Abstract
Eukaryotic cells must contend with a continuous stream of misfolded proteins that compromise the cellular protein homeostasis balance and jeopardize cell viability. An elaborate network of molecular chaperones and protein degradation factors continually monitor and maintain the integrity of the proteome. Cellular protein quality control relies on three distinct yet interconnected strategies whereby misfolded proteins can either be refolded, degraded, or delivered to distinct quality control compartments that sequester potentially harmful misfolded species. Molecular chaperones play a critical role in determining the fate of misfolded proteins in the cell. Here, we discuss the spatial and temporal organization of cellular quality control strategies and their implications for human diseases linked to protein misfolding and aggregation.
Soluble misfolded proteins are concentrated in quality control compartments to enhance their refolding or degradation. Insoluble species that may disrupt protein homeostasis are sequestered to prevent their toxic interactions with the quality control machinery.
ROLE OF PROTEIN QUALITY CONTROL IN CELLULAR INTEGRITY
Maintaining the integrity of the proteome is essential for cell viability. Although energetically favored, the native state of proteins is in a precarious equilibrium (Brockwell and Radford 2007). Proteins often misfold during the life of the cell, as a result of stochastic fluctuations, the presence of destabilizing mutations, stress conditions, or unique metabolic challenges, such as those occurring during cancer or aging (Hartl and Hayer-Hartl 2009). In the cell, misfolded proteins can have deleterious “gain-of-function” activities, in part because of their heightened tendency to aggregate (Dobson 2003). Although the precise mechanisms of toxicity are not well understood, it is clear that misfolded proteins engage in inappropriate interactions with other cellular components and can accumulate in potentially toxic protein inclusions (Lansbury and Lashuel 2006). Protein misfolding is emerging as a major mechanism of human disease, as highlighted by the growing list of “conformational diseases,” which result from the cellular accumulation of misfolded proteins (Muchowski 2002; Sakahira et al. 2002). These include a staggering array of pathologies, ranging from lysosomal storage diseases (Sawkar et al. 2006), cancer (Dai et al. 2007), cystic fibrosis (Koulov et al. 2010) to, most prominently, many neurodegenerative disorders such as Alzheimer (AD), Parkinson’s (PD), and Huntington’s (HD) diseases (Caughey and Lansbury 2003; Cohen and Kelly 2003; Morimoto 2008). It is becoming clear that the cellular capacity to manage the proteome declines during aging and this likely underlies the late onset of neurodegenerative diseases caused by protein misfolding (Cuervo et al. 2005; Ben-Zvi et al. 2009; Demontis and Perrimon 2010).
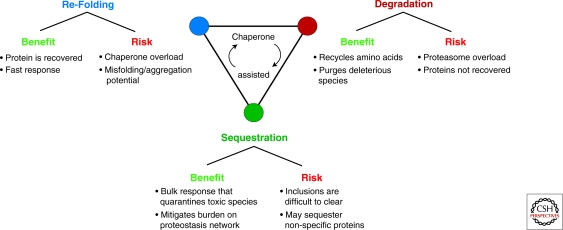
The cell has developed an elaborate machinery that monitors and maintains the health of its proteome (Frydman 2001; Hartl and Hayer-Hartl 2009; Richter et al. 2010). Preserving protein homeostasis, or “proteostasis,” involves several parallel strategies that aim at either refolding, degrading, or sequestering misfolded polypeptides (Fig. 1) (Powers et al. 2009). Central to all these strategies is a network of molecular chaperones that recognizes misfolded proteins (Hartl and Hayer-Hartl 2002; McClellan et al. 2005a). Chaperones can actively promote refolding of the misfolded protein or, if this is not possible, can promote their degradation via the ubiquitin-proteasome pathway (McClellan et al. 2005b). Recent findings have revealed an additional cellular strategy to cope with misfolded proteins that are not refolded or degraded, namely sequestration into specialized quality control compartments (Bagola and Sommer 2008; Kaganovich et al. 2008). The spatial compartmentalization of cellular quality control may help the cell cope with an overload of aberrant proteins, prevent formation of toxic aggregates, and regulate the inheritance of damaged and/or aggregation-prone species (Tyedmers et al. 2010a). Here we review the central mechanisms that maintain protein homeostasis and quality control in eukaryotic cells and highlight the emerging concept that protein quality control is associated with subcellular compartments that sequester and concentrate both soluble and aggregated forms of misfolded proteins.
Figure 1.
Cellular strategies to maintain protein homeostasis. Cells have evolved distinct yet interconnected cellular strategies to maintain protein homeostasis. Each strategy presents advantages and drawbacks. Misfolded proteins can either be refolded, degraded, or delivered to distinct quality control compartments that sequester potentially deleterious species. These strategies are all assisted by molecular chaperones that ensure the system remains balanced. Failure of the cellular strategies can tip the protein homeostasis balance and lead to a decrease in cell viability.
CAUSES AND CONSEQUENCES OF PROTEIN MISFOLDING
Under normal growth conditions, the cell contends with a continuous stream of misfolded proteins arising from inefficient protein biogenesis, expression of mutant proteins, excess unassembled subunits of oligomeric complexes, and inefficiently translocated secretory and mitochondrial precursors (Balch et al. 2008; Voisine et al. 2010). The precise degree to which these processes burden the cellular quality control machinery has been a matter of controversy. In addition to these normal, physiological sources of misfolded proteins, a number of pathological conditions, environmental and metabolic stresses, aging, and cancer, enhance the production of misfolded proteins (Haigis and Yankner 2010). Altogether, these factors can further tax the protein homeostasis machinery. Potential environmental stresses include elevated temperature, exposure to chemicals or heavy metals, viral/bacterial infections, and tissue injury. Potential metabolic stresses are associated with nutrient balance, production of reactive oxygen species (ROS) and mitochondrial dysregulation. These challenges are predominantly met by the activation of the environmental stress response (ESR), which elevates expression of protective cellular components (Voisine et al. 2010). Notably, the high rates of cell division and high mutation rates that accompany cancer lead to a higher load of misfolded proteins (Whitesell and Lindquist 2005). Thus, cancer cells typically overexpress chaperones, and induction of an ESR is an integral part of carcinogenesis (Whitesell and Lindquist 2005). Conversely, aging is associated with a decline of protein homeostasis capacity (Morley et al. 2002; Morley and Morimoto 2004). Presumably, old cells or organisms accumulate deleterious mutations and oxidatively damaged proteins, which overwhelm the capacity of the protein homeostasis network (Gidalevitz et al. 2006). This in turn promotes further protein damage, eventually leading to widespread protein aggregation, toxicity, and cell death (Douglas and Dillin 2010).
Given that increasing the misfolded protein load may tip the balance of the proteostatic network, it is essential for the cell to mount a rapid and robust response to restore cellular homeostasis. The cell relies on two major protein management strategies: (1) the up-regulation of quality control components, and (2) the sequestration of misfolded and/or aggregated proteins. Several transcriptional programs sense stress and enhance expression of molecular chaperones and degradation components. These include the cytosolic ESR, regulated by the transcription factors Hsf1 and Msn2/4 in yeast or HSF-1 and DAF-16 in worms, as well as the UPR in the secretory pathway and the oxidative stress response (Akerfelt et al. 2010). Notably, enhancing chaperone expression via hsf-1 and daf-16 delays aging and protects organisms from neurodegenerative diseases (Morley and Morimoto 2004; Cohen et al. 2006). Little is known about the mechanisms controlling misfolded protein sequestration, thus it is unclear whether and how partitioning into cellular quality control compartments is regulated.
CHAPERONES DICTATE THE BALANCE BETWEEN PROTEIN FOLDING, DEGRADATION, AND AGGREGATION
All aspects of cellular protein homeostasis depend on molecular chaperones (Frydman 2001; Bukau et al. 2006; Schlecht et al. 2011). Chaperones promote the folding of newly synthesized polypeptides, their translocation across membranes, and the refolding of stress-denatured substrates. Chaperones also play a key role in targeting misfolded proteins for degradation as well as preventing aggregation. In eukaryotic cells, these distinct functions are performed by two distinctly regulated chaperone networks: the chaperones linked to protein synthesis (CLIPS), which are functionally and physically linked to the translation machinery and assist folding of newly translated proteins (Albanese et al. 2006), and the heat shock proteins (HSPs), which can be induced by HSF and serve to protect the proteome from stress (Haslbeck et al. 2005).
The cell has several different classes of chaperones, generally classified according to their molecular masses (e.g., hsp100, hsp90, hsp70, hsp60, and hsp40) and small heat shock proteins (smHSPs). Each family is comprised of multiple chaperone isoforms. Certain ATP-driven chaperones, such as Hsp70 and Hsp90 interact with cofactors, which directly influence their ATPase activity and direct them along certain folding pathways (Kampinga and Craig 2010; Taipale et al. 2010). In addition to differences in size and structure, chaperones play different roles spatially and temporarily within the proteostasis network. The ring-shaped chaperonin TRiC/CCT and a subset of Hsp70s assist in de novo protein folding, whereas other chaperones, such as Hsp70s, Hsp90, and the smHSPs appear to assist conformational maturation, oligomeric assembly, and degradation. Importantly, different chaperone systems cooperate functionally to promote folding and degradation (Hartl and Hayer-Hartl 2002). In addition to their well-established role in assisting protein folding and refolding, chaperones can direct the ubiquitination and degradation of misfolded proteins (Youker et al. 2004; McClellan et al. 2005b; Kundrat and Regan 2010). For instance, Hsp70, Hsp110, and Hsp90 are required for degrading misfolded variants of VHL (McClellan et al. 2005a) and other proteins (Hampton 2011). In addition, overexpressing Hsp70 and Hsp40 increases the proteasome-mediated degradation of α-synuclein and polyQ-expanded proteins (Muchowski and Wacker 2005).
Chaperones also prevent the formation of toxic aggregates (Warrick et al. 1999; Muchowski 2002; Morimoto 2008). For example, overexpressing Hsp70 suppresses the toxicity associated with Aβ and tau in AD, α-synuclein in PD, superoxide dismutase (SOD1) in familial Lou Gehrig’s disease (fALS), and polyQ-expanded Huntingtin (Htt) in HD, as well as spinobulbar muscular atrophy (SBMA) and ataxias (Muchowski and Wacker 2005). TRiC/CCT has also been shown to remodel Htt aggregates and appears to be a major cellular modulator of their toxicity (Behrends et al. 2006; Kitamura et al. 2006; Tam et al. 2006). Interestingly, in most of these cases, chaperones do not prevent formation of protein inclusions, but rather likely inhibit formation of toxic species by directing the misfolded species to nontoxic aggregates (Wacker et al. 2004; Muchowski and Wacker 2005; Tam et al. 2009).
Beyond serving as a link between folding and degradation, certain chaperones have the ability to extract and refold proteins from aggregates. The small heat shock proteins (smHSPs) have been observed to tightly associate with aggregates (Haslbeck et al. 2005). A specialized class of chaperones, the ring-shaped hexameric AAA-ATPases can extract misfolded proteins from aggregates in an ATP-dependent manner (Wendler et al. 2007). Chaperones such as ClpB in bacteria and Hsp104 in yeast extract misfolded polypeptides from aggregates by threading them through a central pore lined with aromatic residues (Lum et al. 2004; Haslberger et al. 2008; Wendler et al. 2009). Higher eukaryotes lack a clear Hsp104 homolog, but a “disaggregase” activity has recently been reported (Bieschke et al. 2009; Murray et al. 2010). The extracted protein can then be transferred to Hsp70 and Hsp40 chaperones for refolding or degradation (Buchberger et al. 2010; Lotz et al. 2010; Richter et al. 2010). Interestingly, shifting the balance from refolding to degradation following disaggregation impairs the ability of bacterial cells to withstand stress (Weibezahn et al. 2004).
Although the ability of chaperones to recognize misfolded polypeptides is central to protein quality control, the molecular basis of the triage process by which chaperones decide whether to fold, degrade, or sequester a misfolded protein in an inclusion is perhaps the most critical and least understood question surrounding protein quality control.
PATHWAYS OF MISFOLDED PROTEIN DEGRADATION
Most soluble misfolded proteins are cleared through the ubiquitin-proteasome system (UPS), the major eukaryotic proteolytic pathway (Ciechanover 1998). An enzymatic E1/E2/E3 ubiquitination cascade tightly controls degradation by marking proteins by polyubiquitination, which tags them for destruction by the 26S proteasome. Degradation may require additional factors, such as the p97/Cdc48-Ufd1-Npl4 complex and its various cofactors (Wilkinson 2000). The 26S proteasome in eukaryotes is composed of a 20S proteolytic core and a 19S ATP-dependent regulatory cap (Pickart and Cohen 2004). Different types of polyubiquitin chain linkages can target proteins for degradation (i.e., K48) or specific subcellular localization (i.e., K63) (Xu et al. 2009) and different ubiquitin states (mono- vs. polyubiquitination) can control the activity level and trafficking of proteins (Finley et al. 2004).
The communication between the E3 ubiquitin ligases, which ubiquitinate target proteins and the chaperone machinery is not fully understood. A number of E3 ligases have been shown to ubiquitinate misfolded proteins in an Hsp70-dependent manner. In mammalian cells, the E3 ligase CHIP (carboxy-terminal Hsp70 interacting protein) binds directly to Hsp70 and Hsp90 (Arndt et al. 2007; Vembar and Brodsky 2008). CHIP has been shown to target some Hsp70-bound proteins for degradation, but also can tag substrates with noncanonical ubiquitin chain linkages (e.g., K27 and K63) for functions other than proteasomal degradation (Alberti et al. 2002). In yeast, two E3 ubiquitin ligases anchored to the endoplasmic reticulum (ER) membrane, Doa10 and Hrd1, can mediate Hsp70-dependent ubiquitination of misfolded ER and cytosolic substrates (Nakatsukasa et al. 2008; Hirsch et al. 2009). Recently, two distinct chaperone-mediated quality control pathways were identified whereby misfolded and/or chaperone-bound proteins are ubiquitinated by the E3 ligase Ubr1 in the cytosol and by the E3 ligase San1 in the nucleus (Heck et al. 2010). Future studies should clarify the mechanistic basis for these observations and how Hsp70 and other chaperones control the option of transferring their substrate to an E3 ligase.
Misfolded aggregated proteins can also be degraded by a separate autophagy pathway that involves their ultimate delivery to the lysosome (Klionsky et al. 2010). Autophagy is a nonspecific bulk degradation pathway that was initially described for long-lived cytoplasmic proteins and damaged organelles (He and Klionsky 2009). This process is also a major degradation pathway for many aggregation-prone proteins associated with neurodegenerative disorders (Kroemer et al. 2010). Knockdown of the autophagy genes (e.g., Atg5 and Atg7) leads to aggregation and neurodegeneration in certain mouse models (Hara et al. 2006; Komatsu et al. 2006). Conversely, up-regulation of autophagy can play a protective role in quality control because it can promote the clearance of soluble small-oligomeric aggregates, for instance, in HD models (Rubinsztein et al. 2007; Mizushima et al. 2008). A number of studies suggest that there is a functional relationship between proteasomal and autophagic degradation of misfolded proteins: impairment of the UPS induces compensatory autophagy (Pandey et al. 2007), whereas knockdown of autophagy components leads to the accumulation of proteasomal substrates (Komatsu et al. 2005; Hara et al. 2006). Furthermore, some substrates partition between these two pathways. For instance, the majority of misfolded ER proteins are cleared by the ER-associated degradation (ERAD) pathway that retrotranslocates misfolded proteins to the cytoplasm for degradation by the UPS. If the ERAD pathway does not effectively clear the accumulated misfolded proteins, or if ERAD is impaired otherwise, then the autophagy-lysosome pathway is activated. This acts as a final strategy for ER protection, specifically recognizing and degrading potentially toxic higher molecular weight aggregates (Ishida et al. 2009).
SPATIAL ORGANIZATION OF QUALITY CONTROL PATHWAYS
Accumulation of misfolded proteins in spatially distinct inclusions has been observed in bacterial and eukaryotic cells (Winkler et al. 2010). Their formation is enhanced by various environmental stresses that destabilize folded proteins. Although inclusions often form when the folding and degradation machineries are overwhelmed, there is increasing evidence that inclusion formation is a stereotypical cellular response that aims to spatially and functionally sequester misfolded proteins in specific cellular compartments (Kaganovich et al. 2008). There are also instances of regulated inclusion formation in response to environmental and metabolic stimuli. In yeast, nutrient depletion induces formation of reversible protein inclusions, which are resolubilized after the readdition of nutrients (Narayanaswamy et al. 2009).
Interest into the function and formation of protein inclusions initially arose from the observation that insoluble amyloid aggregates are a hallmark of many neurodegenerative diseases, including Alzheimer, Huntington’s, and prion diseases (Dobson 2006). However, it is clear that many other proteins are found in inclusions, including defective ribosomal products (David et al. 2010), polyubiquitinated proteins (Ross and Poirier 2004), and oxidatively damaged proteins (Nystrom 2005). On heat stress or nutrient stress, sequestration into defined inclusions has been observed for components of the translation machinery (Radonjic et al. 2005), biosynthetic machinery (An et al. 2008), and small HSPs (i.e., Hsp12) (Welker et al. 2010). Furthermore, viruses can also induce a nuclear quality control compartment in infected cells, known as a VICE domain, which contains Hsc70 and the 20S proteasome and degrades nuclear proteins (Livingston et al. 2009).
Several different types of cellular inclusions have been observed in eukaryotic cells. Interestingly, although some inclusions accumulate insoluble aggregated proteins, others contain proteins that exhibit properties of soluble nonaggregated proteins, including the ability to exchange with the cytosolic pool (Kaganovich et al. 2008). Thus, inclusion formation does not solely serve the purpose of sequestering aggregates. As discussed below, these different inclusions likely serve different functions within the quality control network. Insoluble misfolded proteins often accumulate in perinuclear aggregate structures, particularly on proteasome inhibition. One of these structures, termed the aggresome was found to colocalize in mammalian cells with the microtubule-organizing center (MTOC) (Johnston et al. 2000, 2002). Aggresome formation depends on minus-end-directed transport along microtubules (Kopito 2000). In yeast, prions such as Sup35, also accumulate as insoluble amyloid-like aggregates (Tanaka et al. 2006) in distinct, nonperinuclear inclusions (Tyedmers et al. 2010b). Two transiently formed inclusions, termed the ALIS and DALIS, which form in response to immune activation or stress, appear to concentrate soluble proteins targeted for proteasomal or autophagic clearance (Szeto et al. 2006). For instance, the ALIS transiently accumulates polyubiquitinated defective ribosomal products in lipopolysaccharide-stimulated dendritic cells (Lelouard et al. 2002); this compartment also forms in other mammalian cells in response to environmental stresses (Szeto et al. 2006). Formation of the ALIS and DALIS do not depend on microtubule polymerization (Lelouard et al. 2002).
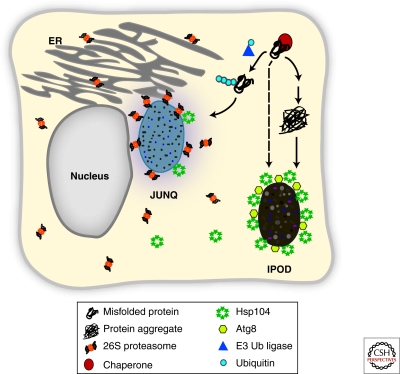
What is the biological significance of such different soluble and insoluble misfolded protein inclusions? Recent work has defined two compartments, called the JUNQ (juxtanuclear quality control) and IPOD (insoluble protein deposit), that sequester distinct types of misfolded proteins and appear to fulfill separate functions within the protein homeostasis network (Fig. 2) (Kaganovich et al. 2008). The JUNQ and the IPOD differ in their spatial and temporal separation of cytosolic inclusions and interface with different protein clearance pathways. The JUNQ compartment is associated with the cytosolic surface of the ER and contains 26S proteasomes and some chaperones such as Hsp104. The JUNQ concentrates soluble misfolded proteins that can either be degraded by the UPS or refolded by cytoplasmic chaperones. The IPOD contains insoluble aggregated proteins, including disease-associated proteins such as huntingtin and prions. The IPOD does not colocalize with proteasomes but does colocalize with the autophagy associated Atg8. The spatial concentration of soluble misfolded proteins into a chaperone and proteasome-rich JUNQ compartment may serve to both enhance clearance rates and enhance the removal of misfolded species from the crowded cellular milieu. In contrast, the spatial sequestration of aggregated proteins in the IPOD compartment may prevent these terminally misfolded aggregates from clogging the proteasome and sequestering soluble chaperones. Additionally, protein inclusions may enhance the efficiency of aggregate clearance, presumably by facilitating interactions with the lysosomal and autophagic pathways (Taylor et al. 2003). Importantly, the JUNQ and IPOD compartments are observed in both yeast and mammalian cells (Kaganovich et al. 2008), pointing to a conservation of these pathways and their functions.
Figure 2.
Distinct quality control compartments in eukaryotic cells. Quality control substrates can be sequestered into two spatial distinct compartments, the JUNQ and the IPOD. Misfolded, ubiquitinated proteins are delivered to the JUNQ, where they can be either degraded by the 26S proteasome or refolded with the assistance of chaperones, such as Hsp104. Insoluble misfolded proteins can also be terminally sequestered in the IPOD. The IPOD may be linked to the autophagy pathway based on its colocalization with Atg8. The molecular chaperone Hsp104 is also sequestered in the IPOD.
The distinct sequestration pathways described above are likely interconnected. For instance, in Caenorhabditis elegans, the HSF-1 transcriptome induces a disaggregation pathway that reduces the aggregation-mediated toxicity of cytosolic Aβ inclusions (Cohen et al. 2006); thus, it is tempting to speculate this enhances their flux through the JUNQ. On overloading of this pathway, the DAF-16 transcriptome enhances sequestration of Aβ into insoluble aggregates that are used for detoxification (Cohen et al. 2006); presumably, these enhance terminal sequestration in an IPOD-like compartment. These observations provide evidence for the cooperation between the two sequestration pathways to ensure maximal cellular protection against toxic aggregates.
How are proteins targeted and delivered to these cellular inclusions? Although it is clear that the processes of protein sequestration into quality control compartments is highly organized, how protein oligomers and aggregates are directed toward these compartments is still poorly understood. Targeting of misfolded proteins to the JUNQ depends on their ubiquitination state; for instance, blocking ubiquitination of misfolded VHL redirects it to the IPOD and renders it insoluble (Kaganovich et al. 2008). It has been proposed that histone deacetylase 6 (HDAC6) helps move ubiquitinated proteins along microtubules toward the MTOC (Johnston et al. 2002). Overexpression of HDAC6 has been shown to suppress toxicity in a fly model of SBMA by inducing a shift from proteasomal to autophagic degradation of the mutant androgen receptor protein (Du et al. 2010). HDAC6 is also implicated in the clearance of aggresomes through selective autophagic turnover and recruits the actin-remodeling machinery for ultimate engulfment (Lee et al. 2010). Another protein that has been proposed to link quality control and autophagic degradation is p62/SQSTM1, which binds to ubiquitinated proteins and autophagic components (Pankiv et al. 2007); although, its function in this process remains controversial (Riley et al. 2010). In yeast, aggregates are transported on actin filaments during mitotic cytokinesis by the polarisome complex and the myosin motor protein Myo2 (Liu et al. 2010). It is currently unclear how nuclear aggregates are formed.
ADVANTAGES OF PROTEIN SEQUESTRATION AS A PROTECTIVE QUALITY CONTROL STRATEGY
Many neurodegenerative diseases are characterized by the formation of intracellular or extracellular inclusions containing amyloid forms of disease-causing misfolded proteins, including huntingtin, α-synuclein, Aβ-peptide, SOD-1, and prion proteins. These proteins do not share primary sequence features, or functional characteristics, but do aggregate into insoluble, β-rich amyloid structures that stain with dyes like Congo Red and Thioflavin T (Wright et al. 2005). However, it is clear that any protein is capable of forming an amyloid under certain conditions (Chiti et al. 2000). Presumably, the ability to interact with the chaperone and quality control machineries distinguishes those disease-causing proteins that accumulate in amyloid inclusions from those that are degraded on misfolding and do not form amyloids.
Although the aggregation of misfolded proteins underlies a range of debilitating diseases (Finkbeiner et al. 2006), formation of insoluble amyloid inclusions can also promote cell survival and may serve as a protective mechanism by sequestering potentially harmful aggregates from the cytosol (Liu et al. 2010). Sequestration of disease-associated proteins into insoluble protein inclusions reduces their cytotoxicity (Arrasate et al. 2004) and reverses proteasomal dysfunction (Bodner et al. 2006). Incorporation of toxic oligomers into protective amyloid-like protein inclusions has been observed to reduce toxicity of Htt and Aß expressed in mammalian cells, C. elegans, and mouse models (Cheng et al. 2007; Bieschke et al. 2009; Cohen et al. 2009). These studies support the hypothesis that sequestration into an IPOD-like structure removes toxic misfolded species from the cellular environment.
The formation of spatially distinct inclusions also affords cells an additional mechanism for misfolded protein clearance, namely through the asymmetric inheritance of inclusions (Kaganovich et al. 2008; Liu et al. 2010). Asymmetric cell divisions entail differential segregation of cellular components between two cells (Henderson and Gottschling 2008). In yeast, asymmetric cytokinesis involves a rejuvenating process in which aged mother cells generate daughters with full replicative potential (Kennedy et al. 1994; Sinclair and Guarente 1997). Notably, whereas normal proteins in yeast diffuse freely between the mother cell and bud (Dobbelaere and Barral 2004), oxidatively damaged proteins are distributed asymmetrically between young mothers and cells in both Saccharomyces cerevisiae and Schizosaccharomyces pombe (Aguilaniu et al. 2003; Shcheprova et al. 2008). Similarly, asymmetric inheritance in dividing mammalian cells has been observed for ubiquitinated proteins (Fuentealba et al. 2008) and protein aggregates (Rujano et al. 2006). Interestingly, during yeast aging the capacity for asymmetric inheritance is lost (Liu et al. 2010), resulting in daughter cells with reduced replicative potential. Thus, the asymmetric inheritance of quality control inclusions may help clear the misfolded protein load in daughter cells in an actively dividing tissue or population.
The observations that formation of large insoluble aggregates can be protective (Arrasate et al. 2004) and that these aggregates do not turn over rapidly (Cohen and Kelly 2003; Balch et al. 2008), together with the finding that they may be inherited asymmetrically (Liu et al. 2010), underscore the advantages of protein sequestration as a quality control strategy.
TOWARD A MODEL FOR MISFOLDED PROTEIN TOXICITY IN AMYLOIDOGENIC DISEASE
Despite the protection afforded by sequestration of aggregates into IPOD-like insoluble inclusions, other inclusions correlate with cellular toxicity. For instance, formation of inclusions of ALS-associated mutant SOD1 (G93A) correlate with cell death, contrary to what is observed for large insoluble Htt inclusions (Arrasate et al. 2004). Indeed, polyQ-expanded Htt and mutant SOD-1 form distinct inclusions on coexpression in mammalian cells (Kitamura et al. 2006; Matsumoto et al. 2006). SOD-1 mutants form mobile soluble inclusions exposed to Hsp70, whereas Htt forms immobile insoluble inclusions impermeable to other cytosolic proteins. Not only do the mobile SOD-1 inclusions recruit and sequester proteasomes, but their appearance also correlates with the onset of toxicity and cell death (Kitamura et al. 2006; Matsumoto et al. 2006). A similar study in C. elegans found that another ALS-associated SOD1 mutant (G85R) causes neuronal toxicity resulting from soluble intracellular inclusions that appear to sequester the Hsp110 chaperone (Wang et al. 2009).
An attractive hypothesis is that misfolded oligomers and soluble aggregates cause toxicity by diminishing the capacity of the protein folding machinery. This hypothesis is supported by a recent study comparing the composition of toxic and nontoxic cellular aggregates in mammalian cells (Olzscha et al. 2011). The toxic aggregates are enriched in cellular metastable proteins compared to the nontoxic ones, suggesting that the toxic aggregates sequester essential chaperones and other factors, which in turn impairs protein homeostasis. Moreover, experiments in yeast, worms, flies, and mammalian cells have all identified a strong connection between misfolding, aggregation, cell death, and dysfunctional protein homeostasis (Lansbury and Lashuel 2006; Brignull et al. 2007). For instance, enhancers of polyQ toxicity include factors involved in protein folding, the stress response, and the UPS (Willingham et al. 2003). Conversely, overexpression or up-regulation of chaperones can delay or suppress aggregation and toxicity of various disease-associated proteins (e.g., polyQ, Aβ, α-synuclein, SOD1) (Fonte et al. 2002; Nollen et al. 2004; van Ham et al. 2008; Roodveldt et al. 2009). These findings support the idea that toxicity arises from adverse interactions between aggregated proteins and the protein homeostasis machinery. This in turn, diverts these essential cellular factors away from their quality control function. A number of studies have identified small oligomeric aggregates as the offending species linked to toxicity (Kayed et al. 2003). For example, in mammalian cell culture and yeast, accumulation of small oligomeric Htt aggregates correlates with toxicity (Kitamura et al. 2006). Moreover, in C. elegans, mutant SOD1 aggregates lead to misfolding of other metastable proteins in the cell (Gidalevitz et al. 2009). It is unclear at this point, which are the key pathways targeted by the toxic aggregates, or even if there is a primary system that is affected. Notably, the deleterious effect of the aggregating species can be modulated by ectopic expression of a variety of different quality control factors. Indeed, overexpression of chaperones, activation of autophagy, and/or activation of the proteasome have all been shown to ameliorate the symptoms of disease-related protein aggregates (Behrends et al. 2006; Fonte et al. 2008) as well as increase organism lifespan (Vacher et al. 2005). These data support a model whereby toxic aggregates sequester or block the function of chaperones and/or other components of the quality control network, leading to a general impairment of the protein homeostasis capacity of the cell.
CONCLUDING REMARKS
The complexity of protein quality control and protein homeostasis likely provides robustness to a process that is critical for cell viability. A surprising emerging concept is that quality control is spatially organized into compartments that manage misfolded proteins hierarchically within the cell. Soluble misfolded proteins are concentrated to enhance their refolding or degradation, whereas insoluble species with the potential to disrupt protein homeostasis are sequestered to prevent their toxic interactions with the quality control machinery. An important question is how the quality control network decides whether an aggregate should be sequestered or engaged in a refolding or degradation process. One possibility is that certain aggregates are simply insoluble and after being recognized as such are delivered for sequestration. Another possibility is that amyloidogenic and potentially toxic species are delivered to a quality control compartment, which sequesters them and allows them to aggregate into a benign amyloid structure. Additional questions for future research include how distinct compartments are organized and maintained and how their substrates are selected. Understanding why cells occasionally fail to compartmentalize misfolded toxic species, allowing them to interfere with normal protein homeostasis, will be instrumental in elucidating the etiology of amyloid diseases.
ACKNOWLEDGMENTS
B.C., M.R., and T.R. contributed equally to this review and are listed alphabetically. The authors thank Drs. Christine Livingston and Raul Andino for comments on the manuscript.
Footnotes
Editors: Richard Morimoto, Jeffrey Kelly, and Dennis Selkoe
Additional Perspectives on Protein Homeostasis available at www.cshperspectives.org
REFERENCES
- Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T 2003. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science 299: 1751–1753 [DOI] [PubMed] [Google Scholar]
- Akerfelt M, Morimoto RI, Sistonen L 2010. Heat shock factors: Integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol 11: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanese V, Yam AY, Baughman J, Parnot C, Frydman J 2006. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell 124: 75–88 [DOI] [PubMed] [Google Scholar]
- Alberti S, Demand J, Esser C, Emmerich N, Schild H, Hohfeld J 2002. Ubiquitylation of BAG-1 suggests a novel regulatory mechanism during the sorting of chaperone substrates to the proteasome. J Biol Chem 277: 45920–45927 [DOI] [PubMed] [Google Scholar]
- An S, Kumar R, Sheets ED, Benkovic SJ 2008. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 320: 103–106 [DOI] [PubMed] [Google Scholar]
- Arndt V, Rogon C, Hohfeld J 2007. To be, or not to be—molecular chaperones in protein degradation. Cell Mol Life Sci 64: 2525–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S 2004. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431: 805–810 [DOI] [PubMed] [Google Scholar]
- Bagola K, Sommer T 2008. Protein quality control: On IPODs and other JUNQ. Curr Biol 18: R1019–R1021 [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW 2008. Adapting proteostasis for disease intervention. Science 319: 916–919 [DOI] [PubMed] [Google Scholar]
- Behrends C, Langer CA, Boteva R, Bottcher UM, Stemp MJ, Schaffar G, Rao BV, Giese A, Kretzschmar H, Siegers K, et al. 2006. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol Cell 23: 887–897 [DOI] [PubMed] [Google Scholar]
- Ben-Zvi A, Miller EA, Morimoto RI 2009. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci 106: 14914–14919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieschke J, Cohen E, Murray A, Dillin A, Kelly JW 2009. A kinetic assessment of the C. elegans amyloid disaggregation activity enables uncoupling of disassembly and proteolysis. Protein Sci 18: 2231–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodner RA, Outeiro TF, Altmann S, Maxwell MM, Cho SH, Hyman BT, McLean PJ, Young AB, Housman DE, Kazantsev AG 2006. Pharmacological promotion of inclusion formation: A therapeutic approach for Huntington’s and Parkinson’s diseases. Proc Natl Acad Sci 103: 4246–4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brignull HR, Morley JF, Morimoto RI 2007. The stress of misfolded proteins: C. elegans models for neurodegenerative disease and aging. Adv Exp Med Biol 594: 167–189 [DOI] [PubMed] [Google Scholar]
- Brockwell DJ, Radford SE 2007. Intermediates: Ubiquitous species on folding energy landscapes? Curr Opin Struct Biol 17: 30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A, Bukau B, Sommer T 2010. Protein quality control in the cytosol and the endoplasmic reticulum: Brothers in arms. Mol Cell 40: 238–252 [DOI] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A 2006. Molecular chaperones and protein quality control. Cell 125: 443–451 [DOI] [PubMed] [Google Scholar]
- Caughey B, Lansbury PT 2003. Protofibrils, pores, fibrils, and neurodegeneration: Separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci 26: 267–298 [DOI] [PubMed] [Google Scholar]
- Cheng IH, Scearce-Levie K, Legleiter J, Palop JJ, Gerstein H, Bien-Ly N, Puolivali J, Lesne S, Ashe KH, Muchowski PJ, et al. 2007. Accelerating amyloid-β fibrillization reduces oligomer levels and functional deficits in Alzheimer disease mouse models. J Biol Chem 282: 23818–23828 [DOI] [PubMed] [Google Scholar]
- Chiti F, Taddei N, Bucciantini M, White P, Ramponi G, Dobson CM 2000. Mutational analysis of the propensity for amyloid formation by a globular protein. EMBO J 19: 1441–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A 1998. The ubiquitin-proteasome pathway: On protein death and cell life. EMBO J 17: 7151–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen FE, Kelly JW 2003. Therapeutic approaches to protein-misfolding diseases. Nature 426: 905–909 [DOI] [PubMed] [Google Scholar]
- Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A 2006. Opposing activities protect against age-onset proteotoxicity. Science 313: 1604–1610 [DOI] [PubMed] [Google Scholar]
- Cohen E, Paulsson JF, Blinder P, Burstyn-Cohen T, Du D, Estepa G, Adame A, Pham HM, Holzenberger M, Kelly JW, et al. 2009. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell 139: 1157–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Bergamini E, Brunk UT, Droge W, Ffrench M, Terman A 2005. Autophagy and aging: The importance of maintaining “clean” cells. Autophagy 1: 131–140 [DOI] [PubMed] [Google Scholar]
- Dai C, Whitesell L, Rogers AB, Lindquist S 2007. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell 130: 1005–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, Kenyon C 2010. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol 8: e1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F, Perrimon N 2010. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 143: 813–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J, Barral Y 2004. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science 305: 393–396 [DOI] [PubMed] [Google Scholar]
- Dobson CM 2003. Protein folding and misfolding. Nature 426: 884–890 [DOI] [PubMed] [Google Scholar]
- Dobson CM 2006. Protein aggregation and its consequences for human disease. Protein Pept Lett 13: 219–227 [DOI] [PubMed] [Google Scholar]
- Douglas PM, Dillin A 2010. Protein homeostasis and aging in neurodegeneration. J Cell Biol 190: 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du G, Liu X, Chen X, Song M, Yan Y, Jiao R, Wang CC 2010. Drosophila histone deacetylase 6 protects dopaminergic neurons against {α}-synuclein toxicity by promoting inclusion formation. Mol Biol Cell 21: 2128–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner S, Cuervo AM, Morimoto RI, Muchowski PJ 2006. Disease-modifying pathways in neurodegeneration. J Neurosci 26: 10349–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D, Ciechanover A, Varshavsky A 2004. Ubiquitin as a central cellular regulator. Cell 116: S29–S32, 22 pp. following S32 [DOI] [PubMed] [Google Scholar]
- Fonte V, Kapulkin V, Taft A, Fluet A, Friedman D, Link CD 2002. Interaction of intracellular β amyloid peptide with chaperone proteins. Proc Natl Acad Sci 99: 9439–9444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonte V, Kipp DR, Yerg J III, Merin D, Forrestal M, Wagner E, Roberts CM, Link CD 2008. Suppression of in vivo β-amyloid peptide toxicity by overexpression of the HSP-16.2 small chaperone protein. J Biol Chem 283: 784–791 [DOI] [PubMed] [Google Scholar]
- Frydman J 2001. Folding of newly translated proteins in vivo: The role of molecular chaperones. Annu Rev Biochem 70: 603–647 [DOI] [PubMed] [Google Scholar]
- Fuentealba LC, Eivers E, Geissert D, Taelman V, De Robertis EM 2008. Asymmetric mitosis: Unequal segregation of proteins destined for degradation. Proc Natl Acad Sci 105: 7732–7737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI 2006. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311: 1471–1474 [DOI] [PubMed] [Google Scholar]
- Gidalevitz T, Krupinski T, Garcia S, Morimoto RI 2009. Destabilizing protein polymorphisms in the genetic background direct phenotypic expression of mutant SOD1 toxicity. PLoS Genet 5: e1000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Yankner BA 2010. The aging stress response. Mol Cell 40: 333–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY 2011. San1-mediated quality control: Substrate recognition “sans” chaperones. Mol Cell 41: 2–3 [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441: 885–889 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M 2002. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science 295: 1852–1858 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M 2009. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol 16: 574–581 [DOI] [PubMed] [Google Scholar]
- Haslbeck M, Franzmann T, Weinfurtner D, Buchner J 2005. Some like it hot: The structure and function of small heat-shock proteins. Nat Struct Mol Biol 12: 842–846 [DOI] [PubMed] [Google Scholar]
- Haslberger T, Zdanowicz A, Brand I, Kirstein J, Turgay K, Mogk A, Bukau B 2008. Protein disaggregation by the AAA+ chaperone ClpB involves partial threading of looped polypeptide segments. Nat Struct Mol Biol 15: 641–650 [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ 2009. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43: 67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck JW, Cheung SK, Hampton RY 2010. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci 107: 1106–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson KA, Gottschling DE 2008. A mother’s sacrifice: What is she keeping for herself? Curr Opin Cell Biol 20: 723–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T 2009. The ubiquitylation machinery of the endoplasmic reticulum. Nature 458: 453–460 [DOI] [PubMed] [Google Scholar]
- Ishida Y, Yamamoto A, Kitamura A, Lamande SR, Yoshimori T, Bateman JF, Kubota H, Nagata K 2009. Autophagic elimination of misfolded procollagen aggregates in the endoplasmic reticulum as a means of cell protection. Mol Biol Cell 20: 2744–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JA, Dalton MJ, Gurney ME, Kopito RR 2000. Formation of high molecular weight complexes of mutant Cu, Zn-superoxide dismutase in a mouse model for familial amyotrophic lateral sclerosis. Proc Natl Acad Sci 97: 12571–12576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JA, Illing ME, Kopito RR 2002. Cytoplasmic dynein/dynactin mediates the assembly of aggresomes. Cell Motil Cytoskel 53: 26–38 [DOI] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J 2008. Misfolded proteins partition between two distinct quality control compartments. Nature 454: 1088–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 11: 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG 2003. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300: 486–489 [DOI] [PubMed] [Google Scholar]
- Kennedy BK, Austriaco NR Jr, Guarente L 1994. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J Cell Biol 127: 1985–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura A, Kubota H, Pack CG, Matsumoto G, Hirayama S, Takahashi Y, Kimura H, Kinjo M, Morimoto RI, Nagata K 2006. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat Cell Biol 8: 1163–1170 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Codogno P, Cuervo AM, Deretic V, Elazar Z, Fueyo-Margareto J, Gewirtz DA, Kroemer G, Levine B, Mizushima N, et al. 2010. A comprehensive glossary of autophagy-related molecules and processes. Autophagy 6: 438–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441: 880–884 [DOI] [PubMed] [Google Scholar]
- Kopito RR 2000. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol 10: 524–530 [DOI] [PubMed] [Google Scholar]
- Koulov AV, Lapointe P, Lu B, Razvi A, Coppinger J, Dong MQ, Matteson J, Laister R, Arrowsmith C, Yates JR 3rd, et al. 2010. Biological and structural basis for Aha1 regulation of Hsp90 ATPase activity in maintaining proteostasis in the human disease cystic fibrosis. Mol Biol Cell 21: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Marino G, Levine B 2010. Autophagy and the integrated stress response. Mol Cell 40: 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundrat L, Regan L 2010. Balance between folding and degradation for Hsp90-dependent client proteins: A key role for CHIP. Biochemistry 49: 7428–7438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbury PT, Lashuel HA 2006. A century-old debate on protein aggregation and neurodegeneration enters the clinic. Nature 443: 774–779 [DOI] [PubMed] [Google Scholar]
- Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, Pandey UB, Kaushik S, Tresse E, Lu J, et al. 2010. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J 29: 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelouard H, Gatti E, Cappello F, Gresser O, Camosseto V, Pierre P 2002. Transient aggregation of ubiquitinated proteins during dendritic cell maturation. Nature 417: 177–182 [DOI] [PubMed] [Google Scholar]
- Liu B, Larsson L, Caballero A, Hao X, Oling D, Grantham J, Nystrom T 2010. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell 140: 257–267 [DOI] [PubMed] [Google Scholar]
- Livingston CM, Ifrim MF, Cowan AE, Weller SK 2009. Virus-induced chaperone-enriched (VICE) domains function as nuclear protein quality control centers during HSV-1 infection. PLoSPathog 5: e1000619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotz GP, Legleiter J, Aron R, Mitchell EJ, Huang SY, Ng C, Glabe C, Thompson LM, Muchowski PJ 2010. Hsp70 and Hsp40 functionally interact with soluble mutant huntingtin oligomers in a classic ATP-dependent reaction cycle. J Biol Chem 285: 38183–38193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum R, Tkach JM, Vierling E, Glover JR 2004. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem 279: 29139–29146 [DOI] [PubMed] [Google Scholar]
- Matsumoto G, Kim S, Morimoto RI 2006. Huntingtin and mutant SOD1 form aggregate structures with distinct molecular properties in human cells. J Biol Chem 281: 4477–4485 [DOI] [PubMed] [Google Scholar]
- McClellan AJ, Scott MD, Frydman J 2005a. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell 121: 739–748 [DOI] [PubMed] [Google Scholar]
- McClellan AJ, Tam S, Kaganovich D, Frydman J 2005b. Protein quality control: Chaperones culling corrupt conformations. Nat Cell Biol 7: 736–741 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ 2008. Autophagy fights disease through cellular self-digestion. Nature 451: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI 2008. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev 22: 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley JF, Morimoto RI 2004. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell 15: 657–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley JF, Brignull HR, Weyers JJ, Morimoto RI 2002. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci 99: 10417–10422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski PJ 2002. Protein misfolding, amyloid formation, and neurodegeneration: A critical role for molecular chaperones? Neuron 35: 9–12 [DOI] [PubMed] [Google Scholar]
- Muchowski PJ, Wacker JL 2005. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci 6: 11–22 [DOI] [PubMed] [Google Scholar]
- Murray AN, Solomon JP, Wang YJ, Balch WE, Kelly JW 2010. Discovery and characterization of a mammalian amyloid disaggregation activity. Protein Sci 19: 836–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL 2008. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell 132: 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanaswamy R, Levy M, Tsechansky M, Stovall GM, O’Connell JD, Mirrielees J, Ellington AD, Marcotte EM 2009. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc Natl Acad Sci 106: 10147–10152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollen EA, Garcia SM, van Haaften G, Kim S, Chavez A, Morimoto RI, Plasterk RH 2004. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci 101: 6403–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystrom T 2005. Role of oxidative carbonylation in protein quality control and senescence. EMBO J 24: 1311–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM 2011. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144: 67–78 [DOI] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, et al. 2007. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 447: 859–863 [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131–24145 [DOI] [PubMed] [Google Scholar]
- Pickart CM, Cohen RE 2004. Proteasomes and their kin: Proteases in the machine age. Nat Rev Mol Cell Biol 5: 177–187 [DOI] [PubMed] [Google Scholar]
- Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE 2009. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem 78: 959–991 [DOI] [PubMed] [Google Scholar]
- Radonjic M, Andrau JC, Lijnzaad P, Kemmeren P, Kockelkorn TT, van Leenen D, van Berkum NL, Holstege FC 2005. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol Cell 18: 171–183 [DOI] [PubMed] [Google Scholar]
- Richter K, Haslbeck M, Buchner J 2010. The heat shock response: Life on the verge of death. Mol Cell 40: 253–266 [DOI] [PubMed] [Google Scholar]
- Riley BE, Kaiser SE, Shaler TA, Ng AC, Hara T, Hipp MS, Lage K, Xavier RJ, Ryu KY, Taguchi K, et al. 2010. Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: A potential role for protein aggregation in autophagic substrate selection. J Cell Biol 191: 537–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roodveldt C, Bertoncini CW, Andersson A, van der Goot AT, Hsu ST, Fernandez-Montesinos R, de Jong J, van Ham TJ, Nollen EA, Pozo D, et al. 2009. Chaperone proteostasis in Parkinson’s disease: Stabilization of the Hsp70/α-synuclein complex by Hip. EMBO J 28: 3758–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Poirier MA 2004. Protein aggregation and neurodegenerative disease. Nat Med 10 (Suppl): S10–S17 [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ 2007. Potential therapeutic applications of autophagy. Nat Rev Drug Discov 6: 304–312 [DOI] [PubMed] [Google Scholar]
- Rujano MA, Bosveld F, Salomons FA, Dijk F, van Waarde MA, van der Want JJ, de Vos RA, Brunt ER, Sibon OC, Kampinga HH 2006. Polarised asymmetric inheritance of accumulated protein damage in higher eukaryotes. PLoS Biol 4: e417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakahira H, Breuer P, Hayer-Hartl MK, Hartl FU 2002. Molecular chaperones as modulators of polyglutamine protein aggregation and toxicity. Proc Natl Acad Sci 99 (Suppl) 4: 16412–16418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawkar AR, D’Haeze W, Kelly JW 2006. Therapeutic strategies to ameliorate lysosomal storage disorders–a focus on Gaucher disease. Cell Mol Life Sci 63: 1179–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlecht R, Erbse AH, Bukau B, Mayer MP 2011. Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat Struct Mol Biol 18: 345–351 [DOI] [PubMed] [Google Scholar]
- Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y 2008. A mechanism for asymmetric segregation of age during yeast budding. Nature 454: 728–734 [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L 1997. Extrachromosomal rDNA circles–a cause of aging in yeast. Cell 91: 1033–1042 [DOI] [PubMed] [Google Scholar]
- Szeto J, Kaniuk NA, Canadien V, Nisman R, Mizushima N, Yoshimori T, Bazett-Jones DP, Brumell JH 2006. ALIS are stress-induced protein storage compartments for substrates of the proteasome and autophagy. Autophagy 2: 189–199 [DOI] [PubMed] [Google Scholar]
- Taipale M, Jarosz DF, Lindquist S 2010. HSP90 at the hub of protein homeostasis: Emerging mechanistic insights. Nat Rev Mol Cell Biol 11: 515–528 [DOI] [PubMed] [Google Scholar]
- Tam S, Geller R, Spiess C, Frydman J 2006. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat Cell Biol 8: 1155–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam S, Spiess C, Auyeung W, Joachimiak L, Chen B, Poirier MA, Frydman J 2009. The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat Struct Mol Biol 16: 1279–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Collins SR, Toyama BH, Weissman JS 2006. The physical basis of how prion conformations determine strain phenotypes. Nature 442: 585–589 [DOI] [PubMed] [Google Scholar]
- Taylor JP, Tanaka F, Robitschek J, Sandoval CM, Taye A, Markovic-Plese S, Fischbeck KH 2003. Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum Mol Genet 12: 749–757 [DOI] [PubMed] [Google Scholar]
- Tyedmers J, Mogk A, Bukau B 2010a. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol 11: 777–788 [DOI] [PubMed] [Google Scholar]
- Tyedmers J, Treusch S, Dong J, McCaffery JM, Bevis B, Lindquist S 2010b. Prion induction involves an ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. Proc Natl Acad Sci 107: 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacher C, Garcia-Oroz L, Rubinsztein DC 2005. Overexpression of yeast hsp104 reduces polyglutamine aggregation and prolongs survival of a transgenic mouse model of Huntington’s disease. Hum Mol Genet 14: 3425–3433 [DOI] [PubMed] [Google Scholar]
- van Ham TJ, Thijssen KL, Breitling R, Hofstra RM, Plasterk RH, Nollen EA 2008. C. elegans model identifies genetic modifiers of α-synuclein inclusion formation during aging. PLoS Genet 4: e1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL 2008. One step at a time: Endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol 9: 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisine C, Pedersen JS, Morimoto RI 2010. Chaperone networks: Tipping the balance in protein folding diseases. Neurobiol Dis 40: 12–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker JL, Zareie MH, Fong H, Sarikaya M, Muchowski PJ 2004. Hsp70 and Hsp40 attenuate formation of spherical and annular polyglutamine oligomers by partitioning monomer. Nat Struct Mol Biol 11: 1215–1222 [DOI] [PubMed] [Google Scholar]
- Wang J, Farr GW, Zeiss CJ, Rodriguez-Gil DJ, Wilson JH, Furtak K, Rutkowski DT, Kaufman RJ, Ruse CI, Yates JR III, et al. 2009. Progressive aggregation despite chaperone associations of a mutant SOD1-YFP in transgenic mice that develop ALS. Proc Natl Acad Sci 106: 1392–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrick JM, Chan HY, Gray B, Chai Y, Paulson HL, Bonini NM 1999. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet 23: 425–428 [DOI] [PubMed] [Google Scholar]
- Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FT, et al. 2004. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119: 653–665 [DOI] [PubMed] [Google Scholar]
- Welker S, Rudolph B, Frenzel E, Hagn F, Liebisch G, Schmitz G, Scheuring J, Kerth A, Blume A, Weinkauf S, et al. 2010. Hsp12 is an intrinsically unstructured stress protein that folds upon membrane association and modulates membrane function. Mol Cell 39: 507–520 [DOI] [PubMed] [Google Scholar]
- Wendler P, Shorter J, Plisson C, Cashikar AG, Lindquist S, Saibil HR 2007. Atypical AAA+ subunit packing creates an expanded cavity for disaggregation by the protein-remodeling factor Hsp104. Cell 131: 1366–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendler P, Shorter J, Snead D, Plisson C, Clare DK, Lindquist S, Saibil HR 2009. Motor mechanism for protein threading through Hsp104. Mol Cell 34: 81–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L, Lindquist SL 2005. HSP90 and the chaperoning of cancer. Nat Rev Cancer 5: 761–772 [DOI] [PubMed] [Google Scholar]
- Wilkinson KD 2000. Ubiquitination and deubiquitination: Targeting of proteins for degradation by the proteasome. Semin Cell Dev Biol 11: 141–148 [DOI] [PubMed] [Google Scholar]
- Willingham S, Outeiro TF, DeVit MJ, Lindquist SL, Muchowski PJ 2003. Yeast genes that enhance the toxicity of a mutant huntingtin fragment or α-synuclein. Science 302: 1769–1772 [DOI] [PubMed] [Google Scholar]
- Winkler J, Seybert A, Konig L, Pruggnaller S, Haselmann U, Sourjik V, Weiss M, Frangakis AS, Mogk A, Bukau B 2010. Quantitative and spatio-temporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO J 29: 910–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CF, Teichmann SA, Clarke J, Dobson CM 2005. The importance of sequence diversity in the aggregation and evolution of proteins. Nature 438: 878–881 [DOI] [PubMed] [Google Scholar]
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J 2009. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137: 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youker RT, Walsh P, Beilharz T, Lithgow T, Brodsky JL 2004. Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol Biol Cell 15: 4787–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]