Abstract
Given the functional interdependencies between the molecular components in a human cell, a disease is rarely a consequence of an abnormality in a single gene, but reflects the perturbations of the complex intracellular network. The emerging tools of network medicine offer a platform to explore systematically not only the molecular complexity of a particular disease, leading to the identification of disease modules and pathways, but also the molecular relationships between apparently distinct (patho)phenotypes. Advances in this direction are essential to identify new diseases genes, to uncover the biological significance of disease-associated mutations identified by genome-wide association studies and full genome sequencing, and to identify drug targets and biomarkers for complex diseases.
Introduction
In humans, as in other organisms, most cellular components exert their functions through interactions with other cellular components, the totality of these interactions representing the human interactome. The potential complexity of this network is daunting: with approximately 25,000 protein-encoding genes, about a thousand metabolites, and an as yet undefined number of distinct proteins (splice variants and more than 300 different post-translationally modified forms1) and functional RNA molecules, the distinct cellular components that serve as the nodes of the interactome easily exceed one hundred thousand. The number of functionally relevant interactions between the components of this network, representing the links of the interactome, is expected to be much larger and remains largely unknown2.
This subcellular interconnectivity implies that the impact of a specific genetic abnormality is not restricted to the activity of the gene product that carries it, but can spread along the links of the network, and alter the activity of gene products that otherwise carry no defects. Therefore, the phenotypic impact of a defect3 is not determined solely by the known function of the mutated gene, but also by the functions of components with which the gene and its products interact and of their interaction partners, i.e., by its network context. Following on this principle, a key hypothesis underlying this review is that a disease is rarely a consequence of an abnormality in a single effector gene product. Instead, the disease phenotype is a reflection of various pathobiological processes that interact in a complex network. A corollary of this hypothesis is that the interdependencies among a cell’s molecular components lead to deep functional, molecular, and causal relationships among apparently distinct phenotypes.
Network-based approaches to human disease can have multiple biological and clinical applications. Indeed, a better understanding of the implications of cellular interconnectedness on disease progression could lead to identification of disease genes and disease pathways, which, in turn, could offer better targets for drug development. These advances could also reshape clinical practice, from the discovery of better and more accurate biomarkers monitoring the functional integrity of the network perturbed by the diseases, to better disease classification, paving the way to personalized therapies and treatment. Our aim here is to present an overview of the organizing principles that govern cellular networks and their role in disease. Indeed, these organizing principles, and the tools and methodologies derived from them, are facilitating the emergence of a body of knowledge that is increasingly referred to as network medicine4–6, offering a quantitative platform to address the complexity of human disease.
The human interactome
Owing to the conservation of biochemical and molecular functions across species, much of our current understanding of cellular networks is derived from model organisms. Yet, in the past decade we witnessed an exceptional growth in human-specific molecular interaction data, helping us understand the interlocking networks that play a key role in human disease7. Most attention is focused on molecular networks, including: protein interaction networks, whose nodes are proteins linked to each other via physical (binding) interactions8, 9; metabolic networks, whose nodes are metabolites linked if they participate in the same biochemical reactions10–12; regulatory networks, whose directed links represent regulatory relationships between a transcription factor and a gene13, or post-translational modifications, such as those between a kinase and its substrates14; and RNA networks, capturing the role of RNA-DNA interactions such as small non-coding microRNAs15 and siRNAs16 in regulating gene expression. In parallel, an increasing number of studies rely on phenotypic networks that include: co-expression networks, in which genes with similar co-expression patterns are linked17; and genetic networks, in which two genes are linked if the phenotype of a double mutant differs from the expected phenotype of two single mutants18, 19. Typically the links of a phenotypic network reflect some pathways in the underlying molecular networks. For example, in yeast, the protein products of gene pairs that display positive genetic interactions often interact directly with each other19.
While current human interactome maps are incomplete and noisy, in the past few years we have witnessed systematic efforts to increase their coverage and accuracy, as well as to estimate the interactome size and correct for known biases2, 20, 21. Yet, in exploring the interplay between networks and human diseases, we first need to assess how comprehensive and accurate the current molecular and phenotypic network maps are, an issue addressed in Box 1.
BOX 1: Biological Network Maps and Interaction Resources.
While the bulk of research on biological networks has focused on E. coli and S. cerevisiae, following the human genome project, the amount of data pertaining to networks in the human cells exceeds in richness and diversity the data available for model organisms. In the following, we briefly discuss the most studied network maps and their limitations, but remind the reader to exercise caution as we are describing a rapidly changing landscape. The links and references to pertinent databases are available online.
Protein-protein interaction networks: In the past five years, significant efforts have been made towards obtaining comprehensive protein interaction maps. High-throughput yeast-two-hybrid maps for humans have been generated by several groups2, 8, 9 yielding over 7,000 binary interactions. The immunoprecipitation and high-throughput mass-spectrometry technique, which identifies co-complexes, has begun to be applied as well to humans93. There have also been major efforts to curate the interactions individually validated in the literature into databases94, such as the Münich Information Center for Protein Sequence (MIPS) protein interaction database, the Biomolecular Interaction Network Database (BIND), the Database of Interacting Proteins (DIP), the Molecular Interaction database (MINT), and the protein Interaction database (IntAct). More recent protein-protein interaction curation efforts, the Biological General Repository for Interaction Datasets (BioGRID), and the Human Protein Reference Database (HPRD) have attempted larger-scale curation of data. Despite these extensive curation efforts, the existing maps are considered incomplete2, and the literature-based datasets, while richer in interactions, are prone to investigative biases21, containing more interactions for the more explored disease proteins36.
Metabolic networks: The metabolic network maps are likely the most comprehensive of all biological networks. Databases such as the Kyoto Encyclopedia of Genes and Genomes (KEGG) and the Biochemical Genetic and Genomics knowledgebase (BIGG) contain the metabolic network of a wide range of species. Recently, Duarte et al.12 published a comprehensive literature-based genome-scale metabolic reconstruction of human metabolism with 2,766 metabolites and 3,311 metabolic and transport reactions. An independent manual construction by Ma et al.95 contains nearly 3,000 metabolic reactions, organized into about 70 human-specific metabolic pathways.
Regulatory networks: Mapping of the human regulatory network is in its infancy, making this network perhaps the most incomplete among all biological networks. Data generated by experimental techniques, such as ChIP-on-chip and ChIP-Sequencing, have started to be collected in databases such as Universal Protein Binding Microarray Resource for Oligonucleotide Binding Evaluation (UniPROBE) and JASPAR. Literature-curated and predicted protein-DNA interactions have been compiled in various databases, such as TRANSFAC and the B-cell interactome (BCI). Human post-translational modifications can be found in databases such as Phospho.ELM, PhosphoSite, and phosphorylation site database (PHOSIDA).
RNA networks: RNA networks can refer to networks containing RNA-RNA or RNA-DNA interactions. Recently, with the increased understanding of microRNAs’ role in disease68, microRNA-gene networks have been constructed using predicted microRNA targets available in databases such as TargetScan, PicTar, microRNA, miRBase, and miRDB. The number of experimentally supported targets is also increasing, which are now compiled in databases such as TarBase and miRecords.
Properties of disease networks
Network medicine relies on a series of advances in network theory22–27, which have provided insights into the properties of biological networks more generally. These studies have indicated that networks emerging in biological, technological, or social systems are not random, but are characterized by a core set of organizing principles, as summarized in Box 2. Understanding diseases in the context of these network principles allows us to answer some fundamental properties of the genes that are involved in disease. Indeed, only about 10% of human genes have a known disease-association28 (Fig. 1a), forcing one to ask: do disease genes have unique, quantifiable characteristics that distinguish them from other, non-disease genes? From a network perspective, this question translates into the following: if we consider the corpus of all known disease genes, are they placed randomly on the interactome, or are there detectable correlations between their location and network topology?
BOX2: Elements of network theory.
An important realization of the past decade is that networks appearing in natural, technological, and social systems are not random, but follow a series of basic organizing principles in their structure and evolution that distinguish them from randomly linked networks. In the following, we summarize the aspects of network theory that pertain to biological networks. For a more detailed exposition see Refs.22–27.While these principles were found to apply to a wide variety of networks, in the context of this review, they refer to biological networks, seen as nodes (e.g., proteins, metabolites, diseases) connected by links (e.g., protein-protein interactions, metabolic reactions, or shared genes) as discussed throughout the review.
Degree distribution and hubs: In a random network, most nodes have approximately the same number of links, and highly connected nodes (hubs) are quite rare. The fraction of links with a given degree, called the degree distribution, follows the well-known Poisson distribution. In contrast, many real networks, including human protein-protein interaction and metabolic networks are scale-free96, which means that the degree distribution has a power-law tail, i.e., the degree distribution P(k) with degree k follows P(k) ~ k−γ, where γ is called the degree exponent. The most noticeable consequence of this property is the presence of a few highly connected hubs that hold the whole network together29. The biological role and dynamical behavior of hubs allowed their classification into “party” hubs, which function inside modules and coordinate specific cellular processes, and “date” hubs, which link together rather different processes and organize the interactome45, 97.
Small world phenomena: Most complex networks (including random networks) display the small world property, which means that there are relatively short paths between any pair of nodes98. This observation means that most proteins (or metabolites) are only a few interactions (or reactions) from any other proteins (metabolites)10, 11, 29. Therefore, perturbing the state of a given node can affect the activity of most nodes in their vicinity as well as of the behavior of the network itself.
Motifs: Some subgraphs (a group of nodes that link to each other forming a small subnetwork within a network) in biological networks appear more (or less) frequently than expected given the network’s degree distribution. Such subgraphs are often called motifs99, and they are likely associated with some optimized biological function (e.g., negative feedback loop, positive feed forward loop, bifan, oscillator).
Modules: Most networks display a high degree of clustering, implying the existence of topological modules, representing highly interlinked local regions in the network. While the identification of such modules can be computationally challenging, a wide array of network clustering tools have emerged in the past few years40–43.
Betweenness centrality: Nodes with a high betweenness centrality (a measure of the number of shortest paths that go through each node) are often called bottlenecks. In networks with directed edges such as regulatory networks, bottlenecks tend to correlate with essentiality100.
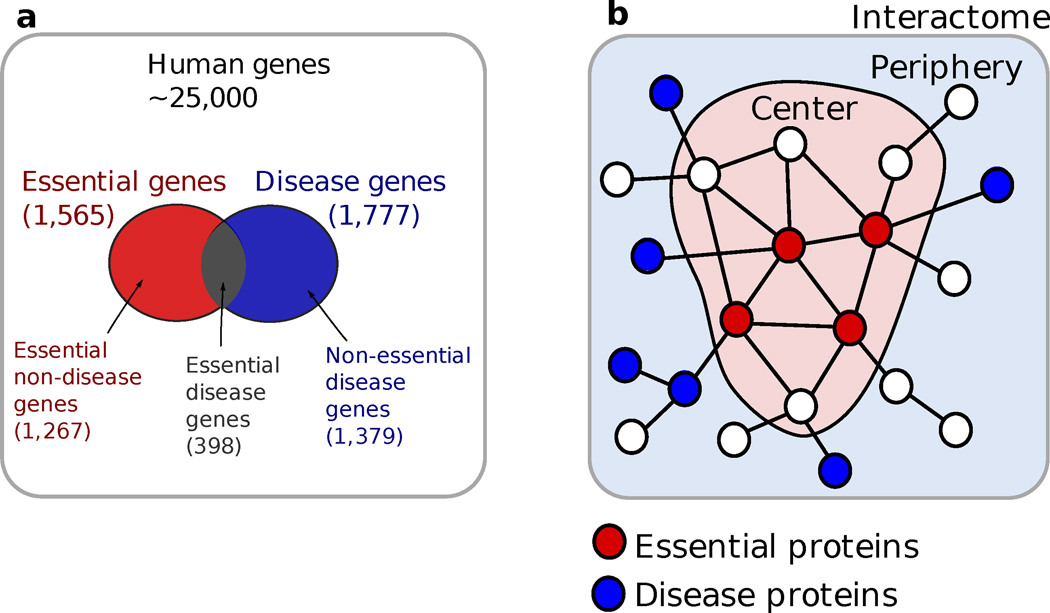
Figure 1. Disease and essential genes in the interactome.
(a) Of the approximately 25,000 genes, only about 1,700 have been associated with specific diseases. In addition, about 1,600 genes are known to be in utero essential, i.e., their absence is associated with embryonic lethality. (b) Schematic illustration of the differences between essential and non-essential disease genes. Non-essential disease genes (illustrated as blue nodes) are found to segregate at the network periphery whereas in utero essential genes (illustrated as red nodes) tend to be at the functional center (encode hubs, expressed in many tissues) of the interactome.
Location of disease genes within networks
An unexpected property of biological networks is the emergence of hub proteins (Box 2), suggesting that hubs must play a special biological role. Indeed, evidence from model organisms indicates that hub proteins tend to be encoded by essential genes29, and that genes encoding hubs are older and evolve more slowly than genes encoding non-hub proteins30–32. The deletion of genes encoding hubs also leads to a larger number of phenotypic outcomes than the deletion of genes encoding less connected proteins21. While the strength of evidence for some of these effects is still debated21, 33, by virtue of the many interactions they have, one expects that the absence of a hub would affect the function of an exceptional number of other proteins. This assumption has led to the hypothesis that, in humans, hubs should typically be associated with disease genes. Indeed, the protein products of up-regulated genes in lung squamous cell carcinoma tend to have a high degree of connectivity34 and 346 proteins implicated in cancer have, on average, twice as many interaction partners as non-cancer proteins35. Moving beyond cancer, one study found that disease proteins in the OMIM Morbid Map28 have more protein-protein interactions than non-disease proteins in a literature-curated protein-protein interaction datase36.
Note, however, that the essential gene concept in simple organisms does not map uniquely into disease genes in humans. Indeed, some human genes are essential in early development, so functional changes in them often lead to first-trimester spontaneous abortions (embryonic lethality). Mutations in such ‘essential’ genes cannot propagate in the population, as individuals carrying them cannot reproduce. In contrast, individuals can tolerate for a long time the disease-causing mutations, often past their reproductive age. The question is, are both (disease and essential) genes associated with hubs? Goh et al37 found that essential genes show a strong tendency to be associated with hubs and expressed in multiple tissues, i.e., they tend to be located at the functional center of the interactome (Fig. 1). Yet, in contrast with our initial hypothesis, non-essential disease genes do not show a tendency to encode hubs and tend to be tissue-specific. That is, from a network perspective, these genes segregate at the functional periphery of the interactome (Fig. 1b). In summary, in human cells it is the essential genes, and not the disease genes, that are encoding hubs. This difference can be understood from an evolutionary perspective: mutations that disrupt hubs have difficulty propagating in the population, as the absence of hubs create so many disruptions that the host may not survive long enough to reproduce. Thus, only mutations that impair functionally or topologically peripheral genes can persist, accounting for the family of heritable diseases, especially those that appear in adulthood.
Local clustering of disease genes – disease modules
If a gene or molecule is involved in a specific biochemical process or disease, its direct interactors might also be suspected to play some role in the same biochemical process. In line with this “local” hypothesis (Box 3), proteins involved in the same disease show a high propensity to interact with each other37, 38. For example, Goh et al.37 observed 290 physical interactions between the products of genes associated with the same disorder, representing a 10-fold increase relative to random expectation (P < 10−6). Furthermore, Gandhi et al.39 and Xu and Li36 found that genes linked to diseases with similar phenotypes have a significantly increased tendency to interact directly with each other. These observations indicate that if we identify a few disease components, the other disease-related components will likely be in their network-based vicinity. That is, we expect that each disease can be linked to a well-defined neighborhood of the interactome, often referred to as a disease module.
BOX 3: Hypotheses of Network Medicine.
Network medicine is based on a series of widely used (and often unspoken) hypotheses and organizing principles that link network structure to biological function and disease. Next, we summarize some of the most frequently utilized hypotheses, their use being discussed in more detail in the main text.
Hubs: Non-essential disease genes (representing the majority of all known disease genes) tend to avoid hubs and segregate at the functional periphery of the interactome. In utero essential genes tend to associated with hubs.
Local hypothesis: Proteins involved in the same disease have an increased tendency to interact with each other.
Corollary of the local hypothesis: Mutations in interacting proteins often lead to similar disease phenotypes.
Disease module hypothesis: Cellular components associated with a specific disease phenotype show a tendency to cluster in the same network neighborhood.
Network parsimony principle: Causal molecular pathways often coincide with the shortest molecular paths between known disease-associated components.
Shared components hypothesis: Diseases that share disease-associated cellular components (genes, proteins, metabolites, miRNAs) show phenotypic similarity and comorbidity.
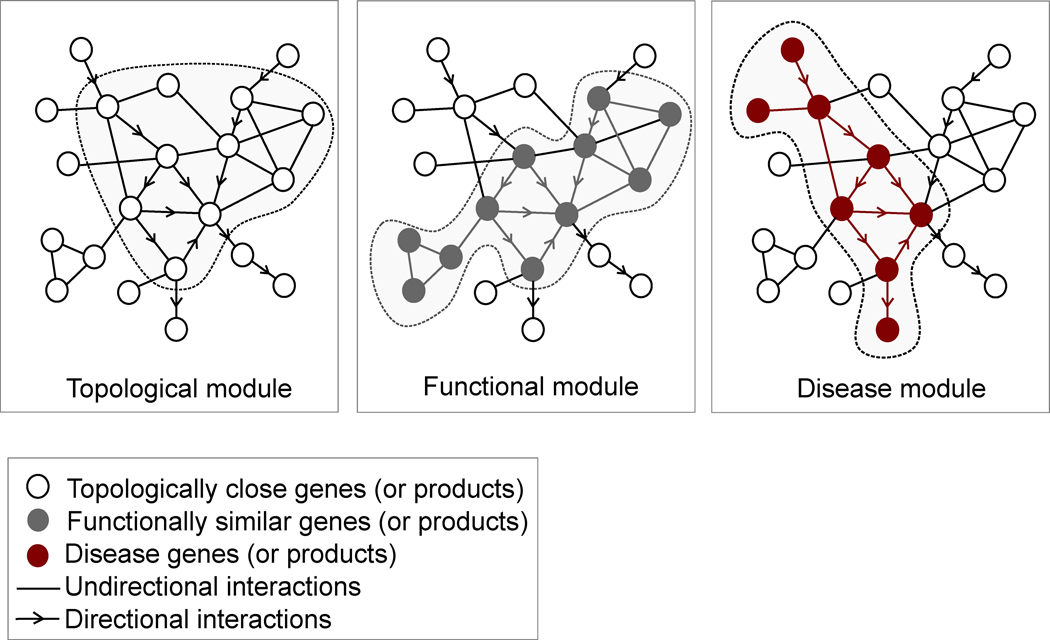
As we try to understand the network-based position of disease genes, we need to distinguish among three distinct phenomena (Fig. 2). A topological module represents a locally dense neighborhood in a network, such that nodes have a higher tendency to link to nodes within the same local neighborhood than to nodes outside of it. It can be identified using various network clustering algorithms40–43 that are blind to the function of individual nodes. By contrast, a functional module represents the aggregation of nodes of similar or related function in the same network neighborhood. Finally, a disease module represents a group of network components that together contribute to a cellular function whose disruption results in a particular disease phenotype.
Figure 2. Disease modules.
Schematic illustration of the three modularity concepts discussed in the review. (a) Topological modules correspond to locally dense neighborhoods of the interactome, such that the nodes of the module show a higher tendency to interact with each other than with nodes outside of the module. As such, topological modules represent a pure network property. (b) Functional modules correspond to network neighborhoods in which there is a statistically significant segregation of nodes of related function. A functional module, thus, requires us to define some nodal characteristics (illustrated as gray nodes), and relies on the hypothesis that nodes involved in closely related cellular functions tend to interact with each other and thus are located in the same network neighborhood. (c) A disease module represents a group of nodes whose perturbation (mutations, deletions, copy number variations, or expression changes) can be linked to a particular disease phenotype, shown as red nodes. The tacit assumption in network medicine is that the topological, functional, and disease modules overlap so that functional modules correspond to topological modules and a disease can be viewed as the breakdown of a functional module.
In the biological literature, there is a tacit assumption that these three concepts are interrelated: cellular components that form a topological module have closely related functions, thus corresponding to a functional module; and a disease is a result of the breakdown of a particular functional module, intimating that a functional module is also a disease module. However, several unique characteristics of disease modules are important to bear in mind. First, a disease module may not be identical to, but likely overlaps with, the topological and/or functional modules. Second, a disease module is defined in relation to a particular disease, and, accordingly, each disease has its own unique module. Finally, a gene, protein, or metabolite can be implicated in several disease modules, which means that different disease modules can overlap.
The disease module hypothesis represents a network-level expansion of the disease gene hypothesis. The emergence of a disease is viewed as a combinatorial problem in which many different defects and perturbations result in a similar disease phenotype, provided they alter the activity of the disease module. Such combinatorial disease mechanisms are well documented in cancer44, but the utility of the disease module hypothesis extends beyond polygenic diseases, and is important even in some monogenic diseases. For example, sickle cell disease, a classic Mendelian disorder, is caused by a single point mutation at position 6 of the beta-chain of hemoglobin. Yet, this simple biochemical phenotype and its corresponding monogenotype do not yield a single pathophenotype: individuals with sickle cell disease can present with painful crises, osteonecrosis, acute chest syndrome, stroke, profound anemia, or mild asymptomatic anemia. Thus, the underlying disease module will likely include all disease modifying genes (e.g., hemoglobin F) that mediate various epigenetic, transcriptional, and post-translational phenomena. An important step of network-based approaches to disease is, therefore, to identify the disease module for the pathophenotype of interest, which, in turn, can guide further experimental work and influence drug development.
Identifying disease modules
Bioinformatics approaches
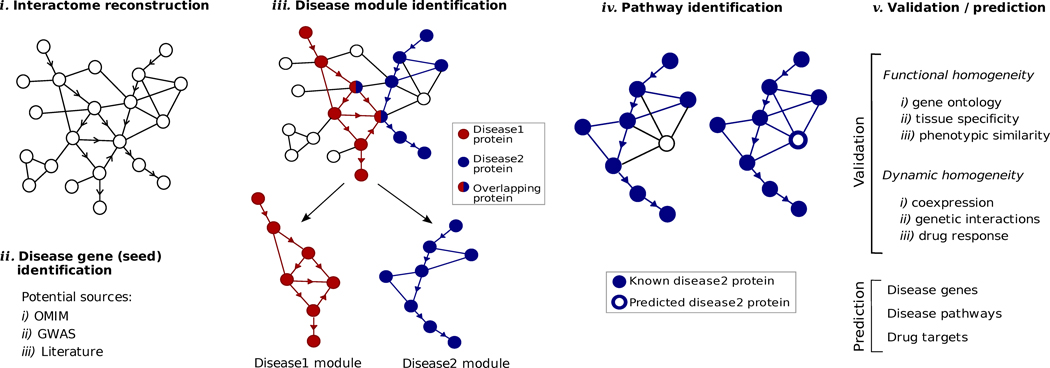
Disease modules can be identified on the basis of currently available data using bioinformatics approaches, whose main steps are described in Figure 3. Variants of this methodology have been applied to a wide range of diseases and pathophenotypes, from cardiovascular disease to various forms of cancer. For example, Taylor et al.45 studied the dynamic modular structure of the protein interaction network in adenocarcinoma of the breast, finding that hub proteins that displayed altered modularity in the human interactome were useful indicators for predicting breast cancer outcome. Similarly, Chen et al.46 relied on co-expression networks constructed from liver and adipose tissues, facilitating the identification of sub-networks associated with genetic loci linked to obesity- and diabetes-related DNA variations. The results confirmed the connection between obesity and a macrophage-enriched metabolic subnetwork, validating three previously unknown genes, LPL, LACTB, and PPM1L, as obesity genes in transgenic mice. An extensive list of diseases to which the disease module has been identified, with the pertinent references, can be found in Table I online.
Figure 3. Identifying and validating disease modules.
- Interactome reconstruction, which merges the most up-to-date information on protein-protein interactions, co-complex memberships, regulatory interactions, and metabolic network maps (Box 1) in the tissue and cell line of interest. These networks are occasionally augmented with phenotypic links, such as coexpression-based relationships, but such phenotypic measures are best utilized later to test the functional homogeneity of the predicted disease module.
- Disease gene (seed) identification, collects the known disease-associated genes obtained from linkage analysis, GWAS, or other sources, serving as the seed of the disease module.
- Disease module identification. The seed genes are placed on the interactome, aiming to identify a subnetwork that contains most of the disease-associated components, exploiting both the functional and topological modularity of the network. If such statistically significant agglomeration is detected, then one can use a combination of clustering tools40–43 to identify the functionally and topologically compact subgraph that contains most disease components, representing the potential disease module. The closer the phenotypic manifestations are of the two diseases (organ, symptoms, drug response), the more significant is the expected overlap between the modules associated with two diseases.
- Pathway identification: Occasionally, the number of components the ascertained disease module contains is so large that it cannot serve as a tractable starting point for further experimental work. In this case it may be necessary to identify the specific molecular pathways whose disruption may be responsible for the disease phenotype. One typically uses the network parsimony principle (Box 3) to select the most likely disease pathways, assuming that causal pathways are the shortest paths connecting the known disease components.
- Validation/prediction: The disease modules are tested for their functional and dynamic homogeneity. The nature of the validation depends on the tools and data available to the investigator; gene expression data can validate the dynamical integrity of the disease module, and GWAS can be used to test the potential links between the SNPs of the predicted cellular components and the disease phenotype. Finally, the predicted disease genes and pathways (serving also as potential drug targets) are tested using the available molecular biology tools and animal models.
The disease module-based approach is useful in exploring pathogen-induced phenotypes, as well. Pathogens often disrupt the cell’s normal activity by interacting with and transferring their genes or proteins to the host cell; thus, a thorough understanding of this process requires a map of the interactions between the molecular components of the virus and the host cell47, 48. A recent study has shown that the array of diseases associated with Epstein-Barr virus and human papillomavirus are linked to genes whose protein products lie in topological proximity to the viral target proteins49, leading to the identification of the viral disease module. In light of these advances, an area ripe for network-based approaches is the bacterial microbiome (and other metagenomes) and its relationship to human disease50.
Experimental mapping of disease modules
Often the rate-limiting step in mapping a disease module is the small coverage of the available cellular interaction maps in the vicinity of the known disease components, requiring additional experimental efforts to identify relevant interactions. This approach was successfully applied to several diseases, including Huntington disease51, spinocerebellar ataxia52, breast cancer53, and schizophrenia54. For example, starting from 23 known ataxia-causing genes, Lim et al.52 used yeast two-hybrid assays to map out their interactions with other human proteins; the interactions of this second group of proteins were then used to build a dense subnetwork two degrees removed from the known ataxia genes. A member of the predicted ataxia disease module, puratrophin-1, a common binding partner to many of the known ataxia genes, which were not previously recognized as having any commonality, was later shown to lead to ataxia-like phenotypes in mice upon its deletion55.
Predicting disease genes
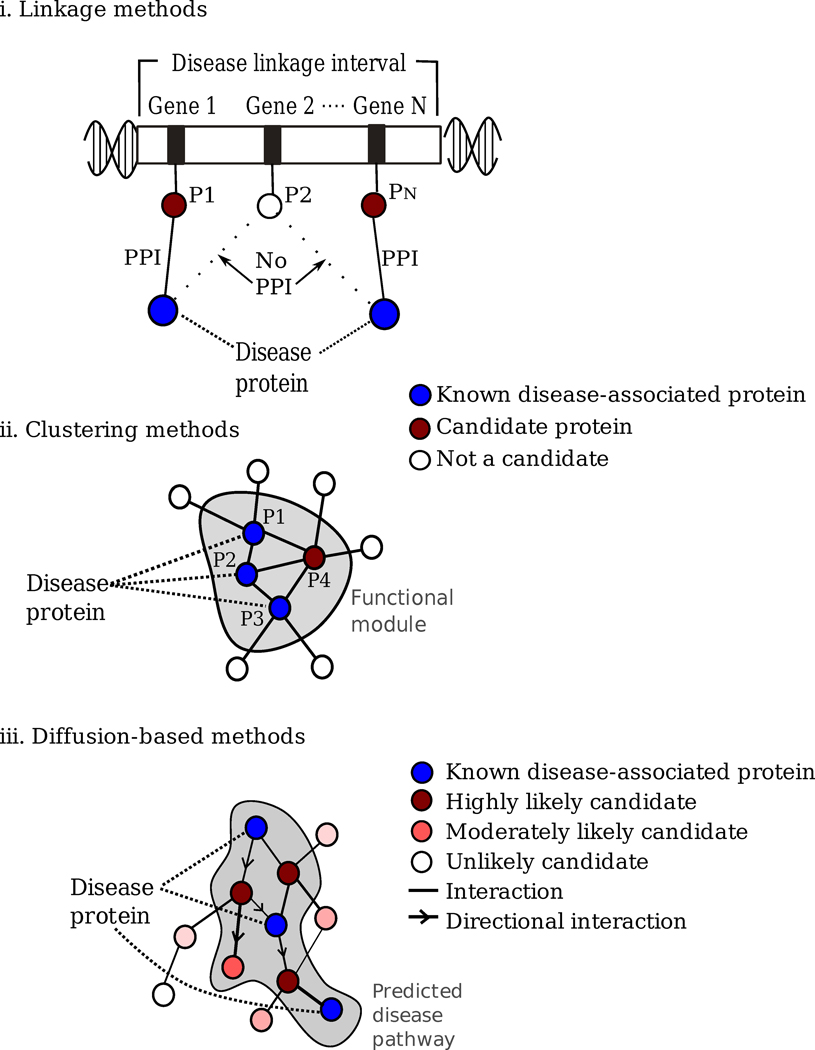
Traditionally, disease-associated genes were discovered by linking genomic intervals containing hundreds of genes to a particular phenotype, or, more recently, with genome-wide association studies (GWAS)56, identifying SNPs that have a statistically significant correlation with the disease. Both methodologies can offer a large number of disease-gene candidates, but identifying the particular gene and the mutation that is causal to the disease remains a difficult undertaking. Recently, a series of increasingly sophisticated network-based tools have been developed to predict potential disease genes, integrating in the network context knowledge about a particular disease, whether it derives from GWAS, full-genome sequencing, linkage methods, or individual studies. The existing tools can be loosely grouped into three categories (Fig. 4):
Linkage methods assume that the direct interaction partners of a disease protein are likely candidates to be associated with the same disease phenotype38, 57, 58. Indeed, Oti et al.38 showed that the set of genes that fell within one of the known disease loci and whose products interacted with a known disease protein were 10-fold enriched in true disease-causing genes; and by considering cellular localization as well, the network information lead to a 1000-fold enrichment over a random selection. Using this feature, they predicted (and confirmed using independent data) that Janus kinase 3 (JAK3) was a candidate protein for severe combined immunodeficiency syndrome, due to its interaction with lymphocyte specific protein-tyrosine (LCK), protein-tyrosine phosphatase (PTPRC), and interleukin 2 receptor (IL2RG), known disease-associated proteins.
Modularity or pathway-based methods assume that all cellular components that belong to the same topological/functional/disease module have a high likelihood of being involved in the same disease59, 60. Thus these methods start with identifying the disease modules (Fig 3), and inspecting their members as potential disease genes. For example, Wu et al.61 showed that many of the recently identified breast cancer susceptibility genes were ranked highly by an algorithm which took advantage of the positive correlation between modularity in the protein interaction network and phenotype-phenotype similarity network.
Diffusion-based methods aim to identify the pathways that are closest to the known disease genes. By releasing hypothetical random walkers from the protein products of the known disease genes, that are then allowed to diffuse along the links of the interactome (moving to any neighboring node with equal probability), one can identify the nodes and links that are closest to the known disease genes, as they will be those most often visited by the random walkers during their random walks. Proteins that interact with several disease proteins will gain a high probabilistic weight, as will those that may not directly interact with any disease proteins but are in close network proximity thereto, helping prioritize proteins and interactions based on their potential involvement in the particular disease. Variants of this methodology have been applied to detect disease genes related to a wide range of diseases, from diabetes mellitus to prostate cancer and Alzheimer disease62, 63.
Figure 4. identifying disease gene candidates.
(i) Linkage methods. Genes located within the linkage interval of a disease whose protein products interact with a known disease-associated protein are considered likely candidate disease genes38, 59. (ii) Clustering methods. Clustering or graph partitioning helps us uncover functional and potential disease modules in the interactome. The members of such modules are considered candidate disease genes59, 61. (iii) Diffusion-based methods: Starting from proteins known to be associated with a disease, a random walker (or a propagator) visits each node in the interactome with a certain probability62, 63. The outcome of the algorithm is a disease-association score assigned to each protein, the likelihood that a particular protein is associated with the disease.
These methodologies (A–C) exploit to an increasing degree the topological and functional information encoded by the interactome. Method A involves only pair-wise linkage information (local hypothesis, Box 3), while the modularity-based method (B) exploits the full network neighborhood of disease genes (disease module hypothesis, Box 3). Finally, diffusion-based methods (C) use the information encoded in the full network topology and the placement of the known disease genes, thereby simultaneously exploiting both topological and functional modularity (together with the parsimony principle, Box 3). It is not surprising, therefore, that a recent comparative study found, that, on the same dataset, linkage-based methods have the least predictive power and that diffusion-based methods offer the best predictive performance59.
Human Diseasome
The highly interconnected nature of the interactome means that at the molecular level, it is difficult, if not counter-intuitive, to consider diseases as being invariably independent of one another. Indeed, different disease modules can overlap, so that perturbations caused by one disease can affect other disease modules. The systematic mapping of such network-based dependencies between the pathophenotypes and their disease modules has culminated in the concept of the diseasome37, representing disease maps whose nodes are diseases and whose links represent various molecular relationships between the disease-associated cellular components. Uncovering such links between diseases not only helps us understand how different phenotypes, often addressed by different medical sub-disciplines, are linked at the molecular level, but can also help us comprehend why certain groups of diseases arise together. The co-morbidity of conditions culled from the diseasome offers insights that may yield novel approaches to disease prevention, diagnosis, and treatment. Diseasome-based approaches could also aid drug discovery, in particular when it comes to the use of approved drugs to treat molecularly linked diseases. Next, we review the construction of such disease maps and the consequences of the observed disease associations.
Shared Gene Hypothesis and the Human Diseasome Network (HDN)
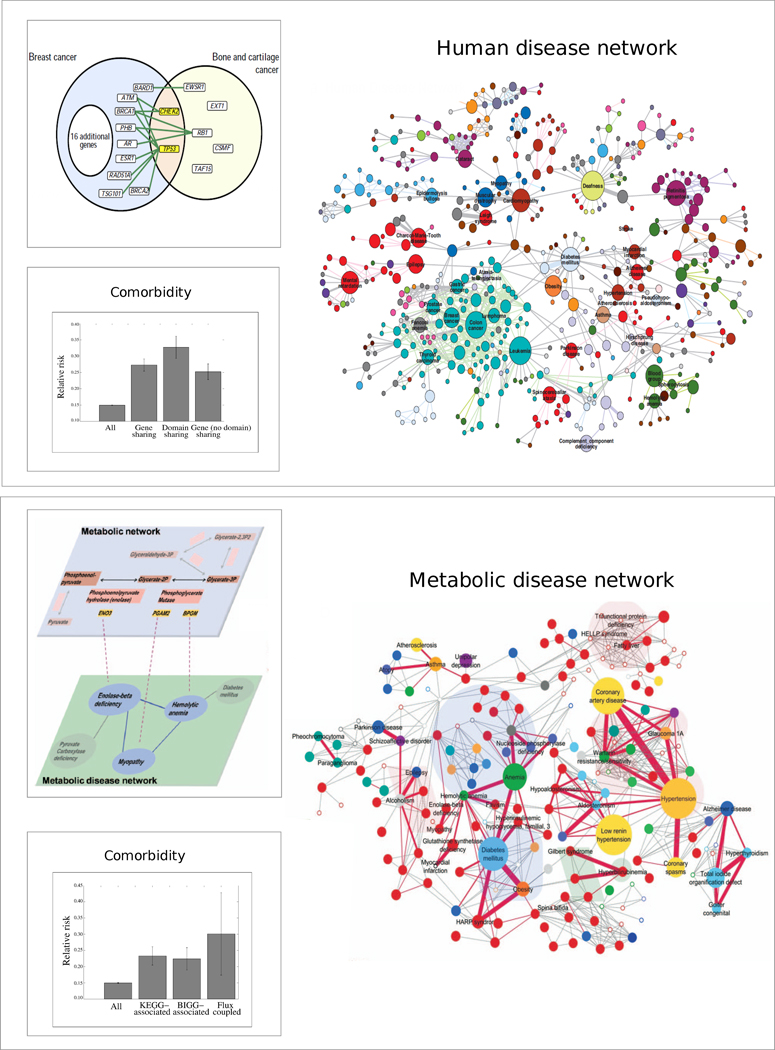
If the same gene is linked to two different disease pathophenotypes, this linkage is often an indication that the two diseases have a common genetic origin. Motivated by this hypothesis, Goh et al.37 used the gene-disease associations collected in the OMIM database to build a network of diseases that are linked if they share one or several genes. In the obtained HDN, 867 of 1,284 diseases with an associated gene are connected to at least one other disease, and 516 of them belong to a single disease cluster (Fig. 5). The clustering of nodes of similar color in Figure 5, denoting the disease class, reflects the fact that similar pathophenotypes have a higher likelihood of sharing genes than pathophenotypes that belong to different disease classes. For example, cancers form a tightly interconnected and easily detectable cluster, held together by a small group of genes associated with multiple cancers, such as P53, KRAS, ERBB2 or NF1.
Figure 5. Disease networks.
(a) Human Disease Network, whose nodes are diseases; two diseases being linked if they share one or several disease-associated genes, as shown in the example involving breast cancer and bone and cartilage cancer64. The large panel shows the giant cluster of the obtained disease network. Not shown are small clusters of isolated diseases37. Node color reflects the disease class of the corresponding diseases to which they belong, cancers appearing as blue nodes and neurological diseases as red nodes. Node size correlates with the number of genes known to be associated with the corresponding disease (after ref.37). The left panel shows the comorbidity between diseases linked in the HDN measured by the logarithm of relative risk, indicating that if the disease-causing mutations affect the same module of the shared disease protein, then the comorbidity is higher64. (b) Metabolic Disease Network, linking two diseases if they are both associated with enzymes and if these enzymes catalyze reactions that share a metabolite (after ref.67). The comorbidity between metabolically linked diseases is higher than those that are not connected, and diseases whose enzymes catalyze reactions that are coupled with each other at the flux level show even higher comorbidity (bottom left panel).
To test if the shared gene relationship has epidemiological consequences in disease occurrence in the population64, we show the comorbidity between linked disease pairs in Figure 5. This analysis indicates that a patient is twice as likely to develop a (comorbid) disease if that disease shares a gene with the primary disease than if that disease does not share a gene with the primary disease. Yet, many disease pairs that share genes do not show significant comorbidity. This lack of comorbidity may occur, in part, because different mutations on the same gene can have different effects on the function of the gene product and on its organ-based expression, therefore, different pathological consequences65 that are context-dependent. Such ‘edgetic’ alleles affect a specific subset of links in the interactome66, and individuals who harbor different mutations in the same gene can develop different disorders. Consistent with this view, disease pairs associated with mutations that affect the same functional domain of a protein show higher comorbidity than disease pairs whose mutations occur in different functional domains64 (Fig. 5).
Shared metabolic pathway hypothesis and the Metabolic Disease Network (MDN)
An enzymatic defect that affects the flux of one reaction may potentially affect the fluxes of all downstream reactions in the same pathway, leading to disease phenotypes that are normally associated with these downstream reactions. Thus, for metabolic diseases, links induced by shared metabolic pathways are expected to be more relevant than the links based on shared genes. In support of this hypothesis, Lee et al.67 constructed a metabolic disease network in which two disorders are connected if the enzymes associated with them catalyze adjacent reactions (Fig. 5). The visually apparent clustering of the MDN mirrors distinct metabolic pathways. For example, purine metabolism consists of 62 reactions associated with 33 diseases, including nucleoside phosphorylase deficiency and congenital dyserythropoietic anemia. These diseases form a visually distinct cluster, highlighted with blue shading in Figure 5. Comorbidity analysis confirms the functional relevance of metabolic coupling: disease pairs linked in the MDN have a 1.8-fold increased comorbidity compared to disease pairs that are not linked metabolically67. Comorbidity is even more pronounced if the fluxes of the reactions catalyzed by the respective disease genes are themselves coupled, i.e., changes in one flux induce significant changes in the other flux, even if the corresponding reactions are not adjacent.
Shared microRNA hypothesis
Prompted by the increasing evidence of the role of miRNAs in human disease, Lu et al.68 connected diseases pairs whose associated genes are targeted by at least one common miRNA molecule. The obtained network displays a disease class-based segregation: for example, cancers share similar associations at the miRNA level, leading to a distinct cancer cluster, which, for example, differs from the cluster associated with cardiovascular diseases, in the miRNA-based disease network.
Phenotypic Disease Network (PDNs)
One can also link disease pairs based on the directly observed comorbidity between them, obtaining a phenotypic disease network. For example, Rzhetsky et al.69 inferred the comorbidity links between 161 disorders from the disease history of 1.5 million patients at the Columbia University Medical Center, and Hidalgo et al.70 built a network involving 657 diseases from the disease history of over 30 million Medicare patients. In these maps, two diseases are connected if their comorbidity exceeds a predefined threshold. The PDN is blind to the mechanism underlying the observed comorbidity, which may be rooted in molecular-level dependencies (as seen for HDN, MDN or mi-RNA based disease networks), or in environmental or treatment-related perturbations of the network. Yet, PDN captures disease progression, as patients tend to develop diseases in the network vicinity of diseases they have already had70. Furthermore, patients who are diagnosed with diseases with more links in the PDN show a higher mortality than those diagnosed with less connected diseases70. Another use of phenotypic information to build a disease network was suggested by Van Driel et al.71, who employed text mining to assign to over 5,000 human phenotypes in the OMIM database a string of phenotypic features from the medical subject heading vocabulary. The overlap of their phenotypic descriptions was used to link various diseases, finding that phenotypic similarity positively correlates with the molecular signatures of two linked diseases, from relatedness at the level of protein sequence to protein motifs and direct protein–protein interactions between the disease-associated proteins.
These studies indicate that the molecular-level links between the known disease components have direct epidemiological consequences, leading to observable comorbidity patterns. While most efforts focused on the role of single molecular or phenotypic measure to capture disease-disease relationships (such as shared genes or metabolites), a comprehensive understanding requires us to inspect multiple sources of evidence, from shared genes to protein-protein interaction based relationships, shared environmental factors, common treatments, affected tissues and organs, and phenotypic manifestations. In line with such integrated approaches, Suthram et al.72 built a disease network by linking two diseases for which the same modules were activated in the specific disease states and Liu et al.73 linked diseases with common environmental influences. While a comprehensive program towards understanding all causal links between diseases is still in its infancy, it will be essential if we seek a deeper understanding of human disease.
Applying network-based knowledge of disease
Network pharmacology
Owing to the often unknown interactions between drug targets and other cellular components, drugs whose efficacy was predicted by specific target-binding experiments may not have the same effect in different clinical settings in which that target is of modified contextual importance (e.g., tissue-specific isoform compensates for the loss of function of the inhibited protein). Furthermore, single-target drugs may, perhaps, correct some dysfunctional aspects of the disease module, but could alter the activity of other network neighborhoods, leading to detectable side effects. This network-based view of drug action implies that most disease phenotypes are difficult to reverse through the use of a single ‘magic bullet,’ i.e., an intervention that affects a single node in the network74. While network-based approaches represent a relatively recent trend in drug discovery, given the intricate network effects drug development must face, the nascent field of network pharmacology75, at the intersection of network medicine and polypharmacology, is poised to become an essential component of drug development strategies.
The utility of network-based approaches in drug discovery has been demonstrated in the search for antibiotic targets against bacterial metabolism. Given the relatively accurate metabolic maps (see Box 1), and that in bacteria flux balance analysis76 and other flux-based methods77 allow the prediction of the flux changes induced by drug-altered enzymatic activity, the metabolic impact of a hypothetical enzyme-blocking drug can be explicitly explored. This process has recently led to the identification and testing of potential new antibacterial targets78. Furthermore, the coupled nature of metabolic fluxes allows for the possibility of rescuing a lost metabolic function through the blocking of additional enzymes, selected to re-route metabolic activity to compensate for the original loss of function, an intriguing alternative to gene therapy79.
There is increasing attention paid to therapies involving multiple targets that may be potentially more effective in reversing the disease phenotype than single drugs80. The efficacy of this approach has been demonstrated by combinatorial therapies of AIDS, cancer, or depression, raising an important question: can one systematically identify multiple drug targets with optimal impact on the disease phenotype? This is an archetypical network problem, leading to methods to identify optimal drug combinations starting either from the metabolic network81, 82, or from the bipartite network linking compounds to their drug-response phenotypes83. Research in this direction has led to potentially safer multi-target combinations for inflammatory conditions, or to the identification of 14 optimal anti-cancer drug combinations81–83.
Equally important, drug-target networks84, 85 that link approved or experimental drugs to their protein targets have helped organize the considerable knowledge base encoding the interplay between diseases and drugs. Its analysis demonstrated the preponderance of palliative drugs, i.e., drugs that do not target the actual source of the disease (i.e., the disease-associated proteins) but proteins in the network neighborhood84 of the disease proteins.
The first step of rational drug design is an understanding of the cellular dysfunction caused by a disease. By definition, this dysfunction is limited to the disease module, which means that one can reduce the search for therapeutic agents to those that induce detectable changes in the particular disease module. This represents a significant reduction of the search space, also aiding the development of biomarkers for disease detection, as changes in the activity of the disease module components are expected to show the strongest correlations with disease progression86.
Disease classification
Contemporary approaches to the classification of human disease are based on observational correlations between pathological analysis and existing knowledge of clinical syndromes. Yet, modern molecular diagnostic tools have shown the shortcomings of this methodology, reflecting both a lack of sensitivity in identifying preclinical disease and a lack of specificity in defining disease unequivocally. For example, hypertrophic cardiomyopathy, an inherited form of heart failure, is caused by a number of mutations in a variety of sarcomeric proteins; however, the clinical phenotype, as well as the anatomic and functional pathophenotypes (via echocardiographic assessment) are essentially indistinguishable from one another87, 88. Similarly, the classification of lymphomas, which has largely relied on histopathology and cell surface marker panels, is recently evolving to molecular-level classification that relies on expression arrays and genomic analysis89, as well as systems approaches90. As a result of this movement toward network-based classification of lymphomas, prognosis can be individualized91 and the promise of individualized therapies more likely to be realized.
Current disease classification, in general, tends to neglect the interconnected nature of many diseases. This failure is partly a response to the focused nature of medical training, as well as the reductionist paradigm that has driven medical diagnosis in the modern era. In an effort to correct this shortcoming, we recently proposed a systems-based network framework for defining human disease92. In this paradigm, the clinical pathophenotype is the systems-driven consequence of a series of linked networks that incorporate the primary disease-causing gene (e.g., the sickle hemoglobin mutation in the 6th position of the beta-chain gene), disease modifying genes (including those that control common (generic) intermediate pathophenotypes (endopathophenotypes) common to all disease (i.e., inflammation, thrombosis/ hemorrhage, fibrosis, immune response, cell proliferation, apoptosis/necrosis) and their network-based determinants, and environmental (and behavioral) determinants (including those that lead modulate gene expression at the transcriptional or epigenetic levels, as well as those that cause post-translational modification of the proteome) and their influence on the functional genome. These subnetwork determinants of the disease network conspire to yield the clinical phenotype in highly individualized ways for simple as well as complex illness92. Clearly, network-based approaches to disease have the potential, therefore, to provide a new and useful framework for classifying disease, defining disease susceptibility, predicting disease outcome, and identifying tailored therapeutic strategies.
Conclusions
In summary, similar to an automotive technician’s inability to fix a car’s electrical problem without an accurate assembly and wiring diagram, a comprehensive understanding of most diseases requires a map of the cell’s intricate wiring diagram, whose breakdown is ultimately responsible for the emergence of a particular disease phenotype. Network medicine seeks to offer this understanding, teaching us that the road towards a reliable network-based approach to disease is currently limited by the incompleteness of the available interactome maps and the limitations of the existing tools to explore the role of networks in disease. For example, investigators are forced to apply traditional statistical tools to network data, assuming that the quantities of interest follow a normal distribution (which they do not—everything from degree distributions to metabolite concentrations are known to be fat tailed), or that the deterministic parameters are independent variables (which, again, they are not—most activity patterns in the cell are correlated). Thus, there is a real need to develop statistical tools that are reliable in the interconnected environment of the cell. Finally, while some principles widely used in network medicine are well documented (like the local hypothesis, Box 3), others like the parsimony principle, or the expected overlap between topological, functional, and disease modules, remain to be quantified and validated.
As helpful as analogies can be, we must realize that there is a fundamental difference between the automotive technician and the physician: the technician can swap the broken component with one that functions correctly. This is a futuristic view of medicine—most drugs do not cure, but only alter the symptoms and signs of the disease. It is also clear, however, that only an integrated understanding of the interactions among the genome, the proteome, the environment, and the pathophenome, mediated by the underlying cellular network, offers a basis for future advances. This perspective has led to a “think globally-act locally” paradigm, fueling advances in network medicine: in order to generate the local network perturbations that may cure a particular disease, we cannot avoid understanding the cells’ global organization.
Supplementary Material
Acknowledgements
We thank A. Sharma, D.-S. Lee and J. Park for useful discussions and suggestions. ALB and NG were supported by National Institutes of Health (NIH) through the Center of Excellence in Genomic Sciences (CEGS), and JL was supported by NIH grants HL061795 (Merit Award), HL81587, HL70819, and HL48743.
Footnotes
Competing interest statement: The authors declare that they have no competing financial interests.
References
- 1.Zhao Y, Jensen ON. Modification-specific proteomics: strategies for characterization of post-translational modifications using enrichments techniques. Proteomics. 2009;9:4632–4641. doi: 10.1002/pmic.200900398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venkatesan K, et al. An empirical framework for binary interactome mapping. Nature Methods. 2008;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 4.Barabási A-L. Network Medicine — From Obesity to the “Diseasome”. NEJM. 2007;357:404. doi: 10.1056/NEJMe078114. [DOI] [PubMed] [Google Scholar]
- 5.Pawson T, Linding R. Network medicine. FEBS Lett. 2008;582:1266–1270. doi: 10.1016/j.febslet.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 6.Zanzoni A, Soler-López M, Aloy P. A network medicine approach to human disease. FEBS Lett. 2009;583:1759–1765. doi: 10.1016/j.febslet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Ideker T, Sharan R. Protein networks in disease. Genome Research. 2008;18:644–652. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rual J-F, et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 9.Stelzl U, et al. A Human Protein-Protein Interaction Network: A Resource for Annotating the Proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 10.Jeong H, et al. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 11.Fell DA, Wagner A. The small world of metabolism. Nature Biotechnology. 2000;18:1121–1122. doi: 10.1038/81025. [DOI] [PubMed] [Google Scholar]
- 12.Duarte NC, et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. PNAS. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 14.Linding R, et al. NetworKIN: a resource for exploring cellular phosphorylation networks. Nucleic Acids Res. 2008;36:D695–D699. doi: 10.1093/nar/gkm902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis BP, Burge CB, Bartel DP. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates that Thousands of Human Genes are MicroRNA Targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 16.Reynolds A, et al. Rational siRNA design for RNA interference. Nature Biotechnology. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 17.Stuart JM, et al. A Gene-Coexpression Network for Global Discovery of Conserved Genetic Modules. Science. 2003;302:249. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 18.Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nature Reviews Genetics. 2007;8:437. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- 19.Beltrao P, Cagney G, Krogan N. Quantitative Genetic Interactions Reveal Biological Modularity. Cell. 2010;141:739. doi: 10.1016/j.cell.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz AS, Yu J, Gardenour KR, Finley RL, Jr, Ideker T. Cost-effective strategies for completing the interactome. Nature Methods. 2009;6:55. doi: 10.1038/nmeth.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu H, et al. High-Quality Binary Protein Interaction Map of the Yeast Interactome Network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barabasi A-L, Oltvai Z. Network biology: Understanding the cell's functional organization. Nature Reviews Genetics. 2004;5:101. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 23.Albert R, Barabasi A-L. Statistical mechanics of complex networks. Rev. Mod. Phys. 2002;74:47–97. [Google Scholar]
- 24.Zhu X, Gerstein M, Snyder M. Getting connected: analysis and principles of biological networks. Genes and Development. 2007;21:1010–1024. doi: 10.1101/gad.1528707. [DOI] [PubMed] [Google Scholar]
- 25.Caldarelli G. Scale free networks. Oxford: Oxford University Press; 2007. [Google Scholar]
- 26.Albert R. Scale-free networks in cell biology. J. Cell Sci. 2005;118:4947–4957. doi: 10.1242/jcs.02714. [DOI] [PubMed] [Google Scholar]
- 27.Newman M, Barabási A-L, Watts DJ. The Structure and Dynamics of Networks. Princeton University Press; 2006. [Google Scholar]
- 28.Amberger J, C A, Scott AF, Hamosh A. McKusick's Online Mendelian Inheritance in Man (OMIM®) Nucleic Acids Res. 2009;37:D793. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeong H, et al. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 30.Fraser HB, et al. Evolutionary rate in the protein interaction network. Science. 2002;296:5568. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- 31.Eisenberg E, Levanon EY. Preferential attachment in the protein network evolution. Physical Review Letters. 2003;91:138701. doi: 10.1103/PhysRevLett.91.138701. [DOI] [PubMed] [Google Scholar]
- 32.Saeed R, Deane CM. Protein protein interactions, evolutionary rate, abundance and age. BMC Bioinformatics. 2006;7:128. doi: 10.1186/1471-2105-7-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jordan IK, Wolf YI, Koonin EV. No simple dependence between protein evolution rate and the number of protein-protein interactions: only the most prolific interactors tend to evolve slowly. BMC Evolutionary Biology. 2003;3:5. doi: 10.1186/1471-2148-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wachi S, Yoneda K, Wu R. Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics. 2005;21:4205–4208. doi: 10.1093/bioinformatics/bti688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jonsson PF, Bates PA. Global topological features of cancer proteins in the human interactome. Bioinformatics. 2006;22:2291–2297. doi: 10.1093/bioinformatics/btl390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu J, Li Y. Discovering disease-genes by topological features in human protein–protein interaction network. Bioinformatics. 2006;22:2800–2805. doi: 10.1093/bioinformatics/btl467. [DOI] [PubMed] [Google Scholar]
- 37.Goh K-I, et al. The human disease network. PNAS. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oti M, et al. Predicting disease genes using protein-protein interactions. J Med Genet. 2006;43:691–698. doi: 10.1136/jmg.2006.041376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gandhi T, et al. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nature Genetics. 2006;38:285–293. doi: 10.1038/ng1747. [DOI] [PubMed] [Google Scholar]
- 40.Girvan M, Newman MEJ. Community structure in social and biological networks. PNAS. 2002;99:7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palla G, Derényi I, Farkas I, Vicsek T. Uncovering the overlapping community structure of complex networks in nature and society. Nature. 2005;435:814–818. doi: 10.1038/nature03607. [DOI] [PubMed] [Google Scholar]
- 42.Ahn Y-Y, Bagrow JP, Lehmann S. Link communities reveal multiscale complexity in networks. Nature. 2010;466:761–764. doi: 10.1038/nature09182. [DOI] [PubMed] [Google Scholar]
- 43.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 45.Taylor IW, et al. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nature Biotechnology. 2009;27:199. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- 46.Chen Y, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uetz P, et al. Herpesviral Protein Networks and Their Interaction with the Human Proteome. Science. 2006;311:239–242. doi: 10.1126/science.1116804. [DOI] [PubMed] [Google Scholar]
- 48.Calderwood MA, et al. Epstein–Barr virus and virus human protein interaction maps. PNAS. 2007;104:7606–7611. doi: 10.1073/pnas.0702332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gulbahce N, et al. Viral perturbations of host networks reflect disease etiology. 2010. doi: 10.1371/journal.pcbi.1002531. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turnbaugh PJ, Gordon JI. An Invitation to the Marriage of Metagenomics and Metabolomics. Cell. 2008;134:708–713. doi: 10.1016/j.cell.2008.08.025. [DOI] [PubMed] [Google Scholar]
- 51.Goehler H, et al. A Protein Interaction Network Links GIT1, an Enhancer of Huntingtin Aggregation, to Huntington’s Disease. Molecular Cell. 2004;15:853–865. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 52.Lim J, et al. A Protein–Protein Interaction Network for Human Inherited Ataxias and Disorders of Purkinje Cell Degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 53.Pujana MA, et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nature Genetics. 2007;39:1338. doi: 10.1038/ng.2007.2. [DOI] [PubMed] [Google Scholar]
- 54.Camargo LM, et al. Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Molecular Psychiatry. 2007;12:74–86. doi: 10.1038/sj.mp.4001880. [DOI] [PubMed] [Google Scholar]
- 55.Amino T, et al. Redefining the disease locus of 16q22.1-linked autosomal dominant cerebellar ataxia. J Hum Genet. 2007;52:643–649. doi: 10.1007/s10038-007-0154-1. [DOI] [PubMed] [Google Scholar]
- 56.Hirschhorn JN. Genomewide Association Studies — Illuminating Biologic Pathways. N Engl J Med. 2009;360:1699. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- 57.Krauthammer M, et al. Molecular triangulation: Bridging linkage and molecular-network information for identifying candidate genes in Alzheimer's disease. PNAS. 2004;101:15148–15153. doi: 10.1073/pnas.0404315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Franke L, et al. Reconstruction of a Functional Human Gene Network, with an Application for Prioritizing Positional Candidate Genes. American Journal of Human Genetics. 2006;78:1011. doi: 10.1086/504300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Navlakha S, Kingsford C. The power of protein interaction networks for associating genes with diseases. Bioinformatics. 2010;26:1057–1063. doi: 10.1093/bioinformatics/btq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lage K, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nature Biotechnology. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 61.Wu X, Jiang R, Zhang MQ, Li S. Network-based global inference of human disease genes. Mol. Syst. Biol. 2008;4:189. doi: 10.1038/msb.2008.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kohler S, et al. Walking the Interactome for Prioritization of Candidate Disease Genes. American Journal of Human Genetics. 2008;82:949–958. doi: 10.1016/j.ajhg.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vanunu O, et al. Associating Genes and Protein Complexes with Disease via Network Propagation. PLoS Computational Biology. 2010;6 doi: 10.1371/journal.pcbi.1000641. e1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park J, et al. The impact of cellular networks on disease comorbidity. Molecular Systems Biology. 2009;5:262. doi: 10.1038/msb.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dudley AM, Janse DM, Tanay A, Shamir R, Church GM. A global view of pleiotropy and phenotypically derived gene function in yeast. Mol. Syst. Biol. 2005;1:1. doi: 10.1038/msb4100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhong Q, et al. Edgetic perturbation models of human inherited disorders. Molecular Systems Biology. 2009;5:321. doi: 10.1038/msb.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee D-S, et al. The implications of human metabolic network topology for disease comorbidity. PNAS. 2008;105:9880–9885. doi: 10.1073/pnas.0802208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu M, et al. An Analysis of Human MicroRNA and Disease Associations. Plos ONE. 2008;3:e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rzhetsky A, et al. Probing genetic overlap among complex human phenotypes. PNAS. 2007;104:11694–11699. doi: 10.1073/pnas.0704820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hidalgo C, et al. A Dynamic Network Approach for the Study of Human Phenotypes. Plos Computational Biology. 2009;5 doi: 10.1371/journal.pcbi.1000353. e1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Driel MA, et al. A text-mining analysis of the human phenome. European Journal of Human Genetics. 2006;14:535–542. doi: 10.1038/sj.ejhg.5201585. [DOI] [PubMed] [Google Scholar]
- 72.Suthram S, et al. Network-Based Elucidation of Human Disease Network-Based Elucidation of Human Disease Enriched for Pluripotent Drug Targets. Plos Computational Biology. 2010;6 doi: 10.1371/journal.pcbi.1000662. e1000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu YI, Wise PH, Butte AJ. The "etiome": identification and clustering of human disease etiological factors. BMC Bioinformatics. 2009;10:S14. doi: 10.1186/1471-2105-10-S2-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nolan GP. What's wrong with drug screening today. Nature Chemical Biology. 2007;3:187. doi: 10.1038/nchembio0407-187. [DOI] [PubMed] [Google Scholar]
- 75.Hopkins AL. Predicting promiscuity. Nature. 2009;462:167. doi: 10.1038/462167a. [DOI] [PubMed] [Google Scholar]
- 76.Fong SS, Palsson BØ. Metabolic gene-deletion strains of Escherichia coli evolve to computationally predicted growth phenotypes. Nat Genet. 2004;36:1056–1058. doi: 10.1038/ng1432. [DOI] [PubMed] [Google Scholar]
- 77.Segrè D, Vitkup D, Church GM. Analysis of optimality in natural and perturbed metabolic networks. PNAS. 2002;99:15112–15117. doi: 10.1073/pnas.232349399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shen Y, et al. Blueprint for antimicrobial hit discovery targeting metabolic networks. PNAS. 2010;10.1073:1–6. doi: 10.1073/pnas.0909181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Motter AE, Gulbahce N, Almaas E, Barabási A-L. Predicting synthetic rescues in metabolic networks. Mol. Syst. Biol. 2008;4:168. doi: 10.1038/msb.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Csermely P, Agoston V, Pongor S. The efficiency of multi-target drugs: the network approach might help drug design. Trends Pharmacol Sci. 2005;26:178–182. doi: 10.1016/j.tips.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 81.Motter AE. Improved network performance via antagonism: From synthetic rescues to multi-drug combinations. Bioessays. 2010;32:236–245. doi: 10.1002/bies.200900128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang K, et al. Finding multiple target optimal intervention in disease related molecular network. Molecular Systems Biology. 2008;4:228. doi: 10.1038/msb.2008.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vazquez A. Optimal drug combinations and minimal hitting sets. BMC Systems Biology. 2009;3 doi: 10.1186/1752-0509-3-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yildirim MA, et al. Drug–target network. Nature Biotechnology. 2007;25:1119. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 85.Keiser MJ, et al. Predicting new molecular targets for known drugs. Nature. 2009;462:175. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schadt EE, Friend SH, Shaywitz DA. A network view of disease and compound screening. Nat. Rev. Drug Disc. 2009;8:286–295. doi: 10.1038/nrd2826. [DOI] [PubMed] [Google Scholar]
- 87.Ho CY, Seidman CE. A contemporary approach to hypertrophic cardiomyopathy. Circulation. 2006;113:e858–e862. doi: 10.1161/CIRCULATIONAHA.105.591982. [DOI] [PubMed] [Google Scholar]
- 88.Morita Y, et al. Shared genetic causes of cadiac hypertrophy in children and adults. N Engl J Med. 2008;358:1899–1908. doi: 10.1056/NEJMoa075463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de Leval L, et al. Molecular classification of T-cell lymphomas. Crit Rev Oncol Hematol. 2009;72:125–143. doi: 10.1016/j.critrevonc.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 90.Basso K. Towards a systems biology approach to investigate cellular networks in normal and malignant B cells. Leukemia. 2009;23:1219–1225. doi: 10.1038/leu.2009.4. [DOI] [PubMed] [Google Scholar]
- 91.Lenz G, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359:2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Loscalzo J, Kohane I, Barabási A-L. Human disease classification in the postgenomic era: A complex systems approach to human pathobiology. Molecular Systems Biology. 2007;3:1–11. doi: 10.1038/msb4100163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ewing RM, et al. Large-scale mapping of human protein–protein interactions by mass spectrometry. Molecular Systems Biology. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cusick ME, et al. Literature-curated protein interaction datasets. Nature Methods. 2009;6:39–46. doi: 10.1038/nmeth.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ma H, et al. The Edinburgh human metabolic network reconstruction and its functional analysis. Molecular Systems Biology. 2007;3:135. doi: 10.1038/msb4100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Barabási A-L, Albert R. Emergence of Scaling in Random Networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 97.Han JD, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, Dupuy D, Walhout AJ, Cusick ME, Roth FP, Vidal M. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 98.Watts DJ, Strogatz SH. Collective dynamics of 'small-world' networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 99.Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network Motifs: Simple Building Blocks of Complex Networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 100.Yu H, Kim PM, Sprecher E, Trifonov V, Gerstein M. The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput Biol. 2007;3:e59. doi: 10.1371/journal.pcbi.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.