Abstract
The adapter protein, Fe65 has been proposed to be the link between the intracellular domains of the amyloid precursor protein, APP (AICD) and the LDL receptor-related protein (LRP-CT). Functional linkage between these two proteins has been established and mutations within LRP-CT affect the amount of Aβ produced from APP. Previous work showed that the AICD binds to the protein interaction domain 2 (PID2) of Fe65. Although the structure of PID1 was solved recently all attempts to demonstrate LRP-CT binding to this domain failed. We used biophysical experiments and binding studies to investigate the binding between these three proteins. Full-length Fe65 bound more weakly to AICD than did N-terminally truncated forms, however the intramolecular domain-domain interactions that had been proposed to inhibit binding could not be observed using amide H/D exchange. Surprisingly, when the LRP-CT is phosphorylated at Tyr4507, it bound to Fe65-PID1 despite the fact that this domain belongs to the Dab-like subclass of PIDs that is not supposed to be phosphorylation dependent. Mutation of a critical arginine abolished binding providing further proof of the phosphorylation-dependence. The Fe65-PID1 domain thus provides a link between the Dab-like class and the IRS-like class of PID domains and is the first Dab-like family member to show phosphorylation-dependent binding.
Keywords: Protein interaction domain (PID), tyrosine phosphorylation, NPXY motif, receptor intracellular domain, H/D exchange, tyrosine phosphorylation
The β-amyloid precursor protein (APP) is a type I membrane protein that can be processed to β-amyloid (Aβ), the main component of amyloid plaques in Alzheimer’s disease (AD). The complicated processes that lead to Aβ production are not well understood. Processing of APP is affected by the expression levels of both the low-density lipoprotein receptor-related protein (LRP) and Fe65 (1–7). The APP intracellular domain (AICD) contains an NPXY motif at position 684–687, a motif known to interact with protein interaction domains (PIDs, also known as phosphotyrosine binding domains or PTBs) (8). Crystal structures have been solved of AICD interacting with PID domains (9, 10), including the C-terminal PID domain of Fe65 (11).
LRP is an endocytic receptor composed of a 515 kD α-subunit and an 85 kD β-subunit that are non-covalently associated. The intracellular domain of LRP (LRP-CT) contains two NPXY motifs at positions 4470–4473 and 4504–4507 that can both be phosphorylated (12, 13). LRP expression levels have been shown to affect Aβ production (1, 5). Pietrzik et al. showed the C-terminal region of LRP, in particular the second NPXY motif, was responsible for LRP’s effect on Aβ production (3). Furthermore, mutation of the tyrosine in this motif abolished these effects.
Fe65 is a 97 kD adapter protein that contains a WW domain followed by two PID domains. Like LRP-CT, Fe65 expression levels have been shown to affect Aβ production (2, 4, 6, 7). In the majority of studies, increased Fe65 expression resulted in decreased Aβ production. Fe65 can also be linked to AD through a splice variant, Fe65a2. This isoform has an altered C-terminus and showed resistance to very late onset Alzheimer’s Disease, possibly through decreased binding to APP (14). The C-terminal PID domain (PID2) of Fe65 interacts with AICD (11, 15, 16) and the crystal structure of this complex reveals a unique binding interface that is much larger than other known PID domain interactions (11). AICD in which the tyrosine in the NPXY motif was mutated to alanine (Y687A) retained the ability to bind Fe65 (16). Both the structure and mutational analysis suggest that PID2 can bind AICD in both the tyrosine-phosphorylated (NPXpY) and non-phosphorylated forms, but the Fe65-AICD NPXpY interaction has not yet been observed.
Fe65 can be processed to a 65 kD form lacking the N-terminal region, and this shorter form was shown qualitatively to have enhanced binding to AICD leading to speculation intramolecular domain interactions may be present in Fe65 (17). Intramolecular interactions were also proposed based on observations of WW domain-PID2 binding in solution (18). One goal of the work presented here was to probe these intramolecular interactions and quantify their effect on binding affinity.
The N-terminal PID domain (PID1) of Fe65 has been reported to interact with the histone acetyltransferase Tip60, the transcription factor CP2, the cytoplasmic domain of the ApoER2 receptor, and LRP-CT (7, 19–21). Interestingly, neither CP2 nor Tip60 contains an NPXY motif. The intracellular domain of ApoER2 does contain an NPXY motif and was shown by co-IP to interact with Fe65 constructs containing the PID1 domain in COS7 cells (7). The Fe65-LRP interaction has been shown in numerous in vivo assays. GST-Fe65 PID1 was able to pull-down LRP-CT in MEF cells (19). In a later study, FRET was observed between the PID1 domain of Fe65 and LRP-CT in an H4 cell line (22). Lastly, co-IP experiments detected Fe65-LRP-CT complexes in HEK293T cells. This interaction could be observed with LRP-CT in which either NPXY motif was deleted, but binding was not observed when both NPXY motifs were deleted (23). The single in vitro experiment showing Fe65 binding to LRP-CT was performed exploiting the trimeric interaction of GST LRP-CT, Fe65, and APP695. GST LRP-CT pull-down of Fe65 was confirmed by detection of APP 35S (23). Radzimanowski et al. were unable to confirm binding of Fe65 PID1 to Tip60, ApoER2, or LRP-CT by isothermal titration calorimetry (ITC), regardless of the phosphorylation state of the NPXY motifs. In vitro binding assays using purified proteins are needed to show direct Fe65 PID1 interactions and the effect the NPXY tyrosine-phosphorylation state has on binding.
Protein interaction domains (PIDs) were formerly called phospho-tyrosine binding domains (PTB), but since phosphorylation is not a binding requirement in all cases, the domain was renamed. PID domains can be divided into 3 families based on structure similarities and binding modes: IRS-like, Shc-like, and Dab-like (24). IRS-like PID domains use two arginine residues to coordinate the phosphate moiety in the NPXpY motif. Shc-like PID domains use two arginine residues and a lysine residue to coordinate the phosphate. Dab-like PID domain binding events are not phosphorylation-dependent. Both PID domains in Fe65 have been classified as Dab-like PID domains. Although PID1 has been classified as a Dab-like PID domain, it contains two key arginine residues that overlap with the IRS binding pocket arginines in structural and sequence alignments (25, 26). These arginines could coordinate a phosphate moiety although binding of a phosphorylated ligand to PID1 has not been observed.
Using a combination of biophysical experiments and binding assays on purified proteins, we provide evidence of the Fe65 PID domains binding to the NPXY motifs in LRP-CT and AICD. Surprisingly, the Fe65 PID1 domain binds the LRP-CT NPXY4507 motif only when it is tyrosine-phosphorylated. LRP-CT only forms a trimeric complex with Fe65 and AICD when this LRP-CT NPXY4507 motif is tyrosine-phosphorylated. The Fe65 PID1 is therefore the first example of a Dab-like PID domain that binds an NPXY motif in a phosphorylation-dependent manner.
Materials and Methods
Protein expression and purification
Human Fe65 constructs containing the PID2 domain (1-662, 236-662, 534-662, 1-708) were expressed with a N-terminal ubiquitin tag to aid in solubility. The ubiquitin fusion vector was generated by cloning the DNA sequence for human ubquitin into the NcoI and BamH1 sites of pHis8 (27). Fe65 proteins were expressed in BL21 (DE3) cells and induced with 0.2 mM IPTG overnight at 18°C in M9ZN media (M9 media supplemented with 10g/L NZ amine). The cell pellet was resuspended in TBS lysis buffer (50 mM Tris-HCl, pH 8, 500 mM NaCl, 1mM BME, 0.5 mM PMSF), sonicated on ice, and centrifuged for 40 minutes at 12,000 rpm. Soluble proteins were captured using Ni-NTA (Qiagen) in TBS pH 8.0 at 4°C eluted with 250mM imidazole, and further purified on a Superdex-75 or Superdex-200 gel filtration column (GE Healthcare) in 50 mM Tris-HCl, pH 8, 150 mM NaCl, 1 mM DTT, 1 mM EDTA. The eluted protein was concentrated in a 10 kD molecular weight cutoff Amicon Ultra centrifugal filter (Millipore).
APP Intracellular Domain (AICD, residues 649-695 using APP695 numbering) with an N-terminal cysteine was cloned into the pMMHb vector modified to include an additional thrombin cleavage site after the TrpLE peptide (27). AICD was expressed in BL21 (DE3) cells and induced with 1 mM IPTG for 4 hours at 37°C. The cell pellet was resuspended in lysis buffer, sonicated on ice, and centrifuged for 40 minutes at 12,000 rpm. After resolubilization of inclusion bodies by sonication in 8M Urea TBS pH 8, protein was loaded onto a 5 mL pre-equilibrated NTA-Ni column. The resulting slurry was washed with 8M Urea TBS pH 8 followed by thrombin cleavage buffer (2 mM CaCl2 TBS pH 8). The slurry was then incubated with 25 ng thrombin in 20 mL cleavage buffer overnight at 25°C. Cleaved protein was washed from the column and purified by reverse phase high performance liquid chromatography (RP-HPLC). The resulting AICD was lyophilized and stored at −20°C.
The human LRP1 gene fragment containing the cytoplasmic domain as well as five residues of the membrane-spanning region (residues 4439-4544) was subcloned into PGEX-4T2. GST fusion proteins were expressed in pLysS cells and induced with 0.2 mM IPTG overnight at 18°C in M9ZN media. The cell pellet was resuspended in lysis buffer, sonicated on ice, and centrifuged for 40 minutes at 12,000 rpm. The supernatant was loaded on a 10 mL pre-equilibrated glutathione sepharose column at 4°C, washed with 3 column volumes of TBS and eluted in 10 mLs of 10 mM glutathione in TBS, pH 7.4. The eluted protein was concentrated in a 10 kD molecular weight cutoff Amicon Ultra centrifugal filter (Millipore) and loaded onto a Superdex-75 gel filtration column in 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM DTT. AICD Y653F, Fe65(R451Q) and LRP-CT Y4478F mutations were introduced using QuickChange mutagenesis (Strategene, La Jolla, CA) and verified by DNA sequencing. Protein concentrations were determined by UV absorbance and identities were confirmed by MALDI-TOF mass spectrometry.
LRP microdomains containing Tyr 4507 (Cys-amino aminohexanoic acid-TNFTNPVY4507ATLY) and Ty 4473 (Cys-amino aminohexanoic acid-VEIGNPTY4473KMYEGGE) in both phosphorylated and non-phosphorylated were prepared as described previously (28).
LRP4488 contains residues 4488-4544 of human LRP1 with an N-terminal cysteine and was cloned into the pMMHb vector modified to include an additional thrombin cleavage site after the TrpLE peptide (27). The expression was carried out as described for AICD.
Phosphorylation of LRP-CT Y4478F, AICD Y653F, and LRP4488
Chicken Src (residues 251-533) was expressed and purified according to previously published protocols (29). Tyrosines in LRP, Y4478F and AICD, Y653F were mutated to ensure exclusive phosphorylation of GST-LRP-CT at Y4507 and AICD at Y687. GST-LRP-CT, AICD, or LRP4488 (230 nmoles) was incubated with 0.7 nmoles purified recombinant c-Src in 2 mLs of 1 mM ATP, 100 mM MnCl2, 100 mM MgCl2, TBS, pH 7.4 for 1 hour at 25°C.
SPR binding experiments
Sensorgrams were recorded on a Biacore 3000 instrument using streptavidin chips as described (30). AICD was biotinylated at the N-terminal cysteine using biotin-maleimide (Pierce). 100 RU, 200 RU, and 300 RU of AICD were immobilized and 50–500 nM Fe65 was injected for 5 min and dissociation was measured for 20 min at 25°C at 50 μL/min. No regeneration step was necessary. Data were fit using the BiaEvaluation 2.0 software and plotted in Origin 7.0.
Amide Exchange Experiments
Native state backbone amide exchange was measured as described previously. Fe65 with or without 2 fold excess AICD was incubated for 0–10 min in deuterated TBS buffer at pH 8. Amide exchange was quenched by tenfold dilution into ice cold 0.1% TFA for a final pH of 2.2. The sample was immediately digested with an excess of immobilized pepsin (Pierce) and the digest mixture was frozen in liquid N2. All samples were analyzed on the same day by MALDI-TOF mass spectrometry (31). The centroids of the mass envelopes were measured and compared to undeuterated controls and corrected for back exchange as described previously (32).
Pull-down assays
AICD and LRP-CT microdomains were immobilized using Sulfolink coupling gel according to the manufacturer’s directions (Pierce, Rockford, IL). Due to the poor solubility of the peptides, all sulfolink immobilization reactions were carried out in 3.5M Guanidine HCl, TBS pH 8.0. The amount of peptide immobilized was quantified by UV absorbance. Efficiency of immobilization was found to be 1–2 mg peptide per mL of beads.
Equal amounts of GST-LRP-CT or GST-LRP-CT (pY4507) were immobilized on glutathione sepharose beads at 1 mg/mL (50% slurry) for 1 hour at 4°C in 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM DTT. The resulting resin was washed 4 times in TBS-X (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM DTT, 2 mM CaCl2, 2 mM MgCl2, 1% Triton-X-100) before being used as bait for Fe65 binding reactions.
Binding reactions were carried out in 1 mL TBS-X containing 15 μL (for AICD and LRP microdomains) or 25 μL (GST-LRP-CT) washed beads and 55 nM–1.2 μM Fe65. Trimeric binding reactions contained 5 μM immobilized AICD, 2.5 μM Fe65, and 260 nM GST-LRP-CT. Fe65 PID2 competition binding reactions contained 650 μM GST LRP-CT, 650 μM AICD, and 24 μM Fe65 PID2 (536-662). Reactions were incubated at 4°C for 1–2 hours, washed 4 times in TBS-X and resuspended in 30μL reducing SDS buffer. Samples were resolved by SDS-PAGE, transferred to PDVF membrane, and probed with Fe65 antibody (1:500, Millipore, 3H6) LRP antibody (1:500, 11H4), anti-HIS antibody (1:1000, Quiagen, Penta-His Ab), or anti-Ubiquitin antibody (1:100, Invitrogen 13-1600).
LRP4488 labeling for fluorescence anisotropy experiments
LRP4488 or LRP4488 phosphorylated at Y4507 (pYLRP4488) was labeled with Oregon Green 488 maleimide (invitrogen, O-6034) at the single cysteine introduced at the N-terminus according to the manufacturers instructions. 1 mL of 100 μM LRP4488 or pYLRP4488 was reduced with immobilized TCEP (Pierce) for 1 hour in TBS, pH 7.5 at room temperature. Reduced LRP4488 was incubated with 20-fold excess Oregon Green 488 and allowed to react for 2 hours at room temperature in the absence of light. The labeled LRP4488 or pYLRP4488 was purified by reverse phase high performance liquid chromatography (RP-HPLC) and lyophilized.
Fluorescence anisotropy experiments
Polarization experiments were performed with 96-well black fluorescence plates (Fluotrac, Greiner Bio-One) on a DTX 880 Multimode Detector Beckman Coulter plate reader with excitation filter at 485 nm and two emission filters at 535 nm equipped with polarizers. Fe65 (0.1–100 μM) with and without 20nM LRP4488 or pYLRP4488 in 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM DTT was added to the wells and incubated for 30 minutes before readings were taken. Anisotropy values at each concentration were determined after subtracting the intensities for Fe65 alone, using the equation r = (I(V,V) – G × I (V,H)) / (I(V,V) – G × 2I (V,H)) where r is the anisotropy, I(V,V) is the fluorescence intensity in the parallel direction, I (V,H is the fluorescence intensity in the perpendicular direction, and G is 0.67, a correction factor for the difference in detection sensitivity for parallel and perpendicular polarized light. Data were fit with KaleidaGraph 4.0 software using the Michaelis-Menten curve fit.
Results
The N-terminal domains of Fe65 weaken binding of Fe65 PID2 to AICD
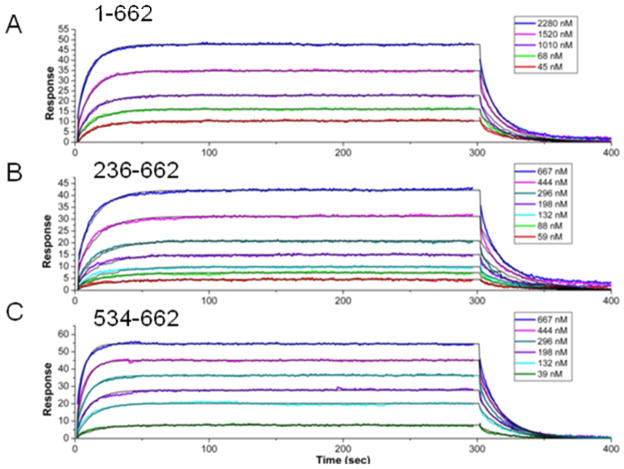
Surface Plasmon Resonance (SPR) binding experiments were performed with AICD (residues 649-695 of the β-amyloid precursor protein) and various Fe65 constructs to explore the inhibitory effects of the N-terminus of Fe65 on the binding of Fe65 PID2 to AICD. Fe65 constructs containing the PID2 domain are unstable in solution and highly prone to aggregation. An ubiquitin tag was introduced to aid solubility, and effectively alleviated these problems. The ubiquitin-Fe65 was able to interact with AICD with similar affinities to those reported in the literature (11). Fe65 containing the full N-terminus (1-662) bound AICD 2.5 fold weaker than Fe65 lacking the N-terminus (236-662) (KD= 2.54+/−0.35 μM and 1.01+/−0.05 μM) (Figure 1A, B). Further truncation of Fe65 to a construct containing only the PID2 domain (534-662) enhanced binding an additional 3 fold (KD= 0.35+/−0.02 μM) (Figure 1C). These results indicate the N-terminal domains do in fact have an inhibitory effect on AICD binding. The N-terminal domains primarily slow the association rate (Table 1).
Figure 1.
SPR binding kinetics of the interaction between AICD and Fe65 N-term-WW-PID1-PID2 1-662 (A), Fe65 WW-PID1-PID2 236-662 (B), or Fe65 PID2 534-662 (C) performed as previously described (30). AICD was biotinylated at the N-terminus through an engineered cysteine and immobilized on a streptavidin sensor chip. Fe65 (50-500 nM) was the flowing analyte. The data were collected on a Biacore 3000 and fit using a simple 1:1 binding model yielding for Fe65 1-662 KD = 2.54 +/− 0.35, for Fe65 236-662 KD = 1.01+/−0.05, and for Fe65 534-662 KD= 0.35 +/− 0.02.
Table 1.
SPR results for the various Fe65 constructs binding to AICD
| Fe65 construct | KD (μM) | ka (M−1s−1) | kd (s−1) | X2 |
|---|---|---|---|---|
| 1-662 | 2.5 ± 0.35 | 2.2 × 104 | 0.055 | 0.78 |
| 236-662 | 1.0 ± 0.05 | 5.7 × 104 | 0.057 | 0.22 |
| 534-662 | 0.35 ± 0.02 | 9.5 × 104 | 0.033 | 0.432 |
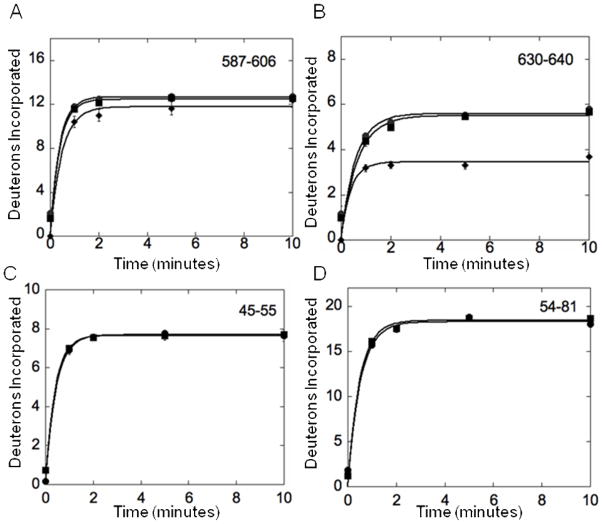
To probe for Fe65 intramolecular interactions, we compared the amide hydrogen/deuterium (H/D) exchange behavior of Fe65 (1-662) to Fe65 (236-662). Peptic fragments covered 44% of the PID2 domain (residues 534-662) including regions of the Fe65 AICD binding interface. H/D exchange analysis showed identical exchange with or without the presence of the N-terminal 235 amino acids. Representative plots are shown in Figure 2A, B (squares and circles) and the data from the 5 minute time point for all peptides from PID2 are summarized in Table 2A. The deuterons incorporated for region 638-649 could not be quantitatively determined due to spectral overlap, but qualitatively showed the same exchange with and without the N terminal region. To ensure that protein interactions would lead to observable changes in H/D exchange, the same experiment was performed on Fe65 (236-662) in the presence of AICD. Significant changes at the AICD binding interface in PID2 were clearly evident (Figure 2B, diamonds).
Figure 2.
Plots of the amide H/D exchange of Fe65. (A) Deuterons incorporated over 10 minutes for Fe65 1-662 (circles), Fe65 236-662 (squares), and Fe65 236-662 with AICD bound (diamonds). Residues 587-606 of the PID2 domain are depicted and show the same solvent accessibility with or without the N-terminal domain. Fe65 236-662 with AICD bound also showed the same solvent accessibility since this region is not in the Fe65:AICD binding interface. (B) Deuterons incorporated over 10 minutes for Fe65 1-662 (circles), Fe65 236-662 (squares), and Fe65 236-662 with AICD bound (diamonds). Residues 630-640 of the PID2 domain are depicted and show the same solvent accessibility with or without the presence of the N-terminal domain. Fe65 236-662 with AICD bound showed decreased solvent accessibility for this region because it is in the binding interface. (C) Plots of the amide H/D exchange of Fe65 236-662 (circles) and Fe65 236-512 (squares) over 10 minutes. Residues 45-55 of the WW domain are depicted and showed the same solvent accessibility with or without the presence of the PID2 domain. (D) Plots of the amide H/D exchange of Fe65 236-662 (circles) and Fe65 236-512 (squares) over 10 minutes. Residues 54-81 of the WW domain are depicted and showed the same solvent accessibility with or without the presence of the PID2 domain.
Table 2.
Amide H/D exchange
| A. Exchange into the PID2 region of Fe65 1-662 vs. 236-662. | ||||
|---|---|---|---|---|
| Fe65 residue | peptide mass | backbone amides | Fe65 1-662 | Fe65 236-662 |
| amides exchanged after 5 min | ||||
| 565-589 | 2555.27 | 21 | 16.1 ± 0.1 | 16.2 ± 0.2 |
| 569-589 | 2127.05 | 18 | 12.7 ± 0.2 | 12.6 ± 0.2 |
| 603-610 | 924.54 | 6 | 2.5 ± 0.1 | 2.5 ± 0.2 |
| 610-621 | 1332.71 | 10 | 5.2 ± 0.1 | 5.3 ± 0.1 |
| 610-620 | 1185.64 | 9 | 5.5 ± 0.1 | 5.5 ± 0.2 |
| 638-649 | 1087.53 | 11 | N/D | N/D |
| B. Exchange into the WW domain region of Fe65 236-702 vs. 236-512. | ||||
|---|---|---|---|---|
| Fe65 residue | peptide mass | backbone amides | Fe65 236-702 | Fe65 236-512 |
| amides exchanged after 5 min | ||||
| 260-270 | 1320.59 | 10 | 7.8 ± 0.1 | 7.6 ± 0.2 |
| 269-296 | 3144.44 | 25 | 18.6 ± 0.1 | 18.8 ± 0.2 |
A second Fe65 intramolecular interaction has been proposed in which the WW domain interacts with the PID2 domain (18). Amide H/D exchange of Fe65 with and without the PID2 domain (236-662 vs. 236-512) showed no changes in the WW domain (residues 253-289, with 81% coverage). Representative plots are shown in Figure 2C, D and the data from the 5 minute time point for all peptides from the WW domain are summarized in Table 2B. The results show no evidence of any strong Fe65 intramolecular interaction involving the WW or PID2 domains.
Fe65 can bind AICD tyrosine phosphorylated at NPXY687
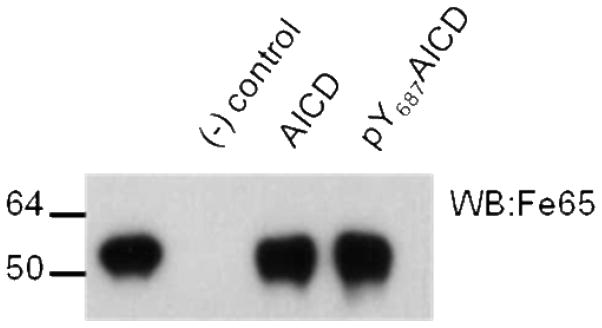
The PID2 domain of Fe65 has been classified as a Dab-like PID domain, the category of PID domains reserved for those that bind non-phosphorylated ligands (24). Dab-like PID domains in some cases highly disfavor tyrosine phosphorylation and in some cases phosphorylation does not affect binding. Fe65 PID2 has not been shown to directly bind any ligand in the NPXpY form. The binding interface of PID2 and AICD is unique in that it is three times larger than known PID domain peptide interactions (11). We sought to determine if Fe65 could bind APP phosphorylated at the tyrosine in the NPXY687 motif. Phosphorylation reactions with purified recombinant Src and AICD resulted in AICD phosphorylation at Y653 and Y687. AICD Y653F was used to ensure phosphorylation exclusively at Y687. AICD or AICDpY687 was coupled to sulfolink resin through an N-terminal cysteine and incubated with Fe65 (236-662). Fe65 binds AICD and AICDpY687 equally well (Figure 3) showing that the PID2 domain of Fe65 can bind ligands in both the phosphorylated and non-phosphorylated states.
Figure 3.
The dependence of binding of Fe65 to AICD on phosphorylation of AICD Y687 was explored using immobilized AICD. 5 μM AICD Y653F (GSKLCKKKQFTSIHHGVVEVDAAVTPEERHLSKMQQNGYENPTYKFFEQMQN) in the phosphorylated or non-phosphorylated form was immobilized using sulfolink coupling gel and incubated with 180 nM Fe65 (236-662) for 1 hour at 4° C. Sulfolink resin alone was used as a negative control. Bound Fe65 was visualized by western blotting with an anti-Fe65 antibody (1:500, Millipore, 3H6). Comparison of the third and fourth lanes reveals that Fe65 binds AICD independent of phosphorylation at Tyr 687.
Tyrosine-phosphorylated and non-phosphorylated LRP-CT NPXY4507 microdomains bind distinct domains in Fe65
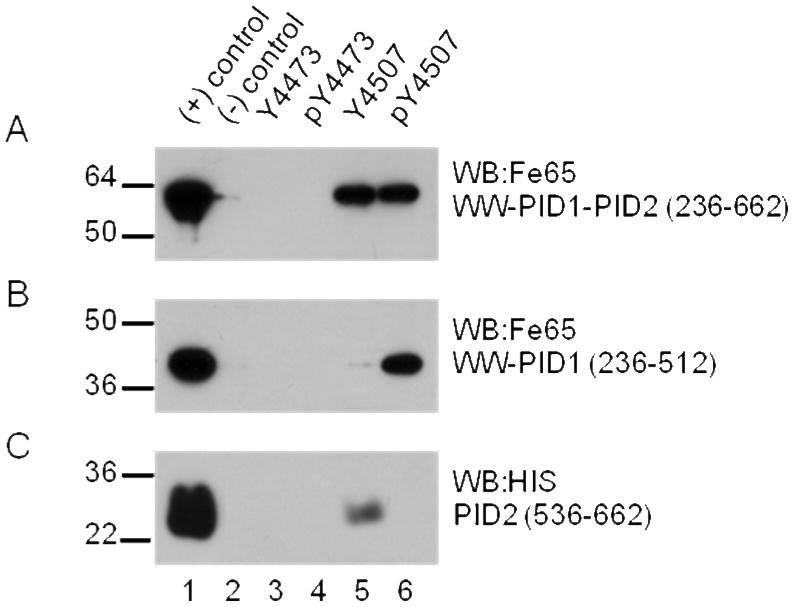
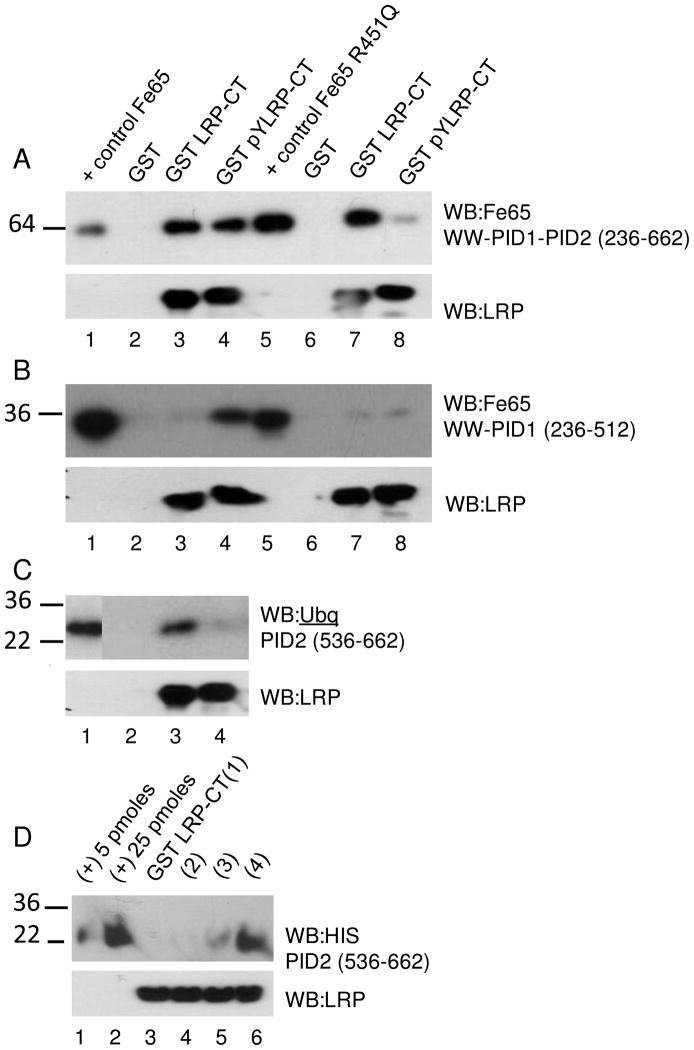
LRP-CT contains two NPXY motifs (4470-4473, 4504-4507) that can both be phosphorylated (13, 33). The phosphorylation state preference of the Fe65 PID domains binding to the LRP NPXY motifs has not been fully explored. Using microdomains centered at each LRP NPXY motif we investigated Fe65 PID domain binding specificity to each LRP NPXY motif in the tyrosine-phosphorylated and non-phosphorylated forms. Each microdomain peptide was coupled to sulfolink resin and incubated with Fe65 WW-PID1-PID2 (236-662), Fe65 WW-PID1 (236-512), or Fe65 PID2 (536-662). All experiments were carried out with freshly prepared, pure Fe65 proteins (Supplementary Figure 1). The LRP microdomains containing Y4473 showed no interaction with Fe65 regardless of their phosphorylation state (Figure 4A, lanes 3 and 4). On the other hand, microdomains containing Y4507 bound Fe65 in both the phosphorylated and non-phosphorylated forms (Figure 4A, lanes 5 and 6). The same assay with Fe65 (236-512) and Fe65 PID2 (536-662) suggests that PID1 interacts with phosphorylated Y4507 while PID2 interacts with the non-phosphorylated Y4507. (Figure 4B, C lanes 5 and 6).
Figure 4.
LRP-CT microdomains binding to (A) Fe65 WW-PID1-PID2 236-662, (B) Fe65 WW-PID1 236-512, or (C) Fe65 PID2 536-662. Microdomains containing Y4473 (Cys-amino aminohexanoic acid-VEIGNPTY4473KMYEGGE) or Y4507 (Cys-amino aminohexanoic acid-TNFTNPVY4507ATLY) in both tryrosine-phosphorylated and non-phosphorylated forms were immobilized using sulfolink coupling gel. Each microdomain (60 μg) was incubated with 180 nM –400 nM Fe65 for 1 hour at 4°C. Sulfolink resin alone was used as a negative control. Bound Fe65 was visualized by western blotting with anti-Fe65 antibody (Millipore, 3H6) or anti-HIS antibody (Quiagen, Penta-His Ab). Fe65 WW-PID1-PID2 bound to both the phospho- and non-phospho-forms of the Y4507-containing microdomain of LRP. Panel B shows that only the phospho-form binds to Fe65 PID1 and Panel C shows that only the non-phospho-form binds to Fe65 PID2.
Recapitulation of the observed FE65 binding preferences in full-length LRP-CT
To determine if the specificity of Fe65 PID1 for LRP-CT in the Y4507 phosphorylated form and Fe65 PID2 for the non-phosphorylated form were also observed in the context of the full LRP-CT, we prepared GST-LRP-CT Y4473F phosphorylated exclusively at Y4507 with recombinant purified Src. GST-LRP-CT (pY4507) or non-phosphorylated GST-LRP-CT was immobilized on glutathione-sepharose and incubated with purified Fe65 constructs (Figure 5). In agreement with the LRP microdomain results, Fe65 WW-PID1-PID2 bound both GST-LRP-CT (pY4507) and non-phosphorylated GST-LRP-CT. Fe65 in which one of the PID1 domain conserved arginines was mutated (R451Q) showed dramatically reduced binding to GST-LRP-CT (pY4507), while the binding of LRP-CT in the non-phosphorylated form was not affected. This effect was also observed using Fe65 WW-PID1. Also in agreement with the microdomain results, Fe65 PID2 binds preferentially to the non-phosphorylated form of LRP-CT (Figure 5C, lane 3).
Figure 5.
LRP-CT binding to (A) Fe65 WW-PID1-PID2 236-662, (B) Fe65 WW-PID1 236-512, or (C) Fe65 PID2 536-662. GST-LRP-CT or GST-LRP-CT (pY4507) was immobilized on glutathione sepharose and 650 nM was incubated with 180–400 nM of each Fe65 construct (lanes 2–4) or Fe65(R451Q) construct (lanes 6–8) for 1 hour at 4°C. GST-coupled sepharose was used as a negative control. Bound Fe65 or Fe65(R451Q) was visualized by western blotting with anti-Fe65 antibody (Millipore, 3H6) or anti-Ubiqitin antibody (Invitrogen 13-1600). Each Fe65 variant (0.05 pmoles) was analyzed as a positive control (lane 1, 5). Equal LRP-CT loading is shown by western blotting with anti-LRP antibody (11H4). (D) LRP-CT binding to Fe65 PID2 536-662. GST-LRP-CT was immobilized on glutathione sepharose and 650 nM was incubated with 3 μM (1), 6 μM (2), 12 μM (3), or 24 μM (4) PID2 for 1 hour at 4°C. Bound Fe65 was visualized by western blotting with anti-HIS antibody (Quiagen, Penta-His Ab). Fe65 PID2 (5 and 25 pmol) is shown as a positive control. Equal LRP-CT loading is shown by western blotting with anti-LRP antibody (11H4).
Fe65 LRP-CT binding affinity is in the micromolar range
In order to show specific, concentration-dependent binding of LRP-CT to Fe65 PID2, GST LRP-CT was incubated with varying amounts of Fe65 PID2 (536-662). GST LRP-CT bound more Fe65 PID2 when incubated with 24 μM as compared to 12 μM (Figure 5D).
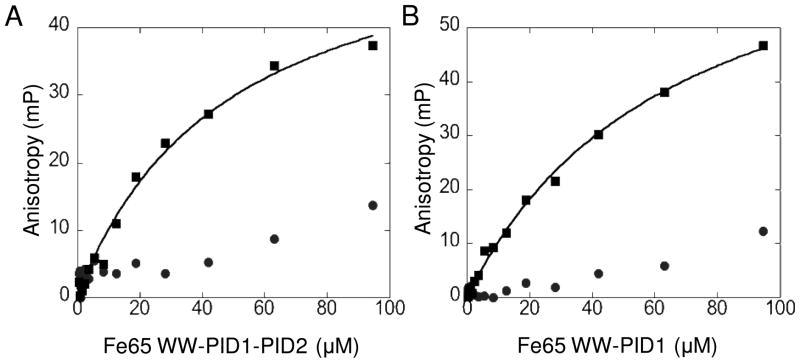
To further establish the interaction between LRP-CT and Fe65, a truncated LRP-CT starting at position 4488 and ending at 4454 (LRP4488) was attached through an N-terminal cysteine to Oregon Green 488 maleimide (Invitrogen) and used in fluorescence anisotropy binding studies. LRP4488 or pYLRP4488 was incubated with varying concentrations of Fe65 WW-PID1-PID2 (236-662) or WW-PID1 (236-512) and the fluorescence anisotropy change upon binding was measured (Figure 6). Both WW-PID1-PID2 and WW-PID1 showed preferred binding to pYLRP4488 with KD values of 48.3+/−7.3 μM and 66.7 +/− 6.2 μM, respectively. Although non-phosphorylated LRP4488 appears to be weakly binding, the affinity was too weak to quantify.
Figure 6.
Fluorescence anisotropy of LRP4488 or pYLRP4488 binding to Fe65. LRP4488 (●) or pYLRP4488 (■) conjugated to Oregon Green 488 maleimide (Invitrogen) was incubated with 0.1–100 μM Fe65 WW-PID1-PID2 236-662 (A) or 0.1–100 μM Fe65 WW-PID1 236-512 (B). Anisotropy values at each Fe65 concentration were determined using a DTX 880 Multimode Dectector Beckman Coulter plate reader with excitation filter at 485 nm and two emission filters at 535 nm equipped with polarizers. Data were fit with KaleidaGraph 4.0 software using a Michaelis-Menten curve fit. pYLRP4488 bound Fe65 WW-PID1-PID2 with KD = 48.3+/−7.3 μM and pYLRP4488 bound Fe65 WW-PID1 with KD = 66.7 +/− 6.2 μM.
LRP-CT and AICD compete for binding to Fe65 PID2
Both non-phosphorylated LRP-CT and AICD bind Fe65 PID2. To determine if these Fe65 ligands share the same binding site, we performed a competition binding experiment. GST-LRP-CT was immobilized on glutathione-sepharose and incubated with Fe65 PID2 (536-662) in the presence and absence of AICD. GST LRP-CT could bind significantly less Fe65 PID2 when an equimolar amount of AICD was present (Figure 7, compare lanes 3 and 4).
Figure 7.
GST LRP-CT (650 nM) was immobilized on glutathione sepharose and allowed to bind 24 μM Fe65 PID2 536-662 in the presence and absence of 650 nM AICD. GST-coupled sepharose was used as a negative control. Bound Fe65 PID2 was visualized by western blotting with anti-HIS antibody (Quiagen, Penta-His Ab). Fe65 PID2 (5 and 25 pmol) is shown as a positive control. Equal LRP-CT loading is shown by western blotting with anti-LRP antibody (11H4).
Fe65 binds preferentially to LRP-CT phosphorylated at tyrosine 4507 when in complex with AICD
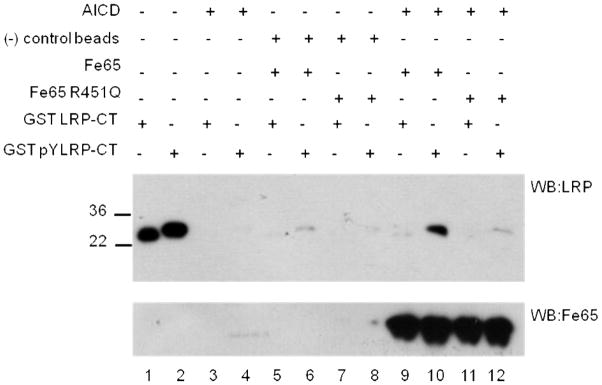
Fe65 has been shown to form a trimeric complex with LRP-CT and AICD (23). We tested whether the phosphorylation state of LRP-CT Y4507 affects the formation of this complex. Immobilized AICD was used as bait to capture purified Fe65 WW-PID1-PID2 (236-662) and LRP-CT or LRP-CT (pY4507). Fe65 and AICD preferentially form a complex with LRP-CT (pY4507) (Figure 8), but not the R451Q mutant, showing that LRP-CT (pY4507) is predominantly interacting with PID1 in the trimeric complex.
Figure 8.
The phosphorylation-dependent binding of LRP-CT to Fe65 in the LRP-CT:Fe65:AICD trimeric complex was explored. Using sulfolink coupling gel, 5 μM AICD was immobilized and incubated with 2.5 μM Fe65 WW-PID1-PID2 (236-662) and 260 nM GST-LRP-CT (pY4507) or GST-LRP-CT for 1 hour at 4° C. Sulfolink resin alone was used as a negative control to ensure LRP-CT was not detected due to aggregation. The reactions were washed and resuspended in reducing sample buffer. Bound GST-LRP-CT was visualized by western blotting with anti-LRP antibody (11H4). Equal Fe65 binding is shown by western blotting with anti-Fe65 antibody (Millipore, 3H6). GST-LRP-CT can only form a complex with Fe65 and AICD when phosphorylated at Y4507 (lane 10).
Discussion
Fe65 contains three protein interaction domains, a WW domain followed by two PID domains. In addition to these defined domains, Fe65 contains an N-terminal region that has been proposed to inhibit interactions with Fe65 ligands (17, 18). Our SPR studies of AICD binding to Fe65 with and without the N-terminal region quantitatively show that the N-terminal region does indeed weaken binding of Fe65 PID2 to AICD, in agreement with the previous studies. It appears that effects from both the far N-terminus and the WW-PID1 region contribute to weakening the binding of AICD to the Fe65 PID2 domain. Although we attempted to use amide H/D exchange to observe the putative intramolecular interactions that might be weakening the AICD binding to the Fe65 PID2 domain, no differences in exchange could be attributed to the presence of the N-terminal region. This indicates that either the intramolecular interactions are too weak to observe by this method, or the interactions take place in regions not covered in the H/D exchange experiment. Clear changes in PID2 were observed upon the addition of AICD, arguing that the first scenario is more likely to be the case. The weaker binding affinity of the Fe65s containing the N-terminal domains was primarily due to slower association rates perhaps indicating that these domains sterically hinder binding.
Phosphorylation of AICD has been observed in Alzheimer’s Disease patient’s brains at several sites including Y687 (34). We report here that AICD can be tyrosine-phosphorylated at Y687 by Src kinase in vitro. Our results show that tyrosine-phosphorylation of the AICD NPXY687 does not affect the binding of AICD to Fe65. The Fe65 PID2-AICD interaction has a large binding interface that does not solely rely on the AICD NPXY binding motif (11). However it is interesting that the addition of the phosphate group to a residue at the interface does not have a significant effect on binding affinity.
LRP-CT contains two NPXY motifs that can both be tyrosine phosphorylated. We found that NPXY4507, but not NPXY4473 bound to Fe65, agreeing with previous binding studies using rat brain lysate (28). All Fe65 interactions occur at the second NPXY4507 in both the tyrosine-phosphorylated and non-phosphorylated forms. Using both the LRP-CT microdomains and full-length LRP-CT, we showed that Fe65 PID1 binds to LRP-CT phosphorylated at Y4507, while Fe65 PID2 prefers non-phosphorylated LRP-CT. Furthermore, we demonstrated that LRP-CT and AICD cannot bind Fe65 PID2 simultaneously, as they compete for binding in pull-down assays. LRP-CT must be phosphorylated at Y4507 in order to form a trimeric complex with Fe65 and AICD, in which AICD binds Fe65 PID2, and LRP-CT (pY4507) binds Fe65 PID1. Although the binding affinities for Fe65-LRP-CT complexes could not be determined by SPR or ITC due to the weak nature of the interaction, fluorescence anisotropy experiments show that Fe65 WW-PID1-PID2 binds pYLRP4488 with a KD of 48.3+/−7.3 μM, and Fe65 WW-PID1 binds pYLRP4488 with a KD of 66.7 +/− 6.2 μM. These experiments further demonstrate that the predominant Fe65-LRP-CT interaction occurs through the PID1 domain of Fe65 and LRP-CT phosphorylated at Y4507. The weak binding affinity of the Fe65 PID2-LRP4488 complex could not be quantified.
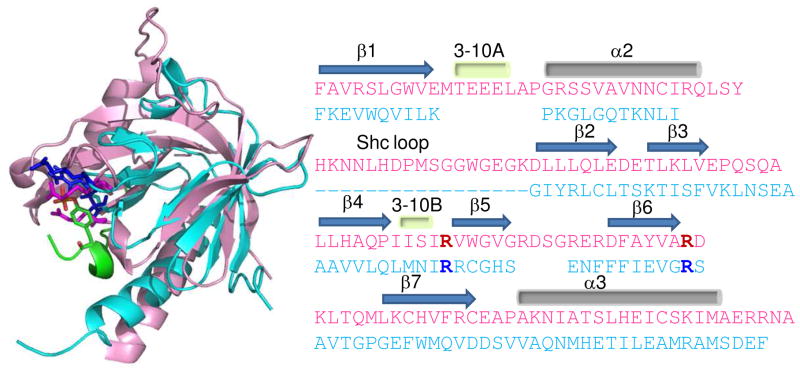
Fe65 PID1 and PID2 have been classified as Dab-like PID domains based on an algorithm using the structures of Dab1, Dab2, NUMB, and X11 (24). IRS-like PID domains were identified based on the structures of IRS, Dok1, and Dok5, and no overlap was found between Dab-like and IRS-like identifications. On the other hand, the Dab-like algorithm identified 16 Shc-like family members showing these two families are close relatives. Although PID1 has been identified as a Dab-like PID domain, it contains features belonging to both the Shc-like and IRS-like PIDs. Shc-like PIDs contain an inserted 22-amino acid loop between α2 and β2, and the Fe65 PID1 also contains a 19-amino acid insertion at this position. Structure and sequence alignments of the Fe65 PID1 with the IRS-like PIDs reveal that Fe65 PID1 residues R451, R470 correspond to the two key phosphate-coordinating arginine residues present in IRS-like PID domains (Figure 9). Remarkably, Fe65 PID1 binds exclusively to LRP-CT in the tyrosine-phosphorylated form and the mutant Fe65(R451Q) loses the ability to bind pYLRP-CT consistent with the functional importance of these arginines. Thus, Fe65 PID1 is the first Dab-like PID domain demonstrated to have phosphorylation-dependent binding.
Figure 9.
Pymol structural alignment of Fe65 PID1 (pink) (PDB accession code 3D8D, (26)) with IRS PID (cyan) bound to the IL-4 receptor NPXY motif (green) (PDB accession code 1IRS, (41)). The phosphotyrosine is colored according to the atoms, the conserved arginines in Fe65 PID1 are red and in IRS PID are blue. Fe65 PID1 R451 and R470 align with the phosphate coordinating arginines of IRS (R212, R227) as shown in the sequence alignment presented with the same color scheme. The alignment was adapted from reference 26.
To explore other Dab-like PID domains that may also be phosphorylation-dependent, we used LALIGN to align Fe65 PID1 with all other Dab-like PID domains found by Unlik et al. in search of aligning the key arginine residues (R451, R470). The Dab-like PID domains belonging to CAPON and GULP were found to have highly basic residues that aligned with the Fe65 PID1 arginine residues. CAPON has been shown to bind dexras and synapsin; both of which do not contain an NPXY motif (35, 36). The PID domain of CAPON is not the only PID domain that has been shown to bind a ligand lacking an NPXY motif, demonstrating the high variability in PID domain binding mechanisms (37). PID domains, including that of Dab, have also been shown to bind phosphoinositide groups at a binding site distinct from the NPXY-peptide binding groove (38). The GULP PID domain has been shown to bind the NPXY motifs of LRP-CT, CED-1, and Stabilin-2 (39, 40). Although these ligands contain NPXY motifs, they have all been shown to bind GULP in the non-phosphorylated form. It will be interesting to see whether GULP will bind more strongly to pYLRP-CT. Our Dab-like PID domain search did not uncover any additional phosphorylation-dependent domains. We suspect that as the PID domain:ligand structure database is expanded, additional binding modes will be revealed and the PID domain classification system may need to be revised.
Supplementary Material
Acknowledgments
This work was funded by NIH grant R01 AG025343. MM was supported by the Hemoglobin and Blood Proteins Training Program (5T32DK007233).
Financial support for this work was provided by NIH grant RO1-AG025343 to EAK.
ABBREVIATIONS
- LRP
Low density lipoprotein receptor-related protein 1
- APP
amyloid precursor protein
- AICD
APP intracellular domain
- CTF
C-terminal fragment
- PID
protein interaction domain
- ITC
isothermal titration calorimetry
- HDX
hydrogen/deuterium exchange mass spectrometry
- TFA
trifluoroacetic acid
- ACN
acetonitrile
Footnotes
An additional figure showing the purity of the proteins is provided as supplementary information. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Ulery PG, Beers J, Mikhailenko I, Tanzi RE, Rebeck GW, Hyman BT, Strickland DK. Modulation of beta-amyloid precursor protein processing by the low density lipoprotein receptor-related protein (LRP). Evidence that LRP contributes to the pathogenesis of Alzheimer’s disease. J Biol Chem. 2000;275:7410–7415. doi: 10.1074/jbc.275.10.7410. [DOI] [PubMed] [Google Scholar]
- 2.Ando K, Iijima KI, Elliott JI, Kirino Y, Suzuki T. Phosphorylation-dependent regulation of the interaction of amyloid precursor protein with Fe65 affects the production of beta-amyloid. J Biol Chem. 2001;276:40353–40361. doi: 10.1074/jbc.M104059200. [DOI] [PubMed] [Google Scholar]
- 3.Pietrzik CU, Busse T, Merriam DE, Weggen S, Koo EH. The cytoplasmic domain of the LDL receptor-related protein regulates multiple steps in APP processing. EMBO J. 2002;21:5691–5700. doi: 10.1093/emboj/cdf568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang B, Hu Q, Hearn MG, Shimizu K, Ware CB, Liggitt DH, Jin LW, Cool BH, Storm DR, Martin GM. Isoform-specific knockout of FE65 leads to impaired learning and memory. J Neurosci Res. 2004;75:12–24. doi: 10.1002/jnr.10834. [DOI] [PubMed] [Google Scholar]
- 5.Zerbinatti CV, Wozniak DF, Cirrito J, Cam JA, Osaka H, Bales KR, Zhuo M, Paul SM, Holtzman DM, Bu G. Increased soluble amyloid-beta peptide and memory deficits in amyloid model mice overexpressing the low-density lipoprotein receptor-related protein. Proc Natl Acad Sci U S A. 2004;101:1075–1080. doi: 10.1073/pnas.0305803101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santiard-Baron D, Langui D, Delehedde M, Delatour B, Schombert B, Touchet N, Tremp G, Paul MF, Blanchard V, Sergeant N, Delacourte A, Duyckaerts C, Pradier L, Mercken L. Expression of human FE65 in amyloid precursor protein transgenic mice is associated with a reduction in beta-amyloid load. J Neurochem. 2005;93:330–338. doi: 10.1111/j.1471-4159.2005.03026.x. [DOI] [PubMed] [Google Scholar]
- 7.Hoe HS, Magill LA, Guenette S, Fu Z, Vicini S, Rebeck GW. FE65 interaction with the ApoE receptor ApoEr2. J Biol Chem. 2006;281:24521–24530. doi: 10.1074/jbc.M600728200. [DOI] [PubMed] [Google Scholar]
- 8.Smith MJ, Hardy WR, Murphy JM, Jones N, Pawson T. Screening for PTB domain binding partners and ligand specificity using proteome-derived NPXY peptide arrays. Mol Cell Biol. 2006;26:8461–8474. doi: 10.1128/MCB.01491-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z, Lee CH, Mandiyan V, Borg JP, Margolis B, Schlessinger J, Kuriyan J. Sequence-specific recognition of the internalization motif of the Alzheimer’s amyloid precursor protein by the X11 PTB domain. EMBO J. 1997;16:6141–6150. doi: 10.1093/emboj/16.20.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yun M, Keshvara L, Park CG, Zhang YM, Dickerson JB, Zheng J, Rock CO, Curran T, Park HW. Crystal structures of the Dab homology domains of mouse disabled 1 and 2. J Biol Chem. 2003;278:36572–36581. doi: 10.1074/jbc.M304384200. [DOI] [PubMed] [Google Scholar]
- 11.Radzimanowski J, Simon B, Sattler M, Beyreuther K, Sinning I, Wild K. Structure of the intracellular domain of the amyloid precursor protein in complex with Fe65-PTB2. EMBO Rep. 2008;9:1134–1140. doi: 10.1038/embor.2008.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boucher P, Liu P, Gotthardt M, Hiesberger T, Anderson RG, Herz J. Platelet-derived growth factor mediates tyrosine phosphorylation of the cytoplasmic domain of the low Density lipoprotein receptor-related protein in caveolae. J Biol Chem. 2002;277:15507–15513. doi: 10.1074/jbc.M200428200. [DOI] [PubMed] [Google Scholar]
- 13.Betts GN, van der Geer P, Komives EA. Structural and functional consequences of tyrosine phosphorylation in the LRP1 cytoplasmic domain. J Biol Chem. 2008;283:15656–15664. doi: 10.1074/jbc.M709514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Q, Cool B, Wang B, Hearn M, Martin G. A candidate molecular mechanism for the association of an intronic polymorphism of FE65 with resistance to very late onset dementia of the Alzheimer type. Hum Mol Genet. 2002;11:465–475. doi: 10.1093/hmg/11.4.465. [DOI] [PubMed] [Google Scholar]
- 15.Fiore F, Zambrano N, Minopoli G, Donini V, Duilio A, Russo T. The regions of the Fe65 protein homologous to the phosphotyrosine interaction/phosphotyrosine binding domain of Shc bind the intracellular domain of the Alzheimer’s amyloid precursor protein. J Biol Chem. 1995;270:30853–30856. doi: 10.1074/jbc.270.52.30853. [DOI] [PubMed] [Google Scholar]
- 16.Borg JP, Ooi J, Levy E, Margolis B. The phosphotyrosine interaction domains of X11 and FE65 bind to distinct sites on the YENPTY motif of amyloid precursor protein. Mol Cell Biol. 1996;16:6229–6241. doi: 10.1128/mcb.16.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Q, Wang L, Yang Z, Cool BH, Zitnik G, Martin GM. Endoproteolytic cleavage of FE65 converts the adaptor protein to a potent suppressor of the sAPPalpha pathway in primates. J Biol Chem. 2005;280:12548–12558. doi: 10.1074/jbc.M411855200. [DOI] [PubMed] [Google Scholar]
- 18.Cao X, Sudhof TC. Dissection of Amyloid-B Precursor Protein-dependent Transcriptional Transavtivation. J Biol Chem. 2004;279:24601–24611. doi: 10.1074/jbc.M402248200. [DOI] [PubMed] [Google Scholar]
- 19.Trommsdorff M, Borg JP, Margolis B, Herz J. Interaction of cytosolic adaptor proteins with neuronal apolipoprotein E receptors and the amyloid precursor protein. J Biol Chem. 1998;273:33556–33560. doi: 10.1074/jbc.273.50.33556. [DOI] [PubMed] [Google Scholar]
- 20.Zambrano N, Minopoli G, de Candia P, Russo T. The Fe65 adaptor protein interacts through its PID1 domain with the transcription factor CP2/LSF/LBP1. J Biol Chem. 1998;273:20128–20133. doi: 10.1074/jbc.273.32.20128. [DOI] [PubMed] [Google Scholar]
- 21.Cao X, Sudhof TC. A transcriptionally active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- 22.Kinoshita A, Whelan CM, Smith CJ, Mikhailenko I, Rebeck GW, Strickland DK, Hyman BT. Demonstration by fluorescence resonance energy transfer of two sites of interaction between the low-density lipoprotein receptor-related protein and the amyloid precursor protein: role of the intracellular adapter protein Fe65. J Neurosci. 2001;21:8354–8361. doi: 10.1523/JNEUROSCI.21-21-08354.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pietrzik CU, Yoon IS, Jaeger S, Busse T, Weggen S, Koo EH. FE65 constitutes the functional link between the low-density lipoprotein receptor-related protein and the amyloid precursor protein. J Neurosci. 2004;24:4259–4265. doi: 10.1523/JNEUROSCI.5451-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uhlik MT, Temple B, Bencharit S, Kimple AJ, Siderovski DP, Johnson GL. Structural and evolutionary division of phosphotyrosine binding (PTB) domains. J Mol Biol. 2005;345:1–20. doi: 10.1016/j.jmb.2004.10.038. [DOI] [PubMed] [Google Scholar]
- 25.Eck MJ, Dhe-Paganon S, Trub T, Nolte RT, Shoelson SE. Structure of the IRS-1 PTB domain bound to the juxtamembrane region of the insulin receptor. Cell. 1996;85:695–705. doi: 10.1016/s0092-8674(00)81236-2. [DOI] [PubMed] [Google Scholar]
- 26.Radzimanowski J, Ravaud S, Schlesinger S, Koch J, Beyreuther K, Sinning I, Wild K. Crystal structure of the human Fe65-PTB1 domain. J Biol Chem. 2008;283:23113–23120. doi: 10.1074/jbc.M800861200. [DOI] [PubMed] [Google Scholar]
- 27.Guttman M, Prieto JH, Croy JE, Komives EA. Decoding of lipoprotein-receptor interactions: properties of ligand binding modules governing interactions with apolipoprotein E. Biochemistry. 2010;49:1207–1216. doi: 10.1021/bi9017208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guttman M, Betts GN, Barnes H, Ghassemian M, van der Geer P, Komives EA. Interactions of the NPXY microdomains of the low density lipoprotein receptor-related protein 1. Proteomics. 2009;9:5016–5028. doi: 10.1002/pmic.200900457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seeliger MA, Young M, Henderson MN, Pellicena P, King DS, Falick AM, Kuriyan J. High yield bacterial expression of active c-Abl and c-Src tyrosine kinases. Protein Sci. 2005;14:3135–3139. doi: 10.1110/ps.051750905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bergqvist S, Croy CH, Kjaergaard M, Huxford T, Ghosh G, Komives EA. Thermodynamics reveal that helix four in the NLS of NF-kappaB p65 anchors IkappaBalpha, forming a very stable complex. J Mol Biol. 2006;360:421–434. doi: 10.1016/j.jmb.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mandell JG, Falick AM, Komives EA. Measurement of amide hydrogen exchange by MALDI-TOF mass spectrometry. Anal Chem. 1998;70:3987–3995. doi: 10.1021/ac980553g. [DOI] [PubMed] [Google Scholar]
- 32.Croy CH, Bergqvist S, Huxford T, Ghosh G, Komives EA. Biophysical characterization of the free IkappaBalpha ankyrin repeat domain in solution. Protein Sci. 2004;13:1767–1777. doi: 10.1110/ps.04731004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barnes H, Ackermann EJ, van der Geer P. v-Src induces Shc binding to tyrosine 63 in the cytoplasmic domain of the LDL receptor-related protein 1. Oncogene. 2003;22:3589–3597. doi: 10.1038/sj.onc.1206504. [DOI] [PubMed] [Google Scholar]
- 34.Lee MS, Kao SC, Lemere CA, Xia W, Tseng HC, Zhou Y, Neve R, Ahlijanian MK, Tsai LH. APP processing is regulated by cytoplasmic phosphorylation. J Cell Biol. 2003;163:83–95. doi: 10.1083/jcb.200301115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaffrey SR, Benfenati F, Snowman AM, Czernik AJ, Snyder SH. Neuronal nitric-oxide synthase localization mediated by a ternary complex with synapsin and CAPON. Proc Natl Acad Sci U S A. 2002;99:3199–3204. doi: 10.1073/pnas.261705799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang M, Jaffrey SR, Sawa A, Ye K, Luo X, Snyder SH. Dexras1: a G protein specifically coupled to neuronal nitric oxide synthase via CAPON. Neuron. 2000;28:183–193. doi: 10.1016/s0896-6273(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 37.Xu H, Lee KW, Goldfarb M. Novel recognition motif on fibroblast growth factor receptor mediates direct association and activation of SNT adapter proteins. J Biol Chem. 1998;273:17987–17990. doi: 10.1074/jbc.273.29.17987. [DOI] [PubMed] [Google Scholar]
- 38.Stolt PC, Vardar D, Blacklow SC. The dual-function disabled-1 PTB domain exhibits site independence in binding phosphoinositide and peptide ligands. Biochemistry. 2004;43:10979–10987. doi: 10.1021/bi049092l. [DOI] [PubMed] [Google Scholar]
- 39.Su HP, Nakada-Tsukui K, Tosello-Trampont AC, Li Y, Bu G, Henson PM, Ravichandran KS. Interaction of CED-6/GULP, an adapter protein involved in engulfment of apoptotic cells with CED-1 and CD91/low density lipoprotein receptor-related protein (LRP) J Biol Chem. 2002;277:11772–11779. doi: 10.1074/jbc.M109336200. [DOI] [PubMed] [Google Scholar]
- 40.Park SY, Kang KB, Thapa N, Kim SY, Lee SJ, Kim IS. Requirement of adaptor protein GULP during stabilin-2-mediated cell corpse engulfment. J Biol Chem. 2008;283:10593–10600. doi: 10.1074/jbc.M709105200. [DOI] [PubMed] [Google Scholar]
- 41.Zhou MM, Huang B, Olejniczak ET, Meadows RP, Shuker SB, Miyazaki M, Trub T, Shoelson SE, Fesik SW. Structural basis for the IL-4 receptor phosphopeptide recognition by the IRS-1 PTB domain. Nat Struct Mol Biol. 1996;3:388–393. doi: 10.1038/nsb0496-388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.