Abstract
Translation elongation factors facilitate protein synthesis by the ribosome. Previous studies identified two universally conserved translation elongation factors EF-Tu/eEF1A and EF-G/eEF2 that deliver aminoacyl-tRNAs to the ribosome and promote ribosomal translocation, respectively1. The factor eIF5A, the sole protein in eukaryotes and archaea containing the unusual amino acid hypusine [Nε-(4-amino-2-hydroxybutyl)lysine]2, was originally identified based on its ability to stimulate the yield (endpoint) of methionyl-puromycin synthesis, a model assay for first peptide bond synthesis thought to report on certain aspects of translation initiation3,4. Hypusine is required for eIF5A to associate with ribosomes5,6, and to stimulate methionyl-puromycin synthesis7. As eIF5A did not stimulate earlier steps of translation initiation8, and depletion of eIF5A in yeast only modestly impaired protein synthesis9, it was proposed that eIF5A function was limited to stimulating synthesis of the first peptide bond or that eIF5A functioned on only a subset of cellular mRNAs. However, the precise cellular role of eIF5A is unknown, and the protein has also been linked to mRNA decay, including the nonsense-mediated mRNA decay (NMD) pathway10,11, and to nucleocytoplasmic transport12,13. Here we show using molecular genetic and biochemical studies that eIF5A promotes translation elongation. Depletion or inactivation of eIF5A in yeast resulted in the accumulation of polysomes and an increase in ribosomal transit times. Addition of recombinant eIF5A from yeast, but not a derivative lacking hypusine, enhanced the rate of tripeptide synthesis in vitro. Moreover, inactivation of eIF5A mimicked the effects of the eEF2 inhibitor sordarin, indicating that eIF5A might function together with eEF2 to promote ribosomal translocation. As eIF5A is a structural homolog of the bacterial protein EF-P14,15, we propose that eIF5A/EF-P is a universally conserved translation elongation factor.
To help clarify the function of eIF5A, encoded by the TIF51A (HYP2) and TIF51B (ANB1) genes in S. cerevisiae, we deleted the non-essential TIF51B gene and constructed a copper-regulated TIF51A degron mutant (tif51a-td) that produces a ubiquitin-Arg codon-eIF5A fusion protein (UBI-R-eIF5A). Under permissive conditions the tif51a-td mutant grew similarly to the isogenic wild type (WT) strain (Figure 1a and Supplementary Figure 1a). Following a shift to nonpermissive conditions, UBI-R-eIF5A was depleted by 6 h and the growth rate of the tif51a-td strain was substantially reduced (Figure 1a and Supplementary Figure 1). This growth defect was complemented by a WT TIF51A plasmid (data not shown). As shown in Figure 1b, depletion of eIF5A following 14 h growth of the tif51a-td mutant under non-permissive conditions, like depletion of the eIF3a (TIF32/RPG1) subunit of the translation initiation factor eIF316, caused a severe reduction in [35S]methionine incorporation into total protein.
Figure 1. eIF5A depletion impairs yeast cell growth and protein synthesis, and causes retention of polysomes.

(a) Isogenic WT (J714) and tif51a-td mutant strains (J702) were serially diluted, spotted on permissive (SCRaf + Cu2+) or non-permissive (SCGal) medium, and incubated 5 days at 25 °C. Total protein synthesis (b) and polysome profiles (c) were analyzed in tif51a-td (J702), tif32-td (YAJ22), and their isogenic WT strains following growth under non-permissive conditions. Incorporation (dpm) of [35S]Met is expressed per A600 unit, and results are representative of triplicate experiments. WT* and tif32-td* strains are isogenic. (c) Cells were treated (+) or untreated (-) with 50 μg/ml cycloheximide (CHX) prior to harvesting, and WCEs were separated on sucrose gradients and fractionated to visualize the indicated ribosomal species. P/M ratios were calculated by comparing areas under the 80S and polysome peaks; ↓polysomes indicates reduced amount of polysomes.
Polyribosome profiles of whole cell extracts (WCEs) from WT and tif51a-td strains grown under nonpermissive conditions were analyzed by velocity sedimentation in sucrose gradients. In WT cells treated with cycloheximide (CHX) to inhibit translation elongation, distinct 40S and 60S ribosomal subunit peaks, as well as 80S monosome and polysome peaks were detected (Figure 1c, upper left panel). Mutations that impair translation initiation, such as the eIF3a degron, reduce the amount of polysomes and cause a corresponding increase in the 80S peak, which consists of both 80S monosomes translating an mRNA and 80S complexes not engaged with an mRNA (note the 5-fold reduction in polysome to monosome (P/M) ratio in the tif32-td versus WT strain, Figure 1c, upper left and middle panels). In contrast, inhibition of translation in the tif51a-td strain resulted in a polysome profile indistinguishable from the WT strain and a similar P/M ratio (Figure 1c, upper panels). Thus, depletion of eIF5A did not appreciably impair translation initiation.
To monitor post-initiation defects, polysome analyses were performed in the absence of CHX. Under these conditions, ribosomes in a WT strain continue elongating during extract preparation, run-off the mRNA, and accumulate as vacant 80S couples (Figure 1c, lower left panel). In contrast, a defect in translation elongation or termination results in slower ribosome run-off and the retention of polysomes, as measured by an increase in the P/M ratio. As shown in Figure 1c (lower panels), polysomes were retained in the tif51a-td strain in the absence of CHX resulting in a 4-fold increase in the P/M ratio; i.e., mimicking the effect of CHX on the WT strain. If eIF5A only functioned during synthesis of the first peptide bond, as was previously proposed8, then post-initiation ribosomes would be expected to continue elongating and run-off the mRNA during extract preparation (like the tif32-td initiation mutant in Figure 1c). The stable polysomes in the tif51a-td strain, which were observed in crude cell extracts and thus include the majority of the mRNAs in the cell, indicate a general translation elongation/termination defect in the absence of eIF5A.
To rapidly inactivate eIF5A, we identified a new temperature-sensitive mutation in TIF51A. The tif51a-D63V mutant exhibited a slow-growth phenotype at permissive temperature (25 °C) that was exacerbated at 37 °C and lethal at 38 °C (Figure 2a), consistent with substantial loss of the protein in vivo (data not shown). Both the tif51a-D63V mutant and the previously characterized tif51a-S149P mutant, isolated as a suppressor of NMD11, stabilized polysomes when incubated for 2 h at the non-permissive temperature (Supplementary Figure 2). Whereas mutations that impair elongation or termination could potentially cause polysome retention in the absence of CHX, our analysis (Supplementary Figure 3a) and previously published results17 of yeast termination factor eRF3 (sup35/sup2) mutants revealed polysome run-off. In contrast, the tif51a-D63V and tif51a-S149P mutants (Supplementary Figure 2), like elongation factor eEF3 and eEF2 mutants18,19, and the eEF2 inhibitor sordarin (Supplementary Figure 3b), caused polysome accumulation. Thus, the primary defect in vivo upon inactivation of eIF5A is impaired translation elongation.
Figure 2. Translation elongation defect in temperature-sensitive tif51a-D63V mutant.
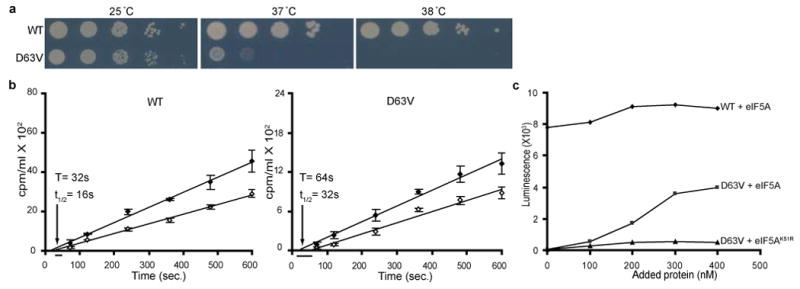
(a) Isogenic WT (J697) and tif51a-D63V mutant (D63V; J698) strains were serially diluted, spotted on SC medium, and incubated 3 days at 25, 37, or 38 °C. (b) WT (left panel) and tif51a-D63V (right) mutant strains were shifted to 36 °C for 2 h, and then labeled with [35S]Met. Fitting lines of total (filled symbols) and completed (open symbols) protein synthesis were obtained by linear regression and used to determine the half-average (t1/2) and average (T) transit time. Data are the average of three independent experiments. (c) In vitro translation activity of heat-treated extracts from WT and tif51a-D63V strains following addition of eIF5A or eIF5AK51R. Results are representative of six independent experiments.
To provide an independent assessment of translation elongation/termination, we determined ribosomal transit times in WT and tif51a-D63V mutant strains cultured at a semi-permissive temperature (36 °C). The ribosomal transit time is the time required for a ribosome, following initiation, to decode an mRNA and release the completed polypeptide chain20,21. Whereas [35S]Met incorporation into total protein (nascent chains + completed polypeptides) increases linearly upon adding the labeled amino acid to cells, there is a lag before [35S]Met incorporation into completed (disengaged from the ribosome and therefore soluble) proteins increases linearly. Plotting the incorporation of [35S]Met into total (nascent + completed) and completed proteins as a function of time yields two lines. The displacement (lag) between the lines reflects the time required to uniformly label the nascent polypeptides; and the average transit time is calculated by doubling the difference in the x-intercepts for the two lines. The average ribosomal transit time in the tif51a-D63V mutant strain at 36 °C (∼64 sec) was ∼2-fold slower than the WT strain (∼32 sec) (Figure 2b), confirming that inactivation of eIF5A impairs translation elongation/termination.
The translation defect in the eIF5A mutant strains could be due to a direct role of eIF5A in translation or an indirect effect of eIF5A on the translational machinery. Following a brief (5 min) incubation at the restrictive temperature (38 °C), extracts from the tif51a-D63V mutant were inactive in translating a luciferase reporter mRNA (Figure 2c, y-intercept). Addition of recombinant WT eIF5A purified from yeast, but not unhypusinated eIF5AK51R, resulted in a dose-dependent restoration of translational activity (Figure 2c). Thus, eIF5A appears to directly stimulate translation and to function as a general translation elongation factor in a manner dependent on its hypusine modification.
To extend the in vitro studies, we examined the effects of eIF5A using an in vitro reconstituted translation system. First, we confirmed previous reports7,8 that eIF5A, but not unhypusinated eIF5AK51R, stimulated methionyl-puromycin synthesis, a model assay of first peptide bond synthesis (data not shown). Next, yeast ribosomes were programmed with a defined mRNA encoding Met-Phe-Phe-Stop (AUG-UUC-UUC-UAA) and initiator [35S]Met-tRNAiMet using purified initiation factors (eIF1, eIF1A, eIF2, eIF5 and eIF5B)22, and these initiation complexes were pelleted over a sucrose cushion. To assay elongation, initiation complexes were reacted with an elongation factor mix containing eEF1A•Phe-tRNAPhe•GTP ternary complex, eEF2, eEF3, GTP and ATP along with eIF5A or the unhypusinated eIF5AK51R mutant. The rate of production of tripeptide product (MFF) was stimulated approximately 2-fold by eIF5A in this purified system whereas the variant eIF5AK51R had no apparent effect (Figure 3a and Supplementary Figure 4a). We also examined the rate of peptide release (termination) using pelleted elongation complexes (carrying an mRNA with a dipeptide ORF and a dipeptidyl-tRNA ([35S]Met-Phe-tRNAPhe) in the P site) to which purified eRF1 and eRF3 were added and the products of the reaction analyzed in a similar fashion. As for the elongation steps, the rate of the termination reaction was stimulated approximately 2-fold by WT eIF5A, but not by the unhypusinated variant (eIF5AK51R) (Figure 3b and Supplementary Figure 4b). In neither case (elongation or termination) were there any effects on the extent (endpoint) of the reaction. Taken together, these data suggest that eIF5A optimizes the ribosome complex for more productive interactions of the mechanistically related tRNA and release factor substrates23.
Figure 3. eIF5A stimulates elongation and termination in vitro.
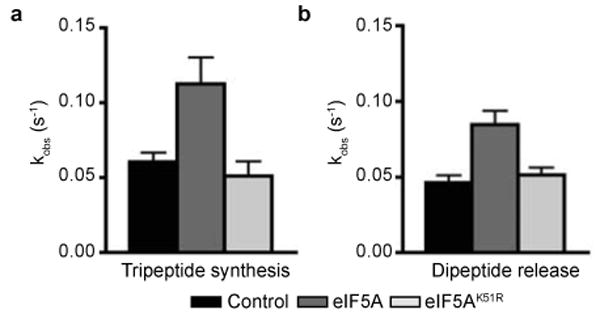
Rate constants for (a) tripeptide synthesis and (b) dipeptide release in the presence of eIF5A, eIF5AK51R, or no added factors. Error bars are the standard error from three independent experiments.
Consistent with eIF5A functioning in translation elongation, the tif51a-D63V and tif51a-S149P mutants impaired programmed ribosomal frameshifting. Interestingly, the eIF5A mutants like treatment of the WT strain with the eEF2 inhibitor sordarin caused a similar and specific inhibition of +1, but not -1, frameshifting (Supplementary Figure 5, see also24). As sordarin also exacerbated the growth defect of the tif51a-D63V mutant (Figure 4a), these data suggest that eIF5A and eEF2 functionally interact in the ribosome elongation cycle. In accordance, eIF5A associated with polysomes in vivo. Following treatment with formaldehyde to freeze polysomes and cross-link factors to the ribosome16, WCEs were subjected to velocity sedimentation in sucrose gradients. As shown in Figure 4b, eIF5A, like elongation factors eEF1A, eEF2, and eEF3, was found at the top of the gradient, in 80S complexes, and in polysomes. In contrast, 3-phosphoglycerate kinase (PGK1), a soluble cytoplasmic protein, was found exclusively at the top of the gradient (Figure 4b), the peripheral ribosomal 40S protein ASC1/RACK1 co-sedimented with the integral 40S protein RPS22 (data not shown), and initiation factors eIF2 (Figure 4b) and eIF3 (data not shown) were distributed across the gradient and did not accumulate on 80S complexes like eIF5A and the elongation factors. As shown in Figure 4c, depletion of eIF5A in the tif51a-td strain resulted in an ∼2-fold accumulation of elongation factor eEF2 on polysomes. These results, combined with the ability of GST-eIF5A to pull-down eEF2 and 80S ribosomes from crude yeast extracts6, support the notion that eIF5A functionally interacts with eEF2 to promote translation elongation.
Figure 4. Functional connection between eIF5A and eEF2.
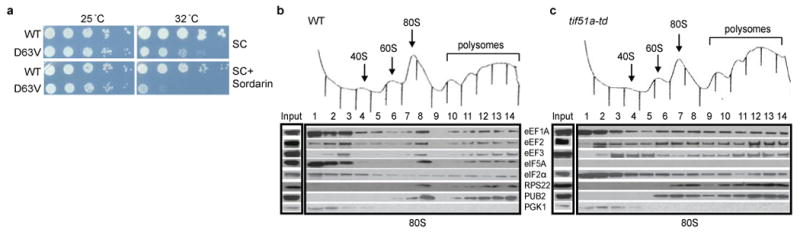
(a) Isogenic WT (J697) and tif51a-D63V (D63V; J698) strains were serially diluted, spotted on SC medium lacking or containing 200 ng/ml sordarin, and incubated 3 days at 25 or 32 °C. (b,c) Cultures of WT or tif51-td strains grown under non-permissive conditions were cross-linked with 1% formaldehyde. WCEs were separated on sucrose gradients, and gradient fractions were subjected to Western analysis using antibodies against the indicated proteins (PUB2 = 60S subunit protein RPL39).
Owing to its unique and essential hypusine modification, eIF5A has been of considerable interest. Our results provide evidence that eIF5A is a translation elongation factor and provide a rationale for its previously proposed roles in translation initiation and mRNA turnover. It is noteworthy that the putative translation initiation function of eIF5A was assigned based on its ability to promote methionyl-puromycin synthesis3,4,8, essentially a translation elongation step. Similarly, as NMD requires ongoing translation, the inhibition of NMD by a mutation in eIF5A11 is consistent with the observation that CHX blocks this form of mRNA turnover25. eIF5A is a structural homolog of the bacterial protein EF-P14,15, which was reported to enhance methionyl-puromycin and poly-Phe synthesis in reconstituted translation assays26,27. As both EF-P and eIF5A alter the Mg2+ optimum for protein synthesis in vitro4,27, the failure to previously detect the critical role of eIF5A/EF-P in translation elongation might stem from the fact that early biochemical screens were performed using in vitro translation reactions with abundant Mg2+ levels. Taken together, our studies reveal that in addition to eEF1A/EF-Tu and eEF2/EF-G a third universal and essential factor, eIF5A/EF-P, is required for translation elongation. Future studies will seek to determine at a molecular level how eIF5A promotes the ribosomal reactions required for translation elongation.
Methods Summary
The tif51a-td degron allele was constructed using methods described previously16 by inserting upstream of and in-frame with the TIF51A open reading frame a DNA cassette carrying a copper-regulated promoter, the protein-destabilizing ubiquitin coding region and an Arg codon (pCUP1-UBI-R)28. To enhance degradation of the UBI-R-eIF5A fusion protein, a galactose-inducible version of the ubiquitin E3-ligase UBR129 was introduced into the tif51a-td strain. The tif51a temperature-sensitive mutants were isolated and growth assays of 10-fold serial dilutions of yeast strains were performed using standard methods. Degron strains were grown under permissive [synthetic complete (SC) medium containing 2% raffinose and 100μM copper sulphate (SCRaf + Cu2+) – where the CUP1 promoter is induced and UBR1 expression is low] or under non-permissive [SC containing 2% galactose lacking copper (SCGal) that represses new synthesis and triggers proteasomal degradation of UBI-R-eIF5A] conditions. Plasmids and yeast strains are listed in Supplementary Table 1 and Supplementary Table 2, respectively. Yeast polysome analyses from formaldehyde cross-linked cells and analysis of ribosomal complexes were performed as described previously16. Anti-yeast eIF5A antiserum was obtained from rabbits immunized with recombinant N-terminal poly-histidine tagged eIF5A produced in bacteria. The assays for programmed ribosomal frameshifting30 were conducted as previously described. The ribosomal transit time measurements21 and the reconstituted translation elongation assays22 were extensions of previously established methods.
Supplementary Material
Acknowledgments
We thank T.G. Kinzy for providing anti-yeast eEF1A, eEF2 and eEF3 antisera, as well as constructs for the purification of eEF2 and eEF3; J. Lorsch for providing constructs for purification of yeast initiation factors; J Dinmann for PRF reporter vectors, and A. Hinnebusch, T.G. Kinzy, A. Jivotovskaya, D. Shelton, C. Grant, J. Lorsch, and members of the Dever and Hinnebusch laboratories for comments and discussion. Salary support provided by HHMI (R.G.) and NIH (D.E.E.). This work was supported in part by the Intramural Research Program of the NIH, NICHD (to T.E.D.).
Footnotes
Full methods and associated references are available in the online version of the paper at www.nature.com/nature.
Author Information: Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Merrick WC, Nyborg J. In: Translational Control of Gene Expression. Sonenberg N, Hershey JWB, Mathews MB, editors. Cold Spring Harbor Laboratory Press; 2000. pp. 89–125. [Google Scholar]
- 2.Wolff EC, Kang KR, Kim YS, Park MH. Posttranslational synthesis of hypusine: evolutionary progression and specificity of the hypusine modification. Amino Acids. 2007;33:341–350. doi: 10.1007/s00726-007-0525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kemper WM, Berry KW, Merrick WC. Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Bα and M2Bβ. J Biol Chem. 1976;251:5551–5557. [PubMed] [Google Scholar]
- 4.Schreier MH, Erni B, Staehelin T. Initiation of mammalian protein synthesis: purification and characterization of seven initiation factors. J Mol Biol. 1977;116:727–753. doi: 10.1016/0022-2836(77)90268-6. [DOI] [PubMed] [Google Scholar]
- 5.Jao DL, Chen KY. Tandem affinity purification revealed the hypusine-dependent binding of eukaryotic initiation factor 5A to the translating 80S ribosomal complex. J Cell Biochem. 2006;97:583–598. doi: 10.1002/jcb.20658. [DOI] [PubMed] [Google Scholar]
- 6.Zanelli CF, et al. eIF5A binds to translational machinery components and affects translation in yeast. Biochem Biophys Res Comm. 2006;348:1358–1366. doi: 10.1016/j.bbrc.2006.07.195. [DOI] [PubMed] [Google Scholar]
- 7.Park MH, Wolff EC, Smit-McBride Z, Hershey JW, Folk JE. Comparison of the activities of variant forms of eIF-4D The requirement for hypusine or deoxyhypusine. J Biol Chem. 1991;266:7988–7994. [PubMed] [Google Scholar]
- 8.Benne R, Hershey JWB. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J Biol Chem. 1978;253:3078–3087. [PubMed] [Google Scholar]
- 9.Kang HA, Hershey JW. Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae. J Biol Chem. 1994;269:3934–3940. [PubMed] [Google Scholar]
- 10.Schrader R, Young C, Kozian D, Hoffmann R, Lottspeich F. Temperature-sensitive eIF5A mutant accumulates transcripts targeted to the nonsense-mediated decay pathway. J Biol Chem. 2006;281:35336–35346. doi: 10.1074/jbc.M601460200. [DOI] [PubMed] [Google Scholar]
- 11.Zuk D, Jacobson A. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J. 1998;17:2914–2925. doi: 10.1093/emboj/17.10.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruhl M, et al. Eukaryotic initiation factor 5A is a cellular target of the human immunodeficiency virus type 1 Rev activation domain mediating trans-activation. J Cell Biol. 1993;123:1309–1320. doi: 10.1083/jcb.123.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zanelli CF, Valentini SR. Is there a role for eIF5A in translation? Amino Acids. 2007;33:351–358. doi: 10.1007/s00726-007-0533-0. [DOI] [PubMed] [Google Scholar]
- 14.Hanawa-Suetsugu K, et al. Crystal structure of elongation factor P from Thermus thermophilus HB8. Proc Natl Acad Sci USA. 2004;101:9595–9600. doi: 10.1073/pnas.0308667101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kyrpides NC, Woese CR. Universally conserved translation initiation factors. Proc Natl Acad Sci USA. 1998;95:224–228. doi: 10.1073/pnas.95.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jivotovskaya AV, Valasek L, Hinnebusch AG, Nielsen KH. Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast. Mol Cell Biol. 2006;26:1355–1372. doi: 10.1128/MCB.26.4.1355-1372.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smirnov VN, et al. Recessive nonsense-suppression in yeast: further characterization of a defect in translation. FEBS Lett. 1976;66:12–15. doi: 10.1016/0014-5793(76)80573-x. [DOI] [PubMed] [Google Scholar]
- 18.Anand M, Chakraburtty K, Marton MJ, Hinnebusch AG, Kinzy TG. Functional interactions between yeast translation eukaryotic elongation factor (eEF) 1A and eEF3. J Biol Chem. 2003;278:6985–6991. doi: 10.1074/jbc.M209224200. [DOI] [PubMed] [Google Scholar]
- 19.Ortiz PA, Kinzy TG. Dominant-negative mutant phenotypes and the regulation of translation elongation factor 2 levels in yeast. Nucleic Acids Res. 2005;33:5740–5748. doi: 10.1093/nar/gki882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan H, Penman S. Regulation of protein synthesis in mammalian cells II Inhibition of protein synthesis at the level of initiation during mitosis. J Mol Biol. 1970;50:655–670. doi: 10.1016/0022-2836(70)90091-4. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen PJ, McConkey EH. Evidence for control of protein synthesis in HeLa cells via the elongation rate. J Cell Physiol. 1980;104:269–281. doi: 10.1002/jcp.1041040302. [DOI] [PubMed] [Google Scholar]
- 22.Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR. Reconstitution of yeast translation initiation. Methods Enzymol. 2007;430:111–145. doi: 10.1016/S0076-6879(07)30006-2. [DOI] [PubMed] [Google Scholar]
- 23.Youngman EM, McDonald ME, Green R. Peptide release on the ribosome: mechanism and implications for translational control. Ann Rev Microbiol. 2008;62:353–373. doi: 10.1146/annurev.micro.61.080706.093323. [DOI] [PubMed] [Google Scholar]
- 24.Harger JW, Meskauskas A, Nielsen J, Justice MC, Dinman JD. Ty1 retrotransposition and programmed +1 ribosomal frameshifting require the integrity of the protein synthetic translocation step. Virology. 2001;286:216–224. doi: 10.1006/viro.2001.0997. [DOI] [PubMed] [Google Scholar]
- 25.Zhang S, et al. Polysome-associated mRNAs are substrates for the nonsense-mediated mRNA decay pathway in Saccharomyces cerevisiae. RNA. 1997;3:234–244. [PMC free article] [PubMed] [Google Scholar]
- 26.Glick BR, Ganoza MC. Identification of a soluble protein that stimulates peptide bond synthesis. Proc Natl Acad Sci USA. 1975;72:4257–4260. doi: 10.1073/pnas.72.11.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganoza MC, Aoki H. Peptide bond synthesis: Function of the efp gene product. Biol Chem. 2000;381:553–559. doi: 10.1515/BC.2000.071. [DOI] [PubMed] [Google Scholar]
- 28.Dohmen RJ, Wu P, Varshavsky A. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science. 1994;263:1273–1276. doi: 10.1126/science.8122109. [DOI] [PubMed] [Google Scholar]
- 29.Labib K, Tercero JA, Diffley JF. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- 30.Harger JW, Dinman JD. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA. 2003;9:1019–1024. doi: 10.1261/rna.5930803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


