Abstract
RNA editing by adenosine deaminases acting on RNAs (ADARs) can be both specific and non-specific, depending on the substrate. Specific editing of particular adenosines may depend on the overall sequence and structural context. However, the detailed mechanisms underlying these preferences are not fully understood. Here, we show that duplex structures mimicking an editing site in the Gabra3 pre-mRNA unexpectedly fail to support RNA editing at the Gabra3 I/M site, although phylogenetic analysis suggest an evolutionarily conserved duplex structure essential for efficient RNA editing. These unusual results led us to revisit the structural requirement for this editing by mutagenesis analysis. In vivo nuclear injection experiments of mutated editing substrates demonstrate that a non-conserved structure is a determinant for editing. This structure contains bulges either on the same or the strand opposing the edited adenosine. The position of these bulges and the distance to the edited base regulate editing. Moreover, elevated folding temperature can lead to a switch in RNA editing suggesting an RNA structural change. Our results indicate the importance of RNA tertiary structure in determining RNA editing.
INTRODUCTION
The most common type of RNA editing in animals involves the conversion of individual adenosine (A) bases to inosine (I) by adenosine deaminases acting on RNA (ADARs) (1,2). Because inosine (I) is read as guanosine (G) during translation, A-to-I conversion in coding sequences leads to amino acid alterations and often entails changes in protein function. The majority of editing events in coding sequences has been identified in the nervous system (3–7).
Any double-stranded region of at least 23 bp may potentially be a substrate for ADARs (8). Such shorter regions of imperfectly paired dsRNA may be precisely edited at one particular adenosine in the midst of dozens of others. In contrast, long regions of perfectly paired dsRNA may be edited non-specifically, and up to 50% of the adenosine residues within an extended, perfect RNA duplex may be edited (8). All known editing substrates of ADARs are predicted to be embedded in extensive duplex structures that are invariably interrupted with loops, bulges and mismatches (9). It is thought that these structural features enable ADARs to recognize specific adenosines (10). Although the structure of the ADAR2 catalytic domain has been determined (11), the specific structural features required for editing have not been fully defined, nor is it understood how sequence and structural variations among editing sites contribute to differences in editing activity (12). Hence, it is still impossible to predict if and to what extent a given RNA might serve as a substrate for the site-specific editing in vivo (9,13).
The mouse GABAA receptor 3 subunit transcript was identified as an ADAR substrate, in which a small exonic RNA duplex is required for I/M site editing (14,15). Editing in Gabra3 RNA transcripts is regulated in a developmental- and tissue-specific manner, reaching a maximum in the adult brain (14–17). Editing at the I/M site of GABAA receptor pre-mRNAs could regulate the function of α3 subunit-containing GABAA receptors, with higher GABA sensitivity, faster activation, slower deactivation and greater outward rectification associated with the non-edited α3 subunit (15,18). Mutagenesis studies indicated that the conserved short duplex structure in Gabra3 RNA is required for editing and might be the minimal natural editing substrate (15). Therefore, this duplex is a well suited model to examine the effect of sequence and structure context on editing choice and efficiency.
In this study, we dissect the role of the stem–loop structure for efficient RNA editing at the Gabra3 I/M site. Unexpectedly, a mimicking stem–loop fails to undergo RNA editing. These results led us to revisit the structural requirement for this editing event by mutagenesis analysis. Point mutations followed by in vivo nuclear injection and editing experiments demonstrated that a non-conserved structure is a determinant for editing. The orientation of bulges at the base of the stem–loop structure seemingly regulates editing efficiency. Moreover, temperature changes can regulate editing by altering the RNA structure. In summary, our results emphasize the role of RNA tertiary structures in modulating RNA editing.
MATERIALS AND METHODS
Materials
Tortoise (Chinemys reevesii), lizard (Aspidoscelis inornata), cattle (Bos taurus), dog and chicken were bought from market. Mouse was kindly provided by the Institute of Cell and Genetics, Zhejiang University. Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany), according to the manufacturers protocol. Genomic DNA was isolated using the Universal Genomic DNA Extraction Kit (TaKaRa, Dalian, China).
RT–PCR
Total RNA was prepared using TRIzol-reagent (Invitrogen, Carlsbad, CA, USA) from different tissues of various animals. Total RNA was reverse transcribed using SuperScript III RTase (Invitrogen) with oligo(dT)15 primer, and the resulting single-stranded cDNA product was treated with RNase H at 37°C for 30 min. The oligonucleotide and primer sequences are listed in Supplementary Table S1. RT–PCR products were gel purified and subjected to direct sequencing with corresponding specific primers. In addition, the products of RT-PCR were purified and cloned into the pGEM-T Easy vector (Promega, Madison, WI, USA) and transformed into competent cells. Sequencing of individual recombinant clones was done using an automatic DNA sequencer.
Chemical probing in vitro
RNA structures were predicted using RNA folding and hybridization software Mfold, version 2.3 (19). Ten pico mole GABRA editing substrates were folded by first denaturing the RNA in the presence of 0.1 M KCl and 50 mM Cacodylate buffer pH 7.0 [for dimethylsulfate (DMS) and Kethoxal probing] or K-borate buffer pH 8.0 [for 1-cyclohexyl-(2-morholinoethy) carbodiimide metho-p-toluene sulfonate (CMCT) probing] at 95°C for 1 min followed by 2 min at room temperature. MgCl2 was then added to 1 mM or 10 mM (final concentration). The samples were incubated at 37°C for 30 min. After folding the chemical modification was performed: (i) DMS probing: DMS was added (final concentration 29 mM) and the samples were incubated at room temperature for 20 min. The reaction was stopped using β-mercaptoethanol (final concentration 280 mM); (ii) CMCT: 10 µl CMCT (42 mg/ml) were added and the samples were incubated at room temperature for 20 min; (iii) Kethoxal probing: 1 µl Kethoxal (7.4 mg/ml) was added and the samples were incubated at 37°C for 20 min. The reaction was stopped using 1 µl 0.5 M K-borate buffer pH 7.0. After precipitation the RNA was used for reverse transcription. Notably, the CMCT-modified RNA was resuspended in 25 mM K-borate buffer pH 8.0 buffer, while the DMS- or Kethoxal-modified RNAs were resuspended in H2O. Each RNA was tested at both Mg2+ concentrations, whereby we observed significant differences only for the Gallus gallus derived substrates, while the modification pattern remained unchanged for the other substrate RNAs (data not shown).
Reverse transcription
One pico mole of RNA was used for reverse transcription, which was essentially performed as described in (20). RT stop controls were obtained from unmodified RNA to detect natural stops of the avian myeloblastosis virus (AMV) reverse transcriptase (Promega) along the RNA template. Analysis of polyacrylamide gels was done with ImageQuant software (GE Healthcare).
Minigene construction, site-directed mutagenesis
Genomic DNA was taken as template, and PCR was performed to attain the corresponding DNA segments encompassing the potentially edited A. Wild-type (WT) minigene DNA, corresponding to the nucleotide positions 1234–1283 (NM_000808), was cloned into the pGEM-T Easy vector (Promega). Site-directed mutagenesis was performed according to the schematic diagrams of minigene constructs by PCR. DNA mutations engineered into expression constructs (M1−M18, M23–M29) were designed to singly disrupt base pairing interactions or, in combination (such as M19–M22), to restore base pairing while changing the identity of base pairing partners. These mutants are identical to WT minigene construct except for the mutated sites. More specific details can be obtained upon request from the authors. All constructs were sequence-verified before in vitro transcription.
In vitro transcription
WT or mutant minigenes were placed downstream of the T7 RNA polymerase promoter by PCR using Pfu Taq DNA polymerase. Transcription was carried out at 37°C for 1 h in the following system: DNA 500 ng, 10XT7 RNA polymerase buffer 2 µl, 50 mM DTT 2 µl, 2.5 mM NTP mix 4 µl, RNase inhibitor (40 U/µl) 0.5 µl, T7 RNA polymerase (Takara Bio, Dalian, China) 10 U and diethylpyrocarbonate (DEPC)-treated water to 20 µl. The template DNA was then degraded with 2 U DNase I (Ambion) after transcription. Synthesized RNAs were collected with Trizol (Invitrogen) and quantified.
RNA folding
The purified RNA samples were equally divided and folded in folding buffer: 95 μl DEPC water and 5 μl 1M MgCl2. After incubating for 2 min at 100°C, four parts RNAs were cooled to 25, 28, 33 and 37°C, respectively, then precipitated with EtOH at −80°C for 2 h. The folded RNAs were dissolved and then microinjected into Xenopus oocytes at 25°C. At temperature-mediated test, the folded RNAs were correspondingly microinjected into oocytes at 28, 33 and 37°C, respectively.
Native and denaturing PAGE
The purified RNA samples were heated for 2 min to 90°C in 50 mM MgCl2, cooled to 25 or 37°C and precipitated upon addition of Ethanol at −80°C. The dissolved RNAs were analysed by 16% native polyacrylamide gel and denaturing polyacrylamide gel, respectively.
Xenopus oocyte nuclei micro-injection
For injection into X. laevis oocyte nuclei, α-32P-ATP labelled RNA was injected in single oocytes. After 2 h oocytes were hand enucleated and the nuclei were homogenized. The RNA was isolated by TRIzol-reagent (Invitrogen). The isolated RNA was heat denatured and placed on ice before adding P1 nuclease. The resulting mononucleotides were separated on cellulose TLC plates as described (21).
RNA editing analysis
Injected X. laevis oocyte nuclei were isolated 2 h after injection and total RNA was prepared using TRIzol-reagent (Invitrogen). In all cases, the RNA samples were treated with DNase (DNA-free, Ambion) to remove contaminating genomic DNA. Total RNA was reverse transcribed using SuperScript III RTase (Invitrogen) with random primers. In cases where not enough nucleotides 3′ of the edited adenosine were present to design a specific primer RNA was polyadenylated at 37°C for 1 h by poly(A) polymerase (Takara) before reverse transcription. RNA editing was quantified as follows: RNA editing produces a HaeIII (GGCC) or Hin1II (CATG) restriction enzyme cutting site. For each RNA sample quantified, three independent PCRs were carried out and the resulting products were cut with HaeIII. In cases where the edited HaeIII site was the target of site-directed mutagenesis, an AluI sites (AGCT) introduced by the mutation was used. Amplified PCR products were separated by electrophoresis through a 12% polyacrylamide gel and then detected by silver staining. Images were captured through a CCD camera, and the quantification of edited and unedited products was done by comparison of the integrated optical density of detected bands measured by the GIS 1D Gel Image System ver. 3.73 (Tanon, Shanghai, China). In addition, the products of RT–PCR were purified and cloned into the pGEM-T Easy vector (Promega). Additionally, sequencing of 20–30 individual clones was done to exclude the appearance of editing events in other portions of the structure. As a result, no additional editing sites were detected except the identified I/M site.
RESULTS
Evolutionary insight into a duplex structure for editing
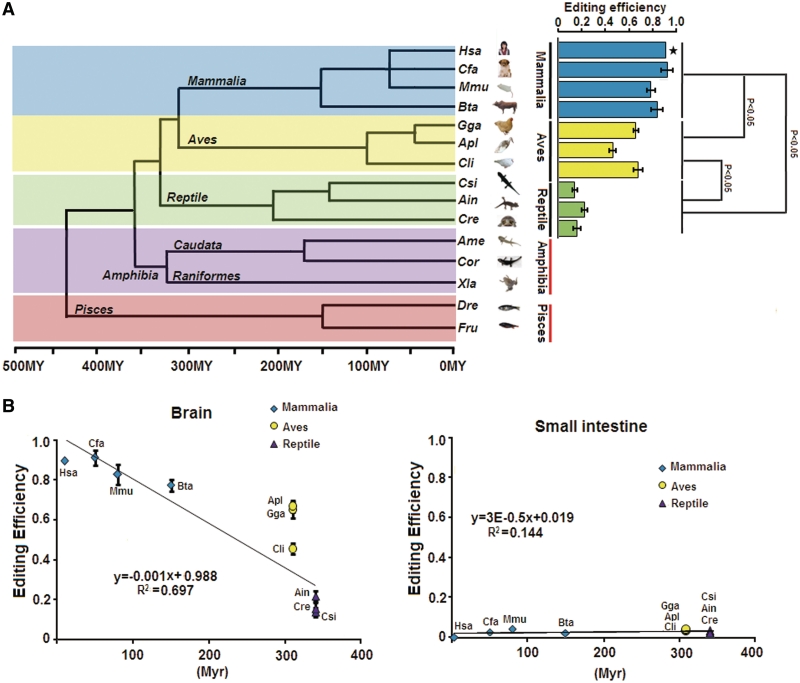
The mouse GABAA receptor α3 subunit transcript (mGabra3) was recently identified as an ADAR substrate, in which a small exonic RNA duplex is required for I/M site editing (14,15). Gabra3 transcripts have a genomically encoded G at the equivalent position in fish (14). To determine the evolutionary origin of I/M RNA editing, we analysed the GABAA receptor 3 subunit from different tissues of three reptiles: Chinese three-keeled pond turtle (Chinemys reevesii), Siamese crocodile (Crocodylus siamensis), little striped whiptail (Aspidoscelis inornata) and two amphibians: Chinese fire-bellied newt (Cynops orientalis), and axolotl (Ambystoma mexicanum). RNA editing could be detected in the three reptilian species but not in Chinese fire-bellied newt and axolotl (Figure 1A and Supplementary Figure S1). Editing could only be detected in the brain but not in the non-neuronal tissues such as small intestine, kidney or adrenal gland (Supplementary Figure S1). Interestingly, editing levels in the brain directly correlated with evolutionary distance from the mammalian lineage (Figure 1). Based on these data, we suggest the following evolutionary scenario for Gabra3 A-to-I RNA editing: G which represents the ancestral state was converted into A with the separation of caudata and raniformes. However, A-to-I RNA editing can immediately revert the G-to-A conversion event, maintaining similarity at the protein level to reduce negative selective pressure (22). During subsequent evolution, editing levels at this site increase, with a maximum in mammalian species.
Figure 1.
Phylogenetic analysis of Gabra3 I/M site. (A) Analysis of Gabra3 editing from mammalian to amphibian. Evolutionary tree showing the phylogenetic relationships among the species. Quantitative analyses of Gabra3 mRNA levels shows increased editing levels in mammalian compared with other classes (P ≤ 0.05). (B) Elevated editing levels in mammalian brain compared with other classes. RNA editing of Gabra3 are low in small intestine from different species. Mammalian (diamonds), Aves (circles), Reptile (triangles).
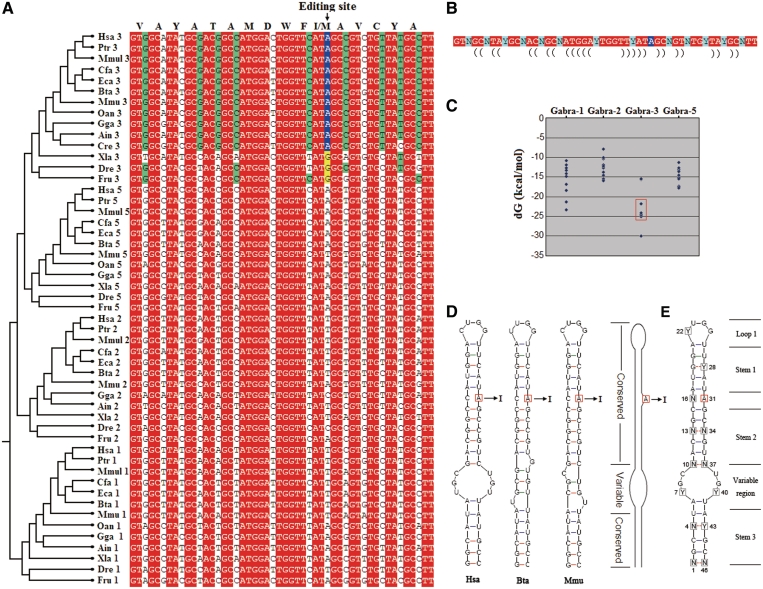
Besides Gabra3, the same 16 amino acid sequences are also encoded in other unedited transcripts, i.e. Gabra1, Gabra2 and Gabra5 (Figure 2A). These Gabra subunits underlie different RNA secondary-structural constraints (Figure 2C). The Gabra3 genes with a highly stable structure can undergo RNA editing; except for one with a genomically encoded G at the equivalent position (Figure 2C). The I/M editing duplex, encoding 16 amino acids, is the smallest natural helix known to be targeted by an ADAR enzyme. Because these sequences have an identical coding capacity, they constitute an interesting and informative model to further investigate the evolutionary determinants of RNA editing.
Figure 2.
Evolutionary and structural analysis of the Gabra subunits of vertebrates. (A) Multiple alignments of nucleotide sequences corresponding to the edited site in Gabra3 from different species and different Gabra isoforms. Notably, the nucleotide sequences encode identical amino acid sequences. Nucleotides showing complete conservation are shaded in red, and synonymous nucleotides showing complete conservation within edited Gabra3 subunits are shaded in green. Residues altered by RNA A-to-I editing are shaded in blue, and genomic substitutions with edited residues at these positions are shaded in yellow. For abbreviations see Supplementary Materials. (B) The degenerate nucleotide sequences encode identical amino acid sequence, where non-synonymous nucleotides (shaded in red) are largely complementary. The edited A is shaded in blue, and synonymous nucleotides are shaded in green. (C) Comparative analysis of the estimated equilibrium free energies (in kcal mol−1) from orthologues of different species from fish to human. The edited genes are boxed. (D) All known edited Gabra3 sequences are predicted to form three types of short duplex structure, with significant difference in shape of the variable region, which was represented by the sequences from H. sapiens (Hsa), M. musculus (Mmu) and B. taurus (Bta), respectively. A schematic picture of the duplexes is shown. (E) Hypothetical architecture of an edited Gabra3 duplex. The potential edited A is squared, and degenerate nucleotides are enclosed in squares. The dotted lines indicate predicted base pairs that depend on complementarities of synonymous nucleotides (G–C, A–U, G–U).
All known edited sequences from human to lizard are predicted to form three types of short duplex structures, with significant differences in shape of a variable region predicted by mFold analysis (Figure 2D and E) (19). This seems to imply that shape variation in this non-conserved region might be tolerated. The dsRNA structure contains a stem with one A–C mismatch that is capped by a tetraloop. Stem 1 of the Gabra3 hairpin is conserved in evolution because G can pair with C or U at Y28 position (Figure 2D and E). Apparently, the N10–N37 mismatch at the synonymous position in cattle Gabra3 (which undergoes RNA editing) can be tolerated in this species (Figure 2D). However, sequence comparative analyses indicate that three pairs (N13–N34; N4–Y43; N1–N46) at the synonymous positions are evolutionarily conserved in all known edited sequences. Except for Gabra3, we failed to find a similar stem–loop structure with pairing state at all N13–N34, N4–Y43 and N1–N46 positions in other genes of Gabra family. This indicates that synonymous changes are under selective constraint to escape RNA editing in these transcripts, which might be unnecessary and even deleterious.
A smart duplex structure for RNA editing
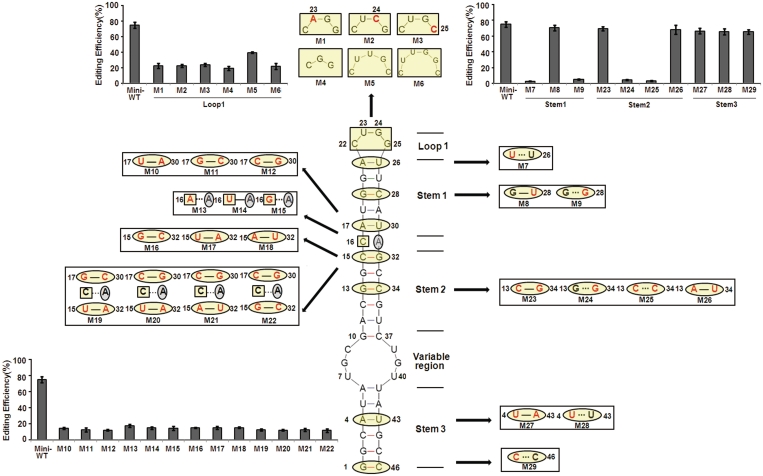
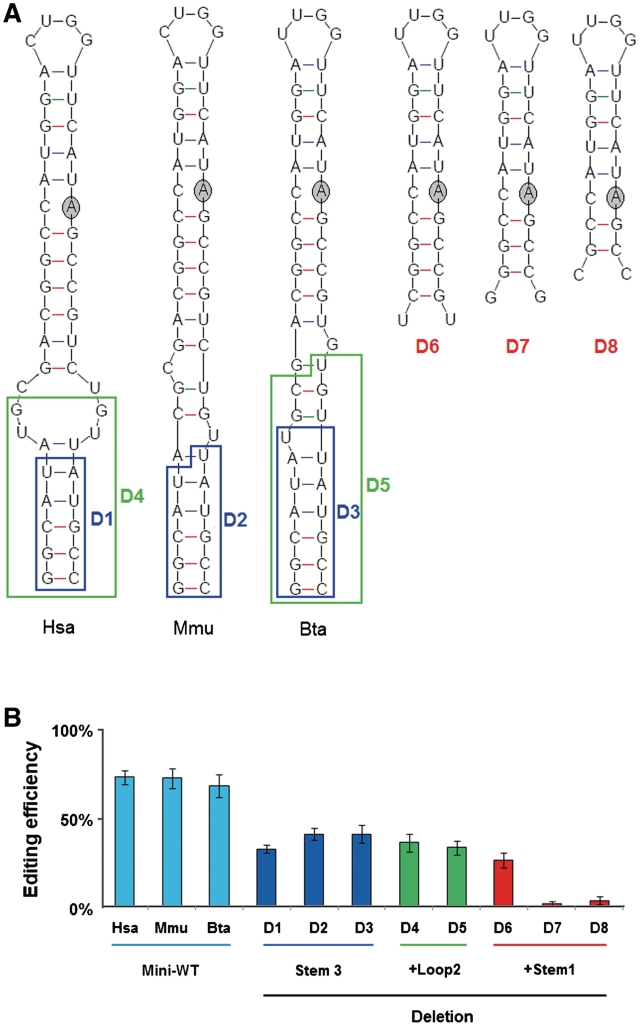
To dissect the role of the stem–loop structure for RNA editing, we synthesized a number of mutant constructs and compared the amount of I/M site editing between the mutants and WT using an in vivo assay (Figure 3). As predicted, most of the mutations that changed 1 bp to another within the stem (i.e. compensatory mutations; Figure 3) had little effect on editing at the I/M site. In contrast, all mutations in Stems 1 or 2 that changed a base pair to a mismatch reduced editing efficiency to almost background levels supporting our prediction (Figure 3). Mutations in Stem 3 did not abrogate editing but only led to a modest reduction (Figure 3). This indicates that the conserved Stem 3 is not obligatory for I/M editing, but can increase editing efficiency. To further elucidate the structural requirements of Stem 3 for selective editing, several constructs were created where either Stem 3 alone or in combination with the variable region was deleted. The resulting 11-bp or 10-bp constructs lacking Stem 3 (and sometimes the variable region) still showed editing, albeit at much reduced efficiency (Figure 4B). Surprisingly, construct D6 only containing 9 bp still showed editing (>25%) (Figure 4B). It is noteworthy that this construct is much shorter than processed siRNAs.
Figure 3.
Sequence and structural elements in hGabra3 required for editing. Predicted RNA secondary structure of the 46-nt hGabra3 substrate. Mutations engineered into expression constructs (M1−M29) were designed to disrupt base pairing interactions or, in combination, to restore base-pairing while changing the identity of base-pairing partners. The mutations (red) and their resultant effects on the predicted duplex structure are shown beside the large structure for each mutant. The edited adenosine is circled in grey. Editing levels of the various constructs was determined by nuclear injection of Xenopus oocytes. Editing levels were quantified by a restriction digestion of the editing site. Data are expressed as mean ± SD from three independent experiments. Mutations in Stems 1 and 2 surrounding the editing site have a strong impact on editing, while mutations in Stem 3 have no effect on editing efficiency.
Figure 4.
Stem 3 is dispensable for RNA editing. The impact of deletions of Stem 3 on RNA editing was tested in various deletion constructs shown in (A). Editing levels of the various WT and mutant transcripts were determined by nuclear injection of Xenopus oocytes followed by RT–PCR and digestion with enzymes diagnostic for the editing site (B). Hsa: Homo sapiens; Mmu: Mus musculus; Bta: Bos taurus. Data are expressed as mean ± SD from three independent experiments.
Comparative analysis indicate that the U/CUCG loop is evolutionarily conserved and C or U is invariantly located at the Y22 position (Figure 2D and E), as confirmed by point mutation. In contrast, the mutations at U23, G24, G25 could markedly reduce editing efficiency (Figure 3). Similarly, deletion or insertion mutations could markedly reduce editing efficiency (Figure 3). Binding of ADAR2 to a GCUMA pentaloop was previously found at the R/G editing site of glutamate receptor subunit B (GluR-B) (23). Our results indicate that U/CUCG terminal loops can play an important role in effective RNA editing. As predicted, sequence context of the edited site is important and changes in the surrounding could markedly reduce editing efficiency (Figure 3). Moreover, introducing complete base pairing in the variable region by mutation reduced editing levels below half of WT levels (Supplementary Figure S3). Together, these results indicate that this smart mini-dsRNA structure, with one A–C mismatch and capped by a tetraloop, is required for efficient editing.
A mimicking duplex structure fails to undergo RNA editing
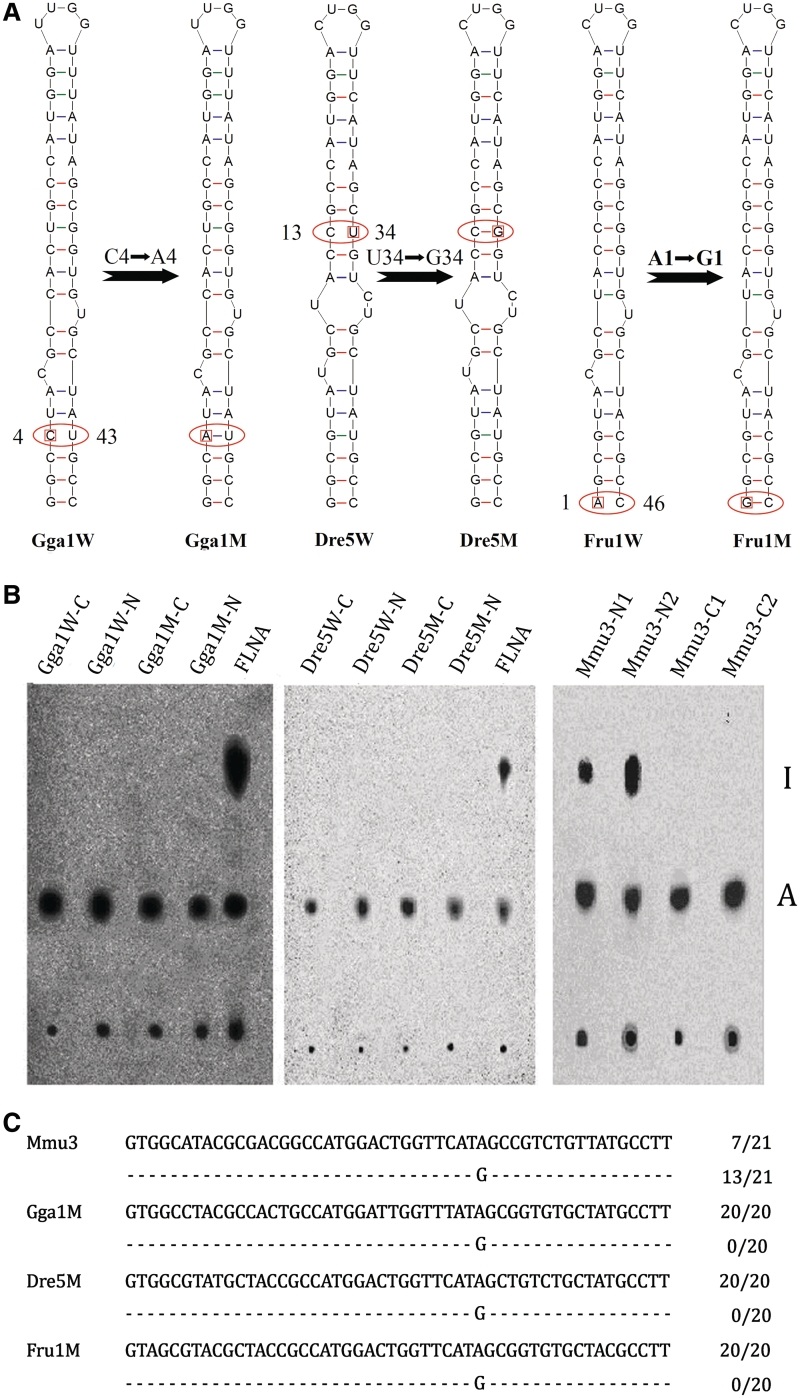
Although we failed to find a stem–loop structure identical to Gabra3 in other Gabra transcripts, a very similar structure is predicted to form in chicken Gabra1 RNA; except for a N4–Y43 mismatch at Stem 3, which represents a natural base pair disruption in Stem 3 (Figure 5A). We also identified naturally occurring structural mutants with a N1–N46 mismatch in Stem 3 of Fugu rubripes Gabra1, and with a N13–N34 mismatch in Stem 2 of Danio rerio Gabra1 (Figure 5A). RNA editing failed to be detected in these transcripts.
Figure 5.
Paired stem regions are not sufficient for editing. (A) Schematic diagrams of WT and mutant (M) minigene constructs tested in oocyte editing assays. A single synonymous change was introduced into chicken Gabra1, D. rerio Gabra5 and F. rubripes Gabra1, converting a mismatch to a paired state. Thus, the mutated stem–loop structures resemble the editing structural rearrangements seen in the Gabra3. The resulting minigenes were evaluated for editing efficiency using Xenopus nuclear injection. (B) Radioactively labeled RNA was injected into X. laevis oocyte nuclei. Injected RNAs were isolated and submitted to P1 digestion and nucleotides were separated on TLC plates. No detectable of editing activity was observed in both Xenopus nuclear (N) and cytoplasm (C), while very strong editing signal was observed in both control constructs (mouse Gabra3 and FLNA). (C) Sequencing further confirmed efficient specific editing at the I/M site in mGabra3 minigene but not in other mutants.
We next investigated whether these natural variants could undergo RNA editing after restoring base pairing by point mutation. A single synonymous change was introduced in chicken Gabra1, converting a C4–U43 mismatch into an A4–U43 paired state (Figure 5A). Thus, the mutated stem–loop structure resembles mammalian Gabra3 (Figure 5A). In vitro synthesized RNA of the resulting minigene was tested for editing by microinjection in Xenopus nuclei. Unexpectedly, no editing was detectable in either Xenopus nuclei or cytoplasms (Figure 5B), while a very strong editing signal was observed in both control constructs (mouse Gabra3 and FLNA). Sequencing confirmed efficient specific editing at the I/M site in the mouse Gabra3 minigene (Figure 2). We also introduced a single synonymous change into D. rerio Gabra5 minigene, converting the C13–U34 unpaired state at Stem 2 into a C13–G34 paired state (Figure 5A). However, similar to the chicken Gabra1 mutant construct we also failed to detect RNA editing in this construct (Figure 5). Also the F. rubripes Gabra1 mutant, converting the A1–C46 unpaired state at Stem 3 into a G1–C46 paired state failed to be edited in the oocyte injection system (Figure 5). To exclude the possibility that sequence preference was responsible for these differences, the conserved stems (Stems 1–3) were further mutated to be nearly or even completely identical to mammalian gabra3 (Supplementary Figure S2). However, the resulting constructs still failed to restore editing. In contrast, only mutations in combination with a substitution in a non-conserved region could raise editing to levels to those of mammalian Gabra3 (Supplementary Figure S2). In fact, such a chicken Gabra1 mutant (Gga–M14) structurally resembles mouse Gabra3. This indicates that chicken Gabra1 might be only edited when the sequence of the variable region totally resembles mouse or human Gabra3. Therefore, these data suggest that the non-conserved region may play an important role in modulating RNA editing.
A structural switch at the non-conserved region for RNA editing
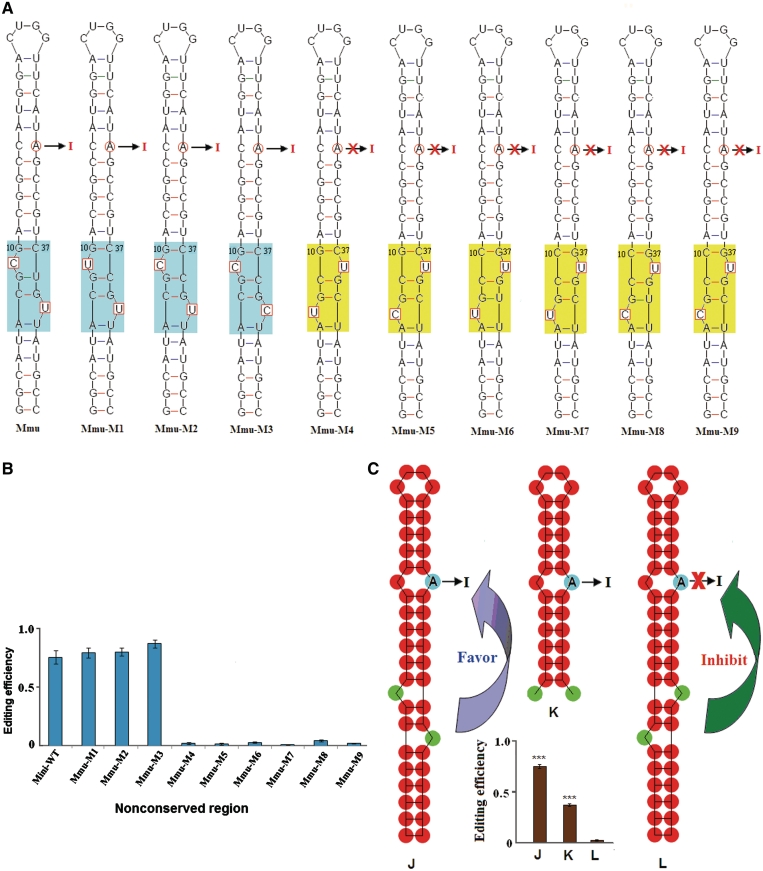
Based on these results the obvious question arose, why no editing could be detected in the transcripts of chicken Gabra1, zebrafish Gabra5 and the mutated constructs (Figure 5). The only difference between Gabra3, and the mutated chicken Gabra1, and zebrafish Gabra5 could be found in the variable region (Figures 2 and 5). To examine whether the sequence at the non-conserved region might affect I/M editing, a series of constructs was analysed (Figure 6A and B). The predicted secondary structures were verified by chemical structure mapping (Figure 7). Surprisingly, only background levels of editing were observed in two constructs where the viable region was altered (Mmu–M4, Mmu–M5) (Figure 6A and B). Comparison of the variable region between editable and uneditable constructs indicates the presence of two bulges, which are separated by 2 bp in most cases, in an asymmetric orientation (Figures 6C and 7). In editable constructs, a bulge (Y9) is located on the 5′ strand in a distance of 6 bp to the editing site A31 and another bulge is observed at position Y40. In contrast, a bulged Y38, which is located on the 3′ strand in a distance of 6 bp to A31, interferes with editing. In these uneditable constructs, Y7 is typically unpaired as well. It is, however, worth mentioning that reducing the distance between the 3′ bulge and the editing site to 5 bp allows efficient editing, as it is the case for Bta Gabra3 (Figures 2D and 4). Interestingly, a relatively open loop, as the one found in the human WT sequence, did not interfere with editing. Moreover, perfect pairing in the variable region introduced by mutation could lead to a decrease in editing below half of WT level (Supplementary Figure S2). Seemingly, the structure of the variable region sets a switch for I/M editing.
Figure 6.
RNA editing is modulated by a structural switch at the non-conserved region. (A) Predicted RNA secondary structure of the 46-nt Gabra3 substrates and mutants. A series of mutants was analysed where nucleotides at the non-conserved region were mutated (enclosed in square). In all cases, the RNA architecture is identical except for the variable region (shaded in colour). The structural shape at the variable region is important for editing. Edited architectures are shaded in blue, while unedited architectures are shaded in yellow. A red cross marks the failure to detect RNA editing (<5%). (B) Editing levels of the various mutant transcripts after injection Xenopus oocyte nuclei. Data are expressed as mean ± SD from three independent experiments. (C) A structural switch at the non-conserved region is a determinant for editing. All examined substrates could be confined to two different shapes. A proximal bulges on the same strand of the edited A inhibits editing, while a distant bulge on the same strand is compatible with editing. ***P < 0.001 compared with each other (using the Student's t-test).
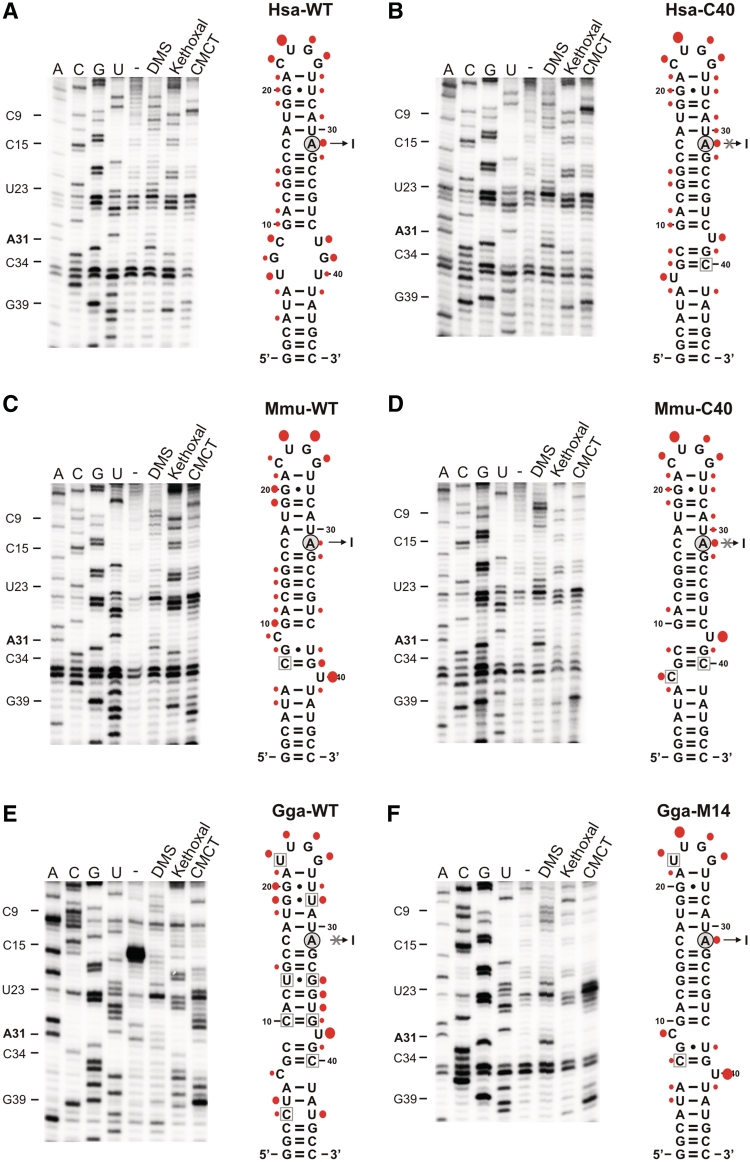
Figure 7.
Structural probing of editing substrates in vitro. Left panels: representative primer extension gels showing the in vitro modification pattern of the GABRA editing substrates of H. sapiens WT (A), H. sapiens mutant C40 (B), Mus musculus WT (C), Mus musculus mutant C40 (D), Gallus gallus WT (the sequence of Gabra1; E) and Gallus gallus mutant 14 (F). A, G, U and C denote sequencing lanes. In the lane ‘–’, natural stops of the reverse transcriptase are shown to detect natural stops of the extension (independent of DMS, CMCT or Kethoxal modification; RNA was folded, but not incubated with either reagent). Lanes labelled DMS: the in vitro DMS modification pattern is shown. DMS methylates A-N1 and C-N3, if they are not involved in H-bonding. Lanes labelled Kethoxal: the in vitro Kethoxal modification pattern is shown. Kethoxal modifies G-N1 and G-N2, if they are not involved in H-bonding. Lanes labelled CMCT: the in vitro CMCT modification pattern is shown. CMCT modifies G-N1 and U-N3, if they are not involved in H-bonding. Right panels: Modifications were plotted onto the secondary-structure maps of respective editing substrates. Red filled circles indicate the detected modification patterns, whereby the size of the filled circles correlates with the relative modification intensity of individual bases. Residues boxed in grey (B–F) indicate base changes relative to the H. sapiens WT editing substrate (A).
Editable WT human and mouse gabra3 and their mutated, uneditable counterparts were tested for structural differences by chemical probing. Also, WT uneditable chicken and the editable mutant 14 of chicken Gabra1 were subjected to chemical probing. As predicted, in mutant and WT mammalian Gabra3, the strongest difference could be observed in the variable region. As predicted a proximal unpaired nucleotide on the strand harbouring the edited adenosine (U39) seemingly interferes with editing. Interestingly, a rather open structure as in the human WT construct does not interfere with editing. In chicken gabra1, the uneditable WT structure showed a very flexible and open conformation at the base of the stem. The editable mutant (Gga–M14), in contrast, was much more protected in this region. Structural probing (Figure 7) confirmed that a 5′ unpaired residue (Y9) is indeed present in the editable substrate RNAs together with a bulged Y40, while base pairing of Y9 and Y40 with nucleotides 39 and 9, respectively, and abolishes editing.
Temperature-mediated switch of RNA editing
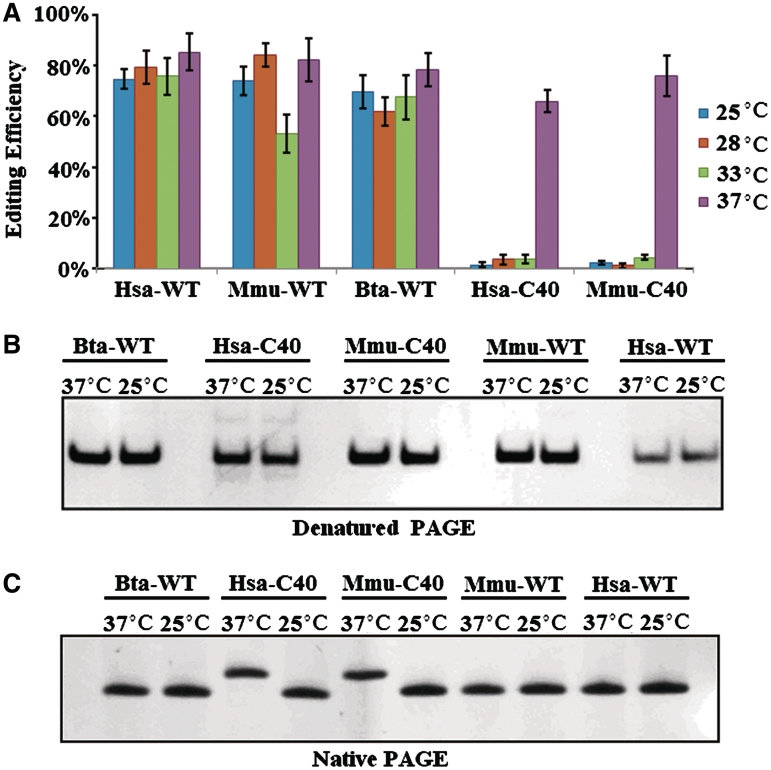
Proper folding of the editing stem–loop might also be affected by temperature. Therefore, to test the role of the variable region on temperature-dependent folding, we performed a temperature control experiment in which editing of the I/M site was monitored. As a result, three WT-constructs could efficiently undergo RNA editing at 25–37°C. Interestingly, mutant minigenes (Hsa–C40, Mmu–C40), which could not undergo RNA editing at 25–33°C, could undergo RNA editing at elevated temperatures (Figure 8A). These results also indicate that editing of WT-constructs is relatively inert to temperature changes.
Figure 8.
Temperature-mediated switch of RNA editing. (A) The editing level under different temperatures. Difference between the secondary structures of folded RNA ranging from 25°C to 37°C. Data are expressed as mean ± SD from three independent experiments. (B and C) RNA conformation influenced by different temperatures. The RNAs folded under different temperature were visualized in denatured (B) and native (C) PAGE (16%) by sliver staining. Mutants (Has–C40, Mmu–C40) exhibited the same mobility in denatured PAGE at 25 and 37°C, while it ran with different mobility under native PAGE.
To further test whether RNA structures change between 25 and 37°C, we first compared electrophoretic mobility of the editing loop in native and denaturing PAGE gels. As expected, three WT-constructs exhibited identical electrophoretic mobility in 25 and 37°C, both in denaturing and native PAGE (Figure 8A). Although Hsa–C40 mutants folded at 37 and 25°C exhibited identical electrophoretic mobility in the denaturing PAGE (Figure 8B), they exhibited different electrophoretic mobility in native PAGE (Figure 8C). Mmu–C40 mutants behaved similarly (Figure 8B and C). This suggests that these mutants can assume two distinct secondary and perhaps tertiary structures from the same sequence at 37 and 25°C. Considering the correlation between structure and RNA editing, this structural alteration might lead to switching RNA editing on and off. Furthermore, these results also indicate that WT-constructs have more evolutionary advantage by resisting changes in editing by temperature.
DISCUSSION
A-to-I editing: beyond RNA secondary structure
Phylogenetic and mutagenesis analyses revealed that duplex structures are required for efficient RNA editing in all substrates such as gluR (24), sytI (25) or ADAR2 (26). Except for the high conservation of the editing site complementary sequence (ECS), there seems to be poor phylogenetic conservation of the primary sequence aside from its ability to form a dsRNA (24). However, the mechanisms underlying selective editing are still largely unknown (27). For example, some gluR-C introns are conserved beyond the sequences required for editing (24). Despite the 40 million years of divergence nucleotides in the edited exon of the sytI genes are invariant within 12 Drosophila species, although not all nucleotides are within the duplex structures required for editing (25). We find that there are much lower GC-content and higher Gibbs free energy in edited exons than in other exons in the Drosophila sytI and other 47 transcripts (28). A previous study also suggests that the 3D structure is important for ADAR substrate recognition and coupling of A-to-I edited sites (29). We analysed and compared the constructs which contained similar RNA pairing structure but tertiary structural variation and found that those have different editing fate. A very recent study reveals structural and sequence requirements of the R/G editing site of GluR-2 for ADAR2 dsRBM binding (30). Similarly, our study indicates that the single nucleotide at Y40 position could play an important role in effective RNA editing by introducing a minor change in shape while leaving the overall stem–loop structure unaltered (Figure 6). Moreover, elevated folding temperature could switch RNA editing, which corresponded with RNA structure change from the same sequence. This finding is interesting as it suggests a different mechanism of editing control for homoiotherms and poikilotherms.
A smart mini-duplex substrate for editing
Previous studies indicate that efficient A-to-I editing occurred with dsRNAs of >23–30 bp in length (8). The crystal structure of a monomeric dsRNA binding domain complexed with dsRNA suggested that a minimum of 16-bp of dsRNA may be sufficient for dsRBD–dsRNA interaction (31). Consistently, a minimal size substrate consisting of a 15-bp stem with a single A–C mismatch could undergo efficient editing (27). Here we define a substrate shorter than 10-bp, with one A–C mismatch and capped by a tetraloop, for proper editing. This represents the shortest substrate for A-to-I editing yet reported in animals. Considering that quite a few mutations not only reduced but turned on/off editing, this duplex is an excellent model to examine the effect of sequence and structure on editing.
Our finding that ADAR can edit such small dsRNA substrates is of particular interest, considering the fact that the RNase III-like ribonuclease Dicer processes long dsRNAs to about 22 bp siRNAs (32). Recent findings point to an intimate interplay between the RNAi and RNA editing pathways (4,33–37). RNA editing greatly reduces the production of siRNAs and consequently antagonizes RNAi effects in vitro (37). Moreover, ADAR certainly can affect RNAi efficacy through A-to-I editing of dsRNA in vivo (35,36). Recent studies identify ADAR1 as a cellular factor that limits the efficacy of siRNA in mammalian cells (38). Notably, ADAR1 is induced by interferon, raising the possibility that this cytokine can modulate RNAi responses (39,40). Our finding therefore suggests that siRNAs could be potential targets for A-to-I editing.
Exon-directed recoding: regulatory hidden layers
Redundancy of the genetic code would allow more than six million DNA isoforms to encode 16 identical Gabra amino acids. The protein-coding region of an mRNA can thus contain one or more overlapping layers of information that modulate gene expression. Apart from the well-known alternative splicing signals, the overlapping RNA secondary structure can contribute to ADAR-mediated recoding of the amino acid sequence. Our results indicate that a single synonymous substitution might result in a nearly complete loss of editing (Figure 6).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China (31071148, 30770469, partial); Natural Science Foundation of Zhejiang Province (No. R3090177); Doctor Foundation of China (J20070724); National Science and Technology Project (2009ZX09103-694); Program for New Century Excellent Talents in University (NCET-04-0531). Austrian Science Foundation (SFB1706 to M.F.J.); Austrian Science Foundation (grant no. Y401 to C.W.). Funding for open access charge: Research grants.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maas S, Rich A, Nishikura K. A-to-I RNA editing: recent news and residual mysteries. J. Biol. Chem. 2003;278:1391–1394. doi: 10.1074/jbc.R200025200. [DOI] [PubMed] [Google Scholar]
- 3.Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 4.Palladino MJ, Keegan LP, O'Connell MA, Reenan RA. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–449. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- 5.Wang Q, Khillan J, Gadue P, Nishikura K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- 6.Tonkin LA, Saccomanno L, Morse DP, Brodigan T, Krause M, Bass BL. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J. 2002;21:6025–6035. doi: 10.1093/emboj/cdf607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoopengardner B, Bhalla T, Staber C, Reenan R. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301:832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- 8.Nishikura K, Yoo C, Kim U, Murray JM, Estes PA, Cash FE, Liebhaber SA. Substrate specificity of the dsRNA unwinding/modifying activity. EMBO J. 1991;10:3523–3532. doi: 10.1002/j.1460-2075.1991.tb04916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong SK, Sato S, Lazinski DW. Substrate recognition by ADAR1 and ADAR2. RNA. 2001;7:846–858. doi: 10.1017/s135583820101007x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lehmann KA, Bass BL. The importance of internal loops within RNA substrates of ADAR1. J. Mol. Biol. 1999;291:1–13. doi: 10.1006/jmbi.1999.2914. [DOI] [PubMed] [Google Scholar]
- 11.Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 2005;309:1534–1539. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linnstaedt SD, Kasprzak WK, Shapiro BA, Casey JL. The fraction of RNA that folds into the correct branched secondary structure determines hepatitis delta virus type 3 RNA editing levels. RNA. 2009;15:1177–1187. doi: 10.1261/rna.1504009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohlson J, Pedersen JS, Haussler D, Ohman M. Editing modifies the GABA(A) receptor subunit alpha3. RNA. 2007;13:698–703. doi: 10.1261/rna.349107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rula EY, Lagrange AH, Jacobs MM, Hu N, Macdonald RL, Emeson RB. Developmental modulation of GABA(A) receptor function by RNA editing. J. Neurosci. 2008;28:6196–6201. doi: 10.1523/JNEUROSCI.0443-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wahlstedt H, Daniel C, Enstero M, Ohman M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009;19:978–986. doi: 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 18.Nimmich ML, Heidelberg LS, Fisher JL. RNA editing of the GABA(A) receptor alpha3 subunit alters the functional properties of recombinant receptors. Neurosci. Res. 2009;63:288–293. doi: 10.1016/j.neures.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liebeg A, Waldsich C. Probing RNA structure within living cells. Methods Enzymol. 2009;468:219–238. doi: 10.1016/S0076-6879(09)68011-3. [DOI] [PubMed] [Google Scholar]
- 21.Hallegger M, Taschner A, Jantsch MF. RNA aptamers binding the double-stranded RNA-binding domain. RNA. 2006;12:1993–2004. doi: 10.1261/rna.125506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian N, Wu X, Zhang Y, Jin Y. A-to-I editing sites are a genomically encoded G: implications for the evolutionary significance and identification of novel editing sites. RNA. 2008;14:211–216. doi: 10.1261/rna.797108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stefl R, Allain FH. A novel RNA pentaloop fold involved in targeting ADAR2. RNA. 2005;11:592–597. doi: 10.1261/rna.7276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aruscavage PJ, Bass BL. A phylogenetic analysis reveals an unusual sequence conservation within introns involved in RNA editing. RNA. 2000;6:257–269. doi: 10.1017/s1355838200991921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reenan RA. Molecular determinants and guided evolution of species-specific RNA editing. Nature. 2005;434:409–413. doi: 10.1038/nature03364. [DOI] [PubMed] [Google Scholar]
- 26.Dawson TR, Sansam CL, Emeson RB. Structure and sequence determinants required for the RNA editing of ADAR2 substrates. J. Biol. Chem. 2004;279:4941–4951. doi: 10.1074/jbc.M310068200. [DOI] [PubMed] [Google Scholar]
- 27.Herbert A, Rich A. The role of binding domains for dsRNA and Z-DNA in the in vivo editing of minimal substrates by ADAR1. Proc. Natl Acad. Sci. USA. 2001;98:12132–12137. doi: 10.1073/pnas.211419898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao J, Wu X, Jin Y. Lower GC-content in editing exons: implications for regulation by molecular characteristics maintained by selection. Gene. 2008;421:14–19. doi: 10.1016/j.gene.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 29.Enstero M, Daniel C, Wahlstedt H, Major F, Ohman M. Recognition and coupling of A-to-I edited sites are determined by the tertiary structure of the RNA. Nucleic Acids Res. 2009;37:6916–6926. doi: 10.1093/nar/gkp731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stefl R, Oberstrass FC, Hood JL, Jourdan M, Zimmermann M, Skrisovska L, Maris C, Peng L, Hofr C, Emeson RB, et al. The solution structure of the ADAR2 dsRBM-RNA complex reveals a sequence-specific readout of the minor groove. Cell. 2010;143:225–237. doi: 10.1016/j.cell.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 33.Heale BS, Keegan LP, McGurk L, Michlewski G, Brindle J, Stanton CM, Caceres JF, O'Connell MA. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J. 2009;28:3145–3156. doi: 10.1038/emboj.2009.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nature Rev. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tonkin LA, Bass BL. Mutations in RNAi rescue aberrant chemotaxis of ADAR mutants. Science. 2003;302:1725. doi: 10.1126/science.1091340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knight SW, Bass BL. The role of RNA editing by ADARs in RNAi. Mol. Cell. 2002;10:809–817. doi: 10.1016/s1097-2765(02)00649-4. [DOI] [PubMed] [Google Scholar]
- 37.Scadden AD, Smith CW. RNAi is antagonized by A-->I hyper-editing. EMBO Rep. 2001;2:1107–1111. doi: 10.1093/embo-reports/kve244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang W, Wang Q, Howell KL, Lee JT, Cho DS, Murray JM, Nishikura K. ADAR1 RNA deaminase limits short interfering RNA efficacy in mammalian cells. J. Biol. Chem. 2005;280:3946–3953. doi: 10.1074/jbc.M407876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iizasa H, Nishikura K. A new function for the RNA-editing enzyme ADAR1. Nature Immunol. 2009;10:16–18. doi: 10.1038/ni0109-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartner JC, Walkley CR, Lu J, Orkin SH. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nature Immunol. 2009;10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.