SUMMARY
Much of the statistical methodology underlying the experimental design of Phase 1 trials in oncology is intended for studies involving a single cytotoxic agent. The goal of these studies is to estimate the maximally tolerated dose, the highest dose that can be administered with an acceptable level of toxicity. A fundamental assumption of these methods is monotonicity of the dose-toxicity curve. This is a reasonable assumption for single agent trials in which the administration of greater doses of the agent can be expected to produce dose-limiting toxicities (DLTs) in increasing proportions of patients. When studying multiple agents, the assumption may not hold since the ordering of the toxicity probabilities could possibly be unknown for several of the available drug combinations. At the same time, some of the orderings are known and so we describe the whole situation as that of a partial ordering. In this article, we propose a new two-dimensional dose-finding method for multiple-agent trials that simplifies to the continual reassessment method (CRM), introduced by O’Quigley, Pepe, and Fisher (1990), when the ordering is fully known. This design enables us to relax the assumption of a monotonic dose-toxicity curve. We compare our approach and some simulation results to a CRM design in which the ordering is known as well as to other suggestions for partial orders.
Keywords: Clinical trial, Continual reassessment method, Dose escalation, Dose finding studies, Maximum tolerated dose, Phase 1 trials, Partial ordering, Toxicity, Drug combination
1. Introduction and background
The primary goal of a typical Phase 1 trial is the estimation of the maximum tolerated dose (MTD) according to some “tolerable” level of toxicity. The MTD is based upon the probability that a patient in the trial experiences a dose-limiting toxicity (DLT). Specifically, it is defined as the dose with a probability of DLT closest to some target toxicity rate. Numerous designs have been proposed for identifying the MTD from a discrete set of preset doses available in which toxicity is described as a binary random variable. The majority of the statistical methods implemented in these designs are based upon the assumption of a monotonic dose-toxicity curve. Using the terminology of Robertson, Wright and Dykstra (1988), the probabilities of toxic response with respect to dose follow a “simple order.” That is, the ordering of the probability of toxicity between any two doses is known and the higher dose corresponds to a greater probability of toxicity. The assumption of a simple order will often appear reasonable in studies that involve a single cytotoxic agent. However, the use of multiple-agent treatments, in which the monotonicity assumption may not hold, is becoming more commonplace in dose-finding studies.
Phase 1 trials combining more than one drug present a bigger challenge in MTD estimation than do single-agent studies. It may be reasonable to assume monotonicity for each drug separately but, when combined, the ordering may not be fully known. We may be able to identify the order of the toxicity probabilities for only a subset of the available treatments, resulting in a partial order. Therefore, it may not be clear which dose should be the next dose assigned in a decision of escalation. Various possible partial order scenarios are described in Robertson et al.(1988), whereas parameter estimation subject to order restrictions is discussed in Dunbar, Conaway, and Peddada (2001), as well as in Hwang and Peddada (1994). In this article, we present a new two-dimensional method for estimating the MTD in Phase 1 trials in which the ordering is not fully known. The proposed design builds off of the continual reassessment method (CRM), which has demonstrated near-optimal properties in trials when the order is known. When the toxicity order is fully known, the proposed method reduces to the CRM.
O’Quigley, Pepe and Fisher (1990) introduced the CRM as an alternative to the traditional Up-and-Down escalation schemes reviewed by Storer (1989). In its original form the CRM is a Bayesian method that uses sequential updating of dose-toxicity information to estimate the dose level at which to treat the next available patient. Suppose that we have a discrete set of k preset dose levels, d1,…, dk. The method begins by assuming a functional dose-toxicity curve, ψ(di, a), that is monotonic in both dose level, di, and the parameter, a; for instance, the logistic curve or power model. After having included j subjects, we can generate an estimate, âj, based on the posterior mean of a. The dose given to the (j + 1) th patient is the level, di, that minimizes Δ(ψ(di, âj), θ), where θ is the target probability of DLT and Δ(v, w) is a measure of distance, such as absolute distance. The procedure continues until some predetermined sample size of patients is exhausted or a stopping rule takes effect.
Although the number of Phase 1 trials that use multiple-agent combinations is increasing, there is relatively little statistical methodology for designing these trials. Korn and Simon (1993) present a graphical method, known as the “tolerable dose diagram”, for guiding the escalation strategy for the combinations. Kramar, Lebecq and Candahl (1999) lay out an a priori ordering for the treatments, and estimate the MTD using a parametric model for the probability of a DLT as a function of the doses of the two agents in combination. Thall et al. (2003) proposed a two-stage Bayesian method for dose-finding with two agents. The dose was first escalated along the diagonal direction in their two-dimensional dose space by increasing the doses of both drugs. Subsequently, two additional treatment combinations were distinguished using a toxicity equivalence contour. Wang and Ivanova (2005) also propose a Bayesian method for two agent combinations where the prior information is based on the single agent toxicity profiles. The allocation of patients to doses is done in a way that is similar to the CRM of O’Quigley, Pepe and Fisher (1990). They lay out an initial grid of points designed to move quickly towards a “solution”, specifically, a dose combination with acceptable toxicity. These initial allocations are essentially a series of individual “standard” phase I trials where the dose-toxicity relationship can be assumed to be monotone. Yin and Yuan (2009) propose a Bayesian dose-finding design that incorporates a copula-type model for estimating the toxicity probabilities of drug combinations. Another design proposed by Yin and Yuan (2009) is a Bayesian design for dose-finding based on latent contingency tables. The approach of Conaway, Dunbar, and Peddada (2004) identifies the simple and partial orders of toxicity probabilities by classifying nodal and nonnodal parameters (see Section 2.2).
The method of this article is developed within the framework of Conaway, Dunbar and Peddada (2004) in that we proceed by laying out all possible simple orders for the probability of toxicity that are consistent with the known orderings among dose combinations. Our strategy is to focus estimation of toxicity probabilities within a small number of simple orders, and allow the properties of the CRM to provide efficient estimates of the MTD within these orders. The remainder of the article is organized as follows: Section 2 gives a general overview of partial order restrictions and the background of the approach of Conaway et al. (2004). Section 3 presents the toxicity probability model and inference associated with the new design, while Section 4 provides simulation results comparing the new method to that of the CRM for fully known orderings, as well as to the approaches of Conaway et al. (2004) and Yin and Yuan (2009). Finally, some discussion on the implications of the new design, along with some topics of interest for further research is presented.
2. Partial ordering of doses
2.1 Example
Consider an example of a Phase 1 study of Samarium Lexidronam / Bortezomib combination therapy for the treatment of relapsed or refractory multiple myeloma. In this particular investigation, a dose-limiting toxicity is defined by a patient experiencing a grade 3+ neutrophenia. This study is described in Berenson et al. (2009) and the treatment combinations are displayed in Table 1. In this example, the ordering of the toxicity probabilities for some of the treatment combinations is not known. Specifically, the trial consists of six drug combinations (doses), d1,…, d6. The toxicity ordering between doses d1 and d2 is known due to the fact that the dose of Bortezomib remains the same while the dose of Sm increases. Such is the case for the ordering between doses d2 and d3. However, the order relationship between doses d3 and d5 and between d3 and d4 is unknown because the dose of Sm decreases while the dose of Bortezomib increases. If we continue to assess the known and unknown toxicity order relationships in this way, it is straightforward to determine that the following order relationships are known: (1) d1 → d2 → d3 → d6, (2) d1 → d4 → d5 → d6 and (3) d2 → d5. In these diagrams, combinations whose orderings are known are connected by arrows. That is, it is known that combination d5 is more toxic than d2 and d4, combination d4 is more toxic than d1, and combination d6 is more toxic than all other combinations. Escalation to a previously untried dose depends on a specification prior to the trial of “possible escalation combinations” associated with each combination. For example, the possible escalation combinations for d1 are d2 and d4, meaning that if d1 was tried and found to be sufficiently non-toxic, then escalation could proceed to the previously untried d2 or d4 Taking into account the subset of drug combinations for which we know the toxicity order, we aim to formulate the possible orderings of the toxicity profile. In the context of the current example, the trial requires the investigation of the following five simple orders.
Table 1.
Drug combinations used in Phase 1 trial of Samarium Lexidronam and Bortezomib
| Drug Combination | ||||||
|---|---|---|---|---|---|---|
| Agent | d1 | d2 | d3 | d4 | d5 | d6 |
| Sm (mCi/kg) | 0.25 | 0.5 | 1.0 | 0.25 | 0.5 | 1.0 |
| Bortezomib (mg/m2) | 1.0 | 1.0 | 1.0 | 1.3 | 1.3 | 1.3 |
d1 → d2 → d3 → d4 → d5 → d6
d1 → d2 → d4 → d3 → d5 → d6
d1 → d2 → d4 → d5 → d3 → d6
d1 → d4 → d2 → d3 → d5 → d6
d1 → d4 → d2 → d5 → d3 → d6
2.2 Dose-escalation in Conaway, Dunbar and Peddada (2004)
Hwang and Peddada (1994) provide a method for estimating parameters subject to partial orders. Their method uses different estimation procedures for “nodal” and “non-nodal” parameters. A nodal parameter is one whose ordering with respect to all other parameters is known. In the Berenson et al. (2009) example, R(d1) is a nodal parameter because it is known that R(d1) ≤ R(di) for i ≥ 2. R(d2) is non-nodal because it is unknown whether R(d2) ≤ R(d4) or R(d4) ≤ R(d2). The estimation procedure of Hwang and Peddada (1994) formed the basis of the method proposed by Conaway, Dunbar and Peddada (2004). In order to eliminate the dependence of the estimates on a single guess at the ordering between non-nodal parameters, Conaway et al.(2004) computed estimates of the parameters under all possible guesses and averaged them. In principle, the next entered patient is assigned to the dose combination with the estimated toxicity probability closest to the target toxicity rate. Once the toxicity status of that patient is observed, the toxicity probabilities are re-calculated and the trial continues. Conaway et al. (2004) apply a stopping rule to their design once enough information about the MTD combination has been attained.
The Hwang-Peddada estimation is carried out only on those dose combinations that have been previously assigned to patients in the trial. The method of Conaway et al. (2004) has the advantage of being a general method for handing partially ordered trials, but the escalation algorithm can be difficult to implement. It is complex. A more straightforward method would be useful, providing the new methodology performs well when compared with Conaway et al (2004). Their estimation method was nonparametric to some degree in that the estimates relied only on monotonicity assumptions. Our approach uses working parametric models, such as the power parameter model of O’Quigley and Shen (1996), with the aim of greater simplicity and possibly greater efficiency. Despite this difference, the approach to be proposed in this article builds from ideas in Conaway et al. (2004). The first is that each of the simple orders consistent with a partial order can be thought of as a model. The second is the importance of dimension reduction. This was a theme of many of the previous papers on dose-finding for combinations (Thall et al. 2003; Conaway, Dunbar and Peddada, 2004; Wang and Ivanova, 2005; Yuan and Yin, 2009). In this article, dimension reduction takes the form of finding, or giving more weight to, a small number of possible orderings. Ideally, the method would quickly identify a few working orderings within which the dose-finding algorithm would then reduce to a standard phase 1 trial. Given the properties of the continual reassessment method (CRM) (O’Quigley, Paoletti and Maccario, 2002), it is reasonable to believe that a method that quickly reduces to the CRM on the correct ordering would have excellent properties in terms of identifying dose combinations with acceptable toxicity.
3. Proposed method for partial ordering
3.1 Modeling toxicity
Suppose that the dose combinations follow a partial order for which there are M possible simple orders consistent with the partial ordering. Consider a trial investigating k treatment combinations, d1,…, dk. The dose for the j th entered patient, Xj, j = 1,…, n can be thought of as random, taking values xj ∈ {d1,…, dk}. Let Y be a binary random variable, where 1 denotes the observation of severe toxic response. For a particular ordering, m, m = 1,…, M, we model the toxicity probability R(xj) via
| (1) |
for some working dose-toxicity model ψm(xj, a) and a ∈ 𝒜. Further, we may wish to take account of any prior information concerning the plausibility of each model and so introduce p (m) = {p(1),…, p(M)}, where p (m) ≥ 0 and where Σm p (m) = 1. Even when there is no prior information available on the possible orderings we can formally proceed in the same way by specifying a discrete uniform for p (m). After inclusion of the first j patients into the trial, we have data in the form of Ωj = {x1, y1,…, xj, yj}. Under model m, we obtain an estimate, âm, for the parameter a. Given the value of âm under model m we can generate estimates R̂(di) = ψm(di, âm), (i = 1,…, k) of the true unknown toxicity probabilities at each of the k treatment combinations. For a target toxicity rate, θ, the i th, i = 1,…, k drug combination, di, assigned to the (j + 1) th patient enrolled in the trial is such that
| (2) |
is minimized. Continuing in this way, the MTD combination is the recommended treatment di for the hypothetical (n+1) th patient such that (2) is minimized after the inclusion of a predetermined sample size of n patients.
3.2 Inference
In the Bayesian framework, we assign a prior probability distribution g(a) for the parameter a of each model, and a prior probability p (m) to each model (simple order). In order to get the trial underway, we will choose the ordering with the largest prior probability, p (m), among the M models. If several, or all, of the models have the same prior probability, then we will choose at random from these orderings. Given the starting ordering, the starting dose, di, is then chosen by the investigators. In order to establish running estimates of the probability of toxicity at the available treatment combinations, we need an expression for the likelihood for the parameter a. After inclusion of the first j patients into the study, the likelihood under m is given by
| (3) |
which, for each ordering, can be used in order to generate a summary value, âm, for a. Given the set Ωj and the likelihood, the posterior density for a is given by
Further, this information can be used to establish the posterior probabilities of the models given the data as
The prior probabilities, p (m), for the ordering are updated by the toxic response data, Ωj. It is expected that the more the data support ordering m, the greater its posterior probability will be. When a new patient is to be enrolled, we choose a single ordering, h, with the largest posterior probability and take the working model, ψh(di, a), associated with this ordering and apply a Bayesian form of the CRM.
Generally, without further randomization, a difficulty can occur in which the ordering corresponding to the greatest posterior probability is chosen over orderings which may never have been tried. Information on these can only ever be obtained through experimentation. Early in the trial, within the posterior distribution of m, there may be only a negligible difference between the largest posterior probability and another value in the distribution. At this point added randomization allows for information to be obtained on competing orderings. Therefore, we do not rely entirely on the maximum of the posterior distribution for guidance as to the most appropriate ordering but rather implement an allocation algorithm in order to obtain information more broadly. Specifically, we generate a random number, h, between 1 and M, weighting each value according to its analogous posterior probability, π(m|Ωj) and implement the working model, ψh(di, a), associated with ordering h. In the latter part of the trial, the accrued information will create a greater gap between the values of posterior probabilities, increasing the likelihood that the ordering with the largest posterior probability will be selected. The randomization algorithm will not be relied upon for the final recommendation of the MTD. For the hypothetical (n+1) th patient, we choose the ordering h with the maximum posterior probability among the M candidate models.
Given the ordering h and the working model, ψh(di, a), we can generate toxicity probability estimates at each level. Beginning with a prior for a, after having included the j th subject, we can compute the posterior probability of a dose-limiting toxicity for di so that
| (4) |
The treatment combination xj+1 ∈ {d1,…, dk} assigned to the (j + 1) th included patient is the dose, di such that (2) is a minimum. The MTD combination is the recommended treatment di such that (2) is minimized after the inclusion of a predetermined sample size of n patients.
3.3 Application to matrix orders
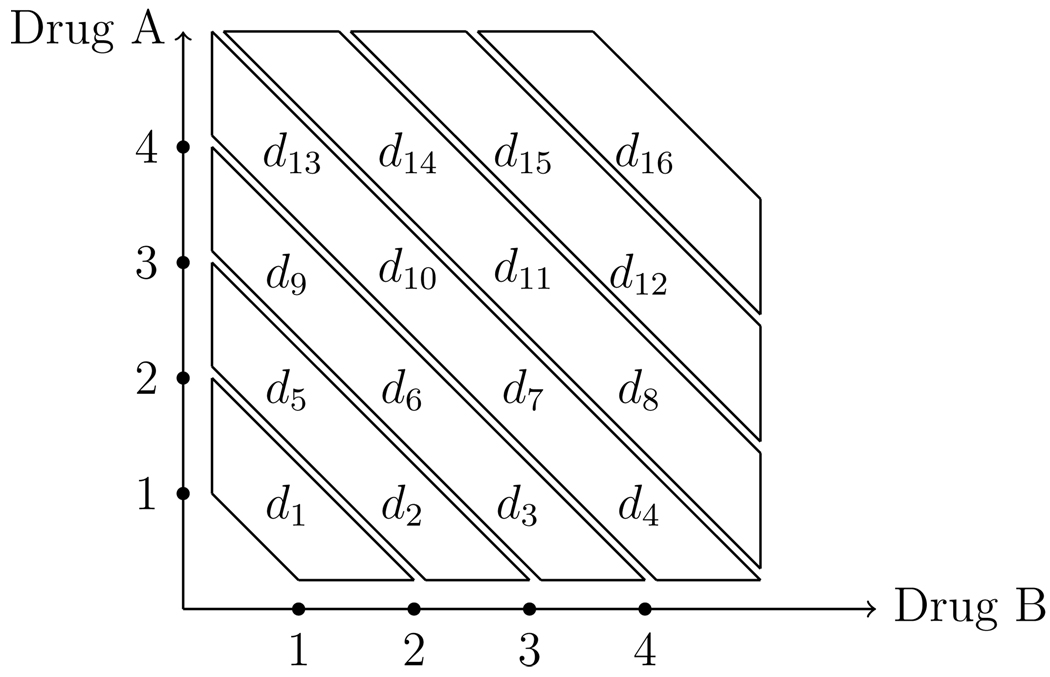
Considering all possible orderings is reasonable in studies like Berenson et al. (2009), but generally, there can be a large number of simple orders. Most of the existing methods address the need to reduce the dimension of the combinations to be studied (Thall et al. 2003; Wang and Ivanova, 2005; Yin and Yuan, 2009). Conaway, Dunbar, and Peddada (2004) propose either choosing, a priori, a small subset of orderings, or randomly selecting a small number of orders. Some approaches selectively eliminate possible combinations as the trial proceeds. In this article, dimension reduction takes the form of specifying a small subset of possible orderings. Consider a two-drug combination trial where drug A has p dose levels and drug B has w dose levels and we assign labels to the combinations di, i = 1,…, k such that k is the total number of treatment combinations and is equal to p × w. For instance, suppose drug A has four dose levels and drug B has four dose levels, with the matrix of treatment combinations given in Figure 1.
Figure 1.
An illustration of zoning a drug combination matrix
It is obvious when looking at the matrix of treatment combinations in Figure 1 that it is not feasible to consider each possible ordering of the dose-toxicity relationship. In order to model toxicity in this setting and have the ability to apply the methods of Sections 3.1 and 3.2, we must select a small subset of simple orders to consider. The only ordering information we have at our disposal, obtained from each agent being investigated individually, is that the toxicity of each agent increases monotonically with dose when the other agent is being held fixed. That is, toxicity increases monotonically across rows and up columns of the matrix of doses in Figure 1. We can use this information to our advantage in selecting the possible simple orders to consider in the trial. Specifically, we are going to use Huang et al. (2007) as a guide in choosing orderings. They label the treatments in a way similar to Figure 1 and place each combination into a “toxicity zone.”
The first zone contains a single combination d1, the second zone contains combinations d2 and d5, and so on. We consider toxicity to increase monotonically with respect to treatment as we move from zone 1 to zone 7. Within each zone, the ordering of the treatments is unknown. Therefore, we are going to formulate the possible simple orders of the toxicity probabilities according to the diagonals of the drug combination matrix. For instance, the three orderings chosen for investigation could be:
m = 1: d1 → d2 → d5 → d3 → d6 → d9 → d4 → d7 → d10 → d13 → d8 → d11 → d14 → d12 → d15 → d16
m = 2: d1 → d5 → d2 → d3 → d6 → d9 → d13 → d10 → d7 → d4 → d8 → d11 → d14 → d15 → d12 → d16.
m = 3: d1 → d5 → d2 → d9 → d6 → d3 → d13 → d10 → d7 → d4 → d14 → d11 → d8 → d15 → d12 → d16.
Subsequent to the possible simple orders being chosen, the methods of Sections 3.1 and 3.2 can be applied in order to model toxicity and estimate the MTD combination. Since, in reality, we do not know the true probability of toxicity at each of the available dose combinations, our objective is to provide some guidance in choosing a proper subset of orderings without knowing whether or not we have included the “correct” ordering in the set. An important idea to this approach is that we do not need to identify the “correct” order, we just need some order which is “close” to the correct one. In fact, at doses far away from the MTD combination, we can work with models which are completely wrong because, in practice, we will never experiment there.
In order to evaluate the performance of this approach, we present simulation studies that compare the methods of Sections 3.1 and 3.2 applied to these three simple orders against existing methodology for matrix orders. Conaway et al. (2004) considered the case in which more than three orderings, in particular six, were chosen but found that choosing a smaller subset of orderings has the advantage of reducing the dimension of the problem and did not significantly reduce the performance of their design. Consequently, we chose to investigate only three possible orderings according to the diagonals of the combination matrix and address the situation of the “correct” ordering not being one chosen for investigation. This topic is discussed in the simulation findings. The simulations compare the performance of the methods proposed in this article with a latent contingency table approach of Yin and Yuan (2009) and the copula regression approach of Yin and Yuan (2009) for a variety of toxicity scenarios in terms of (1) the proportion of trials in which the methods correctly identify the MTD combinations, (2) the proportion of DLT’s induced in arriving at the MTD estimate and (3) the proportion of patients treated at the MTD combinations.
4. Simulations
4.1 Illustration
To illustrate the proposed method, consider the example given in Section 2 involving k = 6 discrete treatment combinations, d1,…, d6. The partial order associated with this trial has associated with it five possible simple orders. Suppose the true toxicity probabilities are R(d1) = 0.04, R(d2) = 0.07, R(d3) = 0.20, R(d4) = 0.35, R(d5) = 0.55 and R(d6) = 0.70, which corresponds to ordering m = 1 being the true dose-toxicity order. The targeted toxicity probability is θ = 0.20, indicating that the “correct” treatment combination for the MTD is given by d3. There is a wide variety of choices for potential working models but the simple power parameter model given by
| (5) |
has shown itself to work well in practice. Here, 𝒜 = (0, ∞) and 0 < αm1 < ⋯ < αmk < 1 is the skeleton representing the prior estimates of the toxicity probabilities. Lee and Cheung (2009) have recently established a systematic approach for selecting the skeleton and, accordingly, the dose levels. For the procedure, we used the optimal value given Table 4 of Lee and Cheung (2009) for a target toxicity probability of θ = 0.20, n = 25 patients, and a prior MTD of l = 3, which is given as δ = 0.08. Implementing the algorithm yielded the skeleton {0.01, 0.07, 0.20, 0.38, 0.56, 0.71} for ordering 1, which were adjusted to correspond to each of the five simple orders consistent with the partial.
Table 4.
Toxicity scenarios 1–10 for the two-drug combinations. The target maximum tolerated dose combinations are in boldface
| Dose Level |
Drug A | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | ||
| Drug B | Scenario 1 | Scenario 2 | |||||||||
| 4 | 0.50 | 0.55 | 0.60 | 0.70 | 0.30 | 0.50 | 0.55 | 0.60 | |||
| 3 | 0.15 | 0.30 | 0.50 | 0.60 | 0.12 | 0.30 | 0.50 | 0.55 | |||
| 2 | 0.10 | 0.12 | 0.30 | 0.45 | 0.10 | 0.15 | 0.30 | 0.45 | |||
| 1 | 0.06 | 0.08 | 0.10 | 0.15 | 0.08 | 0.12 | 0.16 | 0.18 | |||
| Scenario 3 | Scenario 4 | ||||||||||
| 3 | 0.15 | 0.30 | 0.50 | 0.55 | 0.60 | 0.50 | 0.60 | 0.70 | 0.80 | 0.90 | |
| 2 | 0.12 | 0.16 | 0.30 | 0.50 | 0.55 | 0.10 | 0.30 | 0.50 | 0.70 | 0.80 | |
| 1 | 0.06 | 0.08 | 0.10 | 0.30 | 0.50 | 0.06 | 0.10 | 0.15 | 0.30 | 0.50 | |
| Scenario 5 | Scenario 6 | ||||||||||
| 4 | 0.54 | 0.67 | 0.75 | 0.81 | 0.86 | 0.49 | 0.58 | 0.68 | 0.75 | 0.81 | |
| 3 | 0.48 | 0.59 | 0.68 | 0.75 | 0.81 | 0.40 | 0.49 | 0.59 | 0.68 | 0.75 | |
| 2 | 0.40 | 0.45 | 0.59 | 0.67 | 0.74 | 0.27 | 0.40 | 0.45 | 0.59 | 0.67 | |
| 1 | 0.24 | 0.40 | 0.47 | 0.56 | 0.64 | 0.18 | 0.29 | 0.40 | 0.47 | 0.56 | |
For each ordering, we implemented an exponential prior with mean 1 on the parameter a such that g(a) = exp(−a). Before entering the first patient, we choose an ordering by randomly selecting a value from 1 to 5 with each value weighted according to the prior probabilities, p (m), on the ordering. We will assume that, a priori, we believe that each of the five simple orders is equally likely and take p (m) = 1/5. Getting the trial underway, ordering 2 was randomly selected, which corresponds to d1 → d2 → d4 → d3 → d5 → d6. Within this ordering, the first entered patient is treated at a level with α2i as close as possible to the target toxicity rate, θ = 0.20. For m = 2, this dose is d4, on which the first patient is enrolled and experiences toxic side effects. Subsequently, the posterior model probabilities are calculated to be π(1|Ω1) = 0.28, π(2|Ω1) = 0.21, π(3|Ω1) = 0.21, π(4|Ω1) = 0.15 and π(5|Ω1) = 0.15. These values serve as the respective weights for the random selection of a value between 1 and 5, from which ordering 4: d1 → d4 → d2 → d3 → d5 → d6 is chosen. Given m = 4, we take the working model ψ4(di, a) and apply the Bayesian form of the CRM. The estimate for a4 is calculated to be â4 = 0.273, which then allows for the estimation of the dose-limiting toxicity probabilities at each level, yielding R̂(d1) = 0.28, R̂(d4) = 0.48, R̂(d2) = 0.64, R̂(d3) = 0.77, R̂(d5) = 0.85 and R̂(d6) = 0.91. The 2 nd entered patient is then administered combination d1 for which R̂(d1) = 0.28 since, from the available estimates, this is the closest to the target θ = 0.20. This patient does not suffer toxic side effects and the new posterior model probabilities become π(1|Ω2) = 0.31, π(2|Ω2) = 0.21, π(3|Ω2) = 0.21, π(4|Ω2) = 0.13 and π(5|Ω2) = 0.13 from which ordering 2 is chosen. The new estimate, â2, for a2 becomes 0.66. Therefore, d2 becomes the treatment combination with an estimated toxicity probability of R̂(d2) = 0.17, which is closest to the target. The dose that is recommended as the treatment that would have been administered to the 25 th patient, had one been included, is d3. Therefore it is identified (estimated) to be the MTD combination with an estimated probability of toxicity of R̂(d3) = 0.13.
Table 2 shows the results of the simulation for which d3 is chosen as the MTD combination on the basis of all available knowledge.
Table 2.
Sequential trial of 25 patients
| j | h | âh | xj | yj |
|---|---|---|---|---|
| 1 | 2 | 1.00 | d4 | 1 |
| 2 | 4 | 0.27 | d1 | 0 |
| 3 | 2 | 0.66 | d2 | 0 |
| 4 | 1 | 0.65 | d2 | 0 |
| 5 | 3 | 0.91 | d3 | 0 |
| 6 | 1 | 1.05 | d3 | 0 |
| 7 | 2 | 1.06 | d4 | 1 |
| 8 | 1 | 0.91 | d3 | 0 |
| 9 | 1 | 1.00 | d3 | 0 |
| 10 | 1 | 1.08 | d3 | 0 |
| 11 | 1 | 1.14 | d3 | 1 |
| 12 | 1 | 0.89 | d3 | 0 |
| 13 | 2 | 1.00 | d4 | 0 |
| 14 | 1 | 1.02 | d3 | 0 |
| 15 | 1 | 1.07 | d3 | 0 |
| 16 | 1 | 1.11 | d3 | 0 |
| 17 | 1 | 1.15 | d3 | 0 |
| 18 | 1 | 1.19 | d3 | 0 |
| 19 | 1 | 1.23 | d3 | 0 |
| 20 | 1 | 1.27 | d3 | 0 |
| 21 | 1 | 1.30 | d3 | 0 |
| 22 | 1 | 1.33 | d4 | 0 |
| 23 | 1 | 1.38 | d4 | 1 |
| 24 | 1 | 1.25 | d3 | 0 |
| 25 | 1 | 1.28 | d3 | 0 |
4.2 Design comparison
In this section, the set of simulations compare the performance of the CRM for partial orders, which henceforth will be referred to as POCRM, with that of Conaway et al. (2004) for a Phase 1 trial of paclitaxel and carboplatin discussed in Section 2 of their article. The known orderings between doses lead to the investigation of six simple orders consistent with the partial. As previously mentioned, their method incorporates a stopping rule that stipulates that the trial be stopped if nine consecutive patients are enrolled on a single treatment or if the total number of patients on one treatment exceeds eighteen. Because our methods do not implement a stopping rule, but rather exhaust a predetermined sample size of patients, we round the number of patients given for each toxicity scenario in Conaway et al. (2004) to the nearest integer in order to provide a justifiable comparison. The results also compare the proposed method to the original CRM proposed by O’Quigley et al. (1990). The CRM assumes a monotonic dose-toxicity curve, so we judge our method against their approach in order to provide a benchmark of how well the design is performing. It is unreasonable to expect an approach for a partially known toxicity order to perform as well one that assumes a fully known order. Yet, our method reduces to the CRM when the toxicity order is fully known with respect to dose, hence a direct comparison is useful in assessing how much information is lost as a result of not having a fully known dose-toxicity order.
In each set of 1000 simulated trials, the target toxicity rate is θ = 1/3 and we incorporate a uniform prior distribution, p (m). As was noted in the previous section, the probability of dose-limiting toxic response is modeled via the power parameter model with the skeletons generated via the method of Lee and Cheung (2009). In the first toxicity scenario, the approach of Conaway et al. (2004) and POCRM are comparable in terms of correct MTD recommendation. In the second scenario, the POCRM significantly improves the performance of Conaway et al. (2004), correctly recommending the MTD in 13% more trials. In the third scenario, the two methods again perform similarly, while in the fourth scenario, Conaway et al. (2004) does slightly better than POCRM in terms of percent recommendation by a margin of 3%. In the final scenario, the POCRM recommends the “correct” MTD in 8% more of the simulated trials. In terms of the number of toxicities observed in each set of simulations, Conaway et al. (2004) induces less toxicity than the POCRM design in four of the five scenarios.
Table 3 indicates that the simulation results of POCRM are competitive with that of Conaway et al. (2004), making it a practical alternative as a multiple-agent trial design. However, this only gives a snapshot comparison to their method because of the few toxicity scenarios considered. There are advantages to the POCRM in that its methodology and the computation are much less complicated and can be carried out via the use of widely available software. Because it is an extension of the well known CRM, it is believed that it will be more easily understood by clinicians and review boards. At the very least, the method gives the investigator an alternative to his or her design preference when presented with a multiple-agent Phase 1 trial.
Table 3.
Simulation results for partial order continual reassessment method for six possible orderings. The table lists, for each method and set of true toxicity probabilities, the proportion of times each dose is identified as the MTD based on 1000 simulations of the trial
| Dose | d1 | d2 | d3 | d4 | d5 | d6 | n | tox |
|---|---|---|---|---|---|---|---|---|
| R(di) | 0.26 | 0.33 | 0.51 | 0.62 | 0.78 | 0.86 | - | - |
| Conaway et al | 0.35 | 0.52 | 0.11 | 0.02 | 0.00 | 0.00 | 21.3 | 8.5 |
| POCRM | 0.29 | 0.50 | 0.16 | 0.04 | 0.01 | 0.00 | 22.0 | 8.4 |
| CRM | 0.27 | 0.49 | 0.23 | 0.01 | 0.00 | 0.00 | 22.0 | 7.9 |
| R(di) | 0.12 | 0.21 | 0.34 | 0.50 | 0.66 | 0.79 | - | - |
| Conaway et al. | 0.07 | 0.29 | 0.42 | 0.21 | 0.01 | 0.00 | 25.6 | 9.0 |
| POCRM | 0.02 | 0.23 | 0.55 | 0.11 | 0.10 | 0.00 | 26.0 | 10.0 |
| CRM | 0.01 | 0.18 | 0.63 | 0.17 | 0.01 | 0.00 | 25.0 | 7.5 |
| R(di) | 0.04 | 0.07 | 0.20 | 0.33 | 0.55 | 0.70 | - | |
| Conaway et al. | 0.00 | 0.02 | 0.38 | 0.51 | 0.08 | 0.02 | 28.5 | 8.8 |
| POCRM | 0.00 | 0.00 | 0.26 | 0.50 | 0.23 | 0.01 | 29.0 | 10.8 |
| CRM | 0.00 | 0.01 | 0.19 | 0.67 | 0.13 | 0.00 | 28.0 | 8.0 |
| R(di) | 0.01 | 0.04 | 0.05 | 0.17 | 0.33 | 0.67 | - | |
| Conaway et al. | 0.00 | 0.00 | 0.06 | 0.25 | 0.64 | 0.05 | 29.0 | 7.8 |
| POCRM | 0.00 | 0.00 | 0.01 | 0.29 | 0.61 | 0.09 | 29.0 | 9.4 |
| CRM | 0.00 | 0.00 | 0.00 | 0.18 | 0.76 | 0.06 | 28.0 | 6.3 |
| R(di) | 0.01 | 0.02 | 0.05 | 0.15 | 0.20 | 0.33 | - | |
| Conaway et al | 0.00 | 0.00 | 0.01 | 0.04 | 0.37 | 0.59 | 26.2 | 5.8 |
| POCRM | 0.00 | 0.00 | 0.00 | 0.20 | 0.12 | 0.68 | 27.0 | 6.4 |
| CRM | 0.00 | 0.00 | 0.00 | 0.05 | 0.26 | 0.69 | 27.0 | 4.3 |
4.3 Matrix orders
In order to evaluate the operating characteristics and performance of the proposed methods when applied to matrix orders, we ran simulations under six different toxicity scenarios that compare our approach to two existing designs for matrix orders. There are sixteen treatment combinations under investigation in scenarios 1 and 2, fifteen treatment combinations in scenarios 3 and 4, and twenty treatment combinations in scenarios 5 and 6. Table 4 lists the true probability of toxicity for each of these scenarios, with the target MTD combinations indicated in bold type. More specifically, scenarios 1–4 are situations investigated by Yin and Yuan (2009) in their latent contingency table design, whereas scenarios 5 and 6 are two situations examined by Yin and Yuan (2009) using their copula regression design. The scenarios were designed to accommodate the commonly encountered situations in which different numbers and locations of target dose combinations may exist in the treatment combination space. Overall, these simulations provide an assessment of performance across a range of toxicity scenarios and a comparison to a variety of existing approaches. We have also investigated the performance of our approach for higher numbers of combinations, in particular 25 and 36. These simulation results are provided in Web Appendix B.
We began by choosing the three simple dose-toxicity orders given in Section 3.3 according to the diagonals of the matrix of treatments. For each scenario, we consistently chose the orderings in this manner, regardless of the true ordering according to toxicity probabilities presented in Table 4 due to the fact that we wanted to assess the performance of our approach even if we fail to include the “true” ordering in our chosen subset.
For example, in scenario 1, if we label the treatments as in Figure 1, then the true dose-toxicity ordering is d1 → d5 → d2 → d9 → d3 → d6 → d4 → d7 → d10 → d13 → d14 → d11 → d8 → d12 → d15 → d16, which is not one of the three orders chosen by the diagonals given in Section 3.3. In reality, we will never know the true ordering, so choosing the simple orders this way will provide confidence in using the method even if the true ordering is not chosen as one for investigation. Under all scenarios, 2000 trials were simulated. Both of the designs chosen for comparison implemented a stopping rule. Because our method does not incorporate a stopping rule, we use the average sample size of patients (rounded to the nearest integer) that is given in their simulation results. The target toxicity rate is given in bold type in Table 4 and we utilize a uniform prior distribution, p (m). We again generated the skeletons according to the approach of Lee and Cheung (2009).
Table 5 reports the percentage of correct MTD recommendation, the percentage of observed toxicities and the percentage of patients treated at the MTDs for the POCRM and for both designs of Yin and Yuan. Further details concerning the distribution of dose recommendation and allocation are provided in Web Appendix A. In terms of correctly selecting the MTD, the results are comparable. The designs of Yin and Yuan yield a higher percentage in three of the six scenarios, with the gains in performance being slight in scenarios 2, 3, and 6. However, in scenarios 1, 3 and 4, the POCRM improves upon the performance of Yin and Yuan considerably in each case. Table 5 illustrates that the designs of Yin and Yuan can be considered a slightly more conservative approach when examining the percentage of observed toxicities. In each scenario, their designs induce toxic side effects in a smaller percentage of patients when compared with the POCRM. However, POCRM is inducing toxicity in a percentage of patients close to that of the target rate. Finally, in terms of the percentage of patients treated at the MTD combinations, POCRM demonstrates superior performace in each scenario, by consistently treating a higher percentage of patients at the MTD combinations.
Table 5.
Comparison POCRM and existing methodology for matrix orders for 6 toxicity scenarios
| Scenario | ||||||
|---|---|---|---|---|---|---|
| Method | 1 | 2 | 3 | 4 | 5 | 6 |
| % correct recommendation | ||||||
| Yin and Yuan | 41.3 | 53.0 | 62.9 | 55.7 | 44.0 | 48.0 |
| POCRM | 53.0 | 50.0 | 86.0 | 68.0 | 38.0 | 45.0 |
| % of observed toxicities | ||||||
| Yin and Yuan | 27.9 | 27.7 | 28.1 | 29.1 | 36.9 | 35.2 |
| POCRM | 31.1 | 31.1 | 31.3 | 33.4 | 46.0 | 44.0 |
| % patients treated at MTD | ||||||
| Yin and Yuan | 12.8 | 19.2 | 20.5 | 19.0 | 23.4 | 22.0 |
| POCRM | 39.0 | 37.0 | 69.0 | 43.0 | 27.0 | 33.0 |
Overall, the simulation results indicate that, in terms of identifying the MTD, the performance of the new method is comparable to that of existing two-dimensional dose-finding methods (Conaway et al. 2004, Yin and Yuan, 2009). The simulations results support the idea that the “correct” ordering does not need to be one of those chosen for investigation in order for the method to perform well. In parallel with this idea is the notion that we would need to choose more orderings in the hope of increasing our chances of including the true order. Instead, we can choose a small number of orderings, so as to reduce the dimension of the problem, and regardless of whether or not the “correct” order is included, have confidence in the performance of the POCRM in terms of the three criteria used to measure its performance in this section.
5. Discussion
In this article, we present a new design, which leans upon the continual reassessment method, for effectively estimating the MTD in Phase 1 clinical trials when the order of the treatments is only partially known. The goal remains accurate estimation of the MTD, subject to the ethical constraints of treating as many patients as possible at and around the MTD. It is only of indirect interest to establish the correct order and, in as much as the estimate of the MTD itself may remain unaffected by certain orders, then we do not attempt to identify which order is the more correct. An alternative design may be, instead of selecting the ordering with the greatest posterior density and reducing the problem to a standard Phase 1 trial, to estimate the dose to recommend to the next entered patient for each distinct ordering and to use the dose that is most agreed upon across all possible orders. We could work with the cumulative probabilities for some MTD given the set of models. We have investigated this alternative approach and present some simulation results in Web Appendix C.
The simulation results indicate that, in terms of identifying the MTD, the performance of the new method is comparable to that of existing two-dimensional dose-finding methods (Conaway et al. 2004, Yin and Yuan 2009). Because the continual reassessment method for partial orders is a design that relaxes the monotonicity assumption of the CRM, it can be regarded as a multivariate generalization of CRM. Therefore, our approach is compatible with single-agent trials as well, in that it reduces to the CRM when the order of the toxicity probabilities is fully known. Hence, it can serve as a link between single and multiple-agent dose-finding trials. Furthermore, in theoretical investigations or those involving simulations, we can use a CRM with known ordering to provide an upper bound as a gauge on how much information is lost as a result of the partial ordering. Operating characteristics appear to be good although more study, under a broader range of possible situations, may provide more insight into general behavior. It appears possible to find examples which indicate the design to fail Cheung’s (2005) principle of coherence. This is a potential weakness but seems unavoidable in a situation in which there is always some uncertainty as to whether we are, in fact, escalating or de-escalating.
It is natural to ask whether, in large samples, our estimate of the MTD converges to the true MTD. Intuitively this should be so, under suitable conditions. Unfortunately, although we are convinced by intuition and heuristic arguments that our method delivers such large sample consistency, we do not have a formal proof. If a formal proof could be provided, this clearly would bolster the case for our method.
On the other hand, in the context of Phase I combination trials, formal large sample consistency is not a decisive factor, for several reasons: (1) there may not be a unique MTD and a number of drug combinations could conceivably provide the same or similar probabilities of toxicity, (2) in practice sample sizes will always be small, typically no greater than 20 to 30 patients, so that operating characteristics obtained at sample sizes which will be encountered in reality are more important than those obtained at large samples, and these can be obtained via simulations over a wide range of situations, and (3) correct identification of the MTD is only one of several desirable properties - for instance, concentrating experimentation at and around the MTD may be equally or even more important.
We have tested our method in extensive simulation studies, of which only a small part are presented here. The simulation results indicate that our method performs very well. Moreover, MTD prediction based on the true full ordering, regarding the full ordering as perfectly known, yields only a small gain relative to MTD prediction based on our method. Overall, despite the lack of formal large sample theory, the strong showing of our method in extensive simulation studies make us feel confident in recommending it for practical use.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to acknowledge the extensive comments made by two reviewers as well as a detailed critique from the associate editor. These comments have greatly helped us re-focus and sharpen the original submission. The project was supported by Grant Number 1R01CA142859 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Footnotes
Supplementary Materials
Web Appendices A, B and C, referenced in Sections 4.3 and 5, are available under the Paper Information link at the Biometrics website http://www.biometrics.tibs.org.
REFERENCES
- Babb J, Rogatko A, Zacks S. Cancer Phase I clinical trials: Efficient dose escalation with overdose control. Statistics in Medicine. 1998;17:1103–1120. doi: 10.1002/(sici)1097-0258(19980530)17:10<1103::aid-sim793>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Berenson JR, Yellin O, Patel R, Duvivier H, Nassir Y, Mapes R, Abaya CD, Swift RA. A phase I study of Samarium Lexidronam/Bortezomib combination therapy for the treatment of relapsed or refractory multiple myeloma. Clinical Cancer Research. 2009;15(3):1069–1075. doi: 10.1158/1078-0432.CCR-08-1261. [DOI] [PubMed] [Google Scholar]
- Buckland ST, Burnham KP, Augustin NH. Model Selection: An Integral Part of Inference. Biometrics. 1997;53:603–618. [Google Scholar]
- Cheung YK, Chappell R. A Simple Technique to Evaluate Model Sensitivity in the Continual Reassessment Method. Biometrics. 2002;58:671–674. doi: 10.1111/j.0006-341x.2002.00671.x. [DOI] [PubMed] [Google Scholar]
- Cheung YK. Coherence Principles in Dose-finding Studies. Biometrika. 2005;92:863–873. [Google Scholar]
- Chevret S. Statistical Methods in Dose-Finding Experiments. Chichester, U.K: John Wiley and Sons Ltd.; 2006. [Google Scholar]
- Conaway M, Dunbar S, Peddada S. Designs for single- or multiple-agent phase I trials. Biometrics. 2004;60:661–669. doi: 10.1111/j.0006-341X.2004.00215.x. [DOI] [PubMed] [Google Scholar]
- Dunbar S, Conaway M, Peddada S. On improved estimation of parameters subject to order restrictions. Statistics and Applications. 2001;3:121–128. [Google Scholar]
- Huang X, Biswas S, Oki Y, Issa JP, Berry DA. A parallel phase I/II clinical trial design for combination therapies. Biometrics. 2007;63:429–436. doi: 10.1111/j.1541-0420.2006.00685.x. [DOI] [PubMed] [Google Scholar]
- Hwang J, Peddada S. Confidence interval estimation subject to order restrictions. Annals of Statistics. 1994;22:67–93. [Google Scholar]
- Korn EL, Simon R. Using tolerable-dose diagrams in the design of phase I combination chemotherapy trials’. Journal of Clinical Oncology. 1993;11(4):794–801. doi: 10.1200/JCO.1993.11.4.794. [DOI] [PubMed] [Google Scholar]
- Kramar A, Lebecq A, Candalh E. Continual reassessment methods in phase I trials of the combination of two agents in oncology. Statistics in Medicine. 1999;18:1849–1864. doi: 10.1002/(sici)1097-0258(19990730)18:14<1849::aid-sim222>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Lee SM, Cheung YK. Model Calibration in the Continual Reassessment Method. Journal of the Society for Clinical Trial. 2009;6:227–238. doi: 10.1177/1740774509105076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller S. An extension of the continual reassessment method using a preliminary up and down design in a dose finding study in cancer patients in order to investigate a greater number of dose levels. Statistics in Medicine. 1995;14:911–922. doi: 10.1002/sim.4780140909. [DOI] [PubMed] [Google Scholar]
- O’Quigley J, Pepe M, Fisher L. Continual reassessment method: a practical design for Phase I clinical trials in cancer. Biometrics. 1990;46:33–48. [PubMed] [Google Scholar]
- O’Quigley J, Shen LZ. Continual Reassessment Method: A likelihood approach. Biometrics. 1996;52:163–174. [PubMed] [Google Scholar]
- Patnaik A, Warner E, Michael M, Egorin M, Moore M, Siu L, Fracasso P, Rivkin S, Kerr I, Litchman M, Oza A. Phase I dose-finding and pharmacokinetic study of paclitaxel and carboplatin with oral valspodar in patients with advanced solid tumors. Journal of the National Cancer Institute. 2000;18:3677–3689. doi: 10.1200/JCO.2000.18.21.3677. [DOI] [PubMed] [Google Scholar]
- Robertson T, Wright F, Dykstra R. Order Restricted Statistical Inference. New York: Wiley; 1988. [Google Scholar]
- Shen LZ, O’Quigley J. Consistency of continual reassessment method in dose-finding studies. Biometrika. 1996;83:395–406. [Google Scholar]
- Simon R, Korn E. Selecting agent combinations based on total equivalent dose (dose intensity) Journal of the National Cancer Institute. 1990;82:1469–1476. doi: 10.1093/jnci/82.18.1469. [DOI] [PubMed] [Google Scholar]
- Storer BE. Design and analysis of Phase I clinical trials. Biometrics. 1989;45:925–937. [PubMed] [Google Scholar]
- Thall PF, Milliken R, Mueller P, Lee S-J. Dose-finding with two agents in phase I oncology trials. Biometrics. 2003;59:487–496. doi: 10.1111/1541-0420.00058. [DOI] [PubMed] [Google Scholar]
- Wang K, Ivanova A. Two-Dimensional Dose Finding in Discrete Dose Space. Biometrics. 2005;61:217–222. doi: 10.1111/j.0006-341X.2005.030540.x. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Yin G. Sequential continual reassessment method for two-dimensional dose-finding. Statistics in Medicine. 2008;27(27):5664–5678. doi: 10.1002/sim.3372. [DOI] [PubMed] [Google Scholar]
- Yin G, Yuan Y. Bayesian dose finding for drug combinations by copula regression. Journal of the Royal Statistical Society. 2009;58(2):211–224. [Google Scholar]
- Yin G, Yuan Y. A latent contingency table approach to dose-finding for combinations of two agents. Biometrics. 2009;65:866–875. doi: 10.1111/j.1541-0420.2008.01119.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.