Abstract
Restriction–modification systems consist of a modification enzyme that methylates a specific DNA sequence and a restriction endonuclease that cleaves DNA lacking this epigenetic signature. Their gene expression should be finely regulated because their potential to attack the host bacterial genome needs to be controlled. In the EcoRI system, where the restriction gene is located upstream of the modification gene in the same orientation, we previously identified intragenic reverse promoters affecting gene expression. In the present work, we identified a small (88 nt) antisense RNA (Rna0) transcribed from a reverse promoter (PREV0) at the 3′ end of the restriction gene. Its antisense transcription, as measured by transcriptional gene fusion, appeared to be terminated by the PM1,M2 promoter. PM1,M2 promoter-initiated transcription, in turn, appeared to be inhibited by PREV0. Mutational inactivation of PREV0 increased expression of the restriction gene. The biological significance of this antisense transcription is 2-fold. First, a mutation in PREV0 increased restriction of incoming DNA. Second, the presence of the antisense RNA gene (ecoRIA) in trans alleviated cell killing after loss of the EcoRI plasmid (post-segregational killing). Taken together, these results strongly suggested the involvement of an antisense RNA in the biological regulation of this restriction–modification system.
INTRODUCTION
Restriction–modification (R–M) systems are found in many prokaryotes (1). The great abundance of R–M systems in prokaryotes reflects their mobility via transformation, transduction or conjugation (2,3). Many R–M systems are present on plasmids and other mobile elements and can spread from one bacterial host to another, sometimes crossing species boundaries and impacting genome evolution on a global scale (4–6). R–M systems consist of a modification enzyme that methylates a specific DNA sequence in a genome and a restriction endonuclease that cleaves DNA lacking this methylation. They are recognized as bacterial defense systems against invading DNAs, but their biological significance extends far beyond this (7).
R–M systems may represent one form of life, just as other mobile elements (8). Under some conditions, they may switch on bacterial death programs (8,9). Several type II R–M systems can program the death of a cell lineage once it has lost the R–M system (10,11). This is similar to the classical post-segregational host cell killing systems on plasmids (12–14). This post-segregational killing or genetic addiction helps in the maintenance and spread of the R–M system (6,11,15). This suggests that some R–M systems behave as selfish mobile elements, similar to viral genomes and transposons. R–M systems have also been linked with various genome rearrangements (6). Recent studies revealed that modification enzymes methylate many copies of a recognition sequence along the genome and define the epigenetic status of that genome in a combinatorial fashion, which may promote adaptation (16). R–M systems mediate DNA selection during horizontal gene transfer, serving as gene flow ‘gatekeepers’, using epigenetic signs as identification.
R–M systems are thought to possess mechanisms to tightly regulate their gene expression to suppress potentially lethal attacks on their host bacteria. When they enter a new host bacterial cell with a genome lacking proper methylation, they avoid cell killing by expressing the modification enzyme first (17). The restriction endonuclease and modification enzyme activities must be carefully balanced not only at the stage of R–M system establishment in a new host, but also during maintenance of this system. When R–M genes are lost from the cell, the restriction enzyme will attack the chromosome in an act of post-segregational killing.
Their regulatory machinery is expected to be host-independent to bypass differences in host factors affecting their establishment, maintenance and host attack. To date, these regulatory mechanisms have mostly been studied at the transcriptional level. Three main modes of regulation have been distinguished. One employs C-proteins (18), which specifically bind a DNA operator sequence through a helix–turn–helix (HTH) motif to temporally control the expression of the restriction enzyme, modification enzyme or both (19,20). This appears to be a tight, finely tuned regulation mechanism, operating via transcriptional feedback circuits (21). Moreover, C proteins can efficiently delay expression of the restriction enzyme during establishment in a new host cell (17,22). In the second type of regulation, the modification enzyme represses transcription of its own gene and sometimes stimulates expression of a restriction gene through DNA binding via its HTH domain (23–25). In the third mode, the coordinated expression of R–M systems depends on the methylation status of the cognate recognition site(s) within their promoter region (26,27).
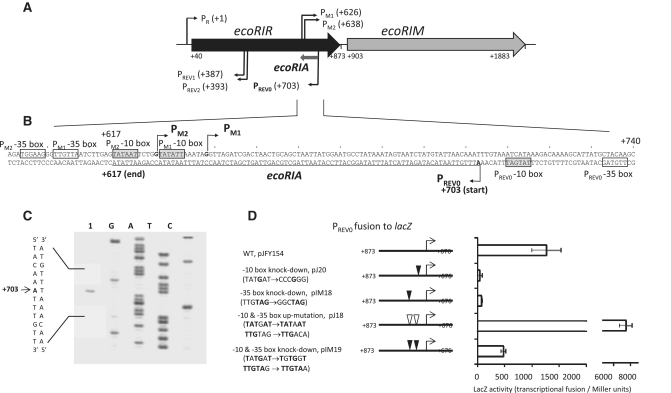
In the EcoRI R–M system, where two genes form a linearly oriented operon, the restriction endonuclease gene (ecoRIR) precedes its modification enzyme gene (ecoRIM) (Figure 1A). We previously identified functional promoters for restriction and modification genes (PR, PM1,M2) (Figure 1A and B) (28,29). We also identified a composite reverse promoter, PREV1,REV2 within the restriction gene (Figure 1A and B), associated with the negative regulation of the ecoRIR gene, and likely the ecoRIM gene (28,29).
Figure 1.
The antisense RNA (Rna0) encoded by the ecoRIA gene. (A) EcoRI restriction–modification system. Coordinates are relative to the transcription initiation site for the bicistronic ecoRIRM mRNA from PR promoter. PM1 and PM2, two overlapping promoters for ecoRIM transcription (28); PREV1 and PREV2, two overlapping reverse promoters associated with negative effects on expression from the PR promoter (29); PREV0, a reverse promoter for an antisense RNA studied in this report. (B) ecoRIA antisense RNA gene. Transcription initiation sites are indicated by arrows. The ecoRIA sequence is underlined. Promoter hexamers are boxed in gray (−10 box) and white (−35 box) (11, this work). (C) Mapping the transcription initiation site by primer extension. Extension from a lacZ gene-specific primer (lacP) (Supplementary Table S1) was carried out using total RNA from E. coli harboring pJFY154 (lane 1, see below). Lanes G, A, T and C represent products of the dideoxy sequencing reactions carried out with the same primer. The initiation site is indicated by an arrow. (D) Promoter activity of PREV0. DNA fragment carrying PREV0 was fused to the lacZ gene (pJFY154). Substitutions were made at the −35 (pIM18) or −10 box (pJ20) of the PREV0 promoter to knock down promoter activity, as well as to increase its activity, by matching them to the consensus sequences for E. coli σ70 RNA polymerase (pJ18). PREV0 activity with the 3 nt changes present in pIM24 (Table 1) was also measured (pIM19) (see Figure 4 below). These changes did not affect the amino acid sequence. Each value represents an average of four measurements, along with their standard deviations.
One of the possible mechanisms underlying the action of the reverse promoter is the transcription of a small antisense RNA. In prokaryotes, small noncoding RNAs are involved in numerous cellular responses to changing environments. The largest group of RNAs studied act by base-pairing with a target RNA to modulate translation initiation or to promote RNA degradation (30–33). A proportion of the small RNAs acting preferentially in cis were discovered mainly in plasmids, phages and transposons (34).
In this work, we discovered another intragenic reverse promoter near the 3′ end of the restriction gene and identified a small antisense RNA transcribed from this region. We demonstrate roles of this antisense RNA gene in the regulation of transcription in this R–M system and in the attack on invading DNA and on host bacteria.
MATERIALS AND METHODS
Bacterial strains and plasmids
Escherichia coli K-12 strains used in this study included MG1655 (35) (from Don Biek, National Cancer Institute, NIH, Bethesda, USA) for the post-segregational cell killing assay and MC1061 [araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)X74 rpsL hsdR2 mcrA mcrB1] (36) for the measurement of LacZ activity. Strain DH10B [F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacΧ74 recA1 DeoR araD139 Δ(ara-leu)7697 galU galK rpsL (StrR) endA1 nupG] (Invitrogen) was used for plasmid construction.
Construction of the promoter-less reporter, pLY2, was reported previously (28,29). All the lacZ transcriptional fusions were constructed in a similar way (Table 1). Corresponding regions of the EcoRI operon from pIK163 (10) were amplified by PCR (primer list in Supplementary Table S1) using KOD plus polymerase (Toyobo), and PCR products were digested with XbaI and cloned into pLY2 linearized with XbaI. All of the substitution mutants were constructed by the megaprimer method (37) and their sequences were confirmed.
Table 1.
Plasmids
| Name | Relevant features/genotype | References |
|---|---|---|
| pIK163 | pBR322 carrying ecoRIRM operon | (10) |
| pHSG415 | pSC101 derivative, thermo-sensitive replication, KanR ApR, CmR | (41) |
| pIK172 | pHSG415 carrying the WT ecoRIRM, ApR, CmR | (11) |
| pIK173 | pHSG415carrying ecoRIR–M+, ApR, CmR | (11) |
| pGEM-T | E. coli TA cloning vector, ApR | Promega |
| pGEM-T2 | pGEM-T carrying an ecoRIRM operon fragment (+431 – +612) | (29) |
| pGEM-T4 | pGEM-T carrying an ecoRIRM operon fragment (+536 – +777) | (29) |
| pLY2 | pACYC184 carrying promoter-less lacZ gene | (29) |
| pLY66 | Transcriptional fusion of a WT ecoRIRM operon fragment with PR promoter (−67 to +400) fused to lacZ in pLY2 | (29) |
| pBLY18 | pJFY161 derivative, double mutations in −10 boxes of PM1M2 promoters (TATAAT to TAGCGG and TATATT to TAAAGC) | This study |
| pJFY35 | Transcriptional fusion of a WT ecoRIRM operon fragment (+873 to +660) to lacZ in pLY2 | This study |
| pJFY47 | Transcriptional fusion of a WT ecoRIRM operon fragment (+873 – +474) to lacZ in pLY2 | This study |
| pJFY153 | Transcriptional fusion of a WT ecoRIRM operon fragment (+873 to +535) to lacZ in pLY2 | This study |
| pJFY154 | Transcriptional fusion of the WT ecoRIRM operon fragment (+873 to +676) to lacZ in pLY2 | This study |
| pJFY185 | Transcriptional fusion of a WT ecoRIRM operon fragment (+873 – +633) to lacZ in pLY2 | This study |
| pJFY161 | Transcriptional fusion of a WT ecoRIRM operon fragment (+873 – +585) to lacZ in pLY2 | This study |
| pJ18 | pJFY154 derivative, up mutations in the −10 and −35 boxes of PREV0 (TATGAT→TATAAT; TTGTAG to TTGACA) | This study |
| pJ20 | pJFY154 derivative, down mutations in the −10 box of PREV0 (TATGAT→CCCGGG) | This study |
| pIM13 | Transcriptional fusion of a WT ecoRIRM operon fragment with promoter PM1,M2 (+585 to +784) fused to lacZ in pLY2 | This study |
| pIM15 | Transcriptional fusion of a WT ecoRIRM operon fragment with promoter PM1,M2 (+406 to +873) fused to lacZ in pLY2 | This study |
| pIM16 | Transcriptional fusion of a ecoRIRM operon fragment with PM1/PM2 promoter (+406 to +709) fused to lacZ in pLY2; PREV0 promoter hexamers are not present | This study |
| pIM18 | pJFY154 derivative, mutations in the −35 box of PREV0 (TTGTAG→GGCTAG) | This study |
| pIM19 | pJFY154 derivative, mutations in the −10 and −35 boxes of PREV0 (TATGAT→TGTGGT and TTGTAG→TTGTAA); no change in R.EcoRI amino-acid sequence | This study |
| pIM11 | Transcriptional fusion of a WT ecoRIR fragment with PR and PM1,M2 promoters (−67–+784) fused to lacZ in pLY2 | This study |
| pIM21 | Derivative of pIM11, mutation as in pJ20 to inactivate PREV0 promoter | This study |
| pIM30 | Derivative of pIM11, mutations as in pBLY18 to inactivate PM1,M2 promoter | This study |
| pIMRM | Entire WT EcoRI system in pACYC184 backbone; CmR | This study |
| pIM24 | pIMRM derivative, 3-nt substitutions as in pIM19 in the −10 and −35 boxes of PREV0 (TATGAT→TGTGGT and TTGTAG→TTGTAA); no change in R.EcoRI amino acid sequence | This study |
| pIM27 | pIMRM derivative; R–M+, deletion of HindIII–BglII fragment of ecoRIR | This study |
| pIM-REV0 | pUC18 derivative carrying ecoRIA gene for the antisense RNA (Rna0) from PREV0 promoter; 4-nt up mutations in PM1,M2 reverse promoter (TGGAAG→GGGATG; TTGTTA→AAGTTA) | This study |
| pIM-ΔRNA | pUC18 derivative with a part of ecoRIR gene (−67 – +130), used as a negative control for testing antisense RNA | This study |
R, restriction; M, modification; WT, wild-type; Cm, chloramphenicol; Ap, ampicillin; Kan, kanamycin.
The p15A replicon plasmids carrying the entire EcoRI R–M system and its variants were constructed as follows. The wild-type EcoRI system in pIMRM was amplified by PCR using pIK163 plasmid template and primers Mr and Rf (Supplementary Table S1), then PCR products were digested with HincII and XbaI and ligated to pACYC184 linearized with the same restriction enzymes. Its R−M+ variant (pIM27) was constructed by cleavage of pIMRM with HindIII and BglII, sites present within the ecoRIR gene, followed by T4 DNA polymerase treatment to blunt the overhangs, and finally plasmid self-ligation. The variant of the EcoRI R–M system, pIM24, containing PREV0 promoter down mutations, was constructed by site-directed mutagenesis (Stratagene).
Plasmids to deliver antisense RNA in trans were generated as pUC18 derivatives as follows. Plasmid pIM-REV0 carrying the antisense gene ecoRIA under the PREV0 promoter was constructed by PCR amplification of an ecoRIR gene fragment (+590–+873) with the 1588bf and 1801r primers, followed by XbaI digestion and cloning into XbaI-linearized pUC18. The 1588bf primer introduced 4 bp substitutions into the composite PM1,M2 promoter (−35 hexamers TGGAAG→GGGATG; TTGTTA→AAGTTA) to overcome transcriptional interference and obtain a higher level of antisense RNA. A negative control plasmid carrying no antisense RNA (pIM-ΔRNA) was constructed by cloning part of the ecoRIR gene (the same length DNA fragment) closer to its 5′ end, into the pUC18 vector. The sequences of all of the constructs were confirmed.
LacZ activity measurement
LacZ activity was determined using exponentially growing cells in LB medium at 37°C as previously described (38). Duplicate measurements were obtained using two clones of each strain. Error bars indicate the standard deviation of four measurements.
Preparation of RNA probes
The oligonucleotides used for probes and primers are listed in Supplementary Table S1. The PCR product amplified from pGEM-T4 using the ‘Colef’ and ‘1800r’ primers was used as a template for ‘probe a’ synthesis by T7 RNA polymerase. ‘Probe b’, previously designated as the ‘right probe’ (29), was complementary to ecoRIR mRNA from +431 to +612. The PCR product amplified from pIK163 (ecoRIRM) by the ‘1128f’ and ‘1748r’ primer pair was used as a template for generating a probe with SP6 RNA polymerase to detect the ecoRIR gene by northern blot analysis. Similarly, the PCR product amplified from pIK163 (ecoRIRM) by the ‘1612f’ and ‘1748r’ primer pair was used as a template for generating a probe with SP6 RNA polymerase to detect the ecoRIM gene by northern blot analysis. In vitro transcription with SP6 or T7 RNA polymerase was performed according to the manufacturer's standard transcription protocol (Promega). After 1 h, 10 U of RQ DNase I (Promega) was added to digest the DNA template. The unincorporated ribonucleotides were removed by passage through a Sephadex G-50 column, and transcribed RNAs of the expected sizes were purified by electrophoresis through an 8% polyacrylamide/8 M urea gel.
Total RNA extraction
Escherichia coli MC1061 cells harboring a plasmid of interest were harvested while growing in exponential phase. For all of the small RNA experiments, total RNA was isolated using Trizol reagent (Invitrogen) according to the manufacturer's instructions. In other cases, total RNA was extracted using the RNeasy Protect Bacteria Mini Kit (Qiagen) according to the manufacturer's instructions. After purification, total RNA was treated with DNase I and its concentration and purity was checked by measuring the absorbance at 260/280 nm and by electrophoresis through a 1.2% formaldehyde-agarose gel.
Primer extension
For the primer extension reaction, 20 µg of total RNA was reverse-transcribed with 1 U of AMV Reverse Transcriptase from the Primer Extension System (Promega) in the presence of 1 pmol of [γ-33P] end-labeled primer, according to the manufacturer's protocol. The sequencing reaction with the fmol ®DNA Cycle Sequencing System (Promega) was performed with the same end-labeled primer and template used for the primer extension, according to the manufacturer's protocol. The reaction products were resolved through an 8% sequencing gel, dried and visualized by exposure to a PhosphorImager screen.
RNase protection
Total RNA annealed to a corresponding complementary RNA probe was digested with E. coli RNase I (Promega), with a minor modification to the manufacturer's protocol (39). Briefly, the same quantity of total RNA was ethanol co-precipitated using an excess of purified RNA probe and the same quantity of loading control (a [γ-33P] end-labeled single-stranded DNA probe, see Supplementary Table S1). The precipitate was dissolved, hybridized overnight at 45°C, and digested according to the manufacturer's protocol. Finally, the ‘RNase-protected’ fragments were resolved in a 10% denaturing polyacrylamide gel, dried and visualized by exposure to a PhosphorImager screen or an X-ray film.
Phage restriction
Restriction activity in vivo was estimated by the ability of cells with an R–M system to restrict plaque formation of λvir bacteriophage. The media used for these experiments contained selective antibiotics. Phage titer was determined by plaque formation using the top agar overlay technique (40).
Post-segregational killing
LB media containing two selective antibiotics was inoculated with a single colony of E. coli MG1655 bearing two plasmids. One plasmid carried a thermo-sensitive replication unit with a wild-type EcoRI R–M system (pIK172), its restriction-negative variant (pIK173) or no EcoRI R–M system (vector control) (pHSG415) (41). The other compatible plasmid was a pUC18 derivative carrying a gene (ecoRIA) for the antisense RNA. After 10–12 h of incubation at 30°C, the cultures were diluted and spread quantitatively either on LB agar containing the appropriate selective antibiotics for both plasmids or on LB agar without any antibiotics, and were incubated at 30°C or 42°C, respectively. Subsequently, the colonies were counted and the ratio of colony-forming units under the two conditions was determined. In parallel, a qualitative spot test was carried out by dropping the same volume of diluted culture on to agar plates.
Bioinformatic analyses
In silico promoter prediction and terminator prediction were performed using the BPROM and FindTerm software, respectively (http://www.softberry.com/all.htm). RNA secondary structure was predicted with RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi).
RESULTS
Novel reverse promoter (PREV0) at the 3′-end of the restriction endonuclease gene
Organization of the EcoRI system along with its promoters is shown in Figure 1A. We previously demonstrated that a down-mutation in the composite reverse promoter, PREV1,REV2 within the restriction gene (Figure 1A) can increase expression of the restriction gene from the PR promoter (29). In the present work, we tested another reverse promoter candidate, revealed by in silico analysis, located at the 3′-terminus of the ecoRIR gene (Figure 1A and B). The corresponding DNA fragment (position +873–+676) was placed in front of the promoter-less lacZ gene. The resulting transcriptional fusion indeed showed high activity (pJFY154, Figure 1D). We designated this reverse promoter as PREV0.
To identify the transcription initiation site of this reverse promoter PREV0, a lacZ gene-specific primer (lacP; Table S1) was end-labeled with [γ-33P]ATP. Extension of this primer hybridized to the total RNA from E. coli harboring pJFY154, revealed that the adenine at coordinate +703 functions as the transcription initiation site (Figure 1B and C).
Based on the location of this transcription initiation site and the consensus sequence of E. coli σ70 RNA polymerase promoters (42,43), we deduced −10 and −35 boxes of PREV0 as TATGAT (consensus TATAAT) and TTGTAG (consensus TTGACA), respectively (Figure 1B). An optimal spacer length of 6 bp was found between the putative −10 box and the transcription initiation site. A 17-bp distance between putative −10 and −35 boxes (Figure 1B) is considered optimal in E. coli promoters (43,44).
To verify the functionality of these promoter elements, we generated a set of transcriptional fusion constructs containing mutations (Figure 1D). Down mutations in the putative −10 or −35 boxes greatly decreased transcriptional activity (pJ20 and pIM18, respectively, Figure 1D), whereas up mutations, matching the consensus sequence in both the −10 and −35 boxes, increased activity about 6-fold (pJ18, Figure 1D).
Antisense RNA (Rna0) from the reverse promoter
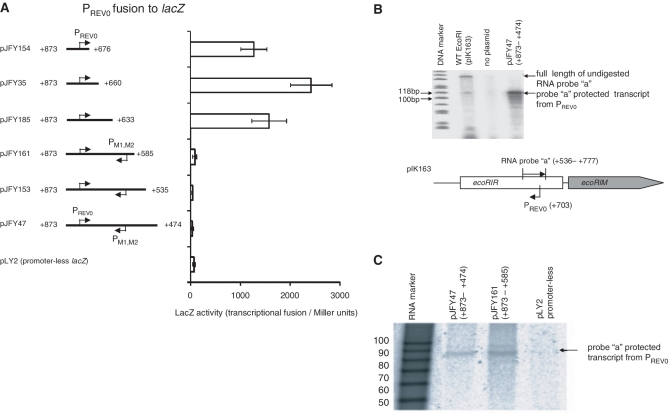
To map the termination site of the antisense RNA from PREV0, PREV0-carrying DNA fragments of increasing downstream length (Figure 2A) were fused to the promoter-less lacZ gene. The sudden drop in lacZ transcription activity between the +873/+633 fragment and the +873/+585 fragment (Figure 2A) suggested termination between +585 and +633, although molecular mechanisms other than termination should also be considered and will be explored in the ‘Discussion’ section.
Figure 2.
Mapping the antisense RNA (Rna0). (A) Mapping by transcriptional fusion. DNA fragments of varying sizes containing PREV0 were cloned upstream of the promoter-less lacZ gene. Each value represents an average of four measurements, along with their standard deviations. (B) RNase protection assay. Lane 1, ΦX174 DNA/HinfI size markers (Promega); lane 2, total RNA from E. coli MC1061 carrying pIK163 (wild-type EcoRI R–M system); lane 3, total RNA from the same strain without a plasmid (negative control); lane 4, total RNA from E. coli MC1061 carrying plasmid pJFY47. The bands below the protected band are likely a result of degradation due to unstable, double-stranded AT-rich ends. A scheme of the RNase protection assay using ‘probe a’, a sense RNA synthesized in vitro, is presented at the bottom of the figure. A map of pJY47 is shown in (A). (C) Length estimation by RNase protection. RNA was detected as in (B). Lane 1, RNA length marker; other lanes, detected RNA in the total RNA of E. coli MC1061 carrying: plasmid pJFY47 (lane 2); plasmid pJFY161 (lane 3); or plasmid pLY2 as the negative control (lane 4). Maps of the promoter fragments are shown in (A).
To detect antisense RNA expressed from the PREV0 promoter in vivo in the context of the EcoRI R–M system, a sensitive RNase protection assay was performed. A 242-nt 32P-labeled RNA probe (complementary from +536 to +777) was synthesized in vitro. The excess probe was hybridized with the total RNA extracted from the cells and cleaved with RNase I to remove RNA overhangs. A specific protected transcript was detected from E. coli harboring a plasmid carrying WT EcoRI R-M system (pIK163) or a plasmid carrying 3′ half of the restriction gene (pJFY47) (Figure 2B).
We compared the lengths of the protected signals in the RNase protection assay between E. coli harboring pJFY161 (+873–+585) and pJFY47 (+873–+470) to define the transcription termination site in detail (Figure 2C). The comparable size of the protected signals revealed that the transcript could not be read through position +585. The estimated length of the antisense RNA was about 88 nt, based on the size of RNA markers (Figure 2C). This result was consistent with the above transcriptional fusion data (Figure 2A) and indicated that the apparent termination site of antisense transcription is around coordinate +617, which is located within the PM1,M2 promoter (Figure 1B).
Neither a start codon nor an open reading frame (ORF) was detected in this RNA, suggesting that this RNA represents a noncoding, small RNA. We designated this antisense RNA gene as ecoRIA (A for antisense) and this RNA as Rna0. We predicted the secondary structure of this RNA (Supplementary Figure S1) and the 5′ terminus of the mRNA transcribed from PM1,M2 (Supplementary Figure S2).
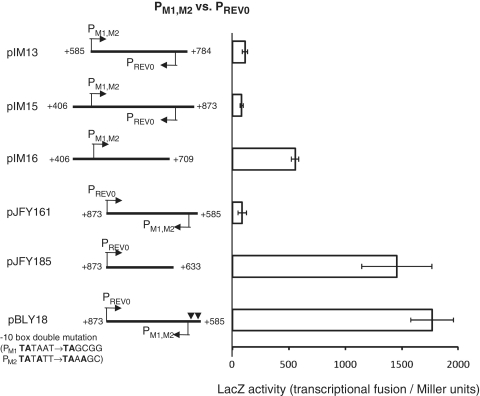
Mutual interference between the PM1,M2 and PREV0 promoters
Termination of antisense transcription from PREV0 occurred at PM1,M2 a composite promoter in the sense orientation (Figure 1B). We did not find any structures resembling Rho-independent-terminators using terminator prediction software. This suggested that the promoter function of PM1,M2 interferes with PREV0-initiated antisense transcription. Indeed, down mutations in the −10 boxes of the PM1,M2 composite promoter increased transcription about 20-fold (pJFY161 versus pBLY18, Figure 3).
Figure 3.
Interference between P M1,2 and PREV0 promoters in transcriptional fusion. Each value represents an average of four measurements, along with their standard deviations.
Because of the symmetry of the two promoters, we examined whether PM1,M2-initiated sense transcription was, in turn, inhibited by the antisense promoter PREV0. When DNA fragments, containing the PM1,M2 promoter, as well as the entire antisense RNA gene, and PREV0 were fused to the promoter-less lacZ gene (pIM13 and pIM15), little activity was detected (Figure 3). Whereas, deletion of the PREV0 promoter (pIM16) increased the activity 7-fold (Figure 3). This suggested that PREV0 function somehow interferes with transcription from PM1,M2.
These in vivo results indicate that the PREV0 reverse promoter action apparently inhibits transcription from the PM1,M2 promoters and vice versa. Possible underlying mechanisms for this will be considered in the ‘Discussion’ section. The net strength of both promoters under inhibition was comparable (pJFY161 versus pIM13, Figure 3), suggesting that they both actively contribute to maintaining fine-tuned coordination of their expression (see ‘Discussion’ section).
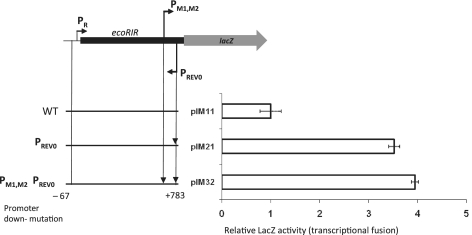
Effect of the reverse promoter mutation on PR activity
To test whether the reverse promoter PREV0 affects ecoRIR expression from PR, we cloned a fragment of WT restriction gene (−67–+783) in front of promoter-less lacZ gene (Figure 4). This restriction endonuclease gene fragment lacked the corresponding codons for 32-amino-acid residues from C-terminus and was inactive (data not shown). The inactivation of PREV0 promoter (plasmid pIM21) increased the net expression by 3.5-fold (Figure 4, 2nd line). PM1,M2 is not responsible for this high expression because a double mutant of PM1,M2 and PREV0 promoters (pIM32) showed a 4-fold increase (Figure 4, 3rd line). These results demonstrated that PREV0 plays some negative role in transcription initiated at PR.
Figure 4.
Effect of PREV0 reverse promoter on PR activity in transcriptional fusion. Each value represents an average of four measurements, along with their standard deviations.
R–M system with a down mutation in the reverse promoter
Next, we investigated the phenotype of the entire EcoRI R–M system with a down mutation in the reverse promoter PREV0. We changed the PREV0 promoter sequence while maintaining the amino acid sequence for the restriction endonuclease.
Based on the PREV0 promoter, we initially designed all possible substitutions (6 bp) for the −10 and −35 promoter hexamers. Despite many attempts, such mutants could not be generated by site-directed mutagenesis using a plasmid harboring the wild-type EcoRI R–M system as a template (pIMRM). Among the screened clones, the majority were found to be restriction-negative, i.e. the restriction gene was disrupted. A small number of restriction-positive clones were identified, which, after sequencing, were found to possess only 2- or 3-nt changes out of the intended 6-bp changes. These changes did not change the amino acid sequence of the restriction endonuclease. We selected one such clone, pIM24, with 3-bp changes in the PREV0 promoter: TATGAT→TGTGGT; TTGTAG→TTGTAA. This 3-bp substitution did not abolish PREV0 promoter activity, but led to a 2.6-fold drop in expression of the lacZ fusion (Figure 1D, pJFY154 versus pIM19).
Reverse promoter mutation strengthens restriction
We examined the effect of the PREV0 promoter down mutation on restriction activity. We compared plaque formation with that of λ bacteriophage, which carries five EcoRI sites (Table 2). The 3-bp substitution mutation conferred almost 9-fold higher restriction than the wild type (Table 2). This result may explain why we could not obtain viable cells with the EcoRI R–M system with mutations that were expected to more severely inactivate PREV0 and lead to high levels of restriction, potentially killing the host bacterial cell. These results clearly indicated the biological significance of this reverse promoter, although the underlying mechanisms are yet to be elucidated (see ‘Discussion’ section).
Table 2.
Reverse promoter mutation increases restriction
| Plasmid | Genotype | Plaque-forming units | Efficiency of plaque formationa | Restriction relative to WT R–M |
|---|---|---|---|---|
| pACYC184 | R− M− (vector) | (2.4 ± 0.4) × 108 | 1 | |
| pIM27 | R− M+ | (2.2 ± 0.3) × 108 | 0.9 | |
| pIMRM | WT (R+ M+) | (1.3 ± 0.3) × 106 | 5.4 × 10−3 | 1 |
| pIM24 | PREV0 downb | (1.5 ± 0.2) × 105 | 6.2 × 10−4 | 8.7 |
R, restriction; M, modification; WT, wild type.
aEfficiency of plaque formation = plaque-forming units on tested plasmid divided by plaque-forming units on pACYC184.
bTATGAT→TGTGGT and TTGTAG→TTGTAA, with no change in R.EcoRI amino acid sequence.
The host bacterium was E. coli MG1655. The standard deviation from four measurements is indicated.
Antisense RNA gene in trans alleviates post-segregational host killing
To further understand the biological role of the antisense RNA in modulation of EcoRI R–M system activity, we examined its role in the attack on host bacterial cells during post-segregational killing (see ‘Introduction’ section).
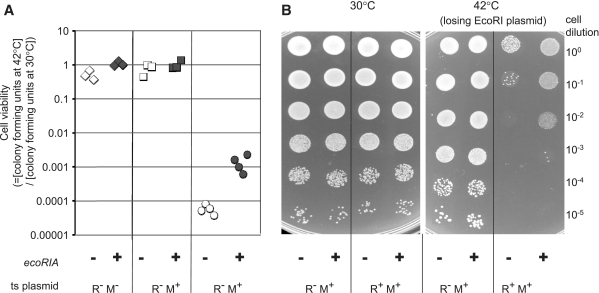
We generated a two-plasmid system in E. coli cells, in which the EcoRI R–M system was carried by a plasmid with a thermo-sensitive replicator, accompanied by a compatible, high-copy number, pUC18-derived plasmid containing the gene (ecoRIA) for the antisense RNA driven from its natural (PREV0) promoter (Table 1). Host cell death was measured after induction of loss of the EcoRI R–M plasmid by blocking its replication through a temperature shift to 42°C. As shown in Figure 5A, the viability of cells with the wild-type EcoRI R–M system (pIK172) in the absence of antisense RNA in trans, showed a significant decrease of 104-fold compared with the undisturbed viability of restriction-negative variants (pHSG415 and pIK173), as reported earlier (10). However, the cells survived the post-segregational killing challenge about 15-fold better in the presence of the antisense RNA gene ecoRIA on a second plasmid (pIM-REV0, Figure 5A, Table 1). These results were confirmed in a qualitative spot assay (Figure 5B).
Figure 5.
Effect of the antisense RNA gene ecoRIA on post-segregational host killing by the EcoRI R–M system. Cell survival after loss of the EcoRI R–M system was measured in the presence of the antisense RNA gene (ecoRIA) in a quantitative (A) and a qualitative assay (B). The E. coli cells carried two plasmids. One was from a series of plasmids harboring EcoRI R–M system variants on a thermo-sensitive pSC101 replicator: vector pHSG415 (diamond); pIK173 (R− M+ square) or pIK172 (R+ M+ circle). The other was from a series of pUC-derivatives to deliver antisense RNA from the PREV0 promoter and ecoRIA gene (pIM-REV0; black) or not, as the negative control (pIM-ΔRNA; white). Dilutions of the culture were spread (A) or spotted (B) on to an agar plate for incubation at a temperature permissive for replication of the RM plasmid (30°C) or at a temperature nonpermissive for its replication (42°C).
A simplest interpretation of these observations is that the antisense RNA (Rna0) transcribed from the PREV0 promoter was delivered in trans and somehow affected the activity of the EcoRI R–M system. However, other possibilities should also be considered (see ‘Discussion’ section).
DISCUSSION
In the present study of the EcoRI R–M system, we identified an antisense RNA (Rna0) from a novel reverse promoter (PREV0) at the 3′ end of the restriction gene and demonstrated the role of this antisense RNA gene in the modulation of gene expression in this R–M system and the attack of incoming and resident DNA. In EcoRI R–M system transcription unit, there are at least six experimentally tested active promoters, where some of them act in overlapping tandems (PREV1,REV2; PM1,M2) and some are arranged convergently (PR versus PREV1,REV2; PREV0 versus PM1,M2) with one promoter facing other promoter within a ecoRIR coding sequence (Figure 1A).
Transcriptional fusion revealed that this reverse promoter (PREV0) and the promoter for the downstream modification gene (PM1,M2) apparently inhibit each other's transcription (Figure 3). The PM1,M2-mediated inhibition of PREV0-initiated transcription resulted in its apparent termination at PM1,M2 (Figures 1 and 2).
Such interference of transcription could take place through various mechanisms. For example (i) transcription termination by a roadblock, in which an RNA polymerase (RNAP) complex bound to a promoter displaces RNAP initiated from an opposite promoter causing termination of transcription; (ii) occlusion, in which a promoter is prevented from RNAP binding due to transient occupation by a passage of elongating RNAP from a facing promoter; (iii) collision of two elongating RNAP complexes leading to premature transcription termination; (iv) sitting duck interference, in which a transcriptional-initiation complex is hit and dislodged by the arrival of an elongation complex from a (stronger) external promoter (45–49); in addition, another possibility is (v) RNA–RNA interference, in which a transcript may be degraded or somehow prevented from being translated after interaction with its complementary RNA formed from the other promoter.
For the inhibition of PREV0-initiated transcription by PM1,M2, possibilities (ii) and (iii) seem unlikely (because the transcript is initiated). Possibility (iv) is also unlikely because the apparent termination of antisense transcription occurs past the transcription initiation site. These leave (i) (termination) and (v) (RNA–RNA) as the most likely explanations. For the inhibition of PM1,M2-initiated transcription by PREV0, however, we cannot exclude any of these five possibilities. A ‘sitting duck’ model has been suggested for regulation of an R–M system (50).
Our investigations have partly depended on the use of promoter mutations. However, we have to remember that other post-transcriptional events such as translation initiation and RNA stability could be affected by sequence changes. For reference, we included a prediction of the secondary structure of Rna0 and the 5′ end of the modification enzyme transcript from PM1,M2 (Supplementary Figures S1 and S2). Because our approach involved transcriptional fusion instead of translational fusion, we do not know the effect, if any, of the reverse promoter on translation from the PM1,M2-initiated transcript.
The presence of a linked reverse promoter might explain the failure to detect strong promoter activity in other R–M systems, such as SalI, with a similar gene organization (51,52).
Convergent promoters have been reported in the regulatory regions of several lambdoid phages, where they control the lysis/lysogeny switch (46,53). Others reported their impact on the switch between replication and conjugational transfer by plasmids (54,55). The net strength of the PM1,M2 promoter and the PREV0 promoter under inhibition was comparable. This suggested that both promoters actively contributed to maintaining coordination of their expression. However, this may represent an unstable equilibrium, whereby activation of one promoter over a threshold may inactivate the other promoter, which would further activate the first promoter (56). This positive feedback loop can form a bi-stable switch, which may be involved in the lifestyle of R–M systems. For example, in the absence of previously synthesized R and M proteins after entry into a new host cell, PM action may be dominant. Accumulation of M protein then might lead to shift toward PREV0 activity. This may be similar to the bi-stable switch in the life style of several lysogenic bacteriophages (57,58).
In SsoII and Ecl18kI R–M systems, the restriction and modification genes are divergent, but their promoters are convergent (R gene – PM – PR – M gene). Their regulation was found to be associated with interference between the two convergent promoters (25,50). In these R–M systems, as well as in MspI (59), two feedback loops have been identified. One, a negative feedback loop, is related to the autoregulation of the modification enzyme and the other, a positive feedback loop, is associated with a boost of restriction transcription after interaction of the modification enzyme with its binding site within the intergenic region.
The effect of reverse promoter mutations on the restriction of incoming bacteriophage DNA indicates the biological relevance of the present study. However, to fully understand the underlying molecular mechanisms, we have to analyze the roles of other promoters, post-transcriptional processes, R–M gene products and host factors, and assemble them into a network of interactions.
The effect of the antisense RNA gene on post-segregational host killing provides another line of evidence for the relevance of our findings to the biology of R–M systems. This trans effect may also provide a clue to understanding the interference between the convergent promoters. Among the four possible mechanisms considered above, (v) namely, degradation or some functional alteration of RNA triggered by its interaction with a complementary RNA, can easily explain this trans effect. However, the five mechanisms were proposed to explain the results of the transcriptional fusions, and this post-segregational killing process includes other processes.
In antisense-RNA-mediated post-segregational killing systems, the antisense RNAs downregulates the toxin gene by basepairing with target toxin mRNA to modulate translation initiation or to promote mRNA degradation (13,60–62), as most small noncoding RNAs in prokaryotes do (30–33). If the antisense RNA (Rna0) alleviates the toxin (R) expression in trans, its action may turn out to be similar to that of these antisense RNAs. Because excess of sequence-specific anti-sense RNA molecules might result in nonspecific down-modulation, further experimental analysis is needed to prove this in the natural context.
In conclusion, our work has demonstrated a novel mechanism of R–M system regulation by noncoding, intragenic, antisense transcription. This novel regulatory system is biologically important in controlling toxic endonuclease genes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Project on Protein Structural and Functional Analyses (Protein 3000); the 21st century COE (Center of Excellence) project of ‘Elucidation of Language Structure and Semantic behind Genome and Life System’; the ‘Grants-in-Aid for Scientific Research’ from the Japan Society for the Promotion of Science (JSPS) (15370099 and 17310113; to I.K.); ‘Postdoctoral Fellowships for Foreign Researchers’ from JSPS (to Y.L.); and by grant N N301 724140 from the Polish Ministry of Science and Higher Education (to I.M.). Funding for open access charge: ‘Grants-in-Aid for Scientific Research’ from JSPS (17310113).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors warmly acknowledge Naofumi Handa and Asao Ichige for their excellent help and advice during the entire study. I.M. is very grateful also to Noriko Takahashi, Ken Ishikawa and Yoshikazu Furuta for useful discussions.
REFERENCES
- 1.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeltsch A, Pingoud A. Horizontal gene transfer contributes to the wide distribution and evolution of type II restriction-modification systems. J. Mol. Evol. 1996;42:91–96. doi: 10.1007/BF02198833. [DOI] [PubMed] [Google Scholar]
- 3.Naderer M, Brust JR, Knowle D, Blumenthal RM. Mobility of a restriction-modification system revealed by its genetic contexts in three hosts. J. Bacteriol. 2002;184:2411–2419. doi: 10.1128/JB.184.9.2411-2419.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kobayashi I, Nobusato A, Kobayashi-Takahashi N, Uchiyama I. Shaping the genome–restriction-modification systems as mobile genetic elements. Curr. Opin. Genet. Dev. 1999;9:649–656. doi: 10.1016/s0959-437x(99)00026-x. [DOI] [PubMed] [Google Scholar]
- 5.Rocha EP, Danchin A, Viari A. Evolutionary role of restriction/modification systems as revealed by comparative genome analysis. Genome Res. 2001;11:946–958. doi: 10.1101/gr.gr-1531rr. [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29:3742–3756. doi: 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fauman EB, Blumenthal RM, Cheng X. Structure and evolution of AdoMet-dependent methyltransferases. In: Cheng X, Blumenthal RM, editors. S-Adenosylmethionine-Dependent Methyltransferases: Structures and Functions. Singapore: World Scientific Publishing; 1999. pp. 1–38. [Google Scholar]
- 8.Asakura Y, Kobayashi I. From damaged genome to cell surface: transcriptome changes during bacterial cell death triggered by loss of a restriction-modification gene complex. Nucleic Acids Res. 2009;37:3021–3031. doi: 10.1093/nar/gkp148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishikawa K, Handa N, Kobayashi I. Cleavage of a model DNA replication fork by a type I restriction endonuclease. Nucleic Acids Res. 2009;37:3531–3544. doi: 10.1093/nar/gkp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kusano K, Naito T, Handa N, Kobayashi I. Restriction-modification systems as genomic parasites in competition for specific sequences. Proc. Natl Acad. Sci. USA. 1995;92:11095–11099. doi: 10.1073/pnas.92.24.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naito T, Kusano K, Kobayashi I. Selfish behavior of restriction-modification systems. Science. 1995;267:897–899. doi: 10.1126/science.7846533. [DOI] [PubMed] [Google Scholar]
- 12.Gerdes K, Rasmussen PB, Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl Acad. Sci. USA. 1986;83:3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerdes K, Thisted T, Martinussen J. Mechanism of post-segregational killing by the hok/sok system of plasmid R1: sok antisense RNA regulates formation of a hok mRNA species correlated with killing of plasmid-free cells. Mol. Microbiol. 1990;4:1807–1818. doi: 10.1111/j.1365-2958.1990.tb02029.x. [DOI] [PubMed] [Google Scholar]
- 14.Hayes F. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science. 2003;301:1496–1499. doi: 10.1126/science.1088157. [DOI] [PubMed] [Google Scholar]
- 15.Mochizuki A, Yahara K, Kobayashi I, Iwasa Y. Genetic addiction: selfish gene's strategy for symbiosis in the genome. Genetics. 2006;172:1309–1323. doi: 10.1534/genetics.105.042895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srikhanta YN, Fox KL, Jennings MP. The phasevarion: phase variation of type III DNA methyltransferases controls coordinated switching in multiple genes. Nat. Rev. Microbiol. 2010;8:196–206. doi: 10.1038/nrmicro2283. [DOI] [PubMed] [Google Scholar]
- 17.Mruk I, Blumenthal RM. Real-time kinetics of restriction-modification gene expression after entry into a new host cell. Nucleic Acids Res. 2008;36:2581–2593. doi: 10.1093/nar/gkn097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tao T, Bourne JC, Blumenthal RM. A family of regulatory genes associated with type II restriction-modification systems. J. Bacteriol. 1991;173:1367–1375. doi: 10.1128/jb.173.4.1367-1375.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knowle D, Lintner RE, Touma YM, Blumenthal RM. Nature of the promoter activated by C.PvuII, an unusual regulatory protein conserved among restriction-modification systems. J. Bacteriol. 2005;187:488–497. doi: 10.1128/JB.187.2.488-497.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vijesurier RM, Carlock L, Blumenthal RM, Dunbar JC. Role and mechanism of action of C. PvuII, a regulatory protein conserved among restriction-modification systems. J. Bacteriol. 2000;182:477–487. doi: 10.1128/jb.182.2.477-487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mruk I, Rajesh P, Blumenthal RM. Regulatory circuit based on autogenous activation-repression: roles of C-boxes and spacer sequences in control of the PvuII restriction-modification system. Nucleic Acids Res. 2007;35:6935–6952. doi: 10.1093/nar/gkm837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakayama Y, Kobayashi I. Restriction-modification gene complexes as selfish gene entities: roles of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion. Proc. Natl Acad. Sci. USA. 1998;95:6442–6447. doi: 10.1073/pnas.95.11.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Som S, Friedman S. Regulation of EcoRII methyltransferase: effect of mutations on gene expression and in vitro binding to the promoter region. Nucleic Acids Res. 1994;22:5347–5353. doi: 10.1093/nar/22.24.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karyagina A, Shilov I, Tashlitskii V, Khodoun M, Vasil'ev S, Lau PC, Nikolskaya I. Specific binding of sso II DNA methyltransferase to its promoter region provides the regulation of sso II restriction-modification gene expression. Nucleic Acids Res. 1997;25:2114–2120. doi: 10.1093/nar/25.11.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fedotova EA, Protsenko AS, Zakharova MV, Lavrova NV, Alekseevsky AV, Oretskaya TS, Karyagina AS, Solonin AS, Kubareva EA. SsoII-like DNA-methyltransferase Ecl18kI: interaction between regulatory and methylating functions. Biochemistry. 2009;74:85–91. doi: 10.1134/s0006297909010131. [DOI] [PubMed] [Google Scholar]
- 26.Beletskaya IV, Zakharova MV, Shlyapnikov MG, Semenova LM, Solonin AS. DNA methylation at the CfrBI site is involved in expression control in the CfrBI restriction-modification system. Nucleic Acids Res. 2000;28:3817–3822. doi: 10.1093/nar/28.19.3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christensen LL, Josephsen J. The methyltransferase from the LlaDII restriction-modification system influences the level of expression of its own gene. J. Bacteriol. 2004;186:287–295. doi: 10.1128/JB.186.2.287-295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Ichige A, Kobayashi I. Regulation of the EcoRI restriction-modification system: identification of ecoRIM gene promoters and their upstream negative regulators in the ecoRIR gene. Gene. 2007;400:140–149. doi: 10.1016/j.gene.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Kobayashi I. Negative regulation of EcoRI restriction enzyme gene associated with intragenic reverse promoters. J. Bacteriol. 2007;189:6928–6935. doi: 10.1128/JB.00127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 31.Majdalani N, Vanderpool CK, Gottesman S. Bacterial small RNA regulators. Crit. Rev. Biochem. Mol. Biol. 2005;40:93–113. doi: 10.1080/10409230590918702. [DOI] [PubMed] [Google Scholar]
- 32.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomason MK, Storz G. Bacterial antisense RNAs: how many are there, and what are they doing? Annu. Rev. Genet. 2010;44:167–188. doi: 10.1146/annurev-genet-102209-163523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 2007;10:102–109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 35.Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 36.Casadaban MJ, Cohen SN. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 37.Sarkar G, Sommer SS. The “megaprimer” method of site-directed mutagenesis. Biotechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 38.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 39.Brewer G, Murray E, Staeben M. RNase OneTM: advantages for nuclease protection assays. Promega Notes Magazine. 1992 [Google Scholar]
- 40.Hendrix RW, Roberts JW, Stahl FW, Weisberg RA. Lambda II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1983. [Google Scholar]
- 41.Hashimoto-Gotoh T, Franklin FC, Nordheim A, Timmis KN. Specific-purpose plasmid cloning vectors. I. Low copy number, temperature-sensitive, mobilization-defective pSC101-derived containment vectors. Gene. 1981;16:227–235. doi: 10.1016/0378-1119(81)90079-2. [DOI] [PubMed] [Google Scholar]
- 42.Oliphant AR, Struhl K. Defining the consensus sequences of E. coli promoter elements by random selection. Nucleic Acids Res. 1988;16:7673–7683. doi: 10.1093/nar/16.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shultzaberger RK, Chen Z, Lewis KA, Schneider TD. Anatomy of Escherichia coli sigma70 promoters. Nucleic Acids Res. 2007;35:771–788. doi: 10.1093/nar/gkl956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen H, Bjerknes M, Kumar R, Jay E. Determination of the optimal aligned spacing between the Shine–Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 1994;22:4953–4957. doi: 10.1093/nar/22.23.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shearwin KE, Callen BP, Egan JB. Transcriptional interference–a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Callen BP, Shearwin KE, Egan JB. Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol. Cell. 2004;14:647–656. doi: 10.1016/j.molcel.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 47.Crampton N, Bonass WA, Kirkham J, Rivetti C, Thomson NH. Collision events between RNA polymerases in convergent transcription studied by atomic force microscopy. Nucleic Acids Res. 2006;34:5416–5425. doi: 10.1093/nar/gkl668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sneppen K, Dodd IB, Shearwin KE, Palmer AC, Schubert RA, Callen BP, Egan JB. A mathematical model for transcriptional interference by RNA polymerase traffic in Escherichia coli. J. Mol. Biol. 2005;346:399–409. doi: 10.1016/j.jmb.2004.11.075. [DOI] [PubMed] [Google Scholar]
- 49.Palmer AC, Ahlgren-Berg A, Egan JB, Dodd IB, Shearwin KE. Potent transcriptional interference by pausing of RNA polymerases over a downstream promoter. Mol Cell. 2009;34:545–555. doi: 10.1016/j.molcel.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Protsenko A, Zakharova M, Nagornykh M, Solonin A, Severinov K. Transcription regulation of restriction-modification system Ecl18kI. Nucleic Acids Res. 2009;37:5322–5330. doi: 10.1093/nar/gkp579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alvarez MA, Chater KF, Rodicio MR. Complex transcription of an operon encoding the SalI restriction-modification system of Streptomyces albus G. Mol. Microbiol. 1993;8:243–252. doi: 10.1111/j.1365-2958.1993.tb01568.x. [DOI] [PubMed] [Google Scholar]
- 52.Alvarez MA, Gomez A, Gomez P, Brooks JE, Rodicio MR. Comparative analysis of expression of the SalI restriction-modification system in Escherichia coli and Streptomyces. Mol. Gen. Genet. 1996;253:74–80. doi: 10.1007/s004380050298. [DOI] [PubMed] [Google Scholar]
- 53.Dodd IB, Egan JB. Action at a distance in CI repressor regulation of the bacteriophage 186 genetic switch. Mol. Microbiol. 2002;45:697–710. doi: 10.1046/j.1365-2958.2002.03038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jagura-Burdzy G, Thomas CM. Dissection of the switch between genes for replication and transfer of promiscuous plasmid RK2: basis of the dominance of trfAp over trbAp and specificity for KorA in controlling the switch. J. Mol. Biol. 1997;265:507–518. doi: 10.1006/jmbi.1996.0747. [DOI] [PubMed] [Google Scholar]
- 55.Sepulveda E, Perez-Mendoza D, Ramirez-Romero MA, Soto MJ, Lopez-Lara IM, Geiger O, Sanjuan J, Brom S, Romero D. Transcriptional interference and repression modulate the conjugative ability of the symbiotic plasmid of Rhizobium etli. J. Bacteriol. 2008;190:4189–4197. doi: 10.1128/JB.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pedersen M, Hammer K. The role of MOR and the CI operator sites on the genetic switch of the temperate bacteriophage TP901-1. J. Mol. Biol. 2008;384:577–589. doi: 10.1016/j.jmb.2008.09.071. [DOI] [PubMed] [Google Scholar]
- 57.Ferrell JE., Jr Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr. Opin. Cell. Biol. 2002;14:140–148. doi: 10.1016/s0955-0674(02)00314-9. [DOI] [PubMed] [Google Scholar]
- 58.Dubnau D, Losick R. Bistability in bacteria. Mol. Microbiol. 2006;61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- 59.Som S, Friedman S. Characterization of the intergenic region which regulates the MspI restriction-modification system. J. Bacteriol. 1997;179:964–967. doi: 10.1128/jb.179.3.964-967.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gerdes K, Wagner EG. RNA antitoxins. Curr. Opin. Microbiol. 2007;10:117–124. doi: 10.1016/j.mib.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 61.Kawano M, Aravind L, Storz G. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 2007;64:738–754. doi: 10.1111/j.1365-2958.2007.05688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fozo EM, Kawano M, Fontaine F, Kaya Y, Mendieta KS, Jones KL, Ocampo A, Rudd KE, Storz G. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol. Microbiol. 2008;70:1076–1093. doi: 10.1111/j.1365-2958.2008.06394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.