Abstract
A gene encoding a putative DNA helicase from Staphylococcus aureus USA300 was cloned and expressed in Escherichia coli. The protein was purified to over 90% purity by chromatography. The purified enzyme, SauUSI, predominantly cleaves modified DNA containing 5mC and 5-hydroxymethylcytosine. Cleavage of 5mC-modified plasmids indicated that the sites S5mCNGS (S = C or G) are preferentially digested. The endonuclease activity requires the presence of adenosine triphosphate (ATP) or dATP whereas the non-hydrolyzable γ-S-ATP does not support activity. SauUSI activity was inhibited by ethylenediaminetetraacetic acid. It is most active in Mg++ buffers. No companion methylase gene was found near the SauUSI restriction gene. The absence of a cognate methylase and cleavage of modified DNA indicate that SauUSI belongs to type IV restriction endonucleases, a group that includes EcoK McrBC and Mrr. SauUSI belongs to a family of highly similar homologs found in other sequenced S. aureus, S. epidermidis and S. carnosus genomes. More distant SauUSI orthologs can be found in over 150 sequenced bacterial/archaea genomes. Finally, we demonstrated the biological function of the type IV REase in restricting 5mC-modified plasmid DNA by transformation into clinical S. aureus strain SA564, and in restricting phage λ infection when the endonuclease is expressed in E. coli.
INTRODUCTION
Restriction enzymes (REases) are grouped into four major types based on subunit architecture, adenosine triphosphate/guanosine triphosphate (ATP/GTP) requirement, sequence specificity and DNA cleavage mechanism (1). Type I restriction–modification (R–M) systems were originally discovered by studying phage plating efficiency among different Escherichia coli hosts (2). Type I restriction enzymes are multi-subunit complexes consisting of M2R2S subunits, and their activity requires ATP hydrolysis (3). Type II REases with 4–8-bp recognition sequences and over 300 unique specificities are widely used in creating recombinant DNA molecules (4). Type III R–M systems are also multi-subunit complexes with R2M2 configuration and require ATP hydrolysis for endonuclease activity (5–7). Two recognition sites in head-to-head or tail-to-tail orientations are essential for type I and type III enzymes to cleave. Modification-dependent REases are presently grouped into type IIM if the target sequence recognition and cleavage are very specific and precise, for example, DpnI, GN6mATC (8) and GlaI, G5mCG5mC (9), MspJI, 5mCNNR (10); or type IV if cleavage is non-specific and variable. Examples of type IV are McrBC, which cleaves 5mC-modified DNA, and GmrSD that attacks glucosylated-hmC (glc-5hmC) T4 DNA (11–15).
Methicillin-resistant Staphylococcus aureus (MRSA) presents a great health threat by hospital-acquired infections (HaMRSA) and more recently by community acquired (CaMRSA) infections characterized by the spread of highly invasive and persistent infections (16,17). Recently, the ability of MRSA strains to acquire vancomycin-resistance genes from Enterococcus species poses a significant threat (18–21). A large number of studies have been carried out to elucidate the mechanism of antibiotic resistance. The general conclusion is that drug resistance genes can be chromosomally encoded or derived from extra chromosomal elements, and acquired by horizontal gene transfer of mobile genetic elements such as conjugative plasmids, transposable elements, integrons or by phage infection and integration (22). The presence of type I, and II R–M systems or type IV restriction systems in clinical S. aureus strains could provide a genetic barrier to the free genetic exchange by transformation, transduction and conjugation (23,24).
A new gene that limits gene transfer was identified in two clinical S. aureus strains (SA564 and UAMS-1) (24). This open reading frame (ORF) contains a super family II DNA helicase domain and was found to be responsible for biological restriction of plasmid transformation when DNA was prepared from E. coli sources and suggested to be a type III restriction endonuclease based on the presence of helicase superfamily II domain. Inactivation of this gene by mutation increased the transformation efficiency by 102–104-fold. However, no companion methylase gene was found adjacent to the newly discovered gene, which is different from the typical gene organization of type III R–M systems (24). There are three possibilities: (i) interference with gene transfer operates by a mechanism other than cleavage [abortive infection, for example, in references (25,26)]; (ii) interference may be due to site-specific cleavage of an unmodified site, with the M gene located somewhere else on the bacterial chromosome; or (iii) cleavage may occur but depend on specific DNA modifications such as 5mC, 5hmC, glc-hmC, N6mA, N4mC or phosphothioation of the DNA backbone (27). Here, we present experimental evidence that indeed SauUSI, a gene nearly identical to those found in S. aureus strains SA564 and UAMS-1, is a modification-dependent REase that requires ATP or dATP for endonuclease activity and that the transformation barrier is due to cleavage activity of 5mC-modified DNA. No type III methylase homolog was found near the SauUSI restriction gene or in the entire sequenced bacterial genome. In addition, we found that divalent cations are required for optimal endonuclease activity. We propose that SauUSI and its homologs are classified as type IV restriction systems. To explore the distribution of SauUSI and its close relatives, we also identified orthologs of this type IV restriction gene in other bacterial species, finding over 150 candidates. In addition, we demonstrate the biological relevance of SauUSI in restricting modified plasmid DNAs in S. aureus and E. coli during transformations and in phage restriction.
MATERIALS AND METHODS
Bacterial strains, genomic DNA and plasmids
ER2566 (C2566, NEB) is an E. coli B strain with the T7 RNA pol gene (T7 gene 1) integrated into the chromosome and deficient in Dcm methylase (Dcm), McrBC and Mrr. The IMPACT protein expression kit and pTYB1 vector were obtained from NEB. Unmodified (Dcm−) λ phage DNA, pUC19 and pBC4 were prepared from ER2566. The backbone vector of pBC4 is pUC19 which carries a BstBI/ClaI fragment of adenovirus-2 DNA. T4gt DNA (deficient in α-and β-glucosyltransferases, containing 5hmC) was a gift from R. Vaisvila (NEB). λ Dcm+Dam−, λ Dcm+Dam+ and pBR322 Dcm+ DNAs were from NEB. Genomic DNA of S. aureus subsp. aureus USA300, FPR3757 was purchased from ATCC. Plasmid DNA was prepared by Qiagen mini-prep spin columns. Transformations of S. aureus cells with plasmid DNA were performed as described in Corvaglia et al. (24).
Protein expression and purification
The SauUSIR gene (2859 bp, 953 aa) flanked by SalI and XhoI restriction sites was amplified by polymerase chain reaction (PCR) using high-fidelity Phusion DNA polymerase (Finnzymes). Following SalI and XhoI restriction digestion, the amplified DNA was ligated to pTYB1 with the same cohesive ends. The ligated DNA was transferred into ER2566 competent cells by transformation. The inserts in two clones with the highest enzyme activity were sequenced and found to contain the wild-type (wt) sequence. For small-scale protein purification, 4 L cells of ER2566 [pTYB1-SauUSIR] were cultured at 37°C in LB supplemented with Amp (100 µg/ml) until late log phase. isopropyl-β-d-thiogalactopyranoside (IPTG) induction was carried out at 0.5 mM final concentration and cell cultures induced at 16°C overnight. Cells were harvested by low-speed centrifugation, resuspended in a sonication buffer (20 mM Tris–HCl, pH 8.5, 0.5 M NaCl, 0.1% Triton X-100) and lysed by repeated sonication. After centrifugation, clarified cell lysate was loaded onto a chitin column (∼21 ml, ∼4 cm × 2.6 cm), which was then extensively washed with 10 column volumes of column buffer (20 mM Tris–HCl, pH 8.5, 0.5 M NaCl). The column was then flushed quickly with 20 ml of column buffer with 50 mM DTT and stored at 4°C overnight to allow the cleavage of SauUSI from the SauUSI–intein–chitin-binding domain (CBD) fusion. Approximately 15 ml of proteins (1 ml × 15) were eluted and analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). To remove the nucleic acids from the eluted proteins, the pooled fractions were loaded onto a Source Q column (GE Healthcare) at 0.25 M NaCl concentration. The flow-through containing the SauUSI protein was then loaded onto a HiTrapTM heparin column (GE Healthcare), and proteins were eluted with a linear gradient of 50 mM to 1 M NaCl. After analysis of the eluted fractions on SDS–PAGE, the fractions with SauUSI were concentrated in a protein concentrator (Amicon/Millipore, 10 000 kDa) by low-speed centrifugation. The protein was diluted in NEB Diluent A buffer and stored at −20°C.
To remove the divalent metal ions bound by the purified SauUSI, the enzyme was supplemented with 20 mM EDTA and then dialyzed against a buffer (50 mM NaCl, 20 mM Tris–HCl, pH 8, 1 mM DTT) for 48 h at 4°C using a dialysis cassette (10 000 Da MWCO; Thermo Scientific Pierce).
For batch purification of wt SauUSI and variants using chitin beads, 10 ml of cells were cultured to late log phase at 37°C. After addition of 0.5 mM IPTG (final concentration), SauUSI protein expression was induced at 16°C overnight. Cell pellets were resuspended in 1 ml of cell lysis buffer (20 mM Tris–HCl, pH 8.5, 0.5 M NaCl, 0.1% Triton X-100), and cells were lysed by sonication. Clarified cell lysates were mixed with 200 µl of chitin beads prewashed with column buffer (20 mM Tris–HCl, pH 8.5, 0.5 M NaCl) and gently mixed on a rocker at 4°C for 1 h. Chitin beads with bound proteins were spun down at low speed centrifugation and washed three times with 1.5 ml of column buffer. The chitin beads were then resuspended in cleavage buffer (20 mM Tris–HCl, pH 8.5, 0.5 M NaCl, 50 mM DTT) and incubated at 4°C overnight. The supernatants containing SauUSI variants were analyzed on SDS–PAGE and assayed for cleavage activity on T4gt DNA.
For constitutive expression of SauUSI endonuclease gene (sauUSIR) in E. coli, a PCR fragment with re-engineered ribosome binding site (GGATCC-GGAGGT-AAATGA-start codon) was cloned into the BamHI site of pBR322 under the TcR promoter. As a control, the sauUSIR gene inserted in the opposite orientation was also constructed.
Site-directed mutagenesis
The catalytic residues H119, K121, N139 and E150 of SauUSI endonuclease were mutated to alanine (Ala) by two-step overlapping PCR using PhusionTM high-fidelity DNA polymerase 2 x master mix (Finnzymes). Full-length amplified DNAs, following DpnI, SalI and XhoI digestion, were gel-purified and ligated to pTYB1 with compatible ends. Desired mutations were confirmed by DNA sequencing. For small-scale affinity purification, cell extracts from 10 ml IPTG-induced overnight culture (16°C) were used for batch purification of the mutant enzymes. For medium-scale purification, cell lysates of 1 L IPTG-induced overnight cell cultures (16°C) were used for chitin column purification. Partially purified SauUSI variant H119A was used for gel filtration chromatography to determine its oligomerization state in NEB buffer 4.
SauUSI digestion and in vitro DNA methylation
SauUSI digestions of various DNA substrates were carried out at 37°C in NEB buffer 4, except specified otherwise. To study the cofactor requirement of SauUSI endonuclease activity, EDTA-treated and extensively dialyzed SauUSI enzyme was incubated with DNA substrates in reaction buffers containing different divalent cations (1 and 5 mM of Mg++, Mn++, Ca++, Co++, Zn++ or Ni++). To understand the NTP or dNTP requirement for the SauUSI enzyme activity, reactions were supplemented with 1 mM ATP, γ-S-ATP (from Sigma–Aldrich), GTP, γ-S-GTP (from Sigma Aldrich), UTP (from Roch Applied Science), dATP, dCTP, dGTP or dTTP. For in vitro DNA modification study, unmodified pBC4 plasmid DNA (Dcm−) was modified by M.MspI (modified base as underlined 5mCCGG), M.HpaII (C5mCGG), M.HhaI (G5mCGC), M.AluI (AG5mCT), M.BamHI (GGAT4mCC) and M.TaqI (TCG6mA), respectively, in the recommended 1 x methylation buffers with S-adenosylmethionine (SAM) at 37°C for 2 h. Methylated plasmid DNAs were purified by spin columns and used as the substrates for SauUSI digestion.
5mC- or 5hmC-modified PCR DNAs with S5mCNGS sites (1, 2, 4, 5 and 6 sites) were produced in PCRs with 5mC (5 m-dCTP) or 5hmC (5hm-dCTP) instead of dCTP. The small amount of unmodified template DNA (<0.1 ng/µl) did not interfere with SauUSI digestion and cleavage site determination. SauUSI-digested PCR DNAs were purified by spin columns using a PCR purification kit (Qiagen) and run-off sequencing was carried out using a Big-Dye® terminator cycle sequencing kit (ABI/Life technologies). Similarly, after digestion of pBR-fnu4HIM (G5mCNGC) plasmid by SauUSI endonuclease, the digested DNA was purified by spin column and used as template for run-off sequencing to determine the cleavage sites.
Gel filtration study of SauUSI H119A variant
Ten microliters of purified SauUSI variant H119A (1 mg/ml) was injected into a Superdex 200 5/150 GL (3 ml bed volume; GE Healthcare). The column was run with NEB buffer 4 at 4°C at a flow rate of 0.2 ml/min. The partition coefficient (Kav) of SauUSI-H119Awas determined by Kav = (ve – vo)/(vt – vo) where ve is the elution volume, vo is the void volume of the column and vt is the total column volume. vo and vt were determined empirically by the addition of blue dextran and DTT, respectively to the sample. A standard curve was created by obtaining the Kav value of the standard proteins ovalbumin (43 kDa), conalbumin (75 kDa) and ferritin (440 kDa) (GE Healthcare) run under the same condition.
Phage plating and restriction of phage by SauUSI
Dcm+ λvir (a gift from A. Fomenkov, Mern Sibley, Lise Raleigh) was prepared from E. coli host ER1793. EcoK Dcm methylase modifies the DNA sequence CCWGG to produce modified sites C5mCWGG (W = A or T). Dcm− λvir was prepared from ER2566, a natural Dcm-deficient B strain. T4gt phage containing 5hmC was prepared from ER2566. Phages were diluted in phage broth or 10 mM MgCl2 solution and used to infect the E. coli hosts.
RESULTS AND DISCUSSION
SauUSI expression and purification
The sauUSIR gene was amplified from genomic DNA of S. aureus subsp. aureus USA300, FPR3757, and inserted into the T7 expression vector pTYB1. This results in a fusion of the ORF to the Sce VMA intein and Bacillus circulans CBD (IMPACT protein purification system, NEB). IPTG-induced ER2566 cell extracts showed endonuclease activity on λ DNA (Dcm+) in the presence of ATP and 10 mM DTT (data not shown). Two clones with the correct-sized insert and the highest endonuclease activity were sequenced and found to carry the wt gene. A small-scale purification procedure was developed. SauUSI was first purified from a chitin column using the DTT-intein cleavage mechanism recommended by the manufacturer. The cleaved and eluted fractions were passed through a Source Q column at moderate salt concentration to remove most of the nucleic acids. The flow-through was then loaded onto a HiTrapTM heparin column and protein eluted with a NaCl gradient. The purification results are shown in Supplementary Figure S1. Two heparin fractions with the highest purity (Supplementary Figure S1, lanes 5 and 6) were concentrated to 0.2 mg/ml and used for further characterization.
SauUSI substrate specificity
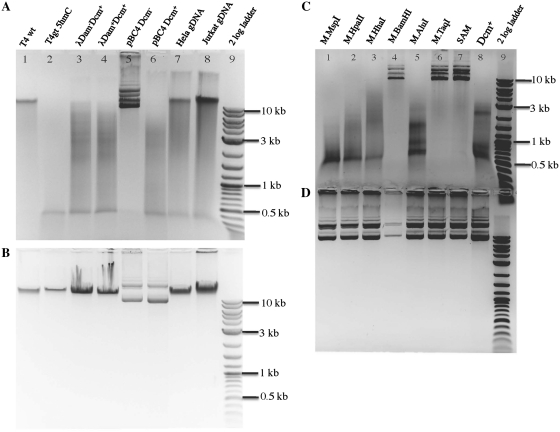
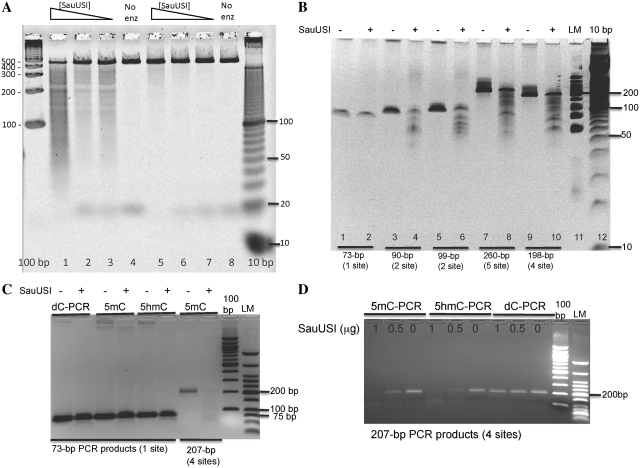
Since SauUSI contains a helicase domain and ATP-binding site (inferred from amino acid sequence similarity to other type I and III REases), we included ATP (1 mM) in all digestions (see below for further data on nucleotide requirement). Figure 1A shows that SauUSI cleaves 5hmC-containing T4gt DNA as well as 5mC-containing DNA from a variety of sources: Dcm-modified λ and pBC4 DNA (pBC4 is a pUC19 derivative carrying a BstBI/ClaI fragment insert from adenovirus DNA), HeLa and Jurkat genomic DNA (human). Escherichia coli Dcm modifies CC(A/T)GG sequences at the internal cytosine, while mammalian DNA carries methylation in some CpG sites. Dam modification does not inhibit cleavage since both Dam+ and Dam− λ DNA were equally cleavable by SauUSI (Figure 1A, lanes 3 and 4). SauUSI displays lower cleavage activity on 5-glc-hmC DNA (glucosylated T4 DNA, wild type) (Figure 1A, lane 1).Unmodified (Dcm−) pBC4 is a poor substrate for SauUSI although a small fraction of DNA was partially linearized (Figure 1A, lane 5), which suggests that SauUSI has a non-specific endonuclease activity at high enzyme concentration, or that there is a low level of contaminating non-specific nuclease. The 0.4–0.5-kb DNA of T4gt digested products was an artifact which dispersed in high percentage agarose gel or PAGE (data not shown). Figure 1B shows the undigested DNA substrates. SauUSI also displays very low endonuclease activity on Dam+ Dcm− pUC19 and pBR322 DNAs (data not shown). The specific activity of SauUSI is determined to be ∼8000 U/mg protein on T4gt phage DNA in the presence of ATP. One unit of SauUSI is defined as the amount of enzyme required to digest 1 µg T4gt DNA into fragments of <500 bp in NEB buffer 4 at 37°C for 1 h.
Figure 1.
SauUSI digestion of modified and unmodified DNA substrates. (A) DNA substrates digested by SauUSI endonuclease in 1X NEB buffer 4 at 37°C for 1 h. (B) Uncleaved DNA substrates. T4 wt DNA contains glucosylated 5-hmC DNA. T4gt deficient in α- and β-glucosyl transfereases contains 5hmC DNA. Lane 9, 2-log DNA size marker. (C) SauUSI digestion of in vitro modified DNA. Plasmid pBC4 was methylated in vitro by the indicated methyltransferase and subsequently digested by SauUSI. Lane 7, mock-modified pBC4 DNA (Dcm−) (only SAM was present, no methylase). Lane 8, in vivo modified pBC4 (Dcm+). (D) In vitro methylated DNA (uncleaved) of the same substrates as in (C).
To further investigate SauUSI’s substrate specificity, Dcm− plasmid pBC4 substrates were modified in vitro by purified methylases and then used as substrates for SauUSI digestion. Figure 1C shows that SauUSI cleaves pBC4 plasmid after in vitro modification by M.MspI (5mCCGG), M.HpaII (C5mCGG), M.HhaI (G5mCGC) or M.AluI (AG5mCT). Modified pBC4 DNAs by M.SssI (5mCG) or M.CviPI (G5mC) also allowed partial cleavage by SauUSI (data not shown). N4mC modified DNA by M.BamHI (GGATm4CC) or N6mA modified by M.TaqI (TCG6mA) are poor substrates for SauUSI (Figure 1C, lanes 4 and 7). Dcm+ pBC4 (C5mCWGG site modified in vivo) was again cleaved by SauUSI. In summary, SauUSI appears to cleave when 5mC is present in the following 3′ sequence contexts: 5mCC, 5mCG, 5mCT and 5mCW (see more sequence requirement below). SauUSI does not cleave N4mC and N6mA-modified DNA (the Dcm- substrates do carry adenine modification in the G6mATC context). This 5mC/hmC-dependent substrate specificity is shared by McrBC, although McrBC activity requires GTP hydrolysis. The digestion pattern, appearing as a ‘smeargram’, suggests non-specific nuclease activity (or strong ‘star’ activity) following incision from (or near) the modified 5mC/5hmC sites. The non-specific nuclease activity might be important to completely destroy the foreign invading DNA (the cleavage products are not easily repaired by simple religation).
We also tested SauUSI digestion of plasmids that have been modified in vivo by C5 methylases. Methylase gene-containing plasmids were co-transformed with pBC4 into a Dcm-deficient host ER2566 and plasmid mixture was prepared and digested with SauUSI in the presence of ATP. All plasmids were found to be mostly resistant to their cognate REases, and thus modifications appear to be complete. The methyltransferases with known modifications that are sensitive to cleavage are M.EagI (CCG5mCCG), M.Fnu4HI (G5mCNGC, confirmed substrate for BisI cleavage) (9), M.BbvI (G5mCAGC, complement strand G5mCTGC) and M.TspRI (5mCASTG). Less sensitive to SauUSI cleavage are M.AclI (AA5mCGTT), M.HpyCH4IV (A5mCGT) and M.NspHI (R5mCATGY). Other methylases are highly likely to create 5mC, but the specific site of modification is not conclusively known. Those that are sensitive to cleavage are M.BssKI (C5mCNGG or 5mCCNGG), M.TseI (G5mCWGC) and M.BsrFI (RC5mCGGY) (Figure 2A). Surprisingly, some methylases with known 5mC modification are nearly resistant. These are M.SacI (GAG5mCTC), M1.BsrDI and M2.BsrDI (G5mCAATG or 5mCATTGC) (Figure 2A). The resistance of bsrDIM1M2-containing plasmids to BsrDI digestion (Figure 2A, lane 20) indicates full modification of BsrDI sites on pBC4. Therefore, incomplete modification does not explain resistance to SauUSI cleavage. Figure 2B shows undigested plasmid DNAs.
Figure 2.
SauUSI digestion of 5mC-modified plasmid DNAs. (A) Plasmids prepared from co-transformants of pACYC-M (or pSYX20-M) and pBC4 were digested by SauUSI for 1 h at 37°C and the digested DNAs were analyzed on a 1% agarose gel. Lanes 1–3, unmodified DNA digested by SauUSI. Lane 4, Dcm-modifed pBC4 digested by SauUSI. Lanes 5, 6, 8–11, 13–17, 19, modified plasmid mixture (plasmid with M gene plus pBC4) digested by SauUSI. Lane 7, M.BbvI-modified plasmid pUC-bbvIM; lane 18, M.TspRI-modified plasmid pUC-tspRIM (M gene under native Thermus promoter). Lane 20, M.BsrDI modified plasmids digested by BsrDI (note: resistance to BsrDI digestion indicates BsrDI site modification). Lane 22, uncut pBC4 plasmid DNA. Lanes 12 and 23, 2-log DNA size ladder. Duplicated DNA samples: lane 5, pACYC-aclIM; lane 6, pSYX20-aclIM; lane 17, pACYC-tspRIM; lane 18, pUC-tspRIM (M gene under Thermus sp. native promoter). (B) Undigested plasmids carrying 5mC methylase genes. Lane 2, pUC-bbvIM; lane 7, pBR-fnu4HIM. Lanes 1, 3–6, 8–12, plasmid with the indicated M gene and pBC4 mixture. (C) SauUSI digestion of N4mC-modified plasmid DNAs. Plasmid pXbaI was purified from a Dcm+ strain (5mC-modified control DNA).
We also tested a number of N4mC-modified plasmids in SauUSI digestion. Plasmids carrying N4mC methylase genes pspGIM, cviSIIIM, TCGA-N4mC methylase (M.TaqI isoschizomer clone from environmental samples, Richard Morgan, unpublished data), bstYIM and bamHIM were poorly digested by SauUSI (Figure 2C), indicating N4mC-modified DNAs are not good substrates for SauUSI. In a control experiment, Dcm+ plasmid pXbaI was cleaved by SauUSI.
The substrate specificity of the in vitro and in vivo modified DNA is listed in Table 1. The relative activity of SauUSI is summarized as follows: S5mCNGS (M.Fnu4HI, M.BssKI, M.Msp, M.Dcm) > G5mCTC (M.SacI), A5mCGT (M.HpyCH4IV) >> 5mCAAT or 5mCATT (M.BsrDI). The best in vitro DNA substrates are T4gt, pBR-fnu4HIM (G5mCNGC) and pACYC-bssKIM (C5mCNGG), which may reflect the frequency of the modified sites. For the limited number of N4mC-modified plasmids that have been tested, SauUSI endonuclease does not appear to cleave modified N4mC DNA efficiently.
Table 1.
Digestion of 5mC-modified plasmid DNAs by SauUSI
| C5 methylase | In vivo/in vitro modification | Modified sites | Relative endonuclease activity |
|---|---|---|---|
| M.HpaII | In vitro | C C GG | ++ |
| M.MspI | In vitro | C CGG | ++ |
| M.EagI | In vivo | CGG C CG | ++ |
| M.BssKI | In vivo | C C NGG (?) | +++ |
| M.BsrFI | In vivo | RCC GGY (?) | ++ |
| M.BbvI | In vivo | G C AGC | ++ |
| M.Fnu4HI | In vivo | G C NGC | +++ |
| M.Dcm | In vivo | C C WGG | ++ |
| M.TseI | In vivo | G C WGC | ++ |
| Consensus sequence | S C NGS | ||
| M.HhaI | In vitro | G C GC | ++ |
| M.SssI | In vivo or in vitro | C G | + |
| M.AclI | In vivo | AAC GTT | + |
| M.HpyCH4IV | In vivo | A C GT | + |
| M.AluI | In vitro | AG C T | ++ |
| M.NspHI | In vivo | R C ATGY | + |
| M.TspRI | In vivo | C ASTG | + |
| M.SacI | In vivo | GAG C TC (?) | +/– |
| M1.BsrDI | In vivo | G C AATG | – |
| M2.BsrDI | In vivo | C ATTGC | – |
(?) indicates possible modified cytosine; “+/–” indicates poor activity; “––” denotes no apparent activity in 1 h digestion, underlined C indicates modified base. R = A/G; S = G/C; W = A/T. The best-modified substrates are G5mCNGC (M.Fnu4HI) and C5mCNGG (M.BssKI).
The density of the 5mC-modified sites also appears to influence SauUSI endonuclease activity. Dcm+ pBR322 or pBC4 additionally modified in vitro by M.MspI (5mCCGG) serves as a better substrate for SauUSI digestion than Dcm+ modified alone (data not shown). The recognition sequence of SauUSI is therefore somewhat similar to BisI (G5mC/NGC) and BlsI (G5mCN/GC) (9). The enzymes are distinct, however: SauUSI cleaves DNA outside of its recognition sequence (see below) and requires ATP or dATP cofactor. Both enzymes require divalent cations, but ATP does not stimulate BisI activity (Priscilla Hiu-Mei Too and Shuang-yong Xu, unpublished data). In addition, the molecular mass of the purified BisI endonuclease was estimated to be ∼21 kDa (±2 kDa) and the apparent molecular mass of SauUSI is ∼100 kDa.
ATP and dATP requirement for endonuclease activity
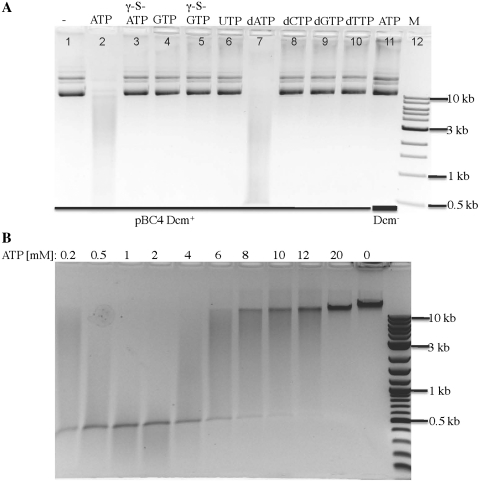
Based on BlastP and amino acid sequence alignment (28) SauUSI contains a predicted ATP binding and DNA helicase domain in the middle part of the protein (approximately at 201–625 aa). To characterize the presumed nucleotide requirement, we tested SauUSI activity on T4gt DNA using different NTP or dNTP in the digestion. Figure 3A shows that only ATP or dATP supports SauUSI endonuclease activity. A non-hydrolyzable ATP analog, γ-S-ATP, does not support SauUSI endonuclease activity, suggesting ATP hydrolysis is necessary for the catalytic reaction. To determine the optimal ATP concentration supporting SauUSI digestion, we tested SauUSI activity at ATP concentrations in the range of 0.2–20 mM on T4gt DNA. SauUSI is optimally active between 1 and 2 mM ATP, with activity detectable between 0.2 and 12 mM ATP under the conditions tested (1 µg T4gt DNA, 1 µg SauUSI, in 50 µl total digestion volume) (Figure 3B). The intracellular ATP concentration was estimated at 2.6 mM when E. coli was grown in culture media (29). No data on intracellular ATP concentration were available for S. aureus USA300 strain. The biological relevance of the preference of ATP/dATP over GTP is not clear. It might be advantageous not competing for the same cofactor if SauUSI-like and McrBC-like type IV restriction systems co-exist in the same bacterium.
Figure 3.
SauUSI digestion of plasmid and T4gt DNAs in the presence of NTP or dNTP. (A) Plasmid DNAs were digested by SauUSI in the presence of NTP or dNTP as indicated. Lanes 1–10, digestion of modified DNA pBC4 (Dcm+). Lane 11, unmodified pBC4 (Dcm−) plus ATP and SauUSI. (B) T4gt DNA was digested by SauUSI at various ATP concentrations (0.2–20 mM).
Salt and metal ion requirement of SauUSI
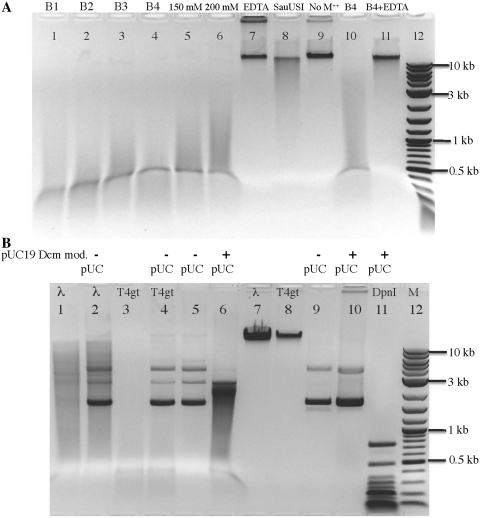
The N-terminus of SauUSI contains a phopholipase D (PLD) family endonuclease catalytic site (the HKD catalytic motif, approximately at 1–200 aa) (30). Some PLD family nucleases are active in the absence of divalent metal ions (in the presence of EDTA) (31,32). We therefore evaluated whether SauUSI requires divalent metal ions for optimal activity. SauUSI is active in the four NEB buffers, all of which contain 10 mM Mg++ (before EDTA treatment and dialysis), with highest activity in buffer 4 (50 mM potassium acetate). It is also active at NaCl concentrations of 50, 100, 150 or 200 mM with 10 mM MgCl2 (Figure 4A, lanes 2 and 3, 5 and 6). SauUSI is partially active in a buffer in the absence of additional metal ions (50 mM NaCl, 10 mM Tris–HCl, pH 8, 1 mM DTT) as shown in Figure 4A, lane 8, but addition of 10 mM EDTA completely inhibited endonuclease activity (Figure 4A, lane 7). To clarify the nature of the requirement, we prepared enzyme depleted of EDTA-chelatable divalent cations. A volume of 20 mM EDTA was added into the SauUI enzyme, which was then dialyzed into a new buffer for 48 h. The metal ion-depleted SauUSI was retested for enzyme activity in buffer with or without divalent cations. Figure 4A, lane 9 shows that SauUSI is inactive after EDTA treatment and the subsequent extensive dialysis. The metal ion-depleted SauUSI regained activity in buffer 4 with Mg++, and its activity was again inhibited if EDTA was added (Figure 4A, lanes 10 and 11). It is not clear whether the metal ion is required for the endonuclease activity or for the ATPase activity or for both. Further experiments are needed to uncouple the two reactions.
Figure 4.
Digestion of T4gt by SauUSI in NEB buffers 1–4, in the presence of EDTA, or in the absence of metal ions (depleted from enzyme preparation). (A) Lanes 1–4, SauUSI digestion in NEB buffers 1 to 4. Lanes 5 and 6, 150 and 200 mM NaCl in a buffer containing 20 mM Tris–HCl, pH 8.0, 1 mM DTT, 10 mM MgCl2. Lane 7, SauUSI digestion in the NTD buffer (50 mM NaCl, 20 mM Tris–HCl, pH 8.0, 1 mM DTT), plus 10 mM EDTA. Lane 8, same as lane 7, except EDTA was left out, no additional metal ions were added to the buffer. Lane 9, T4gt DNA incubated with metal ion-depleted SauUSI in the NTD buffer without additional metal ions. Lane 10, T4gt DNA digested with metal ion-depleted SauUSI in buffer 4. Lane 11, same as lane 10, but supplemented with 10 mM EDTA. (B) SauUSI digestion of modified DNA mixed with unmodified DNA. Lanes 1–6, SauUSI digestion of DNAs. The “–” and “+” on top of pUC indicate unmodified or modified pUC19 by Dcm methylase, respectively. Lane 11, DpnI digestion of pUC19.
We also supplemented the depleted enzyme preparation with Mn++, Ca++, Co++, Zn++, Ni++ or Cu++, finding partial activity in buffers with 1 mM Mn++, Ca++ or Co++. The relative endonuclease activity in different metal ions are Mg++ > Mn++ > Ca++ > Co++ (data not shown). SauUSI is inactive in buffers with metal ions Zn++, Ni++ or Cu++ (1–5 mM, data not shown). The promiscuous use of metal ions for SauUSI activity may provide certain advantage in restricting phage or plasmid DNA if one or two cofactors are in limited supply under certain growth condition. It was interesting to note that glc-hmC-specific endonuclease GmrSD prefers Ca++ metal ion and UTP for optimal activity (15), ScoMcrA prefers Mn++ and Co++ for endonuclease activity (33) and HpyAV, an HNH-type endonuclease, prefers Ni++ for activity (34).
Mixing of unmodified pUC19 DNA with Dcm+ λ (m5C) DNA or T4gt (5hmC) DNA in SauUSI digestion
In principle, SauUSI could have a cryptic endonuclease activity that is activated upon recognition of modified DNA. If so, activation might allow action in trans on a separate unmodified molecule. We find, however, that it acts on 5mC/hmC-modified DNA only in cis. The endonuclease catalytic activity is tightly coupled with 5mC or 5hmC recognition. This was demonstrated with a mixing experiment. We mixed unmodified and modified DNA and digested the mixture with SauUSI. Dam+ Dcm− pUC19 DNA was poorly digested when it was mixed with λ DNA (Dcm+) or T4gt (5hmC) DNA, while λ DNA (Dcm+) or T4gt (5hmC) DNA was preferentially cleaved by SauUSI in the two-DNA mixtures (Figure 4B, lanes 1–4). A small fraction of the unmodified pUC19 DNA (Dcm−) was linearized and nicked by SauUSI endonuclease (see also above). As expected, Dcm+ pUC19 was digested by SauUSI (Figure 4B, lane 6). Similarly, HindIII-digested pUC19 DNA (Dcm−) was not digested when it was mixed with modified λ (Dcm+) or T4gt (5hmC) DNA or mixed with duplex oligos containing two C5mCGG-modified sites (data not shown). This inactivity toward unmodified DNA is important since the native host DNA (self DNA) is presumably not methylated at position 5 of the cytosine (N4mC does not appear to matter). Inspection of the sequenced genome of S. aureus subsp. aureus USA300 does not find any candidate 5mC methylases (two putative Type I N6mA methylases are present, data now shown), consistent with the notion that this type IV enzyme has been evolved to selectively restrict foreign-modified 5mC/5hmC DNA. We changed the classification of SauUSI-like restriction enzymes from type III (24) to type IV, reflecting the fact that SauUSI is a modification-dependent REase, and that the SauUSI-like REase in S. aureus clinical strain SA564 only restricts 5mC-modified plasmid DNA (see below). On the other hand, SauUSI could also be considered as a type IIM enzyme under certain circumstances since its cleavage of short duplex oligos are relatively precise, 2–3 nt outside of its recognition sequence (see below).
Supplementary Figure S2 shows a time course of limited digestion by SauUSI in digesting M.EagI-modified plasmid mixture (pACYC-eagIM and pBC4). Early in the reaction (2–6 min), intermediate DNAs (probably in linear form) were detected. After 20 min digestion, only a smearing pattern was observed. It is likely that the SauUSI endonuclease binds to the M.EagI-modified sites and makes cleavage/nicks on both strands, and may continue digestion along the modified molecules in cis using the DNA helicase activity. SauUSI displays a strong nuclease activity on single-stranded DNA. However, this nuclease activity is not modification dependent (data not shown).
Digestion of 5mC-containing PCR DNA substrates and run-off sequencing of the cleavage products
A ∼430-bp fragment of pACYC184 between the BamHI and SalI site that contains six SCNGS sites was generated by PCR amplification using 5 m-dCTP or regular dCTP. The 5mC-containing PCR product was apparently digested by SauUSI endonuclease, generating a mixture of discrete bands and smearing (Figure 5A). The digestion was incomplete because many of the bands are bigger than the predicted cleavage products of 5mCCGG, C5mCGG, G5mCNGC, C5mCNGG or C5mCWGG generated by virtual digestion using the NEB cutter (http://tools.neb.com/NEBcutter2/). In contrast, the PCR product made using regular dCTP was only much less cleaved at the same enzyme concentrations, confirming the preference of SauUSI to 5mC-modified DNA (Figure 5A).
Figure 5.
SauUSI digestion of 5mC- or 5hmC-containing PCR DNA. (A) Sixty nanograms of a 436-bp substrate DNA was digested with 1, 0.5 and 0.25 µg of SauUSI REase in NEB Buffer 4 with 1 mM ATP for 2 h at 37°C (lanes 1, 2, 3 and 5, 6, 7, respectively). The substrate in lanes 1–4 was made in PCR using 5 m-dCTP. The substrate in lanes 5–8 was made in PCR using regular dCTP. The reactions were analyzed using a 5% TopVision gel (Fermentas) stained with SYBR Gold (Invitrogen). (B) SauUSI digestion of 5mC-containing PCR DNAs (73 bp-1 site, 90 bp-2 sites, 99 bp-2 sites, 260 bp-5 sites and 198 bp-4 sites, respectively). PCR substrates were digested by SauUSI for at 37°C for 2 h and the enzyme was inactivated at 65°C for 20 min. DNA fragments were resolved in a 15% PAGE-Urea gel (Invitrogen) and stained with SYBR Gold. LM, low molecular DNA marker; 10 bp, 10 bp DNA ladder. (C) SauUSI digestion of dC-, 5mC-, 5hmC-containing PCR DNA (73 bp-1 site): 100 bp, 100 bp DNA ladder. DNA was subjected to 2.5% agarose gel electrophoresis. (D) SauUSI digestion of dC-, 5mC-, 5hmC-containing PCR DNA (207 bp-4 sites). Two enzyme concentrations were used in the digestion. DNA fragments were resolved in 2.5% agarose gel electrophoresis.
Similarly, we generated a single site (GCCGC) 73-bp PCR fragment in PCR from pBR322 template and used it as a substrate for SauUSI digestion. The single-site PCR DNA was not cleaved by SauUSI (Figure 5B, lane 2). A two-site PCR substrate (GCTGC and GCAGG) with a length of 90 bp was cleaved by SauUSI; and another two-site substrate (GCGGG, GCCGC) with 99 bp long was also sensitive to SauUSI digestion (Figure 5B, lanes 4 and 6). Longer 5mC-containing PCR products with 5 and 4 sites were also subjected to SauUSI digestion and partially cleaved products were detected (Figure 5B, lanes 8 and 10). Similar results were obtained with 5hmC-containing PCR DNAs. A single site (GCCGC) with 5hmC incorporated in PCR was a poor substrate for SauUSI digestion (Figure 5C), whereas 5hmC-containing PCR DNA with four SauUSI sites were cleavable by SauUSI (Figure 5D). 5hmC-containing PCR DNA with two sites was also cleaved by SauUSI (data not shown). Run-off sequencing from the digested PCR products indicates that SauUSI cleaves DNA 2–3 bp outside of its recognition sequence GCCGC/GCGGC. There are three types of cleavage detected by run-off sequencing. Primer F210 detected two breaks (two double peaks) at the 3′ side of GCGGC recognition site (Supplementary Figure S3, bottom panel) in comparison with undigested pBR322 sequence (top panel) (the cut site distance can be interpreted differently considering the overlapping GCCGC site). Primer F571 detected two breaks (two double peaks) from the 5′ side of GCGGC/GCCGC target site (Supplementary Figure S4, bottom panel) compared to the control DNA sequence using the same primer. Primer F670 detected multiple DNA breaks/double peaks (Supplementary Figure S5), cleavage possibly taking place 3-bp 5′ and 3′ of the GCGGC/GCGGC target site, and another possible cut at 13 bp upstream (one DNA helical turn apart), implying that SauUSI is capable of sliding and cutting at long distance.
We also analyzed the cleavage sites of 5mC-modified plasmid DNA. The fnu4HIM gene was cloned in pBR322 and its expression was driven by the TcR promoter. The plasmid DNA pBR322-fnu4HIM extracted form overnight culture was fully resistant to Fnu4HI digestion (data not shown). Following digestion of pBR322-fnu4HIM by SauUSI for 2 h, the digested DNA was purified and subjected to run-off sequencing. Supplementary Figure S6 (bottom panel) shows that SauUSI cleaves outside of recognition sequence at N3, N10 and N14–18, with complete run-off at N18 from the target site, indicating SauUSI’s ability to travel and cleave at long distance from the G5mCCGC site. Supplementary Figure S6 (top panel) shows the DNA sequencing result from uncut pBR322 DNA. More cleavage sites remain to be analyzed in order to get a complete picture of SauUSI cut sites. The preliminary results indicate that SauUSI endonuclease is most likely to cleave 2–3 bp outside of its recognition sequence on short PCR DNA (∼90–200 bp). For the M.Fnu4HI-modified plasmid DNA (∼6 kb), the SauUSI cleavage sites vary from N3 to N18 (G5mCNGC N3–18, bottom strand). It would be interesting to engineer SauUSI variants to cleave at longer distance at the fixed position.
Site-directed mutagenesis of catalytic residues of SauUSI endonuclease
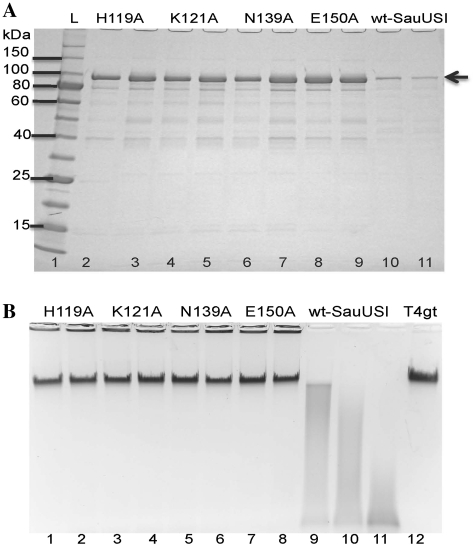
The H119 and K121 residues in SauUSI are highly conserved among PLD family endonucleases; and the N139 and E150 residues are somewhat conserved (N, D, Q, E are sometimes interchangeable at the two positions). We mutated H119, K121, N139 and E150 to Ala by site-directed mutagenesis and partially purified the mutant proteins. Figure 6A shows the purified variant proteins. The mutant protein yields are 4–5-fold higher than the wt enzyme, probably reflecting the reduced toxicity of the mutant endonucleases toward the E. coli expression host. The mutant enzyme variants are inactive in digestion of 5hmC T4gt DNA (Figure 6B, lanes 1–8). In contrast, the wt enzyme (batch-purified SauUSI in lanes 9 and 10, and column-purified SauUSI in lane 11) is active in cleaving T4gt DNA. This mutagenesis experiment confirmed the prediction that H119, K121, N139 and E150 are involved in catalytic activity. Mutations of these four catalytic residues to Ala abolished the endonuclease activity.
Figure 6.
Protein gel analysis of SauUSI variants and activity assay. (A) SDS–PAGE gel analysis of batch purified SauUSI catalytic mutants H119A, K121A, N139A, E150A,and the wt enzyme (two isolates for each mutant are shown). Arrow indicates the SauUSI protein (∼100 kDa). (B) Activity assay of SauUSI mutants and the wt enzyme. Lanes 1–8, T4gt DNA incubated with catalytic mutant proteins. Protein was purified from two isolates for each mutant. Lanes 9 and 10, T4gt digested with two concentration of batch purified wt enzyme; lane 11, wt SauUSI (column purified). Lane 12, uncut T4gt DNA substrate.
Oligomerization state of variant H119A
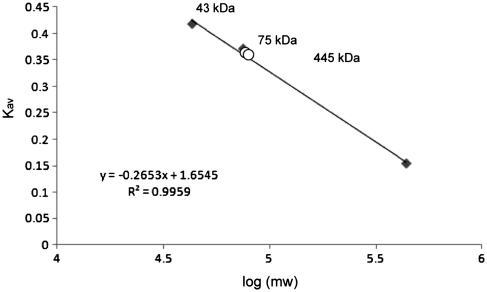
The oligomerization state of SauUSI was investigated using gel-filtration chromatography on an analytical Superdex 200 column. The partition coefficient (Kav) of SauUSI H119A was compared to those of standard proteins to obtain the relative molecular weight (Mr). The elution profile of SauUSI H119A from two separate runs contains a single peak with Mr of 76 kDa (open circles; Figure 7). The absence of peaks that correspond to oligomers of SauUSI indicates that SauUSI exists most likely as monomers in solution in the absence of target DNA. The physical properties of SauUSI enzyme with specific and non-specific DNAs remain to be analyzed.
Figure 7.
Analytical gel filtration chromatography of variant SauUSI-H119A. A Superdex 200 5/150 GL (3 ml bed volume; GE Healthcare) was run with NEBuffer 4 at 4°C at a flow of 0.2 ml/min. The partition coefficient (Kav) of the standard proteins ovalbumin (43 kDa), conalbumin (75 kDa) and ferritin (440 kDa) obtained from two independent runs is plotted against log(mw) (filled diamonds). The Kav of SauUSI H119A (1 mg/ml) obtained from two independent runs under the same running conditions are shown as open circles. A relative molecular weight of 76 kDa was obtained from the standard curve (the insert equation).
Restriction of phage by SauUSI endonuclease
The biological function of type IV restriction systems is to restrict modified foreign DNA (phage or plasmid) (35,15). We tested the restriction of phages by SauUSI endonuclease using Dcm+ or Dcm− λ phages that have been grown on modification proficient or deficient E. coli hosts. The plating efficiency of Dcm+ λ phages is 0.03–0.04% (two independent experiments) on a SauUSI-containing host, compared to the high efficiency on host containing empty vector (Table 2). As expected, SauUSI does not restrict unmodified Dcm− λ phages. A frame-shift mutant of SauUSI (a single base deletion at nt positon 85) abolished the ability to restrict Dcm-modified λ phages. The deletion mutant (frame-shift mutant) was found during screening of sauUSIR gene insert in pBR322.
Table 2.
Phage restriction activity of SauUSI endonuclease
| Cells | Dcm+ λ PFU/ml | Average PFU/ml | Plating efficiency |
|---|---|---|---|
| Exp. 1. ER2566 | 5.8 × 108 | 6.0 × 108 | 100% |
| [pBR322] | 6.1 × 108 | ||
| Exp. 1. ER2566 | 2.0 × 105 | 2.4 × 105 | 0.04% |
| [pBR-sauUSIR] | 2.8 × 105 | ||
| Exp. 1. ER2566 | 5.3 × 108 | 5.6 × 108 | 93.3% |
| [pBR-sauUSIR**] | 5.9 × 108 | ||
| Exp. 2. ER2566 | 3.2 × 108 | 2.9 × 108 | 100% |
| [pBR322] | 2.6 × 108 | ||
| Exp. 2. ER2566 | 9.0 × 104 | 9.5 × 104 | 0.03% |
| [pBR-sauUSIR] | 1.0 × 105 | ||
| Exp. 2. ER2566 | 2.9 × 108 | 3.1 × 108 | 107% |
| [pBR-sauUSIR**] | 3.2 × 108 |
| Cells | Dcm− λ PFU/ml | Average PFU/ml | Plating efficiency |
|---|---|---|---|
| ER2566 | 2.7 × 109 | 2.8 × 109 | 100% |
| [pBR322] | 2.9 × 109 | ||
| ER2566 | 2.4 × 109 | 2.5 × 109 | 89% |
| [pBR-sauUSIR] | 2.5 × 109 | ||
| ER2566 | 2.9 × 109 | 3.0 × 109 | 107% |
| [pBR-sauUSIR**] | 3.0 × 109 |
Plasmid pBR-sauUSIR** carries a single base deletion at nt position 85, causing a frame-shift mutation in the SauUSI endonuclease.
In vivo DNA restriction activity of S. aureus clinical strain SA564 and its restriction-deficient counterpart
Due to the possible existence of other active type I and type IV restriction systems in the S. aureus USA300 strain, we tested the possibility of this type IV (referred to as type III-like in Corvaglia et al., 2010) restriction system restricting methylated DNA in vivo, using S. aureus clinical strain SA564. We measured the transformation efficiencies of plasmid prepared from different E. coli strains. We used strains deficient in adenine methylation (dam−), deficient in cytosine methylation (dcm−), a double mutant (dam− dcm−), or wild-type (wt, dam+ dcm+ DH5α) for both methylation systems. To exclude problems in transformation efficiency other than the restriction of the plasmid DNA, we also prepared plasmid DNA from RN4220, the permissive laboratory S. aureus strain. We have previously shown that plasmid DNA from RN4220 can efficiently be transformed into SA564, independent of the presence or absence of the two restriction systems. Mixtures of plasmids prepared from E. coli and S. aureus were transformed into the clinical strain SA564 and its restriction-deficient counterpart (SA564 hsdR::targetron/type I-deficient, type IV-like::targetron/deficient in SauUSI-like REase) (24). The results presented in Table 3 show that the transformation efficiency of Dcm+-modified DNA is reduced by more than three-order of magnitude in wt SA564 strain. In contrast, plasmid DNA prepared from dcm-deficient strains efficiently transformed wild-type and mutant SA564 strains. In comparison, the absence of dam methylation did not influence the transformation efficiency. These results clearly show that this new restriction system recognizes C-methylated DNA and restricts only Dcm+-modified DNA in vivo.
Table 3.
Transformation efficiency (number of colony forming units) using dcm+ modified plasmid and dam–/dcm– DNA
| Exp.1 | Source of plasmid |
||||
|---|---|---|---|---|---|
| pCN33 DH5α/ pCN38 RN4220 | pCN33 BL21 dcm–/ pCN38 RN4220 | pCN33 LC1015 dam–/ pCN38 RN4220 | pCN33 LC3142 dam–, dcm–/ pCN38 RN4220 | ||
| Recipient strains | 564 | 1/71 100 | 28 500/99 100 | 20/45 500 | 10 200/57 900 |
| 564 type IV– | 8900/30 100 | 8900/53 900 | 7800/32 100 | 7800/40 700 | |
| Exp.2 | Source of plasmid |
||||
|---|---|---|---|---|---|
| pCN38 DH5α/ pCN33 RN4220 | pCN38 BL21 dcm–/ pCN33 RN4220 | pCN38 LC1015 dam–/ pCN33 RN4220 | pCN38 LC3142 dam–, dcm–/ pCN33 RN4220 | ||
| Recipient strains | 564 | 12/17 500 | 78 800/30 900 | 12/15 890 | 23 900/37 600 |
| 564 type IV– | 34 100/7320 | 44 200/10 600 | 64 700/13 300 | 24 900/10 500 | |
Note: Here, we refer to the SauUSI isoschizomer in the S. aureus clinical strain SA564 as a type IV restriction enzyme.
In vivo restriction (toxicity) by constitutive expression of SauUSI endonuclease in Dcm+ E. coli
Initially, we tested plasmid DNA transformation of pTYB1-sauUSIR into Dcm+ or Dcm− E. coli strains. The transformation efficiency of pTYB1-sauUSIR into a Dcm+ strain is only 5–10-fold lower than the empty vector pTYB1 (data not shown). We attributed this low level of restriction to the low expression level of SauUSI from pTYB1 in E. coli under non-inducing condition (or poor in vivo cleavage of the fusion protein SauUSI-intein-CBD to produce active SauUSI). To circumvent this problem, we recloned the sauUSIR gene into pBR322 with an optimal ribosome binding site using a dcm-deficient host so that the sauUSIR gene is constitutively expressed under the TcR promoter. Another clone with the sauUSIR gene inserted in the opposite orientation was also constructed. Both plasmid clones were used to transform Dcm+ E. coli host NEB10β. Table 4 shows the transformation results. The transformation efficiency of pBR-sauUSIR plasmid was reduced by more than four orders of magnitude compared to the empty vector pBR322 or pBR-sauUSIR* (insert in reverse orientation). Although they are not isogenic strains, pBR-sauUSIR plasmid transforms two E. coli B strains deficient in Dcm methylase at high efficiency (data not shown). This experiment has some biological implication in the transfer SauUSI-like restriction genes among bacteria. The results suggest that as long as SauUSI expression level is tightly controlled, sauUSIR-like genes could be acquired by 5mC-methylase carrying hosts.
Table 4.
Transformation efficiency of pBR-sauUSIR, pBR-sauUSIR* and pBR322 plasmids into Dcm+ E. coli strain NEB10β
| Plasmid clone | Comments | CFU/µg DNA | Average CFU/µg |
|---|---|---|---|
| pBR322 | Empty vector | 5.0 × 106 | 4.5 × 106 |
| 4.1 × 106 | |||
| pBR-sauUSIR* | R gene inserted in opposite orientation to TcR promoter | 4.6 × 106 | 4.1 × 106 |
| 3.5 × 106 | |||
| pBR-sauUSIR | R gene insert under TcR promoter | 4.5 × 102 | 3.3 × 102 |
| 3.9 × 102 | |||
| 3.8 × 102 | |||
| 1.0 × 102 | |||
| pBR-sauUSIR** | SauUSI frame-shift mutant | 4.4 × 106 | 3.3 × 106 |
| 2.1 × 106 |
Plasmid pBR-sauUSIR** carries a single base deletion at nt position 85, causing a frame-shift mutation in the SauUSI endonuclease (the mutation was introduced in PCR).
Functional domain organization in SauUSI protein
BlastP analysis of SauUSI to known proteins in Genbank showed that the protein contains at least three functional domains (see Supplementary Figure S7) (36). The N-terminal domain (approximately from 1 to 200 aa) contains the catalytic residues of the phospholipase D (PLD) endonuclease family, specifically at H119-K121-N139-E150. The HxK residues are highly conserved in PLD family endonucleases. N/E/D are also conserved, but can be substituted for each other in some circumstances. The middle part of the protein (approximately from 201 to 625 aa) carries the ATPase/helicase domain that is highly conserved among DEXD superfamily DNA/RNA helicases. There is a conserved putative Mg++ binding site near amino acid 250, suggesting that Mg++ may be required for ATP binding and hydrolysis. The C-terminal region (approximately from 685 to 953 aa) belongs to the conserved domain family DUF3427 found in bacteria and archaea with unknown function. We speculate that this region is the specificity domain that recognizes 5mC or 5hmC. This domain can be found in 255 proteins in the DUF3427 cluster (www.kegg.org) among bacteria and archaea (see Supplementary Table S1 for a list of SauUSI homologs found in many pathogenic G+ bacterial strains). Another conserved motif prediction program found the conserved motif of Type III REases (from 221 to 373 aa in SauUSI, Supplementary Figure S7, bottom panel), explaining why this group of enzymes was originally annotated as putative type III REases containing a DNA/RNA helicase domain in Genbank. We also noted that in some sequenced bacterial genomes, the PLD family endonuclease exists as a separate protein and a DNA/RNA helicase domain in an adjacent protein, possibly regulated by a single operon (data not shown). Site-directed mutagenesis of the conserved residues in the helicase/ATPase and 5mC/5hmC domains will need to be undertaken to confirm these predictions.
Interestingly, there is a conserved gene (SauUSA300_2432) linked to the SauUSI gene (SauUSA300_2431) that has strong amino acid sequence similarity to the pyrophosphohydrolase and MutT/NUDIX family hydrolases. This protein may be involved in the breakdown of pyrophosphate generated by ATP/dATP hydrolysis. The SSDB gene cluster search results from the KEGG server indicated that there are at least 60 SauUSI-like type IV endonucleases linked to the putative pyrophosphohydrolases (data not shown). The function of the latter protein remains to be examined.
Inspection of sequenced Staphylococcus sp. genomes revealed a few type II R-M systems that may create potential substrates for SauUSI following 5mC modification. Staphylococcus aureus RF122 strain contains a putative type II R–M system, whose C5 methylase has significant amino acid sequence similarity to GCNGC methylases (62% aa sequence identity to M.Bsp6I, GCNGC). Staphylococcus aureus ST398 strain carries a putative R–M system with the predicated target sequence of CCNGG (the putative C5 methylase displays 69% aa sequence identity to M1.ScrFI, CCNGG). Staphylococcus aureus TW20 and S. epidermidis RP62A strains carry a putative orphan C5 methylase (possibly prophage encoded multi-specificity C5 methylases) with predicted GGCC specificity. These 5mC-modified DNA at GCNGC, CCNGG and GGCC sites can potentially be cleaved by SauUSI, and 5mC-modified DNA from these strains may be restricted when transferred into S. aureus USA300 strain or Staphylococcus species with active SauUSI-like REases. The susceptibility of these methylase-modified DNAs to SauUSI restriction remains to be tested.
SauUSI displays low activity on short duplex oligos and requires at least two sites for efficient cleavage, possibly SauUSI requiring extensive DNA looping and communication of two distantly bound molecules for optimal activity, as it was observed for type I and type III REases. In SauUSI, the C-terminal region is presumed to contain the 5mC/5hmC recognition domain. Work is underway to clone and express this TRD domain and to characterize its activity. It is possible to select SauUSI mutants that prefer to cleave 5hmC DNA. Such mutants would be useful for epigenetic studies.
CONCLUSIONS
SauUSI is a 5mC/5hmC modification-dependent REase. The endonuclease activity requires ATP or dATP and divalent cations such as Mg++, Mn++, Ca++ or Co++. Although the cleavage appears to be non-specific, the initial incision (nicks or ds breaks) displays certain sequence preferences, with S5mCNGS as the preferred sites. Efficient cleavage requires more than one site; and the cleavage sites outside of its recognition sequence appear to be variable (N2–N18). SauUSI is shown to restrict Dcm+ phage infection and Dcm+ plasmid transformation in E. coli and S. aureus clinical strain SA564. SauUSI endonuclease expression is toxic when it is constitutively expressed in a Dcm+ E. coli strain. SauUSI is a multi-domain protein with a PLD-family endonuclease catalytic site located at the N-terminal domain, a DNA helicase/ATPase located in the middle part of the protein, and a presumed 5mC/hmC TRD domain (DUF3427) located at the C-terminus. Over 150 proteins with 25–100% amino acid sequence identity with a similar functional domain organization are found in sequenced bacterial genomes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
New England Biolabs, Inc. The Swiss National Science Foundation and the Canton of Geneva. Funding for open access charge: New England Biolabs, Inc.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Richard Roberts, Elisabeth Raleigh, and William Jack for their critical comments, and Rick Morgan for discussions. We are grateful to Lise Raleigh, Mern Sibley, Alexey Fomenkov, Rick Morgan, Keith Lunnen, Geoff Wilson, Lucia Greenough, Bill Jack for strains and plasmids. We are appreciative to NEB DNA sequencing lab for sequencing the SauUSI expression clones and SauUSI cleavage products. We express our gratitude to Don Comb and Jim Ellard for their continued support of basic research.
REFERENCES
- 1.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev S, Dryden DT, Dybvig K, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arber W. DNA modification and restriction (Review) Prog. Nucleic Acid Res. Mol. Biol. 1974;14:1–37. doi: 10.1016/s0079-6603(08)60204-4. [DOI] [PubMed] [Google Scholar]
- 3.Wilson GG, Murray NE. Restriction and modification systems. Ann. Rev. Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- 4.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE – a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meisel A, Mackeldanz P, Bickle TA, Kruger DH, Schroeder C. Type III restriction endonucleases translocate DNA in a reaction driven by recognition site-specific ATP hydrolysis. EMBO J. 1995;14:2958–2966. doi: 10.1002/j.1460-2075.1995.tb07296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Humbelin M, Suri B, Rao DN, Hornby DP, Eberle H, Pripfl T, Kenel S, Bickle TA. Type III DNA restriction and modification systems EcoP1 and EcoP15. Nucleotide sequence of the EcoP1 operon, the EcoP15 mod gene and some EcoP1 mod mutants. J. Mol. Biol. 1988;200:23–29. doi: 10.1016/0022-2836(88)90330-0. [DOI] [PubMed] [Google Scholar]
- 7.Szczelkun MD, Friedhoff P, Seidel R. Maintaining a sense of direction during long-range communication on DNA. Biochem. Soc. Trans. 2010;38:404–409. doi: 10.1042/BST0380404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de la Campa AG, Springhorn SS, Kale P, Lacks SA. Proteins encoded by the DpnI restriction gene cassette. J. Biol. Chem. 1988;263:14696–14702. [PubMed] [Google Scholar]
- 9.Tarasova GV, Nayakshina TN, Degtyarev SK. Substrate specificity of new methyl-directed DNA endonuclease GlaI. BMC Mol. Biol. 2008;9:7. doi: 10.1186/1471-2199-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng Y, Cohen-Karni D, Xu D, Chin HG, Wilson G, Pradhan S, Roberts RJ. A unique family of Mrr-like modification-dependent restriction endonucleases. Nucleic Acids Res. 2010;38:5527–5534. doi: 10.1093/nar/gkq327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sutherland E, Coe L, Raleigh EA. McrBC: a multisubunit GTP-dependent restriction endonuclease. J. Mol. Biol. 1992;225:327–358. doi: 10.1016/0022-2836(92)90925-a. [DOI] [PubMed] [Google Scholar]
- 12.Raleigh EA, Murray NE, Revel H, Blumenthal RM, Westaway D, Reith AD, Rigby PWJ, Elhai J, Hanahan D. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. Nucleic Acids Res. 1988;15:1563–1575. doi: 10.1093/nar/16.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulligan EA, Hatchwell E, McCorkle SR, Dunn JJ. Differential binding of Escherichia coli McrA protein to DNA sequences that contain the dinucleotide m5CpG. Nucleic Acids Res. 38:1997–2005. doi: 10.1093/nar/gkp1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mulligan EA, Dunn JJ. Cloning, purification and initial characterization of E. coli McrA, a putative 5-methylcytosine-specific nuclease. Protein Expr. Purif. 2008;62:98–103. doi: 10.1016/j.pep.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bair CL, Black LW. A type IV modification dependent restriction nuclease that targets glucosylated hydroxymethyl cytosine modified DNAs. J. Mol. Biol. 2007;366:768–778. doi: 10.1016/j.jmb.2006.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loomba PS, Taneja J, Mishra B. Methicillin and vancomycin resistant S. aureus in hospitalized patients. J. Glob. Infect. Dis. 2010;2:275–283. doi: 10.4103/0974-777X.68535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deleo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010;375:1557–1568. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stefani S, Goglio A. Methicillin-resistant Staphylococcus aureus: related infections and antibiotic resistance. Int. J. Infect. Dis. 2010;(Suppl. 4):S19–S22. doi: 10.1016/j.ijid.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Zhu W, Clark NC, McDougal LK, Hageman J, McDonald LC, Patel JB. Vancomycin-resistant Staphylococcus aureus isolates associated with Inc18-like vanA plasmids in Michigan. Antimicrob. Agents Chemother. 2008;52:452–457. doi: 10.1128/AAC.00908-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weigel LM, Donlan RM, Shin DH, Jensen B, Clark NC, McDougal LK, Zhu W, Musser KA, Thompson J, Kohlerschmidt D, et al. High-level vancomycin-resistant Staphylococcus aureus isolates associated with a polymicrobial biofilm. Antimicrob Agents Chemother. 2007;51:231–238. doi: 10.1128/AAC.00576-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weigel LM, Clewell DB, Gill SR, Clark NC, McDougal LK, Flannagan SE, Kolonay JF, Shetty J, Killgore GE, Tenover FC. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science. 2003;302:1569–1571. doi: 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- 22.Fluit AC, Schmitz FJ. Resistance integrons and super-integrons. Clin. Microbiol. Infect. 2004;10:272–288. doi: 10.1111/j.1198-743X.2004.00858.x. [DOI] [PubMed] [Google Scholar]
- 23.Waldron DE, Lindsay JA. Sau1: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J. Bacteriol. 2006;188:5578–5585. doi: 10.1128/JB.00418-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corvaglia AR, Francois P, Hernandez D, Perron K, Linder P, Schrenzel J. A type III-like restriction endonuclease functions as a major barrier to horizontal gene transfer in clinical Staphylococcus aureus strains. Proc. Natl Acad. Sci. USA. 2010;107:11954–11958. doi: 10.1073/pnas.1000489107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bidnenko E, Chopin A, Ehrlich SD, Chopin MC. Activation of mRNA translation by phage protein and low temperature: the case of Lactococcus lactis abortive infection system AbiD1. BMC Mol. Biol. 2009;10:4. doi: 10.1186/1471-2199-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GP. The phage abortive infection system, ToxIN, functions as a protein–RNA toxin–antitoxin pair. Proc. Natl Acad. Sci. USA. 2009;106:894–899. doi: 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu G, Ou HY, Wang T, Li L, Tan H, Zhou X, Rajakumar K, Deng Z, He X. Cleavage of phosphorothioated DNA and methylated DNA by the type IV restriction endonuclease ScoMcrA. PLoS Genet. 2010;6:e1001253. doi: 10.1371/journal.pgen.1001253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowry OH, Carter J, Ward JB, Glaser L. The effect of carbon and nitrogen sources on the level of metabolic intermediates in Escherichia coli. J. Biol. Chem. 1971;246:6511–6521. [PubMed] [Google Scholar]
- 30.Ponting CP, Kerr ID. A novel family of phospholipase D homologues that includes phospholipid synthases and putative endonucleases: identification of duplicated repeats and potential active site residues. Protein Sci. 1996;5:914–922. doi: 10.1002/pro.5560050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bao Y, Higgins L, Zhang P, Chan SH, Laget S, Sweeney S, Lunnen K, Xu SY. Expression and purification of BmrI restriction endonuclease and its N-terminal cleavage domain variants. Protein Expr. Purif. 2008;58:42–52. doi: 10.1016/j.pep.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lagunavicius A, Sasnauskas G, Halford SE, Siksnys V. The metal-independent type IIs restriction enzyme BfiI is a dimer that binds two DNA sites but has only one catalytic centre. J. Mol. Biol. 2003;326:1051–1064. doi: 10.1016/s0022-2836(03)00020-2. [DOI] [PubMed] [Google Scholar]
- 33.Liu G, Ou H-Y, Wang T, Li L, Tan H, Zhou X, Rajakumar K, Deng Z, He X. Cleavage of phosphorothioated DNA and methylated DNA by the type IV restriction endonuclease ScoMcrA. PLoS Genetics. 2010;6:e1001253. doi: 10.1371/journal.pgen.1001253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan SH, Opitz L, Higgins L, O'Loane D, Xu SY. Cofactor requirement of HpyAV restriction endonuclease. PLoS ONE. 2010;5:e9071. doi: 10.1371/journal.pone.0009071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raleigh EA. Organization and function of the mcrBC genes of Escherichia coli K-12. Mol. Microbiol. 1992;6:1079–1086. doi: 10.1111/j.1365-2958.1992.tb01546.x. [DOI] [PubMed] [Google Scholar]
- 36.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.