted on a computer using the Linux operating system running the LiteSpeed Web-server [http://www.litespeedtech.com/]. Its architecture follows the model, view, controller pattern used by the Ruby on Rails framework. The models, views, and controllers integrate the functions of the associated sequence query language database, the Web browser, a PACS, and the Web services. Black arrows represent sending metadata to the system and gray arrows depict metadata retrieval using a Web browser. Solid gray arrows show the user’s Web browser requesting the system to select a particular view. The view interacts with the appropriate models representing the different types of information stored and dynamically generate a Web page backed by the database. Hollow gray arrows show sending the appropriate metadata from the database to the user in the form of a Web page generated by the chosen view. Light gray arrows similarly describe how the Web page automatically retrieves appropriate images from the PACS using a Web service. These processes are described in more detail in the following subsections.
Fig 1.
BIMM system architecture and interactions. Black arrows represent the process of sending metadata to the system. Initially, an ontology specifies a lexicon, e.g., the RadLex set of terms and sends it to iPAD, which captures the radiologist’s image annotation. iPAD sends these metadata using the AIM format to the appropriate controller using a Web service. The controller interacts with the models to store the metadata in an SQL relational database. Gray arrows represent the process of retrieving metadata and images from the system. Solid gray arrows indicate requests sent from the client application, e.g., a Web browser. The browser sends an HTTP request to a controller that selects the appropriate view, which generates a Web page and sends it to the user. Hollow gray arrows indicate metadata sent by the system to the user’s Web browser in the form of a Web page. The solid light gray arrow shows the request for image data from the PACS by the Web browser via the WADO protocol. Hollow light gray arrows show the DICOM images from the PACS being sent to the user.
Sending Metadata to BIMM
Inputs to BIMM are encoded using the Annotation and Image Markup (AIM) standard,10–12 which was recently developed for description and storage of image metadata. We used the physician annotation device (iPAD),13 an OsiriX14 plug-in for structuring reports to be consistent with an ontology, capturing ROIs, and generating AIM files, collected metadata from radiologists for each image used in our study (not to be confused with the more recently introduced Apple Inc. tablet device). iPAD provides output of image annotations in the AIM standardized format, which is advantageous because the image content is explicit and machine accessible. iPAD controls the terminology specified by an ontology or lexicon; for our study, we used the RadLex15 controlled terminology. iPAD collects information about image anatomy, imaging observations and their characteristics, and quantitative assessments in AIM-computable format. iPAD sends the AIM annotations to BIMM using a representational state transfer-style Web service (see black arrows in Fig. 1).
This controller parses and stores the information from the AIM file in the database. When a user communicates to the system using a Web browser using a uniform resource locator, a hypertext transfer protocol (HTTP) request is sent to a controller which selects the appropriate view to display to the user through the Web browser. Depending on which view is selected by its uniform resource locator, the appropriate metadata are retrieved from the database and sent to the client. Digital ximaging and communications in medicine (DICOM) images are sent to the Web browser from the PACS using a DICOM Part 18, Web Access to DICOM Persistent Objects service.16 LibXML was used to parse the AIM data [http://xmlsoft.org/]; the parsing and capturing code was written in Ruby using the LibXML-Ruby helper [http://libxml.rubyforge.org].
Data Summary View
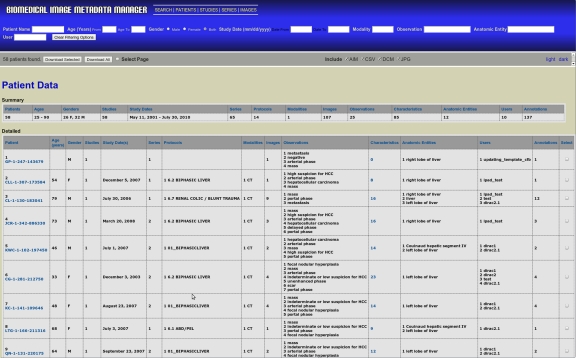
BIMM provides a summary view of the database’s contents, listing groups of annotations organized according to patient, study, series, or image, as selected by the user. Figure 2 shows the patient summary view, which contains two tables; a summary table (top) that contains data describing all of the patients that have had annotations uploaded to the system and a detailed table (bottom) with statistics describing the types of annotations made for each patient. Filtering options, shown in the blue panel at the top of the figure, allow users to select subsets of the database for viewing and downloading.
Fig 2.
Representative summary table page (http://bimm.stanford.edu/patients). The user can generate custom datasets and views using the filtering options available within the blue banner. Customized datasets are downloadable, using buttons and checkboxes within the gray bar below the blue banner, in several formats. Selecting multiple options applies all filters in combination to generate custom datasets. Applying multiple options provides a cumulative effect on the resulting table.
Image Search
Users can query BIMM for images using text entered into a Web browser. The BIMM search page (http://bimm.stanford.edu/search) allows users to enter queries, select a search category (e.g., anatomic entities), and browse or download the search results.
Storing image metadata in a relational database compliant with the AIM schema allows users to search for any image metadata in stored in the AIM files, such as anatomy, imaging observations (IOs), imaging observation characteristics (IOCs), and diagnoses. The image search page allows users to enter search terms which are used to query the BIMM database (Fig. 3). DICOM images stored in the PACS corresponding to the metadata are linked via the DICOM unique identifier (UID) contained in the image metadata and may therefore be retrieved. The image metadata also contains the coordinates of ROIs drawn in the image, which BIMM uses to render the ROI on the images that are displayed with the search results (Fig. 3).
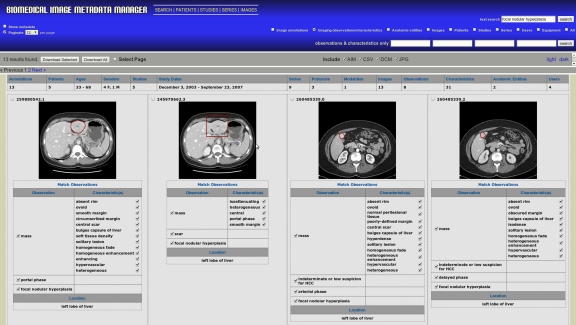
Fig 3.
Sample image search page. Results for a “focal nodular hyperplasia” text query with “imaging observations/characteristics” selected. This query returned 13 images from our database; only the first four, sorted by upload date, are shown. AIM UIDs, unique identifiers defined by the AIM schema and automatically generated by iPAD, are listed above each image.
Two types of search boxes are available: “text search” and “imaging observations/characteristics only” search. The “text search” box finds results matching the query string anywhere in the AIM image metadata. The four “imaging observations/characteristics” boxes allow users to search image metadata using controlled terms (e.g., from RadLex), joined by the logical AND operation. Combining both types of queries together joins the results using the AND operation.
Image search results are sorted by upload date from the most recent; by default, only the anatomic locations and IOs with their characteristics are shown to users (Fig. 3). However, selecting the “show metadata” checkbox displays all of the metadata associated with each image.
Retrieving Similar Images
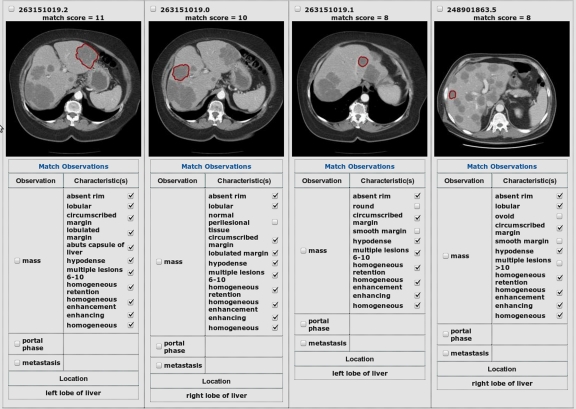
The BIMM application implements a “match observations” feature enabling users to search for images containing similarly described lesions to one selected in the database. This function is implemented by searching BIMM for images having the most IOCs in common with the query image. The user chooses a query image and selects which IOCs are desired for retrieving similar images, i.e., similar images must contain all of the selected IOCs. Then, the user clicks the match observations link. Figure 4 provides some sample results of using this feature. From left to right, the query is shown first, followed by the three most similar images according to matching IOCs.
Fig 4.
Example of similar image retrieval results. User entered a query, metastasis, and clicked “match observations” on one of the text search results. Similar images are retrieved in order of match score, indicated above each image. The matching IOCs that contributed to the computed score are checked. The first image shows the query image, followed by the three most similar images retrieved.
The images returned are ranked according to the scoring function, i.e., the number of matching IOCs, with images having all observations matching the selected image appearing before those having fewer matching observations. In accordance with the AIM schema, for each IO, e.g., “mass,” associated with an annotated image, there can be one or more associated IOCs, e.g., “smooth margins.”
For our evaluation, we considered only the IOCs associated with the “mass” IO and define the similarity between two images in terms of the number of associated IOCs they share. Mathematically, we compute the pairwise image similarity score by using the function
 |
where Ci and Cj are the sets of IOCs corresponding to “mass” in the ith and jth images, respectively, and |·| denotes the counting measure. For example, if a lesion has three IOCs associated with “mass,” then perfectly matching images would have a score of 3, while less accurate matches may have scores of 2, 1, or 0 matching IOCs; if some images have few IOCs, they cannot yield a higher score than images with more IOCs. Since the retrieval results were analyzed on a per-lesion basis, scaling the function to normalize its value has no effect on our evaluation, so it was avoided to maintain the function’s commutativity and to unambiguously show the number of IOCs that are matching from score values.
Image Database Evaluation
Under IRB approval for retrospective analysis of de-identified data, we obtained 74 DICOM images from 34 patients, containing a total of 79 liver lesions. We selected lesions of each type that radiologists would consider typical examples, as follows: cysts were nonenhancing, water density, circumscribed lesions. Hemangiomas showed typical features of discontinuous, nodular peripheral enhancement with fill-in on delayed images. Metastases were hypodense, of soft tissue density, enhanced homogeneously with contrast, and had less well-defined margins than cysts. These three types of lesions are common and span a range of image appearances. In addition to these lesion types, we added five other types to our initial database (Table 1). In Table 1, rows are sorted by the number of lesions (L) that are present for each type. Scans were acquired during the time period February 2007 to August 2008 and used the following range of scan parameters: 120 kVp, 140–400 mAs, and 2.5–5 mm slice thickness.
Table 1.
The Types of Diagnoses and the Number of Lesions (L) of Each Type
| Diagnosis Type | L |
|---|---|
| Cyst | 25 |
| Metastasis | 24 |
| Hemangioma | 14 |
| Hepatocellular carcinoma | 6 |
| Focal nodular hyperplasia | 5 |
| Abscess | 3 |
| Laceration | 1 |
| Fat deposition | 1 |
Using OsiriX, a radiologist manually defined each lesion ROI coordinates and used iPAD to record semantic descriptors for the image (from 161 possible choices). Lesion ROIs were 2D polygons manually defined in OsiriX. The annotations were uploaded to BIMM using the AIM standard.
Performance Evaluation
To evaluate the ability of the matching algorithm to find similar images, we tested the sensitivity and specificity of retrieving images of the same diagnosis in our database of N = 79 lesions belonging to eight different types (see Table 1 for details). There were a total of 72 unique IOCs present among the 79 annotations we studied; the minimum number, maximum number, mean number, and standard deviation of the number of IOCs per annotation were 8, 16, 10.6, and 1.8, respectively. We performed a leave-one-out test on the algorithm by querying each lesion against the remaining lesions in the database and assessed the sensitivity and specificity for retrieving images having the same diagnosis as the query image. The sensitivity and specificity of the top K retrieval results, with 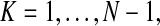 were computed by using a Ruby script to automate the procedure, without using a Web browser to manually perform each query an collect the results. For a fixed value of K, the sensitivity (true-positive rate) was calculated by the number of identical diagnoses, i.e., true positives in the K query results divided by the total number of images in the database with the same diagnosis as the query image. The specificity (the true negative rate) is calculated by the number of non-identical diagnoses in the K query results divided by the total number of non-identical diagnoses in the database. Two of the lesions in the database were represented only as a single image (“fat deposition” and “laceration”); these were excluded from the evaluation since there would be no other examples of these lesions in the database during the leave-one-out testing. The performance of the remaining N − 2 cases were analyzed using mean receiving operator characteristic (ROC) curves. Individual ROC curves for each lesion were defined by the values of the sensitivity and specificity computed for each value of K. We used piecewise linear interpolation to define 11-point curves to compute the mean values at each of the 11 sensitivity coordinates. The area under the ROC curve indicates the potential effectiveness within the framework of a decision support system, with the maximum area of 1 being optimal.
were computed by using a Ruby script to automate the procedure, without using a Web browser to manually perform each query an collect the results. For a fixed value of K, the sensitivity (true-positive rate) was calculated by the number of identical diagnoses, i.e., true positives in the K query results divided by the total number of images in the database with the same diagnosis as the query image. The specificity (the true negative rate) is calculated by the number of non-identical diagnoses in the K query results divided by the total number of non-identical diagnoses in the database. Two of the lesions in the database were represented only as a single image (“fat deposition” and “laceration”); these were excluded from the evaluation since there would be no other examples of these lesions in the database during the leave-one-out testing. The performance of the remaining N − 2 cases were analyzed using mean receiving operator characteristic (ROC) curves. Individual ROC curves for each lesion were defined by the values of the sensitivity and specificity computed for each value of K. We used piecewise linear interpolation to define 11-point curves to compute the mean values at each of the 11 sensitivity coordinates. The area under the ROC curve indicates the potential effectiveness within the framework of a decision support system, with the maximum area of 1 being optimal.
Results
We manually verified that the information stored in the database matched the information from the AIM files for all cases studied by inspecting the metadata elements individually for each case for semantic features and by visually observing the ROI rendered from the 2D spatial coordinates. We manually validated that the “match observations” function correctly implemented the scoring function correctly when constructing similar image retrieval results by counting the number of matching IOCs and comparing this to the reported score. All of the verification and validation procedures were completed successfully.
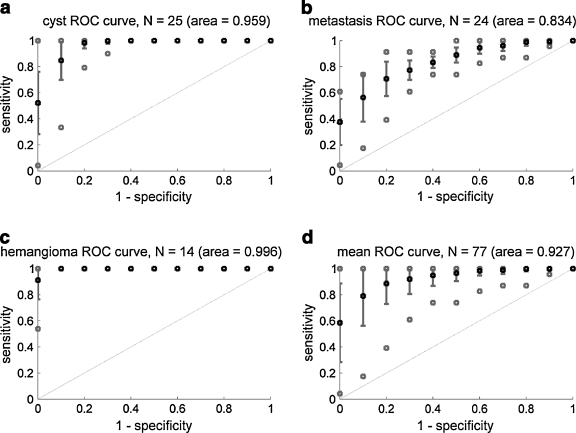
The results of our evaluation using our scoring function are shown in Figure 5. Each 11-point curve represents the effect of varied numbers of similar images retrieved on the sensitivity and specificity of the retrieval results. Average values over the samples of liver lesions are shown in black, error bars and best/worst cases are shown in solid gray lines and open squares, respectively. Panels a through c show the results for the three diagnoses containing more than ten samples; panel d shows the results for all types of lesions combined. The areas under the mean ROC curves are shown parenthetically; these represent the overall effectiveness of the algorithm for classifying lesions by diagnosis based on their IOCs.
Fig 5.
a–d 11-Point ROC curves. Plots show the sensitivities and specificities for retrieving images having the same diagnosis as a query image using a similarity score based on the number of matching IOCs. Points with error bars represent average sensitivity/specificity ±SD using each image of the specified lesion type, i.e., a cyst, b metastasis, c hemangioma, and d all 77 lesions (see Table 1 for lesion types), as the query image. Gray points indicate best and worst cases over all queries of the specified type.
All queries yielded ROC curves above the diagonal (dashed line), indicating that the retrieval results favor lesions with similar diagnoses. For all types of lesions i with a sample size Ni > 10, i.e., cysts, metastases, and hemangiomas, the observed mean ROC curve area was greater than 0.8 (Fig. 5 panels a–c). We also combined the 11-point ROC curves for all queries emcompassing six different lesion image types (cyst, metastasis, hemangioma, hepatocellular carcinoma, focal nodular hyperplasia, and abscess). This mean ROC curve’s area was 0.927, suggesting that the method is highly accurate, overall. However, note that while mean performance was very high, some query lesions produced poor results, as shown by the worst case indications on Figure 5. This is likely caused by poor or incomplete annotation in particular cases and could be improved with more stringent annotation requirements.
Discussion
Managing biomedical image metadata is crucial to enabling researchers and clinicians to use the information in images for investigation and medical care. While an institution’s PACS or, for that matter, a collection of PACS on the internet could potentially be used for decision support by retrieving images containing lesions with a given diagnosis, or having similar appearance to those being reviewed by the radiologist, current PACS implementations do not allow query for images containing particular types of lesions or diagnoses. Our work is relevant in addressing these needs. The query functionality of BIMM might be useful in cases when a radiologist is not confident regarding the diagnosis and could enable a researcher who wishes to develop image-processing algorithms to access particular images using the summary BIMM pages. In addition, research groups from geographically separated locations can combine their data using the globally accessible BIMM application. Searchable databases of image metadata could also be important for radiology research and education by enabling radiologists to find patient cohorts with particular image features.
There are several image databases available, and more under development. Most focus on warehousing images, and generally contain little associated image metadata. For example, the National Biomedical Imaging Archive [NCBI. https://cabig.nci.nih.gov/tools/NCIA] is a public database of biomedical images, but the only image metadata which this resource manages (other than occasionally supplementary data stored in separate files) is derived from the DICOM header. ARRS GoldMiner17 contains images from journal articles and their associated captions, but no other image metadata is available; image search is based only on terms in the associated (unstructured) captions. BIMM is unique in that it can manage a diversity of image metadata, as specified in the AIM standard.
Some existing systems address the problem of utilizing biomedical image metadata for several applications. The Yale Image Finder uses data mining to extract textual metadata that are present in the images themselves.18 ALPHA is a system prototype implementing scalable semantic retrieval and semantic knowledge extraction and representation using ontologies, allowing semantic and content-based queries as well as ontologic reasoning to facilitate query disambiguation and expansion.19 IML, an image markup language, provides a standard for describing image metadata and annotations and allows queries by an image client.20 The MedImGrid system allows semantic and content-based querying using a scalable grid architecture.5 The BIMM system is distinct from the prior work in that it interfaces with a PACS, it supports the use of controlled terminology, it uses a standard for sharing image metadata, it uses metadata describing image features to retrieve similar images based on these features, it provides Web services for communicating with client applications, and it allows sharing of de-identified images and their annotations using a globally accessible Web interface.
In addition to searching image metadata, BIMM also provides image similarity search capabilities, identifying and ranking similar images using a scoring function based on semantic features associated with images. As shown in Figure 5, our approach achieved high sensitivity and specificity for lesion diagnosis when using the IOCs of a query image to retrieve similar images. Our preliminary results appear promising, and they reveal some interesting features about variability in the accuracy of the method depending on the diagnosis. The results were best for hemangiomas and cysts, and not as good for metastases. The features describing hemangiomas and cysts tend to be specific and non-overlapping, whereas the features of metastases can overlap that of other lesions.
One limitation of our approach is that it requires human annotation, currently performed using the iPAD application. At the same time, this is a key attribute of our system, as the human perceptual data captured in BIMM is uniquely informative. While it might be challenging to introduce the current implementation of the iPAD interface into the routine radiology workflow, improvements in the user interface could make this practical in the future. Incorporating voice recognition reporting and controlled terminologies such as RadLex and improving the user interface could streamline structured data collection from images. Another limitation is that our dataset of 79 liver lesions is not very large, contains multiple lesions from the same patients, and contains a limited set of diagnoses; larger databases with more varied image types and diagnoses may be more challenging. We are currently building a larger image database to further validate our results. An alternative to Web-based exchange of image metadata is a distributed, large-scale “grid” of computers from multiple administrative domains, e.g., the National Cancer Institute’s caGrid project.21 It provides a set of services, toolkits for building and deploying of new services, and application programming interfaces for developing client applications.
There are improvements that we could make in our algorithm for finding similar images. Instead of simple frequency of matching IOCs, we could use machine-learning or other optimization algorithms to obtain an optimized set of weights for our scoring function to weight the contributions of each matching IOC in the total score. Such optimization would require a larger set of training data, which we will be pursuing as we expand our database of case material. We plan to optimize our scoring function in the future to maximize the system’s similar image retrieval performance. In addition, our current approach uses semantic features for finding similar images; pixel features are also informative (e.g., texture and lesion boundary22). We will incorporate pixel features into BIMM in the future to improve similar image retrieval results.
Conclusions
We developed a searchable image metadata repository and methods for retrieving similar images using semantic features from their annotations. Preliminary results of the effectiveness of our image retrieval approach appear promising, and future work will focus on increasing the size of our dataset and adding and improving the imaging features included. As our reference dataset grows, we will obtain more clinically relevant evaluation results than those obtained from our single type of images (liver CT), compared to searching an entire PACS database containing multiple modalities and lesion types.
Acknowledgments
This study is supported in part by NIH CA72023.
Contributor Information
Daniel Korenblum, Phone: +1-518-3206414, FAX: +1-866-2335237, Email: dannyko@stanford.edu.
Daniel Rubin, Email: rubin@med.stanford.edu.
Sandy Napel, Email: snapel@stanford.edu.
Cesar Rodriguez, Email: carodriguez@lbl.gov.
Chris Beaulieu, Email: beaulieu@stanford.edu.
References
- 1.Rubin GD. Data explosion: the challenge of multidetector-row CT. Eur J Radiol. 2000;36:74–80. doi: 10.1016/S0720-048X(00)00270-9. [DOI] [PubMed] [Google Scholar]
- 2.Huang HK. Some historical remarks on picture archiving and communication systems. Comput Med Imaging Graph. 2003;27:93–99. doi: 10.1016/S0895-6111(02)00082-4. [DOI] [PubMed] [Google Scholar]
- 3.Lowe HJ, Antipov I, Hersh W, Smith CA: Towards knowledge-based retrieval of medical images. The role of semantic indexing, image content representation and knowledge-based retrieval. Proc AMIA Symp:882–886, 1998 [PMC free article] [PubMed]
- 4.Dina D-F, Sameer A, Mohammad-Reza S, Hamid S-Z, Farshad F, Kost E: Automatically Finding Images for Clinical Decision Support: IEEE Computer Society, 2007
- 5.Hai J, et al: Content and semantic context based image retrieval for medical image grid: IEEE Computer Society, 2007
- 6.Oria V, et al: Modeling Images for Content-Based Queries: The DISIMA Approach, 1997
- 7.Solomon A, Richard C, Lionel B: Content-Based and Metadata Retrieval in Medical Image Database: IEEE Computer Society, 2002
- 8.Warren R, et al. MammoGrid—a prototype distributed mammographic database for Europe. Clin Radiol. 2007;62:1044–1051. doi: 10.1016/j.crad.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 9.Wesley WC, Chih-Cheng H, Alfonso FC, Cardenas AF, Ricky KT. Knowledge-Based Image Retrieval with Spatial and Temporal Constructs. IEEE Trans on Knowl and Data Eng. 1998;10:872–888. doi: 10.1109/69.738355. [DOI] [Google Scholar]
- 10.Channin DS, Mongkolwat P, Kleper V, Sepukar K, Rubin DL. The caBIG Annotation and Image Markup Project. J Digit Imaging. 2010;23:217–25. doi: 10.1007/s10278-009-9193-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubin DL, Mongkolwat P, Kleper V, Supekar K, Channin DS: Medical Imaging on the Semantic Web: Annotation and Image Markup, Stanford University, 2008
- 12.Rubin DL, Supekar K, Mongkolwat P, Kleper V, Channin DS. Annotation and Image Markup: Accessing and Interoperating with the Semantic Content in Medical Imaging. Ieee Intelligent Systems. 2009;24:57–65. doi: 10.1109/MIS.2009.3. [DOI] [Google Scholar]
- 13.Rubin DL, Rodriguez C, Shah P, Beaulieu C: iPad: Semantic annotation and markup of radiological images. AMIA Annu Symp Proc:626–630, 2008 [PMC free article] [PubMed]
- 14.Rosset A, Spadola L, Ratib O. OsiriX: an open-source software for navigating in multidimensional DICOM images. J Digit Imaging. 2004;17:205–216. doi: 10.1007/s10278-004-1014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kundu S, et al. The IR Radlex Project: an interventional radiology lexicon—a collaborative project of the Radiological Society of North America and the Society of Interventional Radiology. J Vasc Interv Radiol. 2009;20:S275–277. doi: 10.1016/j.jvir.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 16.Koutelakis GV, Lymperopoulos DK. PACS through web compatible with DICOM standard and WADO service: advantages and implementation. Conf Proc IEEE Eng Med Biol Soc. 2006;1:2601–2605. doi: 10.1109/IEMBS.2006.260761. [DOI] [PubMed] [Google Scholar]
- 17.Kahn CE, Jr, Thao C. GoldMiner: a radiology image search engine. AJR Am J Roentgenol. 2007;188:1475–1478. doi: 10.2214/AJR.06.1740. [DOI] [PubMed] [Google Scholar]
- 18.Xu S, McCusker J, Krauthammer M. Yale Image Finder (YIF): a new search engine for retrieving biomedical images. Bioinformatics. 2008;24:1968–1970. doi: 10.1093/bioinformatics/btn340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou XS, et al. Semantics and CBIR: a medical imaging perspective. New York, NY, USA: ACM; 2008. [Google Scholar]
- 20.Lober WB, Trigg LJ, Bliss D, Brinkley JM: IML: An image markup language. Journal of the American Medical Informatics Association:403–407, 2001 [PMC free article] [PubMed]
- 21.Oster S, et al. caGrid 1.0: an enterprise Grid infrastructure for biomedical research. J Am Med Inform Assoc. 2008;15:138–149. doi: 10.1197/jamia.M2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Napel S, et al: Automated retrieval of CT images of liver lesions on the basis of image similarity: method and preliminary results. Radiology 256(1): 243–52, 2010 [DOI] [PMC free article] [PubMed]