Abstract
We used a systems biological approach to study innate and adaptive responses to influenza vaccination in humans, during 3 consecutive influenza seasons. Healthy adults were vaccinated with inactivated (TIV) or live attenuated (LAIV) influenza vaccines. TIV induced greater antibody titers and enhanced numbers of plasmablasts than LAIV. In TIV vaccinees, early molecular signatures correlated with, and accurately predicted, later antibody titers in two independent trials. Interestingly, the expression of Calcium/calmodulin-dependent kinase IV (CamkIV) at day 3 was inversely correlated with later antibody titers. Vaccination of CamkIV −/− mice with TIV induced enhanced antigen-specific antibody titers, demonstrating an unappreciated role for CaMKIV in the regulation of antibody responses. Thus systems approaches can predict immunogenicity, and reveal new mechanistic insights about vaccines.
Annual vaccination is one of the most effective methods to prevent influenza1. Currently, two types of seasonal influenza vaccines are licensed in the USA: the trivalent inactivated influenza vaccine (TIV), given by intramuscular injection and the live attenuated influenza vaccine (LAIV), administered intra-nasally. These vaccines contain 3 strains of influenza viruses that need to change annually based on the results of global influenza surveillance data2. The efficacy of influenza vaccine, therefore, depends on the match of antigenicity between vaccine and circulating influenza strains3. Additionally, other factors such as the age and immunocompetence of vaccinees, as well as pre-existing amounts of antibody derived from prior infection or vaccination, contribute to mechanisms that mediate influenza vaccine efficacy1,2,4.
Systems vaccinology has emerged as an interdisciplinary field that combines systems wide measurements and network and predictive modeling applied to vaccinology5. Recently a systems biology approach has been used to identify early gene signatures that correlate with, and predict the later immune responses in humans vaccinated with the live attenuated yellow fever vaccine YF-17D6,7. YF-17D is one of the most successful vaccines ever developed8,9, and stimulates polyvalent innate responses10 and adaptive11 immune responses that can persist for decades after vaccination11. Although systems biological approaches have been used to predict the immunogenicity of YF-17D6,7, a live replicating virus, the extent to which such approaches can be applied to predicting the immunogenicity of inactivated vaccines is unknown. Furthermore, the question of whether systems approaches can predict the immunogenicity of recall responses, as in the case of influenza where the immune response to vaccination is greatly enhanced by the past history of the vaccine recipient, both in terms of prior infections and vaccinations, remains unclear. Importantly, whether such approaches can provide insight into the immunological mechanisms of action of vaccines, and help discover new correlates of protective immunity, is untested. To address these issues, we conducted a series of clinical studies during the annual influenza seasons in 2007, 2008 and 2009, in which healthy young adults were vaccinated with TIV. The goal was to undertake a detailed characterization of the innate and adaptive responses to vaccination with TIV to identify putative early signatures that correlated with or predicted the later immunogenicity, and obtain new insight into the mechanisms underlying immunogenicity.
The results of these studies demonstrate that systems biology approaches can indeed be used to predict the immunogenicity of an inactivated vaccine TIV, with up to 90% accuracy. Interestingly, the expression of one of the genes in the predictive signature at day 3, encoded the Calcium/calmodulin-dependent kinase IV (CaMKIV), and was inversely correlated with plasma hemagglutination inhibition (HAI) antibody titers at day 28. Vaccination of CamkIV −/− mice with TIV induced enhanced antigen-specific antibody titers, demonstrating an unappreciated role for CaMKIV in the regulation of antibody responses. Together, these results demonstrate the utility of systems biology not only in predicting vaccine immunogenicity, but also in offering novel insight into the molecular mechanism of influenza vaccines.
RESULTS
Antibody responses induced by TIV and LAIV
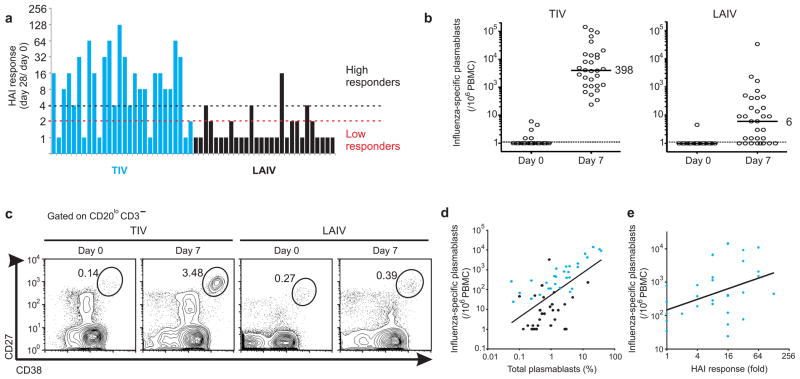
We evaluated the antibody responses in 56 healthy young adults who received either LAIV (28 subjects) or TIV (28 subjects) vaccines, during the 2008 influenza season. Hemagglutination inhibition (HAI) titers for each of the 3 influenza strains contained in LAIV and TIV vaccines were determined using the plasma of vaccinees at baseline (day 0) and at 28 days post-vaccination. The magnitude of antibody responses to influenza vaccine (hereafter referred to as “HAI response”) was measured by the maximum fold-change increase in HAI titers at day 28, relative to the baseline titers, against any of the 3 influenza strains contained in the vaccine (Fig. 1a). The mean HAI response of TIV vaccinees was 6-fold higher than those of LAIV vaccinees (t-test P-value < 0.0001) (Fig. 1a), consistent with numerous previous reports1,12,13. Furthermore, among the TIV vaccinees, there was marked variation (>100 fold) in the magnitude of the HAI response (Fig. 1a). According to FDA’s Guidance for Industry14, seroconversion can be defined by an HAI titer ≥1:40 and a minimum 4-fold increase in antibody titer post vaccination. Thus, we operationally classified the vaccinees into either low HAI responders or high HAI responders, based on whether or not a 4-fold post vaccine increase occurred (Fig. 1a). Most of the TIV vaccinees (22/ 28) were considered high responders and only 6 subjects were classified as low responders. In contrast, most of LAIV vaccinees (24/ 28) were in the low responder group and only 4 were considered high responders (Fig. 1a).
Figure 1.
Analysis of humoral immunity to influenza vaccination. (a) Antibody response determined by HAI titers on the plasma of TIV (blue bars) and LAIV (black bars) vaccinees on day 28 post-vaccination. The bars represent the highest HAI response (fold-change day 28/ day 0) among all 3 influenza strains contained in the vaccine. Subjects were classified as “Low responders” if no increase higher than 2-fold was observed in the HAI response and as “High responders” if the HAI titers on day 28 is ≥4 times higher than the titers at baseline. (b) PBMCs collected from all vaccinees were assayed for influenza-specific IgG secreting plasmablasts by ELISPOT assay at 0 and 7 days after vaccination. Each sample was measured in duplicate, averaged and plotted as plasmablasts per million PBMCs. Median values are shown. (c) Flow cytometry analysis of plasmablasts in blood. The frequency of plasmablast gate (CD3−CD20−/loCD19+CD27hiCD38hi) is shown for a representative TIV (left panel) and LAIV (right panel) vaccinee. (d) Statistically significant positive correlation (Pearson r = 0.58, P-value (two-tail) < 0.0001) between the frequencies of plasmablasts at day 7 determined by flow cytometry and the number of influenza-specific IgG-secreting plasmablasts by ELISPOT on the same day. TIV and LAIV vaccinees are represented by blue and black dots, respectively. (e) Statistically significant positive correlation (Pearson r = 0.43, P-value (two-tail) = 0.02) between influenza-specific IgG secreting plasmablasts at day 7 and the antibody response at day 28 on TIV vaccinees.
Antibodies are produced by antibody-secreting B cells in blood (plasmablasts) or bone marrow and secondary lymphoid organs (fully differentiated plasma cells). High frequencies of antigen-specific plasmablasts in the blood, within a few days of vaccination, reaching a peak at day 7 have been documented15. To determine whether the early plasmablast response to influenza vaccination correlated with the later HAI response, the frequency of influenza-specific plasmablasts at baseline and 7 days post-vaccination was measured (Fig. 1b,c). As previously reported15, rapid clonal expansion of influenza-specific plasmablasts can be observed 7 days after vaccination with TIV vaccine, as measured by the ELISPOT assay (Fig. 1b), and by flow cytometry (Fig. 1c). Here we further show that the expansion of circulating IgG-secreting plasmablasts in TIV vaccinees is also higher than in LAIV vaccinees (Fig. 1b,c). A similar result was observed for IgA-secreting plasmablasts at day 7 post-vaccination (Supplementary Fig. 1a), and a very good correlation between the frequencies of plasmablasts measured by ELISPOT and flow cytometry was evident (Fig. 1d, Supplementary Fig. 1b).
Since very low HAI response was detected after LAIV vaccination, only TIV vaccinees were considered in further correlation analyses. A modest positive correlation was observed between levels of IgG-secreting plasmablasts at day 7 and HAI titer response at day 28 post-vaccination (Fig. 1e). Since the frequencies of plasmablasts return to barely detectable numbers by day 14 post-vaccination15, this correlation suggests that the later antibody response is associated with early circulation of plasmablasts in the blood of vaccinees3. However, given the modest correlation (r = 0.43), there is clearly a need for more robust correlates of immunogenicity.
Molecular signatures of influenza vaccines
We first determined whether TIV and LAIV induced molecular signatures that were detectable in the blood. To identify such signatures of immunogenicity, we first measured the concentrations of key cytokines in the sera of vaccinees on days 0, 3 and 7 post-vaccination using Luminex assays (Supplementary Fig. 2a). Ten cytokines or chemokines were selected based on their importance as key mediators of host immune responses (RANTES, IL1α, IFNα2, MIP1α, Eotaxin, IL12p70, IFN-γ, IL1β, IP-10 and MCP1). Among them, only the chemokine IP-10 (CXCL10) was significantly induced by TIV vaccine at day 3, relative to its level on day 0 (t-test p =0.0189; Supplementary Fig. 2b). None of the cytokines were significantly induced or repressed by LAIV vaccination. The concentration of IP-10 at day 3 compared to baseline were negatively correlated to the HAI response at day 28 post-vaccination (Supplementary Fig. 2c), suggesting a possible involvement of IP-10 in the antibody response. However, the correlation coefficient was modest (r = −0.48), again highlighting the need for more robust correlates of immunogenicity.
To determine in an unbiased way, the expression changes induced by influenza vaccination on a genome-wide scale, a microarray analysis was performed with the peripheral blood mononuclear cells (PBMC) collected from all 56 vaccinees on days 0, 3 and 7 post-vaccination. The expression fold-change of each subject was obtained by subtracting the day 3 or 7 log2 expression values by its corresponding baseline value. Non-informative genes were filtered out if no increase or decrease higher than 25% (1.25-fold) was observed in at least 20% of the vaccinees on day 3 or day 7 compared to baseline. After this step, three independent statistical tests were applied to the remaining genes (see Methods). Only genes identified by all 3 analyses were considered differentially expressed.
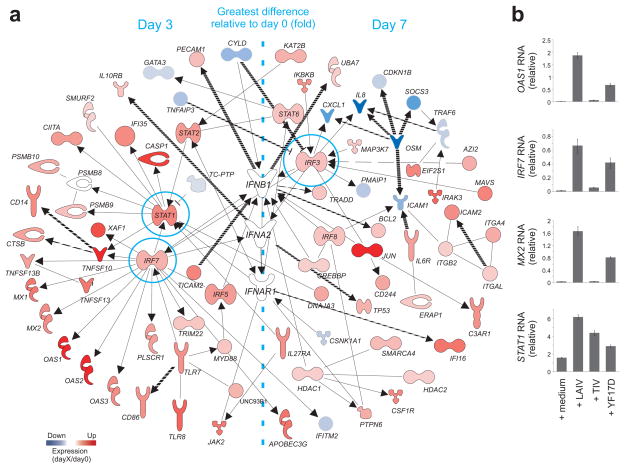
The transcriptome analysis of vaccinees revealed that LAIV and TIV vaccines induce quite different gene signatures (Supplementary Fig. 3a). However the expression of 1,445 probe sets was similarly altered by both vaccines (Supplementary Fig. 3a). Among these common differentially expressed genes (DEG), Ingenuity Pathway Analysis identified a network comprised of several genes related to inflammatory and antimicrobial responses (Supplementary Fig. 3b). These indicated that processes related to innate immunity may influence the immunogenicity of each vaccine. The complete list of DEGs after TIV and LAIV vaccination is shown in Supplementary Table 1. The expression of several interferon (IFN) related genes was altered after LAIV but not TIV vaccination (Fig. 2a,b). Type 1 interferons are central components of the innate immune response to virus16. Therefore the increased expression of type I IFN-related genes may be attributed to the replication competence of the LAIV vaccine. Our analysis identified genes highly associated with the IFN signaling pathways, such as STAT1, STAT2, TLR7, IRF3 and IRF7 (Fig. 2a). It is important to note that the fold-change difference of many IFN-related genes was highest at day 3 post-LAIV immunization (Fig. 2a).
Figure 2.
Molecular signature induced by LAIV vaccination. (a) Interferon (IFN)-related genes differentially expressed after LAIV vaccination. Solid and dashed lines represent respectively, direct and indirect interactions reported for the genes. The colors represent the mean fold-change in gene expression on days 3 or 7 compared to day 0 in all LAIV vaccinees. Genes with expression fold-change highest at day 3 or day 7 post-vaccination are shown on the left or on the right of the network, respectively. (b) Induction of key IFN-related genes was confirmed by quantitative RT-PCR. PBMCs of healthy subjects were stimulated in vitro with different vaccines for 24h. The GAPDH-normalized expression levels of OAS1, IRF7, Mx2 and STAT1 in stimulated PBMCs were compared to those of PBMCs non-stimulated.
The gene signatures of the two influenza vaccines were also compared to another live attenuated vaccine, the yellow fever YF-17D vaccine previously published6. For consistency with the previous publication, the same stringency and criteria were applied to identify the differentially expressed genes in YF17D vaccinees, which is: non-informative genes were filtered out if no increase or decrease higher than 1.41-fold was observed in at least 60% of the vaccinees on day 3 or day 7 compared to baseline; one-way analysis of variance (ANOVA) with Benjamini and Hochberg false-discovery-rate method with a cutoff of 0.05; and genes should be differentially expressed in both YF17D trials6. However, this time we performed the analysis at probe set level, as opposed to defining genes based on UniGene database. Although YF-17D vaccinees have a distinct gene expression profile compared to influenza vaccinees, many IFN-related genes are commonly induced by YF-17D and LAIV vaccines (data not shown). RT-PCR of RNA from PBMCs stimulated in vitro with influenza and YF-17D vaccines confirmed that IFN-related genes are up-regulated 24h after LAIV and YF-17D treatment and not after stimulation with TIV vaccine (Fig. 2b). Taken together, these data demonstrate that vaccination with either TIV or LAIV induces distinct molecular signatures in the blood.
Molecular signatures of sorted cell subsets
Microarray analyses of the gene expression profiles of peripheral blood mononuclear cells (PBMCs) isolated from the blood of vaccinees at baseline, and at days 3 and 7 post vaccination, were performed. One confounding variable here is that the observed transcriptional changes may either result from de novo induction of gene expression, or may simply reflect the changing cellular composition of the PBMC compartment. To overcome this issue, one approach is to isolate and identify genomic signatures of each subset within the PBMC pool. We performed microarray experiments on 4 different FACS-sorted cell types, CD19+ B cells, CD14+ monocytes, CD11chi CD123lo myeloid dendritic cells (mDC) and CD123hi CD11clo plasmacytoid dendritic cells (pDC) from 6 LAIV vaccinees and 6 TIV vaccinees. Total RNA from 96 sorted cell samples at baseline and day 7 was extracted, amplified, labeled and hybridized on Affymetrix chips (see Methods and Supplementary Fig. 4a). SAM17 was performed for each subset, separately comparing the day 7 values with the corresponding baseline values. This approach identified from hundreds to thousands of probe sets differentially expressed after TIV or LAIV vaccination (Supplementary Fig. 4b and Supplementary Table 2), demonstrating that influenza vaccines produce global expression changes on each cell type.
In TIV vaccinees, the highest number of differentially expressed genes (DEGs) was found in mDC and B cells (Supplementary Fig. 4b). However, because a substantial proportion of plasmablasts die upon freeze thawing of cells (data not shown), the DEGs observed in the B cell compartment are likely to be underestimates of the DEGs associated with plasmablasts (Fig. 3d). Nevertheless, we were still able to identify DEGs related to antibody-secreting cells (ASCs) and the unfolded protein response in sorted B cells after TIV immunization (Supplementary Fig. 4c,d). To cope with the large amount of immunoglobulin proteins that are produced, ASCs must greatly increase their secretion machinery, which may lead to an over-expression of mis-folded proteins in the endoplasmic reticulum (ER)18,19. In response to such stress, the cells activate intracellular signal transduction pathways, the unfolded protein response, which protects the cells by enhancing the capacity of the secretory apparatus and by reducing the ER load20. Two transcription factors, XBP1 and ATF6B, which are central orchestrators of the unfolded protein response, were up-regulated in sorted B cells but not in the PBMC analysis after TIV vaccination (Supplementary Fig. 4d).
Figure 3.
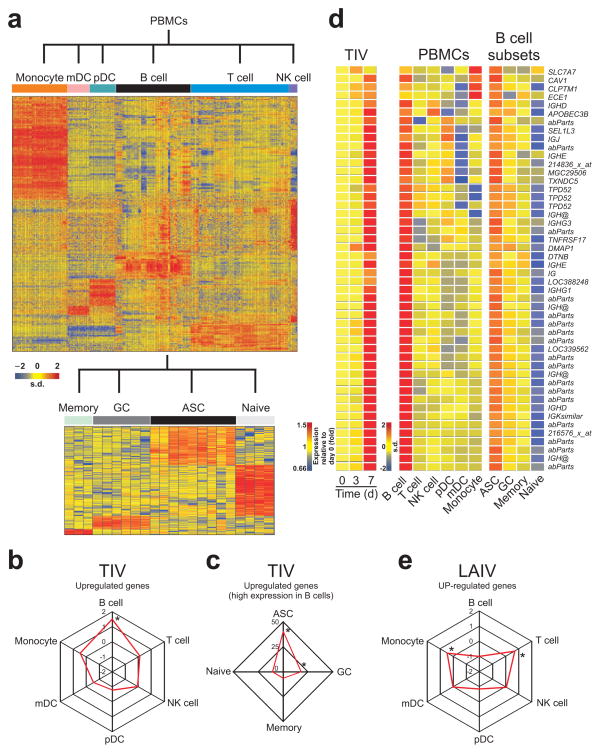
Molecular signatures induced by TIV vaccination. (a) Heat map of gene signatures of immune cells identified by meta-analysis (see Methods). Expression level of each gene (in rows) is represented by the number of standard deviations above (red) or below (blue) the average value for that gene across all samples (in columns). (b) Spider graph showing the fold enrichment of TIV up-regulated genes among the genes highly expressed in any PBMC subset. Fold enrichment is calculated as described in Methods. Cell subsets with statistically significant enrichment (Fisher’s exact test two tailed P-value < 10−10) were marked with asterisks. (c) Spider graph showing the fold enrichment of TIV up-regulated genes among the genes highly expressed in B cells and also highly expressed in a specific B cell subset. (d) Heat map of genes up-regulated by TIV vaccination and also highly expressed in B cells and antibody-secreting cells. The official gene symbol for each probe set is shown on the bottom of the heat map. Probe sets that mapped to antibody variable regions are named ‘abParts’ and those ones not annotated are represented by the Affymetrix probe ID. (e) Spider graph showing the fold enrichment of LAIV up-regulated genes among the genes highly expressed in any PBMC subset.
In LAIV vaccinees, in contrast to what was seen in TIV, the pDC subset generated the highest number of DEGs (Supplementary Fig. 4b). Of the many IFN-related genes induced by LAIV (Fig. 2a), we determined the differential expression of genes in known cell types. In fact, among the 37 IFN-related genes that were differentially expressed in PBMCs and also differentially expressed in at least one FACS-sorted cell subset, 17 and 14 were up-regulated in monocytes and pDC, respectively (Supplementary Fig. 4e, left heat map). However, among the 44 probe sets differentially expressed in at least one cell subset but not in PBMCs, most of them were up-regulated in mDC and pDC (Supplementary Fig. 4e, right heat map). These data suggest that antigen presenting cells may play a significant role in the innate response to LAIV vaccination. The high number of IFN-related genes “missing” in the PBMC analysis may be due to the fact that mDC and pDC represent, together <1% of the total PBMCs21.
This highlights the type of information that can be obtained by examining gene expression profiles in sorted cell types. However, evaluating the gene expression signatures in individual subsets of cells isolated by flow cytometry presents major challenges. The practical use of such approach is very limited, either logistically (the need to use freshly isolated samples to prevent preferential loss of certain cell types such as plasmablasts and effector T cells) or financially (the need for large numbers of gene chips). Therefore, as described below we devised an alternative strategy.
Meta-analysis of cell type specific signatures
Human PBMCs consist of many different cell types, each with a distinct transcriptome. An elegant recent study has highlighted the use of deconvolution analysis to generate cell type-specific gene expression differences in complex tissues22. We devised an independent strategy to discern cell type specific transcriptional signatures from the PBMC microarray analyses. We performed a meta-analysis of publicly available microarray studies, in which the gene expression profiles of isolated individual cell types of PBMCs (e.g. T cells, B cells, monocytes, NK cells etc) or B cells (naïve, memory, germinal center and ASCs from blood or tonsils) had been analyzed (Supplementary Fig. 5a,b). To avoid issues of cross-platform normalization and probe selection, only samples hybridized to Affymetrix Human Genome U133 Plus 2.0 Arrays or Affymetrix Human Genome U133A Arrays were used in our meta-analysis. Additionally, for each study, samples were manually removed based on the severity of the disease or treatment and/or based on the method of cell purification (the list of samples and studies used can be found in Supplementary Table 3). We included in our meta-analysis, microarray data of FACS sorted pDCs and mDCs obtained from PBMCs of TIV and LAIV vaccinees before and after vaccination (Supplementary Fig. 4a). The expression profile of a given cell subset was compared to the expression profile of all other subsets by t-test (P-value < 0.05 and mean fold-change > 2). A gene was designated as highly expressed in a particular cell type by determining the number of times the gene is up-regulated in the cell type in all possible pair-wise comparisons (Supplementary Fig. 5b and Supplementary methods). The genomic signatures of immune cells revealed using this approach (Fig. 3a and Supplementary Table 4) were then compared to the genomic signatures of influenza vaccinees.
The meta-analysis confirmed that TIV up-regulated genes are enriched by genes highly expressed in B cells (Fig. 3b) and among these, genes highly expressed in ASCs (Fig. 3c). The heat map of the ASC genes up-regulated after TIV vaccination can be seen in Fig. 3d. Among the genes up-regulated are “antibody parts”, (which represent rearranged V-D-J immunoglobulin gene segments), and several other genes that encode immunoglobulin parts (IGH@, IGHE, IGHG3, IGHG1 and IGHD), as well as TNFRSF17 (also known as BCMA23, which is a member of the BAFF-BLyS family of receptors, and previously shown to be the best predictor of neutralizing antibody responses to YF-17D6). These results confirm what was seen by flow cytometry and ELISPOT, where an increase in the frequencies of IgG and IgA ASC was observed in the blood of vaccinees at day 7 post-TIV vaccination (Fig. 1 and Supplementary Fig. 1).
In addition to the ASC signature, a signature comprised of several genes orchestrating the unfolded protein response19,20 was observed (data not shown). The high number of XBP-1 target genes differentially expressed after vaccination, is consistent with a role for XBP-1 in orchestrating plasma cell differentiation19. Among them, genes such as ATF6, MANF, CREB3, PDIA4, DNAJB11, HSP90B1, HERPUD1 and DNAJB9 are already known to be involved in the unfolded protein response24–26.
In contrast to TIV, analyses of the transcriptional signature induced by LAIV, using the meta-analyses revealed a significant enrichment of genes highly expressed in T cells and monocytes (Fig. 3e). Many genes highly expressed in NK cells were also found, although these did not have any statistical significance (data not shown). Among the IFN-related genes up-regulated after LAIV vaccination (Fig. 2a), most of them were highly expressed in monocytes and NKs (data not shown). This result is similar to our microarray analyses of FACS-sorted cells obtained from LAIV vaccinees where most of the IFN-related genes differentially expressed in PBMCs and in at least one cell subset were highly expressed in monocytes (Supplementary Fig. 4e, left heat map). These results indicate that the innate and cellular immune responses can play an important role in the mechanism of action of this live attenuated virus vaccine.
Signatures that correlate with antibody response
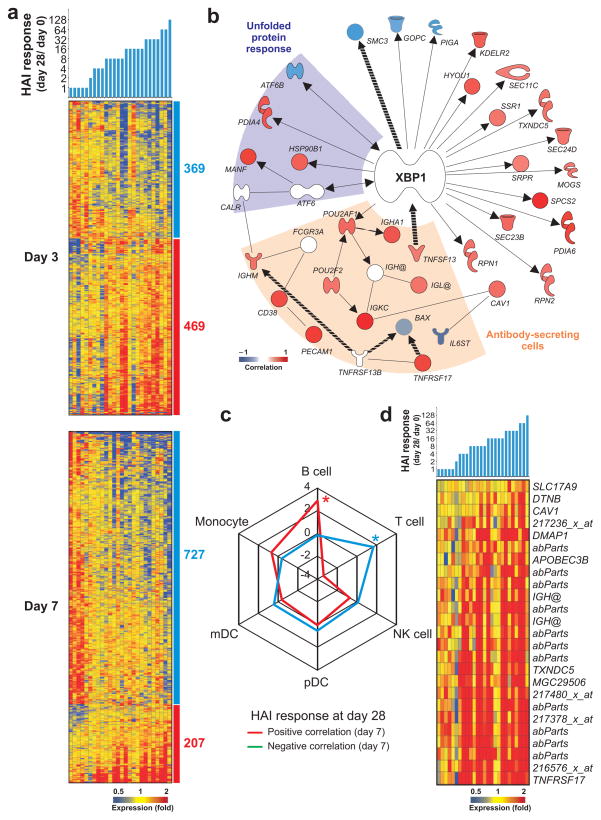
TIV vaccination induces a remarkable variation in the magnitude of the HAI response (Fig. 1a). To gain insight into the potential mechanisms underlying this variation, and to identify gene signatures capable of predicting the magnitude of the HAI response, we searched for early gene signatures that correlate with the B cell responses at day 7 and day 28 post-TIV vaccination. Pearson correlation analysis identified 600–1,100 probe sets that correlated, either directly or inversely, with the magnitude of the HAI antibody response (see Methods and Fig. 4a). Among these correlated genes, there were several genes known to be regulated by the transcription factor XBP-1 and involved in plasma cell differentiation and the unfolded protein response (Fig. 4b). The complete list of genes correlated with the HAI response is available (Supplementary Table 5).
Figure 4.
Molecular signatures that correlate with antibody titers to TIV. (a) Heat map of probe sets (in lines) whose baseline normalized expression at day 3 (top) or day 7 (bottom) correlates (Pearson, P-value < 0.05) to baseline-normalized antibody response at day 28 post-TIV vaccination. Blue and red bars on the right of the heat map indicate the probe sets with negative and positive correlation, respectively (number of probe sets shown). The colors represent the individual fold-change in gene expression at days 3 or 7 compared to day 0 in TIV vaccinees. Probe sets that correlate on both day 3 and 7 to HAI response were counted as “day 7” and the day7-day0 expression was used to represent the expression on heat map. (b) HAI response-correlated genes (Pearson, P-value < 0.05) associated with the Unfolded protein response (purple area), Antibody-secreting cell differentiation (light brown area) and/or regulated by the transcription factor XBP-1. Solid and dashed lines represent, respectively direct and indirect interactions reported for the genes. (c) Spider graph showing the fold enrichment of genes (among those highly expressed in any PBMC subset), whose expression on either day 3 or day 7 post-TIV vaccination is positively (red line) or negatively (blue line) correlated to HAI titers (Pearson, P-value < 0.05). Fold enrichment is calculated as described in Methods. Cell subsets with statistically significant enrichment (Fisher’s exact test two tailed P-value < 10−10) are marked with asterisks. (d) Heat map of probe sets highly expressed in B cells and antibody-secreting cells whose baseline normalized expression correlates (Pearson, P-value < 0.05) to baseline-normalized HAI response.
Ingenuity Pathway Analysis of the genes that were either positively or negatively correlated with the HAI titers, revealed an enrichment of genes related to “cell-mediated immune response” and to “infection mechanism and inflammatory response”, respectively (Supplementary Fig. 6a,b). The identification of genes such as TLR5, CASP1, PYCARD, NOD2 and NAIP suggests new mechanistic links between host innate immunity and humoral responses to influenza vaccination. In fact, different groups have shown that influenza vaccine candidate comprising a recombinant fusion protein linking influenza antigens to the TLR5 ligand (flagellin) may induce potent immunogenicity in mice27 and humans28,29. In addition, canonical pathways, such as “T cell receptor signaling” and “CTLA4 signaling in cytotoxic T lymphocytes” contain many of the genes represented in the “cell-mediated immune response” network and are among the ones with the highest enrichment score in Ingenuity Pathway Analysis (Supplementary Fig. 7a,b). Although further experimentation is necessary, these data indicate a possible association between cellular and humoral responses to TIV vaccination30. Among the top canonical pathways enriched in genes positively correlated to HAI response (Supplementary Fig. 7c), we found networks associated to innate immunity , such as “Natural Killer cell signaling” and “Production of Nitric oxide and reactive oxygen species in macrophages” (Supplementary Fig. 7d). Our analysis also revealed that the expression of interferon related genes (including IFN α and γ receptors) on day 3 post-vaccination is correlated to HAI response, suggesting a link between IFN response and antibody response (Supplementary Fig. 8)31.
Next, we compared the genes whose expression correlated with the HAI response of TIV vaccinees at day 28 with the genomic signatures of immune cells defined by our meta-analysis. This approach revealed that the set of genes positively correlated to HAI response was enriched in genes highly expressed in B cells (Fig. 4c) and, more specifically, in the ASC subset (Fig. 4d). Highly expressed genes in T cells were significantly enriched among the genes with negative correlation to HAI response, supporting the T cell pathways identified by Ingenuity Pathway Analysis as shown in Supplementary Figure 7a,b. Taken together, these data demonstrate identification of early signatures that correlate with later HAI titers induced by TIV.
Molecular signatures that predict antibody responses
Once signatures that correlate with the magnitude of HAI response were delineated, the next step was to identify the minimum sets of genes capable of predicting such a response. Ideally, these sets of genes must be able to accurately classify high versus low responders in additional and independent TIV vaccine trials. For this, we used discriminant analysis via mixed integer programming (DAMIP32,33) which is a very powerful supervised-learning classification method for predicting various biomedical and biobehavioral phenomena32,33.
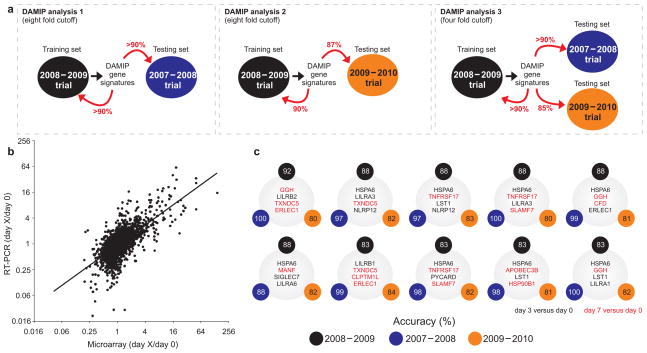
In initial analyses, we classified the TIV vaccinees into 2 extreme groups: very low and very high HAI responders. The former consisted of subjects with 2-fold or lower increase in the HAI titers against any of the 3 influenza strains of the vaccine (Fig. 1a). The latter group consisted of subjects with 8-fold or higher increase in the HAI response for at least one of the 3 influenza strains of the vaccine. Subjects with intermediate HAI response (>= 2-fold and < 8-fold) and subjects for whom microarray data were not available at either day 3 or day 7 post-vaccination (total of 7 subjects) were not analyzed. This trial (named “2008–2009 Trial”) was used to train the DAMIP model to establish an unbiased estimate of correct classification. A second, independent trial was used to evaluate the predictive accuracy of the classification rules identified in the first trial (see Methods and Fig. 5a). The second trial (named “2007–2008 Trial”) consisted of microarray gene expression profile of 9 subjects vaccinated with TIV in the previous year. Using this approach, DAMIP model identified 12 sets of genes containing 2 to 4 genes each (each set associates with one predictive rule) from 2008–2009 trial with 10-fold cross validation accuracy over 90%. The resulting blind prediction accuracy on 2007–2008 trial (predicting low or high antibody responders) is over 90%. Further, some of the 271 set of discriminatory genes offer an accuracy of over 90% in both 10-fold cross validation in the training trial, and in blind prediction accuracy. (Fig. 5a and Supplementary Table 6)
Figure 5.
Signatures that predict the antibody response induced by TIV. (a) Schematic representation of the experimental design used to identify the early gene signatures that predict antibody responses to TIV vaccination. The 2008–2009 Trial was used as a “training set to identify predictive signatures, using the Discriminant Analysis of Mixed Integer Programming (DAMIP) model. These signatures were then tested on the data from the 2007–2008 trial, which represents the “testing set.” The expression of a subset of genes contained within the DAMIP predictive signatures using the 2007–2008 and 2008–2009 trials was then quantified by RT-PCR in a third independent trial (2009–2010 trial). The DAMIP model was again used to confirm the predictive signatures. (b) The expression of a subset of genes contained within the predictive signatures generated by the DAMIP model was validated using RT-PCR. There was a statistically significant positive correlation (2,897 XY pairs, Pearson r = 0.68, P-value < 10−11) between the changes in relative gene expression determined by microarray and RT-PCR analysis. Each point represents a single gene at a given time point. (c) Some of the DAMIP gene signatures identified using 2008–2009 trial as training set and 2007–2008 and 2009–2010 trials as validation sets (i.e. DAMIP model 3). The accuracy represents the number of subjects correctly classified as “low responders” or “high responders” (see legend of Fig. 1a).
We then selected and used real-time RT-PCR experiments to validate 44 genes from the DAMIP gene signatures that had potential biological relevance and/or significance as a predictor of influenza vaccine immunogenicity. A significant positive correlation (R = 0.679, p = 3.25 *10−12) was found between the expression changes on day 3 and day 7 versus baseline detected by microarray and RT-PCR experiments (Fig. 5b and Supplementary Table 7), confirming the microarray data. More importantly, this result gave us confidence to test some of the candidate predictors of immunogenicity in a third and independent influenza vaccine trial (Fig. 5a). RNA from PBMCs of subjects vaccinated with TIV during 2009–2010 influenza season was collected and used in real-time RT-PCR experiments. This trial was comprised of 30 subjects. Real-time RT-PCR expression values of the 44 genes selected from the initial DAMIP gene signatures were then used to validate their capacity for predicting the magnitude of antibody response in this third TIV trial (Fig. 5a). To avoid the identification of over-trained rules, DAMIP program was re-run using the 20082009 trial as training set and the 2007–2008 and 20092010 trials as blind predictive sets. This approach identified 47 sets of genes, some of which can correctly classify > 85% of the vaccinees, as very low or very high antibody responders in any of the 3 trials (Supplementary Table 8).
Since post-vaccination sero-conversion is widely defined as a 4-fold increase in HAI titers34, we decided to run an additional DAMIP analysis using the 4-fold cutoff to classify the vaccinees (Fig. 5a). Thus, subjects with 4-fold or higher increase in the HAI titers post-vaccination were classified as “high” responders and those with 2-fold or lower increase were classified as “low” responders. Using 2008–2009 trial as a training set and 2007–2008 and 2009–2010 trials as blind predictive sets, DAMIP model generated 42 set of gene signatures (Fig. 5c) comprised of 3 to 4 discriminatory genes, some of which had an unbiased estimate of correct classification higher than 85%, as determined by 10-fold cross-validation and blind prediction (see Methods and Supplementary Table 9). One of the genes present in the DAMIP gene signatures, the TNFRSF17 gene, was also identified in the DAMIP models that predict antibody response to YF17D vaccination6.
Among the genes in the TIV vaccine DAMIP models, we found 5 members of the Leukocyte immunoglobulin (Ig)-like receptor (LILR) family (Supplementary Table 9). LILR genes are expressed by immune cell types of both myeloid and lymphoid lineages and are thought to play an immunomodulatory role on innate and adaptive immune systems, by regulating T cells and autoimmunity35–37. Our meta analysis revealed that the LILR genes were most highly expressed in monocytes and myeloid DCs at day 3 after vaccination (data not shown). These results and the presence of 5 members of this family among markers of antibody response to influenza vaccination raises potentially novel roles of these innate immune receptors in regulating antibody responses.
CaMKIV regulates the antibody response
To demonstrate that the gene signatures identified in this study can generate new hypotheses, we selected one gene in the predictive signature, calcium/calmodulin-dependent protein kinase IV (CaMKIV), for functional validation experiments. The CamkIV gene is involved in several processes of the immune system, such as T cell development38–40, inflammatory response41,42 and hematopoietic stem cell maintenance43. However, nothing is known about the possible role of CaMKIV in B cell responses.
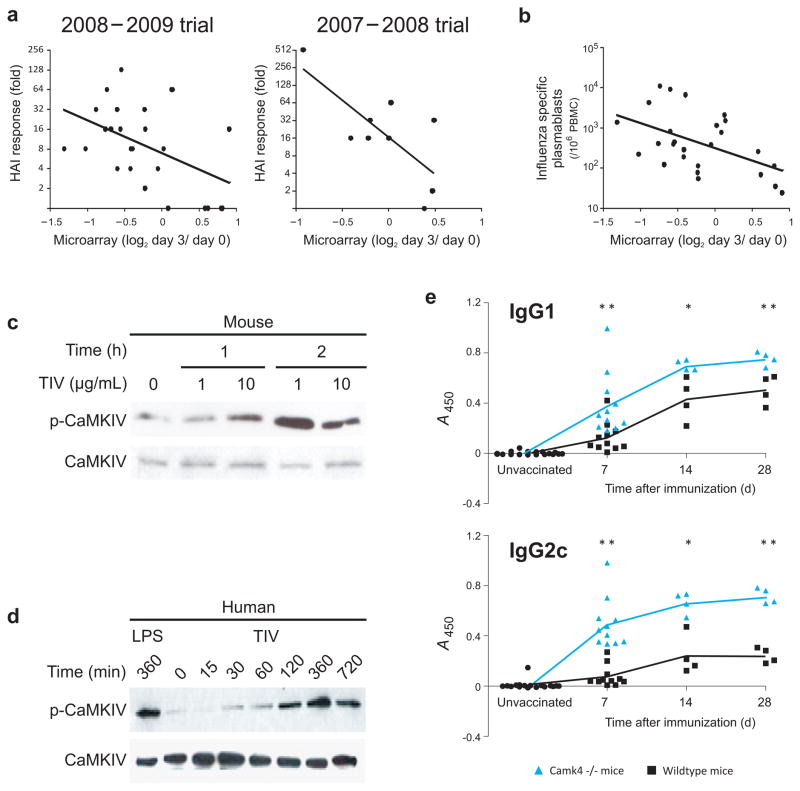
The fold-change in expression of the CaMKIV gene on day 3 post-TIV vaccination is negatively correlated with the antibody response on day 28 post-vaccination in two independent trials (Fig. 6a). Additionally, fold-change expression of CaMKIV is negatively correlated with the expansion of IgG-secreting plasmablasts at day 7 (Fig. 6b), suggesting a possible role for this gene in the regulation of antibody responses to influenza vaccination.
Figure 6.
CAMKIV regulates the antibody response to influenza vaccine. (a) Statistically significant negative correlation between the HAI response at day 28 and the levels of CaMKIV mRNA on PBMCs of vaccinees at day 3 post-vaccination. The left graph represents the TIV vaccinees of 2008–2009 trial (Pearson r = −0.47, P-value (two-tail) = 0.016) and the right graph represents the TIV vaccinees of 2007–2008 trial (Pearson r = −0.73, P-value (two-tail) = 0.024). (b) Statistically significant negative correlation between the number of influenza-specific IgG secreting plasmablasts by ELISPOT at day 7 and the levels of CAMKIV mRNA on PBMCs of vaccinees at day 3 post-vaccination. (c) Phosphorylation of mouse CaMKIV protein after in vitro stimulation of splenocytes with TIV, as determined by western blot. (d) Phosphorylation of CaMKIV protein after in vitro stimulation of human PBMCs treated with TIV for different time points, as determined by western blot. (e) Serum antigen-specific IgG1 (top) and IgG2c (bottom) responses of wild-type (black line) and CamkIV −/− (blue line) mice at day 7, 14 and 28 post-TIV immunization. Student t-test method was used to calculate the statistical significance of each comparison (* = p-value < 0.05, ** = p-value < 0.01). Individual wild type and CamkIV −/− mice are represented by black squares and blue triangles, respectively.
In vitro stimulation of mouse splenocytes with influenza vaccine resulted in phosphorylation of the protein, suggesting that the vaccine may trigger the activation of CaMKIV (Fig. 6c). This finding was further demonstrated in human PBMCs, where in vitro stimulation with influenza vaccine resulted in CaMKIV phosphorylation as early as two hours after stimulation (Fig. 6d). The mechanism by which this occurs is currently being explored.
To check if CaMKIV could regulate the antibody response to influenza vaccine, wild-type (WT) and CamkIV −/− mice were immunized with TIV and serum concentrations of IgG1 and IgG2c were measured on days 7, 14 and 28 post-vaccination (Fig. 6e). After immunization, CamkIV −/− mice showed a significantly higher antibody response compared to WT mice (Fig. 6e). The biggest difference was on day 7, with 3 to 6.5-fold higher antibody levels in knockout mice compared to WT mice (Fig. 6e). These results supported our prediction based on the microarray results and suggest that CaMKIV plays an important role in the regulation of B cell response.
DISCUSSION
Despite their great success, we understand little about the mechanisms by which effective vaccines stimulate protective immune responses. Two exciting new developments are beginning to offer such an understanding: the increasing appreciation of the key roles played by the innate immune system in sensing vaccines and tuning immune responses, and emerging advances in systems biology44. Recently a systems biology approach was used to obtain a global picture of the immune responses in humans to the yellow fever vaccine YF-17D, one of the most successful vaccines ever developed. This approach identified unique biomarkers (molecular signatures) that predicted the magnitude of the antigen-specific CD8+ T cell and antibody responses induced by YF-17D6,7, and the formulation of new hypotheses about the mechanism of action of this vaccine. However whether such an approach will have broad utility in the identification of signatures of immunogenicity of other kinds of vaccines, particularly inactivated vaccines, and whether these signatures would be informative about the underlying mechanisms of immunity, remain unknown. To address these issues, we performed a series of studies over three consecutive influenza seasons. The goal of these studies was to analyze in detail the innate and adaptive responses to vaccination with the influenza vaccines TIV and LAIV, with a view to identifying early molecular signatures that predict the later immune responses, and to obtain insights into the mechanism underlying immunogenicity. According to guidelines established by the FDA14, seroconversion can be defined by an HAI titer ≥1:40 and a minimum 4-fold increase in antibody titer post vaccination. However it often takes several weeks post vaccination to achieve this titer, and therefore the ability to predict seroconversion just a few days following vaccination and identify non responders, would be of great value from a public health perspective. We thus used systems biological approaches to identify early signatures that predicted the HAI titers 4 weeks post vaccination. To accomplish this goal, an interdisciplinary approach was used, including microarray gene expression profiling, RT-PCR, computational methods, combined with cellular and molecular biological approaches as well as experiments involving knockout mice. The data demonstrate that such a systems biological approach can indeed be used not only to identify predictive signatures, but also ascertain new insights about the immunological mechanisms involved.
Although the clinical effectiveness of both vaccines is similar, LAIV induces lower serum antibody response in adults compared to TIV vaccination1,3,45. This likely reflects the lower “take” of the LAIV due to pre-existing mucosal IgA antibodies that can neutralize the virus13. Nevertheless, our microarray analysis identified a large number of differentially expressed genes, most of which were related to the type I IFN response in the PBMCs of LAIV vaccinees. Our next study will focus in analyzing the transcriptomic changes in the nasal mucosa after LAIV vaccination, and how it correlates with or predicts local antibody responses.
Among the genes induced by TIV vaccination, we found an enrichment of genes highly expressed in ASCs. This result may reflect the rapid proliferation of plasmablasts at day 7 post-vaccination15; however, our microarray analysis of B cells sorted from influenza vaccinees indicated that the changes in expression observed in PBMCs can be also derived from real transcriptional changes in B cells. The transcription factor XBP-1, which is essential for the differentiation of ASCs and the unfolded protein response18, and their target genes were up-regulated after TIV vaccination and correlated with IgG and HAI responses. The genes revealed by our study may offer new opportunities to study the complex mechanisms involved in the unfolded protein response and its link to ASC differentiation18.
A key question is whether the signatures that predict the T and B cell response to one vaccine can also predict such responses to another vaccine. Interestingly, of the 133 genes presented in the 271 DAMIP gene signatures that predict the antibody response to TIV vaccination, 7 were also predictors of antibody response to yellow-fever YF-17D vaccination6. Key genes in the predictive signatures are TNFRSF17 (BCMA), a receptor for the B cell growth factor BLyS (known to play a key role in B cell differentiation)23, and CD38, a surface protein that plays important roles in lymphocyte development46,47. TNFRSF17 belongs to a family of molecules (BAFF, APRIL, BAFF- R and TACI), that regulate plasma cell differentiation and antibody production23. Interestingly, there are strong correlations between the expression of genes encoding APRIL, BAFF-R and TACI with the magnitude of the HAI titers to influenza vaccine and with the magnitude of neutralizing antibody response to YF-17D vaccine (data not shown), suggesting that this network of genes may be critically involved in regulating antibody responses to different vaccines. We are currently determining the functional relevance of this network using mouse models. Clearly, it remains to be seen whether this network represents a common predictor of antibody responses induced by many vaccines.
A second question is whether the data generated from such studies will be useful in providing new biological insights into the regulatory mechanisms underlying vaccine immunogenicity. The results of CamkIV −/− mouse experiments demonstrate that such data can indeed reveal unexpected biological targets, which can be mechanistically validated using mouse models. Although the data demonstrate a potent role for CamkIV in regulating antibody responses to influenza vaccine, further work is necessary to delineate the cellular mechanisms involved.
Thirdly, the question of whether signatures that predict immunogenicity also predict efficacy must be considered. Several studies have shown that serum HAI antibody concentrations correlates with protection against influenza48–50. Post-vaccination seroconversion, commonly defined as a 4-fold increase in HAI titers34 represents a useful surrogate for vaccine efficacy when applied to a population. However, this parameter may not provide the optimal prediction of protection in an individual vaccinee, or group of vaccinees. In addition, protective concentrations of antibody may vary accordingly to the prevalent virus subtype and laboratory conducting the assay51. Therefore, we decided to use a more stringent parameter (8-fold or higher increase in the HAI response) to classify subjects with very high antibody responses. Using this cutoff in our analyses, the DAMIP method was able to identify gene signatures that predict the antibody response induced by TIV vaccination. Validation of these gene signatures was performed in a total of 3 independent trials demonstrating the robustness of our approach. To meet the seroconversion definition of FDA’s Guidance for Industry14, i.e. an HAI titer ≥1:40 and a minimum 4-fold increase in antibody titer post vaccination, we re-ran DAMIP using a 4-fold increase as a cutoff to define high HAI responders. Again, DAMIP was able to find sets of 3 to 4 discriminatory genes, with an unbiased estimate of correct classification up to 90% for the three influenza trials. However, the generality of these findings in terms of using gene signatures in PBMCs to predict the immunogenicity and/or efficacy of other vaccines such as mucosal vaccines needs to be tested. Likely, different signatures could be generated from analysis of mucosal tissues.
Finally, although the primary goal of this study was a proof of concept demonstration of the feasibility of this approach in predicting vaccine immunogenicity, (rather than a demonstration of cost effectiveness), it should be noted that in ascertaining the predicting value of our signature in the 2009–2010 trial, in fact we had used a PCR based assay of only a handful of genes (and not on gene expression chips). This demonstrates the feasibility of designing a cost effective, PCR based “vaccine chip” that can predict the immunogenicity of vaccines. Taken together, we reveal how systems biology approaches can be applied to unravel the molecular mechanisms of influenza vaccines. We envision that predictive signatures of influenza vaccine-induced antibody response may have implications in vaccine development, monitoring suboptimal immune responses (in the elderly, infants, or immune compromised populations) or perhaps to identify novel correlates of protection.
METHODS
Clinical study organization
This study is comprised of subjects vaccinated with TIV during the 3 consecutive influenza seasons in, 2008–2009 (trial 1, 28 subjects), 2007–2008 (trial 2, 9 subjects) and 2009–2010 (trial 3, 30 subjects). One trial comprised of subjects immunized with LAIV during 2008–2009 influenza annual season was also included in this study. Young healthy adults, 18 to 50 years old, were vaccinated with one dose of either TIV (Fluarix®, GlaxoSmithKline Biologicals, in 2007–2008 and 2008–2009 seasons and Fluvirin®, Novartis Vaccines and Diagnostics Limited, in 2009–2010 season) or LAIV (FluMist, MedImmune), following current guidelines for influenza vaccination. Supplementary Table 10 shows the influenza virus strains contained on the vaccines. Written informed consent was obtained from each subject with institutional review and approval from the Emory University Institutional Review Board.
Cell, plasma and RNA isolation
Peripheral blood mononuclear cells (PBMC) and plasma were isolated from fresh blood (CPTs; Vacutainer® with Sodium Citrate; BD), following the manufacturer’s protocol. Total RNA from fresh PBMCs (~1.5 × 106 cells) was purified using Trizol® (Invitrogen, Life Technologies Corporation) according to the manufacturer’s instructions. All RNA samples were checked for purity using a ND-1000 spectrophotometer (NanoDrop Technologies) and for integrity by electrophoresis on a 2100 BioAnalyzer (Agilent Technologies).
RT-PCR experiments
Total RNA was reverse-transcribed using a High-Capacity cDNA Archive Kit Protocol (Applied Biosystems Inc.). Custom designed Low Density Array and Quantitative Real-Time PCR Analysis Low Density Arrays for 48 genes were supplied by Applied Biosystems (Supplementary Table 7).
Microarray experiments
Total RNA was hybridized on Human U133 Plus 2.0 Arrays (using GeneTitan platform, Affymetrix, or individual cartridges). Microarray intensity data of probe sets (hereafter referred to “genes”) were normalized by RMA, which includes global background adjustment and quantile normalization. For the microarrays of FACS-sorted immune cell subsets, total RNA was amplified using the WT-Ovation Pico RNA Amplification system (NuGEN) according to the manufacturer’s instructions. Next, cDNA was labeled using the Nugen FL-Ovation cDNA Biotin Module V2 kit, following the manufacturer’s protocol. Hybridization to Affymetrix HT Human Genome U133A Array (Affymetrix, Inc) and washing was performed on the Affymetrix GeneChip Array Station (GCAS) automation platform. The arrays were scanned on the GeneChip HT Array Plate Scanner (Affymetrix 00-0332) and the data were processed using RMA normalization. Additional details can be found in Supplementary Methods.
Hemagglutination inhibition assays
Hemagglutination inhibition (HAI) titers were determined based on the standard WHO protocol, as previously described52. Briefly, plasma samples were treated with receptor destroying enzyme (RDE; Denka Seiken Co.) by adding of 1 part plasma to 3 parts RDE and incubating at 37°C overnight. The following morning, the RDE was inactivated by incubating the samples at 56°C for one hour. The samples were then serially diluted with PBS in 96 well v-bottom plates (Nunc, Rochester, NY) and 4 HA units of either the H1N1, H3N2, or influenza B virus was added to each well. After 30 minutes at room temperature, 50 ul of 0.5% turkey RBCs (Rockland Immunochemicals) suspended in PBS with 0.5% BSA was added to each well and the plates were shaken manually. After an additional 30 minutes at room temperature, the plasma titers were read as the reciprocal of the final dilution for which a button was observed. Negative and positive control plasmas for each virus were used for reference.
B cell ELISPOT experiments and flow cytometry analysis of B cells
Direct ELISPOT to enumerate both total and influenza specific IgG, IgM and IgA plasmablasts present in fresh PBMC samples were essentially done as previously described15,53. Briefly, 96-well ELISPOT filter plates (Millipore, MAHA N4510) were coated overnight with either the influenza vaccine (same vaccine as used for donor vaccination) at a dilution of 1/20 in PBS or with goat anti-human Ig (Caltag). Plates were washed and blocked by incubation with complete RPMI containing 10% FCS at 37°C for 2 hrs. Purified and extensively washed PBMCs were added to the plates in dilution series and incubated for 6 hrs or overnight. Plates were washed with PBS followed by PBS containing 0.05% Tween and then incubated with a biotinylated anti-huIgG, anti-huIgM or anti-huIgA antibody (Caltag) and incubated for 1.5 hrs at room temperature. After washing, the plates were incubated with an avidin-D-HRP conjugate (Vector Laboratories) and finally developed using AEC substrate (3 amino-9 thylcarbozole, Sigma). Developed plates were scanned and analyzed using an automated ELISPOT counter (Cellular Technologies Ltd.). Flow cytometry analysis was performed on whole blood, as previously described15. Briefly, 300–400 μl blood was incubated with the appropriate antibodies for 30 minutes at room temperature. Red blood cells were then lysed by incubation with FACS lysing Solution (Beckton Dickinson) for twice for 4 minutes at RT prior to analysis. Antibodies against CD3, CD20, CD38 and CD19 were purchased from Pharmingen, while anti-CD27 that was purchased from eBiosciences. ASCs were gated and isolated as CD19+CD3−CD20lo/−CD27high CD38high cells. Flow cytometry data was analyzed using FlowJo software.
Meta-analysis of immune cells and Microarray analysis
Full details, including FACS-sorting of immune cell subsets and identification of differentially expressed genes in PBMCs and FACS-sorted cell subsets; the meta-analysis; correlation and predictive analyses can be found in Supplementary Methods.
Analysis of immune responses in CamkIV −/− mice
8–12 week old C57BL6 mice were procured from Jackson Labs (Maine, VM). CamkIV −/−mice were generated as described54 and back bred with C57BL6/J mice for more than 12 generations. Mice were immunized in the right and the left hamstring muscles with a 1:5 diluted human Fluvirin® (Novartis Vaccines and Diagnostics, Cambridge, MA) vaccine dose (in effect, 3ug of HA of each strain included in the vaccine). Mice were bled at time points indicated for analysis of influenza specific antibody responses. All animal procedures were performed in accordance with guidelines established by IACUC at Duke University.
For the antibody ELISA assays, 96 well Nunc maxisorp plates (Nunc, Denmark) were coated with 100μl of 2μg/ml of Fluvirin 2009 or 2010 (Novartis Vaccines and Diagnostics, Cambridge, MA) overnight at 4°C. Plates were washed 3 times with PBS/0.5% Tween 20 using a Biotek auto plate washer and blocked with 200μl of 4% non fat dry milk (Biorad, Hercules, CA) for 2 hours at room temperature. Serum samples from immunized mice at the indicated time points were diluted in 0.1% non-fat dry milk in PBS/0.5% Tween 20 (1:100) and incubated on blocked plates for 2 hours at room temperature. Detection antibodies were purchased from Southern Biotech (Birmingham, AL). The plates were washed 5 times post sample sera incubation, and anti mouse IgG2c-horse radish peroxidase (HRP) conjugate (1:2000) and anti-mouse IgG1-HRP conjugate at (1:5000) in PBS/0.5% Tween 20, were added and plates were incubated for 2 hours at room temperature. Plates were washed 7 times with PBS/0.5% Tween 20, developed using 100μl per well of tetramethylbenzidine (TMB) substrate (BD Biosciences, San Diego, CA) and stopped using 2N H2SO4. Plates were analyzed using a BioRad plate reading spectrophotometer at 450nm with correction at 595nm. Results are represented as O.D values at 450nm at the indicated time points. The details of western blot experiments can be found in Supplementary Methods.
Supplementary Material
Acknowledgments
We thank Barry T. Rouse and Richard Compans for discussion and comments on the manuscript. We thank Herold Oluoch for his excellent technical assistance. The work in the laboratory of B.P. was supported by grants U19AI090023, HHSN266200700006C, U54AI057157, R37AI48638, R01DK057665, U19AI057266, and NO1 AI50025 from the National Institutes of Health and a grant from the Bill & Melinda Gates Foundation. Work in R.A. laboratory was partly funded by grants AI30048 (NIH) and AI057266 (NIH), and CAVD 38645 (Bill and Melinda Gates Foundation Grant). Work in A.R.M. lab was funded by NIH grant DK074701. Work in K.S. laboratory was supported by the Intramural Research Program of the NIAID, NIH. Work in EKL lab was partially support by NSF and CDC grants. The clinical work was supported in part by PHS Grant UL1 RR025008 from the Clinical and Translational Science Award program, National Institutes of Health, National Center for Research Resources.
Footnotes
Accession codes
Gene Expression Omnibus: microarray data, GSE29619.
AUTHOR CONTRIBUTIONS
H.I.N. performed all the experiments and analyses in Figures 2 – 6 and Supplementary Figures 2 – 10; J.W., G.M.L., M.M. and V.K. performed the analyses in Figure 1 and Supplementary Figure 1; E.K.L. performed the DAMIP model analyses in Figure 5; L.R., A.R.M., S.P.K. and N.K. performed the mouse experiments shown in Figure 6; W.N.H. helped in microarray experiments shown in Supplementary Figure 5; S.L. assisted with the bioinformatics analyses of the data in Figure 3; A.A. performed the microarray experiments of samples from 2007 influenza annual season; S.M.K., K.E.K., R.E. and A.K.M. assisted with the collection and processing of samples; K.S. measured the HAI titers; R.A. helped conceive and design the study and supervised the studies in Figure 1 and Supplementary Figure 1. B.P. conceived the study and designed and supervised the experiments and analyses in Figures 1–6 and Supplementary Figures 1–10. B.P. and H.I.N. wrote the paper.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
References
- 1.Sasaki S, et al. Comparison of the influenza virus-specific effector and memory B-cell responses to immunization of children and adults with live attenuated or inactivated influenza virus vaccines. J Virol. 2007;81:215–228. doi: 10.1128/JVI.01957-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fiore AE, et al. Prevention and control of influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2010. MMWR Recomm Rep. 2010;59:1–62. [PubMed] [Google Scholar]
- 3.Sasaki S, et al. Influence of prior influenza vaccination on antibody and B-cell responses. PLoS One. 2008;3:e2975. doi: 10.1371/journal.pone.0002975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeman AM, et al. Humoral and cellular immune responses in children given annual immunization with trivalent inactivated influenza vaccine. Pediatr Infect Dis J. 2007;26:107–115. doi: 10.1097/01.inf.0000253251.03785.9b. [DOI] [PubMed] [Google Scholar]
- 5.Pulendran B, Li S, Nakaya HI. Systems vaccinology. Immunity. 2010;33:516–529. doi: 10.1016/j.immuni.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Querec TD, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaucher D, et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med. 2008;205:3119–3131. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pulendran B. Learning immunology from the yellow fever vaccine:innate immunity to systems vaccinology. Nat Rev Immunol. 2009;9:741–747. doi: 10.1038/nri2629. [DOI] [PubMed] [Google Scholar]
- 9.Monath TP. Yellow fever vaccine. Expert Rev Vaccines. 2005;4:553–574. doi: 10.1586/14760584.4.4.553. [DOI] [PubMed] [Google Scholar]
- 10.Querec T, et al. Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity. J Exp Med. 2006;203:413–424. doi: 10.1084/jem.20051720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrett AD, Teuwen DE. Yellow fever vaccine -how does it work and why do rare cases of serious adverse events take place? Curr Opin Immunol. 2009;21:308–313. doi: 10.1016/j.coi.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 12.Johnson PR, Jr, Feldman S, Thompson JM, Mahoney JD, Wright PF. Comparison of long-term systemic and secretory antibody responses in children given live, attenuated, or inactivated influenza A vaccine. J Med Virol. 1985;17:325–335. doi: 10.1002/jmv.1890170405. [DOI] [PubMed] [Google Scholar]
- 13.Beyer WE, Palache AM, de Jong JC, Osterhaus AD. Cold-adapted live influenza vaccine versus inactivated vaccine: systemic vaccine reactions, local and systemic antibody response, and vaccine efficacy. A meta-analysis. Vaccine. 2002;20:1340–1353. doi: 10.1016/s0264-410x(01)00471-6. [DOI] [PubMed] [Google Scholar]
- 14.Administration, F.a.D. Guidance for Industry: Clinical Data Needed to Support the Licensure of Pandemic Influenza Vaccines. 2007. [Google Scholar]
- 15.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takaoka A, Yanai H. Interferon signalling network in innate defence. Cell Microbiol. 2006;8:907–922. doi: 10.1111/j.1462-5822.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- 17.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc NatlAcad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwakoshi NN, et al. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4:321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- 19.Iwakoshi NN, Lee AH, Glimcher LH. The X-box binding protein-1 transcription factor is required for plasma cell differentiation and the unfolded protein response. Immunol Rev. 2003;194:29–38. doi: 10.1034/j.1600-065x.2003.00057.x. [DOI] [PubMed] [Google Scholar]
- 20.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 21.Ueda Y, et al. Frequencies of dendritic cells (myeloid DC and plasmacytoid DC) and their ratio reduced in pregnant women: comparison with umbilical cord blood and normal healthy adults. Hum Immunol. 2003;64:1144–1151. doi: 10.1016/j.humimm.2003.08.342. [DOI] [PubMed] [Google Scholar]
- 22.Shen-Orr SS, et al. Cell type-specific gene expression differences in complex tissues. Nat Methods. 2010;7:287–289. doi: 10.1038/nmeth.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Avery DT, et al. BAFF selectively enhances the survival of plasmablasts generated from human memory B cells. J Clin Invest. 2003;112:286–297. doi: 10.1172/JCI18025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park SW, et al. The regulatory subunits of PI3K, p85alpha and p85beta, interact with XBP-1 and increase its nuclear translocation. Nat Med. 2010;16:429–437. doi: 10.1038/nm.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu B, Li Z. Endoplasmic reticulum HSP90b1 (gp96, grp94) optimizes B-cell function via chaperoning integrin and TLR but not immunoglobulin. Blood. 2008;112:1223–1230. doi: 10.1182/blood-2008-03-143107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Apostolou A, Shen Y, Liang Y, Luo J, Fang S. Armet, a UPR-upregulated protein, inhibits cell proliferation and ER stress-induced cell death. Exp Cell Res. 2008;314:2454–2467. doi: 10.1016/j.yexcr.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huleatt JW, et al. Potent immunogenicity and efficacy of a universal influenza vaccine candidate comprising a recombinant fusion protein linking influenza M2e to the TLR5 ligand flagellin. Vaccine. 2008;26:201–214. doi: 10.1016/j.vaccine.2007.10.062. [DOI] [PubMed] [Google Scholar]
- 28.Treanor JJ, et al. Safety and immunogenicity of a recombinant hemagglutinin influenza-flagellin fusion vaccine (VAX125) in healthy young adults. Vaccine. 2010;28:8268–8274. doi: 10.1016/j.vaccine.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 29.Talbot HK, et al. Immunopotentiation of trivalentinfluenza vaccine when given with VAX102, a recombinant influenza M2e vaccine fused to the TLR5 ligand flagellin. PLoS One. 2010;5:e14442. doi: 10.1371/journal.pone.0014442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He XS, et al. Cellular immune responses in children and adults receiving inactivated or live attenuated influenza vaccines. J Virol. 2006;80:11756–11766. doi: 10.1128/JVI.01460-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Bon A, et al. Cutting edge: enhancement of antibody responses through direct stimulation of B and T cells by type I IFN. J Immunol. 2006;176:2074–2078. doi: 10.4049/jimmunol.176.4.2074. [DOI] [PubMed] [Google Scholar]
- 32.Lee EK. Large-scale optimization-basedclassification models in medicine and biology. Ann Biomed Eng. 2007;35:1095–1109. doi: 10.1007/s10439-007-9317-7. [DOI] [PubMed] [Google Scholar]
- 33.Brooks JP, Lee EK. Analysis of the consistency of a mixed integer programming-based multi-category constrained discriminant model. Annals of Operations Research. 2010;174:147–168. [Google Scholar]
- 34.Sullivan SJ, Jacobson R, Poland GA. Advances in the vaccination of the elderly against influenza: role of a high-dose vaccine. Expert Rev Vaccines. 2010;9:1127–1133. doi: 10.1586/erv.10.117. [DOI] [PubMed] [Google Scholar]
- 35.Anderson KJ, Allen RL. Regulation of T-cell immunity by leucocyte immunoglobulin-like receptors: innate immune receptors for self on antigen-presenting cells. Immunology. 2009;127:8–17. doi: 10.1111/j.1365-2567.2009.03097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas R, Matthias T, Witte T. Leukocyte immunoglobulin-like receptors as new players in autoimmunity. Clin Rev Allergy Immunol. 2010;38:159–162. doi: 10.1007/s12016-009-8148-8. [DOI] [PubMed] [Google Scholar]
- 37.Brown D, Trowsdale J, Allen R. The LILR family: modulators of innate and adaptive immune pathways in health and disease. Tissue Antigens. 2004;64:215–225. doi: 10.1111/j.0001-2815.2004.00290.x. [DOI] [PubMed] [Google Scholar]
- 38.Krebs J, Wilson A, Kisielow P. Calmodulin-dependent protein kinase IV during T-cell development. Biochem Biophys Res Commun. 1997;241:383–389. doi: 10.1006/bbrc.1997.7823. [DOI] [PubMed] [Google Scholar]
- 39.Wang SL, Ribar TJ, Means AR. Expression of Ca(2+)/calmodulin-dependent protein kinase IV (caMKIV) messenger RNA during murine embryogenesis. Cell Growth Differ. 2001;12:351–361. [PubMed] [Google Scholar]
- 40.Anderson KA, Means AR. Defective signaling in a subpopulation of CD4(+) T cells in the absence of Ca(2+)/calmodulin-dependent protein kinase IV. Mol Cell Biol. 2002;22:23–29. doi: 10.1128/MCB.22.1.23-29.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Illario M, et al. Calmodulin-dependent kinase IV links Toll-like receptor 4 signaling with survival pathway of activated dendritic cells. Blood. 2008;111:723–731. doi: 10.1182/blood-2007-05-091173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato K, et al. Regulation of osteoclast differentiation and function by the CaMK-CREB pathway. Nat Med. 2006;12:1410–1416. doi: 10.1038/nm1515. [DOI] [PubMed] [Google Scholar]
- 43.Kitsos CM, et al. Calmodulin-dependent protein kinase IV regulates hematopoietic stem cell maintenance. J Biol Chem. 2005;280:33101–33108. doi: 10.1074/jbc.M505208200. [DOI] [PubMed] [Google Scholar]
- 44.Pulendran B, Ahmed R. Immunological mechanisms of vaccination. Nat Immunol. 2011;131:509–517. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moldoveanu Z, Clements ML, Prince SJ, Murphy BR, Mestecky J. Human immune responses to influenza virus vaccines administered by systemic or mucosal routes. Vaccine. 1995;13:1006–1012. doi: 10.1016/0264-410x(95)00016-t. [DOI] [PubMed] [Google Scholar]
- 46.Shubinsky G, Schlesinger M. The CD38 lymphocyte differentiation marker: new insight into its ectoenzymatic activity and its role as a signal transducer. Immunity. 1997;7:315–324. doi: 10.1016/s1074-7613(00)80353-2. [DOI] [PubMed] [Google Scholar]
- 47.Deaglio S, Mehta K, Malavasi F. Human CD38: a (r)evolutionary story of enzymes and receptors. Leuk Res. 2001;25:1–12. doi: 10.1016/s0145-2126(00)00093-x. [DOI] [PubMed] [Google Scholar]
- 48.Clements ML, Betts RF, Tierney EL, Murphy BR. Serum and nasal wash antibodies associated with resistance to experimental challenge with influenza A wild-type virus. J Clin Microbiol. 1986;24:157–160. doi: 10.1128/jcm.24.1.157-160.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Potter CW, Oxford JS. Determinants of immunity to influenza infection in man. Br Med Bull. 1979;35:69–75. doi: 10.1093/oxfordjournals.bmb.a071545. [DOI] [PubMed] [Google Scholar]
- 50.Hirota Y, et al. Antibody efficacy as a keen index to evaluate influenza vaccine effectiveness. Vaccine. 1997;15:962–967. doi: 10.1016/s0264-410x(96)00302-7. [DOI] [PubMed] [Google Scholar]
- 51.Belshe RB. Current status of live attenuated influenza virus vaccine in the US. Virus Res. 2004;103:177–185. doi: 10.1016/j.virusres.2004.02.031. [DOI] [PubMed] [Google Scholar]
- 52.Chen GL, Lamirande EW, Jin H, Kemble G, Subbarao K. Safety, immunogencity, and efficacy of a cold-adapted A/Ann Arbor/6/60 (H2N2) vaccine in mice and ferrets. Virology. 2010;398:109–114. doi: 10.1016/j.virol.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crotty S, et al. Cutting edge: long-term B cell memory in humans after smallpox vaccination. J Immunol. 2003;171:4969–4973. doi: 10.4049/jimmunol.171.10.4969. [DOI] [PubMed] [Google Scholar]
- 54.Wu JY, et al. Spermiogenesis and exchange of basic nuclear proteins are impaired in male germ cells lacking Camk4. Nat Genet. 2000;25:448–452. doi: 10.1038/78153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.