Abstract
A workflow is presented that integrates gene expression data, proteomic data, and literature-based manual curation to construct multicellular, tissue-specific models of human brain energy metabolism that recapitulate metabolic interactions between astrocytes and various neuron types. Three analyses are applied for gene identification, analysis of omics data, and analysis of physiological states. First, we identify glutamate decarboxylase as a target that may contribute to cell-type and regional specificity in Alzheimer’s disease. Second, the decreased metabolic rate seen in affected brain regions in Alzheimer’s disease is consistent with a suppression of central metabolic gene expression in histopathologically normal neurons. Third, we identify pathways in cholinergic neurons that couple mitochondrial metabolism and cytosolic acetylcholine production, and subsequently find that cholinergic neurotransmission accounts for ∼3% of brain neurotransmission. Constraint-based modeling can thus contribute to the study and analysis of multicellular metabolic processes in human tissues, and provide detailed mechanistic insight into high-throughput data analysis.
Introduction
Constraint-based reconstruction and analysis of genome-scale microbial metabolic networks has matured over the past decade. This approach has a wide range of applications, such as providing insight into evolution, aiding in metabolic engineering, and providing a mechanistic bridge between genotypes and complex phenotypes1,2. Computational methods3 and a detailed SOP4 have been outlined for the reconstruction of high-quality prokaryotic metabolic networks, and many methods can be deployed for their analysis5,6. Constraint-based modeling of metabolism entered a new phase with the publication of the human metabolic network (HR1)7, based on build-35 of the human genome. Methods allowing tissue-specific model construction have followed8–10.
Many tissue metabolic functions rely on interactions between many cell types. Thus, methods are needed that integrate the metabolic activities of multiple cell types. Here, using HR1, we analyze and integrate omics data with information from detailed biochemical studies to build multicellular constraint-based models of metabolism. We demonstrate this process by constructing and analyzing models of human brain energy metabolism, with an emphasis on central metabolism and mitochondrial function in astrocytes and neurons. Moreover, we provide three detailed examples, demonstrating the use of models to guide experimental work and provide biological insight into the metabolic mechanisms underlying physiological and pathophysiological states in brain.
Results
Building metabolic models of multiple cell types
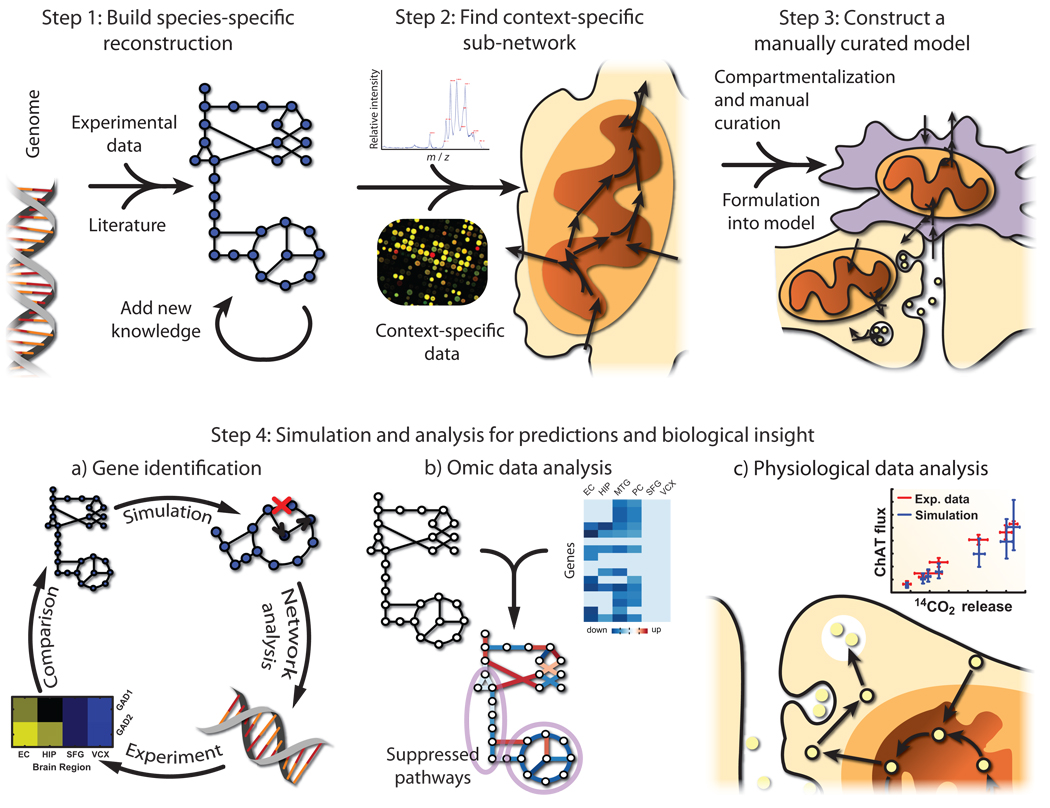
Omics datasets can be difficult to analyze due to their size. However, such datasets can be used to construct large mechanistic models for specific tissues and cell types8,9 that serve as a context for further analysis. The workflow for generating multicellular models, as depicted in Figure 1, consists of the following four steps:
Figure 1. A workflow for bridging the genotype-phenotype gap with the use of high-throughput data and manual curation for the construction of multicellular models of metabolism.
Metabolic models of multicellular tissues can be constructed to gain insight into biology and make testable hypotheses. First, a species-specific reconstruction is built based on genome annotation, experimental data, and knowledge obtained from the literature. Second, high-thoughput data can be mapped to the reconstruction in order to find a context-specific network (e.g., representing a tissue). Third, multicellular models are constructed as the context-specific network is organized into compartments representing different cell types, based on cell-specific knowledge and data. These networks are linked together with the transport of shared metabolites, and then formulated into a model. Fourth, the models are utilized for simulation and analysis to gain insight and generate testable hypotheses. For example, the models can be used to a) predict disease-associated genes, such as glutamate decarboxylase in this work. b) High-thoughput data can be analyzed in the network context to identify sets of genes that change together and affect specific pathways, such as the brain-region-specific suppression of central metabolism in Alzheimer’s disease patients. c) Physiological data can be analyzed in the context of the model, therefore allowing, for example, the calculation of the percentage of the brain that is cholinergic.
Step 1. An organism-specific metabolic network is reconstructed from genome annotation, lists of biomolecular components, and the literature4. Metabolic pathways and associated gene products are not completely known for any species. Thus, a reconstruction is refined through iterations of manual curation, hypothesis generation, experimental validation, and incorporation of new knowledge. HR1 has been through five iterations7.
Step 2. Many gene products are not expressed in all cells at any given time11. Therefore, gene product presence from omic data is mapped to HR1 using the gene-protein-reaction associations to obtain a draft reconstruction for the tissue of interest. This process may be performed manually or algorithmically8,9.
Step 3. Initial context-specific reconstructions are incomplete and may contain false positives due to proximal tissue contamination. Moreover, few high-throughput datasets are cell-type specific. Thus, the initial reconstruction represents the union of metabolic networks from various cell types. To address this problem, the literature is searched to verify enzyme localization and partition the model into compartments representing different cell types and organelles. Upon completion, the reconstruction is converted into a model by specifying inputs, outputs, relevant parameters, and by representing the network mathematically12. See Thiele and Palsson4 for details of proper manual curation.
Step 4. Once the network is accurately reconstructed and converted into an in silico model, it is used for simulation and analysis1,2, for hypothesis generation and to obtain insight into systems-level biological functions.
This workflow was used to build three different multicellular models of brain energy metabolism. Each model represents one canonical neuron type (i.e., glutamatergic, GABAergic, or cholinergic), its interactions with the surrounding astrocytes, and the transport of metabolites through the blood-brain barrier (Fig. 2). This reconstruction focuses on the core of cerebral energy metabolism, including central metabolism, mitochondrial metabolic pathways, and pathways relevant to anabolism and catabolism of three neurotransmitters: glutamate, γ-aminobutarate (GABA), and acetylcholine. Thus, the three models contain the high flux pathways and important reactions in neuron and astrocyte metabolism. These models currently represent the largest and most detailed models of brain energy metabolism 13–15 (see Supplementary Notes). Our models contain 1066, 1067, and 1070 compartment-specific reactions, transformations, and exchanges, involving 983, 983, and 987 metabolite/compartment combinations, for the glutamatergic, GABAergic, and cholinergic models, respectively, and are associated with a total of 403 genes. Lists of reactions, genes, citations, and parameters used to constrain the models are detailed in Supplementary Tables 1– 5. The validity of these models is demonstrated through various tests and comparisons to physiological data. Specifically, our model predicts ATP production rates within 8% of the average published value, and internal flux measurements are consistent with experimentally measured values (see Supplementary Notes). Moreover, three analyses using the models are detailed here. Since most of these analyses cannot be done on previous brain models or HR1 as published, this work provides novel insight into brain energy metabolism.
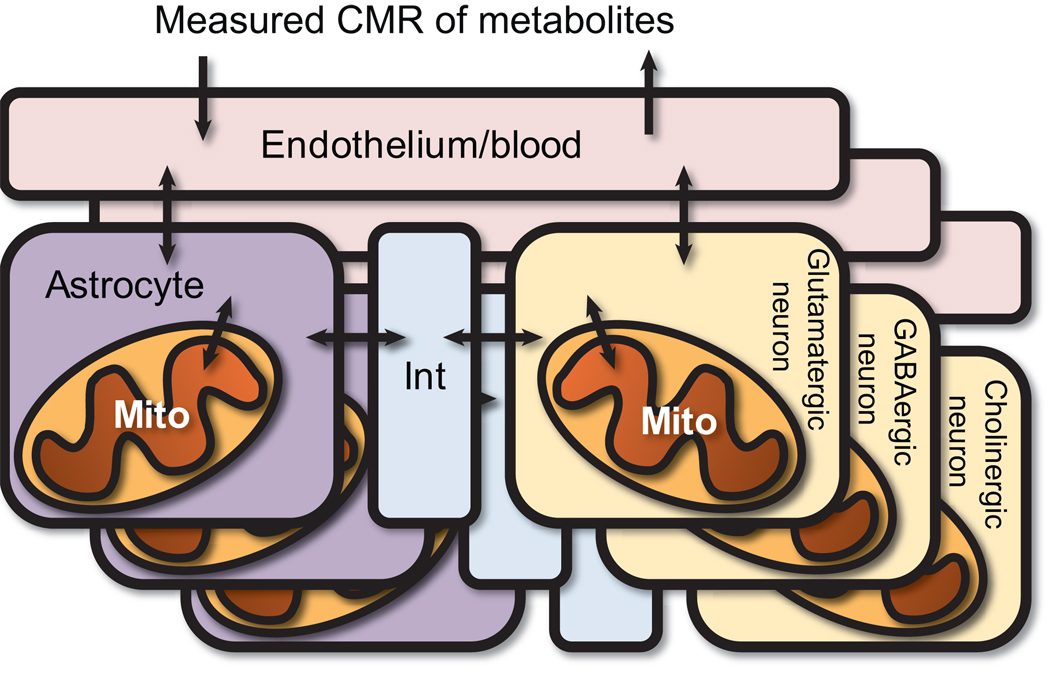
Figure 2. General structure of the models.
Three models were built from the brain reconstruction. Each model consists of various compartments: 1) the endothelium/blood, 2) astrocytes, 3) astrocytic mitochondria, 4) neurons, 5) neuronal mitochondria, and 6) an interstitial space between the cell types. Each neuron metabolic network was tailored to represent a specific neuron type, containing genes and reactions generally accepted to be unique to the neuron type. Mito = mitochondrion, Int = interstitial space, CMR = cerebral metabolic rate.
Identifying a potentially neuroprotective gene in AD
Alzheimer’s disease (AD) is characterized by histopathological features, including neurofibrillary tangles and β-amyloid plaques. Moreover, there is a strong metabolic component, in which metabolic rates of various brain regions decrease years before the onset of dementia16.
Several central metabolic enzymes exhibit altered expression or activity in AD, such as pyruvate dehydrogenase (PDHm), α-ketoglutarate dehydrogenase (AKGDm), and cytochrome c oxidase (CYOO)17–19. The activities of these enzymes are affected by the AD-related proteins β-amyloid and Tau kinase20,21. In silico, as the activity of these enzymes decreases, neurons demonstrate impaired metabolic capacity (Supplementary Fig. 1.a–b), and deficiencies in PDHm activity leads to a decreased cholinergic neurotransmission capacity (Supplementary Fig. 1.c.).
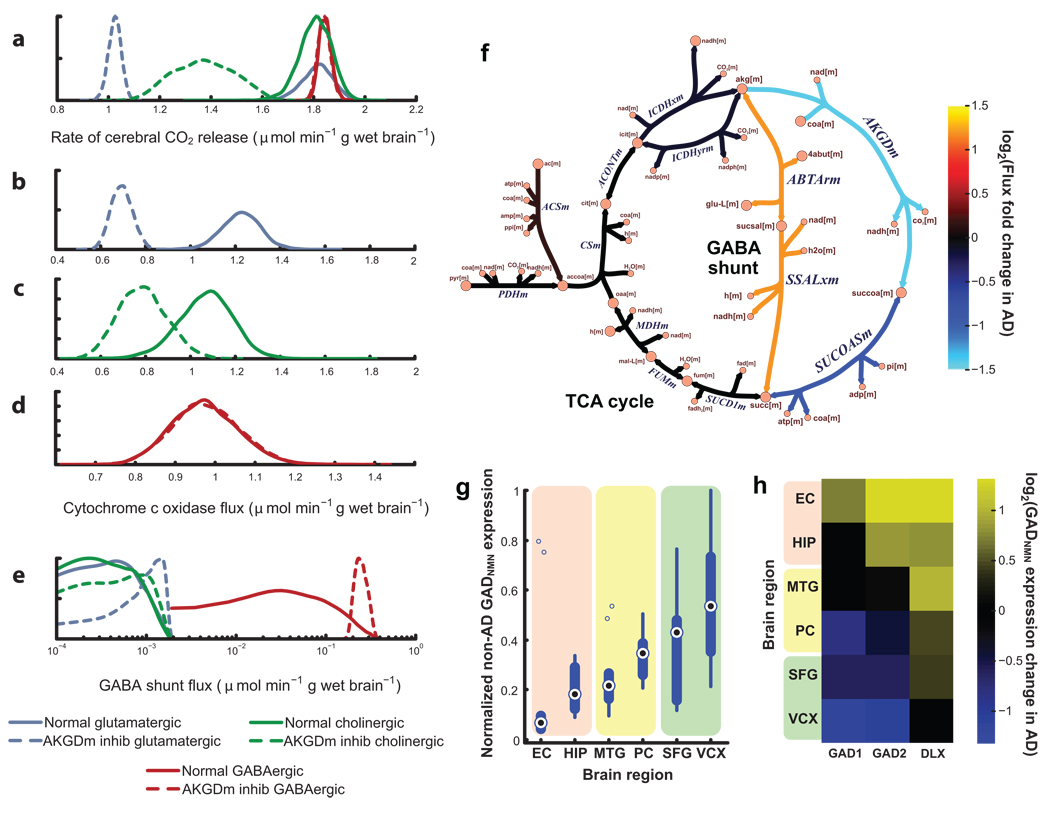
AKGDm deficiency shows the greatest impairment in post-mortem AD brain (57% decrease in activity)18. An in silico analysis shows that this deficiency impairs the metabolic rate in glutamatergic and cholinergic neurons (Fig. 3.a), since it limits oxidative phosphorylation (OxPhos) capacity in neurons (Fig. 3.b–c). Such impairment of OxPhos leads to neuronal apoptosis22. Surprisingly, however, OxPhos is not impaired in the GABAergic neuron model (Fig. 3.d). This model-derived result is consistent with the experimental observation that glutamatergic and cholinergic neurons are more affected in moderate stages of AD23, while most GABAergic cells are relatively unaffected until late stages24. Therefore, the models were further interrogated to identify a mechanism that allows GABAergic neurons to absorb the perturbation, thereby leading to the cell-type-specific effects.
Figure 3. Decrease in α-ketoglutarate dehydrogenase (AKGDm) activity, associated with Alzheimer’s disease (AD), shows cell-type and regional effects in silico consistent with experimental data.
Kernel density plots show the distribution of feasible fluxes for various reactions (a–e). An in silico reduction of AKGDm flux from normal activity (a–e, solid lines) to AD brain activity (a–e, dashed) decreases (a) the oxidative metabolic rate for glutamateric and cholinergic neurons, but not GABAergic neurons. This results from a decrease in the feasible fluxes for oxidative phosphorylation (e.g., cytochrome c oxidase) for both (b) glutamatergic and (c) cholinergic neurons, but not (d) GABAergic cells. This cell-type-specific protection from the AKGDm deficiency results from (e) an increased flux through the GABA shunt in GABAergic cells, by bypassing the damaged AKGDm (f). GABAergic cells maintain a higher GABA shunt flux because of the expression of glutamate decarboxylase (GAD). Neuroprotective properties of GAD are supported by gene expression. (g) Severely damaged brain regions in AD patients have lower GADNMN expression in control brain, while high GADNMN regions (SFG and VCX) show little damage. In AD brain, (h) severely affected regions (HIP and EC) show an increase in GADNMN and the GAD-inducing DLX family, suggesting that non-GAD expressing neurons may be lost in AD. EC = entorhinal cortex, HIP = hippocampus, MTG = middle temporal gyrus, PC = posterior cingulate cortex, SFG = superior frontal gyrus, VCX = visual cortex, NMN = neuron marker normalized, inhib = inhibited. All reaction and metabolite abbreviations are defined in Supplementary Tables 1– 2.
Simulations show that GABAergic neurons absorb the AKGDm perturbation through the GABA shunt (Fig. 3.f), a pathway that uses 4-aminobutyrate transaminase and succinate-semialdehyde dehydrogenase to bypass part of the TCA cycle. However, our models suggest that glutamatergic and cholinergic neurons cannot, despite carrying a small flux through the shunt enzymes (Fig. 3.e). Support for these results includes recent evidence that suggests that cerebellar granule neurons, which have higher levels of GABA, can absorb perturbations to AKGDH through this shunt25.
To identify the mechanism allowing only GABAergic neurons to use the GABA shunt to absorb the AKGDm perturbation, an in silico analysis was performed to identify genes that contribute to this (see Supplementary Notes). This analysis suggests that the two isoforms of glutamate decarboxylase (GAD) could provide the cell-type specific neuroprotection.
GAD allows the GABA shunt to carry a higher flux following the AKGDm perturbation in GABAergic neurons; however, the lack of GAD in other neuron types greatly limits the use of the GABA shunt in silico (Fig. 3.e). Therefore, by fueling the GABA shunt, GAD may play a neuroprotective function, thus contributing to the sparing of GABAergic systems in earlier AD24.
Certain populations of glutamatergic and cholinergic cells tend to be lost earlier in AD, but others survive. Interestingly, while GAD is canonically a GABAergic gene, it occasionally shows low expression in other neuron types, including glutamatergic and cholinergic cells26. Therefore, such populations of non-GABAergic, GAD-expressing neurons would also be protected. Thus, we follow with an analysis of the correlation of GAD expression and AD pathology for further validation.
If GAD has neuroprotective capacity in vivo, two properties of GAD expression are expected. First, brain regions with less GAD per neuron will be more affected in AD, while regions with abundant GAD will be spared. To test this hypothesis, we used a compendium of published microarrays of non-tangle-bearing neurons from six brain regions in Alzheimer patients and age-matched non-Alzheimer controls27. Among control patients, GAD expression levels among the brain regions is consistent with the extent of neuron loss found in AD patients; i.e., brain regions with more neuron loss in AD (e.g., the entorhinal cortex and hippocampus) have lower GAD expression in control patients, while relatively unaffected regions in AD (e.g., superior frontal gyrus and visual cortex) show much higher levels of GAD expression (Fig. 3.g).
For the second property, neurons with low GAD expression should be lost in AD; therefore, GAD expression per neuron should increase in histopathologically affected regions. Consistent with the hypothesis, there is a significant increase in the expression of the brain-specific GAD2 in the entorhinal cortex and hippocampus in AD (p=0.0050 and 0.018, respectively) (Fig. 3.h). As a control, all other neuron-specific genes tested failed to show a correlation with AD pathology (Supplementary Fig. 2), except the Dlx genes, which induce GAD expression in the brain (Fig. 3.h and Supplementary Fig. 3)28. Therefore, these results lend additional support to the possibility that GAD may be providing a neuroprotective effect, and that this effect is correlated with the regional specificity of AD. Moreover, the model was able to guide the identification of a gene and the mechanism for its role in AD, while the subsequent microarray analysis provides experimental support for the model prediction.
Microarray analysis shows pathway suppression in AD
Atrophy alone cannot explain the extent of impaired metabolism in AD in many brain regions29. Therefore, in such regions, there must be metabolic pathway suppression within surviving cells. To test this, we used PathWave30, a method that identifies differentially expressed pathways in omic data based on intra-pathway connectivity in a user-provided metabolic network (see Methods and Supplementary Notes for details).
In this analysis, each brain region demonstrates different amounts of changed metabolic pathways (Supplementary Table 6). The visual cortex and superior frontal gyrus lack any differentially expressed pathways, consistent with previous work that shows little change in metabolic rate in AD in these regions27. However, the posterior cingulate cortex (PC) and middle temporal gyrus (MTG) have the largest numbers of significantly differentially expressed pathways (23 and 18 pathways, respectively). These two regions show significantly decreased metabolic rates in AD, but show fewer histopathological effects27. Both the entorhinal cortex (EC) and hippocampus (HIP) also show decreases in expression of nine metabolic pathways, though the number of suppressed pathways may be lower since these regions suffer a high amount of neuron loss, and only histopathologically healthy neurons were expression profiled. Therefore, more affected neurons may already have been lost or not profiled.
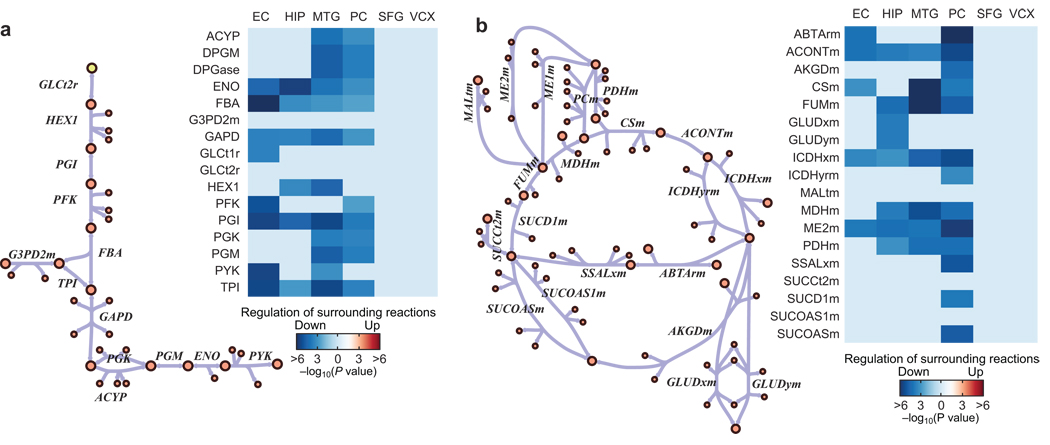
In the PathWave analysis, the four brain regions that show significantly low metabolic rates in AD (PC, MTG, HIP and EC)29 all show a significant suppression of glycolysis and the TCA cycle (Fig. 4). In addition, the HIP, MTG and PCC show a suppression of the malate-aspartate shuttle and OxPhos. Individual regions also show a suppression of other pathways, such as heme biosynthesis (MTG, PC), ethanol metabolism (EC, PC), and several amino acid metabolism pathways. Thus, using PathWave with our models, we find that the decreased metabolic rate in specific regions in AD is associated with the down-regulation of central metabolic gene expression in histopathologically normal neurons (see Supplementary Notes for further details).
Figure 4. Metabolically affected brain regions in AD show significant suppression of central metabolic pathways.
In certain AD brain regions, the metabolic rate of glucose decreases more than can be explained by brain atrophy. PathWave analysis demonstrates that histopathogically normal cells from the metabolically affected brain regions (EC, HIP, MTG, and PC) demonstrate a significant suppression of central metabolic pathways, such as (a) glycolysis and (b) the TCA cycle and surrounding reactions. Metabolically less affected regions (SFG and VCX) show no significant suppression. Reaction suppression shown here is a composite expression of the reaction associated genes and the genes of closely connected reactions. Only significantly changed reactions are shown (FDR = 0.05). EC = entorhinal cortex, HIP = hippocampus, MTG = middle temporal gyrus, PC = posterior cingulate cortex, SFG = superior frontal gyrus, VCX = visual cortex. All reaction and metabolite abbreviations are defined in Supplementary Tables 1– 2.
Models detail properties of cholinergic metabolic coupling
Studies have demonstrated that the use of cytosolic acetyl-CoA for the synthesis of the neurotransmitter acetylcholine comes from the acetyl-CoA formed in the mitochondria. This tight coupling of acetylcholine to mitochondrial metabolism allows treatments that increase glucose uptake in the brain to improve cognitive functions in rats31 and humans with severe cholinergic cognitive pathologies, such as Alzheimer’s Disease and Trisomy-2132. Pathways that transport acetyl-CoA carbon to the cytosol have been suggested; however, the mechanism is still not clear33.
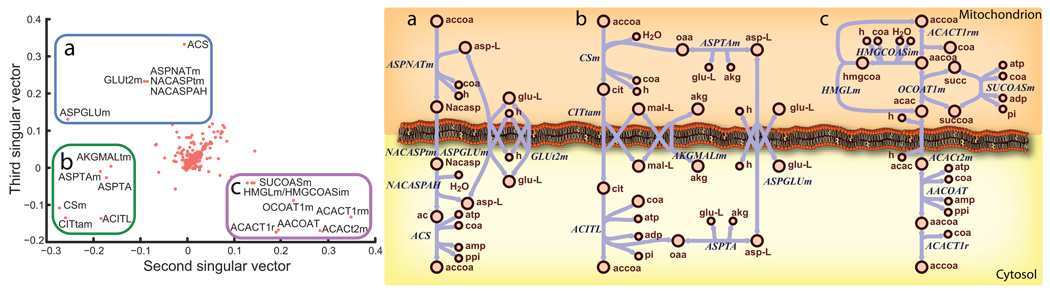
Constraint-based modeling was used to aid in the identification of two pathways that could indirectly transport acetyl-CoA into the cytosol, and provide insight into needed complementary pathways. To identify possible pathways, reaction sets were identified by randomly removing reactions from HR1 until a minimum set was determined that couples the mitochondrial and cytosolic acetyl-CoA pools. This was repeated until more than 21,000 unique minimal reaction sets were identified. Singular value decomposition was then used to identify dominant pathway features that frequently co-occur.
The first singular vector is dominated by reactions that occur most frequently in the reaction sets (e.g., water transport across the cell membrane). However, the second and third singular vectors are dominated by reaction sets that usually co-occur or never co-occur (Fig. 5). These reactions cluster into three distinct pathways, providing hypotheses to aid in the reconstruction process. The omic data used in the reconstruction process, and a thorough literature search eliminated the pathway using cytosolic acetyl-CoA sythetase (Fig. 5.a), and validated the other two, involving the transport and metabolism of acetyl-CoA-derived citrate or acetoacetate, using ATP-citrate lyase (ACITL) or cytosolic acetyl-CoA C-acetyltransferase (ACACT1r), respectively (Fig. 5.b–c; see Supplementary Notes for details).
Figure 5. Singular Value Decomposition (SVD) of feasible pathways elucidates potential pathways that allow for coupling of mitochondria acetyl-CoA metabolism and cytosolic acetylcholine production.
21,000 unique feasible reaction sets were computed, each showing transport of mitochondrial acetyl-CoA carbon to the cytosol in human metabolism. SVD of a matrix of all 21,000 pathways yielded 3 primary pathways that allow this coupling of mitochondrial metabolism to acetylcholine production, by carrying the acetyl-CoA carbon on (a) N-acetyl-L-apartate, (b) citrate, or (c) acetoacetate. As shown by the second singular vector, reactions in the pathway with citrate tend to be missing from pathways when the reactions for the acetoacetate pathway are included. The third singular vector shows a similar relationship of the N-acetyl-L-aspartate pathway. The omic data and known enzyme localization only support the usage of citrate and acetoacetate as potential carriers in neurons.
Our model contains these two pathways and shows a correlation between the flux through mitochondrial pyruvate dehydrogenase and choline acetyltransferase (r = 0.45, p = 3×10−247), consistent with the experimental observation of a tight coupling between mitochondrial metabolism and acetylcholine production. ACITL and ACACT1r also correlate with choline acetyltransferase flux (p < 3 × 10−90). Moreover, it has been reported that the inhibition of ACITL reduces the acetylcholine production rate by 30%33. The in silico inhibition of ACITL reduces acetylcholine production by 7.3%. The in silico decrease is smaller because the model can immediately adapt to the perturbation, while in vivo regulatory responses would take time to adapt. Therefore, it is expected that the model change is smaller. Interestingly, the inhibition of ACACT1r reduces acetylcholine production by 39%. Thus, it seems that cholinergic neurotransmission depends on redundant pathways, and that acetoacetate may play a more dominant role in transporting mitochondrial acetyl-CoA to the cytosol.
The coupling of mitochondrial metabolism to acetylcholine synthesis aids in the treatment of cholinergic disorders. However, knowledge of the abundance of cholinergic neurotransmission also aids in this purpose. It is difficult to identify cholinergic neurons, based solely on cell morphology, since cholinesterases and immunohistochemical markers for cholinergic neurons are also found in non-cholinergic neurons and other tissues34. Therefore, it is unknown what percentage of all neurotransmission is cholinergic.
Using our cholinergic model, we compute the percent contribution of cholinergic neurotransmission based on published data35. The data used for this purpose were obtained from rat brain minces, incubated in solutions containing [1-14C]pyruvate or [2-14C]pyruvate. Both acetylcholine and radiolabeled CO2 were measured at various titrations of several different pyruvate dehydrogenase inhibitors.
The cholinergic model was subjected to similar levels of pyruvate dehydrogenase inhibition. The simulations successfully reproduced the experimentally-witnessed linear relationship between acetylcholine production and metabolic rate, and acetylcholine production was correlated with CO2 release (r = 0.68).
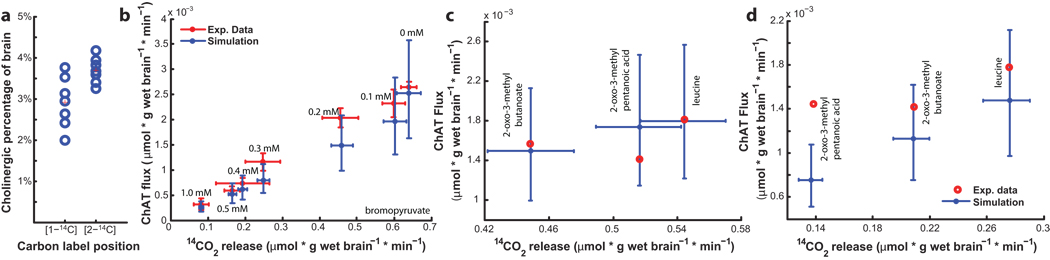
The fraction of cholinergic neurotransmission for the brain was computed by randomly choosing points from both the distributions of experimental data and distributions predicted by the simulations. A scaling factor was subsequently found that reconciles the two. This was repeated for 14 different combinations of pyruvate labeling and pyruvate dehydrogenase inhibitors35, yielding a median predicted cholinergic portion of total brain neurotransmission of 3.3% (Fig. 6.a). After adding this new parameter to the model, the predictions corresponded well with the experimental data sets (Fig. 6.b), including six datasets representing three pyruvate dehydrogenase inhibitors withheld from the previous computations (Fig. 6.c–d). Thus, the model was used in conjunction with experimental data to gain insight into physiological observations and derive important physiological parameters dependent on systems-level activity.
Figure 6. Model-aided prediction of cholinergic contribution is consistent with experimental acetylcholine production.
Percent brain cholinergic neurotransmission was predicted based on 14 sets of experimental data in which brain minces were fed [1-14C]-pyruvate or [2-14C]-pyruvate, followed by measurement of 14C-labeled CO2 and acetylcholine. (a) For each experiment, the feasible amount of the brain that can generate the experimental response was computed, centering at 3.3%. (b) This parameter was employed in the analysis, and the updated model predictions were consistent with experimental data, such as seen in the case of treating the brain minces with [1-14C]-pyruvate and increasing levels of the pyruvate-dehydrogenase inhibitor bromopyruvate. Moreover, the updated model predictions were consistent with measured 14C-labeled CO2 and acetylcholine production for brain minces that were treated with three PDHm inhibitors withheld from previous computations for both supplementation with (c) [1-14C]-pyruvate and (d) [2-14C]-pyruvate. Error bars on the simulation results represent 25th and 75th percentiles. ChAT = choline acetyltransferase.
Discussion and Conclusions
In this study we presented a workflow for generating tissue-specific, multicellular metabolic models. Through the analysis and integration of omic data, followed by manual curation, we used this workflow to build a first-draft manually-curated multicellular metabolic reconstruction of brain energy metabolism. Three models were generated from this reconstruction, representing different types of neurons coupled to astrocytes. We employed these models in three distinct analyses, in which we made predictions and gained systems-level insights into Alzheimer’s disease and cholinergic neurotransmission.
As experimental methods and data resolution improve, the accuracy of these models and their predictions should also improve. Improvements in neuroimaging and metabolomics will allow for more precise quantification of metabolite flow through the blood-brain-barrier, which is of interest since dysfunction of this system accompanies many neurological disorders and injuries36. In addition, improvements in transcriptomics and proteomics will provide higher-resolution quantification of cell- and organelle-specific genes and proteins. This data will allow models to account for neuron groups in specific brain regions, subcellular heterogeneity within cells, and the inclusion of less abundant glial cells. For example, higher-resolution models may provide insight into metabolic changes that occur in specific cell populations, such as the structures closely related to the olfactory system, which are affected in the early stages of Alzheimer’s disease37.
As seen in this work, novel insight into mammalian tissue-specific metabolism may be gained as more multicellular models are constructed. Our models demonstrate metabolic coupling and synergistic activities that more coarse-grained models miss, since the three analyses presented here were not possible using HR1 or the previous models of brain metabolism. The compartmentalization of metabolic processes within cells38, between cells39, and in host-pathogen interactions has an important role in normal physiology. Therefore, such models may provide greater insight and more accurately predict true cellular functions.
Lastly, while this work stands as an example of how one may construct such models, it also demonstrates a few types of analyses that can be done with these models. Scores of additional methods have been developed for genome-scale reconstructions ranging from constraint-based analyses5 to topological studies40.
In a broad sense, this study serves as an example of how mechanistic genotype-phenotype relationships can be built. From the genotype one can begin to reconstruct the network for an organism. The integration of high-throughput data and careful manual curation can add context-specific mechanistic network structure to genomic information. Thus, this network becomes a representation of the complex genetic interactions and biochemical mechanisms underlying observed phenotypes. This complex, but mechanistic, relationship between the genotype and phenotype can be used as a foundational structure upon which additional high-throughput data can be analyzed and predictive simulations can be conducted, thus leading to improved understanding, testable hypotheses, and increased knowledge1,2,40.
Methods
Reconstruction of iNL403
This work focuses on the core of cerebral energy metabolism and the pathways that play a critical role in cell-type specific functions in the brain. The pathways included in this work include mitochondrial metabolic pathways, central metabolic pathways closely tied to mitochondrial function, and additional pathways that are needed for modeling neuron and astrocyte functions. To reconstruct these pathways, a list of known human mitochondrial, glycolytic, and transport reactions were extracted from the manually-curated human metabolic reconstruction (HR1)7. From this list, reactions were directly added to the brain reconstruction if brain-localized protein or gene expression was suggested by the Human Protein Reference Database (release 5) (HPRD)41 or H-inv (version 4.3) (HINV)42, both of which provide tissue expression presence calls for each gene. Proteomic data from live human brain, acquired for the HUPO brain proteome project (BPP) (www.ebi.ac.uk/pride)43, were also used (See Supplementary Table 7 for accession numbers). Additional reactions were added as dictated by biochemical data from the literature (Supplementary Table 1). This reconstruction is designated iNL403, since it contains 403 genes. Reactions and pathways were manually curated to verify presence in the human brain and to determine cell-type localization, thus yielding a first-draft metabolic reconstruction of the brain metabolic network. Reactions unique to the different neuron types were determined from the literature (see notes in Supplementary Table 1), and consist largely of the reactions needed to make and metabolize their associated neurotransmitters. A list of all reactions, supporting data, citations, and a comparison with previous brain metabolism models can be found in Supplementary Tables 1– 4 and 8. Models in SBML format and model updates can be obtained from http://systemsbiology.ucsd.edu/In_Silico_Organisms/Brain.
Constraint-based modeling
Constraint-based modeling and analysis of metabolic networks has been previously described5, 12. Briefly, all of the reactions are described mathematically by a stoichiometric matrix, S, of size m x n, where m is the number of metabolites and n is the number of reactions, and each element is the stoichiometric coefficient of the metabolite in the corresponding reaction. The mass balance equations at steady state are represented as
where v is the flux vector12. Maximum and minimum fluxes and reaction reversibility, when known, are placed on each reaction, further constraining the system as follows:
At this point the model can then be used with many constraint-based methods5 to study network characteristics.
The S matrix was constructed with the mass and charge balanced reactions from the reconstruction. Select metabolites, known to cross the blood-brain barrier, were added as exchange reactions, allowing those metabolites to leave or enter the extracellular space in the model. A few metabolites from network gaps were allowed to enter or leave the system from the cytosol or mitochondria. This was only used when transporter mechanisms or subsequent pathway steps where not known, and when their entrance or removal from the system was necessary for model function. When available, cerebral metabolic rates were used from published data to constrain the upper and lower bounds of the exchange reactions44, 45. All parameters are detailed in Supplementary Tables 1 and 5.
Monte Carlo sampling
Monte Carlo sampling was used to generate a set of feasible flux distributions (points). The method is based on the artificially centered hit and run algorithm with slight modifications. Initially, a set of non-uniform pseudo-random points, called warm-up points, is generated. In a series of iterations, each point is randomly moved, always remaining within the feasible flux space. This is done by 1) choosing a random direction, 2) computing the limits of how far one can travel in that direction, and 3) choosing a new random point along this line. After many iterations, the set of points is mixed and approach a uniform sample of the solution space, thus providing a distribution for each reaction that represents the range and probability of the flux for each reaction, given the network topology and model constraints. For more detail, see the Supplementary Notes.
Simulating enzyme deficiencies
Enzyme deficiencies were obtained from the literature18. To simulate each deficiency, the distribution for all candidate flux states was determined using Monte Carlo sampling. From this distribution, the most probable flux was found and reduced by the fraction reported in the literature. All candidate states were then recomputed and compared with normal candidate flux states.
Alzheimer’s disease microarray analysis
Microarrays were obtained from the Gene Expression Omnibus (GSE5281). Arrays consist of 161 Affymetrix Human Genome U133 Plus 2.0 Arrays that profile the gene expression from laser-capture microdissected histopathologically normal neurons from six different brain regions of Alzheimer’s disease patients and age-matched controls. These arrays were not used in model construction.
Arrays were gcrma normalized using the bioconductor package for R. Pearson’s correlation coefficients were computed for all array pairs, and arrays with r < 0.8 were discarded (i.e., GSM119643, GSM119661, GSM119666, and GSM119676).
Different arrays had different levels of glial contamination. Therefore, to assess the amount of GAD (neuron-specific), the GAD1 and GAD2 levels on each array were normalized as follows. For each array, the relative amount of neuron material was determined by computing a ratio for four neuron-specific genes to the median level across all arrays. Neuron-specific genes were chosen to represent different neuron parts, including the soma, axon, and synaptic bouton (TUBB346, NeuN47, SYN148, and ACTL6B49). These were summed to compute a relative amount of neuron material (NM) for each array, j,
for each neuron marker gene gi. Since GAD genes are neuron-specific in the central nervous system, these were normalized for each array by the associated relative amount of neuron material, thus termed GADNMN for neuron-marker normalized GAD. It is assumed in this study that the neuron markers used here do not change their expression level per neuron between Alzheimer’s patients and age-matched control, since no published studies have demonstrated that these genes change expression in healthy cells through the progression of Alzheimer’s disease. It is possible that there is down-regulation of some neuron markers among neurons bearing neurofibrillary tangles, since synapse loss is a hallmark of Alzheimer’s disease23. However, the arrays used in this study profile histopathologically normal neurons and the surrounding glial cells. Therefore, it is not expected that there will be significant changes in the expression of these key neuronal genes in the data used here. Lastly, the inclusion of multiple genes from different cell regions helps to minimize the effects from expression changes not attributable to glial cell contamination. The results presented in this work are robust to the removal of each neuron marker gene (Supplementary Fig. 4).
PathWave analysis
PathWave allows for the elucidation of pathways that significantly change together. Its advantage over other methods, such as Gene Set Enrichment Analysis, is that it takes metabolic network connectivity into account in order to identify changes in pathways.
PathWave was used as published previously30. The reactions in each model were subdivided into biologically relevant functional pathways. Reactions that were involved in multiple pathways were added to each associated pathway. Reactions and their pathways are shown in Supplementary Table 9.
Since PathWave analyzes microarray data based on closely connected reactions, pathways were simplified by removing all metabolites with connectivity greater than eight in the metabolic network. Exceptions are listed in Supplementary Table 10.
For each of these simplified metabolic pathways, reactions were laid into a 2-dimensional, regular square lattice grid. To optimally preserve neighborhood relations of the reactions, adjacent nodes of the network were placed onto the grid as close to each other as possible. We mapped each expression data set, obtained from the Gene Expression Omnibus (GSE5281), onto the corresponding reactions of the transcribed enzymes. If a reaction was catalyzed by a complex of proteins, the average expression was taken. The resulting expression values of each reaction were z-transformed. Haar wavelet transforms on the optimized grid representation of each pathway were performed to explore every possible expression pattern of neighboring reactions and to define groups of reactions within a pathway that showed significant differences between samples of different conditions.
To obtain significance values, the sample labels were permutated (n = 10,000) and scores were calculated for each wavelet and permutation. The scores represent the absolute value of the logarithm of the p-value for each wavelet feature, calculated by t-tests. For the best hit (highest score of the non-permutated wavelet features) a p-value was obtained from the reference distributions and represented the significance for the corresponding pathway. The p-value for each pathway was corrected for multiple testing (FDR = 0.05)50. Only pathways with more than three significantly differentially regulated reactions were further considered (FDR = 0.05). To obtain local patterns in the pathways, all wavelet features were statistically tested applying t-tests and corrected for multiple testing. Statistically significant features contained those sub-graphs of the metabolic network that showed differentially regulated patterns. Reconstructing these sub-graphs allowed us to directly detect the regions of interest in the metabolic network (Supplementary Table 11).
Identifying pathways for acetylcholine synthesis
An FBA-derived approach was employed to identify all possible pathways coupling the mitochondrial and cytosolic acetyl-CoA pools using known reactions in human metabolism. First, the potential pathways were identified using HR17. A reaction that supplies mitochondrial acetyl-CoA was added to the model. A second reaction was added to remove cytosolic acetyl-CoA from the model. Lastly, all other metabolite uptake and secretion constraints were opened. Reactions were randomly removed until a minimum pathway was identified, capable of carrying flux between mitochondrial and cytosolic acetyl-CoA. This was repeated until more than 21,000 unique sets of reactions were identified. An r x p binary matrix was then built with the p unique reaction sets consisting of r reactions. Each element (i,j) of this matrix was 0 if reaction i was absent from pathway j or 1 if reaction i was in pathway j. Rows for all reactions that were never necessary were subsequently removed from the matrix. Singular value decomposition was then used, followed by varimax factor rotation of the first five singular vectors. Singular vector loadings demonstrated the dominant sets of reactions, and their major dependencies that could be used to couple mitochondrial acetyl-CoA metabolism and cytosolic acetylcholine metabolism.
Predicting cholinergic neurotransmission
The percentage cholinergic neurotransmission was computed based on published data35. The previously published data were obtained from rat brain minces that were incubated in solutions containing [1-14C]pyruvate or [2-14C]pyruvate. Both acetylcholine and radiolabeled CO2 were measured at various titrations of several different inhibitors of pyruvate dehydrogenase (PDHm) (see Supplementary Notes for all inhibitors).
Simulations were conducted using the cholinergic model. The models were allowed to take up the same substrates provided experimentally35, at rates consistent with the data (see Supplementary Table 5). Monte Carlo sampling was used to identify all feasible flux states. This was done for various levels of PDHm inhibition, ranging from 0 to 90% inhibition. The percentage cholinergic neurotransmission was computed by randomly selecting a feasible flux state from each level of PDHm inhibition and computing the slope of the sum of labeled CO2-producing fluxes and choline acetyltransferase for the different simulations. A similar slope was computed from randomly sampled points from the reported experimental distributions. The ratio of these slopes represents a feasible percentage cholinergic neurotransmission. This was repeated 1000 times and the median value was reported. Comparisons with the experimental data were done by suppressing the in silico pryruvate dehydrogenase flux until the measured CO2 release rate was obtained. At this level of suppression, the resulting predicted acetylcholine production rate was compared with the experimentally measured rates. See Supplementary Notes for more detailed methods.
Supplementary Material
Acknowledgments
The authors thank Dr. Gary Gibson at Cornell University, Dr. Ines Thiele at the University of Iceland, and Marc Abrams, Dr. Monica Mo and Dr. Christian Barrett at UCSD for suggestions pertaining to this work. This work was funded in part by a Fulbright fellowship, an NSF IGERT Plant Systems Biology training grant (# DGE-0504645), NIH grant 2R01GM068837_05A1, and the Helmholtz Alliance on Systems Biology and the BMBF by the NGFN+ neuroblastoma project ENGINE.
Footnotes
Author contributions
N.E.L., J.C., A.Y., M.P.A. and B.Ø.P. conceived and designed model. N.E.L., J.C., G.S., R.K., R.E., J.S., A.B. and R.A.L. performed data analyses. The manuscript was written by N.E.L., G.S., J.S., A.B. and B.Ø.P.
References
- 1.Feist AM, Palsson BO. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat. Biotechnol. 2008;26:659–667. doi: 10.1038/nbt1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oberhardt MA, Palsson BO, Papin JA. Applications of genome-scale metabolic reconstructions. Mol. Syst. Biol. 2009;5:320. doi: 10.1038/msb.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breitling R, Vitkup D, Barrett MP. New surveyor tools for charting microbial metabolic maps. Nat. Rev. Microbiol. 2008;6:156–161. doi: 10.1038/nrmicro1797. [DOI] [PubMed] [Google Scholar]
- 4.Thiele I, Palsson BO. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protocols. 2010;5:93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis NE, Jamshidi N, Thiele I, Palsson BØ. Encyclopedia of Complexity and Systems Science. Vol. 5535. New York: Springer; 2009. [Google Scholar]
- 6.Orth JD, Thiele I, Palsson BO. What is flux balance analysis? Nat. Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duarte NC, et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc. Natl. Acad. Sci. U. S. A. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker SA, Palsson BO. Context-specific metabolic networks are consistent with experiments. PLoS Comput. Biol. 2008;4:e1000082. doi: 10.1371/journal.pcbi.1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shlomi T, Cabili MN, Herrgard MJ, Palsson BO, Ruppin E. Network-based prediction of human tissue-specific metabolism. Nat. Biotechnol. 2008 doi: 10.1038/nbt.1487. [DOI] [PubMed] [Google Scholar]
- 10.Jerby L, Shlomi T, Ruppin E. Computational reconstruction of tissue-specific metabolic models: application to human liver metabolism. Mol. Syst. Biol. 2010;6:401. doi: 10.1038/msb.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ponten F, et al. A global view of protein expression in human cells, tissues, and organs. Mol. Syst. Biol. 2009;5:337. doi: 10.1038/msb.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palsson Bin. Systems biology : properties of reconstructed networks. Vol. 322. New York: Cambridge University Press, Cambridge; 2006. [Google Scholar]
- 13.Chatziioannou A, Palaiologos G, Kolisis FN. Metabolic flux analysis as a tool for the elucidation of the metabolism of neurotransmitter glutamate. Metab. Eng. 2003;5:201–210. doi: 10.1016/s1096-7176(03)00029-6. [DOI] [PubMed] [Google Scholar]
- 14.Cakir T, Alsan S, Saybasili H, Akin A, Ulgen KO. Reconstruction and flux analysis of coupling between metabolic pathways of astrocytes and neurons: application to cerebral hypoxia. Theor. Biol. Med. Model. 2007;4:48. doi: 10.1186/1742-4682-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Occhipinti R, Puchowicz MA, LaManna JC, Somersalo E, Calvetti D. Statistical analysis of metabolic pathways of brain metabolism at steady state. Ann. Biomed. Eng. 2007;35:886–902. doi: 10.1007/s10439-007-9270-5. [DOI] [PubMed] [Google Scholar]
- 16.Reiman EM, et al. Functional brain abnormalities in young adults at genetic risk for late-onset Alzheimer's dementia. Proc. Natl. Acad. Sci. U. S. A. 2004;101:284–289. doi: 10.1073/pnas.2635903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukui H, Diaz F, Garcia S, Moraes CT. Cytochrome c oxidase deficiency in neurons decreases both oxidative stress and amyloid formation in a mouse model of Alzheimer's disease. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14163–14168. doi: 10.1073/pnas.0705738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bubber P, Haroutunian V, Fisch G, Blass JP, Gibson GE. Mitochondrial abnormalities in Alzheimer brain: mechanistic implications. Ann. Neurol. 2005;57:695–703. doi: 10.1002/ana.20474. [DOI] [PubMed] [Google Scholar]
- 19.Gibson GE, et al. Alpha-ketoglutarate dehydrogenase in Alzheimer brains bearing the APP670/671 mutation. Ann. Neurol. 1998;44:676–681. doi: 10.1002/ana.410440414. [DOI] [PubMed] [Google Scholar]
- 20.Casley CS, Canevari L, Land JM, Clark JB, Sharpe MA. Beta-amyloid inhibits integrated mitochondrial respiration and key enzyme activities. J. Neurochem. 2002;80:91–100. doi: 10.1046/j.0022-3042.2001.00681.x. [DOI] [PubMed] [Google Scholar]
- 21.Hoshi M, et al. Regulation of mitochondrial pyruvate dehydrogenase activity by tau protein kinase I/glycogen synthase kinase 3beta in brain. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2719–2723. doi: 10.1073/pnas.93.7.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorman AM, Ceccatelli S, Orrenius S. Role of mitochondria in neuronal apoptosis. Dev. Neurosci. 2000;22:348–358. doi: 10.1159/000017460. [DOI] [PubMed] [Google Scholar]
- 23.Ginsberg SD, Che S, Counts SE, Mufson EJ. Single cell gene expression profiling in Alzheimer's disease. NeuroRx. 2006;3:302–318. doi: 10.1016/j.nurx.2006.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai MKP, Ramirez MJ, Tsang SWY, Francis PT. In: Neurobiology of Alzheimer's Disease. Dawbarn D, Allen SJ, editors. New York: Oxford; 2007. pp. 245–282. [Google Scholar]
- 25.Santos SS, et al. Inhibitors of the alpha-ketoglutarate dehydrogenase complex alter [1-13C]glucose and [U-13C]glutamate metabolism in cerebellar granule neurons. J. Neurosci. Res. 2006;83:450–458. doi: 10.1002/jnr.20749. [DOI] [PubMed] [Google Scholar]
- 26.Hassel B, Johannessen CU, Sonnewald U, Fonnum F. Quantification of the GABA shunt and the importance of the GABA shunt versus the 2-oxoglutarate dehydrogenase pathway in GABAergic neurons. J. Neurochem. 1998;71:1511–1518. doi: 10.1046/j.1471-4159.1998.71041511.x. [DOI] [PubMed] [Google Scholar]
- 27.Liang WS, et al. Altered neuronal gene expression in brain regions differentially affected by Alzheimer's disease: a reference data set. Physiol. Genomics. 2008;33:240–256. doi: 10.1152/physiolgenomics.00242.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stuhmer T, Anderson SA, Ekker M, Rubenstein JL. Ectopic expression of the Dlx genes induces glutamic acid decarboxylase and Dlx expression. Development. 2002;129:245–252. doi: 10.1242/dev.129.1.245. [DOI] [PubMed] [Google Scholar]
- 29.Ibanez V, et al. Regional glucose metabolic abnormalities are not the result of atrophy in Alzheimer’s disease. Neurology. 1998;50:1585–1593. doi: 10.1212/wnl.50.6.1585. [DOI] [PubMed] [Google Scholar]
- 30.Schramm G, et al. PathWave: discovering patterns of differentially regulated enzymes in metabolic pathways. Bioinformatics. 2010;26:1225–1231. doi: 10.1093/bioinformatics/btq113. [DOI] [PubMed] [Google Scholar]
- 31.Ragozzino ME, Pal SN, Unick K, Stefani MR, Gold PE. Modulation of hippocampal acetylcholine release and spontaneous alternation scores by intrahippocampal glucose injections. J. Neurosci. 1998;18:1595–1601. doi: 10.1523/JNEUROSCI.18-04-01595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watson GS, Craft S. Modulation of memory by insulin and glucose: neuropsychological observations in Alzheimer’s disease. Eur. J. Pharmacol. 2004;490:97–113. doi: 10.1016/j.ejphar.2004.02.048. [DOI] [PubMed] [Google Scholar]
- 33.Cooper JR. Unsolved problems in the cholinergic nervous system. J. Neurochem. 1994;63:395–399. doi: 10.1046/j.1471-4159.1994.63020395.x. [DOI] [PubMed] [Google Scholar]
- 34.Karczmar AGin. Exploring the vertebrate cholinergic nervous system. Vol. 686. New York, N.Y.: Springer; 2006. [Google Scholar]
- 35.Gibson GE, Jope R, Blass JP. Decreased synthesis of acetylcholine accompanying impaired oxidation of pyruvic acid in rat brain minces. Biochem. J. 1975;148:17–23. doi: 10.1042/bj1480017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abbott NJ, Ronnback L, Hansson E. Astrocyte-endothelial interactions at the blood-brain barrier. Nat. Rev. Neurosci. 2006;7:41–53. doi: 10.1038/nrn1824. [DOI] [PubMed] [Google Scholar]
- 37.Thompson MD, Knee K, Golden CJ. Olfaction in persons with Alzheimer's disease. Neuropsychol. Rev. 1998;8:11–23. doi: 10.1023/a:1025627106073. [DOI] [PubMed] [Google Scholar]
- 38.Schryer DW, Peterson P, Paalme T, Vendelin M. Bidirectionality and compartmentation of metabolic fluxes are revealed in the dynamics of isotopomer networks. Int. J. Mol. Sci. 2009;10:1697–1718. doi: 10.3390/ijms10041697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serres S, Raffard G, Franconi JM, Merle M. Close coupling between astrocytic and neuronal metabolisms to fulfill anaplerotic and energy needs in the rat brain. J. Cereb. Blood Flow Metab. 2008;28:712–724. doi: 10.1038/sj.jcbfm.9600568. [DOI] [PubMed] [Google Scholar]
- 40.Lee DS, et al. The implications of human metabolic network topology for disease comorbidity. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9880–9885. doi: 10.1073/pnas.0802208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mishra GR, et al. Human protein reference database--2006 update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujii Y, Imanishi T, Gojobori T. H-Invitational Database: integrated database of human genes. Tanpakushitsu Kakusan Koso. 2004;49:1937–1943. [PubMed] [Google Scholar]
- 43.Reidegeld KA, et al. The power of cooperative investigation: summary and comparison of the HUPO Brain Proteome Project pilot study results. Proteomics. 2006;6:4997–5014. doi: 10.1002/pmic.200600305. [DOI] [PubMed] [Google Scholar]
- 44.Lying-Tunell U, Lindblad BS, Malmlund HO, Persson B. Cerebral blood flow and metabolic rate of oxygen, glucose, lactate, pyruvate, ketone bodies and amino acids. Acta Neurol. Scand. 1980;62:265–275. doi: 10.1111/j.1600-0404.1980.tb03035.x. [DOI] [PubMed] [Google Scholar]
- 45.Lying-Tunell U, Lindblad BS, Malmlund HO, Persson B. Cerebral blood flow and metabolic rate of oxygen, glucose, lactate, pyruvate, ketone bodies and amino acids. Acta Neurol. Scand. 1981;63:337–350. doi: 10.1111/j.1600-0404.1981.tb00788.x. [DOI] [PubMed] [Google Scholar]
- 46.Tischfield MA, et al. Human TUBB3 Mutations Perturb Microtubule Dynamics, Kinesin Interactions, and Axon Guidance. Cell. 2010;140:74–87. doi: 10.1016/j.cell.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim KK, Adelstein RS, Kawamoto S. Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors. J. Biol. Chem. 2009;284:31052–31061. doi: 10.1074/jbc.M109.052969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Camilli P, Cameron R, Greengard P. Synapsin I (protein I), a nerve terminalspecific phosphoprotein. I. Its general distribution in synapses of the central and peripheral nervous system demonstrated by immunofluorescence in frozen and plastic sections. J. Cell Biol. 1983;96:1337–1354. doi: 10.1083/jcb.96.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olave I, Wang W, Xue Y, Kuo A, Crabtree GR. Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes Dev. 2002;16:2509–2517. doi: 10.1101/gad.992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benjamini Y, Yekutieli D. The Control of the False Discovery Rate in Multiple Testing under Dependency. The Annals of Statistics. 2001;29:1165–1188. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.