Abstract
Aim
To identify gene-expression signatures predicting cytarabine response by an integrative analysis of multiple clinical and pharmacological end points in acute myeloid leukemia (AML) patients.
Materials & methods
We performed an integrated analysis to associate the gene expression of diagnostic bone marrow blasts from acute myeloid leukemia (AML) patients treated in the discovery set (AML97; n = 42) and in the independent validation set (AML02; n = 46) with multiple clinical and pharmacological end points. Based on prior biological knowledge, we defined a gene to show a therapeutically beneficial (detrimental) pattern of association of its expression positively (negatively) correlated with favorable phenotypes such as intracellular cytarabine 5´-triphosphate levels, morphological response and event-free survival, and negatively (positively) correlated with unfavorable end points such as post-cytarabine DNA synthesis levels, minimal residual disease and cytarabine LC50.
Results
We identified 240 probe sets predicting a therapeutically beneficial pattern and 97 predicting detrimental pattern (p ≤ 0.005) in the discovery set. Of these, 60 were confirmed in the independent validation set. The validated probe sets correspond to genes involved in PIK3/PTEN/AKT/mTOR signaling, G-protein-coupled receptor signaling and leukemogenesis. This suggests that targeting these pathways as potential pharmacogenomic and therapeutic candidates could be useful for improving treatment outcomes in AML.
Conclusion
This study illustrates the power of integrated data analysis of genomic data as well as multiple clinical and pharmacologic end points in the identification of genes and pathways of biological relevance.
Keywords: acute myeloid leukemia, cytarabine, gene-expression profiles, genetic signatures
The most widely used front-line chemotherapeutic regimens for the treatment of acute myeloid leukemia (AML) is cytarabine (Ara-C) combined with an anthracycline. Although these regimens have been demonstrated to achieve complete remission in approximately 90% of AML patients, the clinical outcome is unsatisfactory, with 50–60% of young patients having long-term disease-free survival [1–5]. Similarly, among adults 60 years or older of age, the clinical outcome is dismal, with only 40–55% of patients achieving complete remission [1]. Therefore, further improvements in the therapy are needed. Improving the understanding of factors that influence the efficacy of cytarabine in the context of its use in contemporary therapy could suggest ways to improve treatment outcomes.
The development of resistance to first-line chemotherapy remains a common reason for treatment failure with adverse side effects contributing to morbidity and mortality in AML [6–8]. As cytarabine is one of the mainstays in AML chemotherapy treatment, a better understanding of the factors contributing to cytarabine resistance could potentially improve treatment outcome, as well as associated adverse side effects.
The cytotoxic activity of cytarabine depends on its intracellular conversion to cytarabine 5´-triphosphate (Ara-CTP). Pharmacokinetic genes including deoxycytidine kinase, cytidine deaminase, cytosolic 5´-nucleotidases and human equilibrative nucleoside transporter have been implicated in influencing cytarabine response [9]. In addition, DNA polymerase, topoisomerases, bcl-2 and TP53 have been associated with Ara-C sensitivity and clinical response [10]. However, the exact molecular mechanisms and pathways responsible for cytarabine-mediated leukemic cell death are still not clear. Recent efforts to identify gene-expression signatures associated with cytarabine resistance have mostly used human or murine leukemia cell lines and/or expression arrays that do not represent the entire genome [11–15]. We have previously interrogated the association of cytarabine cytotoxicity with genome-wide gene expression, as well as genetic variation in lymphoblast cell lines from healthy individuals [16].
However, the identification of a true genetic signature predicting cytarabine response requires comprehensive evaluation in AML patients. In the last two decades, gene-expression profiles have provided valuable information on the new biologically and prognostically relevant risk groups in AML [17]. In the present study, we have taken a unique approach in order to identify the gene-expression signatures predicting response in AML patients by the comprehensive analysis of multiple pharmacokinetic, pharmacodynamic (cytarabine dependent) and clinical end points in patients enrolled in two clinical trials. The unique feature of our study is the simultaneous and integrative analysis of multiple in vivo and in vitro pharmacological (cytarabine dependent) and clinical end points in AML patients to identify gene-expression signatures that exhibit a therapeutically meaningful pattern of associations with all the end points. Our study is the first to explore genome-wide gene expression in order to predict meaningful patterns of association with multiple pharmacologic and clinical end points. The identification of gene-expression signatures predicting response that are driven by cytarabine-related end points may be useful in designing more effective chemotherapeutic regimens.
Materials & methods
Patients
This study included 42 subjects from the AML97 clinical trial [18,19] and 46 subjects from the AML02 clinical trial [5] (clinicaltrials.gov identifier NCT00136084 [101]) with previously collected microarray gene-expression data.
For the AML97 trial, patients aged 21 years or younger with all subtypes of AML, except acute promyelocytic leukemia with the t(15;17) PML-RARα fusion, were eligible for enrollment. Patients were randomly assigned to receive either a short daily infusion of cytarabine (arm A) or a continuous infusion of cytarabine (arm B). Patients in arm A received five daily 2-h infusions of cytarabine (500 mg/m2/day) and five daily 30 mini-infusions of cladribine (9 mg/m2), which began 24 h after the start of the first cytarabine infusion. There was a 2-h intervals between the end of each cladribine infusion and the start of each cytarabine infusion. Patients in arm B received cytarabine (500 mg/m2/day) as a 120-h continuous infusion and five daily 30 mini-infusions of cladribine (9 mg/m2), which began 24 h after the start of the cytarabine infusion.
The AML02 trial enrolled AML patients aged 22 years or younger excluding acute promy-elocytic leukemia or Down syndrome patients, but those with all other subtypes of de novo or secondary AML, as well as patients with mixed-lineage leukemia, were eligible. Patients were randomized to receive induction I therapy containing either high-dose cytarabine (3 g/m2 intravenously over 3 h, given every 12 h on days 1, 3 and 5) or low-dose cytarabine (100 mg/m2 intravenously over 30 min, given every 12 h on days 1–10) plus daunorubicin (50 mg/m2 intravenously over 6 h on days 2, 4 and 6) and etoposide (100 mg/m2 intravenously over 4 h on days 2–6) (Figure 1). Subsequent therapy was adapted based on minimal residual disease (MRD) as assessed by flow cytometry and diagnostic risk features. Details of the study outcome of this protocol are described elsewhere [5].
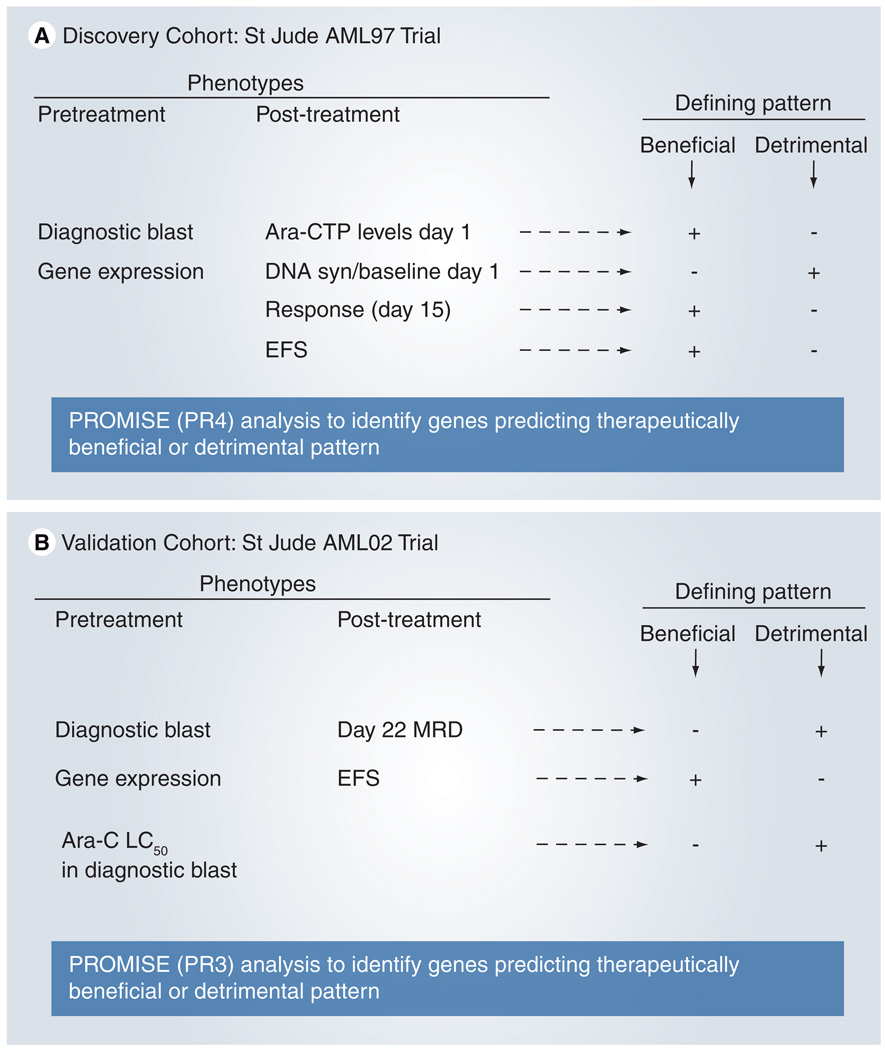
Figure 1. Treatment schema and our strategy for using PROMISE to identify genes associated with beneficial or detrimental patterns of association.
(A) AML97 test set, (B) AML02 validation set.
Ara-C: Cytarabine; Ara-CTP: Cytarabine 5´-triphosphate; DNA syn: DNA synthesis; EFS: Event-free survival; MRD: Minimal residual disease; PROMISE: Projection onto the Most Interesting Statistical Evidence.
The study designs were approved by the institutional review boards of participating institutions. Written informed consent was obtained from patients, parents or guardians, as well as assent from the patients, as appropriate, before enrollment in the study.
Pharmacology & clinical end points
For the AML97 trial, four pharmacological and clinical end points were assessed. Intracellular concentrations of Ara-CTP were measured in leukemia cells from bone marrow samples obtained on the first day (ara-CTP1; after cytarabine alone), as previously described [18]. In addition, the rate of DNA synthesis in leukemia cells from the bone marrow was determined at diagnosis and on day 1 of therapy, as previously described [18]. From these measurements of DNA synthesis rates, we computed the log-ratio of the day 1 rates to the baseline rate [20]. Response was determined by morphologic examination of a bone marrow aspirate collected after the first course of chemotherapy (day 15 after initiation of chemotherapy). Complete response was defined as less than 5% blasts in the marrow and no definitive evidence of leukemia by morphology (e.g., Auer rods) or karyotype. No response was defined as 15% or more blasts in the marrow. In this era when morphology was the only modality to evaluate response, all other cases were considered undefined or as partial responses. To more accurately define response to therapy, bone marrow evaluation was repeated weekly as necessary to classify the undefined cases. The duration of event-free survival (EFS) was defined as the time from diagnosis until the date of failure (induction failure, relapse, death or the development of a second malignancy) or until the date of last contact of all event-free survivors. Patients who did not attain a complete remission after two courses of therapy were considered to have failed at time zero.
For the AML02 trial, three pharmacologic and clinical end points were assessed. On day 22 after the start of therapy, MRD was measured by flow cytometry, as previously described [21]. The sensitivity of leukemic cells to cytarabine was determined in vitro with the use of the 4-day 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) drug-resistance assay [22]. Briefly, bone marrow was obtained at diagnosis, and mononuclear cells were isolated using Ficoll-Hypaque density-gradient centrifugation within 24 h. Cells were resuspended in modified RPMI-1640 medium, which was supplemented with 20% fetal calf serum, penicillin (100 IU/ml), streptomycin (100 µg/ml) and Fungizone™ (Invitrogen; 0.125 µg/ml), as well as ITS medium supplement containing insulin (5 ng/ml), transferring (5 µg/ml), and sodium selenite (5 ng/ml), as previously described [22]. If necessary, samples were further enriched to achieve more than 80% blasts by the use of magnetic cell sorting (Miltenyi Biotech, Germany). The cells were treated with varying concentrations of cytarabine (range 0.002–2.5 ng/µl) to determine the LC50 value. In addition, EFS was defined as described above.
Gene-expression profiling
Gene-expression profiling of leukemia cells from the diagnostic bone marrow of 42 patients in the test set (AML97) and 46 in the validation set (AML02) was performed using Affymetrix U133 gene chips [23]. Details regarding RNA isolation, the labeling of cRNA and the scanning of Affymetrix arrays have been published previously [23]. Primary data for the AML97 subjects are available [102].
Statistical analysis
A novel statistical procedure, Projection onto the Most Interesting Statistical Evidence (PROMISE [20]), was applied to the expression, pharmacological and clinical data from the AML97 clinical trial in order to identify candidate probe sets (genes) with therapeutically beneficial or detrimental patterns of association. For the discovery analysis of AML97 data, a probe set was defined to have a therapeutically beneficial pattern of association if its expression levels positively associated with day 1 Ara-CTP levels, morphological response and the duration of EFS and negatively associated with the rate of DNA synthesis on day 1 relative to that at baseline. Positive associations with day 1 Ara-CTP levels, morphological response and EFS are defined as therapeutically beneficial because higher Ara-CTP levels in leukemic cells should improve the prospects of achieving remission. In addition, morphological response is a desirable clinical outcome and a long EFS is the ultimate objective of therapy, respectively. A negative association with the DNA synthesis rate relative to baseline is defined as therapeutically beneficial as the DNA synthesis rate is an indicator of leukemic proliferation and the pharmacodynamic objective of delivering cytarabine is to induce apoptosis in leukemic cells by interfering with DNA synthesis. A therapeutically detrimental pattern was defined as the opposite of the beneficial pattern. To account for therapy, the analysis was stratified by arm and the amendment of intrathecal therapy. P-values were computed using 10,000 permutations. Additional details are provided in the Supplementary Tables 1–4 & Supplementary Figures 1 & 2; www.futuremedicine. com/doi/suppl/10.2217/pgs.10.191.
As a validation analysis, PROMISE was used to examine the association between candidate probe sets’ expression and pharmacological and clinical end point data in the AML02 clinical trial. In this analysis, a probe set was defined as having a therapeutically beneficial pattern of association if its expression levels negatively associated with LC50 values, day 22 MRD levels and if it was positively associated with the duration of EFS. Negative associations with LC50 and day 22 MRD were defined as therapeutically beneficial because lower LC50 values indicate the leukemia is more sensitive to Ara-C, whereas lower MRD levels indicate greater efficacy of clinical treatment, respectively. A positive association with EFS duration was defined as therapeutically beneficial as EFS is the ultimate objective of therapy. To account for therapy, the analysis was stratified by arm. P-values were computed using 10,000 permutations.
Gene-set enrichment analyses were incorporated into each PROMISE analysis, as previously described. The molecular function, pathway and biological process gene-set definitions were used in these analyses [103]. For each gene set, the enrichment statistic was defined as the average of the absolute values of the PROMISE statistics of member probe sets. For each gene set, a p-value was determined using the same set of 10,000 permutations utilized in the PROMISE analysis.
The associations between EFS and other end points were measured by Jung’s statistics, and associations between other end points were measured by Spearman’s correlation. The significance was determined by 10,000 permutations, as stratified by treatment arms. The rank-sum test was used to identify probe sets with differential expression, according to the presence or absence of a core-binding karyotype (inv[16] or t[8;21]). All statistical analyses were performed using R software [104].
Results
Patient characteristics
Table 1 lists the demographic features of the patients enrolled in the discovery (n = 42) and validation (n = 46) cohorts. No significant differences were observed between the two arms, as well as the two cohorts in age, gender and white blood cell count. The cytogenetic features were also distributed similarly in the two cohorts. The clinical outcomes for discovery (AML97) [19] and validation cohorts [5] are published elsewhere. Table 2 lists the various pharmacokinetic, pharmacodynamic and clinical end points for AML patients included in our study. The initial administration of cytarabine before other agents allowed us to perform pharmacology studies from samples obtained at day 1, without the potentially confounding effects of other remission-induction agents.
Table 1.
Demographic data for 42 AML97 patients used as a discovery cohort and 46 AML02 patients used as the validation cohort for this study.
| Clinical features | Discovery set induction I arm |
Validation set randomized arm |
|||||
|---|---|---|---|---|---|---|---|
| Arm A | Arm B | Total | HDAC | LDAC | Total | ||
| Gender | Female | 8 | 16 | 24 | 14 | 12 | 26 |
| Male | 11 | 7 | 18 | 7 | 13 | 20 | |
| Age | <10 years | 8 | 14 | 22 | 11 | 14 | 25 |
| ≥10 years | 11 | 9 | 20 | 10 | 11 | 21 | |
| WBC | <50 | 11 | 16 | 27 | 17 | 11 | 28 |
| ≥50 | 8 | 7 | 15 | 4 | 14 | 18 | |
| Race | Black | 4 | 6 | 10 | 3 | 6 | 9 |
| White | 12 | 9 | 21 | 14 | 17 | 31 | |
| Other | 3 | 8 | 11 | 4 | 2 | 6 | |
| Cytogenetics | inv(16) | 2 | 3 | 5 | 3 | 7 | 10 |
| t(8;21) | 4 | 3 | 7 | 1 | 2 | 3 | |
| t(9;11) | 2 | 0 | 2 | 2 | 2 | 4 | |
| 11q23 | 2 | 4 | 6 | 5 | 1 | 6 | |
| Normal | 0 | 5 | 5 | 2 | 7 | 9 | |
| Others | 9 | 8 | 17 | 8 | 6 | 14 | |
Distribution of demographics was not statistically different within a cohort for arms or between the discovery and validation cohort (p > 0.05) for all parameters.
HDAC: High-dose cytarabine; LDAC: Low-dose cytarabine; t: Translocation; WBC: White blood cell.
Table 2.
Pharmacological and outcome variables used in the PROMISE analysis in AML97 (discovery cohort) PR4 and AML02 (validation cohort) PR3 analysis.
| Variable | Median (range) | Median (range) |
|---|---|---|
| Discovery cohort AML97 | Arm A | Arm B |
| Day 1 Ara-CTP | 0.28 (0.09–2.43) | 0.22 (0.01–0.88) |
| DNA synthesis (100† day1/baseline) | 8.06 (1.27–62.59) | 19.7 (2.4–148) |
| OS† | 0.368 (0.104) | 0.565 (0.10) |
| EFS† | 0.368 (0.104) | 0.522 (0.10) |
| Validation cohort AML02 | HDAC arm | LDAC arm |
| IC50 | 0.28 (0.01–5.0) | 0.394 (0.0055–5.0) |
| Day 22 MRD† | 12 neg, 4 int, 5 high | 15 neg, 2 int, 8 high |
| OS‡ | 0.905 (0.068) | 0.706 (0.102) |
| EFS‡ | 0.857 (0.081) | 0.640 (0.111) |
Intermediate: 0.1% ≤ MRD ≤ 1%; High: MRD >1%.
5-year OS or EFS estimates with Peto and Pike standard error.
3-year OS or EFS estimates with Peto and Pike standard error.
Ara-CTP: Cytarabine 5´-triphosphate; EFS: Event-free survival; HDAC: High-dose cytarabine; Int: Intermediate;
LDAC: Low-dose cytarabine; MRD: Minimal residual disease; Neg: Negative; OS: Overall survival; PROMISE: Projection onto the Most Interesting Statistical Evidence.
Associations among end points
Statistical associations among clinical and pharmacologic end points in our study were generally concordant with the previously established biological relationships used to define the beneficial and detrimental patterns for PROMISE analysis (Table 3). Each statistically significant (p < 0.05) correlation among end point variables agreed with the biological relationship used to define the PROMISE statistic. Only DNA synthesis in AML97 demonstrated some statistically insignificant discordant associations (p > 0.07 in all cases). Moreover, the directions of these discordant associations were inconsistent across the four therapy-defined strata (data not shown). Thus, because the biological relationship of DNA synthesis and Ara-CTP is well-established, and our data did not provide any statistically compelling evidence to the contrary, we opted to include DNA synthesis as a ‘detrimental’ pharmacologic end point in our PROMISE analysis.
Table 3.
Associations among pharmacologic and clinical end points in the AML97 and AML02 clinical cohorts.
| Cohort | Clinical end points | Association statistics |
Consistency of trend |
p-value |
|---|---|---|---|---|
| AML97 | Day 1 Ara-CTP and EFS | 0.117 | Yes | 0.386 |
| DNA synthesis and EFS | 0.238 | No | 0.073 | |
| Response and EFS | 0.368 | Yes | 0.001 | |
| Day 1 Ara-CTP vs response | 0.277 | Yes | 0.105 | |
| DNA synthesis vs response | 0.171 | No | 0.321 | |
| Day 1 Ara-CTP vs DNA synthesis | 0.111 | No | 0.56 | |
| AML02 | LC50 vs EFS | 0.089 | Yes | 0.531 |
| MRD vs EFS | −0.286 | Yes | 0.022 | |
Ara-CTP: Cytarabine 5´-triphosphate; EFS: Event-free survival; MRD: Minimal residual disease.
Identification of genes predicting therapeutically beneficial or detrimental patterns
We first applied the PROMISE procedure to identify genes with therapeutically beneficial or detrimental patterns of association with the end points in the AML97 discovery cohort. In this analysis, a gene was considered to have a therapeutically beneficial pattern of association (Figure 1A) if its expression demonstrated a positive arm-adjusted correlation with Ara-CTP concentrations (higher Ara-CTP levels increase the probability of clearing leukemic cells), morphological response (complete response is better than partial response, which is better than no response) and EFS duration, as well as a negative arm-adjusted correlation with day 1 DNA synthesis rates relative to baseline. The PROMISE analysis with four end points (PR4) identified 240 probe sets (annotated to 228 known genes) with a therapeutically beneficial pattern, and 97 probe sets (annotated to 94 known genes) with a detrimental pattern as being significant at the p ≤ 0.005 level (for complete list please see Supplementary Table 1).
Validation of gene-expression signatures predicting cytarabine response
To validate the findings in the discovery set, we performed a PROMISE analysis of data collected from the independent AML02 validation cohort consisting of 46 patients. For this cohort, a gene was considered to demonstrate a therapeutically beneficial pattern (Figure 1B) if its expression demonstrated negative arm-adjusted correlations with the LC50 (greater LC50 indicates greater resistance) of cytarabine for diagnostic leukemia cells and categorized levels of MRD (low, intermediate and high, <0.1%, 0.1–1% and >1%, respectively) after one course of therapy, and a positive arm-adjusted correlation with EFS. A gene with opposite correlations was considered to have a detrimental pattern of association. This analysis of three end points in the AML02 study was denoted PR3. A probe set initially identified by the PR4 analyses at the 0.005 level (by a two-sided test) was considered to be validated if the PR3 analysis indicated the same association pattern (beneficial or detrimental) and gave a two-sided p-value of less than 0.10 (which is equivalent to a significance at the 0.05 level in an appropriately defined one-sided test).
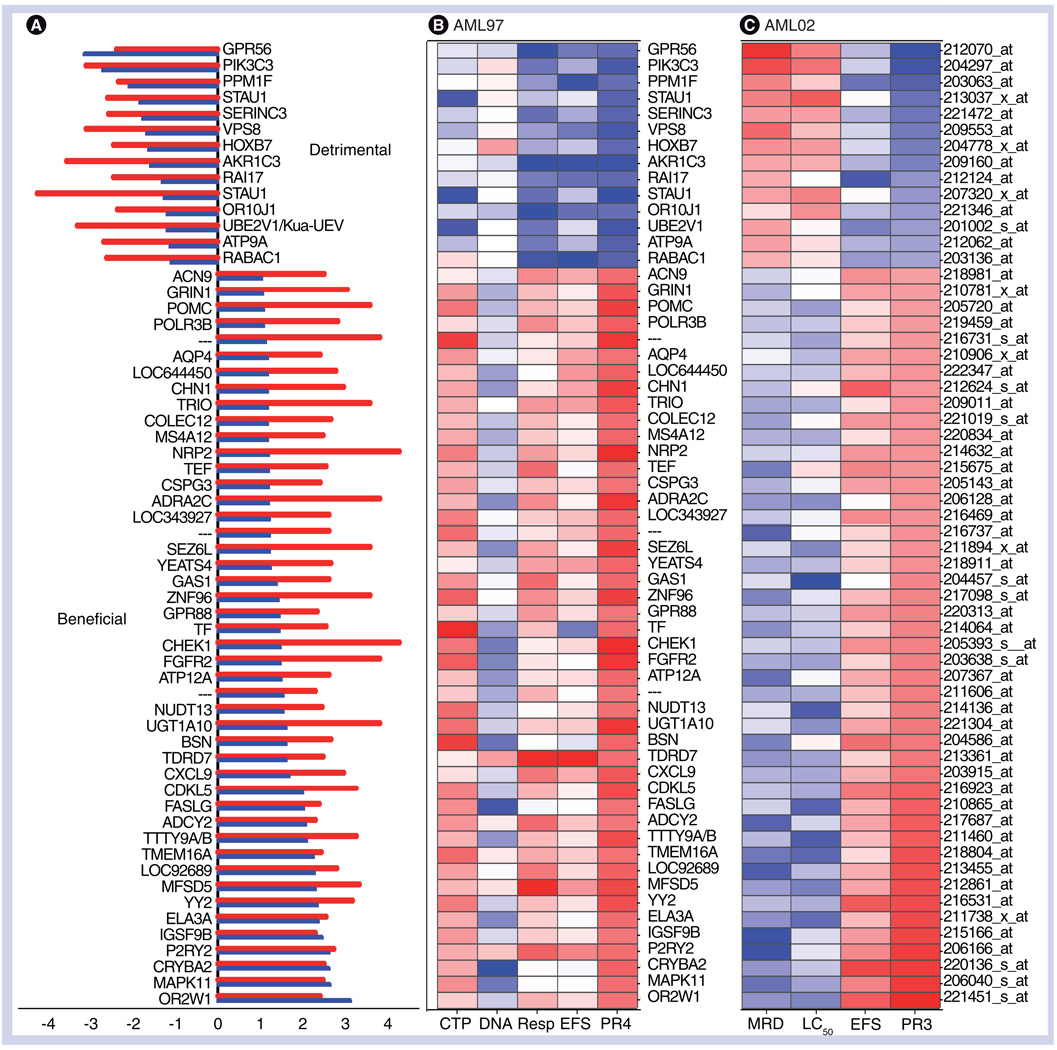
The PR3 analysis validated 60 unique probe sets (53 known genes) initially identified in the PR4 discovery analysis (Supplementary Table 2). Of these, 46 probes demonstrated a therapeutically beneficial pattern (5% of 240 = 12 probe sets expected to be erroneously selected by chance) and 14 demonstrated a detrimental pattern (5% of 97 = 5 probe sets expected to be erroneously selected by chance) with multiple end points (Figure 2A). The association of each probe with an individual phenotype is depicted in Figure 2B & 2C for discovery (AML97) and validation sets (AML02), respectively.
Figure 2. Therapeutically beneficial and detrimental patterns of association detected by the PROMISE procedure.
(A) PR3-validated results of the PR4 analysis included 46 probe sets associated with beneficial, and 14 with detrimental patterns in both datasets. The x-axis values give a log10 p-value with sign defined by the pattern (negative for detrimental and positive for beneficial). The result from AML97 is shown by the red bar and the result from AML02 is shown by the blue bar. (B & C) Heat maps depicting the statistical significance of the correlation of 60 validated probe sets with each end point in the AML97 test set and AML02 validation set. Each row represents a gene corresponding to 2A, and each column represents phenotypes, such as intracellular cytarabine 5´-triphosphate levels; DNA synthesis relative to baseline; Resp and EFS (time-related censor for EFS); LC50 (LC50 of cytarabine in diagnostic leukemia cells); MRD; and EFS. PR4 and PR3 indicate the stat values corresponding to the PROMISE analysis in the discovery and validation sets, respectively. Colors are assigned according to the signed log10 p-value.
EFS: Event-free survival; MRD: Minimal residual disease; PROMISE: Projection onto the Most Interesting Statistical Evidence; Resp: Response after the first course of therapy.
Our validated list included several genes with potential biological relevance. Notable genes exhibiting a therapeutically detrimental pattern included PIK3C3, which is involved in the PI3K/PTEN/Akt/mTOR signaling cascade, plays a crucial role in cell growth and survival and is activated in various cancers, including leukemia [24]. The expression of HOXB7 has a detrimental pattern of association. Earlier studies have demonstrated that the overexpression of HOX genes, HOXB5 and HOXB2, are associated with poor outcome [25], and proposed a role of homeobox genes in leukemogenesis [26]. AKR1C3, an aldo-keto reductase, has been demonstrated to regulate myeloipoiesis owing to its ability to metabolize prostaglandin D2. Genetic variants in AKR1C3 have been associated with a risk of childhood leukemia [27] and an inhibition of AKR1C3 expression in myeloid leukemia cell lines promotes cell differentiation [28]. Recently, a combination of benzafibrate and medroxyprogesterone acetate, which both inhibit AKR1C3, has been suggested for the treatment of AML [29]. Other detrimental genes of interest included UB2EV1 (encoding ubiquitin conjugating enzyme that has been shown to inhibit apoptosis through NF-κB activation [30]), GPR56 (a G-protein-coupled receptor that has been implicated in tumorigenesis), PPM1F and RAI17.
Genes with a therapeutically beneficial pattern included: FASLG (codes for the ligand of Fas, interaction between Fas and Faslg is critical for triggering apoptosis), MAPK11 (a Map kinase involved in multiple cellular processes such as apoptosis, proliferation, migration, cell death, and differentiation) and FGFR2 (influences Bax and Bcl; earlier studies have demonstrated overexpression of FGFR1 to be associated with good outcome in AML [25]). In addition, genes associated with beneficial patterns are involved in the G-protein-coupled receptor signaling pathway (GPR88, P2RY2, CXCL9, POMC, FASLG, ADRA2C, GRIN1 and ADCY2), the cell cycle (CHEK1, GAS1 and UBE2V1) and transcription (YY2).
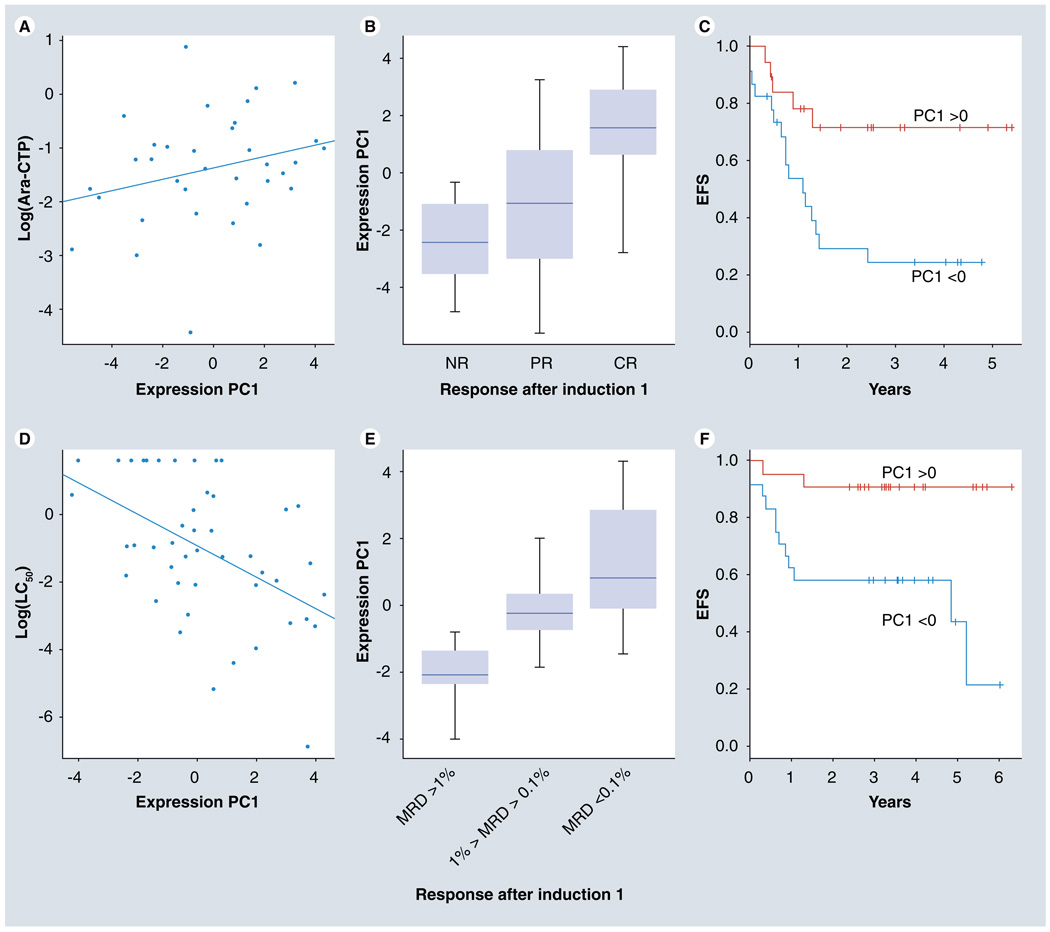
To visually depict our results, we determined principal components for the log-transformed expression values of these 60 probe sets for the combined cohort (Supplementary Figure 2). In the AML97 cohort, the first principal component (PC1) demonstrated a positive association with Ara-CTP levels (Figure 3A), a negative association with DNA synthesis rate relative to baseline (data not shown), a positive association with improved morphological response (Figure 3B) and a positive association with EFS (Figure 3C). In the AML02 cohort, PC1 demonstrates a strong negative association with LC50 (Figure 3D), a negative association with MRD (Figure 3E), and a positive association with EFS (Figure 3F).
Figure 3. Association of principal component of expression of 60 validated probe sets with multiple phenotypes from the AML97 discovery set and AML02 validation set.
In the discovery set (AML97 cohort), the PC1 shows positive association with: (A) Ara-CTP levels; (B) improved morphological response; and (C) EFS. Within the validation set (AML02 cohort), PC1 shows: (D) negative association with LC50; (E) MRD; and (F) positive association with EFS.
Ara-CTP: Cytarabine 5´-triphosphate; CR: Complete response; EFS: Event-free survival; MRD: Minimal residual disease; NR: No response; PC1: First principal component; PR: Partial response.
Functional analysis by gene-set enrichment analyses
To link the therapeutically meaningful association patterns with specific biological processes and molecular functions, we incorporated gene set enrichment analyses into each PROMISE analysis. Our analysis identified 40 biological process gene sets, 41 molecular function gene sets and 12 genomic location gene sets as being significant at the p = 0.05 level in both the PROMISE analyses (Supplementary Table 4). The identified gene sets suggest roles of disease pathology (cell cycle, proliferation, differentiation and maturation), cytarabine pharmacology (nucleotide binding, nucleoside triphosphate activity and other nucleotide processes), cell–cell signaling (G-protein-coupled receptor signaling pathway, cytokine and chemokine mediated signaling pathway, cyclic nucleotide and phosphoinositide-mediated signaling, MAK kinase and RAS GTPase activation). Of interest, genes located at 7q 22 and 7q31 were predominantly associated with beneficial gene pattern, and loss of chromosome seven has been associated with a poor prognosis.
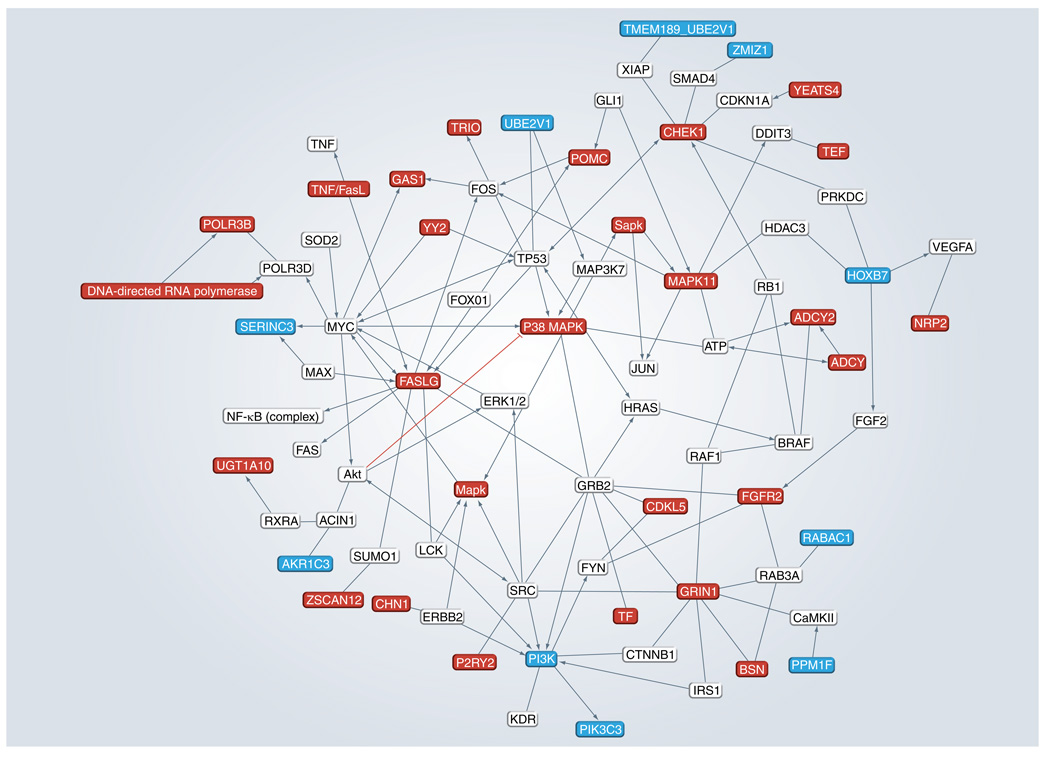
Using the ingenuity pathway analysis tool (literature-based annotations) we generated an interactome map for the validated genes. As shown in Figure 4, there is significant interaction between the validated genes, as well as with other genes of biological interest: TP53, FOS, FOXO1, Jun, VEGF, ERK1/2, MAPK and the NF-κB complex.
Figure 4. Interactome map generated by the Ingenuity Pathway Analysis tool.
A total of 60 validated genes were analyzed for any reported association with genes belonging to apoptosis signaling and/or cell cycle and proliferation networks in Ingenuity Pathway Analysis. Only those genes that were directly associated with our gene(s) of interest were retained in the network shown. For the sake of simplicity the resulting network was additionally trimmed to represent relationships with validated genes.
Discussion
In this study, we identified and validated genes that demonstrated patterns of association with multiple clinical and pharmacological end points of potential therapeutic relevance. The identified genes play important roles in biological processes of known relevance to cancer therapy. PIK3C3 demonstrated a strong detrimental pattern of association. The PIK3 family of proteins are well accepted as oncogenes and are involved in PIK3/PTEN/AKT/mTOR signaling by activating Akt [31,32], Erk [33] and Raf [34,35]. Our results support the observations that PI3K and its downstream signaling contribute to the development of drug resistance in myeloid leukemia. In addition our results have identified multiple genes (GPR88, GPR56, P2RY2, CXCL9, POMC, FASLG, ADRA2C, GRIN1 and ADCY2) involved in G-protein-coupled receptor signaling, which interacts with PI3K signaling and MAP kinases. These findings suggest that the introduction of PI3K inhibitors might be beneficial for a subset of patients with specific expression profiles.
Of interest, identification of aldo-keto reductase (AKR1C3) as a detrimental gene further strengthens the argument for its involvement in leukemogenesis [36], as well as its role in interfering with drug response. AKR1C3 has been identified as a suppressor of cell differentiation and potential novel combination therapies with AKR1C3 inhibitors have the potential to improve AML chemotherapy [28,29]. Ubiquitin conjugating enzyme UBE2V1, is a potential proto-oncogene that has been implicated in the activation of the NF-κB signaling pathway, as well as in an increased expression of its target antiapoptotic protein, Bcl-2 [30,37]. Its overexpression has been demonstrating to confer prolonged cell survival and protects cells from stress-induced apoptosis [30]. The observed association of UBE2V1 expression with a therapeutically detrimental pattern is in accordance with these observations, and requires further investigation.
These gene-expression signatures, coupled with other prognostic factors such as cytogenetics/demographics, could be used to predict response and provide an opportunity to adapt therapies to best match individual patient’s leukemia profile. One such approach is currently being evaluated to improve the breast cancer prognosis, whereby gene-expression based commercially available prognostic assays (e.g., Oncotype Dx® and Mammaprint®) are being used to predict reoccurrence of cancer [38]. In addition targeted therapies that enhance the activity of beneficial genes (and associated pathways) or inhibit the activity of detrimental genes (and associated pathways) may lead to improved clinical outcomes for children with AML.
In addition, none of the major cytarabine metabolic pathway genes were identified. This finding may be attributed to a variety of biological or statistical factors. For some of these genes, genotype may be more important than expression. For example, we have identified some inactivating polymorphisms in DCK [39]. In addition, across the cohort, the pattern of expression levels of genes with opposing effects on Ara-C metabolism may preclude them from being statistically identified. For example, we observed that DCK and NT5C2 expression were positively correlated (Spearman correlation = 0.2, p = 0.068), and it is now known that these genes have opposing effects on Ara-C metabolism. Finally, the sample size of our study was determined by the number of available samples, and thus, may not have been of adequate power to detect the known Ara-C metabolism genes [40].
Our study illustrates the power of integrated data analysis. We used PROMISE to perform an integrated data analysis that increases the statistical power in order to identify genes demonstrating therapeutically meaningful patterns of association with multiple end points. This was achieved by more effectively leveraging all available information in a simultaneous integrative analysis. Traditional approaches to identify genomic variables associated with specific patterns of multiple end points include screening the association of genomic variables with each end point individually and then identifying genes that are significant in each analysis and have the desired pattern of association. One of the problems with the traditional approach is the lack of statistical power and that the results are difficult to interpret as each end point involves multiple testing. PROMISE avoids the problems associated with traditional approaches and performs one test that directly addresses the question of whether a gene demonstrates the association pattern of interest, and thus, improves the statistical power and simplifies interpretation. In this study, we overcame the limitation of small sample sizes in both clinical trials by leveraging data from multiple end point variables in a statistically robust and biologically meaningful way to successfully identify and validate 60 probe sets with clear relevance to AML and its treatment. The improvements in statistical power will likely be more dramatic in larger studies with multiple end point variables. Analogous approaches with other genomic variables (e.g., SNPs and genomic copy number) and end point variables can be used to identify relevant genes in other contexts [41]. Our analyses lead to many novel insights, with only 20% of the 60 validated probe sets being associated with the cytogenetic risk group.
We were able to perform this integrative analysis because we collected four pharmacologic and clinical end points for each patient in the AML97 study and three pharmacologic and clinical end points for each patient in the AML02 study. Thus, for each cohort, we were able to compute a statistic for the association of each probe set’s expression with each end point. In addition, for each cohort, we used the PROMISE procedure to combine the association statistics for each probe set into an association pattern statistic and determine its p-value. To our knowledge, this is the first study to use such an integrative analysis procedure in order to identify pleiotropic genes with effects on multiple clinical and pharmacological end points. As such, we cannot meaningfully compare our validation success rate to that of other published studies. Regardless of this, the validation success rate of any study depends on the state of the biological system under consideration, experimental design and sample size.
Conclusion & future perspective
In conclusion, we used prior biological knowledge to define therapeutically beneficial and detrimental patterns of association with multiple pharmacological and clinical end points for cohorts of AML patients. Althrough PROMISE-based integrated analysis we identified and validated biologically interesting gene-expression signatures predicting therapeutically beneficial or detrimental response. The identification of unique gene-expression signatures and genetic variation responsible for interpatient variation in these candidate genes can be applied clinically to identify patients at risk of reduced efficacy or increased risk of side effects and to tailor therapy to achieve maximum clinical benefit.
Executive summary.
Background on cytarabine
-
▪
Inadequate efficacy of cytarabine-containing first-line chemotherapy remains a major therapeutic challenge in acute myeloid leukemia (AML), and a better understanding of the factors contributing to cytarabine resistance could improve treatment outcome.
Merits of current study
-
▪
Efforts to identify gene-expression signatures associated with cytarabine resistance in the last few years have mostly used human or murine leukemia cell lines and/or expression arrays that do not represent the entire genome.
-
▪
In the last decade, multiple studies have also used gene-expression profiling for classification of most of the known genetic subclasses in AML.
-
▪
Integrative genome-wide expression analysis with multiple end points can identify a genetic signature predicting cytarabine response in AML patients.
-
▪
The unique feature of our study is the simultaneous and integrative analysis of multiple in vivo and in vitro pharmacological and clinical end points in AML patients to define a therapeutically meaningful pattern and to identify gene-expression signatures predicting this pattern.
-
▪
These signatures, coupled with other factors such as cytogenetics/demographics, could be used predict response and provide an opportunity to design most effective combination regimen using available standard-of-care agents and targeted therapies that best match the leukemia profile.
-
▪
Finally, the pharmacodynamic genes identified in our study are potential candidates for future pharmacogenomic studies. Genetic variation within these genes could be applied clinically to identify patients and tailor therapy to achieve maximum clinical benefit.
Supplementary Material
Acknowledgments
This work was supported in part by grants CA57629, CA21765 and R01CA132946 (Jatinder K Lamba) from the NIH (US Department of Health and Human Services) and by the American Lebanese Syrian Associated Charities (ALSAC).
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Contributor Information
Jatinder K Lamba, Department of Experimental & Clinical Pharmacology, University of Minnesota, MN, USA.
Kristine R Crews, Department of Pharmaceutical Sciences, St Jude Children’s Research Hospital, Memphis, TN, USA.
Stanley B Pounds, Department of Biostatistics, St Jude Children’s Research Hospital, Memphis, TN, USA.
Xueyuan Cao, Department of Biostatistics, St Jude Children’s Research Hospital, Memphis, TN, USA.
Varsha Gandhi, Department of Experimental Therapeutics & Leukemia, The University of Texas, MD Anderson Cancer Center, Houston, TX, USA.
William Plunkett, Department of Experimental Therapeutics & Leukemia, The University of Texas, MD Anderson Cancer Center, Houston, TX, USA.
Bassem I Razzouk, Children’s Center for Cancer & Blood Diseases, Peyton Manning Children ’s Hospital at St Vincent, Indianapolis, IN 46260, USA.
Vishal Lamba, Department of Genetics & Cell Biology, University of Minnesota, Minneapolis, MN, USA.
Sharyn D Baker, Department of Pharmaceutical Sciences, St Jude Children’s Research Hospital, Memphis, TN, USA.
Susana C Raimondi, Department of Pathology, St Jude Children’s Research Hospital, Memphis, TN, USA.
Dario Campana, Department of Oncology, St Jude Children’s Research Hospital, Memphis, TN, USA.
Ching-Hon Pui, Department of Oncology, St Jude Children’s Research Hospital, Memphis, TN, USA.
James R Downing, Department of Pathology, St Jude Children’s Research Hospital, Memphis, TN, USA.
Jeffrey E Rubnitz, Department of Oncology, St Jude Children’s Research Hospital, Memphis, TN, USA.
Raul C Ribeiro, Department of Oncology, St Jude Children’s Research Hospital, Memphis, TN, USA.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Shipley JL, Butera JN. Acute myelogenous leukemia. Exp. Hematol. 2009;37:649–658. doi: 10.1016/j.exphem.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Creutzig U, Zimmermann M, Lehrnbecher T, et al. Less toxicity by optimizing chemotherapy, but not by addition of granulocyte colony-stimulating factor in children and adolescents with acute myeloid leukemia: results of AML-BFM 98. J. Clin. Oncol. 2006;24:4499–4506. doi: 10.1200/JCO.2006.06.5037. [DOI] [PubMed] [Google Scholar]
- 3.Lange BJ, Smith FO, Feusner J, et al. Outcomes in CCG-2961, a children’s oncology group Phase 3 trial for untreated pediatric acute myeloid leukemia: a report from the children’s oncology group. Blood. 2008;111:1044–1053. doi: 10.1182/blood-2007-04-084293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsukimoto I, Tawa A, Horibe K, et al. Risk-stratified therapy and the intensive use of cytarabine improves the outcome in childhood acute myeloid leukemia: the AML99 trial from the Japanese Childhood AML Cooperative Study Group. J. Clin. Oncol. 2009;27:4007–4013. doi: 10.1200/JCO.2008.18.7948. [DOI] [PubMed] [Google Scholar]
- 5. Rubnitz JE, Inaba H, Dahl G, et al. Minimal residual disease-directed therapy for childhood acute myeloid leukaemia: results of the AML02 multicentre trial. Lancet Oncol. 2010;11(6):543–552. doi: 10.1016/S1470-2045(10)70090-5. ▪ Reports clinical outcome results from the AML02 study.
- 6. Galmarini CM, Thomas X, Calvo F, et al. Potential mechanisms of resistance to cytarabine in AML patients. Leuk. Res. 2002;26:621–629. doi: 10.1016/s0145-2126(01)00184-9. ▪▪ Comprehensive review of mechanisms of resistance to cytarabine.
- 7.Galmarini CM, Thomas X, Calvo F, et al. In vivo mechanisms of resistance to cytarabine in acute myeloid leukaemia. Br. J. Haematol. 2002;117:860–868. doi: 10.1046/j.1365-2141.2002.03538.x. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez-Calotti P, LJordheim P, Giordano M, Dumontet C, Galmarini CM. Substrate cycles and drug resistance to 1-β-d-arabinofuranosylcytosine (araC) Leuk. Lymphoma. 2005;46:335–346. doi: 10.1080/10428190400015683. [DOI] [PubMed] [Google Scholar]
- 9. Cros E, Jordheim L, Dumontet C, Galmarini CM. Problems related to resistance to cytarabine in acute myeloid leukemia. Leuk. Lymphoma. 2004;45:1123–1132. doi: 10.1080/1042819032000159861. ▪▪ Comprehensive review of mechanisms of resistance to cytarabine response.
- 10. Lamba JK. Genetic factors influencing cytarabine therapy. Pharmacogenomics. 2009;10:1657–1674. doi: 10.2217/pgs.09.118. ▪▪ Comprehensive review of pharmacogenetic factors influencing cytarabine.
- 11.Abe S, Funato T, Takahashi S, et al. Increased expression of insulin-like growth factor i is associated with Ara-C resistance in leukemia. Tohoku J. Exp. Med. 2006;209:217–228. doi: 10.1620/tjem.209.217. [DOI] [PubMed] [Google Scholar]
- 12.Kanno S, Higurashi A, Watanabe Y, Shouji A, Asou K, Ishikawa M. Susceptibility to cytosine arabinoside (Ara-C)-induced cytotoxicity in human leukemia cell lines. Toxicol. Lett. 2004;152:149–158. doi: 10.1016/j.toxlet.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 13. Takagaki K, Katsuma S, Horio T, et al. cDNA microarray analysis of altered gene expression in Ara-C-treated leukemia cells. Biochem. Biophys. Res. Commun. 2003;309:351–358. doi: 10.1016/j.bbrc.2003.08.009. ▪ Gene-expression study to identify genes associated with Ara-C resistance.
- 14.Yin B, Kogan SC, Dickins RA, Lowe SW, Largaespada DA. Trp53 loss during in vitro selection contributes to acquired Ara-C resistance in acute myeloid leukemia. Exp. Hematol. 2006;34:631–641. doi: 10.1016/j.exphem.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 15.Yin B, Tsai ML, Hasz DE, Rathe SK, Le Beau MM, Largaespada DA. A microarray study of altered gene expression after cytarabine resistance in acute myeloid leukemia. Leukemia. 2007;21:1093–1097. doi: 10.1038/sj.leu.2404595. [DOI] [PubMed] [Google Scholar]
- 16.Hartford CM, Duan S, Delaney SM, et al. Population-specific genetic variants important in susceptibility to cytarabine arabinoside cytotoxicity. Blood. 2003;113:2145–2153. doi: 10.1182/blood-2008-05-154302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller BG, Stamatoyannopoulos JA. Integrative meta-analysis of differential gene expression in acute myeloid leukemia. PLoS ONE. 2010;5(3):e9466. doi: 10.1371/journal.pone.0009466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crews KR, Gandhi V, Srivastava DK, et al. Interim comparison of a continuous infusion versus a short daily infusion of cytarabine given in combination with cladribine for pediatric acute myeloid leukemia. J. Clin. Oncol. 2002;20:4217–4224. doi: 10.1200/JCO.2002.10.006. [DOI] [PubMed] [Google Scholar]
- 19. Rubnitz JE, Crews KR, Pounds S, et al. Combination of cladribine and cytarabine is effective for childhood acute myeloid leukemia: results of the St Jude AML97 trial. Leukemia. 2009;28(8):1410–1416. doi: 10.1038/leu.2009.30. ▪ Reports clinical outcome data from the AML97 study.
- 20. Pounds S, Cheng C, Cao X, et al. PROMISE: a tool to identify genomic features with a specific biologically interesting pattern of associations with multiple end point variables. Bioinformatics. 2009;25:2013–2019. doi: 10.1093/bioinformatics/btp357. ▪▪ Describes the details of the PROMISE method for integrative analysis of multiple end points.
- 21.Coustan-Smith E, Ribeiro RC, Rubnitz JE, et al. Clinical significance of residual disease during treatment in childhood acute myeloid leukaemia. Br. J. Haematol. 2003;123:243–252. doi: 10.1046/j.1365-2141.2003.04610.x. [DOI] [PubMed] [Google Scholar]
- 22.Holleman A, Cheok MH, den Boer ML, et al. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N. Engl. J. Med. 2004;351:533–542. doi: 10.1056/NEJMoa033513. [DOI] [PubMed] [Google Scholar]
- 23.Ross ME, Mahfouz R, Onciu M, et al. Gene expression profiling of pediatric acute myelogenous leukemia. Blood. 2004;104:3679–3687. doi: 10.1182/blood-2004-03-1154. [DOI] [PubMed] [Google Scholar]
- 24.McCubrey JA, Steelman LS, Abrams SL, et al. Targeting survival cascades induced by activation of Ras/Raf/MEK/ERK, PI3K/PTEN/Akt/mTOR and Jak/STAT pathways for effective leukemia therapy. Leukemia. 2008;22:708–722. doi: 10.1038/leu.2008.27. [DOI] [PubMed] [Google Scholar]
- 25.Bullinger L, Dohner K, Bair E, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N. Engl. J. Med. 2004;350:1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- 26.van Oostveen J, Bijl J, Raaphorst F, Walboomers J, Meijer C. The role of homeobox genes in normal hematopoiesis and hematological malignancies. Leukemia. 1999;13:1675–1690. doi: 10.1038/sj.leu.2401562. [DOI] [PubMed] [Google Scholar]
- 27.Liu CY, Hsu YH, Pan PC, et al. Maternal and offspring genetic variants of AKR1C3 and the risk of childhood leukemia. Carcinogenesis. 2008;29:984–990. doi: 10.1093/carcin/bgn071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desmond JC, Mountford JC, Drayson MT, et al. The aldo-keto reductase AKR1C3 is a novel suppressor of cell differentiation that provides a plausible target for the non-cyclooxygenase-dependent antineoplastic actions of nonsteroidal anti-inflammatory drugs. Cancer Res. 2003;63:505–512. [PubMed] [Google Scholar]
- 29.Khanim FL, Hayden RE, Birtwistle J, et al. Combined bezafibrate and medroxyprogesterone acetate: potential novel therapy for acute myeloid leukaemia. PLoS ONE. 2009;4:e8147. doi: 10.1371/journal.pone.0008147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Syed NA, Andersen PL, Warrington RC, Xiao W. Uev1A, a ubiquitin conjugating enzyme variant, inhibits stress-induced apoptosis through NF-κB activation. Apoptosis. 2006;11:2147–2157. doi: 10.1007/s10495-006-0197-3. [DOI] [PubMed] [Google Scholar]
- 31.Gold MR, Ingham RJ, McLeod SJ, et al. Targets of B-cell antigen receptor signaling: the phosphatidylinositol 3-kinase/Akt/glycogen synthase kinase-3 signaling pathway and the Rap1 GTPase. Immunol. Rev. 2000;176:47–68. doi: 10.1034/j.1600-065x.2000.00601.x. [DOI] [PubMed] [Google Scholar]
- 32.Ong SH, Hadari YR, Gotoh N, Guy GR, Schlessinger J, Lax I. Stimulation of phosphatidylinositol 3-kinase by fibroblast growth factor receptors is mediated by coordinated recruitment of multiple docking proteins. Proc. Natl Acad. Sci. USA. 2001;98:6074–6079. doi: 10.1073/pnas.111114298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marlowe JL, Puga A. Aryl hydrocarbon receptor, cell cycle regulation, toxicity, and tumorigenesis. J. Cell Biochem. 2005;96:1174–1184. doi: 10.1002/jcb.20656. [DOI] [PubMed] [Google Scholar]
- 34.Juliano RL. Signal transduction by cell adhesion receptors and the cytoskeleton: functions of integrins, cadherins, selectins, and immunoglobulin-superfamily members. Annu. Rev. Pharmacol. Toxicol. 2002;42:283–323. doi: 10.1146/annurev.pharmtox.42.090401.151133. [DOI] [PubMed] [Google Scholar]
- 35.Knall C, Young S, Nick JA, Buhl AM, Worthen GS, Johnson GL. Interleukin-8 regulation of the Ras/Raf/mitogen-activated protein kinase pathway in human neutrophils. J. Biol. Chem. 1996;271:2832–2838. doi: 10.1074/jbc.271.5.2832. [DOI] [PubMed] [Google Scholar]
- 36.Birtwistle J, Hayden RE, Khanim FL, et al. The aldo-keto reductase AKR1C3 contributes to 7,12-dimethylbenz(a)anthracene-3,4-dihydrodiol mediated oxidative DNA damage in myeloid cells: implications for leukemogenesis. Mutat. Res. 2009;662:67–74. doi: 10.1016/j.mrfmmm.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 37.Xia ZP, Sun L, Chen X, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim C, Paik S. Gene-expression-based prognostic assays for breast cancer. Nat. Rev. Clin. Oncol. 2010;7(6):340–347. doi: 10.1038/nrclinonc.2010.61. [DOI] [PubMed] [Google Scholar]
- 39.Pounds S, Cheng C. Sample size determination for the false discovery rate. Bioinformatics. 2005;21:4263–4271. doi: 10.1093/bioinformatics/bti699. [DOI] [PubMed] [Google Scholar]
- 40.Lamba JK, Crews K, Pounds S, et al. Pharmacogenetics of deoxycytidine kinase: identification and characterization of novel genetic variants. J. Pharmacol. Exp. Ther. 2007;323:935–945. doi: 10.1124/jpet.107.128595. [DOI] [PubMed] [Google Scholar]
- 41.Pounds S, Cao X, Cheng C, et al. Integrated analysis of pharmacokinetic, clinical, and snp microarray data using Projection onto the Most Interesting Statistical Evidence with adaptive permutation testing; Presented at: IEEE International Conference on Bioinformatics and Biomedicine; Washington, DC, USA. 2009. Nov 1–4, [DOI] [PMC free article] [PubMed] [Google Scholar]
Websites
- 101.Treatment of Patients With Newly Diagnosed Acute Myeloid Leukemia or Myelodysplasia. http://clinicaltrials.gov/ct2/show/ NCT00136084.
- 102.St Jude Children’s Research Hospital. www.stjuderesearch.org/data/AML1.
- 103.Gene set enrichment analysis. www.broadinstitute.org/gsea/msigdb/collections.jsp.
- 104.The R Project for Statistical Computing. www.r-project.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.