Abstract
Background:
To identify in vivo new cardiac binding sites of serum response factor (SRF) in genes and to study the response of these genes to mild over-expression of SRF, we employed a cardiac-specific, transgenic mouse model, with mild over-expression of SRF (Mild-O SRF Tg).
Methodology:
Microarray experiments were performed on hearts of Mild-O-SRF Tg at 6 months of age. We identified 207 genes that are important for cardiac function that were differentially expressed in vivo. Among them the promoter region of 192 genes had SRF binding motifs, the classic CArG or CArG-like (CArG-L) elements. Fifty-one of the 56 genes with classic SRF binding sites had not been previously reported. These SRF-modulated genes were grouped into 12 categories based on their function. It was observed that genes associated with cardiac energy metabolism shifted toward that of carbohydrate metabolism and away from that of fatty acid metabolism. The expression of genes that are involved in transcription and ion regulation were decreased, but expression of cytoskeletal genes was significantly increased. Using public databases of mouse models of hemodynamic stress (GEO database), we also found that similar altered expression of the SRF-modulated genes occurred in these hearts with cardiac ischemia or aortic constriction as well.
Conclusion and significance:
SRF-modulated genes are actively regulated under various physiological and pathological conditions. We have discovered that a large number of cardiac genes have classic SRF binding sites and were significantly modulated in the Mild-O-SRF Tg mouse hearts. Hence, the mild elevation of SRF protein in the heart that is observed during typical adult aging may have a major impact on many SRF-modulated genes, thereby affecting cardiac structure and performance. The results from our study could help to enhance our understanding of SRF regulation of cellular processes in the aged heart.
Keywords: SRF modulated genes, SRF binding sites, mouse heart, mild-SRF over-expression, gene expression, striated muscle
Introduction
It is well appreciated that the mammalian adult heart undergoes a number of changes with advancing age.1–3 Recent studies indicate that one of the key transcription factors, serum response factor (SRF), plays an important role in the regulation of cardiac genes during development and adult aging.4–7 SRF is a member of the MADS (MCM1, Agamous, Deficiens, SRF) family of transcription factors that regulates the genes that are usually considered to be immediate-early genes and muscle-related genes.8,9 SRF also serves to regulate cell proliferation, cell size, and cell survival.7,10–12
SRF modulates genes by binding to the cognate response element, the serum response element (SRE), which contains a consensus sequence of CC(A/T)6GG also known as the CArG box.13–16
In addition, SRF can also regulate the gene promoters containing the CArG-like (CArG-L) elements, which have only a single base mismatch from the classic CArG box.8,17 It has been estimated that hundreds of SRF-modulated genes that contain CArG and/or CArG-L motifs may exist in both the mouse and human genomes.18 However, to identify those genes with CArG or CArG-L elements remains a challenge.
The level of SRF expression increases by approximately 20% from the age of 3 months to 20 months (from young adulthood to early senescence) in rodent hearts.4,6 It is plausible that this increased SRF might contribute to altered expression of SRF-modulated genes, thereby affecting cardiac function in aged mice. In our previous study, we reported the generation and characterization of transgenic mice with mild cardiac-specific SRF overexpression of approximately 40%–50%.6 Mild overexpression of SRF produced cardiac changes in young adult transgenic mice similar to that observed in normal senescence. By 6 months of age, the hearts of these young adult transgenic mice had changes that usually appear later, at around 20 months or older, which include mild cardiomyocyte hypertrophy, cardiac fibrosis and mildly increased left ventricular wall thickness. The cardiac functional changes, including a 20% reduction in early diastolic LV filling (peak E) and a 35% decline in peak E-to-peak-A (late diastolic filling) ratio, are similar to those seen clinically in late life as part of typical human adult myocardial aging.6,19,20 It appeared likely that SRF-modulated genes may have contributed to the cardiac phenotype in this model of myocardial aging.6
To determine the response in vivo of potential SRF-modulated genes, we examined the cardiac gene profile of the Mild-O-SRF Tg. We found that the expression of 207 cardiac genes was significantly altered in the transgenic mice compared to their non-transgenic littermates. Bioinformatics analysis of the promoter of 207 genes showed that 93% of the genes in the 6 month old Mild-O-SRF Tg hearts had CArG or CArG-L elements. These genes encoded a broad spectrum of proteins involved in multiple functions including metabolism, cytoskeleton, transcription and translational regulation, extracellular matrix, ion transport, stress response, as well as protease and protease inhibitors.
Experimental Procedures
Transgenic mouse with mild cardiac-specific overexpression of SRF (Mild-O-SRF Tg)
The generation and characterization of transgenic mice with mild cardiac-specific overexpression of SRF was previously reported.6 At 6 months of age, the Mild-O-SRF Tg mice manifested cardiac changes with mild diastolic impairment, suggestive of an “aged heart”.6 These alterations included mild fibrosis and hypertrophy, with preserved systolic function, but impaired diastolic function. Therefore, 6-monthold transgenic and non-transgenic mice were used in this study. The studies were conducted with Institutional Review Board approval from the University of Arkansas for Medical Sciences, and in accordance with the NIH Guiding Principles for Research Involving Animals.
Total RNA isolation, GeneChip hybridization and preliminary data analysis
Total RNA was isolated from the cardiac ventricles of the transgenic and non-transgenic mice as previously described.21 The total RNA preparations were then subjected to a purification procedure using RNeasy Mini Spin Columns (Qiagen). The total RNA preparations from five animals were pooled per group. Each sample from one group was hybridized to an independent GeneChip MGU74Av2 (Affymetrix). The GeneChip hybridization and preliminary data analysis were performed according to the standard procedures at the Genomic Center at Beth Israel Deaconess Medical Center in Boston.22
| Gene | Forward sequence | Reverse sequence |
|---|---|---|
| B4Galnt1 | TCGCTGACGACAGTGACAAAC | TGCCGAAGGGCATGAAGTAG |
| Apod | TGGCCACCGATTATGAAAACT | CCACATGGAAGAGCCAGAAGA |
| Pfkp | TCAGCCCTGCACCGAATTAT | CATCACCTCCAGGACGAAGGT |
| Acot9 | CATATGAACTTGCATGGGCTACTG | AACCAACTTCAACAGGCTTCTGA |
| Slc27a1 | GGACGCCAGTAGTGGTATATGCT | GCGGAACAGGTTGGCTACAG |
| Lipc | CCTCAGCACCCGGAAACA | CACCCGTGGATGATCATGATAA |
| Coasy | CCGTGGGTGGCACTTTTG | GCTCCTGGGCCAGTACACAT |
| Pafah1b3 | GTTGTGCTGGGCCTGCTT | CCTGTCGGTTTTTCTCTCGAA |
| Pgk2 | TGCTAAAGGAACCAAAGCTCTCAT | GGCGCAGCAAGTAGCAGTATCT |
| Apoa1 | AGAGCAACCCTACCTTGAACGA | TGGCTTTCTCGCCAAGTGT |
| Maoa | CCCATTCCGTGGTGCATT | CATCCATTGTCCTCCACAGGTT |
| Cyp2b10 | GAAAGTCCAAAAGGAGATTGATCAG | CATTTTGGTGCGGTCATCAAG |
| Cyp3a16 | CTGCAGGAGGAGATCGATGAG | CCATCGCCATCACGGTATC |
| Cyp3a11 | AACTGCAGGATGAGATCGATGAG | TTCATTAAGCACCATATCCAGGTATT |
| Uox | TCAAACACGTCCATGCATTCA | TTCTCATCTGCTCCACCTCACA |
| Myl1 | TGAACAGTTTCTGCCCATGATG | CAGACCCTCAACGAAATCTTCATA |
| Anxa10 | GACATGCTGATTGACATCCTAACAC | CATGCTCTGATAGGTCCCAGCTA |
| Rab8b | GGCGAAGACGTACGATTATCTGT | GTTGAAGGCGTCCTCTGAGAA |
| Acta2 | TCCTGACGCTGAAGTATCCGATA | GGTGCCAGATCTTTTCCATGTC |
| Casq1 | TTCTTAGATCCCAGCCCAGTTC | GTGGCGGGAAGAGAAACAGA |
| Actn1 | CCTCCCGGATGCAGACAA | TGGACGATCTTGGACACTTCAT |
| Epb4.2 | TCACGGCAATGGCAAGGT | CGGTTTACCAATGGCCATATTT |
| Dstn | TGAAGCAGGTACCATGGATGAC | GCCATGCCTGTGATGTTAACA |
Microarray data analysis
The microarray data analysis and data interpretation were performed using ArrayTrack.23 A list of differentially expressed genes (DEGs) were identified using a t-test with a combination of cut off P-value (P < 0.05) and fold change (FC > 2). The Array-Track Gene Ontology (GO) tool Gene Ontology for Function Analysis (GOFFA) was subsequently applied to the DEGs for biological interpretation.24,25 The statistical significance of a GO term was determined using Fisher’s Exact Test. Furthermore, the GOPath and TreePrune in GOFFA were also used to identify significant biological functions based on the DEGs. In addition to GOFFA analysis, DEGs were also analyzed in canonical pathway maps using GeneGo MetaCore. Experimental data are visualized as red/blue thermometers pointing up/down, and signifying up/down-regulation of the map objects.
The microarray data is MIAME compliant and has been deposited in GEO database with ACCESSION number GSE19874.
Validation of the array data: real time PCR
Validation of the Affymetrix data was performed by qPCR analysis with the ABI PRISM 7700 Sequence Detection System (Applied Biosystem, CA) using standard procedures. All microarray results that were changed ≥2 fold were validated by real-time RT PCR. The primers used in experiments are enumerated in the table above.
To allow comparison of qRT-PCR values, the concentration of cDNA in each sample was adjusted to yield similar amounts of PCR product when amplified by primers for 18S. The 18S reaction was performed using standard curves representing 5, 1.25, 0.31, and 0.08 ng/μl of the pooled cDNA. Standard curves were generated for all other targets using pooled RT-PCR products at 80, 20, 5 and 1.25 ng/μl. Relative standard curve method is used to calculate the amplification difference between the samples.
Analysis of SRF binding sites: the classic CArG and CArG-like motifs in the gene promoter
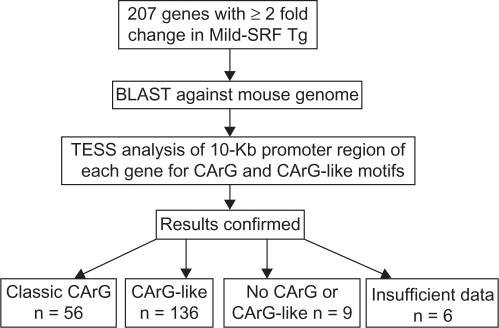
The criteria for the classic CArG motif is a 10-bp element that has the sequence CC(A/T) 6GG and CArG-like element has a single base mismatch from its classic counterpart.8 Briefly, the mRNA sequences were obtained from the RefSeq database for most of the 207 genes; the mRNA sequences were also obtained from GenBank database for several genes that did not have reference sequences in the RefSeq database as of September 2007. The reference mRNA sequences were submitted to BLAST for comparison with the mouse genomic DNA sequence in the mouse genome database. After noting the orientation of the alignment, the appropriate 10-Kb genomic DNA sequence upstream from the transcription start point corresponding to the promoter region of each gene was isolated and analyzed using a web-based bioinformatics tool TESS at http://www.cbil.upenn.edu/cgi-bin/tess/tess. Because TESS results are model-based, the potential CArG and CArG-like sequences were verified by both using LALIGN (http://www.ch.embnet.org/software/LALIGN_form.html) and visual confirmation (Fig. 1).
Figure 1.
Flow diagram showing the method of defining classic CArG and CArG-like elements in 207 SRF-modulated genes in response to mild-SRF overexpression in vivo. The sequence of each gene was BLAST-ed against the mouse genome and the promoter of each gene was identified. The classic CArG and/or CArG-like sequences in the promoter region were identified by Transcription Element Search (TESS) software and confirmed by LALIGN software and visual comparison (see text in methods section for details).
Comparison of differentially expressed genes in the Mild-O-SRF Tg mice with other mouse models
To ascertain if our data compared well with other models of aging or cardiac ischemia, we looked at 27 other GEO microarray data bases of aging and heart available online at the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/). The GEO database was searched for expression data from experiments using mouse hearts on the Affymetrix MGU74aV2 chip. The 27 gene expression data sets were also analyzed for differential expression by fold-change and P-value (two-tailed T-test). The lists of differentially expressed genes were combined and filtered with our list of 207 differentially expressed SRF-modulated genes. Fold-change in gene expression was examined at 2-fold change and P-value was tested at P < 0.05.
Results
Mild overexpression of SRF altered many cardiac genes in vivo
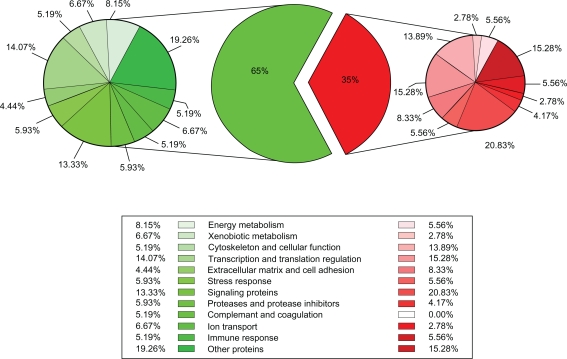
To examine the impact of mild overexpression of SRF on cardiac gene expression and to explore the potential mechanism underlying the functional changes resembling cardiac aging, microarray analysis was performed using mouse hearts from the 6-month-old transgenic mice and non-transgenic littermates. The expression of 207 cardiac genes was significantly altered in the transgenic mice compared to non-transgenic littermates (Tables 1 and 2). We consider all 207 of these genes as “SRF-modulated genes”. Among them, 65% (135 of 207) of the genes were down-regulated, whereas 35% (72 of 207) of the genes were up-regulated, indicating that increased SRF expression repressed the expression of a majority of cardiac genes (Fig. 2).
Table 1.
List of the 56 SRF-modulated genes containing “classic CArG” element that are altered in response to mild-SRF overexpression in vivo. The down-regulated genes are in the left column, whereas up-regulated genes are in the right column. Genes marked with a “*” are those which were found in this study to contain previously unreported CArG elements. Genes marked with a “+” are genes which contain both “classic CArG” and non-classic “CarG-like” elements.
| Downregulated | Upregulated | ||||||
|---|---|---|---|---|---|---|---|
| Classic CArG (56 genes) | 8430410A17Rik* | Cyp3a16*+ | II1f5*+ | Osteonectin*+ | Anxa10* | Epb4.2*+ | Postn+ |
| Aadac *+ | Dsc2*+ | Jag2*+ | Pcsk5*+ | Aqr*+ | Fh1l+ | Rab8b* | |
| Akr1c6*+ | E2f3*+ | Lancl2*+ | Rrm1*+ | R3hcc1*+ | Hspa1b*+ | Serpine1*+ | |
| Aqp4*+ | Elovl2* | LOC547428*+ | Serpina1a/b/c/d*+ | BB001228* | Icsbp1* | Slc20a1*+ | |
| Brcal*+ | Es1*+ | Mup1*+ | Slc4a8*+ | Cyp2b10*+ | II4* | Tbp*+ | |
| C77681* | Hoxb8* | Mup2*+ | Spink3*+ | Dlg7*+ | Myh7+ | Tpm2+ | |
| Cdca5*+ | Hpx*+ | Myh4*+ | Ugt2b5*+ | Emp1*+ | Myl1*+ | Usp29*+ | |
| Crk*+ | Ift81*+ | Ofa* | Uox* | Emr1*+ | Nppa+ | ||
| Cwf19I1* | |||||||
| CArG-like (136 genes) | 1100001G20Rik* | Dmrtb1* | Kcnq2* | Rps12* | A430107D22Rik* | Chx10* | Iqgap1* |
| AI042964* | Efnb3* | Kng1* | Rps17* | AA222883* | Col1a1* | Maoa* | |
| Apc2* | Eif2s3y* | Lgals2* | S100a9* | Acot9* | Ctgf | Ndrg4* | |
| Apoa1* | F10 | Lhx8* | Scg2* | Acta2 | Cxcl12* | Rrp12* | |
| Atp1a1* | Fdft1* | Lifr* | Six3* | Actn1 | Dnase1l2* | Pfkp* | |
| BphI* | Fgb* | Lipc* | Skb1* | Apod* | Dpp7 | Plagl1* | |
| Bub1* | Fmnl1* | Loc380988 | Slc10a1* | Arhgdig* | Dstn | Ptpns1* | |
| C77691* | Gabra6* | Mug1/2* | Slc27a1* | Atf3* | Fbln1* | Pus1* | |
| C8g* | Gabrb2* | Mup3* | Sp4* | Avpi1* | Gdf15* | Rap2ip* | |
| Camk2a* | Gm1418* | Mup5* | Stard10* | B4galnt1* | Gipc1* | Rho* | |
| Capn2* | Gm1419* | Itga8 | Sult1a1* | Bdkrb1* | H2-DMb1* | Rps4y2* | |
| Cdh15* | Gnmt* | Nsun2* | Syt3* | Bgn* | Hnedc2* | Srxn1* | |
| Cfi* | Gria2* | Oat* | Tecta* | Bmp10* | Hspa1a* | Tgfb3* | |
| Cldn11* | Gtf3c4* | Olfr18* | Tesp2* | Casq1 | Id2* | Wbp5* | |
| Cpa3* | H2-M10.1* | Orm1* | Tk1* | Ccl8* | II3ra | Znrf4* | |
| Csh1* | Hba-x* | Pafah1b3* | Tnfaip3* | ||||
| Cyp3a11* | Hcrt* | Pgk2* | Trpm7* | ||||
| Dcc* | Homer1* | Plekhh1* | Tspan8* | ||||
| Dclre1a* | Hrc* | Ppil2* | Tyms-ps* | ||||
| Ddb1* | II10ra* | Prlpf* | Ubd* | ||||
| Ddx3x* | II15 | Pzp | Vax2* | ||||
| Defcr5* | Inhbc* | Rpl34 | Vps13c* | ||||
| Diablo* | Insm1* | ||||||
| No CArG or CArG-like (9 genes) Insuff data (6) | 1500003O03Rik | Coasy | Hmx1 | Plg | Av169168 | DIx5 | |
| Arhgap9 | D8ertd738e | Plekha 1 | |||||
| Cr1 | Gm1060 | Igh-V | Igh-VX24 | Birc1c | H2-TL-T17-c | ||
Table 2.
Functional categories of SRF-modulated genes. Each of the 207 SRF-modulated genes was assigned to one of 12 categories. 56 genes contained classic CArG elements, 136 genes contained CArG-like elements, 15 genes did not have both elements, or there are not sufficient data to show that they had either element.
| Category | CArG | CArG-like | Without CArG/CArG-like*, or insufficient data |
|---|---|---|---|
| 1. Energy metabolism | 4 | 10 | 1 |
| 2. Xenobiotic metabolism | 4 | 7 | |
| 3. Cytoskeleton | 8 | 9 | |
| 4. Transcription and translation | 7 | 20 | 3 |
| 5. Extracellular matrix | 2 | 10 | |
| 6. Stress response | 2 | 10 | |
| 7. Signaling proteins | 7 | 24 | 2 |
| 8. Proteases, and protease inhibitors | 5 | 6 | |
| 9. Complement and coagulation | 0 | 6 | 1 |
| 10. Ion transport | 3 | 8 | |
| 11. Immune response | 1 | 6 | 4 |
| 12. Other proteins | 13 | 20 | 4 |
| Total | 56 | 136 | 15 |
Figure 2.
A Pie chart schematic showing percentage distribution of 207 SRF-modulated genes in response to mild-SRF overexpression in vivo. The pie chart in the center depicts percent distribution of all the 207 genes: 35% (72 of 207) of the genes are increased (in red), and 65% (135 of 207) of the genes are decreased (in green). The smaller pie charts on the right (shades of red) and left (shades of green) show distribution of the functional categories of genes within the increased or decreased subdivision. The percentage distribution is marked next to each category on the pie chart as well as in the legend.
Most of the genes that were significantly impacted by mild overexpression of SRF contained CArG and/or CArG-like elements
To examine whether SRF activated or repressed genes that contain CArG and/or CArG-like elements within their promoter regions, we further analyzed transcriptional sites within the 10-Kb promoter region in each of the 207 genes. The classic CArG element was defined as a 10-bp DNA sequence with “CC(A/T)6GG”. The “CArG-like” element was defined as a 10-bp sequence with only one substitution from the consensus sequence. As shown in the flow diagram in Figure 1, CArG and CArG-like elements within the 10-Kb promoter region of each gene was analyzed and verified by using software program TESS and LALIGN.26,27 The existence of one or more of either a CArG or a CArG-like element in the promoter sequence of each of the 207 genes was then confirmed by visual inspection.
Most of the genes that were significantly impacted by mild overexpression of SRF contained CArG and/or CArG-like elements. As shown in Tables 1 and 2, approximately 93% (192 of 207) of the cardiac genes that responded significantly to SRF overexpression contained CArG and/or CArG-like elements. Roughly 29% (56 of 192) of the SRF-modulated genes contained at least one classic CArG element, while 71% (136 of 192) of them had at least one CArG-like element. Of those 56 genes with a classic CArG element, approximately 79% (44 of 56) also contained at least one CArG-like element. Many of these genes have not been previously reported as SRF-modulated genes (Table 1 and Table S1). In addition, mild over-expression of SRF repressed 65% (124/192) and activated 35% (68/192) of the SRF-modulated genes (Fig. 2). These data indicate that SRF both represses and activates genes containing CArG and/or CArG-like elements.
Mild overexpression of SRF affected SRF-modulated genes in multiple functional categories
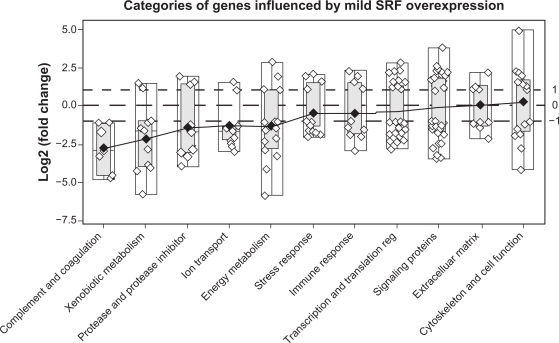
To assess the significance of altered expression of SRF-modulated genes on cardiac function, the 207 SRF-modulated genes were grouped into 12 categories according to their function and Gene Ontology (GO) term. They are energy metabolism, xenobiotic metabolism, cytoskeleton, transcription and translation regulation, extracellular matrix (ECM), stress response, signaling proteins, protease and protease inhibitors, complement and coagulation, ion transport, immune response as well as other proteins (Table 2, Figs. 2, 3 and 4). The 192 genes that contain CArG and/or CArG-like elements were distributed among all of the 12 categories (Table 2). The 56 genes that contain at least one classic CArG element were also found in 11 of the 12 categories (Table 2). Mild over-expression of SRF down-regulated a majority of the genes in 10 categories, but up-regulated most of the genes in the category of cytoskeleton and cellular function (Table S1).
Figure 3.
A box-plot of the magnitude of change in gene expression (log2) in the Mild-SRF transgenic mice distributed among 12 different categories (category of “other proteins” not shown). Each gene is represented by a small circle. White boxes depict the range limits, and the light gray boxes, the inter-quartile range. The diamonds represent the means and are joined by a solid line. The categories are displayed from the most down-regulated (on the left) to the most up-regulated (on the right). Note that in the cytoskeletal category more genes are upregulated than downregulated.
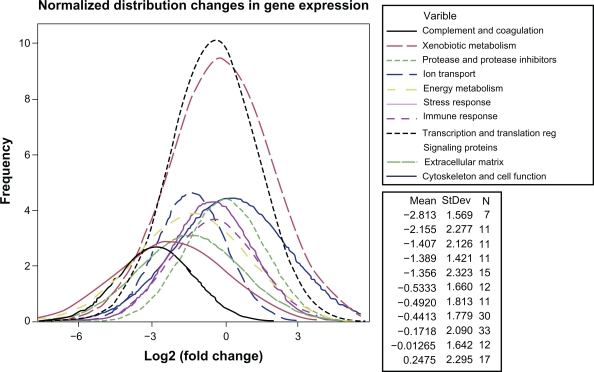
Figure 4.
Normalized distribution curves of SRF-modulated genes in response to mild-SRF overexpression in vivo. The legend at the top right shows the functional categories as variables. The legend at the bottom right, shows the mean fold change in gene expression, the standard deviation (SD) and the number (N) of genes in each category.
It was found that mild overexpression of SRF changed the expression of genes regulating energy metabolism. For example, phosphofructokinase, a key enzyme that controls the pace of glycolysis was elevated over 2-fold. The SRF-modulated genes that regulate fatty acid metabolism were down-regulated. Solute carrier family 27 (slc27a1), which catalyzes the transfer of long-chain fatty acids across the plasma membrane, was down 2-fold.28 Lipase, which catalyzes the rate-limiting step in adipose tissue lipolysis, was down 2-fold.29 Elongase 2 (Elovl2), which performs the first regulatory step (condensation) in the elongation cycle in fatty acid synthesis, was down 5-fold.30 Esterase 1 (Es-1), which hydrolyzes a variety of esters including fatty acid esters of estradiol, was down 17.5-fold. Coenzyme A synthase was decreased 2.2-fold.
Our data also revealed that SRF impacted the genes involved in transcriptional and translational regulation. The genes that were up-regulated include distal-less homeobox 5 (4.8-fold), activating transcription factor 3 (ATF3, up 4.5-fold), TATA box binding protein (TBP, up 3.8-fold), and four and a half lim domains 1 (Fhl1, up 3-fold), Id2, which forms heterodimer with other HLH proteins, was up 2.3-folds. The down-regulated genes include Six3 (down 2-fold), Gtf3c4 (down 2.6-fold), Lhx8 (down 2-fold), Hmx1 (down 2.2-fold), Sp4 (down 2.6-fold), and E2F3 (down 5-fold). The proprotein convertase (PCSK5), which mediates post-translational endoproteolytic processing for several integrin alpha subunits, was down 2.7-fold.
An equal number of genes in the ECM category were up-regulated and down-regulated. Mild SRF overexpression not only changed the expression of cytoskeletal genes but also changed the expression of many genes in other functional categories. For instance, several ECM genes were up-regulated, including type I collagen, fibulin and biglycan. Serpine1/PAI-1, which inhibits the degradation of ECM proteins, was also up 2.9-fold. Changes were also observed in genes that play a regulatory role in cardiac hypertrophy and fibrosis, such as TGF-beta3 (up 3-fold); connective tissue growth factor (CTGF), which promotes fibroblast proliferation and myocyte growth, was increased by 4-fold. GDF15, a member of TGF-beta superfamily and a potential biomarker for cardiac disease, was increased 14-fold.31 Periostin, which regulates collagen I fibrillogenesis, was elevated over 4-fold.32,33 Annexins (ANXs) are a large group of calcium-binding proteins participating in diverse important biological processes.34,35 In the mild-SRF transgenic mouse heart, annexin a10 was increased by more than 30-fold.
Alteration was also observed in the expression of genes involved in proteolysis. Ubiquitin specific peptidase 29 (USP29) was increased 3.7-fold. Dipeptidyl peptidase 7 (DPP7), was increased 2.7-fold. Spink3, a Kazal type 1 serine peptidase inhibitor, was decreased 2.5-fold. Serpina1a, a member of serine/cysteine peptidase inhibitors, was reduced 16-fold. Alpha-2-thiol proteinase inhibitor (Kng1) was decreased 29-fold.
Interestingly, several ion regulation genes were found to be SRF-modulated genes. For instance, several ion transport genes were down-regulated. ATP1a1 was down 3-fold, slc4a8 was down 2.5-fold, TRPM7 was down 3-fold, KCNQ2 was down 3-fold, and Sodium/bile acid co-transporter (slc10a) was down 8-fold. Aqp4, a gene involved in water transport, was down 5-fold. Casq1, a calcium handling protein, was up 2.8-fold. The histidine rich calcium binding protein (HRC), which interacts with SERCA2, was down 2.8-fold.36
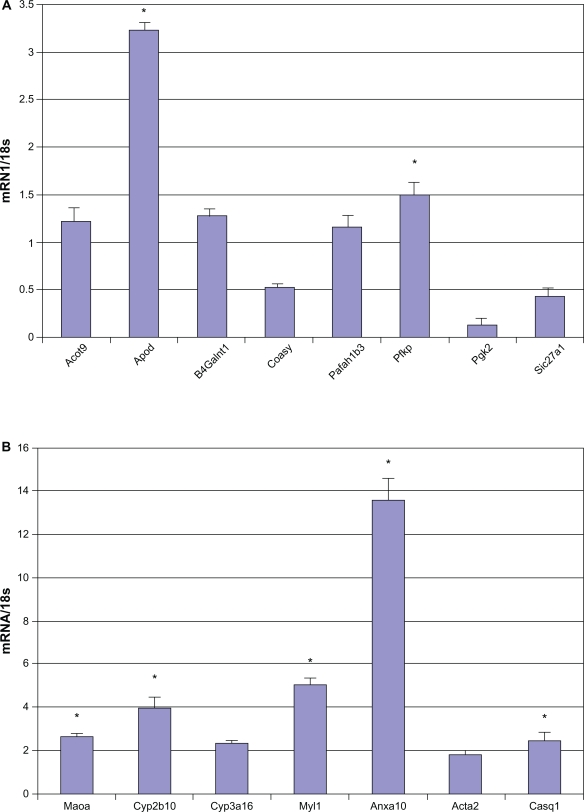
All genes that were differentially expressed ≥2 fold were validated with real-time RT-PCR and representative results are depicted in Figure 5A and 5B. B4galnt1 (beta-1,4-n-acetyl-galactosaminyl transferase 1), important in energy metabolism and the synthesis of gangliosides, was elevated 7 fold in the Mild-O-SRF Tg mouse hearts. Abnormalities in ganglioside metabolism maybe associated with Type I diabetes mellitus.37 Expression of Apod (ApolipoproteinD) a soluble lipid carrier is found in most human tissues, but especially in glia of the nervous system.38–41 Its expression was increased 3.6 fold in the Mild-O-SRF Tg (Fig. 5A). Acot9 was increased 2 fold in Mild-O-SRF Tg. Acyl CoA is important in triglyceride synthesis and its increase can potentially increase cardiovascular risk profile (Fig. 5A).42 CoasyCoA synthase (CoASy) was decreased in the Mild-O-SRF Tg hearts (Fig. 5A). Coasyis a mitochondria-associated enzyme which mediates two final stages of de novo CoA biosynthesis.43 Pafah1b3 (platelet activating factor acetylhydrolase) and Pfkp (phosphofructokinase) which is involved in adiposity were also confirmed with qRT-PCR (Fig. 5A).44,45 Anxa10 was elevated more than 13 fold on q-RT-PCR (Fig. 5B).34,35
Figure 5.
The results of real-time RT-PCR are given as a relative expression of mRNA normalized to the 18S housekeeping gene, fold change in gene expression. n = 5 mice in Mild-O SRF Tg (trangenic mouse with mild over-expression of SRF) and non-transgenic group. Results are provided as means ± SD.
Note: *P < 0.05 Non-Tg, vs. Mild-O SRF Tg using the Mann Whitney U test.
Altered expression of SRF-modulated genes was also observed in other mouse models of stress
SRF is a delayed immediate early response gene.46 To examine whether SRF-modulated genes might be differentially regulated in other mouse models of stress, we obtained cardiac gene expression data from Gene Expression Omnibus (GEO) database. Analysis of 27 microarray data sets representing various mouse models of cardiac stress revealed that many SRF-modulated genes are indeed differentially regulated in response to stress, including the 194 SRF-modulated genes reported in the present study. Further analysis revealed that most of the classic CArG SRF-modulated genes (57 of 65) were also observed to be differentially expressed in these other mouse models (Table 3).
Table 3.
Comparison of expression of the SRF-modulated genes containing classic CArG elements between mild overexpression SRF transgenic mouse and other mouse models. The cardiac gene microarray data of 27 experiments were downloaded from the Gene Expression Omnibus (GEO) database. The expression of SRF-modulated genes was compared among mild-SRF transgenic (Tg) mice and other mouse models in the GEO database that used the same Affymetrix gene chip as in our experiment. The majority of these mouse models used in comparison also had hemodynamic, ischemic or exercise induced stress. The results indicate that 13 genes (in pink) in other mouse models were either up-regulated or down-regulated in the same direction as that in the mild-SRF Tg mouse (labeled as “A” for agreement in the direction of change with the SRF Tg mouse). There were 20 genes (in purple) that showed a mixed pattern of being either “up- or down-regulated” (labeled as “A” for agreement or “D” for disagreement in the direction of change with the SRF Tg mouse) in the various models compared with the SRF Tg mouse. The other 15 genes (in light blue) were allaltered in the opposite direction to that which was observed in the mild-SRF Tg mouse (“D” for disagreement).
| Gene identifier | 6Mo SRF Tg vs. nTg | GDS488 7 day MI | GDS794 21 day TAC | GDS446 dnPI3K | GDS488 1 hr MI | GDS446 caPI3K | GDS144 5 wk exercise | GDS591 Doxy. (-), (tTA)+ | GDS591 Doxy. (+), (tTA)+ | GDS488 4 hr MI | GDS488 24 hr MI | GDS496 3 wk TNFa+ | GDS488 8 wk MI | GDS794 2 day TAC | GDS2258 female TAC | GDS1001 ATP1a1+/– | GDS2258 male TAC | GDS1302 3 ug/kg TCDD | GDS144 10 min exercise | GDS794 10 day TAC | GDS1247 Dysf –/– | GDS648 IGF1R+, caPI3K | GDS1302 6 ug/kg TCDD | GDS1302 1.5 ug/kg TCDD | GDS144 4 wk exercise | GDS2335 diabetic Endu | GD S2335 healthy Endu | GDS648 IGF1R+, dnPI3K | Percent agreement |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cwf19l1* | ↓ | A ↓ | A ↓ | 100 | |||||||||||||||||||||||||
| AA930519 | ↓ | A ↓ | 100 | ||||||||||||||||||||||||||
| Mup1* | ↓ | A ↓ | 100 | ||||||||||||||||||||||||||
| Myh4* | ↓ | A ↓ | A ↓ | A ↓ | A ↓ | A ↓ | 100 | ||||||||||||||||||||||
| Nppa | ↑ | A ↑ | A ↑ | A ↑ | A ↑ | 100 | |||||||||||||||||||||||
| Jag2* | ↓ | A ↓ | 100 | ||||||||||||||||||||||||||
| Fhl1 | ↑ | A ↑ | A ↑ | A ↑ | A ↑ | A ↑ | A ↑ | 100 | |||||||||||||||||||||
| Il15 | ↓ | A ↓ | A ↓ | A ↓ | A ↓ | A ↓ | 100 | ||||||||||||||||||||||
| Serpine1* | ↑ | A ↑ | A ↑ | A ↑ | 100 | ||||||||||||||||||||||||
| Pcsk5* | ↓ | A ↓ | 100 | ||||||||||||||||||||||||||
| Hspa1b* | ↑ | A ↑ | A ↑ | 100 | |||||||||||||||||||||||||
| Cyp2b10* | ↑ | A ↑ | 100 | ||||||||||||||||||||||||||
| Es1* | ↓ | A ↓ | 100 | ||||||||||||||||||||||||||
| Ift81* | ↓ | A ↓ | A ↓ | A ↓ | A ↓ | A ↓ | A ↓ | D ↑ | D ↑ | 75 | |||||||||||||||||||
| Ofa* | ↓ | A ↓ | A ↓ | D ↑ | 67 | ||||||||||||||||||||||||
| Cdca5* | ↓ | A ↓ | D ↑ | 50 | |||||||||||||||||||||||||
| Lancl2* | ↓ | A ↓ | D ↑ | D ↑ | 33 | ||||||||||||||||||||||||
| LOC547428* | ↓ | A ↓ | D ↑ | D ↑ | 33 | ||||||||||||||||||||||||
| R3hcc1 | ↑ | D ↓ | A ↑ | D ↓ | D ↓ | 25 | |||||||||||||||||||||||
| Tpm2 | ↑ | A ↑ | A ↑ | A ↑ | A ↑ | D ↓ | 80 | ||||||||||||||||||||||
| Myh7* | ↑ | A ↑ | A ↑ | A ↑ | D ↓ | 75 | |||||||||||||||||||||||
| Myl1* | ↑ | A ↑ | A ↑ | D ↓ | 67 | ||||||||||||||||||||||||
| Rab8b* | ↑ | A ↑ | D ↓ | D ↓ | 33 | ||||||||||||||||||||||||
| Emp1* | ↑ | A ↑ | A ↑ | A ↑ | A ↑ | A ↑ | D ↓ | D ↓ | 71 | ||||||||||||||||||||
| Emr1* | ↑ | A ↑ | A ↑ | D ↓ | A ↑ | D ↓ | 60 | ||||||||||||||||||||||
| Irf8 | ↑ | A ↑ | A ↑ | D ↓ | D ↓ | A ↑ | 60 | ||||||||||||||||||||||
| Tbp* | ↑ | A ↑ | D ↓ | 50 | |||||||||||||||||||||||||
| Postn # | ↑ | A ↑ | A ↑ | A ↑ | A ↑ | A ↑ | D ↓ | A ↑ | D ↓ | 75 | |||||||||||||||||||
| Il1f5* | ↓ | A ↓ | D ↑ | 50 | |||||||||||||||||||||||||
| Aqp4* | ↓ | A ↓ | D ↑ | A ↓ | A ↓ | 75 | |||||||||||||||||||||||
| Slc20a1* | ↑ | A ↑ | A ↑ | D ↓ | 67 | ||||||||||||||||||||||||
| Hpx* | ↓ | A ↓ | D ↑ | 50 | |||||||||||||||||||||||||
| Ugt2b5* | ↓ | A ↓ | D ↑ | 50 | |||||||||||||||||||||||||
| C77681* | ↓ | D ↑ | D ↑ | 0 | |||||||||||||||||||||||||
| Mup1* | ↓ | D ↑ | 0 | ||||||||||||||||||||||||||
| Anxa10* | ↑ | D ↓ | 0 | ||||||||||||||||||||||||||
| Dsc2* | ↓ | D ↑ | D ↑ | 0 | |||||||||||||||||||||||||
| Epb4.2* | ↑ | D ↓ | 0 | ||||||||||||||||||||||||||
| Crk* | ↓ | D ↑ | D ↑ | 0 | |||||||||||||||||||||||||
| Il4* | ↑ | D ↓ | 0 | ||||||||||||||||||||||||||
| Aqr* | ↑ | D ↓ | 0 | ||||||||||||||||||||||||||
| Brca1* | ↓ | D ↑ | 0 | ||||||||||||||||||||||||||
| E2f3* | ↓ | D ↑ | D ↑ | 0 | |||||||||||||||||||||||||
| Slc4a8* | ↓ | D ↑ | 0 | ||||||||||||||||||||||||||
| Serpina1d* | ↓ | D ↑ | D ↑ | 0 | |||||||||||||||||||||||||
| Cyp3a16* | ↓ | D ↑ | 0 | ||||||||||||||||||||||||||
| Akr1c6* | ↓ | D ↑ | 0 | ||||||||||||||||||||||||||
| Elovl2* | ↓ | D ↑ | D ↑ | 0 | |||||||||||||||||||||||||
| 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 83 | 80 | 76 | 75 | 75 | 67 | 57 | 50 | 50 | 50 | 45 | 45 | 43 | 33 | 0 | 0 | 0 | 0 | 0 |
Note: Columns from left to right: The 1st column shows the gene symbol. The 2nd column (in yellow) shows the direction of alteration of genes in the mild over-expression SRF Tg hearts. Columns 3–29 are a list of mouse models in the GEO database used for comparison. Experimental systems (columns) and genes (rows) were sorted so that those with the highest percentage agreement with the SRF Tg mouse in differentially expressed genes were toward the upper-left corner of the table (Table 3). The last column of the right and the last row at the bottom provides the percentage agreement of differentially expressed genes in the various models with the SRF Tg model.
Of those SRF-modulated genes that were differentially expressed in both the myocardial infarction (MI) model (GEO number: GDS488) and in the mild SRF transgenic model, the direction of change of these genes was similar at time points from 1 hr to 8 wks after MI (Table 3). At 1 hour after myocardial infarction, the expression of periostin increased and expression of LOC547428 (predicted to be similar to T-cell receptor alpha chain) decreased, both in agreement with the changes seen in SRF-transgenic mice. At four hours, the expression of four and a half LIM domains 1 (Fhl1), serine (or cysteine) peptidase inhibitor clade E member 1 (Serpine1), heat shock protein 1B (Hspa1b), epithelial membrane protein 1 (Emp1), and solute carrier family 20 member 1 (Slc20a1) were increased, as they were also in SRF-transgenic mice. At one day post MI, natriuretic peptide precursor type A (Nppa) and Emp1 expression were increased as they were in SRF-transgenic. Expression of esterase 1 (Es1), intraflagellar transport 81 homolog (Ift81) were similarly decreased between MI and SRF-Tg models. At seven days after MI, expression levels of Nppa, Fhl1, Serpine1, Emp1, interferon regulatory factor 8 (Irf8), and periostin were increased, while those of interleukin 15 (Il-15) and Ift81 were decreased; all of these changes were in the same direction as those in the SRF-Tg model. At eight weeks after MI, the expression levels of myosin heavy polypeptide 4 (Myh4) and UDP glucuronosyltransferase 2 family polypeptide B5 (UGT2B5) were both down, and that of Emr1 was up, all of which were in agreement with those in the SRF-Tg model. 47 Thus, the SRF transgenic model shared certain similarities in ischemic stress signaling with that of the MI model. In the transverse aortic constriction (TAC) model (GEO number: GDS794), there were also interesting similarities.48 After two days of TAC, expression levels of Fhl1, RAB8B (a member of the RAS oncogene family) were increased, and that of Il-15 was decreased, all in the same direction as in the SRF-Tg model (Table 3). After ten days of TAC, jagged 2 (Jag2), Il-15, Ift81, oncofetal antigen (Ofa), and aquaporin 4 (Aqp4) were down-regulated in the same direction as in the SRF-Tg model. After twenty-one days of TAC, the expression levels of Fhl1, Serpine1, Myh7, Emp1, and Periostin were up, and that of Ift81 was down, all of which were in agreement with the SRF-Tg model.48 Thus, the SRF transgenic model also shared certain similarities with the TAC model. These data suggest that common signaling pathways mediating stress response may be involved in the regulation of SRF-modulated genes (Table 3).
Discussion
This study has several major findings. A set of 207 SRF-modulated genes and their in vivo response to SRF regulation in the heart has been identified. Among them, 192 genes had CArG and/or CArG-like elements in their promoter regions. The 179 genes (51 of the 56 genes with classic CArG elements, and 123 of the 136 genes with CArG-like elements) were not previously reported to have SRF binding sites.
The 207 SRF-modulated genes could be assigned to 12 categories based on their function and Gene Ontology term, suggesting that SRF regulates multiple cellular functions via its target genes. Overexpression of SRF repressed 65% of the SRF-modulated genes. The gene profile revealed that in mild-SRF transgenic hearts, genes associated with cardiac energy metabolism shifted toward that of carbohydrate metabolism and away from that of fatty acid metabolism. It also revealed decreased expression of many genes that regulate transcriptional activity, stress response, protein turnover and ion regulation. However, the expression of cytoskeletal genes was considerably increased. Overall, the changes in cardiac gene expression are similar to those that are observed during adult aging. These findings support the notion that an elevation of SRF protein level in typical adult aging may contribute to the altered cardiac structure and function observed during senescence.6
It is likely that SRF overexpression contributed to the altered energy metabolism in the mild-SRF transgenic heart. In the heart, energy usually comes from beta-oxidation of fatty acids and glycolysis, the proportion of which changes during different stages of life. During fetal life, myocardial ATP is derived predominantly from glycolysis and lactate oxidation. After birth, a rapid increase in fatty acid oxidation occurs along with a decline in glycolytic and lactate oxidative rates.49 In the healthy adult heart, about 60%–90% of the ATP generation in the mitochondria comes from beta-oxidation of fatty acids, and the rest comes from pyruvate that is derived from glycolysis and lactate.50 A decline in fatty acid oxidation together with an increase in carbohydrate metabolism has been observed in the senescent heart.51–53 The changes in gene expression of the aged heart include an up-regulation of phosphofructokinase, an allosteric enzyme that controls the rate of glycolysis by converting fructose 6-phosphate to fructose 1,6-bisphosphate, and down-regulation of solute carrier family 27 (slc27a1), which catalyzes the transfer of long-chain fatty acids across the plasma membrane.28,51 In the mild-SRF transgenic heart, the up-regulation of phosphofructokinase together with down-regulation of several proteins, including solute carrier family 27 (slc27a1),28 carnitine acetyltransferase,54 long-chain acyl-CoA synthetase 1 (ASCL1),55 lipase, and elongase 2 (Elovl2),30 indicate that a reduction in fatty acid metabolism also occurred in the present SRF model. This shift away from fatty acid toward carbohydrate metabolism also occurs at certain disease stages and/or in some experimental models of diabetes and heart failure.56,57 SRF has also been reported to mediate glucose response via their binding sites in the human nuclear receptor LXRbeta gene.58
Aging is associated with altered transcriptional and translational regulation.3,53,59 During the typical aging process, most components of the cardiovascular system undergo gradual change, including a progressive loss of myocytes with subsequent hypertrophy of the remaining viable myocytes. The net result is a change in the ratio of myocytes to fibroblasts. As myocytes are lost and fibroblasts continue to divide and produce collagen, the physical properties of the aging heart become altered.3 From the point of view of molecular biology, a “selective” decline in gene expression is a common feature of aging in various tissues across species.59 A number of studies have demonstrated that during adult aging, a majority of the genes are downregulated in various tissues, including heart, kidney, prostate, oocytes and monocytes.52,53,60,61,63–65 However, expression of a minority of the genes is actually increased with age. For instance, a number of cytoskeletal and ECM proteins are usually increased in both the old human and rodent hearts.53 The altered gene expression observed during aging is dynamic and well-regulated.59 SRF overexpression in the heart also changed the expression of a group of genes involved in transcriptional and translational regulation. Increased expression of four and a half lim domains 1 (FHL1) indicates the presence of cardiac stress as seen in other mouse model.66 Altered expression of TATA box binding protein and the subunit of general transcription factor are likely to affect the transcriptional initiation and efficiency, while altered expression of the subunit 3 of eukaryotic translation initiation factor 2 is likely to affect the translational process. Altered expression of ribosomal proteins S4, S12, S17, and L34, are likely to affect mRNA splicing, ribosome assembly, translational fidelity and protein synthesis.67,68 Altered expression of ribonucleotide reductase m1 and thymidine kinase 1 could affect DNA synthesis and repair, nucleotide metabolism as well as cell-cycle progression.69
Comparing our data from mild-SRF transgenic mice with those in aged mice in the literature, it is found that they share similarities in terms of cardiac gene expression.51,53 Similar to that in aged mice, the majority of cardiac genes in most of the functional categories were decreased in the transgenic mice, whereas cytoskeletal genes were increased in the transgenic mice. Some well studied ECM proteins, including collagen I, were also significantly increased in the mild-SRF transgenic mouse model as well as in old age. Genes such as Apod, increased in our transgenic mouse, have also been shown to be increased in Alzheimer’s disease, Parkinson’s, stroke and schizophrenia. In Alzhemer’s disease, it can be found in amyloid plaques within the brains of patients.38–41 It is possible that an increase in Apod is a compensatory mechanism in degenerative diseases as well as aging as it is involved in membrane lipids and degradation pathways.38–41
The most interesting comparison was between our SRF Tg model with mild over-expression of SRF and the dominant negative PI3kinase (dnPI3K) model (Table 3, second to the last column on the right). The hearts of the dnPI3K model had a 100% disagreement in the direction of gene change when compared with the differentially expressed genes in hearts of the SRF Tg model (Table 3, last column and bottom row). This is notable because the dnPI3K mouse model has been reported by various investigators as being anti-hypertrophic and even anti-cardiac aging,70,71 whereas the SRF Tg model with mild over-expression of SRF is an model of cardiac aging.6 Hence, the data in Table 3, especially as it pertains to PI3K, further validates our model of cardiac aging with mild over-expression of SRF in the heart. Although suppression of PI3K has been reported to prevent cardiac aging, a recent study suggests that dominant negative PI3K mice are more susceptible to congestive heart failure after myocardial infarction.72 However, variation in these studies could be explained by differences in the temporal expression of the transgene during development and adulthood, compensatory gene responses as well as strain differences in mice.73
The present in vivo mouse model revealed many previously unreported SRF-modulated genes. The effect of SRF on gene expression has been studied both in vitro and in vivo. The in vitro transfection assay using cell lines has been frequently utilized to study the effect of SRF on the expression of a number of SRF-modulated genes.74,75 Since many cell lines that are used in the assay may not have the same SRF cofactors as that in cardiac myocytes and fibroblasts, the experimental data in cell lines may differ from that in the intact heart. The cardiac-specific mild-SRF transgenic mouse provides us with an excellent tool to identify new SRF-modulated genes and to study the response of these novel genes to cardiac SRF overexpression. Interestingly, most of the cardiac genes that responded significantly to SRF overexpression contained CArG and/or CArG-like elements in their promoters. The down-regulation of a majority of SRF-modulated genes and up-regulation of a minority of SRF-modulated genes in the present study support the notion that SRF-dependent gene regulation is complex. SRF-modulated genes are regulated by multiple transcription regulators including SRF, SRF cofactors, SRF isoforms and a number of microRNAs.5,11,75–79 Other mechanisms of gene regulation include nonsense-mediated mRNA decay.80
Taken together, our findings demonstrate that a mild elevation of the SRF protein level that is observed in the rodent heart during typical adult aging may have a major impact on many cardiac genes, thereby affecting multiple aspects of cardiac structure, metabolism and performance in old age.
Supplementary Material
Acknowledgments
This study was supported in part by NIH grants AG-018388, and AG-026091 from the Department of Health and Human Services, The Geriatric Research, Education, and Clinical Center (GRECC), and the Central Arkansas Veterans Healthcare System. We thank Sophie Wang and Drs. Joel Lawitts, Marjorie Beggs and Richard Dennis for their helpful input, and John Williams for expert assistance with manuscript preparation.
Footnotes
Authors’ Contributions
XMZ participated in the design of the study, carried out part of the experiments and drafted the manuscript. GA participated in the coordination of the study and the writing of the manuscript. SH participated in the bioinformatics analysis and drawing the figures.
BB and CH participated in the coordination of the study. YZ participated in the most of the experiments. XG participated in the Chip hybridization. HF and WT participated in the microarray data analysis. JYW led the study and participated in the design of the study, the statistical analysis and overall interpretation of results. All authors have read and approved the final manuscript.
Disclosure
This manuscript has been read and approved by all authors. The views presented in this article do not necessarily reflect those of the US Food and Drug Administration. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers of this paper report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Pugh KG, Wei JY. Clinical implications of physiological changes in the aging heart. Drugs Aging. 2001;18:263–76. doi: 10.2165/00002512-200118040-00004. [DOI] [PubMed] [Google Scholar]
- 2.Wei JY. Age and the cardiovascular system. N Engl J Med. 1992;327:1735–9. doi: 10.1056/NEJM199212103272408. [DOI] [PubMed] [Google Scholar]
- 3.Wei JY. Understanding the aging cardiovascular system. Geriatrics and Gerontol International. 2004;4:S298–303. [Google Scholar]
- 4.Lu XG, Azhar G, Liu L, Tsou H, Wei JY. SRF binding to SRE in the rat heart: influence of age. J Gerontol A Biol Sci Med Sci. 1998;53:B3–10. doi: 10.1093/gerona/53a.1.b3. [DOI] [PubMed] [Google Scholar]
- 5.Wang D, Passier R, Liu ZP, et al. Regulation of cardiac growth and development by SRF and its cofactors. Cold Spring Harb Symp Quant Biol. 2002;67:97–105. doi: 10.1101/sqb.2002.67.97. [DOI] [PubMed] [Google Scholar]
- 6.Zhang X, Azhar G, Furr MC, Zhong Y, Wei JY. Model of functional cardiac aging: young adult mice with mild overexpression of serum response factor. Am J Physiol Regul Integr Comp Physiol. 2003;285:R552–60. doi: 10.1152/ajpregu.00631.2002. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Chai J, Azhar G, et al. Early postnatal cardiac changes and premature death in transgenic mice overexpressing a mutant form of serum response factor. J Biol Chem. 2001;276:40033–40. doi: 10.1074/jbc.M104934200. [DOI] [PubMed] [Google Scholar]
- 8.Miano JM. Serum response factor: toggling between disparate programs of gene expression. J Mol Cell Cardiol. 2003;35:577–93. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- 9.Treisman R, Ammerer G. The SRF and MCM1 transcription factors. Curr Opin Genet Dev. 1992;2:221–6. doi: 10.1016/s0959-437x(05)80277-1. [DOI] [PubMed] [Google Scholar]
- 10.Poser S, Impey S, Trinh K, Xia Z, Storm DR. SRF-dependent gene expression is required for PI3-kinase-regulated cell proliferation. EMBO J. 2000;19:4955–66. doi: 10.1093/emboj/19.18.4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schratt G, Philippar U, Hockemeyer D, Schwarz H, Alberti S, Nordheim A. SRF regulates Bcl-2 expression and promotes cell survival during murine embryonic development. EMBO J. 2004;23:1834–44. doi: 10.1038/sj.emboj.7600188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinhold B, Schratt G, Arsenian S, et al. Srf(−/−) ES cells display non-cell-autonomous impairment in mesodermal differentiation. EMBO J. 2000;19:5835–44. doi: 10.1093/emboj/19.21.5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boxer LM, Prywes R, Roeder RG, Kedes L. The sarcomeric actin CArG-binding factor is indistinguishable from the c-fos serum response factor. Mol Cell Biol. 1989;9:515–22. doi: 10.1128/mcb.9.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hautmann MB, Madsen CS, Mack CP, Owens GK. Substitution of the degenerate smooth muscle (SM) alpha-actin CC(A/T-rich)6GG elements with c-fos serum response elements results in increased basal expression but relaxed SM cell specificity and reduced angiotensin II inducibility. J Biol Chem. 1998;273:8398–406. doi: 10.1074/jbc.273.14.8398. [DOI] [PubMed] [Google Scholar]
- 15.Taylor M, Treisman R, Garrett N, Mohun T. Muscle-specific (CArG) and serum-responsive (SRE) promoter elements are functionally interchangeable in Xenopus embryos and mouse fibroblasts. Development. 1989;106:67–78. doi: 10.1242/dev.106.1.67. [DOI] [PubMed] [Google Scholar]
- 16.Treisman R. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell. 1986;46:567–74. doi: 10.1016/0092-8674(86)90882-2. [DOI] [PubMed] [Google Scholar]
- 17.Zilberman A, Dave V, Miano J, Olson EN, Periasamy M. Evolutionarily conserved promoter region containing CArG*-like elements is crucial for smooth muscle myosin heavy chain gene expression. Circ Res. 1998;82:566–75. doi: 10.1161/01.res.82.5.566. [DOI] [PubMed] [Google Scholar]
- 18.Miano JM, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am J Physiol Cell Physiol. 2007;292:C70–81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- 19.Grandi AM, Venco A, Barzizza F, Scalise F, Pantaleo P, Finardi G. Influence of age and sex on left ventricular anatomy and function in normals. Cardiology. 1992;81:8–13. doi: 10.1159/000175770. [DOI] [PubMed] [Google Scholar]
- 20.Lakatta EG. Cardiovascular aging in health. Clin Geriatr Med. 2000;16:419–44. doi: 10.1016/s0749-0690(05)70021-5. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Azhar G, Chai J, et al. Wei, Cardiomyopathy in transgenic mice with cardiac-specific overexpression of serum response factor. Am J Physiol Heart Circ Physiol. 2001;280:H1782–92. doi: 10.1152/ajpheart.2001.280.4.H1782. [DOI] [PubMed] [Google Scholar]
- 22.Jones J, Otu H, Spentzos D, et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res. 2005;11:5730–9. doi: 10.1158/1078-0432.CCR-04-2225. [DOI] [PubMed] [Google Scholar]
- 23.Tong W, Cao X, Harris S, et al. ArrayTrack--supporting toxicogenomic research at the US. Food and Drug Administration National Center for Toxicological Research. Environ Health Perspect. 2003;111:1819–26. doi: 10.1289/ehp.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun H, Fang H, Chen T, Perkins R, Tong W. GOFFA: Gene Ontology For Functional Analysis—A FDA Gene Ontology Tool for Analysis of Genomic and Proteomic Data. BMC Bioinformatics. 2006;7(Suppl 2):S23. doi: 10.1186/1471-2105-7-S2-S23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang XQ, Miller W. A time-efficient, linear-space local similarity algorithm. Adv Appl Math. 1991;12:337–57. [Google Scholar]
- 27.Schug J, Overton GC. TESS: transcription element search system on the WWW Computational Biology and Informatics Laboratory, School of Medicine, University of Pennsylvania. 1997.
- 28.Schaffer JE, Lodish HF. Expression cloning and characterization of a novel adipocyte long chain fatty acid transport protein. Cell. 1994;79:427–36. doi: 10.1016/0092-8674(94)90252-6. [DOI] [PubMed] [Google Scholar]
- 29.Li Z, Sumida M, Birchbauer A, Schotz MC, Reue K. Isolation and characterization of the gene for mouse hormone-sensitive lipase. Genomics. 1994;24:259–65. doi: 10.1006/geno.1994.1614. [DOI] [PubMed] [Google Scholar]
- 30.Jakobsson A, Westerberg R, Jacobsson A. Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog Lipid Res. 2006;45:237–49. doi: 10.1016/j.plipres.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Kempf T, Horn-Wichmann R, Brabant G, et al. Circulating concentrations of growth-differentiation factor 15 in apparently healthy elderly individuals and patients with chronic heart failure as assessed by a new immunoradiometric sandwich assay. Clin Chem. 2007;53:284–91. doi: 10.1373/clinchem.2006.076828. [DOI] [PubMed] [Google Scholar]
- 32.Norris RA, Damon B, Mironov V, et al. Periostin regulates collagen fibrillogenesis and the biomechanical properties of connective tissues. J Cell Biochem. 2007;101:695–711. doi: 10.1002/jcb.21224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oka T, Xu J, Kaiser RA, et al. Genetic Manipulation of Periostin Expression Reveals a Role in Cardiac Hypertrophy and Ventricular Remodeling. Circ Res. 2007 doi: 10.1161/CIRCRESAHA.107.149047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Camors E, Monceau V, Charlemagne D. Annexins and Ca2+ handling in the heart. Cardiovasc Res. 2005;65:793–802. doi: 10.1016/j.cardiores.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 35.Liu SH, Lin CY, Peng SY, et al. Down-regulation of annexin A10 in hepatocellular carcinoma is associated with vascular invasion, early recurrence, and poor prognosis in synergy with p53 mutation. Am J Pathol. 2002;160:1831–7. doi: 10.1016/S0002-9440(10)61129-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arvanitis DA, Vafiadaki E, Fan GC, et al. The Histidine-Rich Calcium Binding Protein Interacts With Sarcoplasmic Reticulum Ca-ATPase. Am J Physiol Heart Circ Physiol. 2007 doi: 10.1152/ajpheart.00278.2007. [DOI] [PubMed] [Google Scholar]
- 37.Boraska V, Torlak V, Skrabić V, et al. Glycosyltransferase B4GALNT1 and type 1 diabetes in Croatian population: clinical investigation. Clin Biochem. 2009 Jun;42(9):819–22. doi: 10.1016/j.clinbiochem.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 38.Terrisse L, et al. Increased levels of apolipoprotein D in cerebrospinal fluid and hippocampus of Alzheimer’s patients. J Neurochem. 1998;71:1643–50. doi: 10.1046/j.1471-4159.1998.71041643.x. [DOI] [PubMed] [Google Scholar]
- 39.Navarro A, Del Valle E, Astudillo A, Gonzalez del Rey C, Tolivia J. Immunohistochemical study of distribution of apolipoproteins E and D in human cerebral beta amyloid deposits. Exp Neurol. 2003;184:697–704. doi: 10.1016/S0014-4886(03)00315-7. [DOI] [PubMed] [Google Scholar]
- 40.Kalman J, McConathy W, Araoz C, Kasa P, Lacko AG. Apolipoprotein D in the aging brain and in Alzheimer’s dementia. Neurol Res. 2000;22:330–6. doi: 10.1080/01616412.2000.11740678. [DOI] [PubMed] [Google Scholar]
- 41.Morais Cabral JH, et al. Arachidonic acid binds to apolipoprotein D: implications for the protein’s function. FEBS Lett. 1995;366:53–6. doi: 10.1016/0014-5793(95)00484-q. [DOI] [PubMed] [Google Scholar]
- 42.Breus O, Panasyuk G, Gout IT, Filonenko V, Nemazanyy I. CoA synthase is in complex with p85alphaPI3K and affects PI3K signaling pathway. Biochem Biophys Res Commun. 2009 Aug 7;385(4):581–5. doi: 10.1016/j.bbrc.2009.05.102. Epub 2009 May 29. [DOI] [PubMed] [Google Scholar]
- 43.Assadi AH, Zhang G, McNeil R, Clark GD, D’Arcangelo G. Pafah1b2 mutations suppress the development of hydrocephalus in compound Pafah1b1; Reln and Pafah1b1; Dab1 mutant mice. Neurosci Lett. 2008 Jul 4;439(1):100–5. doi: 10.1016/j.neulet.2008.04.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scott BT, Olson N, Long GL, Bovill EG. Novel isoforms of intracellular platelet activating factor acetylhydrolase (PAFAH1b2) in human testis; encoded by alternatively spliced mRNAs. Prostaglandins Other Lipid Mediat. 2008 Mar;85(3–4):69–80. doi: 10.1016/j.prostaglandins.2007.10.005. Epub 2007 Nov 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malzahn D, Balavarca Y, Lozano JP, Bickeböller H. Tests for candidate-gene interaction for longitudinal quantitative traits measured in a large cohort. BMC Proc. 2009 Dec 15;3(Suppl 7):S80. doi: 10.1186/1753-6561-3-s7-s80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsou H, Azhar G, Lu XG, Kovacs S, Peacocke M, Wei JY. Age-associated changes in basal c-fos transcription factor binding activity in rat hearts. Exp Cell Res. 1996;229:432–7. doi: 10.1006/excr.1996.0388. [DOI] [PubMed] [Google Scholar]
- 47.Tarnavski O, McMullen JR, Schinke M, Nie Q, Kong S, Izumo S. Mouse cardiac surgery: comprehensive techniques for the generation of mouse models of human diseases and their application for genomic studies. Physiol Genomics. 2004;16:349–60. doi: 10.1152/physiolgenomics.00041.2003. [DOI] [PubMed] [Google Scholar]
- 48.Zhao M, Chow A, Powers J, Fajardo G, Bernstein D. Microarray analysis of gene expression after transverse aortic constriction in mice. Physiol Genomics. 2004;19:93–105. doi: 10.1152/physiolgenomics.00040.2004. [DOI] [PubMed] [Google Scholar]
- 49.Makinde AO, Kantor PF, Lopaschuk GD. Maturation of fatty acid and carbohydrate metabolism in the newborn heart. Mol Cell Biochem. 1998;188:49–56. [PubMed] [Google Scholar]
- 50.Stanley WC, Chandler MP. Energy metabolism in the normal and failing heart: potential for therapeutic interventions. Heart Fail Rev. 2002;7:115–30. doi: 10.1023/a:1015320423577. [DOI] [PubMed] [Google Scholar]
- 51.Lee CK, Allison DB, Brand J, Weindruch R, Prolla TA. Transcriptional profiles associated with aging and middle age-onset caloric restriction in mouse hearts. Proc Natl Acad Sci U S A. 2002;99:14988–93. doi: 10.1073/pnas.232308999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sample J, Cleland JG, Seymour AM. Metabolic remodeling in the aging heart. J Mol Cell Cardiol. 2006;40:56–63. doi: 10.1016/j.yjmcc.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 53.Volkova M, Garg R, Dick S, Boheler KR. Aging-associated changes in cardiac gene expression. Cardiovasc Res. 2005;66:194–204. doi: 10.1016/j.cardiores.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 54.Jogl G, Tong L. Crystal structure of carnitine acetyltransferase and implications for the catalytic mechanism and fatty acid transport. Cell. 2003;112:113–22. doi: 10.1016/s0092-8674(02)01228-x. [DOI] [PubMed] [Google Scholar]
- 55.Durgan DJ, Smith JK, Hotze MA, et al. Distinct transcriptional regulation of long-chain acyl-CoA synthetase isoforms and cytosolic thioesterase 1 in the rodent heart by fatty acids and insulin. Am J Physiol Heart Circ Physiol. 2006;290:H2480–97. doi: 10.1152/ajpheart.01344.2005. [DOI] [PubMed] [Google Scholar]
- 56.Tuunanen H, Ukkonen H, Knuuti J. Myocardial fatty acid metabolism and cardiac performance in heart failure. Curr Cardiol Rep. 2008;10:142–8. doi: 10.1007/s11886-008-0024-2. [DOI] [PubMed] [Google Scholar]
- 57.van der Vusse GJ, van Bilsen M, Glatz JF. Cardiac fatty acid uptake and transport in health and disease. Cardiovasc Res. 2000;45:279–93. doi: 10.1016/s0008-6363(99)00263-1. [DOI] [PubMed] [Google Scholar]
- 58.Nilsson M, Dahlman-Wright K, Karelmo C, Ebeling M, Gustafsson JA, Steffensen KR. Elk1 and SRF transcription factors convey basal transcription and mediate glucose response via their binding sites in the human LXRB gene promoter. Nucleic Acids Res. 2007;35:4858–68. doi: 10.1093/nar/gkm492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Helfand SL, Inouye SK. Rejuvenating views of the ageing process. Nat Rev Genet. 2002;3:149–53. doi: 10.1038/nrg726. [DOI] [PubMed] [Google Scholar]
- 60.Ashton KJ, Willems L, Holmgren K, Ferreira L, Headrick JP. Age-associated shifts in cardiac gene transcription and transcriptional responses to ischemic stress. Exp Gerontol. 2006;41:189–204. doi: 10.1016/j.exger.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 61.Hamatani T, Falco G, Carter MG, et al. Age-associated alteration of gene expression patterns in mouse oocytes. Hum Mol Genet. 2004;13:2263–78. doi: 10.1093/hmg/ddh241. [DOI] [PubMed] [Google Scholar]
- 62.Lau KM, Tam NN, Thompson C, Cheng RY, Leung YK, Ho SM. Age-associated changes in histology and gene-expression profile in the rat ventral prostate. Lab Invest. 2003;83:743–57. doi: 10.1097/01.lab.0000069519.06988.24. [DOI] [PubMed] [Google Scholar]
- 63.Maes OC, Xu S, Yu B, Chertkow HM, Wang E, Schipper HM. Transcriptional profiling of Alzheimer blood mononuclear cells by microarray. Neurobiol Aging. 2007;28:1795–809. doi: 10.1016/j.neurobiolaging.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 64.Melk A, Mansfield ES, Hsieh SC, et al. Transcriptional analysis of the molecular basis of human kidney aging using cDNA microarray profiling. Kidney Int. 2005;68:2667–79. doi: 10.1111/j.1523-1755.2005.00738.x. [DOI] [PubMed] [Google Scholar]
- 65.Rysa J, Leskinen H, Ilves M, Ruskoaho H. Distinct upregulation of extracellular matrix genes in transition from hypertrophy to hypertensive heart failure. Hypertension. 2005;45:927–33. doi: 10.1161/01.HYP.0000161873.27088.4c. [DOI] [PubMed] [Google Scholar]
- 66.Sheikh F, Raskin A, Chu PH, et al. An FHL1-containing complex within the cardiomyocyte sarcomere mediates hypertrophic biomechanical stress responses in mice. J Clin Invest. 2008;118:3870–80. doi: 10.1172/JCI34472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Herr AJ, Atkins JF, Gesteland RF. Coupling of open reading frames by translational bypassing. Annu Rev Biochem. 2000;69:343–72. doi: 10.1146/annurev.biochem.69.1.343. [DOI] [PubMed] [Google Scholar]
- 68.Winkelmann DA, Kahan L, Lake JA. Ribosomal protein S4 is an internal protein: Localization by immunoelectron microscopy on protein-deficient subribosomal particles. Proc Natl Acad Sci U S A. 1982;79:5184–8. doi: 10.1073/pnas.79.17.5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Welin M, Kosinska U, Mikkelsen NE, et al. Structures of thymidine kinase 1 of human and mycoplasmic origin. Proc Natl Acad Sci U S A. 2004;101:17970–5. doi: 10.1073/pnas.0406332102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McMullen JR, Shioi T, Huang WY, et al. The insulin-like growth factor 1 receptor induces physiological heart growth via the phosphoinositide 3-kinase(p110alpha) pathway. J Biol Chem. 2004 Feb 6;279(6):4782–93. doi: 10.1074/jbc.M310405200. [DOI] [PubMed] [Google Scholar]
- 71.Inuzuka Y, Okuda J, Kawashima T, et al. Suppression of phosphoinositide 3-kinase prevents cardiac aging in mice. Circulation. 2009 Oct 27;120(17):1695–703. doi: 10.1161/CIRCULATIONAHA.109.871137. [DOI] [PubMed] [Google Scholar]
- 72.Lin RC, Weeks KL, Gao XM, et al. PI3K(p110 alpha) protects against myocardial infarction-induced heart failure: identification of PI3K-regulated miRNA and mRNA. Arterioscler Thromb Vasc Biol. 2010 Apr;30(4):724–32. doi: 10.1161/ATVBAHA.109.201988. [DOI] [PubMed] [Google Scholar]
- 73.Lu Z, Jiang YP, Wang W, et al. Loss of cardiac phosphoinositide 3-kinase p110 alpha results in contractile dysfunction. Circulation. 2009 Jul 28;120(4):318–25. doi: 10.1161/CIRCULATIONAHA.109.873380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang D, Chang PS, Wang Z, et al. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–62. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 75.Zhang SX, Garcia-Gras E, Wycuff DR, et al. Identification of direct serum-response factor gene targets during Me2SO-induced P19 cardiac cell differentiation. J Biol Chem. 2005;280:19115–26. doi: 10.1074/jbc.M413793200. [DOI] [PubMed] [Google Scholar]
- 76.Chen JF, Mandel EM, Thomson JM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–33. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kemp PR, Metcalfe JC. Four isoforms of serum response factor that increase or inhibit smooth-muscle-specific promoter activity. Biochem J. 2000;345(Pt 3):445–51. [PMC free article] [PubMed] [Google Scholar]
- 78.Xiao J, Luo X, Lin H, et al. MicroRNA miR-133 represses HERG K+ channel expression contributing to QT prolongation in diabetic hearts. J Biol Chem. 2007;282:12363–7. doi: 10.1074/jbc.C700015200. [DOI] [PubMed] [Google Scholar]
- 79.Zhang X, Azhar G, Zhong Y, Wei JY. Identification of a novel serum response factor cofactor in cardiac gene regulation. J Biol Chem. 2004;279:55626–32. doi: 10.1074/jbc.M405945200. [DOI] [PubMed] [Google Scholar]
- 80.Zhang X, Azhar G, Huang C, Cui C, Zhong Y, Huck S, Wei JY. Alternative splicing and nonsense-mediated mRNA decay regulate gene expression of serum response factor. Gene. 2007;400:131–9. doi: 10.1016/j.gene.2007.06.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.