Abstract
During evolution, organisms have gained functional complexity mainly by modifying and improving existing functioning systems rather than creating new ones ab initio. Here we explore the interplay between two processes which during evolution have had major roles in acquisition of new functions: gene duplication and protein domain rearrangements. We consider four possible evolutionary scenarios: gene families that have undergone none of these event types; only gene duplication; only domain rearrangement; or both events. We characterize each of the four evolutionary scenarios by functional attributes. Our analysis of ten fungal genomes indicates that at least for the fungi clade, species significantly appear to gain complexity by gene duplication accompanied by expansion of existing domain architectures via rearrangements. We show that paralogs gaining new domain architectures via duplication tend to adopt new functions compared to paralogs that preserve their domain architectures. We conclude that evolution of protein families through gene duplication and domain rearrangement is correlated with their functional properties. We suggest that in general, new functions are acquired via the integration of gene duplication and domain rearrangements rather than each process acting independently.
1. Introduction
The increasing complexity of organisms during evolution has been attributed to duplications and recombinations of existing genes rather than ab initio formation of new functional units in the genome (Chothia et al., 2003). Duplication of entire genes has a key role in the emergence of new functions, with three possible fates (Force et al., 1999): (i) one of the duplicates degenerates by accumulating deleterious mutations while the other preserves the original function; (ii) the original function is divided and maintained by the two copies, a phenomenon called subfunctionalization; and (iii) one of the duplicates retains the ancestral function while the other acquires a new function (neofunctionalization). In some cases, the duplicates exhibit a combination of the above functional scenarios, where both duplicates lose parts of the original function and gain one or more new functions (He and Zhang, 2005b). Typically, the maintenance of several copies of a gene leads to functional specialization and provides a greater chance to adapt to new environmental conditions. For example, it has been shown that gene duplication results in a faster divergence between species when compared to single copy genes (Gu et al., 2004).
An additional driving force for organisms’ complexity is protein domain rearrangements. Domains are highly conserved units from which proteins are composed. Their length ranges from ~35 to 250 amino acids; they have a conserved sequence and usually fold independently of other such units in the protein (Ponting and Russell, 2002). Most eukaryotic proteins are composed of more than one domain (Apic et al., 2001). Although the potential number of domain combinations is enormous, the actual number is limited, since the creation of new genes is the result of expansion of existing domain architectures, a process in which (i) a genomic interval that codes for one or more domains is duplicated; (ii) the duplicated region selectively diverges by mutations, deletions or insertions; and, sometimes, (iii) a recombination or fusion with other genes occurs (Vogel et al., 2005; Patthy, 2003; Bork, 1991; Moore et al., 2008). The prevalence of this evolutionary scenario results in many domain architectures that have a common ancestor; the vast majority of similar domain architectures have emerged from a common descent and furthermore, more than 90% of domains from the same superfamily are duplicates. (Przytycka et al., 2006; Gough, 2005; Chothia and Gough, 2009). Another outcome of the duplication process is the dominance of simple domain rearrangements, with the addition or removal of domains usually occurring at the gene termini (Fong et al., 2007). When the addition of a domain into an existing domain architecture of a protein increases the protein functionality it is termed domain accretion (Koonin et al., 2000). The presence of additional domains allows the protein to interact with more proteins (or, nucleic acids); thus, it is not surprising that many instances of domain accretion were detected in signal transduction proteins and in regulatory processes, and that it is more widespread in the evolution of eukaryotes than in prokaryotes (Koonin et al., 2000). Domain rearrangements have a key role in the emergence of typical features of vertebrates and chordates such as cartilage and the inner ear, and the complicated craniofacial structures (Kawashima et al., 2009), in the evolution of the metazoan signaling system (King et al., 2008), and in the development of innate immune systems in both vertebrates and invertebrates (Zhang et al., 2008). Analysis of domain recombination in the yeast mating response pathway showed that some recombination events lead to functional variations such as increased mating efficiency (Peisajovich et al., 2010). Domain recombination also resulted in greater diversity in pathway response dynamics than gene duplication and it was also observed that some domains progress in evolutionary patterns that correlate with biological processes (Jin et al., 2009).
Gene duplication and domain rearrangement are key factors for gaining organism complexity (Chothia and Gough, 2009). However, little is known about how they actually combine during evolution. Several studies have shown that in many cases there is either a strong selection against one of the processes or a preference for the other. A selection against gene duplication is the basis of the ‘gene balance hypothesis’ (Papp et al., 2003; Yang et al., 2003), which suggests that duplications of genes encoding proteins that are subunits in a complex can be deleterious due to dosage problems. Ciccarelli et al. (Ciccarelli et al., 2005) have shown that in some cases, a selection against duplication can result in changes in the protein architecture. In addition, housekeeping genes were shown to be less likely to be duplicated than others (Hooper and Berg, 2003), while membrane transporters tend to be duplicated (Saier, 2003).
It has been shown that the frequency of changes in domain gain and loss is higher after duplication (Buljan and Bateman, 2009) and yeast duplicate genes have more domains than singletons (He and Zhang, 2005a; Lin et al., 2007). It was suggested that multi-domain genes are more likely to be retained after duplications since the relatively large number of independent units, i.e. the domains, facilitates the survival of the duplicate by subfunctionalization and subsequent neofunctionalization events. An example for a neofunctionalization is the SH2 domain. The SH2 domain has a major role in signaling proteins by binding phosphorylated tyrosine. It was suggested that in the amoeba Dictyostelium and in yeast, the SH2 domain occurs without its phosphotyrosine-binding function. During the evolution of multicellular organisms, the SH2 and kinases were merged into a single protein that was undergoing a mutation and selection for tyrosine phosphorylation and recognition. At the final stage, the domain diversified to carry out its known function in phosphotyrosine signaling (Apic and Russell, 2010). However, a similar domain architecture does not necessarily indicate functional conservation; comparison of the evolution in S. cerevisiae after whole-genome duplication (WGD) with non-WGD paralogs indicated that even if the protein domain architectures are maintained, functions, cellular processes and localizations can vary (Grassi et al., 2010).
To what extent evolution of gene duplication and domain rearrangements are correlated? In this work we explore the range of complexity of gene duplication and domain architectures which has evolved over more than 300 million years, in 10 fungal species. The species represent a wide range of diversified genomes of single celled fungal organisms. Among these species are the genomes of S. cerevisiae and C. glabrata which share common whole genome duplication in their ancestry. The well characterized S. cerevisiae genome enables us to explore functional characteristics of a fungal genome in light of domain rearrangements and duplications. Here, we characterize functional attributes that are related to evolution by gene duplication and domain rearrangements. We show that complexity acquired by gene duplication and domain rearrangements can fall into four predominant evolutionary scenarios, which are distinguished from each other by the functions of the proteins they span, the way these proteins interact and their phylogenetic history.
2. Materials and Methods
2.1 Data acquisition
Clusters of orthologous groups were obtained from the eggNOG database (Jensen et al., 2008), containing genes from ten fungal genomes (in parentheses is the percentage of a proteome of each studied species (Letunic and Bork, 2007)): Kluyveromyces lactis (92.2%), Ashbya gossypii (96.1%), Saccharomyces cerevisiae (81.5%), Candida glabrata (93.9%), Debaryomyces hansenii (86.6%), Yarrowia_lipolytica (80.4%), Aspergillus_fumigatus (83.5%), Schizosaccharomyces pombe (86.6%), Cryptococcus neoformans (78.8%) and Encephalitozoon cuniculi (63.4%). In total, our data set contained 4,815 clusters of orthologs spanning 34,725 genes. The fungal phylogeny is as represented in (James et al., 2006). We used the Interpro (Mulder et al., 2007) domain annotation for UniProt proteins (UniProt, 2008). We considered only clusters having at least two genes with known domain composition. Manually-curated protein complexes were obtained from the MIPS database (Mewes et al., 1999). Protein-protein interaction data were assembled from recent publications and public databases (DIP, BioGRID) (Xenarios et al., 2000; Gavin et al., 2006; Krogan et al., 2006; Reguly et al., 2006) with a total of 24,140 interactions. The interactions were assigned reliability estimates which were computed using a logistic regression model that takes into account the experimental techniques through which each of the interactions was detected (Sharan et al., 2005). The list of S. cerevisiae essential genes was downloaded from the Saccharomyces Genome Deletion Project (Winzeler et al., 1999). Essential ORF deletions were defined as those that survived only as heterozygous diploids.
2.2 Functional coherency analysis
Functional coherency of protein sets was based on the Gene Ontology (GO) (Ashburner et al., 2000) biological process annotation. As the majority of fungal species lack functional annotation with GO terms, the GO terms used in this work are those of S. cerevisiae. For each class, we used a hypergeometric score to evaluate its functional coherency with respect to each of the biological process terms. The resulting p-values were further corrected for multiple testing using the false discovery rate (FDR) procedure (Benjamini and Hochberg, 1995).
2.3 Enrichment in essential S. cerevisiae genes and protein complexes
For the analyses of essential genes we counted how many orthologous groups have at least one essential S. cerevisiae gene. The analysis of proteins participating in protein complexes was performed by counting how many orthologous groups contain at least one gene whose protein product is known to be part of a protein complex in S. cerevisiae (Mewes et al., 1999).
3. Results and Discussion
3.1 Gene duplication and domain rearrangements during fungi evolution
We explored 4,815 clusters of orthologous genes spanning the evolution of ten fungal genomes (Figure 1A). Each cluster contains both orthologous and paralogous genes, as defined by the eggNOG database (Jensen et al., 2008). This database was selected as it provides data on both orthologous groups and protein domain architectures. Genes within these clusters exhibit a variety of evolutionary changes. Here we focused on the patterns of domain architectures, as defined by the sequential order of the domains in a protein, and gene duplication events. We found that most orthologous groups (73.3%) have a single architecture for all their member genes. A similar fraction (71.8%) of orthologous groups contain no duplicates, i.e., each of their member genes has at most a single copy in each of the ten species.
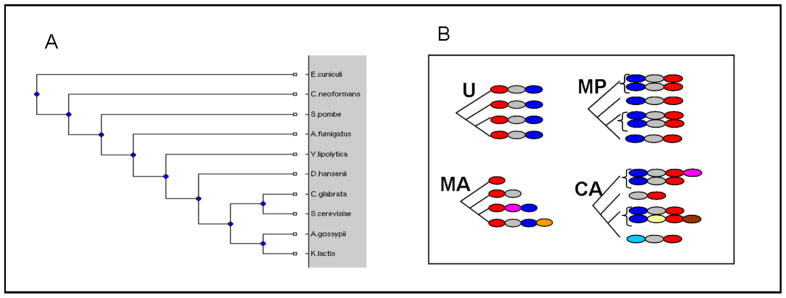
Figure 1.
A. Tree of fungi species used in this work. B. Illustration of orthologous groups’ classification: U uniform, MA multiarch, MP multipar, CA complexarch. Leaves correspond to proteins existing in contemporary species. Colors represent different protein domains.
The above findings motivated us to partition the 4,815 orthologous groups into four classes, according to their predominant evolutionary scenarios (see Figure 1B). The first set, named Uniform, is composed of all orthologous groups (2,607) having a single domain architecture for all genes, and no duplicates. The second set, Multiarch, is composed of orthologous groups (849) having no duplicates and at least two different domain architectures. The third set, Multipar, is composed of orthologous groups (923) with single domain architecture and at least one duplicated gene. The fourth, Complexarch, consists of orthologous groups (436) having at least two different domain architectures and at least one duplicated gene. The families composing the Complexarch set are heterogeneous and include evolutionary scenarios involving domain rearrangements and gene duplications in the different species. For example, a Complexarch orthologous group may be composed of 2 or more paralogs with the same domain architecture in a particular species and an ortholog with different domain architecture in other species. Another possible Complexarch orthologous group may be composed of single (rather than paralogous) proteins with the same domain architecture across all species except for one species where a paralog with different domain architecture is found.
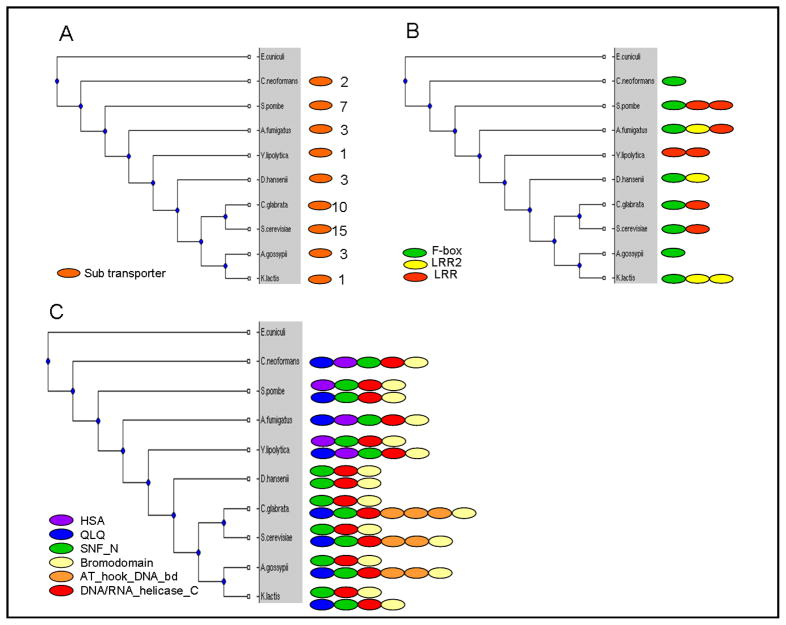
Examples of multipar, multiarch and complexarch orthologous groups are shown in Figure 2. Figure 2A presents a cluster of orthologs having more than two paralogs in most species. All of these proteins belong to a large family of monosaccharide transporters, which are known to transport various types of hexose sugars. Each protein consists of exactly one domain and the multiple duplicates in a species allow specification for various hexose types. Some of the transporters of this family recognize glucose and others recognize galactose or fructose. Indeed, it has been shown that multiple types of hexose transporters in yeast have a role in increasing the fitness in a low-glucose environment (Brown et al., 1998). Figure 2B shows a cluster of orthologs having at most one copy in each species, and different domain architectures. These proteins are part of the SCF E3 ubiquitin ligase complex that has a key role in the regulation of cell cycle progression. These proteins contain the F-box domain that mediates protein-protein interaction and interacts directly with the Skp1 protein in the SCF complex. In addition, these proteins contain one or more variable domains that are thought to mediate interactions with SCF substrates. Domain variability of these proteins is important for substrate recognition and hence in substrate-specific ubiquitination pathways (Nakayama and Nakayama, 2005). In this example, F-box and LRR domains are found both in ancestral single-domain genes as well as in a fused form. However, this is not a general feature of the analyzed clusters; these fusions occur in only 2.1% of the clusters. Furthermore, only 14.9% of the domains are found in more than a single cluster. Figure 2C presents a cluster of orthologous proteins involved in transcriptional regulation. These proteins are part of the SWI/SNF complex. By changing the contacts between the DNA and histones, chromatin structure is altered, which enables the binding of transcription factors to their response elements. In all species but D. hansenii, selection against duplication resulted either in no duplicates (C. neoformans and A. fumigatus) or in new domain architectures in the duplicated gene, including subfunctionalization (S. pombe).
Figure 2.
Examples of orthologous groups exhibiting different evolutionary scenarios. A. Multipar family of monosaccharide transporters. Multiple duplicates enable different types of hexoses. Shown are the numbers of duplicates for each species. B. A Multiarch family of proteins that are part of the SCF E3 ubiquitin ligase complex. C. A Complexarch family of proteins that are part of the SWI/SNF complex.
In order to check whether domain rearrangement and gene duplication events occur in an independent manner, we applied a Chi-square test to the four classes. We found that the number of clusters across the four sets deviated significantly from the random expectation (p<8.2e-10), with the Uniform and Complexarch groups being significantly more populated than expected (p<9.23e-8, hypergeometric test). The expected number of each class is the product of the frequencies of its characteristics among all families divided by the total number of families; for example, to calculate the expected number of the Uniform class we multiplied the number of all singledomain families by the number of all single-copy families and divided by the total number of families.
Next, we sought to determine whether the phylogenetic profile of a cluster influence its assignment to any of the four classes. We measured the fraction of each of the four classes in the 10 fungal genomes. A genome is represented in a partition if it has at least one copy of a gene in a cluster participating in that partition. The distribution of the four classes across the genomes is shown in Figure S1. We found that Uniform, Multiarch, Multipar and Complexarch are equally distributed across species; namely there is no tendency in any of the species to adopt one of the above four evolutionary scenarios over the other (p<0.05, Spearman correlation).
3.2 Functional-based characteristics
We characterized the different partition classes in terms of their functional attributes. A summary of the following results is presented in Table 1. First, we tested whether any of the sets is enriched in essential S. cerevisiae genes. To this end, we counted how many orthologous groups have at least one essential S. cerevisiae gene. We used the hypergeometric test to evaluate the significance of the results, using for each class the corresponding parameters: the number of orthologous groups having essential genes in the class, the number of orthologous groups in the class, the total number of ourthologous groups and the total number of orthologous groups having essential genes. We found that the Uniform and Multiarch sets are enriched for essential genes (p<0.006 and p<4.4e-8, respectively, hypergeometric score), while an opposite trend was observed in the Multipar set. These results are in congruence with previously published phenotypic results, showing that a possible mechanism of compensation for gene deletion is the existence of duplicate genes (Gu et al., 2003). By definition, essential genes are obligatory for the survival of the organism so a possible explanation may be that they do not have a duplicate gene for compensation and therefore the fraction of essential genes in the Multipar set is lower than in the other sets. Next, we examined the number of orthologous groups containing at least one gene whose protein product is known to be part of a protein complex in S. cerevisiae. The statistical analysis was similar to that used for essential genes, replacing the parameter of essential genes by participation in protein complexes according to MIPS (Mewes et al., 1999). We found that the Uniform and Multiarch sets are enriched in genes coding for proteins which participate in these complexes (p<0.001 and p<0.0005, respectively, hypergeometric score). These findings are in congruence with the ‘gene balance hypothesis’, suggesting that genes coding for proteins that are part of complexes have a strong selection against duplication. Thus, these genes would either present a conserved architecture (Uniform) or progress in an evolutionary path that does not include duplications (Multiarch). These results may also be linked to works on phenotypic effects of one copy and of duplicated genes in yeast. It has been shown that duplication of a single gene participating in certain complexes is expected to be harmful to S. cerevisiae and that large families of proteins are rarely involved in complexes (Papp et al., 2003).
Table 1.
Functional attributes of evolutionary classes.
| Analysis | Uniform | Multiarch | Multipar | Complexarch |
|---|---|---|---|---|
| Fraction of yeast proteins that participate in complexes (%) | 7.78* | 8.39* | 5.85§ | 6.36 |
| Fraction of essential yeast proteins (%) | 19.79* | 25.2* | 10.51§ | 14.44 |
| Average number of domains in a protein (including repetitives) | 1.2 | 2.46 | 1.22 | 2.37 |
| Average number of domains (counting only unique domains) | 1.14 | 1.57 | 1.15 | 1.73 |
| Fraction of fungi-specific domains (%) | 5.23* | 1.68§ | 3.9 | 4.38* |
| Fraction of ancient clusters (%) | 62.14§ | 73.73* | 75.5* | 83.71* |
| Mean degree in S. cerevisiae protein-protein interaction network | 11.12§ | 15.2 | 13.93 | 15.18 |
| Mean clustering coefficient in S. cerevisiae protein-protein interaction network | 0.154* | 0.156* | 0.102§ | 0.116 |
significantly high (hypergeometric score, p-value < 0.05)
significantly low (hypergeometric score, p-value < 0.05)
To investigate the protein interaction characteristics within the four classes, we evaluated the average number of interactors of S. cerevisiae proteins in a protein-protein interaction network. For each class, we pooled all its S. cerevisiae proteins and calculated their average number of interactors (degree) in the network. By applying the same algorithm to random, size preserving classes, we were able to assign an empirical p-value to each class. Randomized classes were created by pooling all clusters of orthologous groups and randomly assigning these clusters for each of the four classes, preserving their original size. We found that Multiarch proteins had the highest degree, with 15.2 different interaction partners on average. Complexarch proteins had 15.1 interactions on average. The Multipar and Uniform classes had 13.9 and 11.1 interactions on average, respectively. The latter was significantly low (p<0.01, empirical p-value) compared to the average number of interactions in the other classes. We also characterized the network modularity of the classes. For each S. cerevisiae protein, we calculated its clustering coefficient in the protein-protein interaction network, indicating how many of its interacting partners also interact with each other. For each partition, we pooled all S. cerevisiae proteins and calculated their average clustering coefficient. We found that Multiarch and Uniform proteins had the highest clustering coefficient (0.156 and 0.154, respectively, p<0.01, empirical p-value). Complexarch proteins had a clustering coefficient of 0.11 while the lowest clustering coefficient was measured in the Multipar proteins (0.102, p<0.01, empirical p-value).
We used the gene ontology (GO) (Ashburner et al., 2000) ‘biological process’ annotation to test whether the four classes could be characterized by certain biological processes. Our analysis indicated that the four partitions were distinguished from each other by their functions. We found that the Uniform orthologous groups were enriched in metabolic and biosynthetic processes and catalytic activity (p<10e-15). Basic functions such as metabolic and biosynthetic processes are expected to be highly conserved among species and indeed we found that the Uniform proteins are enriched in these essential functions. Multiarch orthologous groups were enriched in metabolic processes, response to stress, cell division and cell cycle processes and mitosis (p<7e-12). Multipar orthologous groups were enriched in transport processes (p<2e-11) and metabolic and biosynthetic processes, movement and cellular homeostasis (p<3.2e-7). The observation that Multipar proteins were enriched in functions that are related to transport is supported by the previous findings that the evolution of transporter families is mostly mediated by gene duplications rather than by other processes (Saier, 2003). Complexarch orthologous groups were mainly enriched in regulatory and signal transduction functions (p<10e-15). The acquisition of new domains by domain manipulations enabled Multiarch and Complexarch proteins to acquire new functions and to increase their connectivity in signaling systems; thus we find that these partitions are mostly enriched by regulatory and signal transduction functions. The characterization of the four classes in terms of GO biological processes reveals that the attributes of the classes are correlated with their functional enrichment. While the Uniform class, presenting the most conservative evolutionary scenario was enriched in basic, essential functions such as biosynthesis and metabolic processes, the Complexarch class, which is permissive in terms of both domain shuffling and duplication, enables proteins participating in this group to acquire advanced, complex functions such as signal transduction.
3.3 Age-based characteristics
We explored the age of domains composing the genes in the different classes. We classified the domains into new, which are specific to fungi and ancient, which can be found also in other eukaryotes, in archaea or bacteria (Cohen-Gihon et al., 2007). Surprisingly, we found that Uniform orthologous groups are enriched in new domains (p<0.01, hypergeometric score) while Multiarch orthologous groups are enriched in ancient domains (p<0.01). This result suggests either that proteins in the Uniform clusters have fungi-specific roles or that they are non-ancient. To decide which of these two alternatives is the likely explanation we analyzed also the ages of the clusters: we defined the age of a cluster according to the lowest common ancestor of its members. We classified the clusters of the Ascomycota phylum as new or Ascomycota-specific. This monophyletic group of organisms, the largest phylum of Fungi, has diverged about 300 million years ago and its members are known as the sac-fungi (James et al., 2006). Accordingly, ancient clusters include common ancestors that diverged before the divergence of Ascomycota. On the corresponding tree in our work, an ancient cluster includes the root and its direct son while a new cluster includes common ancestors that diverged later. We found that the Uniform class has the lowest fraction of ancient clusters and is enriched in new clusters, while the Complexarch class has the highest fraction of ancient clusters. These findings suggest that Uniform clusters contain Ascomycota-specific proteins, or some protein families that were not detected in the non-Ascomycota species, perhaps due to lower percentage of proteome covered in non-Ascomycota species compared to Ascomycota species (see Methods). The distribution of cluster ages in the four classes is shown in Figure S2.
3.4 Domain number characteristics
Next, we measured the average number of domains composing a protein within the different orthologous groups either by counting the total number of domains within a protein or by counting only the unique domains in a protein (excluding repeated domains). Both measures indicated that Complexarch and Multiarch proteins have on average more domains in their proteins comparing to other groups (Table 1). Interestingly, while the average number of the total and unique domains in Multipar and Uniform proteins is similar, in Complexarch and Multiarch proteins the average total number is greater by 50% as compared to the average unique number, indicating that approximately half of the domains gained by a protein were duplications of already existing domains. These results are in congruence with previous studies (Bjorklund et al., 2005; Bjorklund et al., 2006), suggesting that repetitions of duplicated domains have important binding properties and are involved in protein-protein interactions and support our findings that proteins of the Multiarch and Complexarch classes are involved in protein complexes and protein-protein interactions more than proteins composing the two other classes.
3.5 Paralogs in Complexarch families tend to adopt different architectures and different functions
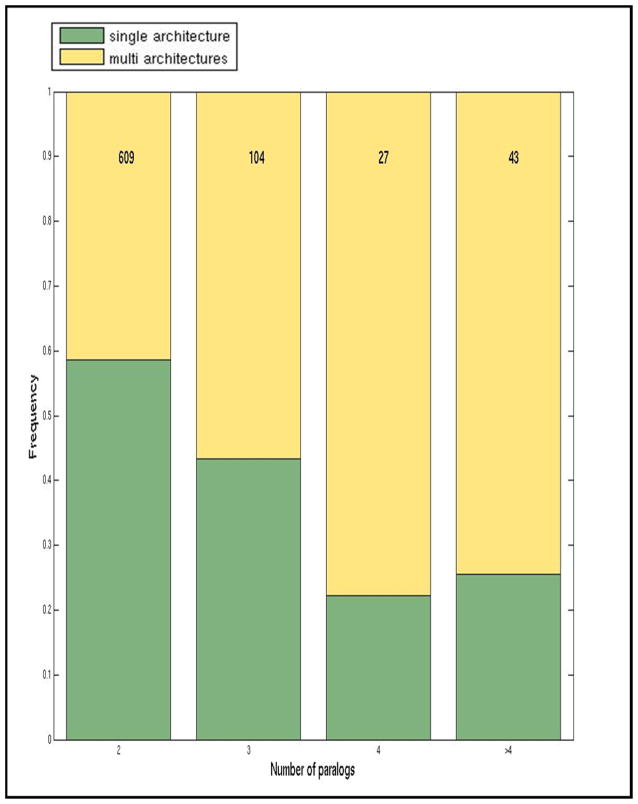
To further explore the interplay between gene duplication and domain rearrangements, we focused on protein families that evolved using both processes, that is, the Complexarch families. We focused on protein families with at least two paralogs and sought to investigate the relationship between the number of paralogs and the number of different architectures that they adopt. For each set of paralogs in the Complexarch class, we counted how many unique domain architectures were present. Interestingly, we found that almost half (42%) of the proteins that present two paralogs formed different domain architectures. The fraction of multi architectures increases with the number of paralogs: 75% of genes having 5 or more paralogs had more than one domain architecture (Figure 3). The distribution of the fractions of single- and multi- architectures across different number of paralogs is significantly different from the random expectation (p-value < 1.28e-7, Chi-square test). The expected distribution was calculated by multiplying the frequencies of each characteristic (single- or multi-domain and number of paralogs) among all proteins and dividing by the total number of proteins. Thus, for example, to calculate the expected value of the number of multi-domain proteins with 2-paralogs we multiplied the number of all multi-domain proteins by the number of all proteins having 2 paralogs and divided the product by the total number of proteins of the analysis. It should be mentioned that in some cases the observed frequent changes in domain compositions may result from an imperfect assignment of domains caused by the changes in the number of domain repeats.
Figure 3.
Analysis of the number of unique domain architectures within paralogous proteins of the Complexarch group. X-axis: the number of paralogs of a gene family. Y-axis: the fraction of families that have a single architecture (green) or more than a single architecture (yellow). Numbers on top represent the number of protein families in each category.
The Complexarch class is composed of heterogeneous families in terms of evolutionary scenarios. It includes protein families in which the domains were rearranged after duplication as well as families in which there are identical paralogs (in terms of domain architectures) but a single protein in another species undergone rearrangement. To create more homogeneous groups, we divided the Complexarch protein families into (i) high complexity families, i.e. families having two or more paralogs in at least one species, with each paralog presenting different domain architecture from the other paralogs (30.3% of the families), (ii) low complexity families, i.e families having two or more paralogs in at least one species, with each paralogous group of proteins presenting an identical domain architecture in at least two paralogs (48.6%) and (iii) families that do not fall into any of these categories, that is, families with two identical (in terms of domain architecture) paralogs in one species and different paralogs in other species (21.1%). Then, we identified the function of the paralogs in each of the three categories, using their GO ‘biological process’ terms (Ashburner et al., 2000). We defined the function of each paralog as its most specific GO biological process term. A pair of paralogs was defined as having a similar function if both paralogs have the same most specific GO term, and similarly, having different functions if their most specific annotations are different. We found that in high complexity families, 26% of the paralogs had different functions while in low complexity groups only 7% changed their function. In group (iii) 5.4% of the paralogs changed their function. On the other hand, while implementing a similar analysis on paralogs in Multipar clusters, we found that only 6% of the paralogs had different functions. These findings indicate that domain rearrangement is coupled to a change in function. These results are in congruence with the duplication-degeneration-complementation model (Force et al., 1999) that was discussed above; moreover, they emphasize the importance of domain rearrangement events in the evolution of duplicates, particularly in large protein families.
4. Conclusions
In this work we defined and studied four evolutionary classes of protein families with respect to gene duplication and domain rearrangements. We have shown that some protein families were under selection against duplication (Multiarch); others against domain rearrangements (Multipar). In congruence with previous studies (Papp et al., 2003; Yang et al., 2003), we have shown that Multipar proteins are depleted in protein complexes probably since duplicates can interrupt the balance among the gene products in the complexes; however, they are enriched in transporter families. Evolution of proteins participating in complexes has therefore occurred via the Multiarch proteins, by acquiring new domains; in support of this conclusion, our results indicate that this class is enriched with proteins that take part in complexes. Families that present a conserved profile of gene evolution (Uniform) are also enriched in proteins participating in complexes; these proteins also do not maintain duplicates. Interestingly, the fraction of fungi-specific domains (5.23%) in this class is 3-fold larger than in the Multiarch class. On the other hand, the fraction of prokaryotic domains in families of proteins that have undergone either duplication and/or domain rearrangements is larger than in other families. Moreover, we found that in about half of the cases the acquisition of new domains by a protein occurs through the duplication of one or more of its existing domains.
Notably, we found that more than 400 families, called Complexarch families in this work, have undergone both duplications and domain manipulations during the course of evolution. Some of these families contain proteins that have paralogs with different domain architectures. Protein families that survived such complicated evolutionary changes are of special interest. Their genes have the ability to survive a duplication event and to acquire new functions by gaining additional domains. In this regard, genes with a large number of domains are more likely to survive a duplication event (He and Zhang, 2005a; Lin et al., 2007). Their large number of domains may help the duplicates undergo subfunctionalization, where both copies still maintain the original function. An alternative way to survive a duplication event is the acquisition of new domains by the duplicate. In these cases, one duplicate maintains the original function while the other acquires new properties. This can happen right after the duplication or following other events, such as change in expression or neofunctionalization by sequence divergence which played a role in the retention of the duplicated gene (He and Zhang, 2005b). To further substantiate this point, we performed an analysis of domain architectures in paralogous proteins with the Complexarch families. We observed that the greater the number of paralogs a species has, the greater is the variety of domain architectures they adopt. This increase in the number of architectures in paralogs is significant. However, such an increase in changes in domain composition may result from an imperfect assignment of domains caused by changing the number of repetitive domains. Moreover, we explored the functions of high-complexity protein families, which are about one third of all Complexarch families. In these families, all the paralogs with a particular species adopt different domain architectures. We found that high-complexity protein families tend to adopt different functions compared to families in which paralogs maintain two or more duplicates with the same domain architecture. These results serve to validate that the functional variety is greater when gene duplication is accompanied by domain rearrangements. Obviously, the organism benefits from such duplications followed by domain manipulations, since merging both processes increases organism complexity considerably, increasing both the protein connectivity and the number of gene copies. On top of the domain accretion scenario (Koonin et al., 2000) where existing architectures tend to gain complexity by the acquisition of new domains, we find that in many cases this occurs mainly in duplicated genes. The abundance of proteins that were targets of both gene duplication and domain manipulations, along with higher level functionality, suggest that combining these two processes is synergistic and highly advantageous during evolution.
Supplementary Material
Figure S1 Distribution of the four evolutionary classes across species. A genome is represented in a class if it has at least one copy of a gene in a cluster participating in that class.
Figure S2 Distribution of the four evolutionary classes across cluster ages. Cluster age was defined according to the lowest common ancestor of its members and is represented by the number of the corresponding internal node on the phylogenetic tree. Accordingly, the root of the tree is represented by the age of 10.
Acknowledgments
We thank Nir Yosef for providing us with the protein-protein interaction network data. We thank Itai Yanai for helpful advices. This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract numberHHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. ICG is a fellow of the Edmond J. Safra Bioinformatics Program and of the Ela Kodesz Research and Scholarship Fund at Tel-Aviv University. RS was supported by a research grant from the Israel Science Foundation (no. 385/06).
References
- Apic G, Gough J, Teichmann SA. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J Mol Biol. 2001;310:311–25. doi: 10.1006/jmbi.2001.4776. [DOI] [PubMed] [Google Scholar]
- Apic G, Russell RB. Domain recombination: a workhorse for evolutionary innovation. Sci Signal. 2010;3:pe30. doi: 10.1126/scisignal.3139pe30. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B. 1995;57:289–300. [Google Scholar]
- Bjorklund AK, Ekman D, Elofsson A. Expansion of protein domain repeats. PLoS Comput Biol. 2006;2:e114. doi: 10.1371/journal.pcbi.0020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorklund AK, Ekman D, Light S, Frey-Skott J, Elofsson A. Domain rearrangements in protein evolution. J Mol Biol. 2005;353:911–23. doi: 10.1016/j.jmb.2005.08.067. [DOI] [PubMed] [Google Scholar]
- Bork P. Shuffled domains in extracellular proteins. FEBS Lett. 1991;286:47–54. doi: 10.1016/0014-5793(91)80937-x. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Todd KM, Rosenzweig RF. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol Biol Evol. 1998;15:931–42. doi: 10.1093/oxfordjournals.molbev.a026009. [DOI] [PubMed] [Google Scholar]
- Buljan M, Bateman A. The evolution of protein domain families. Biochem Soc Trans. 2009;37:751–5. doi: 10.1042/BST0370751. [DOI] [PubMed] [Google Scholar]
- Chothia C, Gough J. Genomic and structural aspects of protein evolution. Biochem J. 2009;419:15–28. doi: 10.1042/BJ20090122. [DOI] [PubMed] [Google Scholar]
- Chothia C, Gough J, Vogel C, Teichmann SA. Evolution of the protein repertoire. Science. 2003;300:1701–3. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- Ciccarelli FD, von Mering C, Suyama M, Harrington ED, Izaurralde E, Bork P. Complex genomic rearrangements lead to novel primate gene function. Genome Res. 2005;15:343–51. doi: 10.1101/gr.3266405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Gihon I, Nussinov R, Sharan R. Comprehensive analysis of cooccurring domain sets in yeast proteins. BMC Genomics. 2007;8:161. doi: 10.1186/1471-2164-8-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong JH, Geer LY, Panchenko AR, Bryant SH. Modeling the evolution of protein domain architectures using maximum parsimony. J Mol Biol. 2007;366:307–15. doi: 10.1016/j.jmb.2006.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–45. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–6. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Gough J. Convergent evolution of domain architectures (is rare) Bioinformatics. 2005;21:1464–71. doi: 10.1093/bioinformatics/bti204. [DOI] [PubMed] [Google Scholar]
- Grassi L, Fusco D, Sellerio A, Cora D, Bassetti B, Caselle M, Lagomarsino MC. Identity and divergence of protein domain architectures after the yeast whole-genome duplication event. Mol Biosyst. 2010 doi: 10.1039/c003507f. [DOI] [PubMed] [Google Scholar]
- Gu Z, Rifkin SA, White KP, Li WH. Duplicate genes increase gene expression diversity within and between species. Nat Genet. 2004;36:577–9. doi: 10.1038/ng1355. [DOI] [PubMed] [Google Scholar]
- Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421:63–6. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- He X, Zhang J. Gene complexity and gene duplicability. Curr Biol. 2005a;15:1016–21. doi: 10.1016/j.cub.2005.04.035. [DOI] [PubMed] [Google Scholar]
- He X, Zhang J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005b;169:1157–64. doi: 10.1534/genetics.104.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper SD, Berg OG. On the nature of gene innovation: duplication patterns in microbial genomes. Mol Biol Evol. 2003;20:945–54. doi: 10.1093/molbev/msg101. [DOI] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsch HT, Rauhut A, Reeb V, Arnold AE, Amtoft A, Stajich JE, Hosaka K, Sung GH, Johnson D, O’Rourke B, Crockett M, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schussler A, Longcore JE, O’Donnell K, Mozley-Standridge S, Porter D, Letcher PM, Powell MJ, Taylor JW, White MM, Griffith GW, Davies DR, Humber RA, Morton JB, Sugiyama J, Rossman AY, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, Spotts RA, Serdani M, Crous PW, Hughes KW, Matsuura K, Langer E, Langer G, Untereiner WA, Lucking R, Budel B, Geiser DM, Aptroot A, Diederich P, Schmitt I, Schultz M, Yahr R, Hibbett DS, Lutzoni F, McLaughlin DJ, Spatafora JW, Vilgalys R. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–22. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Julien P, Kuhn M, von Mering C, Muller J, Doerks T, Bork P. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2008;36:D250–4. doi: 10.1093/nar/gkm796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Xie X, Chen C, Park JG, Stark C, James DA, Olhovsky M, Linding R, Mao Y, Pawson T. Eukaryotic protein domains as functional units of cellular evolution. Sci Signal. 2009;2:ra76. doi: 10.1126/scisignal.2000546. [DOI] [PubMed] [Google Scholar]
- Kawashima T, Kawashima S, Tanaka C, Murai M, Yoneda M, Putnam NH, Rokhsar DS, Kanehisa M, Satoh N, Wada H. Domain shuffling and the evolution of vertebrates. Genome Res. 2009;19:1393–403. doi: 10.1101/gr.087072.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, Marr M, Pincus D, Putnam N, Rokas A, Wright KJ, Zuzow R, Dirks W, Good M, Goodstein D, Lemons D, Li W, Lyons JB, Morris A, Nichols S, Richter DJ, Salamov A, Sequencing JG, Bork P, Lim WA, Manning G, Miller WT, McGinnis W, Shapiro H, Tjian R, Grigoriev IV, Rokhsar D. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–8. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Aravind L, Kondrashov AS. The impact of comparative genomics on our understanding of evolution. Cell. 2000;101:573–6. doi: 10.1016/s0092-8674(00)80867-3. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St Onge P, Ghanny S, Lam MH, Butland G, Altaf-Ul AM, Kanaya S, Shilatifard A, O’Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–43. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–8. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Lin YS, Hwang JK, Li WH. Protein complexity, gene duplicability and gene dispensability in the yeast genome. Gene. 2007;387:109–17. doi: 10.1016/j.gene.2006.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewes HW, Heumann K, Kaps A, Mayer K, Pfeiffer F, Stocker S, Frishman D. MIPS: a database for genomes and protein sequences. Nucleic Acids Res. 1999;27:44–8. doi: 10.1093/nar/27.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AD, Bjorklund AK, Ekman D, Bornberg-Bauer E, Elofsson A. Arrangements in the modular evolution of proteins. Trends Biochem Sci. 2008;33:444–51. doi: 10.1016/j.tibs.2008.05.008. [DOI] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, Copley R, Courcelle E, Das U, Daugherty L, Dibley M, Finn R, Fleischmann W, Gough J, Haft D, Hulo N, Hunter S, Kahn D, Kanapin A, Kejariwal A, Labarga A, Langendijk-Genevaux PS, Lonsdale D, Lopez R, Letunic I, Madera M, Maslen J, McAnulla C, McDowall J, Mistry J, Mitchell A, Nikolskaya AN, Orchard S, Orengo C, Petryszak R, Selengut JD, Sigrist CJ, Thomas PD, Valentin F, Wilson D, Wu CH, Yeats C. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–8. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama KI, Nakayama K. Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin Cell Dev Biol. 2005;16:323–33. doi: 10.1016/j.semcdb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–7. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Patthy L. Modular assembly of genes and the evolution of new functions. Genetica. 2003;118:217–31. [PubMed] [Google Scholar]
- Peisajovich SG, Garbarino JE, Wei P, Lim WA. Rapid diversification of cell signaling phenotypes by modular domain recombination. Science. 2010;328:368–72. doi: 10.1126/science.1182376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Russell RR. The natural history of protein domains. Annu Rev Biophys Biomol Struct. 2002;31:45–71. doi: 10.1146/annurev.biophys.31.082901.134314. [DOI] [PubMed] [Google Scholar]
- Przytycka T, Davis G, Song N, Durand D. Graph theoretical insights into evolution of multidomain proteins. J Comput Biol. 2006;13:351–63. doi: 10.1089/cmb.2006.13.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguly T, Breitkreutz A, Boucher L, Breitkreutz BJ, Hon GC, Myers CL, Parsons A, Friesen H, Oughtred R, Tong A, Stark C, Ho Y, Botstein D, Andrews B, Boone C, Troyanskya OG, Ideker T, Dolinski K, Batada NN, Tyers M. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J Biol. 2006;5:11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH., Jr Tracing pathways of transport protein evolution. Mol Microbiol. 2003;48:1145–56. doi: 10.1046/j.1365-2958.2003.03499.x. [DOI] [PubMed] [Google Scholar]
- Sharan R, Suthram S, Kelley RM, Kuhn T, McCuine S, Uetz P, Sittler T, Karp RM, Ideker T. Conserved patterns of protein interaction in multiple species. Proc Natl Acad Sci U S A. 2005;102:1974–9. doi: 10.1073/pnas.0409522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–5. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Teichmann SA, Pereira-Leal J. The relationship between domain duplication and recombination. J Mol Biol. 2005;346:355–65. doi: 10.1016/j.jmb.2004.11.050. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M’Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–6. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Xenarios I, Rice DW, Salwinski L, Baron MK, Marcotte EM, Eisenberg D. DIP: the database of interacting proteins. Nucleic Acids Res. 2000;28:289–91. doi: 10.1093/nar/28.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lusk R, Li WH. Organismal complexity, protein complexity, and gene duplicability. Proc Natl Acad Sci U S A. 2003;100:15661–5. doi: 10.1073/pnas.2536672100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zmasek CM, Dishaw LJ, Mueller MG, Ye Y, Litman GW, Godzik A. Novel genes dramatically alter regulatory network topology in amphioxus. Genome Biol. 2008;9:R123. doi: 10.1186/gb-2008-9-8-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Distribution of the four evolutionary classes across species. A genome is represented in a class if it has at least one copy of a gene in a cluster participating in that class.
Figure S2 Distribution of the four evolutionary classes across cluster ages. Cluster age was defined according to the lowest common ancestor of its members and is represented by the number of the corresponding internal node on the phylogenetic tree. Accordingly, the root of the tree is represented by the age of 10.