Abstract
Systemic lupus erythematosus (SLE) is an autoimmune disease characterized by the production of an array of pathogenic autoantibodies, including high-affinity anti-dsDNA IgG antibodies. These autoantibodies are mutated and class-switched, mainly to IgG, indicating that immunoglobulin (Ig) gene somatic hypermutation (SHM) and class switch DNA recombination (CSR) are important in their generation. Lupus-prone MRL/faslpr/lpr mice develop a systemic autoimmune syndrome that shares many features with human SLE. We found that Ig genes were heavily mutated in MRL/faslpr/lpr mice and contained long stretches of DNA deletions and insertions. The spectrum of mutations in MRL/faslpr/lpr B cells was significantly altered, e.g., increased dG/dC transitions, and increased targeting of the RGYW/WRCY mutational hotspot and the WGCW AID-targeting hotspot. We also showed that MRL/faslpr/lpr greatly upregulated CSR, particularly to IgG2a and IgA in B cells of the spleen, lymph nodes and Peyer’s patches. In MRL/faslpr/lpr mice, the significant upregulation of SHM and CSR was associated with significantly increased expression of AID, which mediates DNA lesion, the first step in SHM and CSR, and translesion DNA synthesis (TLS) polymerase (pol) θ, pol η and pol ζ, which are involved in DNA synthesis/repair process associated with SHM and, possibly, CSR. Thus, in lupus-prone mice, SHM and CSR are dysregulated, as a result of enhanced AID expression and, therefore, DNA lesions, and dysregulated DNA repair factors, including TLS polymerases, which are involved in the repair process of AID-mediated DNA lesions.
Keywords: activation-induced cytidine deaminase (AID), antibody, autoantibody, B cell, class switch DNA recombination (CSR), DNA deletion, DNA insertion, lupus, somatic hypermutation (SHM)
Introduction
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease characterized by the production of autoantibodies with multiple specificities, mainly for nuclear antigens. These autoantibodies cause lesion to specific organs, including kidneys, central nervous system and heart [1, 2]. Despite the evidence for the pathogenic role of these autoantibodies, the mechanisms that lead to their production remain largely unknown. An important feature of pathogenic autoantibodies in lupus is that they are heavily mutated [1, 3–11], emerging from unmutated V(D)J templates of natural autoantibody-producing cell precursors in the primary B cell repertoire [12–18]. Indeed, reversion of hypermutated V(D)J sequences of anti-dsDNA autoantibodies to germline led to loss of dsDNA-binding [6, 17, 19, 20], indicating that somatic hypermutation (SHM) is critical in the generation of these autoantibodies. Not only are pathogenic autoantibodies hypermutated to acquire a high-affinity for the selecting self-antigen(s), but are also class-switched, mostly to IgG, through class switch DNA recombination (CSR). IgG autoantibodies can gain access to the extracellular space, thereby mediating tissue/organ injury.
MRL/faslpr/lpr mice develop a systemic autoimmune syndrome that shares many features with human SLE. These mice are characterized by the spontaneous development of a severe systemic autoimmune disease, exhibiting autoantibody production, hypergammaglobulinemia, lymphadenopathy and immune complex glomerulonephritis [21, 22]. In lupus-prone MRL/faslpr/lpr mice, the serum level of the major pathogenic Ig isotypes, IgG2a and IgG1, is increased by more than 10-fold, as compared to normal mouse controls [23]. Consistent with the role of SHM and CSR in the generation of pathogenic autoantibodies in MRL/faslpr/lpr mice, ablation of these two processes through knockout of Aicda gene, which encodes activation-induced cytidine deaminase (AID), led to abrogation of lupus nephritis [24].
SHM and CSR are highly regulated. In the germinal centers (GC) of secondary lymphoid organs, antigen-primed B cells are activated in a T cell-dependent fashion to undergo SHM and CSR. SHM is initiated by transcription of the targeted V(D)J region DNA and eventually inserts mismatches in both DNA strands. The SHM machinery preferentially targets the RGYW/WRCY (R = A or G, Y = C or T, W = A or T) mutational hotspot [25–28]. CSR entails germline IH-S-CH transcription of the upstream and downstream CH genes that will be involved in the recombination event, generation of DSBs in the S regions located 5′ of the respective CH genes and, eventually, ligation of the upstream switch (S) region DNA end with the downstream S region DNA end, and deletion of the intervening DNA. Such a recombination event brings a rearranged VHDJH segment initially upstream of Cμ or Cδ exons into close proximity of Cγ, Cα or Cε, thereby preserving antibody specificity [29].
SHM and CSR would entail two sequential stages: generation of DNA lesions and DNA lesion repair [30–35]. The first stage of both SHM and CSR require the intervention of AID. AID preferentially deaminates dC within WRC in single-stranded DNA [36–40], as existing in the transcription bubble of V(D)J and S region DNA, in SHM and CSR, respectively [36, 38, 41–44]. The resulting dU:dG mispair can be “replicated over”, yielding dC → dT or dG → dA transition (Phase 1a). Alternatively, dU can be deglycosylated by the uracil DNA glycosylase Ung, yielding an abasic site, a lesion that can be bypassed by error-prone TLS polymerases during DNA replication, thereby leading to insertion of transition and transversion mutations, or can be processed by the base excision repair (BER) pathway (Phase 1b). This entails excision by apurinic/apyrimidinic endonuclease (APE) or the Mre11-Rad50 lyase [45, 46] to create DNA nicks. Proximal nicks on opposite strands give rise to double-stranded DNA breaks (DSBs), which are obligatory intermediates in CSR [30–33, 41, 42, 44, 47–54]. Alternatively, the dU:dG mispair can recruit the Msh2-Msh6 heterodimer, MutSα, to initiate the MMR pathway (Phase 2), leading to DNA nicks, single-strand DNA gaps and DSBs through the intervention of a yet to be identified endonuclease(s) and exonuclease I (Exo I) [55, 56]. In SHM, completion of MMR through intervention of error-prone TLS polymerases, instead of the high-fidelity pol δ and pol ε, would lead to the introduction of mutations during DNA re-synthesis [33, 43, 54, 57]. In CSR, repair of DSBs, as occurring in both Phase 1b and Phase 2, likely by a non-classical non-homologous end-joining (NHEJ) pathway [58], leads to S-S region synapsis [59].
Mutation frequency is significantly higher in B cells of SLE patients than in healthy subjects [60, 61]. Accordingly, both SLE patients and lupus mice display high levels of circulating IgGs, including pathogenic autoantibodies. Prolonged T:B cell contact centered on CD154:CD40 engagement, dysregulated production of B cell-stimulating T cell-derived cytokines and/or B cell apoptosis may contribute to abnormal SHM or CSR in lupus B cells [62]. However, the molecular mechanisms that are responsible for enhanced SHM or CSR remain to be identified [62]. It has been suggested that Aicda mRNA level is greatly increased in GC B cells of lupus-prone BXD2 mice, as compared to C57BL/6 mice [63, 64], and, conversely, lupus nephritis is abrogated in AID-deficient MRL/faslpr/lpr mice [24].
We contend that in the lupus, the expression of AID and TLS polymerases and, therefore, SHM and CSR are dysregulated, resulting in the generation of highly mutated and class-switched autoantibodies. We show here that lupus-prone MRL/faslpr/lpr mice display high rates of SHM and CSR in association with extensive DNA lesions, including deletions and insertions, in the Ig heavy chain (Igh) locus. In these autoimmune mice, high SHM and CSR rates, and extensive DNA lesion are associated with upregulation of AID expression and TLS DNA polymerases, suggesting that these AID-mediated DNA lesions and TLS polymerase-mediated DNA repair play an important role in the emergence of lupus autoantibodies.
Materials and Methods
Autoimmune and non-autoimmune mice
MRL/faslpr/lpr and C57BL/6 mice were purchased from the Jackson Laboratory (Bar Harbor, ME). The mice were housed in pathogen-free facilities, and were provided with autoclaved food and deionized water. All animal protocols were approved by the Institutional Animal Care and Use Committee of University of California, Irvine, CA.
Ig CSR analysis
The numbers of (B220+) B cells or GC (PNAhi B220+) B cells expressing IgM, IgG1, IgG2a, IgG2b, IgG3 or IgA were determined by FACS analysis. Single cell suspensions were prepared from spleen, axillary lymph nodes, brachial lymph nodes, inguinal lymph nodes or Peyer’s patches of 11-week-old MRL/faslpr/lpr and non-autoimmune C57BL/6 mice and stained with PE-conjugated anti-mouse CD45R (B220) mAb (clone RA3-6B2) (eBioscience, San Diego, CA) or Alexa Fluor® 647-conjugated PNA (Invitrogen Corp., Carlsbad, CA), as well as rat anti-mouse IgM (clone II/41), anti-mouse IgG1 (clone A85-1)-, anti-mouse IgG2a (clone R19-15)-, anti-mouse IgG2b (clone R12-3)-, anti-mouse IgG3 (clone R40-82)-, or anti-mouse IgA-FITC (clone C10-3) mAb (BD Biosciences, San Jose, CA). Cells were fixed with 1% paraformaldehyde in PBS and analyzed using a FACSCalibur™ flow cytometer (BD Biosciences).
Amplification and sequence analysis of the intronic VJ558DJH4-iEμ DNA
Peyer’s patches PNAhi B cells were obtained from 11-week-old MRL/faslpr/lpr and non-autoimmune C57BL/6 mice and used to analyze SHM in the intronic DNA downstream of rearranged VJ558 genes. Phusion™ DNA polymerase (New England Biolabs Inc., Ipswich, MA) was utilized for chromosomal DNA amplifications. Primers for PCR amplifications were synthesized by Operon (Huntsville, AL). The intronic heavy chain region downstream of rearranged VHDJH was amplified using nested PCR involving primer pairs, JX13F: 5′-AGCCTGACATCTGAGGAC-3′ and JX4R: 5′-TCTGATCGGCCATCTTGACTC-3′ for the first PCR round and JX15F: 5′-CATCTGAGGACTCTGCNGTCTAT-3′ and JX5R: 5′-CCTCACTCCCATTCCTCGGTTAAA-3′ for the second, yielding an about 900 bp fragment if rearrangement involves JH4. Reaction conditions for the first round were 94°C for 1 min, 58°C for 1.5 min, 68°C for 1.45 min for 35 cycles, and for the second-round PCR were 94°C for 45 sec, 58°C for 1 min and 68°C for 1.45 min for 35 cycles. PCR products were cloned into the pCR-TOPO™ vector (Invitrogen Corp.) and sequenced. In the case of the JH-iEμ intronic DNA, only JH4-iEμ sequences were analyzed. Sequences were analyzed using the MacVector 7.2 software (Accelrys Inc., San Diego, CA). The differences in frequency and spectrum of mutations in MRL/faslpr/lpr and C57BL/6 mice were analyzed using χ2 tests.
Quantitative real-time PCR (qRT-PCR) analysis of Aicda, TLS pol θ, pol η, and pol ζ (rev3) mRNA
Lymph nodes, thymus, spleen, Peyer’s patches, and liver cells were isolated from MRL/faslpr/lpr and non-autoimmune C57BL/6 mice. Total RNA was extracted from 2–10 × 106 cells using Trizol (Invitrogen Corp.) according to manufacturer’s instructions. Residual DNA was removed from the extracted RNA with DNase I (New England Biolabs Inc.). The first strand cDNAs were synthesized from equal amounts of total RNA (2 μg) using the SuperScript™ Preamplification System (Invitrogen Corp.) and Oligo dT-primer. For real-time quantification of transcripts, the cDNA in test samples was diluted 10 fold in distilled, deionized water and 2 μl was used as a template in a volume of 25 μl containing each primer at 0.3 mM. RT-PCR primers were as follows: Aicda forward 5′-TGCTACGTGGTGAAGAGGAG-3′ and reverse 5′-TCCCAGTCTGAGATGTAGCG-3′, pol θ forward 5′-AGCCCTCAGCTCCGGTGTGGAC-3′ and reverse 5′-GGAAACACGGAAGAAAGCGTTG-3′, pol η forward 5′-GCCCAACCGCCAAACCCTGGTCTCAC-3′ and reverse 5′-GGTAGCGCAGAGGAAGAGCATTGTG-3′, pol ζ (rev3) forward 5′-TCTCAGTCTCCCACAGGCAAAC-3′ and reverse 5′-CTTTTCTGGCACATCCGAAGG-3′, gapdh forward 5′-TTCACCACCATGGAGAAGGC-3′ and reverse 5′-GGCATGGACTGTGGTCATGA-3′. Quantitative real-time qRT-PCR analysis was performed using an DNA Engine Opticon 2 Real-Time PCR Detection System (Bio-Rad Laboratories, Inc., Hercules, CA) to measure SYBR-green (DyNAmo HS SYBR Green, New England Biolabs Inc.) incorporation with the following protocol: 50°C for 2 min, 95°C for 10 min, 40 cycles of 95 °C for 10 sec, 60°C for 20 sec, 72°C for 30 sec, 80°C for 1 sec, and data acquisition at 80°C, and 72°C for 10 min. Melting curve analysis was done from 72°C–95°C and samples were incubated for another 5 min at 72°C. The ΔΔCt method was used for data analysis.
Results
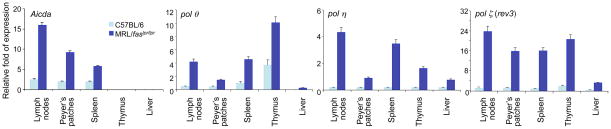
Aicda, and TLS pol θ, pol η, and pol ζ (rev3) are upregulated in lymph nodes, Peyer’s patches and spleen of lupus-prone MRL/faslpr/lpr mice
AID is essential for the first stage of SHM and CSR (the generation of DNA lesions). TLS pol θ, pol η and pol ζ play important roles in the second stage (DNA repair), thereby contributes to the overall frequency and spectrum of mutations. We hypothesized that a significantly increased expression of AID and TLS polymerases underlie the dysregulated SHM and CSR in lupus B cells. As shown by us and others, expression of AID, TLS pol θ, pol η and pol ζ is induced in B cells activated by the same stimuli that induce SHM and CSR, including CD154, LPS or BCR crosslinking [26–28, 42, 65–67]. Consistent with our hypothesis, we found that in lupus-prone MRL/faslpr/lpr mice, the expression, as measured by real-time qRT-PCR, of Aicda, pol θ, pol η and pol ζ (rev3) in lymph nodes, Peyer’s patches and the spleen, which contained a large proportion of hypermutating and class switching GC B cells, was significantly higher than that in non-autoimmune C57BL/6 mice (Figure 1). Thus, the transcription regulation program of Aicda and TLS polymerases is significantly upregulated in B cells of lupus-prone MRL/faslpr/lpr mice.
Figure 1.
Aicda, polθ, pol η,and pol ζ (rev3) are preferentially expressed in lymph nodes, Peyer’s patches and spleen of lupus-prone MRL/faslpr/lpr mice. Total RNA was prepared from spleen, lymph nodes, Peyer’s patches, thymus and liver of non-immunized 8-week old MRL/faslpr/lpr or age-matched non-autoimmune C57BL/6 mice. The levels of Aicda, polθ, pol η, and pol ζ (rev3) transcripts were analyzed by real-time qRT-PCR using SYBR green and normalized to gapdh expression. Data are mean values ± SD from 3 independent experiments.
Lupus-prone MRL/faslpr/lpr mice show increased loads of somatic point-mutations in the Igh locus
In SLE patients, peripheral B cells show a marked increased mutation frequency [62], displaying an overall frequency of mutations in nonproductive Ig VλJλ or VκJκ DNA that is 4- to 7-fold that of healthy subjects [60–62]. To analyze SHM in lupus-prone mice, we analyzed mutations in the intronic JH4-iEμ hi) B cells from 11-week old female sequence downstream of rearranged VJ558DJH4 DNA in GC (PNA MRL/faslpr/lpr and age-matched female non-autoimmune C57BL/6 mice, using methods reported by us and others [26–28, 66, 68]. As we previously argued, mutations in this intronic region are not subjected to positive or negative selection, therefore, reflecting the inherent nature and bias of the SHM machinery [28]. In addition, their frequency is still relatively high, even though they account for the tail of the V(D)J hypermutation wave.
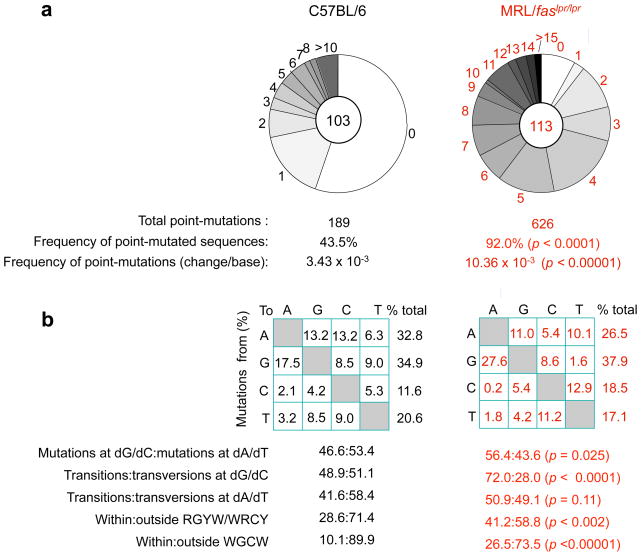
VJ558DJH4-iEμ DNA was amplified using a nested PCR involving two VJ558 FR3 forward primers and two different JH4 intronic reverse primers. Analysis of 113 and 103 JH4-iEμ intronic DNA sequences (534 bp) from 3 11-week-old MRL/faslpr/lpr and 3 age-matched non-autoimmune C57BL/6 mice showed that in MRL/faslpr/lpr mice, 92% of JH4-iEμ DNA sequences carried point-mutations, while only about 43% from C57BL/6 mice did so (p < 0.0001) (Figure 2). The overall frequency of mutations in MRL/faslpr/lpr mice was at least 3-fold that of C57BL/6 mice (p < 0.00001): 10.36 × 10−3 versus 3.43 × 10−3 change/base, respectively. Thus, SHM is significantly upregulated in lupus-prone MRL/faslpr/lpr mice.
Figure 2.
Mutation frequency is greatly increased and mutation spectrum is significantly altered in lupus-prone MRL/faslpr/lpr mice. (a) Pie charts depict the proportions of sequences that carry 1, 2, 3, etc. point-mutations over the 534 bp intronic JH4-iEμ DNA in GC (PNAhi) B cells from Peyer’s patches of three 11-week old MRL/faslpr/lpr and C57BL/6 mice. The numbers of sequences analyzed are at the center of the pies. (b) Compilations, with the numbers indicating percentages of all mutations scored in the pool of all point-mutations from MRL/faslpr/lpr and C57BL/6 mice. Below the compilations, the ratio of mutations at dG/dC to those at dA/dT, the ratio of transition:transversion substitutions at both dC/dG and dA/dT, and the ratio of mutations within and outside RGYW/WRCY mutational hotspots and WGCW AID hotspots are indicated. The significance of differences in the mutation frequency/spectrum between MRL/faslpr/lpr and C57BL/6mice was analyzed with the χ2 test. p < 0.05 were considered statistically significant.
Somatic point-mutations in lupus-prone MRL/faslpr/lpr mice are altered in spectrum
In MRL/faslpr/lpr mice, the greatly increased frequency of somatic point-mutations was associated with significantly altered spectrum of mutations. These autoimmune mice showed a preference in mutations at dG/dC compared to dA/dT, and exhibited a dominance of dG/dC transitions, which increased by over 47.2% compared to the non-autoimmune C57BL/6 mice (Figure 2), while the proportion of transition mutations at dA/dT was not significantly changed (p = 0.11). The increase in transition mutations at dG/dC resulted from increased transitions at both dG and dC, by 57.7% and 143.4%, respectively (p < 0.0001).
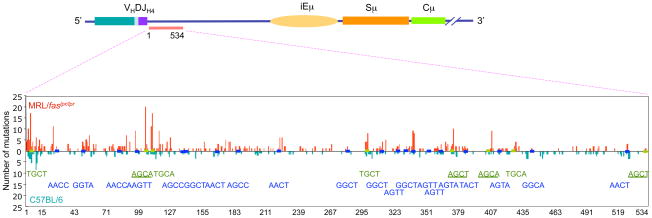
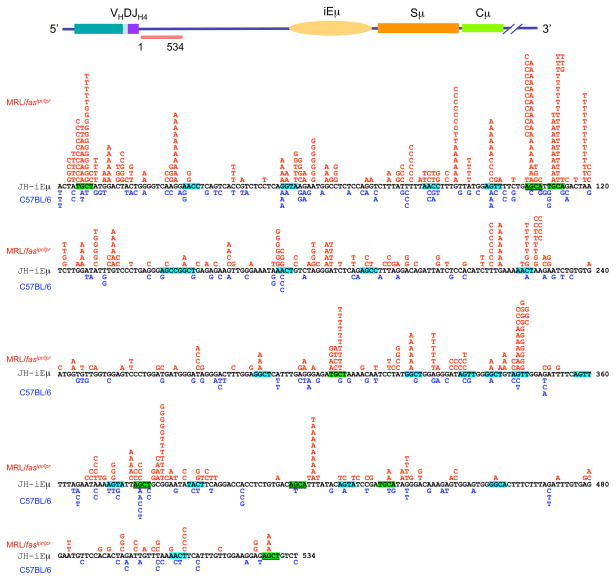
MRL/faslpr/lpr mice also displayed an increased targeting of mutations to the RGYW/WRCY hotspot (Figure 2). In these mice, the proportion of mutations that segregated with RGYW/WRCY was more than 44% higher than in C57BL/6 mice: 41.2% versus 28.6% (p < 0.002). RGYW/WRCY contains the WRC motif and its complement GYW, which are preferentially targeted by AID in vitro, indicating that the intrinsic SHM hotspot owes much to the inherent sequence specificity of AID itself [40]. Since AID can form dimers in vivo [69, 70], partially overlapping WRC on opposite strands would facilitate binding by two subunits of an AID dimer. The ensuing AID target hotspot in vivo would then be WGCW, which represents the most frequent embodiment of the hotspot of SHM [40]. The number of mutations targeting WGCW was significantly increased in MRL/faslpr/lpr mice (Figures 2, 3 and 4). In these mice, the mutations segregating within WGCW were more than 26.5% of the overall JH4-iEμ mutations, as compared to 10.1% in C57BL/6 mice (p < 0.0001). In MRL/faslpr/lpr mice, 82% of WGCW mutations targeted dG/dC, while only 47% did so in C57BL/6 mice. At least 70% of dG/dC mutations within WGCW in MRL/faslpr/lpr mice were transitions. Thus, most point-mutations in MRL/faslpr/lpr mice consisted of dG/dC mutations, mainly dG/dC transitions, and segregated within the WGCW AID hotspot.
Figure 3.
SHM is upregulated and preferentially targets the WGCW AID hotspot in lupus-prone mice. The 534 bp intronic JH4-iEμ DNA of Peyer’s patch PNAhi B cells of 11-week-old lupus-prone MRL/faslpr/lpr and age-matched non-autoimmune C57BL/6 mice were sequence analyzed. Mutations in MRL/faslpr/lpr mouse DNA are shown in red, mutations in C57BL/6 mouse DNA are shown in blue. WGCW AID hotspots are shown in green. RGYW/WRCY mutational hotspots are shown in blue. AGCT and AGCA motifs, which are iterations of both WGCW and RGYW are underlined.
Figure 4.
SHM is significantly upregulated in the lupus-prone mice. The 534 bp intronic JH4-iEμ DNA of Peyer’s patch B cells of 11-week-old lupus-prone MRL/faslpr/lpr and non-autoimmune C57BL/6 mice were sequence analyzed. Mutations in MRL/faslpr/lpr mouse DNA are shown in red, mutations in C57BL/6 mouse DNA are shown in blue. WGCW AID hotspots are highlighted green. RGYW/WRCY mutational hotspots are highlighted blue. AGCT and AGCA motifs, which are iterations of both WGCW and RGYW are underlined.
Lupus-prone MRL/faslpr/lpr mice undergo increased CSR
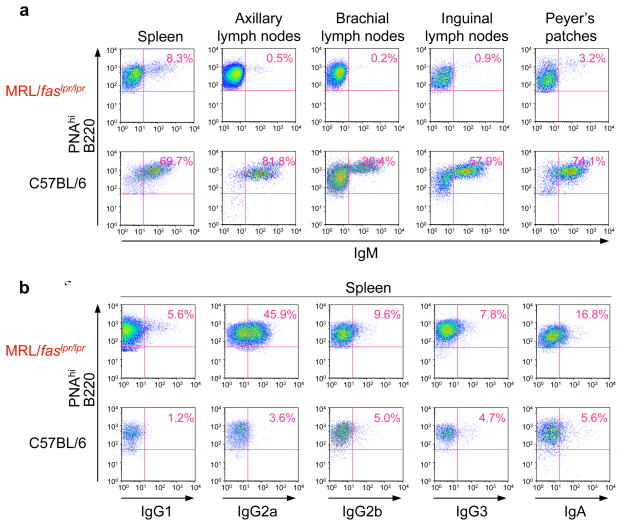
The important feature of the pathogenic autoantibodies found in patients with SLE is that they are IgG produced by an oligoclonal expansion of B cells. In individual SLE patients and lupus mice, the onset of clinical disease is associated with switching of autoantibodies from IgM to IgG. The predominance of IgG over IgM among pathogenic autoantibodies in human SLE and murine lupus indicates that CSR is an important mechanism underlying the development of disease. In our analysis of B220+ cells in spleen, axillary lymph nodes, brachial lymph nodes, inguinal lymph nodes and Peyer’s patches of lupus-prone MRL/faslpr/lpr mice, as few as 2.3% and 2.4% of total B cells (axillary and brachial lymph nodes) were surface IgM+ (Figure 5a), while in non-autoimmune control C57BL/6 mice, IgM+ B cells accounted for as many 86.0% and 54.9% of total B cells (spleen and axillary lymph nodes). The reduced surface IgM+ B cells in MRL/faslpr/lpr mice reflected the increased proportions of B cells that underwent CSR. In these mice, more than 21% of spleen B cells switched to IgG2a, the dominant isotype of autoantibodies in murine lupus, a percentage near 7-fold higher than that in C57BL/6 mice, and up to 25% of B cells switched to IgA in the spleen (Figure 5b and 5c). The increased proportion of switched B cells in MRL/faslpr/lpr mice did not merely result from an overall increased number of GCs (not shown), but also stemmed from an increased proportion of B cells that underwent CSR among GC (PNAhi) B cells in these autoimmune mice as compared to their non-autoimmune C57BL/6 counterparts (Figure 6). Almost 46% of splenic GC (PNAhi) B cells in MRL/faslpr/lpr mice underwent CSR to IgG2a, as compared with only 3.6% in non-autoimmune C57BL/6 control mice. Thus, in lupus-prone MRL/faslpr/lpr mice, B cells significantly upregulate CSR to all secondary Ig isotypes, with a predominance of CSR to IgG2a.
Figure 5.
CSR is significantly upregulated in lupus-prone MRL/faslpr/lpr mice. (a) Cells from spleen, lymph nodes or Peyer’s patches of MRL/faslpr/lpr or non-autoimmune C57BL/6 mice were stained with PE-labeled anti-B220 mAb, and FITC-labeled anti-IgM mAb, the IgM- B220+ cells are considered as switched B cells; (b) Cells from spleen, lymph nodes or Peyer’s patches of MRL/faslpr/lpr or non-autoimmune C57BL/6 mice were stained with PE-labeled anti-B220 mAb, and FITC-labeled anti-IgA mAb; (c) Spleen cells from MRL/faslpr/lpr or non-autoimmune C57BL/6 mice were stained with PE-labeled anti-B220 mAb, and FITC-labeled anti-IgG1, anti-IgG2a, anti-IgG2b, or anti-IgG3 mAb. Stained cells were then analyzed by FACS.
Figure 6.
Significant increased CSR to all isotypes in GC B cells of lupus-prone mice. (a) GC (PNAhi) B cells from spleen, lymph nodes or Peyer’s patches of lupus-prone MRL/faslpr/lpr or non-autoimmune C57BL/6 mice were stained with PE-labeled anti-B220 mAb, FITC-labeled anti-IgM mAb and then analyzed for surface fluorescence by FACS. (b) GC (PNAhi) B cells from the spleens of MRL/faslpr/lpr or C57BL/6 mice were stained with PE-labeled anti-B220 mAb, FITC-labeled anti-IgM, anti-IgG1, anti-IgG2a, anti-IgG2b, anti-IgG3 or anti-IgA mAb, and then analyzed for surface fluorescence by FACS.
Lupus-prone MRL/faslpr/lpr mice accumulate DNA deletions and insertions in the Igh locus
We found that, in addition to the greatly increased loads of point-mutations and significantly higher CSR rates, MRL/faslpr/lpr mice displayed a high frequency of deletions and insertions in the Igh locus. We chose to analyze the intronic JH4-iEμ sequence for DNA lesions, as this area is targeted by the SHM machinery. We reasoned that the impact of AID-mediated dC deamination could give rise single- or double-stranded DNA breaks, eventually leading to losses (deletions) or additions (insertions) of DNA sequences.
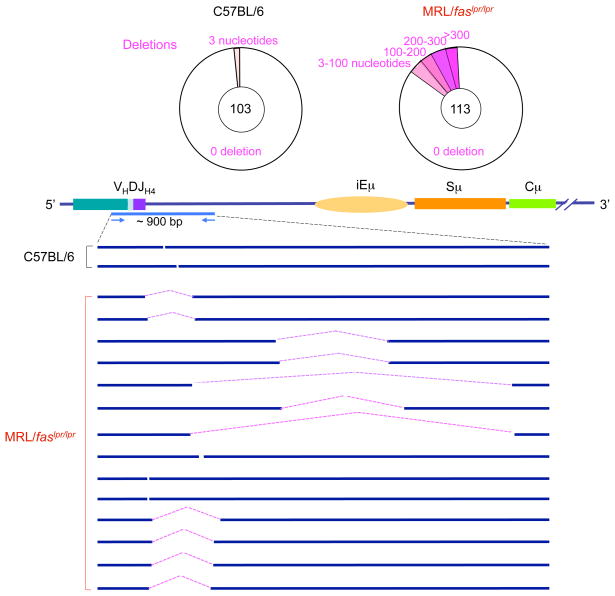
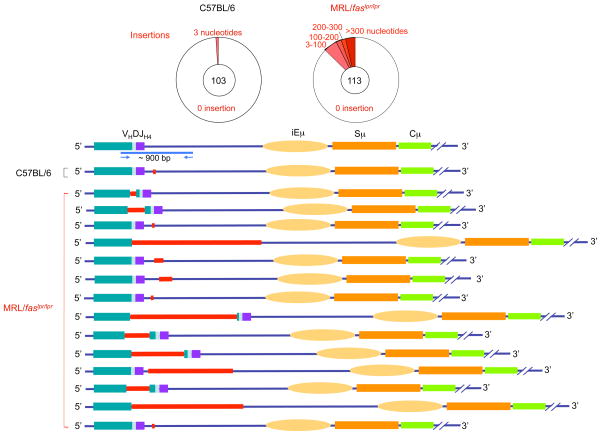
We amplified and analyzed VJ558DJH4-iEμ DNA sequences from Peyer’s patch GC (PNAhi) B cells of 3 11-week-old MRL/faslpr/lpr and 3 age-matched C57BL/6 mice. While only 3 of 103 VJ558DJH4-iEμ sequences from the Peyer’s patch B cells of C57BL/6 mice bore deletions or insertions (2 deletions and 1 insertion), all of which were only 3-bp in length, 28 of 113 VJ558DJH4-iEμ sequences from Peyer’s patch B cells of MRL/faslpr/lpr mice contained DNA deletions or insertions, respectively. Over 50% of these deletions and insertions consisted of more than 100 bp (Figures 7, 8, and 9). With the exception of one of the insertions containing a 419 bp sequence of chromosome 2, the nature and origin of the insertions were not identified. Thus, in autoimmune MRL/faslpr/lpr mice, the Igh locus accumulates multiple lesions, including both wide DNA deletions and long DNA insertions.
Figure 7.
The Igh locus in the B cells of lupus-prone MRL/faslpr/lpr mice contains a high frequency of deletions. Pie charts depict the proportions of sequences that contain different numbers of deletions (pink) over the about 900 bp intronic VJ558DJH4-iEμ DNA in GC (PNAhi) B cells from Peyer’s patches of three 11-week-old MRL/faslpr/lpr and three 11-week-old C57BL/6 mice. Depicted are locations and sizes of deletions.
Figure 8.
The Igh locus in the B cells of lupus-prone MRL/faslpr/lpr mice contains a high frequency of insertions. Pie charts depict the proportions of sequences that contain different numbers of insertions (red) over the about 900-bp VJ558DJH4-iEμ DNA in GC (PNAhi) B cells from Peyer’s patches of three 11-week old MRL/faslpr/lpr and three 11-week-old C57BL/6 mice. Depicted are locations and sizes of insertions.
Figure 9.
The junction sequences of deletions and deletions in the Igh locus of MRL/faslpr/lpr mice. VJ558DJH4-iEμ DNA of GC (PNAhi) B cells from Peyer’s patches of MRL/faslpr/lpr or C57BL/6 mice were amplified and sequenced. Each sequence is compared with germline Igh sequence. (a) The deleted nucleotides are indicated by pink dots. The numbers on top of each aligned sequence indicate upstream and downstream breakpoints of recombined sequences. (b) The inserted sequences are in red and underlined. The upstream and downstream VJ558DJH-iEμ sequences linked to the inserted DNA are in blue.
Discussion
The importance of somatically hypermutated and class switched, mainly IgG, autoantibodies in the pathological manifestations of systemic lupus is well established, but little is known about the precise molecular mechanisms underlying the generation of such autoantibodies. In these studies, we have shown markedly enhanced SHM and CSR as well as significant loads of DNA lesions in the Igh locus of lupus-prone MRL/faslpr/lpr mice. We also shown that in these autoimmune mice, Igh locus DNA lesions and increased SHM and CSR were associated with upregulation of AID and TLS pol θ, pol η and pol ζ. Our findings suggest that in lupus B cells, enhanced AID expression and dysregulation of TLS polymerases lead to aberrant SHM and CSR, which likely play a role in the generation of autoantibodies. They also provide a first evidence that DNA deletions and insertions occur at a high rate in the Ig locus of lupus B cells, likely as a result of the overall dysregulation of AID and other important elements of the SHM and CSR machineries.
Our findings imply that the dysregulation of SHM and CSR in lupus B cells is mediated by a significantly increased AID expression and, possibly, an enhanced recruitment of TLS polymerases to V(D)J and S region DNA, respectively. This is supported by our demonstration that somatic mutations in lupus B cells preferentially targeted the WGCW AID hotspot and were mainly dG/dC mutations, namely, dG → dA and dC → dT transitions. It has been suggested that the level of AID affects SHM and CSR, and, therefore, the affinity maturation and the class switching of autoantibodies in autoimmunity [64, 71]. The significantly higher Aicda expression in BXD2 autoimmune mice, as compared to their non-autoimmune mouse counterparts, is dependent on CD4+ T cells and is particularly apparent in B cells expressing a high level of CD86 [64]. Soluble CTLA-4-mediated inhibition of the CD28:CD86 co-engagement results in an inhibition of the “spontaneous” GC reaction, a reduction in Aicda expression to a level comparable to that in normal mice, and decreased production of IgG autoantibodies [64], strongly suggesting that AID induction is critical to SHM and CSR in lupus B cells, and the generation of autoantibodies. Indeed, lupus nephritis is abrogated in AID-deficient MRL/faslpr/lpr mice, in which circulating anti-dsDNA IgGs are barely detectable [24].
The spontaneous splenic GC reaction in autoimmune MRL/faslpr/lpr, as well as NZBxNZW F1, BXSB or BXD2 mice [64, 72], but not in age-matched, non-autoimmune mice, coincides with the appearance of autoantibodies [21, 22], indicating that the SHM and CSR processes that lead to generation of high affinity IgG autoantibodies unfold mainly, although possibly not exclusively, in GCs. The intrinsic hyperactivities of lupus B cells, likely compounded by a dysregulated interaction of these B cells with other immune cells, would result in not only a higher frequency of GCs [72], but also a sequence of molecular events leading to aberrant SHM and CSR. Increased AID expression in lupus B cells perhaps reflects an aberrant regulation of these lymphocytes by other immune cells in the GC, likely through CD154:CD40 engagement and co-stimulatory receptors, and/or their intrinsic hyperactivity. Accordingly, these events would be reflected in the higher frequency of mutations in B cells from SLE patients, as compared to healthy individuals [60, 61], and the high proportion of IgM+IgD+ B cells spontaneously undergoing CSR in SLE patients [73].
In CSR, AID deaminates dCs in S regions, leading to the emergence of DSBs. In addition to their role in CSR, AID-induced DSBs can promote Igh-c-Myc chromosomal translocations [74], suggesting that AID also mediates the generation of DNA lesions, such as DSBs, outside S regions. The frequency of DSBs in S region and non-S region DNA has been thought to correlate with the level of AID expression. In an Aicda+/− plasmacytoma mouse model, reduced AID expression levels impaired S region DSBs and Igh-c-Myc translocations [75]. Accordingly, in Aicda+/− MRL/faslpr/lpr mice, a 25–40% reduction of Aicda expression leads to significantly reduced high-affinity anti-dsDNA IgGs [71], further suggesting that a high level of AID expression is critical for the generation of pathogenic autoantibodies. Conversely, the high frequency of deletions and insertions in the Igh locus we have reported here likely resulted from DSBs effected by AID that we have shown to be significantly upregulated in lupus-prone MRL/faslpr/lpr mice. Reported Igh-oncogene translocations involves in general the 3′ portion of JH-iEμ and S region DNA [76], which was not included in the DNA sequence we analyzed here for deletions and insertions. Therefore, is reasonable to assume that the actual frequency of deletions and insertions in the Igh locus of MRL/faslpr/lpr mice was much higher than what we have shown here. The overall increased deletions, insertions, point-mutations and altered nature of mutations in MRL/faslpr/lpr mice strongly point at overall increased DNA lesions by increased AID level and altered DNA repair in these autoimmune mice.
TLS polymerases play a major role in SHM [32, 33]. In the first stage of SHM and CSR, AID deaminates dC to mediate DNA lesion generation. In the second stage, DNA repair, error-prone TLS polymerases insert mutations not only in the immediate vicinity of deaminated dC, but also as far as 15- and 10-nucleotides 5′ and 3′ from the deamination site, respectively [32, 33], thereby affecting the distribution, frequency and spectrum of mutations. Upregulated AID and TLS polymerases would explain the greatly increased frequency and altered distribution and spectrum of mutations we have shown here in lupus-prone MRL/faslpr/lpr mice. TLS pol θ, pol η and pol ζ play important yet different roles in SHM. As we have shown, knockdown of the Rev3 subunit of pol ζ or deficiency in pol θ results in a significantly reduced overall mutation frequency, at both dC/dG and dA/dT [27, 28]. By contrast, deficiency in pol η results in significantly decreased dA/dT mutations with a virtually normal mutation frequency [77, 78]. Pol θ, pol η and pol ζ are preferentially expressed in hypermutating B cells [27, 28, 79] and are induced in B cells by the stimuli that induce AID expression and/or SHM, such as CD154, LPS or BCR crosslinking [26–28, 42, 65, 66, 80]. In autoimmune B cells, enhanced AID expression and AID recruitment to Igh locus would lead to enhanced recruitment of TLS polymerases. Upregulated AID expression in autoimmune mice coincides with an upregulation of TLS, which may or may not keep up with the increased load of DNA lesions effected by the upregulated AID. In lupus B cells, the higher expression of AID would yield more DNA lesions, resulting in enhanced recruitment of TLS polymerases to Igh locus and increased occurrence of DSBs, and leading to a high loads of deletions and insertions, as well as SHM and CSR.
The association of SLE with B cell malignancies has been widely reported [81–84]. Findings in cohort studies provide support for an increased risk of B cell lymphoma in SLE, but their meaning and pathogenic implications are of difficult interpretation [85]. B cell lymphomas frequently bear chromosomal translocations that juxtapose Ig loci with an oncogene [76]. These translocations represent one of the first steps in the neoplastic transformation of a B cell. Their frequent involvement of the Ig locus is thought to stem directly from the DNA lesions occurring in V(D)J recombination, SHM and CSR [51, 86]. The significantly increased DNA deletions and insertions in the Igh locus of lupus-prone mice suggest that translocations are also increased in lupus B cells, possibly playing a significant role in the lymphomagenesis associated with this disease. Our current experiments aim at defining the frequency, nature and generation of DNA lesions in SLE patents and their relative contribution to the origin of translocations.
Acknowledgments
This work was supported by the NIH grants AI 045011, AI 060573 and AI 079705 to P.C.
References
- 1.Shlomchik MJ, Madaio MP. The role of antibodies and B cells in the pathogenesis of lupus nephritis. Springer Semin Immunopathol. 2003;24:363–375. doi: 10.1007/s00281-003-0119-1. [DOI] [PubMed] [Google Scholar]
- 2.Diamond B. Autoimmunity. Immunol Rev. 2005;204:5–8. doi: 10.1111/j.0105-2896.2005.00258.x. [DOI] [PubMed] [Google Scholar]
- 3.Shlomchik MJ, Aucoin AH, Pisetsky DS, Weigert MG. Structure and function of anti-DNA autoantibodies derived from a single autoimmune mouse. Proc Natl Acad Sci USA. 1987;84:9150–9154. doi: 10.1073/pnas.84.24.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shlomchik M, Mascelli M, Shan H, Radic MZ, Pisetsky D, Marshak-Rothstein A, Weigert M. Anti-DNA antibodies from autoimmune mice arise by clonal expansion and somatic mutation. J Exp Med. 1990;171:265–292. doi: 10.1084/jem.171.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Es JH, Gmelig Meyling FH, van de Akker WR, Aanstoot H, Derksen RH, Logtenberg T. Somatic mutations in the variable regions of a human IgG anti-double-stranded DNA autoantibody suggest a role for antigen in the induction of systemic lupus erythematosus. J Exp Med. 1991;173:461–470. doi: 10.1084/jem.173.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winkler TH, Fehr H, Kalden JR. Analysis of immunoglobulin variable region genes from human IgG anti-DNA hybridomas. Eur J Immunol. 1992;22:1719–1728. doi: 10.1002/eji.1830220709. [DOI] [PubMed] [Google Scholar]
- 7.Radic MZ, Mackle J, Erikson J, Mol C, Anderson WF, Weigert M. Residues that mediate DNA binding of autoimmune antibodies. J Immunol. 1993;150:4966–4977. [PubMed] [Google Scholar]
- 8.William J, Euler C, Christensen S, Shlomchik MJ. Evolution of autoantibody responses via somatic hypermutation outside of germinal centers. Science. 2002;297:2066–2070. doi: 10.1126/science.1073924. [DOI] [PubMed] [Google Scholar]
- 9.Wellmann U, Letz M, Herrmann M, Angermuller S, Kalden JR, Winkler TH. The evolution of human anti-double-stranded DNA autoantibodies. Proc Natl Acad Sci USA. 2005;102:9258–9263. doi: 10.1073/pnas.0500132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atassi MZ, Casali P. Molecular mechanisms of autoimmunity. Autoimmunity. 2008;41:123–132. doi: 10.1080/08916930801929021. [DOI] [PubMed] [Google Scholar]
- 11.Elkon K, Casali P. Nature and functions of autoantibodies. Nat Clin Pract Rheumatol. 2008;4:491–498. doi: 10.1038/ncprheum0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casali P. Polyclonal B cell activation and antigen-driven antibody response as mechanisms of autoantibody production in SLE. Autoimmunity. 1990;5:147–150. doi: 10.3109/08916939009002973. [DOI] [PubMed] [Google Scholar]
- 13.Chai SK, Mantovani L, Kasaian MT, Casali P. Natural autoantibodies. Adv Exp Med Biol. 1994;347:147–159. doi: 10.1007/978-1-4615-2427-4_15. [DOI] [PubMed] [Google Scholar]
- 14.Kasaian MT, Ikematsu H, Balow JE, Casali P. Structure of the VH and VL segments of monoreactive and polyreactive IgA autoantibodies to DNA in patients with systemic lupus erythematosus. J Immunol. 1994;152:3137–3151. [PMC free article] [PubMed] [Google Scholar]
- 15.Casali P, Schettino EW. Structure and function of natural antibodies. Curr Top Microbiol Immunol. 1996;120:167–179. doi: 10.1007/978-3-642-85226-8_17. [DOI] [PubMed] [Google Scholar]
- 16.Putterman C, Diamond B. Immunization with a peptide surrogate for double-stranded DNA (dsDNA) induces autoantibody production and renal immunoglobulin deposition. J Exp Med. 1998;188:29–38. doi: 10.1084/jem.188.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z, Schettino EW, Padlan EA, Ikematsu H, Casali P. Structure-function analysis of a lupus anti-DNA autoantibody: central role of the heavy chain complementarity-determining region 3 Arg in binding of double- and single-stranded DNA. Eur J Immunol. 2000;30:2015–2026. doi: 10.1002/1521-4141(200007)30:7<2015::AID-IMMU2015>3.0.CO;2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wardemann YS, ASJWYEM, MCN Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 19.Ichiyoshi Y, Casali P. Analysis of the structural correlates for antibody polyreactivity by multiple reassortments of chimeric human immunoglobulin heavy and light chain V segments. J Exp Med. 1994;180:885–895. doi: 10.1084/jem.180.3.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ichiyoshi Y, Casali P. Analysis of the structural correlates for self-antigen binding by natural and disease-related autoantibodies. In vitro expression of recombinant and/or mutagenized human IgG. Ann N Y Acad Sci. 1995;764:328–341. doi: 10.1111/j.1749-6632.1995.tb55844.x. [DOI] [PubMed] [Google Scholar]
- 21.Andrews BS, Eisenberg RA, Theofilopoulos AN, Izui S, Wilson CB, McConahey PJ, Murphy ED, Roths JB, Dixon FJ. Spontaneous murine lupus-like syndromes. Clinical and immunopathological manifestations in several strains. J Exp Med. 1978;148:1198–1215. doi: 10.1084/jem.148.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theofilopoulos AN, Dixon FJ. Murine models of systemic lupus erythematosus. Adv Immunol. 1985;37:269–390. doi: 10.1016/s0065-2776(08)60342-9. [DOI] [PubMed] [Google Scholar]
- 23.Murphy ED, Roths JB. A new congenic inbred strain, MRL/Mp Mouse News Lett. 1978;58:51–52. [Google Scholar]
- 24.Jiang C, Foley J, Clayton N, Kissling G, Jokinen M, Herbert R, Diaz M. Abrogation of lupus nephritis in activation-induced deaminase-deficient MRL/lpr mice. J Immunol. 2007;178:7422–7431. doi: 10.4049/jimmunol.178.11.7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang B, Casali P. The CDR1 sequences of a major proportion of human germline Ig VH genes are inherently susceptible to amino acid replacement. Immunol Today. 1994;15:367–373. doi: 10.1016/0167-5699(94)90175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zan H, Li Z, Yamaji K, Dramitinos P, Cerutti A, Casali P. B cell receptor engagement and T cell contact induce Bcl-6 somatic hypermutation in human B cells: identity with Ig hypermutation. J Immunol. 2000;165:830–839. doi: 10.4049/jimmunol.165.2.830. [DOI] [PubMed] [Google Scholar]
- 27.Zan H, Komori A, Li Z, Cerutti A, Schaffer A, Flajnik MF, Diaz M, Casali P. The translesion DNA polymerase ζ plays a major role in Ig and bcl-6 somatic hypermutation. Immunity. 2001;14:643–653. doi: 10.1016/s1074-7613(01)00142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zan H, Shima N, Xu Z, Al-Qahtani A, Evinger AJI, Zhong Y, Schimenti JC, Casali P. The translesion DNA polymerase θ plays a dominant role in immunoglobulin gene somatic hypermutation. EMBO J. 2005;24:3757–3769. doi: 10.1038/sj.emboj.7600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stavnezer J, Amemiya CT. Evolution of isotype switching. Semin Immunol. 2004;16:257–275. doi: 10.1016/j.smim.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Diaz M, Casali P. Somatic immunoglobulin hypermutation. Curr Opin Immunol. 2002;14:235–240. doi: 10.1016/s0952-7915(02)00327-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu X, Feng J, Komori A, Kim EC, Zan H, Casali P. Immunoglobulin somatic hypermutation: double-strand DNA breaks, AID and error-prone DNA repair. J Clin Immunol. 2003;23:235–246. doi: 10.1023/a:1024571714867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Z, Fulop Z, Zhong Y, Evinger AJ, Zan H, Casali P. DNA lesions and repair in immunoglobulin class switch recombination and somatic hypermutation. Ann NY Acad Sci. 2005;1050:146–162. doi: 10.1196/annals.1313.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casali P, Pal Z, Xu Z, Zan H. DNA repair in antibody somatic hypermutation. Trends Immunol. 2006;27:313–321. doi: 10.1016/j.it.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Noia JM, Neuberger MS. Molecular Mechanisms of Antibody Somatic Hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 35.Neuberger MS, Rada C. Somatic hypermutation: activation-induced deaminase for C/G followed by polymerase η for A/T. J Exp Med. 2007;204:7–10. doi: 10.1084/jem.20062409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 37.Dickerson SK, Market E, Besmer E, Papavasiliou FN. AID mediates hypermutation by deaminating single stranded DNA. J Exp Med. 2003;197:1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 39.Sohail A, Klapacz J, Samaranayake M, Ullah A, Bhagwat AS. Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucleic Acids Res. 2003;31:2990–2994. doi: 10.1093/nar/gkg464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beale RCL, Oetersen-Mahrt SK, Watt IN, Harris RS, Rada C, Neuberger MS. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J Mol Biol. 2004;337:585–596. doi: 10.1016/j.jmb.2004.01.046. [DOI] [PubMed] [Google Scholar]
- 41.Papavasiliou FN, Schatz DG. Somatic hypermutation of immunoglobulin genes; merging mechanisms for genetic diversity. Cell. 2002;109:s35–s44. doi: 10.1016/s0092-8674(02)00706-7. [DOI] [PubMed] [Google Scholar]
- 42.Zan H, Wu X, Komori A, Holloman WK, Casali P. AID-dependent generation of resected double-strand DNA breaks and recruitment of Rad52/Rad51 in somatic hypermutation. Immunity. 2003;18:727–738. doi: 10.1016/s1074-7613(03)00151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rada C, Di Noia JM, Neuberger MS. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell. 2004;16:163–171. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 44.Rush JS, Fugmann SD, DGS Staggered AID-dependent DNA double strand breaks are the predominant DNA lesions targeted to Sμ in Ig class switch recombination. Int Immunol. 2004;16:549–557. doi: 10.1093/intimm/dxh057. [DOI] [PubMed] [Google Scholar]
- 45.Yabuki M, Fujii MM, Maizels N. The MRE11-RAD50-NBS1 complex accelerates somatic hypermutation and gene conversion of immunoglobulin variable regions. Nat Immunol. 2005;6:730–736. doi: 10.1038/ni1215. [DOI] [PubMed] [Google Scholar]
- 46.Larson ED, Cummings WJ, Bednarski DW, Maizels N. MRE11/RAD50 cleaves DNA in the AID/UNG-dependent pathway of immunoglobulin gene diversification. Mol Cell. 2005;20:367–375. doi: 10.1016/j.molcel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 47.Bross L, Fukita Y, McBlane FCD, Rajewsky K, Jacobs H. DNA double-strand breaks in immunoglobulin genes undergoing somatic hypermutation. Immunity. 2000;13:589–597. doi: 10.1016/s1074-7613(00)00059-5. [DOI] [PubMed] [Google Scholar]
- 48.Papavasiliou FN, Schatz DG. Cell-cycle-regulated DNA double-stranded breaks in somatic hypermutation of immunoglobulin genes. Nature. 2000;408:216–221. doi: 10.1038/35041599. [DOI] [PubMed] [Google Scholar]
- 49.Papavasiliou FN, Schatz DG. The activation-induced deaminase functions in a postcleavage step of the somatic hypermutation process. J Exp Med. 2002;195:1193–1198. doi: 10.1084/jem.20011858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bross L, Muramatsu K, Kinoshita K, Honjo H, Jacobs H. DNA double-strand breaks: prior to but not sufficient in targeting hypermutation. J Exp Med. 2002;195:1187–1192. doi: 10.1084/jem.20011749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Unniraman S, Zhou S, Schatz DG. Identification of an AID-independent pathway for chromosomal translocations between the Igh switch region and Myc. Nat Immunol. 2004;5:1117–1123. doi: 10.1038/ni1127. [DOI] [PubMed] [Google Scholar]
- 52.Casali P, Zan H. Class switching and Myc translocation: how does DNA break? Nat Immunol. 2004;5:1101–1103. doi: 10.1038/ni1104-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 54.Neuberger MS, Di Noia JM, Beale RC, Williams GT, Yang Z, Rada C. Somatic hypermutation at A. T pairs: polymerase error versus dUTP incorporation. Nat Rev Immunol. 2005;5:171–178. doi: 10.1038/nri1553. [DOI] [PubMed] [Google Scholar]
- 55.Bardwell PD, Woo CJ, Wei K, Li Z, Martin A, Sack SZ, Parris T, Edelmann W, Scharff MD. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat Immunol. 2004;5:224–229. doi: 10.1038/ni1031. [DOI] [PubMed] [Google Scholar]
- 56.Vallur AC, Maizels N. Activities of human exonuclease 1 that promote cleavage of transcribed immunoglobulin switch regions. Proc Natl Acad Sci U S A. 2008;105:16508–16512. doi: 10.1073/pnas.0805327105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neuberger MS, Harris RS, Di Noia J, Petersen-Mahrt SK. Immunity through DNA deamination. Trends Biochem Sci. 2003;28:305–312. doi: 10.1016/S0968-0004(03)00111-7. [DOI] [PubMed] [Google Scholar]
- 58.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, Rajewsky K, Alt FW. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 59.Chaudhuri J, Basu U, Zarrin A, Yan C, Franco S, Perlot T, Vuong B, Wang J, Phan RT, Datta A, Manis J, Alt FW. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- 60.Dorner T, Heimbacher C, Farner NL, Lipsky PE. Enhanced mutational activity of Vkappa gene rearrangements in systemic lupus erythematosus. Clin Immunol. 1999;92:188–196. doi: 10.1006/clim.1999.4740. [DOI] [PubMed] [Google Scholar]
- 61.Dorner T, Kaschner S, Hansen A, Pruss A, Lipsky PE. Perturbations in the impact of mutational activity on Vlambda genes in systemic lupus erythematosus. Arthritis Res. 2001;3:368–374. doi: 10.1186/ar329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dorner T, Lipsky PE. Molecular basis of immunoglobulin variable region gene usage in systemic autoimmunity. Clin Exp Med. 2005;4:159–169. doi: 10.1007/s10238-004-0051-2. [DOI] [PubMed] [Google Scholar]
- 63.Hsu HC, Zhou T, Kim H, Barnes S, Yang P, Wu Q, Zhou J, Freeman BA, Luo M, Mountz JD. Production of a novel class of polyreactive pathogenic autoantibodies in BXD2 mice causes glomerulonephritis and arthritis. Arthritis Rheum. 2006;54:343–355. doi: 10.1002/art.21550. [DOI] [PubMed] [Google Scholar]
- 64.Hsu HC, Wu Y, Yang P, Wu Q, Job G, Chen J, Wang J, Accavitti-Loper MA, Grizzle WE, Carter RH, Mountz JD. Overexpression of activation-induced cytidine deaminase in B cells is associated with production of highly pathogenic autoantibodies. J Immunol. 2007;178:5357–5365. doi: 10.4049/jimmunol.178.8.5357. [DOI] [PubMed] [Google Scholar]
- 65.Zan H, Cerutti A, Schaffer A, Dramitinos P, Casali P. CD40 engagement triggers switching to IgA1 and IgA2 in human B cells through induction of endogenous TGF-β. Evidence for TGF-β but not IL-10-dependent direct Sμ→Sα and sequential Sμ→Sγ, Sγ→Sα DNA recombination. J Immunol. 1998;161:5217–5225. [PMC free article] [PubMed] [Google Scholar]
- 66.Zan H, Cerutti A, Dramitinos P, Schaffer A, Li Z, Casali P. Induction of Ig somatic hypermutation and class switching in a human monoclonal IgM+ IgD+ cell line in vitro: Definition of the requirements and the modalities of hypermutation. J Immunol. 1999;162:3437–3447. [PMC free article] [PubMed] [Google Scholar]
- 67.Zan H, Casali P. AID- and Ung-dependent generation of staggered double-strand DNA breaks in immunoglobulin class switch DNA recombination: A post-cleavage role for AID. Mol Immunol. 2008;46:45–61. doi: 10.1016/j.molimm.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu X, Tsai CY, Patam MB, Zan H, Chen JP, Lipkin SM, Casali P. A role for the MutL mismatch repair Mlh3 protein in immunoglobulin class switch DNA recombination and somatic hypermutation. J Immunol. 2006;176:5426–5437. doi: 10.4049/jimmunol.176.9.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dickerson SK, Market E, Besmer E, Papavasiliou FN. AID mediates hypermutation by deaminating single stranded DNA. J Exp Med. 2003;197:1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prochnow C, Bransteitter R, Klein MG, Goodman MF, Chen XS. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 2007;445:447–451. doi: 10.1038/nature05492. [DOI] [PubMed] [Google Scholar]
- 71.Jiang C, Zhao ML, Diaz M. Activation-induced deaminase heterozygous MRL/lpr mice are delayed in the production of high-affinity pathogenic antibodies and in the development of lupus nephritis. Immunology. 2008 doi: 10.1111/j.1365–2567.2008.02882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luzina IG, Atamas SP, Storrer CE, daSilva LC, Kelsoe G, Papadimitriou JC, Handwerger BS. Spontaneous formation of germinal centers in autoimmune mice. J Leukoc Biol. 2001;70:578–584. [PubMed] [Google Scholar]
- 73.Liu S, Cerutti A, Casali P, Crow MK. Ongoing immunoglobulin class switch DNA recombination in lupus B cells: analysis of switch regulatory regions. Autoimmunity. 2004;37:431–443. doi: 10.1080/08916930400010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, Honjo T, Nussenzweig A, Nussenzweig MC. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 75.Takizawa M, Tolarova H, Li Z, Dubois W, Lim S, Callen E, Franco S, Mosaico M, Feigenbaum L, Alt FW, Nussenzweig A, Potter M, Casellas R. AID expression levels determine the extent of cMyc oncogenic translocations and the incidence of B cell tumor development. J Exp Med. 2008;205:1949–1957. doi: 10.1084/jem.20081007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kuppers R, Dalla-Favera R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 2001;20:5580–5594. doi: 10.1038/sj.onc.1204640. [DOI] [PubMed] [Google Scholar]
- 77.Delbos F, De Smet A, Faili A, Aoufouchi S, Weill JC, Reynaud CA. Contribution of DNA polymerase η to immunoglobulin gene hypermutation in the mouse. J Exp Med. 2005;201:1191–1196. doi: 10.1084/jem.20050292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martomo SA, Yang WW, Wersto RP, Ohkumo T, Kondo Y, Yokoi M, Masutani C, Hanaoka F, Gearhart PJ. Different mutation signatures in DNA polymerase η- and MSH6-deficient mice suggest separate roles in antibody diversification. Proc Natl Acad Sci USA. 2005;102:8656–8661. doi: 10.1073/pnas.0501852102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zeng X, Winter DB, Kasmer C, Kraemer KH, Lehmann AR, Gearhart PJ. DNA polymerase η is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat Immunol. 2001;2:537–541. doi: 10.1038/88740. [DOI] [PubMed] [Google Scholar]
- 80.Xu Z, Zan H, Pal Z, Casali P. DNA replication to aid somatic hypermutation. Adv Exp Med Biol. 2007;596:111–127. doi: 10.1007/0-387-46530-8_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cibere J, Sibley J, Haga M. Systemic lupus erythematosus and the risk of malignancy. Lupus. 2001;10:394–440. doi: 10.1191/096120301678646128. [DOI] [PubMed] [Google Scholar]
- 82.Bernatsky S, Boivin JF, Joseph L, Rajan R, Zoma A, Manzi S, et al. An international cohort study of cancer in systemic lupus erythematosus. Arthritis Rheum. 2005;52:1481–1490. doi: 10.1002/art.21029. [DOI] [PubMed] [Google Scholar]
- 83.Hansen A, Lipsky PE, Dorner T. B-cell lymphoproliferation in chronic inflammatory rheumatic diseases. Nat Clin Pract Rheumatol. 2007;3:561–569. doi: 10.1038/ncprheum0620. [DOI] [PubMed] [Google Scholar]
- 84.Niller HH, Wolf H, Minarovits J. Regulation and dysregulation of Epstein-Barr virus latency: Implications for the development of autoimmune diseases. Autoimmunity. 2008;41:298–328. doi: 10.1080/08916930802024772. [DOI] [PubMed] [Google Scholar]
- 85.Bernatsky S, Clarke A, Ramsey-Goldman R. Malignancy and systemic lupus erythematosus. Curr Rheumatol Rep. 2002;4:351–358. doi: 10.1007/s11926-002-0045-6. [DOI] [PubMed] [Google Scholar]
- 86.Unniraman S, Schatz DG. AID and Igh switch region-Myc chromosomal translocations. DNA Repair. 2006;5:1259–1264. doi: 10.1016/j.dnarep.2006.05.019. [DOI] [PubMed] [Google Scholar]