Abstract
Copper isotopes may prove to be a useful tool for investigating bacteria–metal interactions recorded in natural waters, soils, and rocks. However, experimental data which attempt to constrain Cu isotope fractionation in biologic systems are limited and unclear. In this study, we utilized Cu isotopes (δ65Cu) to investigate Cu–bacteria interactions, including surface adsorption and intracellular incorporation. Experiments were conducted with individual representative species of Gram-positive (Bacillus subtilis) and Gram-negative (Escherichia coli) bacteria, as well as with wild-type consortia of microorganisms from several natural environments. Ph-dependent adsorption experiments were conducted with live and dead cells over the pH range 2.5–6. Surface adsorption experiments of Cu onto live bacterial cells resulted in apparent separation factors (Δ65Cusolution–solid = δ65Cusolution – δ65Cusolid) ranging from +0.3‰ to +1.4‰ for B. subtilis and +0.2‰ to +2.6‰ for E. coli. However, because heat-killed bacterial cells did not exhibit this behavior, the preference of the lighter Cu isotope by the cells is probably not related to reversible surface adsorption, but instead is a metabolically-driven phenomenon. Adsorption experiments with heat-killed cells yielded apparent separation factors ranging from +0.3‰ to –0.69‰ which likely reflects fractionation from complexation with organic acid surface functional group sites. For intracellular incorporation experiments the lab strains and natural consortia preferentially incorporated the lighter Cu isotope with an apparent Δ65Cusolution–solid ranging from ~+1.0‰ to +4.4‰. Our results indicate that live bacterial cells preferentially sequester the lighter Cu isotope regardless of the experimental conditions. The fractionation mechanisms involved are likely related to active cellular transport and regulation, including the reduction of Cu(II) to Cu(I). Because similar intracellular Cu machinery is shared by fungi, plants, and higher organisms, the influence of biological processes on the δ65Cu of natural waters and soils is probably considerable.
1. INTRODUCTION
The stable isotopes of copper (Cu) can be used to investigate linkages between the geosphere and biosphere. Copper isotopes are substantially fractionated during abiotic chemical reactions such as surface adsorption (Balistrieri et al., 2008; Pokrovsky et al., 2008), aqueous complexation (e.g., Zhu et al., 2002; Vance et al., 2008; Bigalke et al., 2010), and oxidation or reduction (e.g., Zhu et al., 2002; Ehrlich et al., 2004). The latter mechanisms are reflected in the isotopic variation reported among Cu°-, Cu(I)- and Cu(II)-bearing minerals in nature (e.g., Larson et al., 2003; Markl et al., 2006; Asael et al., 2007; Mathur et al., 2009). Vance et al. (2008) also reported natural variations in δ65Cu for rivers, estuaries, and sea water. Copper present as dissolved organic-ligand complexes in rivers and estuaries was isotopically heavier than Cu associated with the particulate load (Vance et al., 2008).
Reactions with biological components also fractionate Cu isotopes; however, the magnitudes and directions of biologic fractionations and the processes that underpin them remain unclear. For example, Pokrovsky et al. (2008) found that in several cases Cu isotopes were not fractionated substantially during surface adsorption reactions with bacteria, but in others the lighter Cu isotope was preferentially sorbed. In contrast, Mathur et al. (2005) and Kimball et al. (2009) suggested that the heavier Cu isotope was preferentially sequestered by Acidithiobacillus ferrooxidans cells coated by iron-(oxy)hydroxides. It was not possible in this case, however, to distinguish between the adsorption of Cu onto cell surfaces versus co-precipitation and adsorption onto the mineral coatings. Zhu et al. (2002) measured the isotopic changes of Cu(II) during its incorporation into proteins synthesized by bacteria and yeast. They found that the selected proteins preferentially incorporated the lighter Cu isotope and that in some cases this incorporation involved the reduction of Cu(II) to Cu(I). Buchl et al. (2008) demonstrated that inactivation of a metal-binding prion protein in the brains of mice resulted in preferential uptake of the heavier Cu isotope relative to brain tissue with unaltered proteins.
Part of the existing ambiguity associated with Cu isotope results from biological experiments is attributable to the fact that individual biochemical reactions are not easily isolated. Specifically, the fractionation of Cu isotopes during reversible cellular–surface adsorption has not been adequately distinguished from reactions involving intra-cellular incorporation of Cu (a problem also identified by Wasylenki et al. (2007) for Fe and Mo interactions with bacteria). Bacterial cells contain negatively-charged organic acid functional groups (carboxyl, phosphoryl, hydroxyl, etc.) that readily form complexes with aqueous metal cations like Cu (e.g., Beveridge and Fyfe, 1985; Beveridge, 1989). As the pH of a system increases, more surface functional groups become deprotonated, allowing the bacteria to adsorb more metal cations from solution (e.g., Beveridge and Murray, 1980; Fein et al., 1997). These surface reactions are thought to be fully reversible and have been described using surface complexation models (e.g., Fein et al., 1997). Bacterial surface–metal complexation reactions are analogous in many ways to the complexation of Cu by dissolved humic substances, as the metal–organic complexes that form in both cases are chemically and structurally similar (e.g., Meador, 1991; Ma et al., 1999; Borrok and Fein, 2004; Ashworth and Alloway, 2007). In addition to forming surface complexes with bacterial cells, Cu can be transported from the surface and into the cell where it is used in small amounts for metabolic processes and enzyme function (e.g., Brand et al., 1983; Peña et al., 1999; Finney and O’Halloran, 2003). At higher concentrations, Cu is toxic to the cells and can lead to the production of hydroxyl radicals and/or the structural displacement of necessary metals like calcium and magnesium (Beveridge and Koval, 1981; Koch et al., 1997).
In this study, we evaluated the Cu isotopic fractionation behavior for arguably two of the most important and ubiquitous bacterial interactions with metals in natural systems; surface adsorption and intracellular uptake. We performed batch Cu-adsorption and Cu-uptake experiments using individual Gram-positive (Bacillus subtilis) and Gram-negative (Escherichia coli) bacteria, as well as natural consortia of microorganisms isolated from natural waters. The laboratory strains of bacteria, B. subtilis and E. coli, were chosen because their copper-binding mechanisms have been previously investigated (Arnesano et al., 2003; Bertini et al., 2010) and they have different cell wall configurations. B. subtilis, a Gram-positive bacteria, has a thick cell wall comprised of layers of peptidoglycan intertwined with teichoic acids. E. coli, a Gram-negative bacteria, has a thin cell wall with limited peptidoglycan surrounded by a thick outer-membrane rich in lipopolysaccarides.
2. METHODS
All media and reagents were prepared using >18 MΩ ultra-pure water, and were sterilized in an autoclave. Reaction vessels, sample bottles, and pipette tips were acid-washed overnight in a sub-boiling solution of 10% HCl and rinsed three times with pure water prior to use. Ultra-pure trace metal-grade acids and bases were used for experiments and sample preparation.
2.1. Cu–bacteria surface adsorption experiments
Escherichia coli and B. subtilis, Gram-negative and Gram-positive species, respectively, were the sole focus of the surface adsorption experiments with Cu. These bacteria were grown in Trypticase Soy Broth (TSB) with 0.5% yeast extract for a period of 24 h under aerobic conditions in an incubator/shaker at 25 °C. After the cells reached the mid-log phase of growth, they were harvested by centrifugation at 3800g and then washed five times in 0.01 M NaClO4 (the experimental electrolyte) to remove any residual media and metals (e.g., Borrok and Fein, 2005). Each washing step involved suspension of the bacteria in fresh electrolyte followed by separation of the electrolyte from the cells via centrifugation. After the fifth and final wash the bacterial pellet was centrifuged at 5000g for 1 h (supernatant was decanted during this process several times) and the moist mass of the pellet was determined. The moist mass to dry mass ratio for both cell types was about 15–1; however, the moist mass is presented here. The TSB media itself contained 0.34 mg/L Cu (average of triplicate samples analyzed using the ICP-OES as described below). This level of Cu was acceptable for the adsorption experiments, because the amount of experimental Cu added to the system was large by comparison and Cu from the media was removed from the surfaces of the bacteria by washing. The supernatant from the final wash was analyzed using the ICP-OES technique and Cu was not present in the solution above the 8 µg/L detection limit. In hindsight, the choice of a sodium-rich electrolyte was a poor one for isotope work, as the 23Na40 Ar polyatomic combination directly interferes with 63Cu. This complexity was overcome by the isotopic preparation steps described below.
Five types of adsorption experiments were conducted: (1) pH-dependent adsorption at different Cu/bacteria ratios using live cells, (2) pH-dependent adsorption at constant Cu/bacteria ratio using dead cells, (3) Cu loading at a constant pH with live cells, (4) adsorption kinetics and reversibility with live cells, and (5) control experiments without bacteria. All adsorption experiments were conducted below pH 6.5 so aqueous Cu(II) did not appreciably hydrolyze or reach saturation with respect to Cu-oxide or -oxyhydroxide phases. This stable pH range was confirmed using Visual MINTEQ version 2.53 (Gustafsson, 2007). Copper(II) was diluted from an in-house atomic adsorption standard for use in the adsorption experiments.
2.1.1. pH-dependent adsorption experiments
Batch adsorption experiments were conducted with E. coli and B. subtilis in stock solutions of 0.01 M NaClO4 with component ratios of moist bacteria (in g) to Cu(II) (in mg/L) of 5:10 and 15:2. These component ratios were chosen to highlight possible differences in the Cu–organic complexation chemistries of the different functional group sites. Thirty milliliter samples of the homogeneous bacteria–Cu stock suspensions were collected and placed in batch reaction vessels. The pH of each vessel was individually adjusted to values between 2.5 and 6.5 using 0.2 mL aliquots of 0.1 N HNO3 or 0.2 N NaOH. The experimental batches were allowed to interact on a shaker table for 1 h after which the solution pH was measured. This equilibration time for adsorption reactions was determined using the kinetic information described below. The reaction vessels were centrifuged and filtered with a pre-cleaned 0.45 µm nylon syringe filter to separate the bacterial pellets from the supernatant. Control experiments were conducted using the same methodologies, but without the bacterial component.
The supernatant was preserved through acidification by adding 0.5 mL of concentrated ultrapure HNO3 and then analyzed for its Cu content using an ICP-OES method (described below). A split from each sample was used for Cu isotopic analysis. Adsorption of Cu onto the bacterial pellet was determined by the difference between the initial Cu concentration of the stock solution and the concentration of Cu remaining in solution after filtration.
The cells used in adsorption experiments were thought to be “non-metabolizing” because they were carefully washed to remove nutrients and were suspended in an electrolyte solution that provided no energy. Despite these precautions we decided to test the possibility that the cells retained some stored energy for metabolic function that impacted Cu isotopes. To this end, we performed a set of adsorption experiments with intact but heat-killed E. coli cells. The same experimental procedures described above were followed for adsorption of Cu onto dead E. coli cells. Cells were heat-killed by placing the bacteria-electrolyte stock solution in a hot water bath where the temperature was slowly raised. After extensive testing, we found that the minimum effective temperature and time necessary to kill but not lyse the E. coli cells was 40 °C for 15 min. After subjecting the cells to this treatment we could not achieve bacterial growth after 48 h of incubation on TSB agar plates. Cell growth did occur after treatments of 35 and 38 °C. The cellular integrity of the heat-killed E. coli was verified through microscopic examination of Gram-stained cells.
2.1.2. Metal loading experiments
Metal loading experiments were conducted to test whether isotopic fractionation of Cu changed as available functional group sites become saturated with Cu at a single pH. First, a stock solution of 5 g/L of E. coli or 1 g/L B. subtilis in a 0.01 M NaClO4 was prepared and adjusted to pH 4 using a few mL of 0.1 N HNO3. This experiment was conducted at pH 4 to assure that the isotopic differences measured were attributable to surface adsorption only and not from precipitation. The solution was stirred using a magnetic stirring rod (raised 1 in. from the bottom of the vessel with a plastic mounting device) and the pH was held constant with a Radiometer TIM 856 autotitrator/pH-stat instrument. Copper (from the AA standard or a Cu-perchlorate solid in the case of the experiment with B. subtilis) was added to the stock solution in 1 h steps to achieve concentrations of approximately 5, 10, 25, 50, and 100 mg/L. Each addition of Cu was followed by a 1 h reaction time after which 30 mL samples of the homogeneous suspension were collected using a sterile, pre-rinsed serological pipette. As described above, the samples were filtered, the filtrate was acidified, the preserved samples were analyzed for their Cu content, and sample splits were processed for isotopic analysis.
2.1.3. Kinetics and equilibrium experiments
Kinetics experiments were conducted with both laboratory bacterial strains. A stock solution of 5 g/L bacteria and 10 mg/L of Cu in 0.01 M NaClO4 was held at a constant pH of 4.3 using the pH-stat instrument. Thirty milliliters of the homogeneous mixture was collected after about 5, 15, 30, 60, and 80 min, 25 h, and 48 h. Each sample was filtered through a pre-cleaned 0.45 µm nylon syringe filter; the filtrate was acidified and analyzed for its metal content on the ICP-OES and a split of the sample was prepared for isotopic analysis. Reversibility experiments were conducted to test whether Cu adsorption onto surface functional group sites was an equilibrium process. A stock solution of 5 g/L bacteria and 10 mg/L of Cu in 0.01 M NaClO4 was held at a constant pH of 4.0 for 3 h before being adjusted to pH 5.0 (using 0.2 N NaOH). After 2 h at pH 5, the solution was adjusted back to pH 4.0 using 0.2 N HNO3 where it was held constant for two more hours. At the beginning of the experiment and after each pH change, 30 mL samples were collected at approximately 1, 5, 15, 30, 60, 90, 120 and 180 min. One additional sample was collected after 24 h at the final pH of 4.
2.1.4. Control experiments without bacteria
All procedures followed for the pH-dependent adsorption experiments at constant Cu/bacteria ratio were duplicated for the control experiment with the exception that bacteria were not added. The objectives of this experiment were to verify that Cu did not sorb to the filter or experimental apparatus and that Cu was soluble over the pH range predicted by thermodynamic modeling.
2.2. Metabolic uptake experiments
2.2.1. Cell washing experiments
In order to verify that we could successfully isolate intracellular Cu from surface-bound Cu, we performed a series of washing experiments. The first set of washing experiments focused on the removal of surface adsorbed Cu(II) from E. coli and B. subtilis. The second set of experiments focused on the removal of Cu(II)-citrate from solutions in contact with these same bacteria. The wash experiments were carried out using a series of solutions of varying pH and ionic strength designed to remove Cu but not damage the integrity of the cell walls. Each experiment was subjected to five washing cycles consisting of centrifugation and then re-suspension in fresh wash solution. The supernatant from the individual wash cycles was collected and analyzed using an ICP-OES and Total Organic Carbon (TOC) analyzer to determine Cu and dissolved OC concentrations, respectively. The latter analyses confirmed that cells were not lysed during the washing experiments (data not shown). Optical microscopy and TEM images also confirmed that the cells remained intact after the washes.
2.2.2. Intracellular incorporation experiments
For the intracellular incorporation experiments we additionally used natural consortia of microorganisms collected from the Rio Grande Reservoir (RGR) and Cement Creek (CC) in Southwest Colorado. The RGR was characterized by high pH (~10), moderate human and water fowl activity, high DOC content, and turbid water. Cement Creek is a mountain stream rich in Fe and other metals derived from acid rock drainage. The CC site was distinguished by low pH (~3.7), high conductivity (>1600 µS m−1) and high levels of dissolved Fe(II), Cu(II), and Zn(II). Both samples of wild-type microorganisms were characterized by Gram-staining with light microscopy. The RGR consortia included fungi, diatoms, and protozoa, as well as Gram-positive and Gram-negative bacteria of differing morphologies (cocci, bacilli, vibrio). The CC consortia contained Gram-negative rods, Gram-positive cocci, and some fungi.
Experiments were performed by growing bacteria aerobically at 25 °C in an incubator/shaker in a specially-formulated basal medium amended with Cu(II)-citrate. The media recipe followed that for “FeDM” described by Hersman et al. (2001) and contained the following per L of 18 MΩ pure water: 1g glucose, 0.5 g K2HPO4, 1.0 g NH4Cl, 0.2 g MgSO4·7H2O, 0.2 g CaCl2·2H2O, 8.33 g succinate disodium salt, 30 mM Fe-EDTA, 4.77 g HEPES buffer, and 0.125 mL of additional trace elements (5 mg MnSO4·H2O, 6.5 mg CoSO4·7H2O, 3.3 mg ZnSO4 and 2.4 mg MoO3 per 100 mL of water). Prior to the addition of Cu(II)-citrate, 6 mL of the basal media was evaporated to dryness and dissolved in 3 mL of 2% HNO3 for analysis of its trace element content. It contained no measurable Cu (i.e., there was less than the 4 µg/L of Cu in the basal media). The basal media was amended with 3 mg/L Cu(II)-citrate (about 1 mg/L Cu) for each bacterial experiment and control experiments were performed with bacteria but no Cu and with Cu but no bacteria. Cu(II)-citrate was chosen because the aqueous citrate complex keeps Cu(II) from reacting with the cell surface functional group sites but is still fully bioavailable for the uptake of Cu(II) into the cells (Nybroe et al., 2008). Control experiments verified the aqueous stability of the Cu(II)-citrate complex and that this complex did not react with the bacterial surface (see Section 3).
Experimental solutions were either inoculated directly from cultures grown on agar plates using a sterilized metal loop (in the case of B. subtilis and E. coli) or were inoculated through the addition of 20 µL (per 1 L of experimental solution) of the original water sample (in the case of the bacterial consortia). This inoculation contributed less than 5 ng of Cu to the experimental solutions. Both bacterial strains and consortia were grown in the basal media amended with Cu(II)-citrate until they reached mid-log phase (determined by UV–vis measurements). At this stage the cells were harvested via centrifugation. Harvested cells were washed three times with a solution of 0.2 M MgSO4 at pH 1.5 (similar to the washing method developed by Cheng et al. (1970)). Cells were digested with a mixture of ultra-pure 70% HNO3 and 30% H2O2, evaporated to dryness, and re-dissolved in 2% HNO3. This solution was analyzed for its Cu content using an ICP-OES and sample splits were prepared for isotopic analysis.
2.3. ICP-OES and UV–vis measurements
The supernatant from all adsorption and control experiments, as well as the bacterial digests from the intracellular uptake experiments, were analyzed for their Cu concentrations using a Perkin-Elmer Optima 5300DV ICP-OES instrument. Standards were diluted from a CertiSPEX® multi-element standard. Uncertainty was quantified through replicate analyses and was found to be less than ±5%. USGS standard reference water T-143 was used as an external check to verify accuracy. Concentrations of Cu for this sample fell within the certified range of 20.4–24.2 µg/L. Optical density measurements for bacterial growth were conducted using a Hach 2300 UV–vis photo-spectrometer at a light wavelength of 600 nm. Uncertainty was quantified through replicate analysis of samples and was found to be ±2%.
2.4. TEM analysis
Transmission electron microscope (TEM) images of bacteria before and after exposure to Cu were collected using a Zeiss EM-10 TEM in the Department of Biological Sciences Analytical Cytology Core Facility at the University of Texas at El Paso. Cells from the selected experiments were pelleted through centrifugation and then suspended in warm agar. A glutaraldehyde-osmium tetroxide fixation technique was used to prepare the specimens. These methods are similar to those described previously by Kimball et al. (2009). Agar specimens were minced with a razor blade to a size of ~1 mm3 and transferred to fixation vials were they were filled 1/3 full with 3% glutaraldehyde in 0.12 M Millonig’s phosphate buffer (pH 7.4). Vials were placed in a rotary agitator at room temperature for 1 h. Glutaraldehyde was removed from the vials and the samples were washed three times with fresh cold 0.06 M phosphate buffer (5–15 min per wash). A solution of 1% OsO4 in 0.12 M phosphate buffer was added to cover the specimens in the vials. Vials were placed in an ice bucket (dark, 0–4 °C) for 1.5 h before the OsO4 was removed and the specimens washed with cold 0.06 M phosphate buffer 3 more times followed by three rinses of double distilled water. Uranyl acetate (0.5%) was added to the specimens and they were wrapped with aluminum foil and left to incubate at room temperature for 1 h. The uranyl acetate was then removed and the sample rinsed three times with distilled water. Samples were dehydrated by washing with an ethanol and acetone series. Poly/Bed 812 plastic (Poly-sciences, Inc., Warrington, PA) was allowed to infiltrate into the samples for 6 h, after which a drop of the plastic was placed at the bottom of a BEEM capsule. Specimens were transferred to the BEEM capsules and then 100% plastic was used to fill the remaining capsule volume. The capsule was placed in an embedding tray for at least 48 h in a 60 °C oven. Thick sections (1 µm) were stained with Tolui-dine Blue and Fuschin to determine suitable areas to be thin sectioned with a Leica Ultracut ultramicrotome (60–90 nm). Hardened specimens were sectioned into ~90 nm sections and mounted on 200 mesh copper grids for analysis. Thin sections were post stained with uranyl acetate followed by Reynolds lead citrate. Grids were examined and photographed in a Zeiss EM-10 transmission electron microscope (Carl Zeiss SMT, Inc. Peabody, MA) operating at an accelerating voltage of 60 or 80 kV. Digital images were obtained with a Gatan 385C CCD camera.
2.5. Isotopic analysis
Selected samples from the adsorption, control, and intracellular uptake experiments were prepared for isotopic analysis. First, Cu was isolated from the solution matrix through anion exchange chromatography as described by Borrok et al. (2007). Savillex® Teflon beakers, pipette tips and sample storage containers were all acid-washed overnight in a sub-boiling 10% HCl bath and washed three times in ultra-pure water prior to use. BDH Aristar® ultra pure trace metal grade HCl (<1 ppt Cu) was used for column chemistry. Column separations were performed in a Class 100 HEPA-filtered laminar flow clean bench. Column separation of Cu proved to be a relatively simple process because Cu was the only element of interest in the experiments; however, in several cases Na (from the NaClO4 electrolyte) broke through into the Cu fraction during the initial separation. Hence, we passed the Cu fraction (about 1.5 µg of Cu) through the column procedure a second time for additional purification (e.g., Pribil et al., 2010). One hundred percent (±5%) of the Cu was recovered from the columns, as the fractions before and after Cu elution had no detectable Cu (given the detection limits less than 0.04 µg of Cu were present). Procedural blanks for every column separation event were evaluated on the MC-ICP-MS, and in all cases the 63Cu voltage signal for the blanks was less than 0.1% of the voltage of the samples, suggesting that the blanks contained less than about 1.5 ng of Cu.
For the adsorption experiments we prepared only supernatant samples for analysis and not bacterial pellets. Because bacterial pellets remain “wet” after centrifugation, the adsorbed Cu is allowed to co-mingle with the free Cu that remains in solution. Therefore, the Cu isotopic signature of the bacterial pellet is compromised (e.g., Barling and Anbar, 2004). Instead, we calculated the δ65Cu for the solid fraction through isotopic mass balance according to the following equation:
| (1) |
Isotopic analyses were conducted using a Nu Instruments multicollector ICP-MS at the Center for Environmental and Earth Isotopic Research at UTEP. The NIST SRM 976 Cu standard was used as a reference material. The SRM standard was also passed through the column chromatography process to eliminate differences in sample matrix (e.g., Archer and Vance, 2004). All samples were evaporated to dryness after column separations and 1 mL of 2% ultrapure HNO3 was added and evaporated to dryness to remove residual Cl from the column chemistry. Samples, standards, and blanks were reconstituted in ultra-pure 0.2% HNO3 in preparation for delivery into the MC-ICP-MS via a de-solvating nebulizer (Nu DSN 100) system. Each sample was bracketed by the SRM standard and results are presented in standard delta notation (parts per thousand) relative to the average of the two bracketing standards,
| (2) |
Experimental results are also reported as isotopic separation factors, which are defined by the following equation:
| (3) |
Long term analytical precision was determined by replicate analyses of two in-house Cu standards, a J.T. Baker® AA standard and a digested Cu-rod obtained from a nearby Cu refinery. Each of these standards were prepared as unknowns and purified through the column chemistry procedure. Over a period of 8 months, the 2σ precision for the Cu rod standard was ±0.16‰, (n = 12) and the 2σ precision of the AA standard was ±0.14‰ (n = 15). The uncertainties for column duplicates fell within these ranges. Error bars reported in figures reflect calculated 2σ uncertainties of replicates or in cases where fewer replicates were performed the average uncertainty is reported. The complete isotope dataset is presented in Electronic Annex-1.
3. RESULTS AND DISCUSSION
3.1. Adsorption experiments
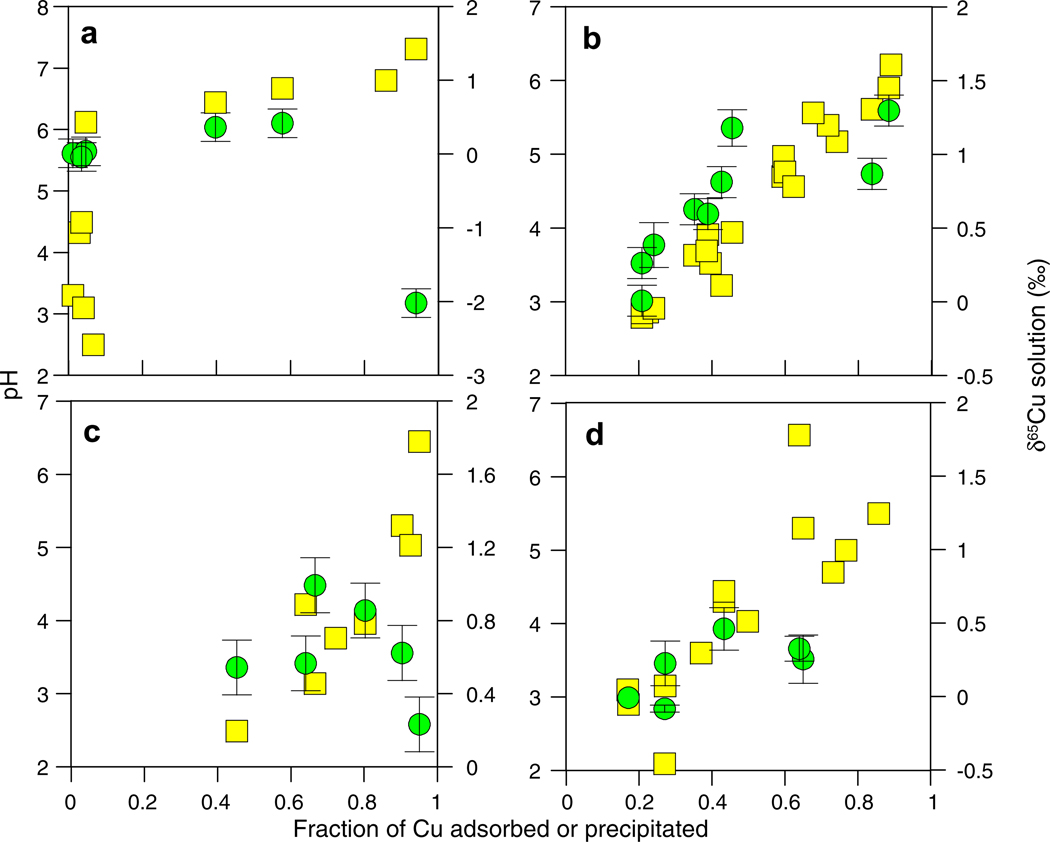
Concentration and isotopic data for the pH-dependent adsorption experiments are presented in Fig. 1a–d. In all cases the fraction of Cu adsorbed from solution increased with increasing pH over the tested pH range of 2–6.5. This is an expected result, because as pH increases, more bacterial-surface functional group sites become deprotonated and available to react with free Cu(II) (Beveridge and Fyfe, 1985; Beveridge, 1989; Fein et al., 1997). Cells exposed to Cu during the adsorption experiments remained intact (see TEM images below) and were able to grow when streaked onto nutrient agar after a 24 h incubation period.
Fig. 1.
Cu concentration (yellow squares) and δ65Cu (green circles) plotted as a function of the fraction of Cu adsorbed and pH for (a) an abiotic precipitation experiment, (b) adsorption of 10 mg/L of Cu by 5 g/L E. coli, (c) adsorption of 2 mg/L Cu by 15 g/L E. coli, and (d) adsorption of 10 mg/L Cu by 5 g/L B. subtilis. Error bars for concentration data fall within the symbols. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The control experiment without bacteria (Fig. 1a) demonstrated that no Cu precipitation or adsorption onto the reaction vessel occurred over the pH range of 2.5–6, but Cu did start to precipitate at pH >6.5. E. coli and B. subtilis adsorbed similar amounts of Cu at the same pH and experimental conditions (Fig. 1b and d), while an increase in the E. coli: Cu(II) ratio resulted in more Cu(II) binding at lower pH (Fig. 1c).
The δ65Cu of the starting solution for all adsorption experiments was +0.71‰ ± 0.16; however, for illustration purposes we have normalized the adsorption data to a starting point of 0‰ in all figures by subtracting +0.71‰ from each sample. The control experimental data (Fig. 1a) demonstrate that the heavier Cu isotope is preferentially incorporated into Cu oxy-hydroxide precipitates (leaving the solution depleted in the heavy isotope) during abiotic precipitation at pH >~6.5. During the initial stages of precipitation, the δ65Cu of the solution increased slightly suggesting the possibility of a modest kinetic isotope effect followed by equilibrium fractionation.
The δ65Cu of the solution in the bacterial adsorption experiments became heavier relative to the starting δ65Cu (Fig. 1b–d). This demonstrates that the bacteria became enriched in the lighter isotope of Cu. This observation holds for experimental systems with variable bacteria:Cu ratios and for both strains of bacteria. The apparent separation factor for this process, Δ65Cusolution–solid (Eq. (3)), was as large as +2.6‰ for the E. coli adsorption experiments and +1.4‰ for the B. subtilis experiment. The preferential sequestration of the lighter Cu isotope during bacterial surface adsorption experiments was an unexpected result, as adsorption experiments with mineral surfaces have shown that the heavier Cu isotope is preferentially incorporated into the Cu–surface complex (e.g., Balistrieri et al., 2008; Pokrovsky et al., 2008). The initial shift to heavier δ65Cu in solution occurs at low levels of adsorption and low pH; however, the δ65Cu of the solution does not change much after about 40% adsorption (Fig. 1b–d). Because the fraction adsorbed does not correlate with the δ65Cu in a consistent manner, it appears that either (1) the observed fractionations are attributable to a process unrelated to adsorption, or (2) that the observed fractionations are the result of summing of fractionations from several individual reactions governed by the differences in chemical bonding environments active over the tested pH range.
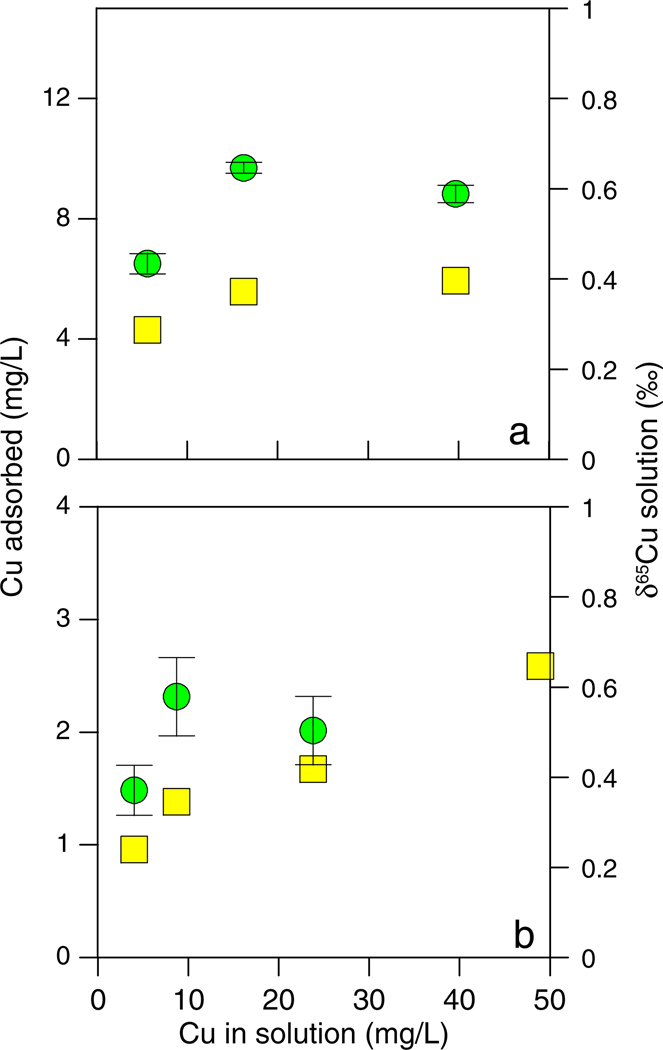
The results of the metal loading experiments are presented in Fig. 2a and b for E. coli and B. subtilis, respectively. In the metal loading experiments, the pH is held constant to isolate its impact on the isotopic changes. The adsorption of Cu onto both bacterial species increased with increased concentrations of Cu in solution, but too few points were tested to effectively evaluate the data using a Langmuir model. Under low Cu loads, the δ65Cu of the solutions became enriched in the heavier Cu isotope relative to the parent solutions, and the magnitude of the fraction-ation was similar to that measured for the pH-dependent experiments. The δ65Cu of the experimental solutions remained relatively consistent as more Cu was added to them. However, because the δ65Cu was still elevated while smaller and smaller fractions of the total Cu were adsorbed, the calculated Δ65Cusolution–solid, increased with increasing Cu load, reaching values greater than +4.0‰ in both systems. This isotopic shift is probably not related to changes in surface adsorption, because the amount of adsorbed Cu did not substantially change in the B. subtilis experiment despite the large isotopic shift. In order to estimate fractionation behavior at the highest metal:bacteria ratios, we analyzed the δ65Cu of several digested bacteria pellets. The results (not shown) indicate that the lighter isotope of Cu is indeed enriched in the bacteria at high metal:bacteria ratios, but quantification of the magnitude of the fractionation was difficult because aqueous Cu was not fully removed from the cells (see Section 2.5).
Fig. 2.
Cu concentration (yellow squares) and δ65Cu (green circles) data for Cu-loading experiments at pH 4 with (a) 5 g/L E. coli and (b) 1 g/L B. subtilis. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
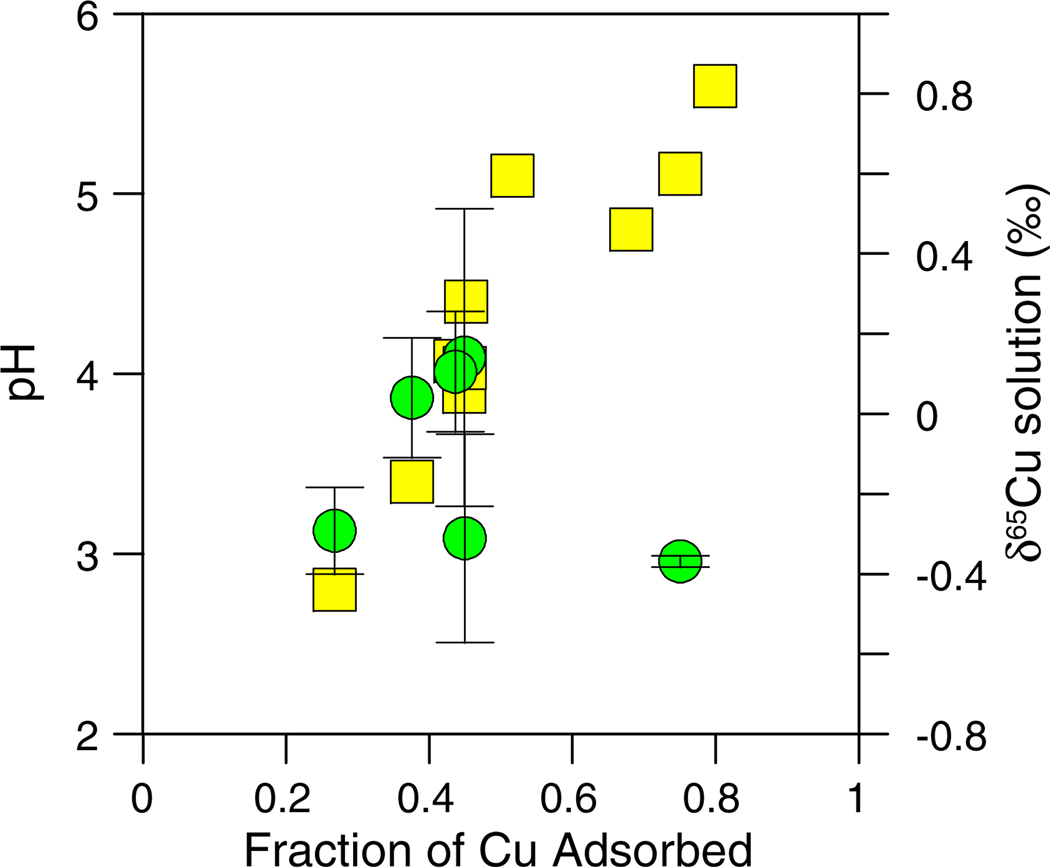
Data from the adsorption experiment with intact but heat-killed E. coli cells are shown in Fig. 3. The extent of Cu adsorption onto the dead cells increased with increasing pH and the extent of Cu adsorption was similar to that of the corresponding live cell experiment (Fig. 1b). The δ65Cu of the solution, however, showed almost no fractionation (within analytical uncertainties) for samples collected at pH 3.4, 3.9, and 4.0. However, at pH 4.1 and 5.1 there was a slight preference for adsorption of the heavier Cu isotope. At pH 4.1 (45% adsorption) the Δ65Cusolution–solid was –0.69 ± 0.25‰, while at pH 5.1 (75% adsorption) the Δ65Cusolution–solid was –0.49 ± 0.02‰. This behavior is the opposite of that observed for live cells, suggesting that preferential incorporation of light Cu isotopes is an active cellular process, presumably triggered using stored energy. At pH 2.7 (initially pH 2.3 before the 1 h reaction time) the bacteria also preferentially sequestered the heavier Cu isotope. However, because of the low pH, this sample may have been impacted by cell lysis. This could have facilitated exposure of Cu to different cellular components and may have allowed some Cu bound to these components to pass through the filter.
Fig. 3.
Cu concentration (yellow squares) and δ65Cu (green circles) data plotted as a function of the fraction of Cu adsorbed for the experiment with 10 mg/L Cu and 5 g/L of dead (heat-killed) E. coli. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. Kinetics and reversibility experiments
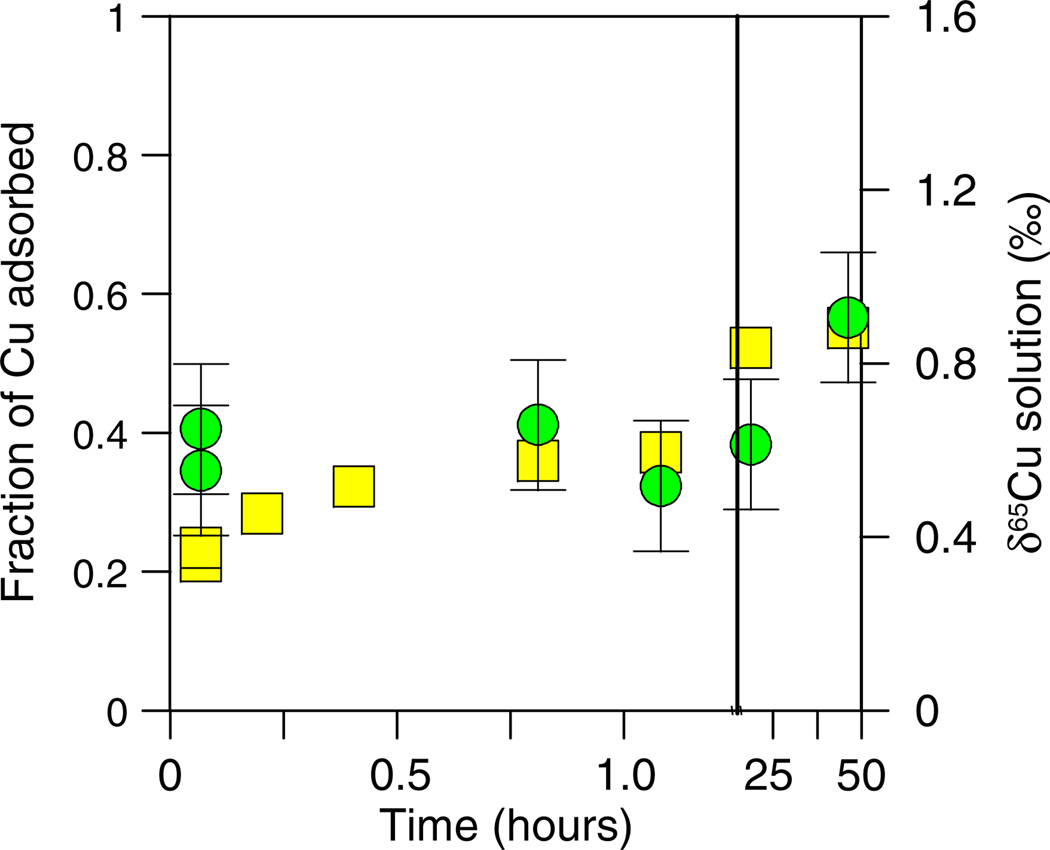
The results of the kinetics and reversibility experiments are presented in Figs. 4–6. As observed in the pH-dependent adsorption experiments, the δ65Cu of the solution initially increased relative to the starting δ65Cu during the kinetic experiments. Figs. 4 and 5 demonstrate that the bulk of Cu(II) adsorption onto E. coli occurs in the first several minutes; however, some additional Cu(II) adsorbs slowly over a period of about 48 h (Fig. 4). The δ65Cu of the experimental solutions, however, did not appreciably change despite the fluctuations in the adsorbed concentrations of Cu that occurred as time increased, with the possible exception of the final point at 48 h where a small increase was observed (Fig. 4).
Fig. 4.
Cu concentration (yellow squares) and δ65Cu (green circles) data plotted as a function of time for the kinetics experiment with E. coli (10 mg/L Cu and 5 g/L bacteria). The solid line distinguishes a break in the scale of the X-axis. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
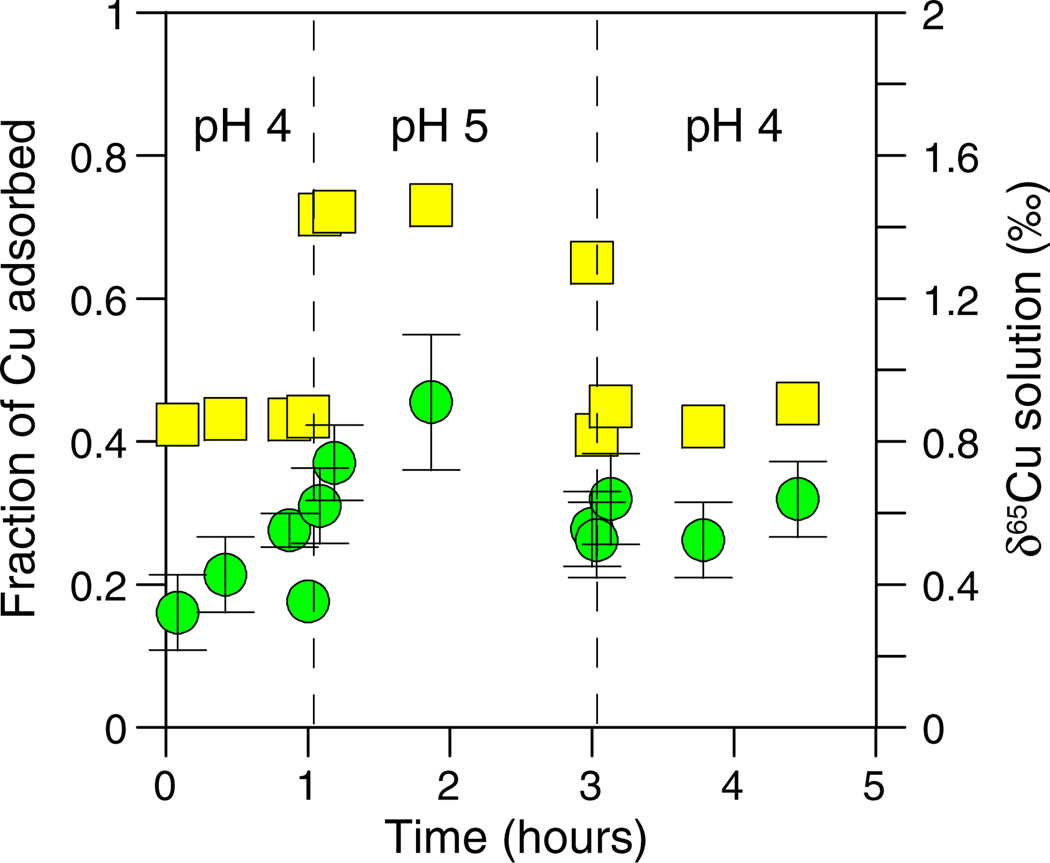
Cu concentration (yellow squares) and δ65Cu (green circles) data plotted as a function of time for the reversibility experiment with B. subtilis (10 mg/L Cu and 5 g/L bacteria). The pH of the experiment was changed from 4 to 5 and back to 4 at the times identified by the dashed lines. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
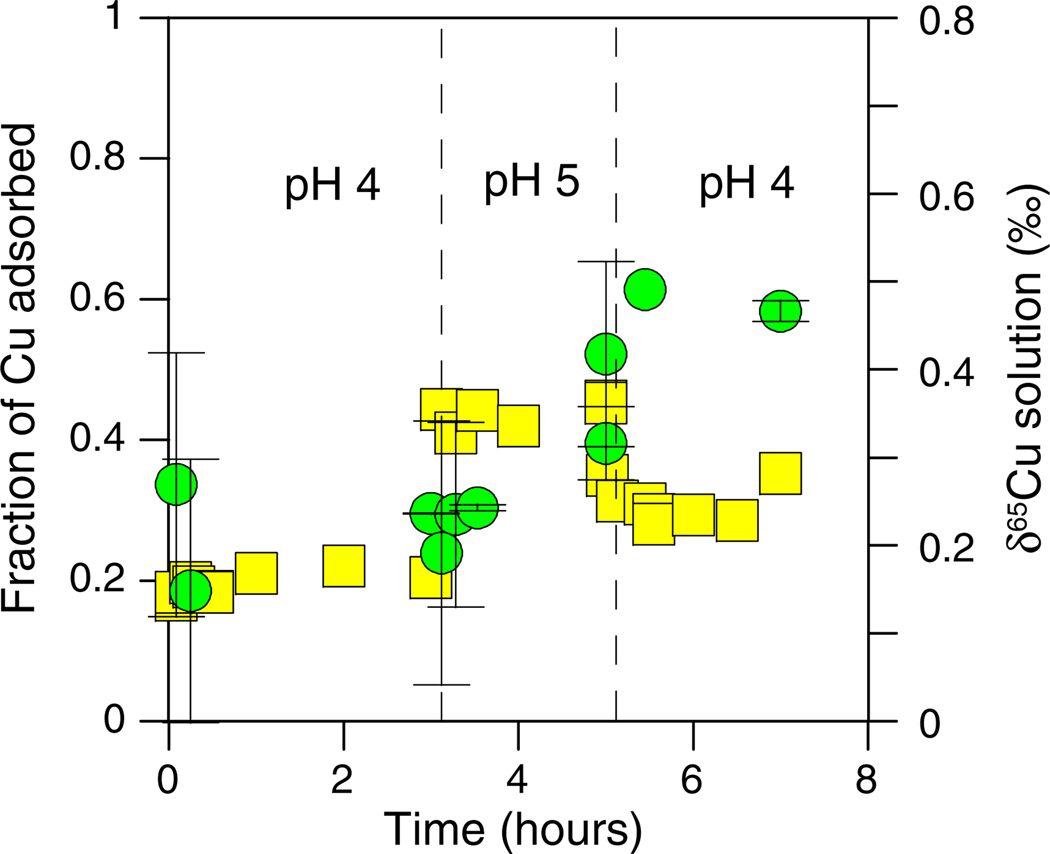
Cu concentration (yellow squares) and δ65Cu (green circles) plotted as a function of time for the kinetics and reversibility experiment with E. coli (10 mg/L Cu and 5 g/L bacteria). The pH of the experiment was changed from 4 to 5 and back to 4 at the times identified by the dashed lines. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The extent of Cu adsorption onto E. coli was not fully reversible as ~20% of the Cu was adsorbed initially at pH 4, while ~30% of the Cu was adsorbed upon returning to pH 4 after the intermediate adsorption step at pH 5 (Fig. 5). The irreversible sequestration of a small amount of Cu in the E. coli cells was confirmed in our washing experiments (see below).
The adsorption of Cu onto B. subtilis cells was rapid, did not increase as a function of time (for the 2 h period tested), and appeared to be fully reversible (Fig. 6). However, the δ65Cu of the solution did not equilibrate as quickly. At the start of the experiment, the δ65Cu of the solution immediately increased relative to the starting solution and increased further as a function of time during the initial pH step. This may indicate that Cu isotopes were being fractionated via a kinetic isotope affect, perhaps related to surface processes for B. subtilis which sequester and reduce Cu or transport it across the cell membrane (see Section 3.6). The δ65Cu of the solution did not appreciably change at the pH 5 step and remained relatively consistent as pH was changed back to 4.0 (Fig. 6). Because the δ65Cu for each of the reversibility experiments did not significantly change in response to changes in the amount of Cu adsorbed (Figs. 5 and 6), surface adsorption was probably not the primary control on the isotopic changes.
3.3. Washing experiments
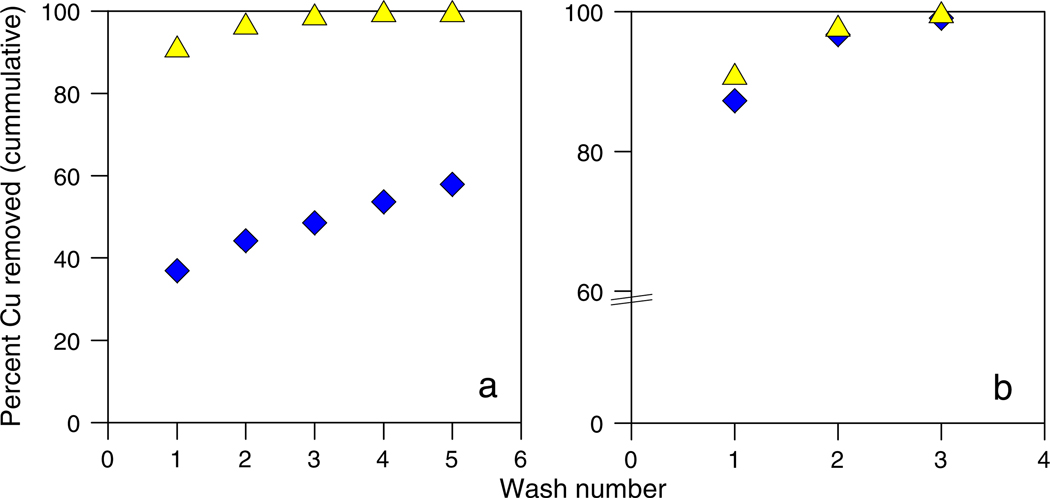
Washing experiments were conducted to identify the most effective method(s) for the removal of adsorbed Cu from bacterial surfaces while keeping intracellular Cu intact (Fig. 7). We used six different chemical solutions to wash E. coli and B. subtilis after exposure to Cu(II) at pH ~4.5. At this condition ~5 mg/L of Cu(II) were adsorbed to the bacterial surfaces prior to washing. Washes with EDTA were not attempted because there is some evidence that EDTA treatments can disrupt the integrity of the cell membranes of Gram-negative bacteria (Wilkinson, 1967; Alakomi et al., 2003). The most effective chemical agent for Cu removal was a simple pH 1.5 HNO3 solution with or without a 0.2 M MgSO4 electrolyte (Cheng et al., 1970). The other electrolyte solutions were slightly less effective. The pH 1.5 washes of the B. subtilis cells yielded 100 ± 5% recovery of Cu, while washes of E. coli were only effective for the recovery of 60 ± 5% of Cu (Fig. 7a). This finding is confirmation of the reversibility experimental results (Fig. 5), which demonstrated that adsorption of Cu(II) onto E. coli was not a fully reversible process.
Fig. 7.
The cumulative percentage of adsorbed Cu removed from B. subtilis (yellow triangles) and E. coli (blue diamonds) using washes of 0.2 M MgSO4 at pH 1.5. Panel (a) depicts washes from experiments conducted with Cu(II) and panel (b) depicts washes from experiments conducted with Cu(II)-citrate. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Largely because of this irreversible binding behavior, we evaluated whether we could be successful in removing Cu(II)-citrate associated with the bacterial surfaces through washing. Because Cu(II)-citrate forms a strong aqueous complex in pH-circumneutral solutions we expected that (1) Cu(II)-citrate would not bind strongly with the bacterial surface and (2) what Cu(II)-citrate did associate with the bacterial surface would be easily removed. Fig. 7b presents the results of washing experiments for Cu(II)-citrate and shows that 100 ± 5% of the Cu was recovered from both bacterial species in the first three washes.
3.4. Intracellular incorporation experiments
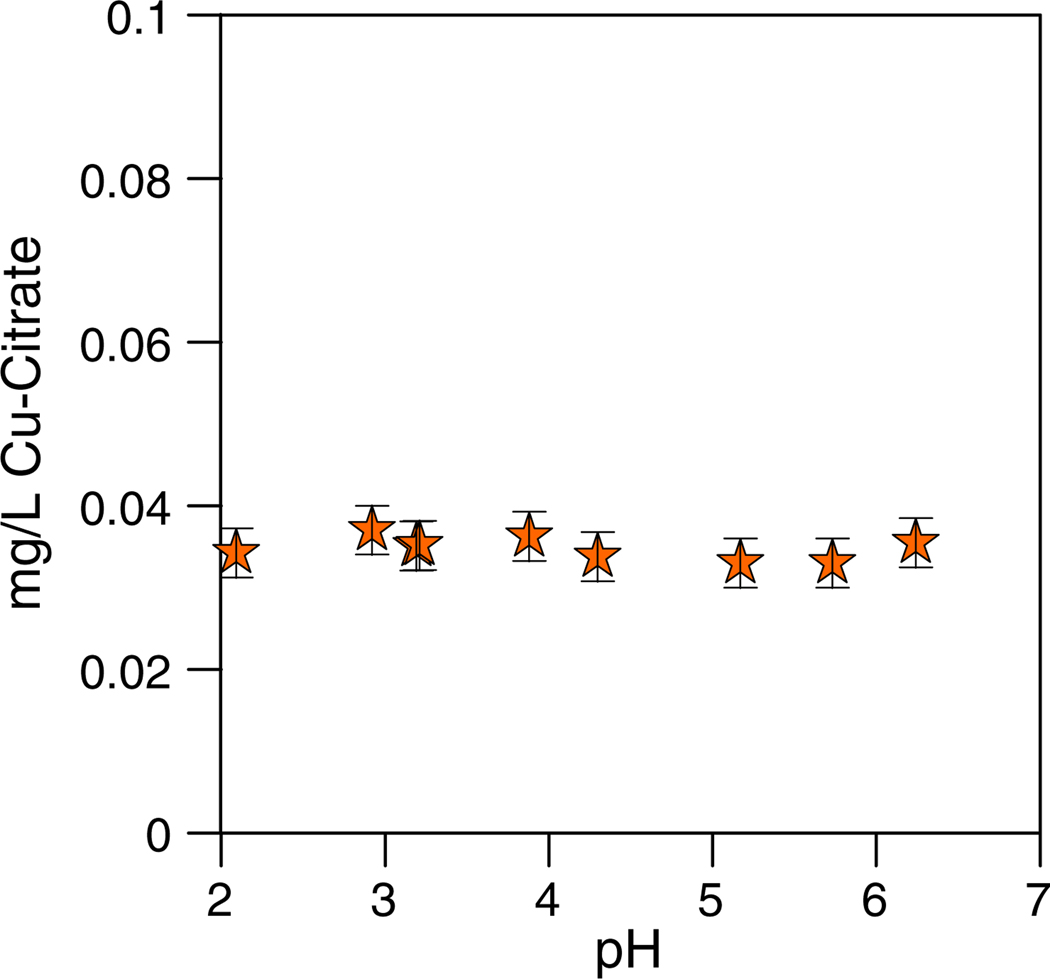
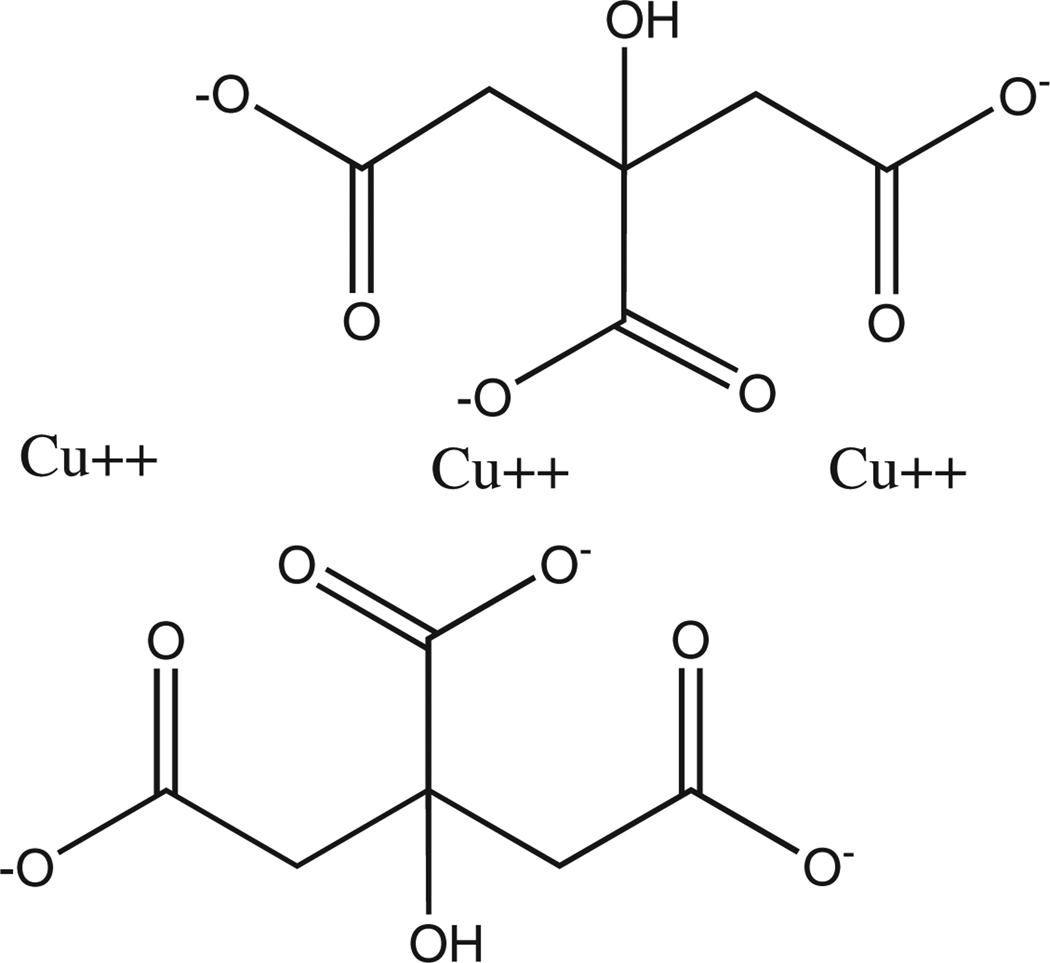
Cu(II)-citrate was used for the intracellular incorporation experiments because it did not bind appreciably with the bacterial surface (see above) and we needed additional flexibility in the pH range for the growth experiments (i.e., Cu not complexed as Cu(II)-citrate would mostly precipitate under circumneutral pH conditions). Control experiments confirmed that the Cu(II)-citrate complex did not appreciably precipitate, or adsorb to the reaction vessels under the experimental pH conditions (Fig. 8). Cu(II)-citrate [Cu3(C6H5O7)2] contains 3 Cu atoms bound to deprotonated carboxylic functional group locations, as illustrated in Fig. 9 (e.g., Green et al., 1998; Ivanov and Tsakova, 2002). It is possible that the choice of Cu(II)-citrate over other possible Cu–organic complexes might have some influence on the bacterial metabolisms. However, we believe that Cu(II)-citrate is a suitable analog for Cu in natural environments because (1) free Cu(II) is all but absent in natural waters at circumneutral pH because of its propensity to hydrolyze and precipitate (e.g., Smith and Martell, 1978) and (2) most Cu that is present is bound to organic ligands associated with dissolved humic substances that are dominated by carboxylic functional group sites (Gauthier et al., 1987).
Fig. 8.
Concentration of Cu in solution as a function of pH for the control experiment with Cu(II)-citrate. Copper did not precipitate or adsorb onto the reaction vessel over this pH range.
Fig. 9.
Molecular structure of Cu(II)-citrate.
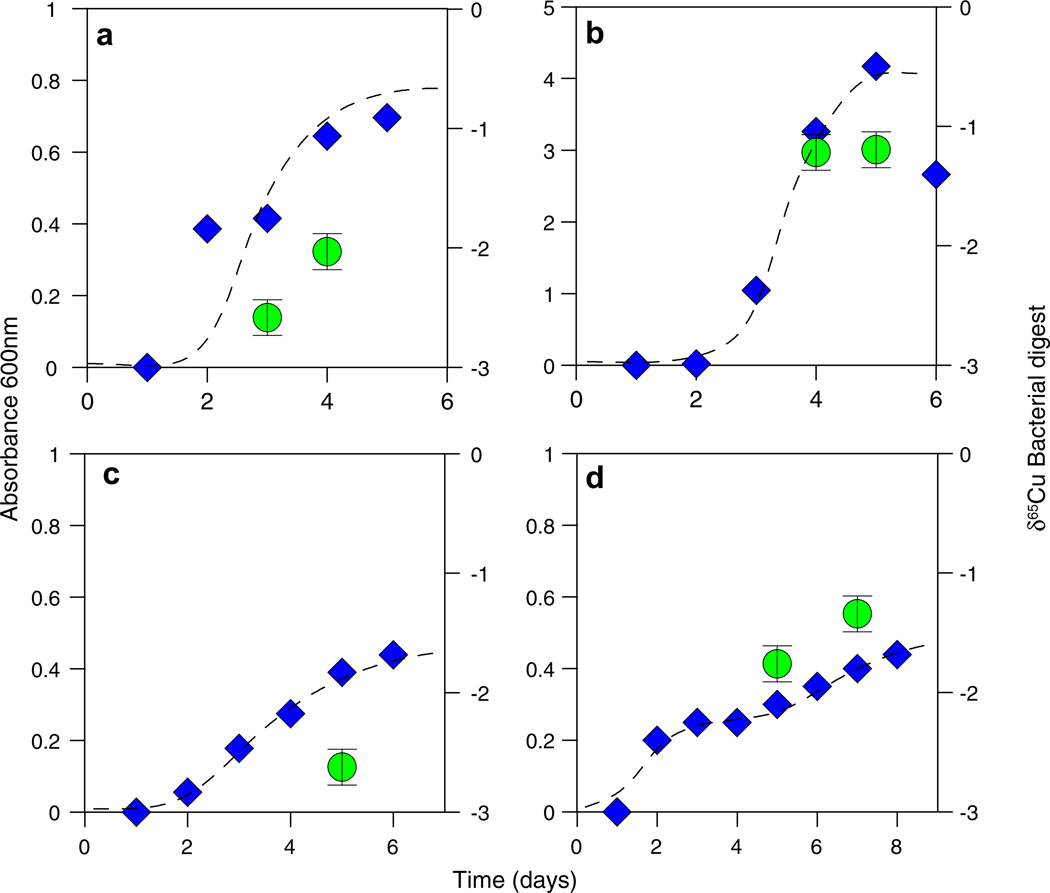
Results for the intracellular incorporation experiments are presented in Fig. 10a–d. All of the bacterial species and consortia reached exponential growth phase by day 4 of the experiments. E. coli (Fig. 10a), B. subtilis (Fig. 10b), the RGR bacterial consortium (Fig. 10c), and the CC bacterial consortium (Fig. 10d) all preferentially incorporated the lighter isotope of Cu during growth. Although the direction of the isotopic fractionation was consistent for all experiments, the magnitude of the fractionation (relative to the starting solution) was dependent on the species and consortium. This may be attributable to the fact that some Cu enzymes are specific to certain families of organisms (Bertini et al., 2010).Wecalculated the apparent Δ65Cusolution–solid for each of the metabolic experiments based on the fraction of the total Cu pool incorporated during growth and the δ65Cu of the washed and digested bacterial pellets. B. subtilis and E. coli incorporated only a small fraction of the total Cu (<2%) and fractionated the Cu isotopes by an average of +1.2‰ and +2.3‰ (Δ65Cusolution–solid, Eq. (2)), respectively. The RGR consortium incorporated almost 40% of the Cu in solution and fractionated the isotopes by +4.4‰ (Δ65Cusolution–solid). The CC consortium incorporated 20% of the Cu in the solution and fractionated the isotopes by an average of +1.9‰ (Δ65Cusolution–solid). The RGR and CC consortia contained several Gram-positive and Gram-negative bacterial species, but the RGR also contained large amounts of other microorganisms, including fungi and protozoa. The large fractionations observed in the RGR consortia might be attributable in part to the different types of microorganisms present.
Fig. 10.
Absorbance at 600 nm (blue diamonds) and δ65Cu (green circles) data plotted as a function of time for intracellular incorporation experiments with (a) E. coli, (b) B. subtilis, (c) RGR consortium, and (d) CC consortium. Growth behavior is illustrated by dashed curves. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5. Sites of Cu accumulation and impact on cell morphology
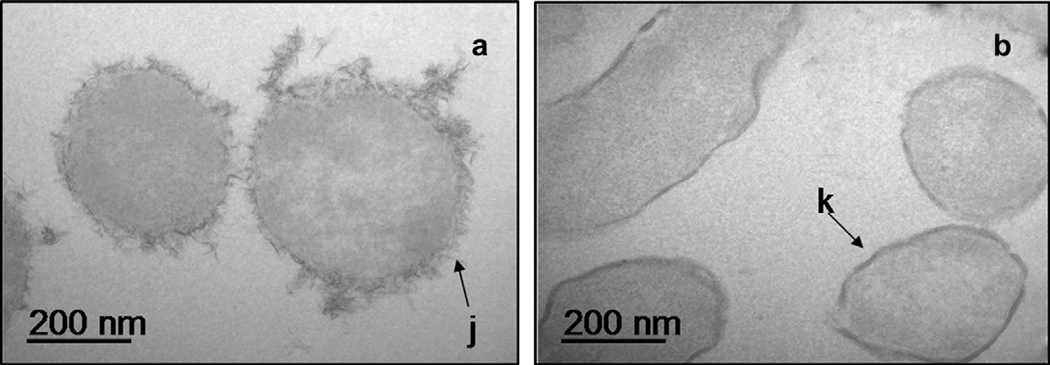
We collected TEM images of E. coli cells used in the adsorption and intracellular uptake experiments to evaluate the impact of Cu on the cells and to help identify the sites of Cu-binding (Fig. 11a and b). Prior to Cu adsorption, E. coli cells looked “healthy” and had an intact lipopolysaccharide (LPS) layer (Fig. 11a). After exposure to Cu, the LPS layers appear to be absent (Fig. 11b). The polysaccharide chains of LPS extend outward for a distance of about 10 nm from the surface of the outer-membrane, and the hydrophobic barrier made by the LPS is stabilized by structural cations such as Ca2+ and Mg2+ that link adjacent molecules via salt bridging (Beveridge and Koval, 1981; Caroff and Karibian, 2003). Copper can displace the weakly bonded salt bridges, disrupting the LPS and outer membrane while leaving the integrity of the cell wall intact (e.g., Borrok et al., 2004). The location of adsorbed Cu is distinguished by the electron dense outer ring of the E. coli cells (Fig. 11b) that was not present in the control specimen (Fig. 11a). It appears that most Cu is associated with the E. coli surface and/or periplasmic space, and lesser amounts of Cu probably penetrated inside the cell.
Fig. 11.
Transmission electron micrographs of (a) E. coli with intact LPS (arrow “j”) grown in TSB with 0.5% yeast extract and washed five times with 0.01 M NaClO4 to remove residual media and (b) E. coli after adsorption with 10 mg/L Cu(II) and 5 g/L bacteria. The electron dense region (arrow “k”) reflects Cu accumulation at the outer rim of cells.
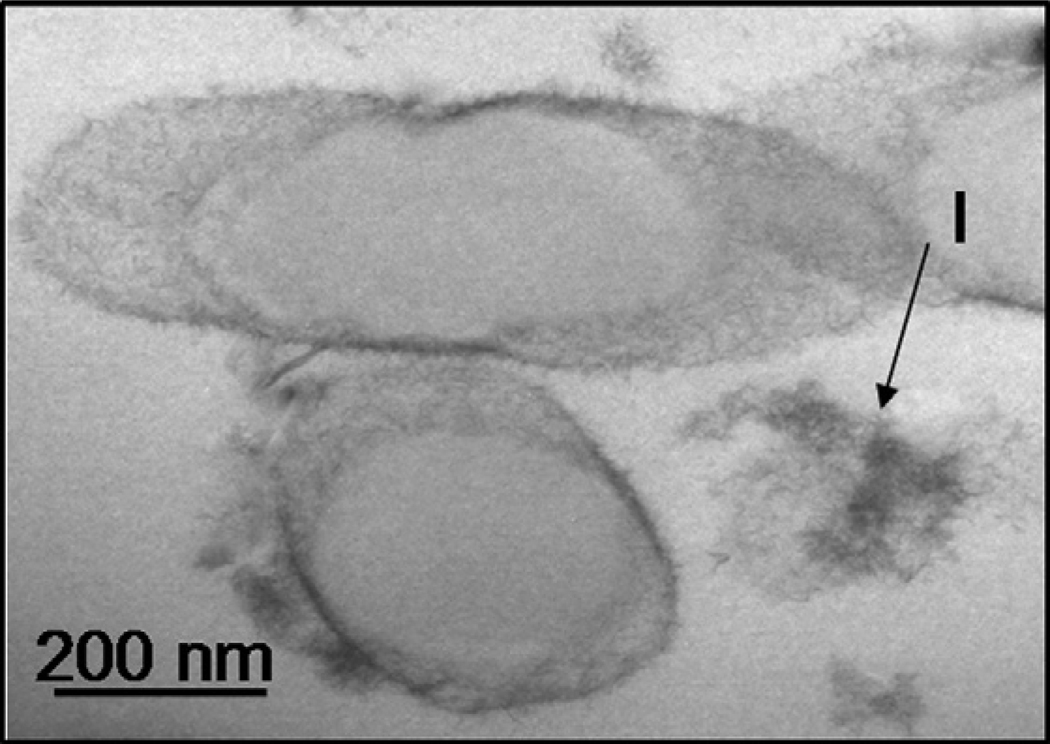
Prior to Cu adsorption, B. subtilis cells had thick, intact cell walls coated with some EPS residue. Exposure to Cu apparently stressed the cells and in some cases of prolonged exposure induced sporulation. Fig. 12 shows a TEM image of B. subtilis cells that were sporulating after exposure to Cu. Cu-induced sporulation of Bacillus cells has been documented previously by Kolodziej and Slepecky (1964). Copper (as indicated by the darker electron dense regions) appears to be sequestered between the outer coat of the spore and the inner membrane of the parent cell (Fig. 12). Ghosal et al. (2010) reported similar behavior for several monovalent metal cations which accumulated in the outer structures of Bacillus spores.
Fig. 12.
Transmission electron micrograph of B. subtilis cells in the process of sporulation after prolonged exposure (>72 h) to Cu(II) (experiment with 10 mg/L Cu(II) and 5 g/L bacteria). The electron dense region between the outer structure of the spores and the inner membrane of the parent cells reflects Cu accumulation (arrow “l” identifies lysed cell components).
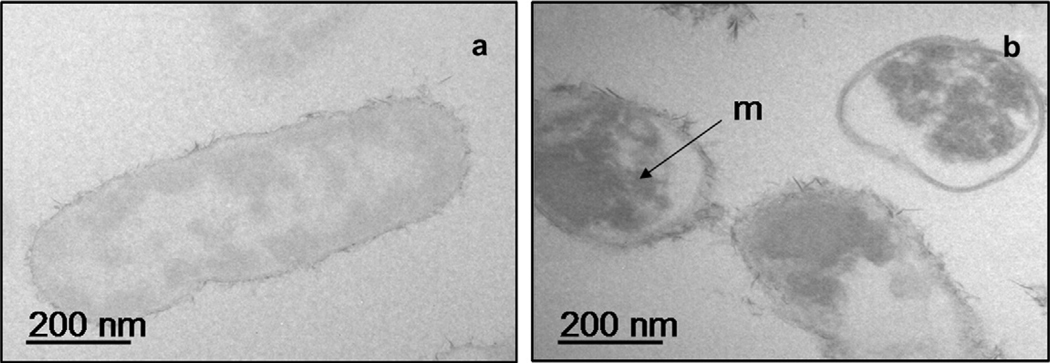
TEM images of E. coli cells before and after the intracellular uptake experiments (grown in basal media or basal media + Cu(II)-citrate), are presented in Fig. 13a and b. Copper, represented by the electron dense material can be seen inside all of the bacterial cells (Fig. 13b). This supports the suggestion that isotopic changes in these experiments were related to intracellular uptake and not to surface reactions.
Fig. 13.
Transmission electron micrographs of (a) washed E. coli cells grown in basal media and (b) E. coli grown in basal media amended with Cu-citrate. The image shows distinct electron-dense regions of Cu accumulation within the cells (arrow “m”).
3.6. Mechanisms of Cu isotope fractionation
Although the distribution of Cu in the adsorption experiments is largely controlled by cell surface reactions, the fractionation of Cu isotopes appears to be controlled by active metabolic processes. This is best demonstrated through our control adsorption experiment using dead but intact bacteria cells. Because metabolic functions were disabled in this case, we assume that the fractionation of Cu isotopes is attributable to complexation reactions with deprotonated organic-acid surface functional groups. Previous chemical and spectroscopic investigations (e.g., Beveridge and Murray, 1980; Kelly et al., 2002; Boyanov et al., 2003) suggest that deprotonated phosphoryl and carboxylic functional group sites are the dominant metal binding moieties over the pH range of 2–6. However above pH 5, metal-carboxyl complexes (often bi-dentate in structure) are the dominant metal surface species (Boyanov et al., 2003; Kretschmer et al., 2004). In our experiment, Cu isotopes were not fractionated beyond analytical uncertainties for most points below pH 4, but the bacterial cells became measurably enriched in the heavy Cu isotope at pH 4.1 and 5.1 (Fig. 3). This behavior is similar to that described by Bigalke et al. (2010) for the complexation of Cu(II) with insoluble humic acid. In the Bigalke et al. (2010) investigation, the humic acid complex was enriched in 65Cu by +0.27‰. Both observations are consistent with equilibrium isotopic fractionation where the heavier Cu isotope is preferentially incorporated into the stronger bonding environment of the surface complex. Like bacterial surfaces, natural organic matter (e.g., humic acid) is rich in carboxylic functional group sites (Dube et al., 2001; Kudeyarova, 2007). Cu binds strongly to these sites, often forming inner-sphere complexes with shorter Cu–O bond distances than those for the Cu(H2O)62+ aqueous complex (Korshin et al., 1998; Karlsson et al., 2006; reviewed in Bigalke et al. (2010)). This view is also consistent with recent reports of dissolved Cu in river waters (presumably bound to organic ligands) enriched in the heavier Cu isotope relative to bulk igneous rocks (Vance et al., 2008).
Pokrovsky et al. (2008) also reviewed available literature on the bonding distances of Cu–organic ligand complexes, but came to a different conclusion. They suggested that the bonding environments for Cu–organic complexes were similar enough to Cu(H2O)62+ that isotopic fractionation was negligible or that the lighter isotope of Cu would be preferentially incorporated into the surface complex (at least under acidic pH conditions where phosphoryl sites dominate). This explanation provided support for their Cu isotopic data collected during a set of bacteria adsorption experiments (Pokrovsky et al., 2008). In these experiments (with live bacterial cells), no systematic Cu fractionation was observed except under acidic pH conditions where the lighter Cu isotope was preferentially associated with the bacteria. A possible alternate interpretation of these results is that Cu isotopes were impacted by active metabolic transport reactions in addition to surface complexation.
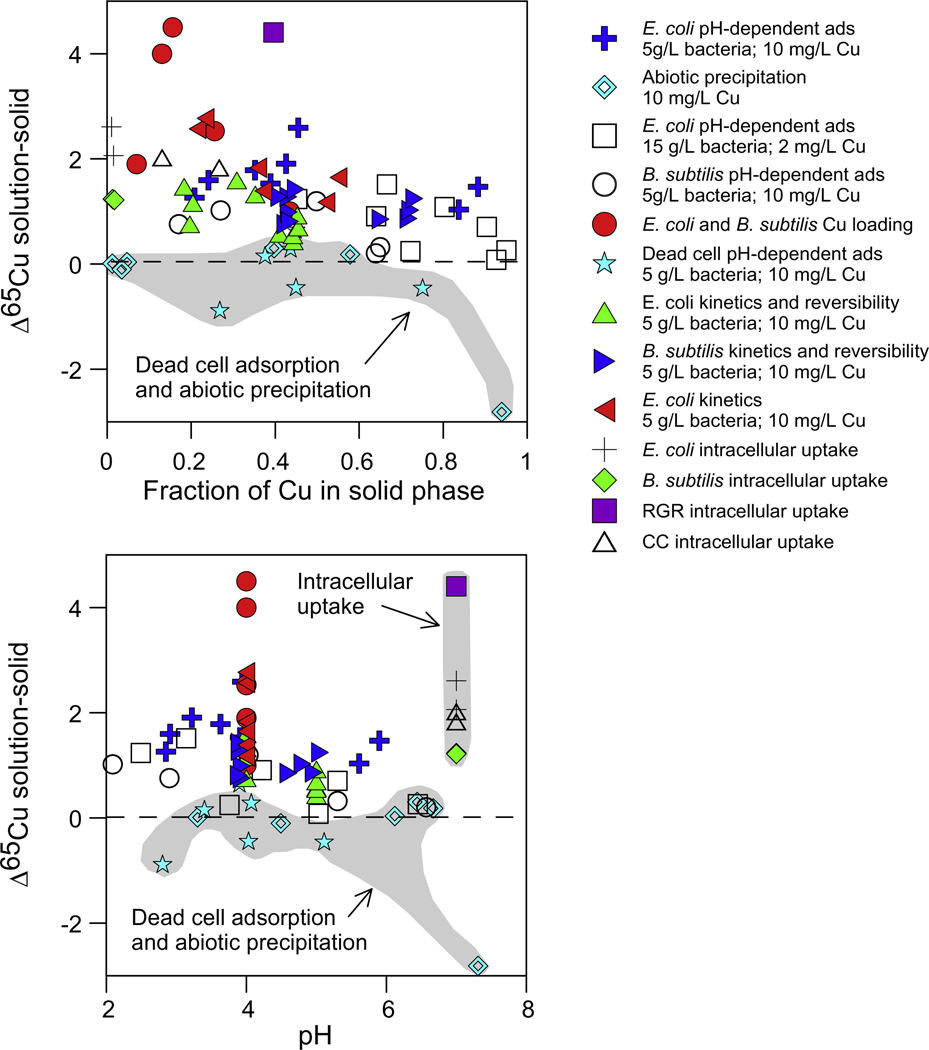
In our study, Cu adsorption experiments using live cells, as well as active growth experiments with Cu(II)-citrate, resulted in substantial isotopic fractionation of Cu. In all cases, the lighter Cu isotope was preferentially sequestered by the bacteria. For comparison, the apparent separation factors (Δ65Cusolution–solid, Eq. (2)) for all experimental data are presented in Fig. 14. The lack of correlation between changes in δ65Cu and the extent of adsorption, the similarity of the results among the adsorption and growth experiments, and the preference for incorporation of the lighter Cu isotope all suggest that the mechanisms of fractionation involve active regulation of Cu in the cells. Both E. coli and B. subtilis contain proteins to traffic, deliver, and regulate Cu. Most Cu which is incorporated into proteins and enzymes is reduced from Cu(II) to Cu(I) or is bound to enzymes used in catalysis where the Cu(I)–Cu(II) redox gradient is exploited (Waldron and Robinson, 2009). Active transport of Cu was possible in the adsorption experiments in the absence of growth media, because the cells were able to use stored energy reserves. The internal cycling of Cu in this case may have been triggered by the large amounts of free Cu(II) in these experiments.
Fig. 14.
Δ65Cusolution–solid calculated via isotopic mass balance versus (a) the fraction of Cu sequestered and (b) pH for all experiments.
Once Cu traverses the outer membrane of E. coli (probably via porins), it is regulated by 4 homeostatic mechanisms involving specific Cu binding proteins housed in the periplasmic space, and is controlled in the cytoplasm by three additional Cu proteins (Nikaido and Vaara, 1985; Rensing and Grass, 2003; Bertini et al., 2010). The periplasmic copper proteins found in E. coli are: copper-resistance protein (CopC) which binds both Cu(I) and Cu(II) at two distinct sites, Cu-transport protein (CusF) which binds Cu(I), Cu–Zn superoxide dismutase (SOD) and amine oxidase catalytic domain (amine oxidase) which both have sites to change the oxidation state of Cu when coupled to electron transfer, catalysis or oxygen binding (Arnesano et al., 2003; Banci et al., 2005; Bertini et al., 2010). Two of the three Cu cytoplasmic proteins, CueR and CsoR, are transcription factors which serve in transcription regulation and contain Cu(I) sites with two Cys ligands and an additional His in the case of CsoR (Bertini et al., 2010). The third cytoplasmic protein, YhcH-like, has a role in sialic acid catabolism (Teplyakov et al., 2005), however the oxidation state of the incorporated Cu is unknown. Combined NMR and EXAFS examination by Banci et al. (2005) has shown that the CopC protein binds Cu(II) via two histidine residues and two oxygen ligands at bond distances of 1.99 and 1.97 Å, respectively. Cu(I) in the same protein is bound to three S atoms and one histidine at distances of 2.30 and 1.9 Å, respectively.
Bacillus subtilis similarly regulates Cu through Cu chaperone proteins and efflux systems (e.g., Smaldone and Helmann, 2007) and contains CueR, CsoR and membrane-bound CopC. Additionally, B. subtilis has copper transport proteins (e.g., CopZ) with a heavy metal-associated domain in the cytoplasm and membrane as well as the membrane anchored protein, glutathione peroxidase-like (Sco1) (Banci et al., 2005; Bertini et al., 2010). Gram positive bacteria, such as B. subtilis, appear to prevent possible damage to the cell by using membrane-anchored chaperones outside the cell, such as Sco1, to load Cu into the cytoplasm (Bertini et al., 2010).
Differences in the locations of the Cu transport proteins that serve as the initial holding places for Cu before it is transferred inside the cellular membrane may explain why in adsorption experiments some Cu was irreversibly bound to E. coli but not B. subtilis. Copper-transport proteins in Gram-positive bacteria are anchored to the cell membrane, but extend to the outer reaches of the cell wall (Mattatall et al., 2000). Copper-transport proteins in Gram-negative bacteria like E. coli are housed within the periplasmic space (Finney and O’Halloran, 2003; Bertini et al., 2010). Copper associated with chaperones in the periplasmic space would not be easily removed by washing, whereas Cu associated with proteins near the cell wall/solution interface is likely to be easily removed.
Much of the intracellular Cu machinery described above appears to be “universal” among bacterial species. Bertini et al. (2010) recently used the Protein Data Bank to systematically analyze copper binding sites for all proteins that typically carry at least one Cu atom. Out of the 57 representative organisms investigated, only five were found lacking in Cu machinery; however, these five organisms were obligate host-associated microbes capable of obtaining the needed molecules from their hosts (Bertini et al., 2010). Many of the Cu binding proteins associated with bacteria are shared by plants and higher organisms, including humans (Harrison et al., 2000; Solioz, 2002).
The preference for the lighter Cu isotope by bacteria likely reflects the combination of kinetic affects rising from the rapid and sometimes irreversible incorporation of Cu and the reduction of Cu(II) to Cu(I). Cu isotopes are additionally impacted in the adsorption experiments by cell surface complexation reactions; however, in most cases this affect is probably small relative to the kinetic and redox mechanisms. The biologic redox affect is similar in many ways to the abiotic redox cycling of Cu in nature. Cu(II)-bearing minerals are typically isotopically heavier than co-evolved Cu(I)-bearing minerals by 1–3‰ (e.g., Asael et al., 2007). Similarly, experimental reduction of Cu(II) to form a Cu(I) sulfide mineral resulted in a 3‰ shift where the lighter Cu isotope was enriched in the sulfide phase (Ehrlich et al., 2004). This affect is even seen during the oxidative leaching of Cu(I) minerals in that aqueous Cu(II) becomes enriched in the heavier Cu isotope (Mathur et al., 2005; Fernandez and Borrok, 2009; Kimball et al., 2009). For bacterial cells, however, the redox fractionation mechanism cannot explain our data unless the isotopic changes of Cu inside the cell are translated to isotopic changes in the bulk solution. In other words, the Cu(I) accumulated in the cell must equilibrate with the Cu(II) in bulk solution. This could occur through transfer of isotopically heavy Cu out of the cell through efflux pumps or via atom exchange reactions on the cell wall or in the periplasmic space. Our Cu-loading experiments may suggest that the degree of Cu stress plays an important part in the process. As more Cu was loaded the bacteria cells became further enriched in the lighter Cu isotopes. Bacteria often contain sensing mechanisms that respond to increasing levels of Cu stress (Rouch et al., 1985; Cobine et al., 1999). When extracellular concentrations of Cu reach about 10 µM, the internal Cu machinery (cop operon transcription) is up-regulated (Pécou et al., 2006). As Cu concentrations increase, greater efflux of Cu from the cell is expected (Rensing and Grass, 2003).
Our findings also support the earlier work by Zhu et al. (2002) which demonstrated that specific Cu-binding proteins isolated from bacteria and yeast incorporated the lighter Cu isotope relative to the starting solution. Furthermore, Pokrovsky et al. (2008) described similar isotopic fractionation during Cu adsorption onto Pseudomonas aurofaciens over the pH range 2–4; however, they attributed this to differences in bonding environment and not to the metal trafficking machinery of the cells. These findings could also be consistent with a biological control for Cu isotopic signatures in natural waters. Vance et al. (2008) suggested that Cu–organic ligand complexes are responsible for the enrichment of heavy Cu isotopes in dissolved riverine water; however, preferential incorporation of the lighter Cu isotope into microorganisms and plants could produce the same affect.
The apparent separation factors for all experimental data (Fig. 14) show a large degree of non-systematic variation in the magnitude of Cu isotope fractionation as they appear to be independent of pH and fraction adsorbed. This variability was also documented in the Pokrovsky et al. (2008) investigation and for different proteins in the Zhu et al. (2002) investigation. Although the temptation with a highly-variable dataset is to dismiss its relevance, we believe that in this case the variability of the data is relevant. Once we understand that the observed fractionations are not related to simple cell surface adsorption (this is why we refer to them as “apparent” separation factors), but are a reflection of active cellular function, variation in the magnitude of fractionation seems logical. The δ65Cu is impacted not only by the types and abundances of bacterial Cu-binding proteins, but also by the time and extent of Cu exposure, the pH, and the degree of cell death, cell lysis, and in some cases sporulation (without a redox change, Fig. 12). Additional differences between adsorption and intracellular incorporation experiments include the localities of Cu binding (see Figs. 11–13) and the availability of nutrients. Collectively, these observations suggest Cu isotopes are sensitive probes of complex intracellular Cu machinery and could be used as tools to better understand these reactions.
4. CONCLUSIONS
The fractionation of Cu isotopes by bacteria is a complex process that can be only partly constrained through bulk adsorption and intracellular uptake experiments. Additional experiments are needed that isolate Cu transport through the cell membrane, Cu regulation in the periplasmic space (for Gram-negative species), and transfer of Cu into the cytoplasm. Future work should focus on linking Cu isotopic fractionation to these specific reactions and on spectroscopic work to constrain the bonding environments of internalized and surface-bound Cu. With this initial work, however, we can conclude the following:
Cu complexation with organic acid functional groups present on dead bacterial surfaces exhibits a preference for the heavier Cu isotope at pH 4.1 and 5.1 (and presumably at higher pHs). This observation is consistent with a recent study by Bigalke et al. (2010) where the heavier Cu isotope was preferentially incorporated into insolubilized humic acid. However, because the magnitude of the fractionation is small relative to active metabolic processes, it seems likely that intracellular biological processes also exert an important control on the δ65Cu of natural waters.
The behavior of Cu during bacterial surface interactions is more complex than previously thought based on Cu concentration data alone. Although the distribution of Cu in these experiments is controlled by surface adsorption, the isotopic changes suggest that some Cu enters the cells. In the case of E. coli, some of this Cu becomes irreversibly bound. Metal isotopic techniques can provide new insights into the mechanisms and binding environments associated with bacterial surface adsorption that were previously obscured in the metal concentration data.
The magnitude and time-dependence of the Cu isotopic fractionation measured for the adsorption experiments with live cells probably reflects the metal transfer rate across the intracellular membrane and the reduction of Cu(II) to Cu(I) by the available proteins within the internal Cu regulatory systems. If true, Cu isotopes might be used as a chemical tool to gather valuable toxicological information and to learn about the mechanisms of Cu-bearing enzymes and proteins. Because there is structural and functional conservation among bacteria, yeast and humans (Peña et al., 1999; Solioz, 2002), this tool could provide insights for several important biological systems.
Regardless of the experimental conditions (i.e., growth, media, species, form of Cu, etc.), live bacteria cells preferentially incorporated the lighter Cu isotope and left the solution enriched in the heavy Cu isotope. Because abiotic reactions such as adsorption (e.g., Balistrieri et al., 2008; Pokrovsky et al., 2008) and precipitation (without a redox change, Fig. 1a) preferentially incorporate the heavier Cu isotope into the solid phase, we can identify a “Cu isotope biosignature” in these simple systems. Because the Cu binding proteins associated with bacteria in this study are shared by many other bacterial species, fungi, plants, and higher organisms (Peña et al., 1999; Harrison et al., 2000; Solioz, 2002), Cu isotopes appear to be well-suited tools for the discovery of biological markers on Earth. It is also likely, however, that a variety of abiotic chemical reactions play a complicating role.
Supplementary Material
ACKNOWLEDGMENTS
This publication was made possible through funding by the National Science Foundation (NSF) Grant 0745345, the Center for Earth and Environmental Isotope Research (NSF MRI Grant 0820986) and by Grant No. 2G12RR008124-16A1 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). Transmission electron microscopy was provided by the Analytical Cytology Core Facility at the Border Biomedical Research Center at UTEP funded by NIH Grant # 2G12RR08124-16A1. The publication contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH. Graduate funding for J. Navarrete was partly provided through the University of Texas System Louis Stokes Alliance for Minority Participation (LSAMP), NSF Grant HRD-0832951. We would like to thank Jasper G. Konter and Minghua Ren for their editorial and analytical support, respectively. The manuscript was improved through insightful suggestions from three anonymous reviewers and Associate Editor, Clark Johnson.
Footnotes
APPENDIX A. SUPPLEMENTARY DATA
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.gca.2010.11.011.
REFERENCES
- Alakomi HL, Saarela M, Helander I. Effect of EDTA on Salmonella enterica serovar Typhimurium involves a component not assignable to lipopolysaccharide release. Microbiology. 2003;149:2015–2021. doi: 10.1099/mic.0.26312-0. [DOI] [PubMed] [Google Scholar]
- Archer C, Vance D. Mass discrimination correction in multiple-collector plasma source mass spectrometry: an example using Cu and Zn isotopes. J. Anal. At. Spectrom. 2004;19:656–665. [Google Scholar]
- Arnesano F, Banci L, Bertini I, Mangani S, Thompsett AR. A redox switch in CopC: an intriguing copper trafficking protein that binds copper(I) and copper(II) at different sites. Proc. Nat. Acad. Sci. USA. 2003;100:3814–3819. doi: 10.1073/pnas.0636904100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asael D, Matthews A, Bar-Matthews M, Halicz L. Copper isotope fractionation in sedimentary copper mineralization (Timna Valley, Israel) Chem. Geol. 2007;243:238–254. [Google Scholar]
- Ashworth DJ, Alloway BJ. Complexation of copper by sewage sludge-derived dissolved organic matter: effects on soil sorption behavior and plant uptake. Water Air Soil Pollut. 2007;187:187–196. [Google Scholar]
- Balistrieri LS, Borrok DM, Wanty RB, Ridley WI. Fractionation of Cu and Zn isotopes during adsorption onto amorphous Fe(III) oxyhydroxide: experimental mixing of acid rock drainage and ambient river water. Geochim. Cosmochim. Acta. 2008;72:311–328. [Google Scholar]
- Banci L, Bertini I, Mangani S. Integration of XAS and NMR techniques for the structure determination of metallo-proteins. Examples from the study of copper transport proteins. J. Synchrotron Radiat. 2005;12:94–97. doi: 10.1107/S090904950402789X. [DOI] [PubMed] [Google Scholar]
- Barling J, Anbar AD. Molybdenum isotope fractionation during adsorption by manganese oxides. Earth Planet. Sci. Lett. 2004;217:315–329. [Google Scholar]
- Bertini I, Cavallaro G, McGreevy KS. Cellular copper management – a draft user’s guide. Coord. Chem. Rev. 2010;254:506–524. [Google Scholar]
- Beveridge TJ. Role of cellular design in bacterial metal accumulation and mineralization. Rev. Microbiol. 1989;43:147–171. doi: 10.1146/annurev.mi.43.100189.001051. [DOI] [PubMed] [Google Scholar]
- Beveridge TJ, Fyfe WS. Metal fixation by bacterial cell walls. Can. J. Earth Sci. 1985;22:1893–1898. [Google Scholar]
- Beveridge TJ, Koval SF. Binding of metals to cell envelopes of Escherichia coli K-12. Appl. Environ. Microbiol. 1981;42:325–335. doi: 10.1128/aem.42.2.325-335.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge TJ, Murray RG. Sites of metal deposition in the cell wall of Bacillus subtilis. J. Bacteriol. 1980;141:876–887. doi: 10.1128/jb.141.2.876-887.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigalke M, Weyer S, Wilcke W. Copper isotope fractionation during complexation with insolubilized humic acid. Environ. Sci. Technol. 2010;44:5496–5502. doi: 10.1021/es1017653. [DOI] [PubMed] [Google Scholar]
- Borrok D, Fein JB. Distribution of protons and Cd between bacterial surfaces and dissolved humic substances determined through chemical equilibrium modeling. Geochim. Cosmochim. Acta. 2004;68:3043–3052. [Google Scholar]
- Borrok DM, Fein JB. The impact of ionic strength on the adsorption of protons, Pb, Cd, and Sr onto the surfaces of Gram negative bacteria: testing non-electrostatic, diffuse and triple-later models. J. Colloid Interface Sci. 2005;286:110–126. doi: 10.1016/j.jcis.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Borrok D, Fein JB, Tischler M, O’Loughlin E, Meyer H, Liss M, Kemner K. The effect of acidic solutions and growth conditions on the adsorptive properties of bacterial surfaces. Chem. Geol. 2004;207:107–119. [Google Scholar]
- Borrok DM, Wanty RB, Ridley WI, Wolf R, Lamothe PJ, Adams M. Separation of copper, iron, and zinc from complex aqueous solutions for isotopic measurement. Chem. Geol. 2007;242:400–414. [Google Scholar]
- Boyanov MI, Kelly SD, Kemner KM, Bunker BA, Fein JB, Fowle DA. Adsorption of cadmium to Bacillus subtilis bacterial cell walls: a pH-dependent X-ray absorption fine structure spectroscopy study. Geochim. Cosmochim. Acta. 2003;67:3299–3311. [Google Scholar]
- Brand LE, Sunda WG, Guillard RL. Limitation of marine phytoplankton reproductive rates by zinc, manganese and iron. Limnol. Oceanogr. 1983;28:1182–1198. [Google Scholar]
- Buchl A, Hawkesworth CJ, Ragnarsdottir KV, Brown DR. Re-partitioning of Cu and Zn isotopes by modified protein expression. Geochem. Transact. 2008;9:11. doi: 10.1186/1467-4866-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caroff M, Karibian D. Structure of bacterial lipopoly-saccharides. Carbohydr. Res. 2003;338:2431–2447. doi: 10.1016/j.carres.2003.07.010. [DOI] [PubMed] [Google Scholar]
- Cheng KJ, Ingram JM, Costerton JW. Release of alkaline phosphatase from cells of Pseudomonas aeruginosa by manipulation of cation concentration and of pH. J. Bacteriol. 1970;104:748–753. doi: 10.1128/jb.104.2.748-753.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobine P, Wickramasinghe WA, Harrison MD, Weber T, Solioz M, Dameron CT. The Enterococcus hirae copper chaperone CopZ delivers copper(I) to the CopY repressor. FEBS Lett. 1999;445:27–30. doi: 10.1016/s0014-5793(99)00091-5. [DOI] [PubMed] [Google Scholar]
- Dube A, Zbytniewski R, Kowalkowski T, Cukrowska E, Buszewski B. Adsorption and migration of heavy metals in soil. Pol. J. Environ. Stud. 2001;10:1–10. [Google Scholar]
- Ehrlich S, Butler I, Halicz L, Richard D, Oldroyd A, Matthews A. Experimental study of the copper isotope fractionation between aqueous Cu(II) and covellite, CuS. Chem. Geol. 2004;209:259–269. [Google Scholar]
- Fein JB, Daughney CJ, Yee N, Davis TA. A chemical equilibrium model for metal adsorption onto bacterial surfaces. Geochim. Cosmochim. Acta. 1997;61:3319–3328. [Google Scholar]
- Fernandez A, Borrok DM. Fractionation of Cu, Fe, and Zn isotopes during the oxidative weathering of sulfide-rich rocks. Chem. Geol. 2009;264:1–12. [Google Scholar]
- Finney LA, O’Halloran TV. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science. 2003;300:931–936. doi: 10.1126/science.1085049. [DOI] [PubMed] [Google Scholar]
- Gauthier TD, Seitz WR, Grant CL. Effects of structural and compositional variations of dissolved humic materials on pyrene Koc values. Environ. Sci. Technol. 1987;21:243–248. doi: 10.1021/es00157a003. [DOI] [PubMed] [Google Scholar]
- Ghosal S, Leighton TJ, Wheeler KE, Hutcheon ID, Weber PK. Spatially resolved characterization of water and ion incorporation in Bacillus spores. Appl. Environ. Microbiol. 2010;76:3275–3282. doi: 10.1128/AEM.02485-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green TA, Russell AE, Roy S. The development of a stable citrate electrolyte for the electrodeposition of copper – nickel alloys. J. Electrochem. Soc. 1998;145:875–881. [Google Scholar]
- Gustafsson JP. Visual MINTEQ, version 2.53. 2007 Available from: < http://www.lwr.kth.se/English/OurSoftware/vminteq/index.htm>.
- Harrison MD, Jones CE, Solioz M, Dameron CT. Intracellular copper routing: the role of copper chaperones. Trends Biol. Sci. 2000;25:29–32. doi: 10.1016/s0968-0004(99)01492-9. [DOI] [PubMed] [Google Scholar]
- Hersman LE, Forsythe JH, Ticknor LO, Maurice PA. Growth of Pseudomonas mendocina on Fe(III)(hydr)-oxides. Appl. Environ. Microbiol. 2001;67:4448–4453. doi: 10.1128/AEM.67.10.4448-4453.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S, Tsakova V. Influence of copper anion complexes on the incorporation of metal particles in polyaniline. Part I: copper citrate complex. J. Appl. Electrochem. 2002;32:701–707. [Google Scholar]
- Karlsson T, Persson P, Skyllberg U. Complexation of copper(II) in organic soils and in dissolved organic matter – EXAFS evidence for chelate ring structures. Environ. Sci. Technol. 2006;40:2623–2628. doi: 10.1021/es052211f. [DOI] [PubMed] [Google Scholar]
- Kelly SD, Kemner KM, Fein JB, Fowle DA, Boyanov MI, Bunker BA, Yee N. X-ray absorption fine structure determination of pH-dependent U-bacterial cell wall interactions. Geochim. Cosmochim. Acta. 2002;66(2):3855–3871. [Google Scholar]
- Kimball BE, Mathur R, Dohnalkova AC, Wall AJ, Runkel RL, Brantley SL. Copper isotope fractionation in acid mine drainage. Geochim. Cosmochim. Acta. 2009;73:1247–1263. [Google Scholar]
- Koch KA, Marjorette M, Penña MMO, Thiele DJ. Copper-binding motifs in catalysis, transport, detoxification and signaling. Chem. Biol. 1997;4:549–560. doi: 10.1016/s1074-5521(97)90241-6. [DOI] [PubMed] [Google Scholar]
- Kolodziej B, Slepecky RA. Trace metal requirements for sporulation of Bacillus megaterium. J. Bacteriol. 1964;88:821–830. doi: 10.1128/jb.88.4.821-830.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshin GV, Frenkel AI, Stern EA. ESAFS study of the inner shell structure in copper(II) complexes with humic substances. Environ. Sci. Technol. 1998;32:2699–2705. [Google Scholar]
- Kretschmer XC, Meitzner G, Gardea-Torresday JL, Webb R. Determination of Cu environments in the cyanobacterium Anabaena flos-aquae by X-ray absorption spectroscopy. Appl. Environ. Microbiol. 2004;70:771–780. doi: 10.1128/AEM.70.2.771-780.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudeyarova AY. Application of basic chemical concepts to understanding the formation and transformation mechanisms of humic substances: a revue of publication and own experimental data. Soil Chem. 2007;40:934–948. [Google Scholar]
- Larson PB, Maher K, Ramos FC, Chang Z, Gaspar M, Meinert LD. Copper isotope ratios in magmatic and hydrothermal ore-forming environments. Chem. Geol. 2003;201:337–350. [Google Scholar]
- Ma H, Kim SD, Cha DK, Allen HE. Effect of kinetics of complexation by humic acid on toxicity of copper to Ceriodaphnia dubia. Environ. Toxicol. Chem. 1999;18:28–837. [Google Scholar]
- Markl G, Lahaye Y, Schwinn G. Copper isotopes as monitors of redox processes in hydrothermal mineralization. Geochim. Cosmochim. Acta. 2006;70:4215–4228. [Google Scholar]
- Mathur R, Ruiz J, Titley S, Liermann L, Buss H, Brantley S. Cu isotopic fractionation in the supergene environment with and without bacteria. Geochim. Cosmochim. Acta. 2005;69:5233–5246. [Google Scholar]
- Mathur R, Titley S, Barra F, Brantley S, Wilson M, Phillips A, Munizaga F, Maksaev V, Vervoort J, Hart G. Exploration potential of Cu isotope fractionation in porphyry copper deposits. J. Geochem. Explor. 2009;102:1–6. [Google Scholar]
- Mattatall NR, Jazairi J, Hill BC. Characterization of YpmQ, an accessory protein required for the expression of cytochrome c oxidase in Bacillus subtilis. J. Biol. Chem. 2000;275:28802. doi: 10.1074/jbc.M002741200. [DOI] [PubMed] [Google Scholar]
- Meador JP. The interaction of pH, dissolved organic carbon, and total copper in the determination of ionic copper and toxicity. Aquat. Toxicol. 1991;19:13. [Google Scholar]
- Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybroe O, Brandt KK, Ibrahim YM, Tom-Petersent A, Holm PE. Differential bioavailability of copper complexes to bioluminescent Pseudomonas fluorescens reporter strains. Environ. Toxicol. Chem. 2008;27:2246–2252. doi: 10.1897/08-025.1. [DOI] [PubMed] [Google Scholar]
- Pécou E, Maass A, Remenik D, Briche J, Gonzalez M. A mathematical model for copper homeostasis in Enterococcus hirae. Math. Biosci. 2006;203:222–239. doi: 10.1016/j.mbs.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Peña MMO, Lee J, Thiele DJ. A delicate balance: homeostatic control of copper uptake and distribution. J. Nutr. 1999;129:1251–1260. doi: 10.1093/jn/129.7.1251. [DOI] [PubMed] [Google Scholar]
- Pokrovsky OS, Viers J, Enmova EE, Kompantseva EI, Freydier R. Copper isotope fractionation during its interaction with soil and aquatic microorganisms and metal oxy(hydr)oxides: possible structural control. Geochim. Cosmochim Acta. 2008;72:1742–1757. [Google Scholar]
- Pribil MJ, Wanty RB, Ridley WI, Borrok DM. Influence of sulfur-bearing polyatomic species on high precision measurements of Cu isotopic composition. Chem. Geol. 2010;272:49–54. [Google Scholar]
- Rensing C, Grass G. Escherichia coli mechanisms of copper homeostasis in a changing environment. Microbiol. Rev. 2003;27:197–213. doi: 10.1016/S0168-6445(03)00049-4. [DOI] [PubMed] [Google Scholar]
- Rouch D, Camakaris J, Lee BTO, Luke RKJ. Inducible plasmid-mediated copper resistance in Escherichia coli. J. Bacteriol. 1985;154:1263–1268. [Google Scholar]
- Smaldone GT, Helmann JD. CsoR regulates the copper efflux operon copZA in Bacillus subtilis. Microbiology. 2007;153:4123–4128. doi: 10.1099/mic.0.2007/011742-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RM, Martell AE. Critical Stability Constants. V. 1. New York and London: Plenum Press; 1978. [Google Scholar]
- Solioz M. Role of proteolysis in copper homoeostasis. Biochem. Soc. Trans. 2002;4:688–691. doi: 10.1042/bst0300688. [DOI] [PubMed] [Google Scholar]
- Teplyakov A, Obmolova G, Toedt J, Galperin MY, Gilliland GL. Crystal structure of the bacterial YhcH protein indicates a role in sialic acid catabolism. Am. Soc. Microbiol. 2005;187:5520–5527. doi: 10.1128/JB.187.16.5520-5527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance D, Archer C, Bermin J, Perkins J, Statham PJ, Lohan MC, Ellwood MJ, Mills RA. The copper isotope geochemistry of rivers and the oceans. Earth Plant. Sci. Lett. 2008;274:204–213. [Google Scholar]
- Waldron KJ, Robinson NJ. How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol. 2009;7:25–35. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
- Wasylenki LE, Anbar AD, Liermann LJ, Mathur R, Gordon GW, Brantley SL. Isotope fractionation during microbial metal uptake measured by MC-ICP-MS. J. Anal. At. Spectrom. 2007;22:905–910. [Google Scholar]
- Wilkinson SG. The sensitivity of Pseudomonads to ethylenediaminetetra-acetic acid. J. Gen. Microbiol. 1967;47:67–76. doi: 10.1099/00221287-47-1-67. [DOI] [PubMed] [Google Scholar]
- Zhu XK, Guo Y, Williams RJP, O’Nions RK, Matthews A, Belshaw NS, Canters GW, de Waal EC, Weser Ul, Burgess BK, Salvato B. Mass fractionation processes of transition metal isotopes. Earth Plant. Sci. Lett. 2002;200:47–62. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.