Abstract
Pilus assembly in Caulobacter crescentus occurs during a short period of the cell cycle and pili are only present at the flagellar pole of the swarmer cell. Here we report a novel assay to visualize pili by light microscopy that led to the purification of Caulobacter pili and the isolation of a cluster of seven genes, including the major pilin subunit gene pilA. This gene cluster encodes a novel group of pilus assembly proteins. We have shown that the pilA promoter is activated late in the cell cycle and that transcription of the pilin subunit plays an important role in the timing of pilus assembly. pilA transcription is regulated by the global two-component response regulator CtrA, which is essential for the expression of multiple cell cycle events, providing a direct link between assembly of the pilus organelle and bacterial cell cycle control.
Keywords: assembly/bacteriophage/Caulobacter/cell cycle/pili
Introduction
Pili are extracellular filaments, found on a wide variety of bacteria, that play an important role in adhesion of pathogenic bacteria to their host, biofilm formation, conjugative DNA transfer, non-flagellar motility and bacteriophage infection (Clewell, 1993; O’Toole and Kolter, 1998; Soto and Hultgren, 1999; Wall and Kaiser, 1999). Pilus filaments range from 2 to 10 nm in diameter and from 0.2 to 6 µm in length (Strom and Lory, 1993; Hultgren et al., 1996). Some bacteria have pili localized to the poles of the cells, and others have pili covering the entire cell surface. These structures are formed by the polymerization of pilin subunits at the base of the growing filament (Lowe et al., 1987). In Gram-negative bacteria, the pilin subunit must be secreted across both inner and outer membranes before being assembled into an extracellular filament. Although several distinct mechanisms of pilus assembly have been described, they all share common requirements: peptidases that process the signal peptide found on prepilin, energy provided by ATP hydrolysis for transport of pilin across the inner membrane, and specialized outer membrane proteins that form channels that allow the pilin subunit to reach the cell surface (Christie, 1997; Soto and Hultgren, 1999).
In many bacteria, pili are receptors for bacteriophage, which bind either to the tip or the sides of the pilus filament (Clewell, 1993). Studies of infection of Escherichia coli by the filamentous DNA phage fd led to the idea that pili may be dynamic structures (Brinton, 1971; Jacobson, 1972). According to this model, phage infection is a two-step process: first, fd binds to the tip of the F-pilus, then the pilus retracts, bringing the phage to the cell surface where interaction with a secondary receptor facilitates DNA injection (Click and Webster, 1997). Subsequent studies of Pseudomonas aeruginosa infection by the DNA phage PO4 also support this model (Bradley, 1974).
Caulobacter crescentus is a non-pathogenic, α-purple bacterium that divides asymmetrically (Brun et al., 1994) to generate two progeny cells: a non-motile stalked cell and a motile swarmer cell with several pili and a single flagellum (see Figure 9B). The stalked cell grows and replicates its chromosome, producing a predivisional cell with a new flagellum at the pole opposite the stalk. Pilus assembly occurs later than the assembly of the flagellum; the flagellum is assembled at the incipient swarmer pole of the predivisional cell, while pili are found only on progeny swarmer cells (Sommer and Newton, 1988). When the swarmer cell later differentiates into a stalked cell, the flagellum is released from the pole to be replaced by a new stalk. Pili are also lost at this transition, and it has been suggested that pilus retraction may be responsible for this loss (Lagenaur and Agabian, 1977; Sommer and Newton, 1988).
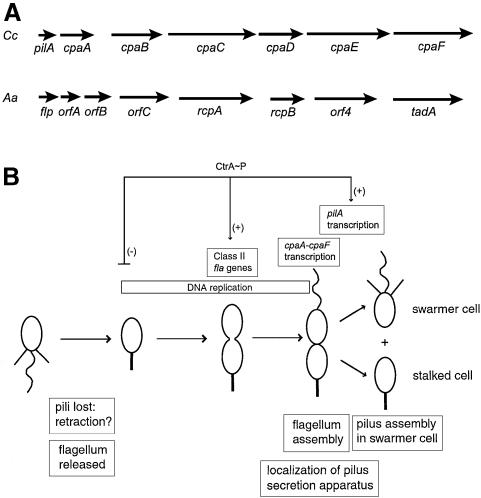
Fig. 9. Cell cycle control of a novel cluster of pilus biogenesis genes. (A) The Caulobacter pilus assembly gene cluster is similar to the flp–rcp–tadA region found in the human oral pathogen A.actinomycetemcomitans. The gene order and predicted function of the Caulobacter (Cc) and Actinobacillus (Aa) clusters are conserved. (B) Model for the cell cycle control of Caulobacter pilus assembly. DNA replication and temporally controlled transcription events are shown above a schematic of the cell cycle. Morphological changes are indicated below. CtrA∼P represses the initiation of DNA replication, activates the expression of the Class II flagellar (fla) genes, and late in the cell cycle, activates the transcription of the pilA gene. The genes encoding putative components of the pilus secretion machinery (cpaA–cpaF) are transcriptionally induced before the pilA gene.
Pilus biogenesis in Caulobacter provides a unique system to study the temporal control of organelle assembly and to understand the mechanisms that restrict its assembly to a specific cellular location. Although evidence is accumulating that many proteins are localized to specific sites in the bacterial cell (Shapiro and Losick, 2000), the mechanisms involved are unknown. In this paper, we report the identification and temporal regulation of a cluster of genes involved in pilus biogenesis in Caulobacter. The pilin subunit was purified using a novel fluorescence assay for visualizing bacterial pili, allowing the isolation of the gene encoding the pilin subunit, pilA. Six adjacent genes required for Caulobacter pilus assembly, cpaA–cpaF, were identified. Although the Caulobacter pilin subunit shares some features with Type IV pilins, the entire cluster of pilus assembly genes most closely resembles a gene cluster recently identified in the human oral pathogen, Actinobacillus actinomycetemcomitans (Inoue et al., 1998; Haase et al., 1999). Transcription of the pilA gene is regulated by the same response regulator, CtrA, that controls multiple cell cycle events, such as DNA replication, DNA methylation and flagellar biogenesis (Quon et al., 1996). The timing of pilus assembly can be altered by expressing the pilA gene from a constitutive promoter, demonstrating that transcriptional regulation of the pilin subunit gene is an important factor in determining the cell cycle pattern of pilus assembly. The identification of CtrA as a positive regulator of pilus biogenesis provides a direct link between pilus assembly and the genetic network that controls cell cycle progression.
Results
Bacteriophage φCbK binds to the polar pili on Caulobacter swarmer cells
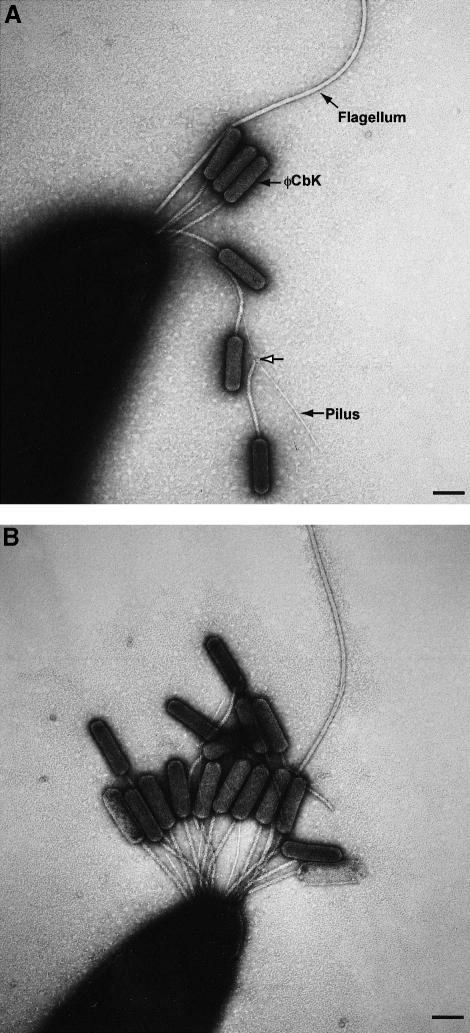
Caulobacter-specific bacteriophage include a generalized transducing phage φCr30, which binds to the S-layer surface protein, a small spherical RNA phage φCb5, which binds to the sides of pili, and a large DNA phage, φCbK, which binds specifically to the flagellar pole of swarmer cells (Schmidt, 1966; Agabian-Keshishian and Shapiro, 1971; Edwards and Smit, 1991). Mutants selected for φCbK resistance were found always to be resistant to φCb5, suggesting a relationship between φCbK infection and pili (Lagenaur et al., 1977). We have extended previous studies of φCbK infection by examining short adsorption times by electron microscopy. Cells fixed immediately after the addition of phage (∼15 s adsorption time) showed φCbK bound to the sides of pili, in addition to the cell pole (Figure 1A). After 15 min of adsorption, φCbK was bound primarily to the cell pole (Figure 1B). φCbK infection may be a two-step process in which initial attachment to pili is followed by pilus retraction and subsequent binding to a cell surface receptor. Thus, φCbK appears to be a pili-specific Caulobacter phage, and provides an explanation for the co-selection of φCbK and φCb5 resistance. We used the fact that pili are an adsorption site for phage φCbK both to purify the pilin subunit and as a genetic selection to identify genes involved in pilus biogenesis.
Fig. 1. Electron microscopy of φCbK infection of C.crescentus swarmer cells showing that φCbK is a pili-specific bacteriophage. The flagellar pole of two swarmer cells is shown. (A) φCbK infection after 15 s adsorption. φCbK attaches to pili by its long non-contractile tail (attachment site marked with a white arrow). Some of the phage are bound to the cell pole. (B) φCbK infection after 15 min adsorption. φCbK is bound predominantly to the cell pole. Scale bars ∼120 nm.
Purification of Caulobacter pili using a novel fluorescence assay and identification of the pilA locus encoding the pilin subunit
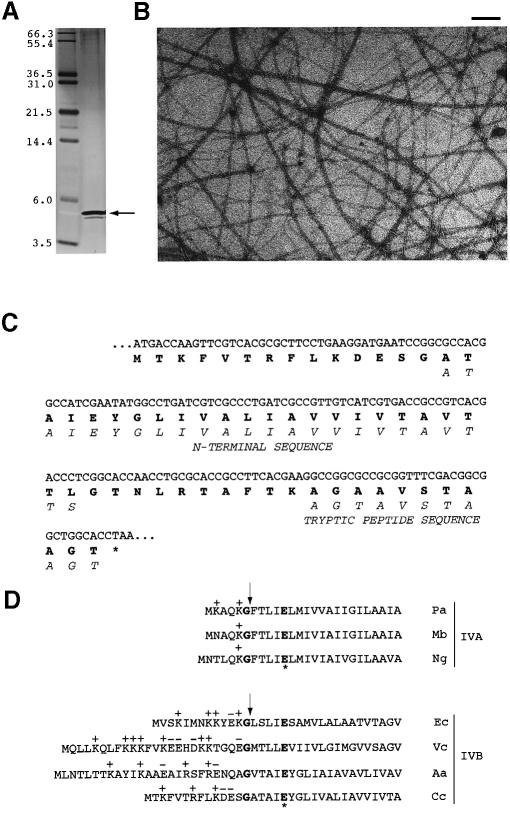
Aided by a novel fluorescence assay (described in Materials and methods) that allows the visualization of pili by light microscopy (see QuickTime movie in the Supplementary data at The EMBO Journal Online), we purified pili filaments. Pure pili allowed us to obtain the N-terminal amino acid sequence and then the gene encoding the major pilin subunit. We used a hyperpiliated strain (bNY30a), closely related to NA1000, as our source of pili and a modification of the purification protocol described by Lagenaur and Agabian (1977). Examination of the purified pili fraction by PAGE revealed one major band at ∼4.8 kDa (Figure 2A) and intact filaments by electron microscopy (Figure 2B). We obtained both N-terminal and internal tryptic peptide sequences of this 4.8 kDa protein and used this information to search the TIGR Caulobacter genome sequence for the gene encoding the pilin subunit. A single open reading frame (ORF) was identified encoding a predicted protein of 59 amino acids, which we named pilA, for the pilin subunit gene (Figure 2C). The predicted start methionine is 14 residues upstream from the first residue obtained by N-terminal sequencing, suggesting that pilin has a leader peptide that is processed after Gly14 to yield a mature protein of 45 amino acids (4360 Da). The size of the purified pilin protein (4.8 kDa) determined by gel electrophoresis closely matches that of the predicted mature pilin subunit.
Fig. 2. Purification of bNY30a pili and identification of the NA1000 pilA gene. (A) Purified pili (∼2 µg protein) visualized by silver stain. Pili were purified from a hyperpiliated C.crescentus strain bNY30a and examined by 16.5% Tris-tricine SDS–PAGE. One major band (marked with an arrow) of ∼4.8 kDa was identified as the pilin subunit. Molecular weight markers are labeled in kilodaltons. (B) Electron micrograph of purified pili, stained with uranyl acetate. This fraction contains intact pili filaments, of a single diameter. Scale bar ∼100 nm. (C) Comparison of the predicted NA1000 PilA sequence with protein sequence data obtained from purified bNY30a pili. The predicted sequence of NA1000 PilA is shown in bold. The N-terminal sequence data and tryptic peptide sequences are shown in italics. (D) Caulobacter PilA has a Type IV-like leader peptide. Shown are the N-terminal sequences for three Type IVA pilin subunits: Pseudomonas aeruginosa PAK pilin (Pa; DDBJ/EMBL/GenBank accession No. X02402), Moraxella bovis pilin (Mb; DDBJ/EMBL/GenBank accession No. M92155) and Neisseria gonorrhoeae PilE (Ng; DDBJ/EMBL/GenBank accession No. X66834). Two Type IVB pilin subunits, EPEC E.coli BfpA (Ec; DDBJ/EMBL/GenBank accession No. Z12295) and V.cholerae TcpA (Vc; DDBJ/EMBL/GenBank accession No. U09807), are aligned with C.crescentus PilA (Cc) and A.actinomycetemcomitans Flp (Aa; DDBJ/EMBL/GenBank accession No. AB005741) because they have a longer leader peptide and the N-terminal residue is not a phenylalanine. Cleavage of Type IV leader peptides occurs after a conserved glycine (marked with an arrow). A conserved glutamate (asterisk) is found at position +5.
Identification of a cpa gene cluster
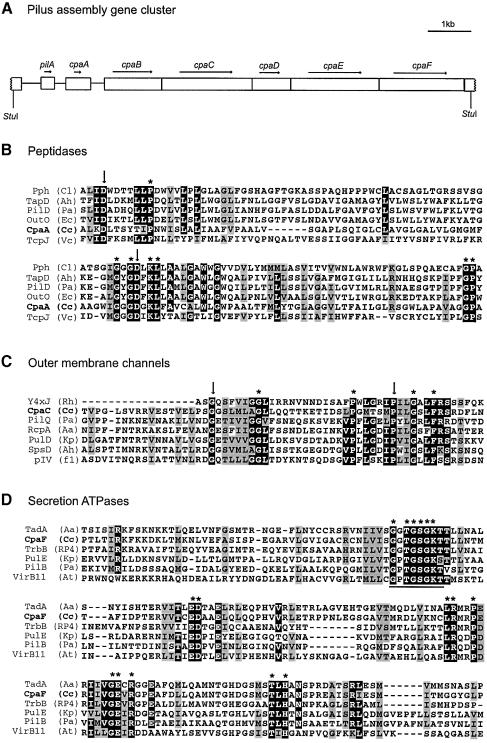
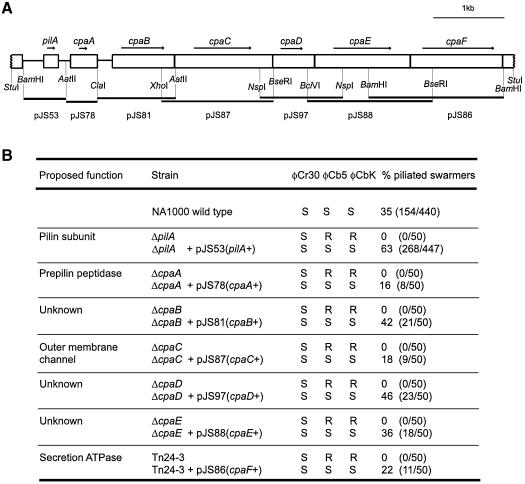
Phage φCbK resistance was used as a genetic tool to identify Caulobacter genes required for pilus assembly. The RNA phage φCb5 produces very cloudy plaques on the wild-type host NA1000, making it difficult to use in a genetic screen (Schmidt and Stanier, 1965). In contrast, φCbK produces clear plaques, which allows easy detection of phage-resistant mutants. We specifically looked for φCbK-resistant mutants that retained motility, to avoid the pleiotropic class of genes that affect multiple polar structures (Fukuda et al., 1976; Wang et al., 1993). From a collection of mini-Tn5lacZ2 insertion mutants, we identified one mutant, Tn24-3, with the appropriate phenotype. The region flanking the transposon insertion site was used to isolate an 8 kb StuI complementing fragment. Sequence analysis of this region identified seven ORFs with appropriate Caulobacter codon bias (Figure 3A). The pilA gene in this cluster encodes the pilin subunit that we identified biochemically, as described in the previous section.
Fig. 3. Sequence analysis of a pilus assembly gene cluster. (A) Diagram of an 8 kb StuI fragment that complements transposon mutant Tn24-3. Seven open reading frames were identified on this fragment, including the pilA locus encoding the pilin subunit and three other genes with homology to known pilus assembly proteins. (B) Alignment of the deduced amino acid sequence of Caulobacter CpaA with several members of the prepilin peptidase family. Only the region of the protein that is thought to contain the active site of the peptidase has been aligned. Putative active site residues are marked with an arrow. Invariant residues are marked with an asterisk. The sequences used in this alignment are Chlorobium limicola plasmid pCL1, Pph (Cl; DDBJ/EMBL/GenBank accession No. U77780), Aeromonas hydrophila TapD (Ah; DDBJ/EMBL/GenBank accession No. U20255), P.aeruginosa PilD (Pa; DDBJ/EMBL/GenBank accession No. M32066), Erwinia caratovora OutO (Ec; DDBJ/EMBL/GenBank accession No. X70049), C.crescentus CpaA (Cc) and V.cholerae TcpJ (Vc; DDBJ/EMBL/GenBank accession No. M74708). (C) Caulobacter CpaC is similar to the PulD/pIV family of outer membrane channels. Only the most highly conserved region of this protein family has been aligned. Invariant residues are marked with an asterisk and two functionally important residues are indicated with an arrow. The sequences used in this alignment are Rhizobium sp. NGR234 Y4×J (Rh; DDBJ/EMBL/GenBank accession No. AE000106), C.crescentus CpaC (Cc), P.aeruginosa PilQ (Pa; DDBJ/EMBL/GenBank accession No. L13865), A.actinomycetemcomitans RcpA (Aa; DDBJ/EMBL/GenBank accession No. AF139249), Klebsiella pneumoniae PulD (Kp; DDBJ/EMBL/GenBank accession No. M32613), A.hydrophila SpsD (Ah; DDBJ/EMBL/GenBank accession No. L41682) and coliphage f1 pIV (f1; DDBJ/EMBL/GenBank accession No. V00606). (D) Caulobacter CpaF is a member of the TrbB/VirB11 family of secretion ATPases. Only the most highly conserved region, surrounding the Walker Box (underlined), has been aligned. Invariant residues are marked with an asterisk. The sequences used in this alignment are A.actinomycetemcomitans TadA (Aa; DDBJ/EMBL/GenBank accession No. AF152598), C.crescentus CpaF (Cc), plasmid RP4 TrbB (RP4; DDBJ/EMBL/GenBank accession No. M93696), K.pneumoniae PulE (Kp; DDBJ/EMBL/GenBank accession No. M32613), P.aeruginosa PilB (Pa; DDBJ/EMBL/GenBank accession No. M32066) and Agrobacterium tumefaciens plasmid pTiC58, VirB11 (At; DDBJ/EMBL/GenBank accession No. X53264).
Downstream of pilA are six genes, which we named cpaA–cpaF for Caulobacter pilus assembly. The protein encoded by cpaA is similar to the C-terminal region of prepilin peptidases (Figure 3B), which are bifunctional enzymes that process and methylate substrates required for Type II protein secretion or Type IV pilus assembly (Nunn and Lory, 1991; Russel, 1998). Downstream of cpaA is a group of five closely spaced genes, which most likely form a single transcriptional unit. Three of these genes, cpaB, cpaD and cpaE, encode proteins of unknown function. The protein encoded by cpaC is related to the PulD/pIV family of proteins, refered to as secretins (Nouwen et al., 1999). These proteins are required for extracellular secretion, filamentous phage assembly and pilus biogenesis, and are thought to function as a gated channel in the outer membrane (Russel et al., 1997). The most highly conserved portion of these secretins (Russel, 1994), containing an invariant glycine and proline residue, has been aligned with CpaC (Figure 3C).
The fifth gene in this cluster, cpaF, encodes a protein related to the TrbB/VirB11 family of Walker Box-containing proteins. The TrbB/VirB11 family of proteins are involved in DNA uptake, extracellular secretion and pilus assembly (Hobbs and Mattick, 1993). Mutations in the Walker Box motif abolish their function in vivo, suggesting that ATP binding or hydrolysis is critical to their function (Turner et al., 1993; K.M.Stephens et al., 1995). The biochemical function of the TrbB/Virb11 protein family is not understood, but it is thought that the energy from ATP hydrolysis is used to export proteins across the inner membrane (Russel, 1998). An alignment of CpaF with other members of the TrbB/VirB11 protein family is shown in Figure 3D.
pilA, cpaA, cpaB, cpaC, cpaD, cpaE and cpaF are required for Caulobacter pilus assembly
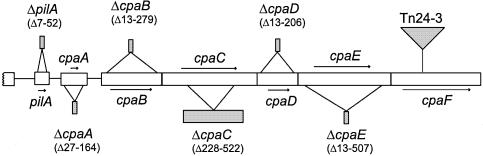
To test directly the function of the genes in the pilA–cpa cluster, we constructed in-frame deletion mutants of pilA, cpaA, cpaB, cpaC, cpaD and cpaE in wild-type C.crescentus strain NA1000 (Figure 4). All six chromosomal in-frame deletion strains, as well as the transposon insertion in the cpaF gene, were found to be resistant to the pili-specific phages φCbK and φCb5, but remained sensitive to φCr30 (Figure 5B). These mutant strains all exhibited normal morphology and motility (data not shown) and were found to be pili-less by electron microscopy. Phage sensitivity and piliation could be restored for each mutant strain by a plasmid carrying the corresponding wild-type allele (Figure 5A), demonstrating that the phenotype was due to the effect of a single gene mutation. The degree of complementation, as determined by the percentage of piliated swarmer cells, varied among the different constructs, perhaps due to inappropriate levels of expression of the particular gene product. Taken together, these data indicate that the seven genes contained on the 8 kb StuI complementing fragment are required for pilus assembly in Caulobacter.
Fig. 4. Schematic of the mutations used in this study. Six in-frame deletions were made in the Caulobacter pilA–cpaA cluster. The amino acids deleted in each gene are indicated. The mutation in cpaF was a mini-Tn5lacZ2 insertion (Tn24-3).
Fig. 5. Complementing clones and electron microscopy data. (A) Diagram of the plasmid clones used to complement each of the mutants shown in Figure 4. (B) Summary of complementation data. All seven mutants were found to be sensitive (S) to the generalized transducing phage φCr30, and resistant (R) to the pili-specific phages φCbK and φCb5, and pili-less as determined by electron microscopy. Phage sensitivity and piliation were restored by a plasmid carrying the corresponding wild-type allele, whereas the vector alone had no effect (not shown). The number of swarmer cells examined for each strain by electron microscopy is shown in parentheses.
Pilin transcription is activated by the CtrA response regulator and CtrA∼P binds the pilA promoter
We examined Caulobacter piliation by electron microscopy using the negative stain uranyl acetate. Consistent with previous work (Sommer and Newton, 1988), we found that 35% of swarmer cells (n = 440) were piliated and that predivisional cells were rarely piliated (1.4%, n = 278). Piliated swarmer cells had 1–7 pili, with an average of 2.6 pili per cell (data not shown). Pili are lost at the swarmer to stalked cell transition and reappear only in the progeny swarmer cell, after cell division (Sommer and Newton, 1988). To understand the mechanism that controls the timing of pilus assembly, we have examined the promoter of the pilA gene and its time of expression during the cell cycle.
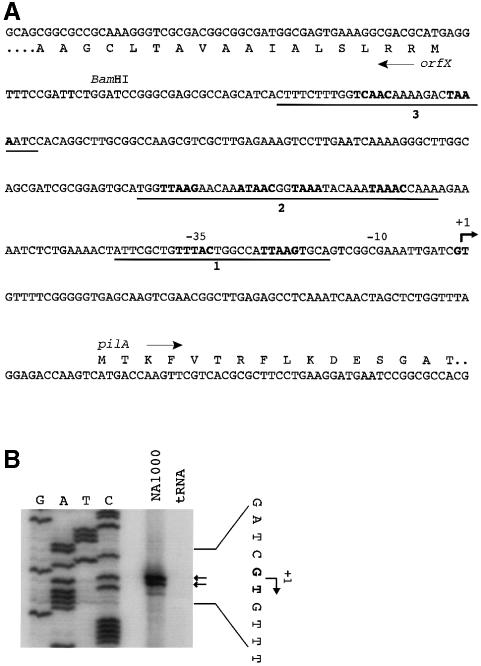
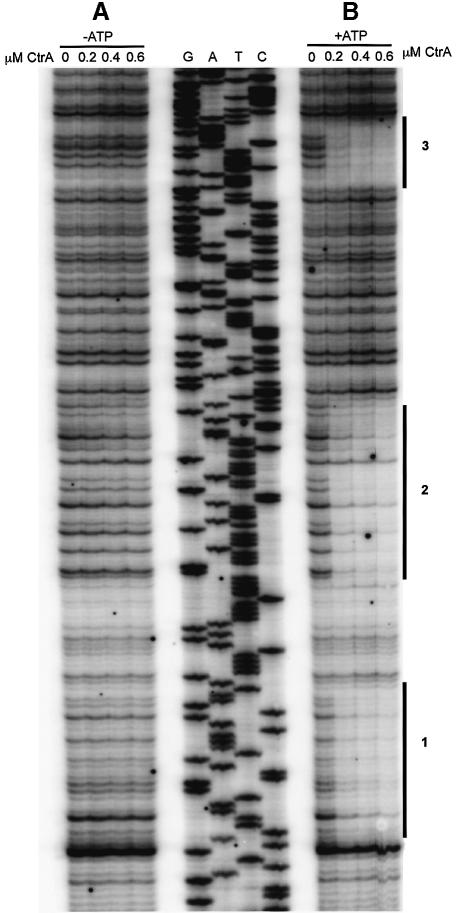
The start site of pilA transcription was identified by primer extension analysis using a primer located 30 nucleotides downstream of the start codon of the pilA gene (Figure 6A). The start site was localized to two adjacent nucleotides, a G and T (Figure 6B). Inspection of the promoter region revealed a potential CtrA binding site overlaping the –35 region (Figure 6A), a configuration observed in many genes known to be regulated by CtrA (Quon et al., 1996). To test directly whether transcription of the pilA gene is regulated by CtrA, we constructed a pilA promoter fusion to lacZ (PpilA–lacZ). This construct (pJS70) extended from a BamHI site (–230 nt) to just downstream of the ATG start codon (+90 nt). An ORF in the opposite direction to pilA is just upstream of the BamHI site (orfX) so we believe that this fragment contains the entire pilin promoter (Figure 6A). β-galactosidase activity was measured at 28°C in wild-type C.crescentus strain NA1000, and in ctrA401ts (LS2195), a temperature-sensitive lethal allele of ctrA, each carrying pJS70. At the permissive temperature (28°C), ctrA401ts is a partial loss of function mutation (Quon et al., 1996). Promoter activity of PpilA–lacZ in ctrA401ts (882 ± 136 Miller units) is 17% of wild type (5294 ± 1200 Miller units), demonstrating that CtrA is a positive regulator of pilA transcription. Using a similar promoter fragment, we performed DNase I protection analysis using purified CtrA protein, which was phosphorylated in vitro (Reisenauer et al., 1999). We found that CtrA∼P, but not unphosphorylated CtrA, binds to the pilA promoter in three regions (Figure 7), coinciding with four consensus CtrA binding sites (Figure 6A). Taken together, these results demonstrate that CtrA directly regulates pilA transcription.
Fig. 6. Identification of the pilA transcription start site. (A) Sequence of the pilA promoter region. The pilA transcription start site is marked with a +1 and an arrow. Three regions which are protected by CtrA∼P (see Figure 7) are underlined, and CtrA binding motifs are marked in bold. A divergently transcribed gene, orfX, is 316 nt upstream of the predicted PilA methionine. (B) Start site of pilA transcription determined by primer extension. Ten micrograms of total RNA obtained from NA1000 or 10 µg of yeast tRNA were hybridized with the pilinrev2 primer and transcribed with Superscript II at 42°C. A sequence ladder generated with the same primer was used to determine the start site of transcription. The sequencing ladder corresponds to the non-coding strand. The doublet start site indicated with small arrows, and designated in bold, was observed with several different RNA preparations and two additional primers (data not shown).
Fig. 7. DNase I protection of the pilin promoter with purified CtrA∼P. Purified CtrA was phosphorylated with MBP–EnvZ fusion protein in vitro in a reaction mixture containing ATP. A sequence ladder generated using the pilinrev2 primer was used to identify the protected bases. (A) In the absence of ATP, no protected regions were observed. CtrA concentrations in the footprint reactions are indicated. (B) Three distinct regions of the promoter were protected with CtrA∼P and are marked with a solid black line. These protected regions coincide with CtrA binding motifs in the pilA promoter shown in Figure 6A.
Transcription of the pilA gene occurs only in late predivisional and swarmer cells
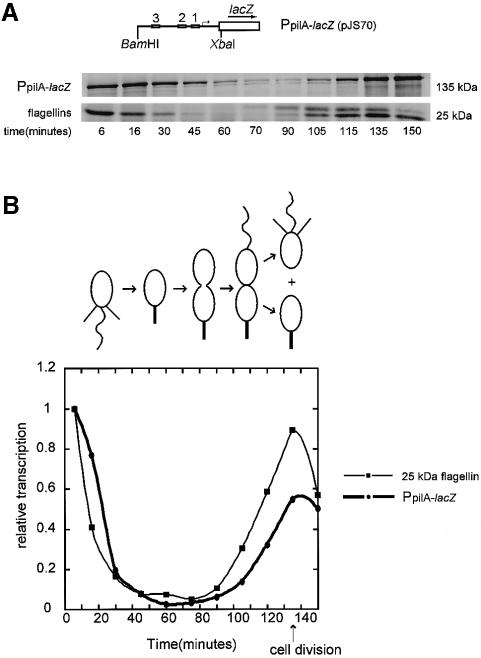
The cell cycle pattern of pilA transcription was determined in synchronized wild-type cultures containing a PpilA–lacZ fusion on a low copy number plasmid (pJS70). The cells were pulse labeled with [35S]methionine during the cell cycle. Immunoprecipitation with anti-β-galactosidase antibodies was used to determine the timing of pilA transcription (Figure 8A). We compared pilA transcription to the time of expression of the well-characterized flagellin genes (Figure 8B). We found that pilA transcription is cell cycle regulated and its pattern of expression is similar to the timing of pilus assembly determined by electron microscopy.
Fig. 8. Transcription of pilA is cell cycle controlled. (A) Diagram of the PpilA–lacZ fusion construct, pJS70. Wild-type C.crescentus strain NA1000 carrying pJS70 was synchronized and allowed to proceed through the cell cycle. The synthesis of β-galactosidase was monitored by pulse labeling cells with [35S]methionine for 5 min at the times indicated and immunoprecipitating with anti-β-galactosidase antibody. Labeled β-galactosidase was resolved on a 10% SDS–PAGE gel. As a control, the flagellin subunits were immunoprecipitated from the same labeled cell extracts. (B) The β-galactosidase and the 25 kDa flagellin bands were quantitated using a phosphoimager and were normalized to the value of the most intense band in each series. A diagram of the Caulobacter cell cycle is shown. The pattern of pilA transcription coincides with the pattern of 25 kDa flagellin gene transcription and is similar to the timing of pilus assembly observed by electron microscopy.
To determine whether the time of pilA transcription affects the timing of pilus assembly, the pilA gene was expressed throughout the cell cycle by replacing its native promoter with the constitutive Ptac promoter. This Ptac–pilA construct (pJS96), on a high copy plasmid (∼20 copies per cell), was mated into wild-type C.crescentus strain NA1000 and the percentage of piliated swarmers and predivisional cells was determined by electron microscopy (Table I). Constitutive expression of pilA resulted in a higher percentage of swarmer cells with visible pili. In addition, we observed a significant increase in the percentage of piliated predivisional cells, indicating a shift in the timing of pilus assembly to an earlier point in the cell cycle. These results demonstrate that the time of transcription of the pilin subunit gene plays an important role in controlling the time of pilus assembly.
Table I. Constitutive transcription of pilA alters timing of pili formation.
| NA1000+ vector (pJS71) | NA1000+ Ptac–pilA (pJS96) | |
|---|---|---|
| Swarmers | 38%a | 78% |
| Predivisionals | 4% | 21% |
Wild-type C.crescentus NA1000 carrying either vector only (pJS71) or Ptac–pilA (pJS96) were examined by electron microscopy and the percentage of piliated swarmer cells and piliated predivisional cells (with a flagellum) was determined.
aPercentage piliation (n = 100).
Discussion
In this paper, we provide both biochemical and genetic data demonstrating that a 4.8 kDa protein (4360 Da predicted) is the major subunit of Caulobacter pili. The pilin subunit, encoded by the pilA gene, has a leader peptide similar to that found in Type IV pilin (see Figure 2D), which is characterized by a net positive charge, cleavage after a glycine residue, and a conserved glutamate at position +5 (Strom and Lory, 1993). The presence of a Type IV-like leader peptide suggests that Caulobacter PilA is processed by a specific prepilin peptidase. The gene downstream of pilA, cpaA, encodes a protein with homology to the C-terminal region of prepilin peptidases. Prepilin peptidase PilD, first identified in P.aeruginosa, cleaves prepilin after a conserved glycine residue and methylates the resulting N-terminal phenylalanine (Nunn and Lory, 1991). Based on our N-terminal sequence data, the N-terminal residue of mature PilA is an alanine. It is known that prepilin peptidase can methylate an alanine residue (Strom and Lory, 1991); however, we do not know whether PilA is methylated. Structure–function studies of P.aeruginosa PilD and the Vibrio cholerae homolog TcpJ suggest that the methylase and peptidase activities reside in separate parts of the protein (Strom et al., 1993; Pepe and Lory, 1998; LaPointe and Taylor, 2000). Caulobacter CpaA is similar only to the domain required for peptidase activity, and lacks a conserved cytoplasmic loop implicated in methyltransferase function. We propose that CpaA is a functional peptidase, but not a methylase, and is required for the processing of PilA prepilin. A direct role of CpaA in the processing of PilA awaits the production of an anti-pilin antibody.
There is a striking similarity in gene order and predicted function between the Caulobacter pilA–cpa cluster and the flp–rcp–tadA genomic region of A.actinomycetemcomi tans (Figure 9A). Three genes in the Actinobacillus cluster, flp, rcpA and tadA, have been implicated in pilus assembly (Inoue et al., 1998; Haase et al., 1999; D.Figurski, personal communication). Flp was purified from Actinobacillus pili and is likely to be the major pilin subunit (Inoue et al., 1998). Flp is a small protein that has a Type IV-like leader sequence and the predicted mature protein is 40% identical to Caulobacter PilA. Interestingly, the gene orfB, which is just downstream of flp, encodes a protein that is similar to the C-terminal region of prepilin peptidases. OrfB may be a functional peptidase, required for processing the Flp subunit, analogous to that proposed for the Caulobacter proteins CpaA and PilA. The genes orfC, rcpB and orf4 encode proteins that have similar hydropathy profiles to Caulobacter CpaB, CpaD and CpaE, and may also be required for Actinobacillus pili formation. Both Caulobacter CpaC and Actinobacillus rough colony protein A (RcpA) are members of the PulD/pIV family of outer membrane secretins. Several of these secretins have been purified and shown to form a toroid shaped channel, with an inner diameter of ∼5–10 nm (Linderoth et al., 1997; Bitter et al., 1998; Nouwen et al., 1999). The size of this channel is sufficiently large that an intact pilus filament could exit through it (Bitter et al., 1998). It is likely that RcpA is required for Actinobacillus pilus assembly, analogous in function to CpaC, but a ΔrcpA strain has not yet been reported. CpaF and TadA are both related to the TrbB/VirB11 family of putative secretion ATPases, and are likely to serve analogous functions in Caulobacter and Actinobacillus, respectively. In fact, a transposon insertion in the tadA gene is defective in pilus assembly (D.Figurski, personal communication). Recently, gene clusters similar to those found in Caulobacter and Actinobacillus have been identified, and the gene order has been shown to be largely conserved in a group of diverse bacteria, suggesting that this is a novel family of pilus biogenesis genes (S.Kachlany, P.Planet, M.Bhattacharjee, E.Kollia, R.DeSalle, D.Fine and D.Figurski, personal communication).
Here, we report the first molecular genetic study of a chromosomally encoded pilus system in the α-purple subdivision of bacteria. It is likely that the Caulobacter pilus system will be present and conserved in other α-purple bacteria. In support of this, we have identified predicted proteins similar to cpaB–cpaF in Sinorhizobium meliloti, a member of the α-group, but the contigs are currently too short to determine whether they are derived from a single genomic region (our unpublished data). In addition, we have identified potential cpaF genes in various α-purple bacteria by Southern blot analysis, including Asticcacaulis biprosthecum, Asticcacaulis excentricus, Caulobacter bacteroides and Hyphomicrobium zavarzinii (data not shown).
Using C.crescentus as a model system, we have a unique opportunity to understand the connection between the bacterial cell cycle and the control of pilus assembly. We have identified the pilA promoter and shown that it is activated by the response regulator CtrA. CtrA is a global regulator, which controls multiple cell cycle events, including the time of initiation of DNA replication, DNA methylation, cell division and flagellar biogenesis (Quon et al., 1996, 1998; Kelly et al., 1998). The proper turnover and phosphorylation of CtrA are necessary for cell cycle progression (Domian et al., 1997). This study demonstrates that CtrA controls pilus biogenesis, and therefore provides a direct link between pilus assembly and the regulation of cell cycle progression. Induction of pilA gene transcription occurs later in the cell cycle than all other known direct targets of CtrA (Stephens and Shapiro, 1993; C.M.Stephens et al., 1995; Quardokus et al., 1996; Quon et al., 1996, 1998; Reisenauer et al., 1999). Reisenauer et al. (1999) have proposed that differences in the affinity of CtrA for its DNA binding sites and interaction with additional regulatory proteins may be responsible for the differential induction of CtrA target genes. Clearly, the complex structure of the pilin promoter region, which contains four CtrA binding sites, needs to be dissected further in order to understand how CtrA binding affinity, and possibly additional factors, controls the timing of pilA transcription.
The cell cycle transcription profile of all the genes in the pilA–cpa cluster has recently been determined using DNA microarray analysis of synchronous populations of cells (M.Laub and L.Shapiro, personal communication). The genes cpaB–cpaF are co-induced 15 min before the pilA gene. The gene encoding the putative Caulobacter prepilin peptidase, cpaA, is induced slightly after the cpaB–cpaF group. Thus, these six genes, likely to be encoding components of the pilin secretion apparatus, are all induced prior to pilA gene transcription and pilus assembly. We propose a model for the cell cycle control of pilus assembly in which the secretion machinery is localized to the flagellar pole of the late predivisional cell, but pili filaments are not assembled until after cell division when a sufficient amount of processed pilin subunits has accumulated (Figure 9B). Consistent with our model, we observed pili on predivisional cells when the pilA gene was constitutively transcribed from the Ptac promoter, demonstrating that a functional secretion apparatus is present in the predivisional cell. Expression of the pilin subunit appears to be the final step in pilus assembly.
Our results contrast with previous reports, which identified an 8.0 kDa protein as the pilin subunit (Smit et al., 1981) that led to a post-transcriptional model for pilus assembly (Smit and Agabian, 1982; Smit, 1987). In order to resolve this issue, we cloned the gene encoding the reported 8.0 kDa protein and generated an in-frame deletion. Strains lacking the 8.0 kDa protein are still piliated and sensitive to φCbK infection (J.Skerker and L.Shapiro, unpublished data), demonstrating that the 8.0 kDa protein is not required for Caulobacter pilus assembly and is not the major pilin subunit. The identification of the genes encoding the pilin subunit and pilus assembly proteins presented here have allowed experiments that revealed transcriptional regulation as the basis of temporally controlled pilus biogenesis.
Materials and methods
Bacterial strains, phage and growth conditions
The bacterial strains, phage and plasmids used in this study are listed in Table II. Escherichia coli strains were grown at 37°C in Luria–Bertani broth supplemented with ampicillin (50 µg/ml), tetracycline (10 µg/ml), kanamycin (50 µg/ml) or spectinomycin (50 µg/ml) as necessary. Caulobacter crescentus strains were grown at 30°C in peptone yeast extract (PYE) or minimal medium (M2G) supplemented with spectinomycin (25 µg/ml), kanamycin (20 µg/ml) or tetracycline (1 µg/ml) as necessary. For solid media, PYE-agar plates were supplemented with spectinomycin (100 µg/ml), kanamycin (20 µg/ml) or tetracycline (2 µg/ml). Plasmids were transferred into Caulobacter by conjugation or electroporation (Ely, 1991). Preparations of φCbK and φCr30 were routinely obtained by plate lysis using NA1000 as the host as previously described (Ely, 1991). Strain bNY30a served as the host for φCb5, because it yielded higher titer lysates than NA1000 (Lagenaur and Agabian, 1977).
Table II. Bacterial strains, phage and plasmids.
| Relevant characteristic(s) | Reference or source | |
|---|---|---|
| Caulobacter crescentus | ||
| NA1000 | CB15 syn-1000; synchronizable wild-type derivative | Evinger and Agabian (1977) |
| bNY30a | hyperpiliated derivative of CB13B1a, originally SW16-Pil 200 | J.Poindexter |
| LS2195 | NA1000 ctrA401ts | Quon et al. (1996) |
| LS2549 | NA1000 cpaF::miniTn5lacZ2; Tn24-3, KanR | this work |
| LS3118 | NA1000 ΔpilA | this work |
| LS3214 | NA1000 ΔcpaA | this work |
| LS3215 | NA1000 ΔcpaB | this work |
| LS3216 | NA1000 ΔcpaC | this work |
| LS3217 | NA1000 ΔcpaD | this work |
| LS3218 | NA1000 ΔcpaE | this work |
| Escherichia coli | ||
| S17-1 | conjugal transfer of plasmids to C.crescentus | Simon et al. (1983) |
| DH10B | host for cloning and DNA sequencing | Gibco-BRL |
| Bacteriophage | ||
| φCr30 | generalized transducing phage | Ely and Johnson (1977) |
| φCb5 | pili-specific RNA phage | Bendis and Shapiro (1970) |
| φCbK | swarmer-specific DNA phage | Lagenaur et al. (1977) |
| Plasmids | ||
| pKS+ | pBluescript II KS+ Cloning vector, AmpR | Stratagene |
| pCR 2.1-TOPO | cloning vector for PCR products, AmpR, KanR | Invitrogen |
| pNPTS138 | pLitmus 38-derived vector with added nptI, sacB, and RK2 oriT, KanR | M.R.K.Alley |
| pRK290-20R | pRK290 with pIC20R polylinker, TetR | M.R.K.Alley |
| pRKlac290 | lacZ transcriptional fusion vector, pRK290 derivative, TetR | Gober and Shapiro (1992) |
| p10-1 | Tn24-3 complementing cosmid; pLAFR5 based,TetR | this work |
| pJS2 | 8kb StuI fragment derived from p10-1 in pKS+, AmpR | this work |
| pJS53 | BamHI–AatII fragment containing pilA in pRK290-20R, TetR | this work |
| pJS56 | in-frame deletion construct for pilA in pNPTS138, KanR | this work |
| pJS70 | pilA promoter–lacZ fusion in pRKlac290, TetR | this work |
| pJS71 | SpecR derivative of pBBR1MCS | this work |
| pJS78 | AatII–ClaI from pJS2 containing cpaA in pJS71, SpecR | this work |
| pJS81 | ClaI–AatII from pJS2 containing cpaB in pJS71, SpecR | this work |
| pJS86 | BamHI–BamHI from pJS2 containing cpaF in pJS71, SpecR | this work |
| pJS87 | XhoI–BseRI from pJS2 containing cpaC in pJS71, SpecR | this work |
| pJS88 | BciVI–BseRI from pJS2 containing cpaE in pJS71, SpecR | this work |
| pJS89 | in-frame deletion construct for cpaA in pNPTS138, KanR | this work |
| pJS90 | in-frame deletion construct for cpaB in pNPTS138, KanR | this work |
| pJS91 | in-frame deletion construct for cpaC in pNPTS138, KanR | this work |
| pJS92 | in-frame deletion construct for cpaD in pNPTS138, KanR | this work |
| pJS93 | in-frame deletion construct for cpaE in pNPTS138, KanR | this work |
| pJS96 | Ptac–pilA construct in pJS71, SpecR | this work |
| pJS97 | NspI–NspI from pJS2 containing cpaD in pJS71, SpecR | this work |
Electron microscopy of φCbK infection complexes and Caulobacter piliation
Mid-log phase cultures were fixed with an equal volume of pili fix (5% glutaraldehyde in 50 mM cacodylate–HCl buffer pH 7.4). To examine φCbK adsorption, 100 µl of cell culture and phage lysate (1010 p.f.u./ml) were mixed and either immediately fixed with pili fix solution or incubated for 15 min at room temperature prior to fixation. After fixation (10 min at 25°C), the cells were collected by centrifugation, washed with 1 ml of PYE and resuspended in 100 µl of PYE. Samples were stained with 1% uranyl acetate and examined at 40 000× magnification on a Philips EM 300 operated at 80 kV. The same fixation and staining procedure was used to prepare samples for examining Caulobacter piliation, except that no φCbK was added.
Fluorescent labeling of φCbK and visualization of Caulobacter pili
Phage lysates were concentrated by centrifugation in a TLA100.2 rotor at 60 000 r.p.m. for 2 h. The pellet was resuspended in PYE to a final titer of 1011–1012 p.f.u./ml. Phage were stained with YO-PRO-1 iodide (Molecular Probes, Inc.) by mixing 20 µl of phage with 1 µl of dye stock (1 mM in DMSO) and incubated at 4°C in the dark for 48 h (Hennes and Suttle, 1995). To visualize Caulobacter pili, 1 µl each of fluorescent phage and purified pili were mixed on a glass slide, and observed using a Nikon Eclipse E800 microscope and a HQ-FITC filter set (Chroma Technology Corporation). Free phage were easily distinguished from the ordered lines of phage that are pili coated with fluorescent φCbK. Real-time imaging of fluorescently labeled pili was performed with an intensified CCD camera (VideoScope International, ICCD-1000F) at 250–500× final magnification (see the Supplementary data at The EMBO Journal Online). The video signal was recorded on S-VHS tapes with a VCR (Panasonic AG-6740), digitized with a Pentium based PC (P3; 500 MHz) using a Matrox Marvel G200 frame grabber card (30 f.p.s.), and converted into a QuickTime movie using Adobe Premiere 5.0.
Pili purification and N-terminal sequencing
Caulobacter pili were purified from a spontaneous hyperpiliated mutant bNY30a, originally called SW16-Pil 200 (Lagenaur and Agabian, 1977). The purification procedure was a modification of the method described previously (Lagenaur and Agabian, 1977). The fluorescent assay described above was used to monitor fractions throughout the purification. One liter of C.crescentus bNY30a was grown to stationary phase and the cells were pelleted by centrifugation at 6000 g for 30 min. The pili-rich supernatant was adjusted to 0.5 M NaCl, 2% w/v polyethylene glycol 8000 (PEG 8000) and the solution was incubated overnight at 4°C. Pili were collected by centrifugation at 15 000 g for 30 min. The pellet was resuspended in 40 ml of PYE, centrifuged at 2500 g for 1 h, and the cleared supernatant was spun for 20 h at 38 000 r.p.m. (SW41 Ti rotor) to collect the pili. The pellet was resuspended in 5 ml TMC (Tris–HCl pH 7.5, 2 mM MgCl2, 1 mM CaCl2), centrifuged at 15 000 g for 15 min to remove any insoluble material, and then CsCl was added to a final density of 1.236 g/ml. After 64 h of centrifugation (SW 50.1 rotor, 60 000 r.p.m.), a pilus band, visible at the approximate center of the gradient, was collected and dialyzed against distilled water. Purified pili were analyzed by electrophoresis on 16.5% Tris–tricine gels, and the separated proteins were transferred to Immobilon-PSQ membrane (Millipore, Inc.). After staining with Coomassie Blue, the pilin band was excised and the N-terminal sequence was determined by Edman degradation (Stanford PAN facility). In addition, tryptic peptides derived from an in-gel digest were isolated by HPLC, and N-terminal sequence obtained (Stanford PAN facility).
Isolation of φCbK-resistant mutant Tn24-3 and complementing clones
We screened a library of insertion mutants generated with the transposon mini-Tn5lacZ2 (de Lorenzo et al., 1990; M.R.K.Alley, unpublished collection) to identify strains that were φCbK resistant but had no motility defects. Mutant Tn24-3 was φCbKR, had no motility defects, and kanamycin resistance was 100% linked to φCbKR as determined by co-transduction analysis with φCr30. We cloned the genomic DNA flanking the insertion site and used a 300 bp PstI–BamHI fragment to probe a pLAFR5 cosmid library (Alley et al., 1991). We identified cosmid p10-1, which restored phage sensitivity to mutant Tn24-3. An 8 kb StuI complementing fragment was then isolated from p10-1 using the same 300 bp probe. Sequence analysis revealed that Tn24-3 has an insertion in the cpaF gene.
Sequence analysis
The 8 kb StuI fragment was sequenced using Thermo Sequenase (USB Corp.) and BigDye Terminators (PE Biosystems). Preliminary sequence data were obtained from the C.crescentus genome project at The Institute for Genomic Research (http://www.tigr.org). The sequence of the 8 kb StuI fragment has been deposited in the DDBJ/EMBL/GenBank database under accession No. AF229646.
Allelic replacement experiments
We constructed in-frame deletions of pilA and cpaA–cpaE to test directly their function in Caulobacter pilus assembly. Two PCR products extending ∼500 bp from either side of the deletion point were synthesized using Advantage HF polymerase (Clontech). The resulting products were first cloned into pCR2.1-TOPO (Invitrogen) and sequenced. An EcoRI–XbaI fragment of PCR product ‘A’ and an XbaI–HindIII fragment of PCR product ‘B’ were ligated into pNPTS138 (see Table II). The engineered XbaI site produces two new in-frame codons (Ser, Arg). This strategy was used to generate in-frame deletions for pilA, cpaA, cpaB, cpaD and cpaE. An in-frame deletion of cpaC was constructed by removing an internal PstI fragment. Allelic replacement was performed by a two-step sacB counterselection procedure (Stephens et al., 1996). Colony PCR, using a set of primers flanking each deletion point, was used to screen for the loss of the wild-type allele and the presence of the in-frame deletion.
DNase I protection experiments
Native CtrA was overexpressed in E.coli using the pET expression system (Novagen), solubilized from inclusion bodies with 6 M guanidine–HCl and purified by Q-Sepharose chromatography with a continuous NaCl gradient. CtrA eluted at ∼280 mM NaCl and was 95% pure by SDS–PAGE (K.Ryan, unpublished data). Template DNA for the pilA promoter was generated by PCR using the foward primer spefw (5′-GGACTAGTGACCGGCCGAGACCGGCAGG-3′) and the reverse primer xbarev (5′-GCTCTAGACGTGACGAACTTGGTCATGAC-3′). In vitro phosphorylation of purified CtrA and DNase I protection experiments was performed as described (Reisenauer et al., 1999).
Primer extension and cell cycle analysis of pilA transcription
RNA was isolated from mid-log phase NA1000 cultures using a hot phenol procedure (Sambrook et al., 1989) and primer extensions performed using the pilinrev2 primer (5′-gccgtggcgcggattcatc-3′) as described (Ausubel et al., 1989). A sequencing ladder using the pilinrev2 primer and pJS2 as template was used to determine the start site nucleotide. The pilA promoter–lacZ fusion (PpilA–lacZ) was generated using the primers spefw and xbarev. The resulting product was digested with BamHI (∼300 nt from the pilA start codon) and XbaI, and cloned into pRKlac290, to give pJS70. Wild-type strain NA1000 carrying pJS70 was synchronized by Ludox density gradient centrifugation (Evinger and Agabian, 1977). Analysis and quantitation of cell cycle transcription were performed as described (Stephens and Shapiro, 1993).
Supplementary data
Supplementary data to this paper (QuickTime movie) are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank M.R.K.Alley for his collection of φCbK-resistant mutants, J.Poindexter for providing strains and phage, J.Smit for providing DNA sequence and antibodies, F.Thomas and N.Ghori for instruction and advice in electron microscopy, D.Winant for performing N-terminal and tryptic peptide sequencing, M.Laub for providing unpublished microarray data, K.Ryan for supplying purified CtrA and E.Haase for helpful discussion regarding the flp-rcp-tad locus. Preliminary sequence data were obtained from The Institute for Genomic Research (http://www.tigr.org). Sequencing of C.crescentus was accomplished with support from the Department of Energy. We particularly thank David Figurski and members of his laboratory for access to their unpublished material and informing us of the widespread nature of the Actinobacillus gene cluster. We thank members of the Shapiro laboratory for critical reading of this manuscript. This work was supported by NIH grant GM32506/5120MZ and J.M.S. was supported by a NSF Predoctoral Fellowship.
References
- Agabian-Keshishian N. and Shapiro,L. (1971) Bacterial differentiation and phage infection. Virology, 44, 46–53. [DOI] [PubMed] [Google Scholar]
- Alley M.R., Gomes,S.L., Alexander,W. and Shapiro,L. (1991) Genetic analysis of a temporally transcribed chemotaxis gene cluster in Caulobacter crescentus.Genetics, 129, 333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds) (1989) Current Protocols in Molecular Biology. John Wiley and Sons, New York, NY. [Google Scholar]
- Bendis I. and Shapiro,L. (1970) Properties of Caulobacter ribonucleic acid bacteriophage fCb5. J. Virol., 6, 847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitter W., Koster,M., Latijnhouwers,M., de Cock,H. and Tommassen,J. (1998) Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol. Microbiol., 27, 209–219. [DOI] [PubMed] [Google Scholar]
- Bradley D.E. (1974) The adsorption of Pseudomonas aeruginosa pilus-dependent bacteriophages to a host mutant with nonretractile pili. Virology, 58, 149–163. [DOI] [PubMed] [Google Scholar]
- Brinton C.C. Jr (1971) The properties of sex pili, the viral nature of ‘conjugal’ genetic transfer systems and some possible approaches to the control of bacterial drug resistance. CRC Crit. Rev. Microbiol., 1, 105–160. [DOI] [PubMed] [Google Scholar]
- Brun Y.V., Marczynski,G. and Shapiro,L. (1994) The expression of asymmetry during Caulobacter cell differentiation. Annu. Rev. Biochem., 63, 419–450. [DOI] [PubMed] [Google Scholar]
- Christie P.J. (1997) Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J. Bacteriol., 179, 3085–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D.B. (1993) Bacterial Conjugation. Plenum Press, New York, NY. [Google Scholar]
- Click E.M. and Webster,R.E. (1997) Filamentous phage infection: required interactions with the TolA protein. J. Bacteriol., 179, 6464–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V., Herrero,M., Jakubzik,U. and Timmis,K.N. (1990) Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing and chromosomal insertion of cloned DNA in Gram-negative eubacteria. J. Bacteriol., 172, 6568–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domian I.J., Quon,K.C. and Shapiro,L. (1997) Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell, 90, 415–424. [DOI] [PubMed] [Google Scholar]
- Edwards P. and Smit,J. (1991) A transducing bacteriophage for Caulobacter crescentus uses the paracrystalline surface layer protein as a receptor. J. Bacteriol., 173, 5568–5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B. (1991) Genetics of Caulobacter crescentus. Methods Enzymol., 204, 372–384. [DOI] [PubMed] [Google Scholar]
- Ely B. and Johnson,R.C. (1977) Generalized transduction in Caulobacter crescentus.Genetics, 87, 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evinger M. and Agabian,N. (1977) Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol., 132, 294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda A., Miyakawa,K., Iida,H. and Okada,Y. (1976) Regulation of polar surface structures in Caulobacter crescentus: pleiotropic mutations affect the coordinate morphogenesis of flagella, pili and phage receptors. Mol. Gen. Genet., 149, 167–173. [DOI] [PubMed] [Google Scholar]
- Gober J.W. and Shapiro,L. (1992) A developmentally regulated Caulobacter flagellar promoter is activated by 3′ enhancer and IHF binding elements. Mol. Biol. Cell, 3, 913–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase E.M., Zmuda,J.L. and Scannapieco,F.A. (1999) Identification and molecular analysis of rough-colony-specific outer membrane proteins of Actinobacillus actinomycetemcomitans. Infect. Immun., 67, 2901–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennes K.P. and Suttle,C.A. (1995) Direct counts of viruses in natural waters and laboratory cultures by epifluorescence microscopy. Limnol. Oceanogr., 40, 1050–1055. [Google Scholar]
- Hobbs M. and Mattick,J.S. (1993) Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol., 10, 233–243. [DOI] [PubMed] [Google Scholar]
- Hultgren S.J., Jones,C.H. and Normark,S. (1996) Bacterial adhesins and their assembly. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella Cellular and Molecular Biology, Vol. 2. ASM Press, Washington, DC, pp. 2730–2756. [Google Scholar]
- Inoue T., Tanimoto,I., Ohta,H., Kato,K., Murayama,Y. and Fukui,K. (1998) Molecular characterization of low-molecular-weight component protein, Flp, in Actinobacillus actinomycetemcomitans fimbriae. Microbiol. Immunol., 42, 253–258. [DOI] [PubMed] [Google Scholar]
- Jacobson A. (1972) Role of F pili in the penetration of bacteriophage fl. J. Virol., 10, 835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A.J., Sackett,M.J., Din,N., Quardokus,E. and Brun,Y.V. (1998) Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev., 12, 880–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagenaur C. and Agabian,N. (1977) Caulobacter crescentus pili: structure and stage-specific expression. J. Bacteriol., 131, 340–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagenaur C., Farmer,S. and Agabian,N. (1977) Adsorption properties of stage-specific Caulobacter phage fCbK. Virology, 77, 401–407. [DOI] [PubMed] [Google Scholar]
- LaPointe C.F. and Taylor,R.K. (2000) The type 4 prepilin peptidases comprise a novel family of aspartic acid proteases. J. Biol. Chem., 275, 1502–1510. [DOI] [PubMed] [Google Scholar]
- Linderoth N.A., Simon,M.N. and Russel,M. (1997) The filamentous phage pIV multimer visualized by scanning transmission electron microscopy. Science, 278, 1635–1638. [DOI] [PubMed] [Google Scholar]
- Lowe M.A., Holt,S.C. and Eisenstein,B.I. (1987) Immunoelectron microscopic analysis of elongation of type 1 fimbriae in Escherichia coli.J. Bacteriol., 169, 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouwen N., Ranson,N., Saibil,H., Wolpensinger,B., Engel,A., Ghazi,A. and Pugsley,A.P. (1999) Secretin PulD: association with pilot PulS, structure and ion-conducting channel formation. Proc. Natl Acad. Sci. USA, 96, 8173–8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D.N. and Lory,S. (1991) Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc. Natl Acad. Sci. USA, 88, 3281–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole G.A. and Kolter,R. (1998) Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol., 30, 295–304. [DOI] [PubMed] [Google Scholar]
- Pepe J.C. and Lory,S. (1998) Amino acid substitutions in PilD, a bifunctional enzyme of Pseudomonas aeruginosa. Effect on leader peptidase and N-methyltransferase activities in vitro and in vivo.J. Biol. Chem., 273, 19120–19129. [DOI] [PubMed] [Google Scholar]
- Quardokus E., Din,N. and Brun,Y.V. (1996) Cell cycle regulation and cell type-specific localization of the FtsZ division initiation protein in Caulobacter. Proc. Natl Acad. Sci. USA, 93, 6314–6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quon K.C., Marczynski,G.T. and Shapiro,L. (1996) Cell cycle control by an essential bacterial two-component signal transduction protein. Cell, 84, 83–93. [DOI] [PubMed] [Google Scholar]
- Quon K.C., Yang,B., Domian,I.J., Shapiro,L. and Marczynski,G.T. (1998) Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl Acad. Sci. USA, 95, 120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisenauer A., Quon,K. and Shapiro,L. (1999) The CtrA response regulator mediates temporal control of gene expression during the Caulobacter cell cycle. J. Bacteriol., 181, 2430–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel M. (1994) Mutants at conserved positions in gene IV, a gene required for assembly and secretion of filamentous phages. Mol. Microbiol., 14, 357–369. [DOI] [PubMed] [Google Scholar]
- Russel M. (1998) Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J. Mol. Biol., 279, 485–499. [DOI] [PubMed] [Google Scholar]
- Russel M., Linderoth,N.A. and Sali,A. (1997) Filamentous phage assembly: variation on a protein export theme. Gene, 192, 23–32. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schmidt J.M. (1966) Observations on the adsorption of Caulobacter bacteriophages containing ribonucleic acid. J. Gen. Microbiol., 45, 347–353. [DOI] [PubMed] [Google Scholar]
- Schmidt J.M. and Stanier,R.Y. (1965) Isolation and characterization of bacteriophages active against stalked bacteria. J. Gen. Microbiol., 39, 95–107. [DOI] [PubMed] [Google Scholar]
- Shapiro L. and Losick,R. (2000) Dynamic spatial regulation in the bacterial cell. Cell, 100, 89–98. [DOI] [PubMed] [Google Scholar]
- Simon R., Priefer,U. and Puhler,A. (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. BioTechnology, 1, 784–790. [Google Scholar]
- Smit J. (1987) Localizing the subunit pool for the temporally regulated polar pili of Caulobacter crescentus. J. Cell Biol., 105, 1821–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit J. and Agabian,N. (1982) Caulobacter crescentus pili: analysis of production during development. Dev. Biol., 89, 237–247. [DOI] [PubMed] [Google Scholar]
- Smit J., Hermodson,M. and Agabian,N. (1981) Caulobacter crescentus pilin. Purification, chemical characterization and NH2-terminal amino acid sequence of a structural protein regulated during development. J. Biol. Chem., 256, 3092–3097. [PubMed] [Google Scholar]
- Sommer J.M. and Newton,A. (1988) Sequential regulation of developmental events during polar morphogenesis in Caulobacter crescentus: assembly of pili on swarmer cells requires cell separation. J. Bacteriol., 170, 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto G.E. and Hultgren,S.J. (1999) Bacterial adhesins: common themes and variations in architecture and assembly. J. Bacteriol., 181, 1059–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens C.M. and Shapiro,L. (1993) An unusual promoter controls cell-cycle regulation and dependence on DNA replication of the Caulobacter fliLM early flagellar operon. Mol. Microbiol., 9, 1169–1179. [DOI] [PubMed] [Google Scholar]
- Stephens C.M., Zweiger,G. and Shapiro,L. (1995) Coordinate cell cycle control of a Caulobacter DNA methyltransferase and the flagellar genetic hierarchy. J. Bacteriol., 177, 1662–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens C., Reisenauer,A., Wright,R. and Shapiro,L. (1996) A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc. Natl Acad. Sci. USA, 93, 1210–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens K.M., Roush,C. and Nester,E. (1995) Agrobacterium tumefaciens VirB11 protein requires a consensus nucleotide-binding site for function in virulence. J. Bacteriol., 177, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom M.S. and Lory,S. (1991) Amino acid substitutions in pilin of Pseudomonas aeruginosa. Effect on leader peptide cleavage, amino-terminal methylation and pilus assembly. J. Biol. Chem., 266, 1656–1664. [PubMed] [Google Scholar]
- Strom M.S. and Lory,S. (1993) Structure–function and biogenesis of the type IV pili. Annu. Rev. Microbiol., 47, 565–596. [DOI] [PubMed] [Google Scholar]
- Strom M.S., Bergman,P. and Lory,S. (1993) Identification of active-site cysteines in the conserved domain of PilD, the bifunctional type IV pilin leader peptidase/N-methyltransferase of Pseudomonas aeruginosa. J. Biol. Chem., 268, 15788–15794. [PubMed] [Google Scholar]
- Turner L.R., Lara,J.C., Nunn,D.N. and Lory,S. (1993) Mutations in the consensus ATP-binding sites of XcpR and PilB eliminate extracellular protein secretion and pilus biogenesis in Pseudomonas aeruginosa. J. Bacteriol., 175, 4962–4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall D. and Kaiser,D. (1999) Type IV pili and cell motility. Mol. Microbiol., 32, 1–10. [DOI] [PubMed] [Google Scholar]
- Wang S.P., Sharma,P.L., Schoenlein,P.V. and Ely,B. (1993) A histidine protein kinase is involved in polar organelle development in Caulobacter crescentus. Proc. Natl Acad. Sci. USA, 90, 630–634. [DOI] [PMC free article] [PubMed] [Google Scholar]