Abstract
The large number of candidate genes made available by comprehensive genome analysis requires that relatively rapid techniques for the study of function be developed. Here, we report a rapid and convenient electroporation method for both gain- and loss-of-function studies in vivo and in vitro in the rodent retina. Plasmid DNA directly injected into the subretinal space of neonatal rodent pups was taken up by a significant fraction of exposed cells after several pulses of high voltage. With this technique, GFP expression vectors were efficiently transfected into retinal cells with little damage to the operated pups. Transfected GFP allowed clear visualization of cell morphologies, and the expression persisted for at least 50 days. DNA-based RNA interference vectors directed against two transcription factors important in photoreceptor development led to photoreceptor phenotypes similar to those of the corresponding knockout mice. Reporter constructs carrying retinal cell type-specific promoters were readily introduced into the retina in vivo, where they exhibited the appropriate expression patterns. Plasmid DNA was also efficiently transfected into retinal explants in vitro by high-voltage pulses.
Recent advances in comprehensive expression profiling, such as microarray analysis (1, 2) and serial analysis of gene expression (SAGE, ref. 3), have enabled the identification of a large number of genes that might play a role in mammalian retinal development and disease. It is now more important than ever that a rapid and convenient method for the analysis of function be developed, particularly for application in intact tissue in vivo or in vitro.
We have previously used retroviral vectors, based on Moloney murine leukemia virus (MLV) to deliver genes to the developing retina (4–6). Such vectors can enable expression of virally transduced genes in the retina without the time and expense of transgenic animals. However, there are disadvantages inherent in the use of such vectors. First, the use of an MLV vector is limited to gene transfer into mitotic retinal progenitor cells, as it requires proliferation of the target cells for integration. Second, it is time consuming to prepare high titer virus stocks to achieve efficient gene transfer. Third, MLV vectors have a size limitation for insert DNA (typically <7 kb). Fourth, such vectors do not readily allow introduction of more than two genes into the same cells. Some of these problems can be bypassed by using other types of viral vectors, such as lentivirus (7) and adeno-associated virus (8), although size and efficiency still limit certain applications and one still must make viral stocks.
To overcome these problems, we have applied a relatively new technique for the introduction of DNA into neonatal mouse and rat retinae, that of in vivo electroporation, which has recently been applied to several tissues of various animal species (reviewed in ref. 9). A solution of DNA is directly injected into the subretinal space of neonatal mouse/rat pups, and electric pulses are applied by using tweezer-type electrodes. This method is faster than other viral and transgenic gene transfer methods. The gene transduction efficiency of this method is also higher than that of MLV viral vectors. We have also applied an electroporation technique for gene introduction into retinal explants by using a micro electroporation chamber (in vitro electroporation).
In this article, we report several types of experiments performed with these gene transfer methods. These include (i) promoter analysis, (ii) gain-of-function analysis using a bicistronic expression vector, and (iii) loss-of-function analysis using a DNA-based RNA interference (RNAi) vector.
Materials and Methods
Animals. Timed pregnant Sprague–Dawley rats and Swiss–Webster mice were purchased from Taconic Farms, and CD1 mice were from Charles River Breeding Laboratories. All of the animal experiments in this study were approved by the Institutional Animal Care and Use Committee at Harvard University.
DNA Construction. For construction of pCAG-GFP and pCAG-DsRed, cDNAs encoding enhanced GFP (EGFP) and DsRed2 were excised from pEGFP-N1 (Clontech) and pDsRed2-N1 (Clontech), respectively, and cloned into pCAGGS (10) with modified multiple cloning sites. For construction of pCMV-GFP, pEF-GFP, and pUB-GFP, the promoter region of pCAG-GFP was replaced by the cytomegalovirus (CMV) promoter excised from pEGFP-N1, the human elongation factor (EF) 1α promoter excised from pCE-EGFP-1 (11), and human ubiquitin C promoter excised from pFUGW (12), respectively.
For construction of pRho-2.2K-DsRed, pCABP5–4.7K-DsRed, and pCRALBP-4K-DsRed, the promoter region of pCAG-DsRed was replaced by bovine rhodopsin promoter (from –2174 to +70), mouse calcium binding protein 5 (CABP5) promoter (from –4534 to +148, National Center for Biotechnology Information locus ID 29865), and mouse cellular retinaldehyde binding protein (CRALBP) promoter (from –3932 to +71, National Center for Biotechnology Information locus ID 19771), respectively. The bovine rhodopsin promoter was excised from the –2174-lacZ construct (13), and the mouse CABP5 and CRALBP promoters were amplified by genomic PCR. The following PCR primers were used to amplify the promoters: mouse CABP5 promoter, 5′-CCCTTTGCTAGCACTGAGACCCTTTAATACG-3′ and 5′-CCTGGATTGCAATGCTGTCTCTCACACTTGC-3′; mouse CRALBP promoter, 5′-TCCCT T TCTCCTATGAGA AGCGGGAGGCCC-3′ and 5′-CCA AGCTCTGATGTCA AGATGGCCCCTCCT-3′. pRho-2.2K-CFP was constructed by replacing the coding region of DsRed2 in pRho-2.2K-DsRed with enhanced cyan fluorescent protein (ECFP) excised from pECFP-N1 (Clontech).
For construction of pCAGIG, an internal ribosome entry site (IRES)-GFP cassette excised from pMX-IRES-GFP (14) was ligated into the pCAGGS vector. A cDNA encoding mouse Rax (15) was amplified by PCR to trim the 5′ and 3′ UTRs and cloned into pCAGIG to make pCAGIG-mRax. RNAi vectors for mouse Crx, Nrl, and GAPDH were constructed by inserting the annealed oligonucleotides into pBS/U6 (16) digested with ApaI (blunted) and EcoRI. Oligonucleotides were as follows: mouse Crx (coding region 408–428), 5′-GGCATCTCAGATTCTTACAGAAGCTTCTGTAAGAATCTGAGATGCCCTTTTTG-3′ and 5′-AATTCAAAAAGGGCATCTCAGATTCTTACAGAAGCTTCTGTAAGAATCTGAGATGCC-3′; mouse Nrl (306–326), 5′-GGTCCTGTCTCTATGGAAGGAAGCTTCCTTCCATAGAGACAGGACCCTTTTTG-3′ and 5′-AATTCAAAAAGGGTCCTGTCTCTATGGAAGGAAGCTTCCTTCCATAGAGACAGGACC-3′; mouse GAPDH (393–413), 5′-GGTGTGAACCACGAGAAATAAAGCTTTATTTCTCGTGGTTCACACCCTTTTTG-3′ and 5′-AATTCAAAAAGGGTGTGAACCACGAGAAATAAAGCTTTATTTCTCGTGGTTCACACC-3′.
In Vivo Electroporation and Virus Infection. Newborn rat or mouse pups were anesthetized by chilling on ice, and a small incision was made in the eyelid and sclera near the lens with a 30-gauge needle. DNA solutions (3∼6 μg/μl) in PBS containing 0.1% fast green as a tracer were injected into the subretinal space through the incision by using a Hamilton syringe with a 32- or 33-gauge blunt-ended needle under a dissecting microscope. For newborn rat pups, ≈1 μl of DNA was injected, and for mouse newborn pups, ≈0.5 μl of DNA was injected. After DNA injection, tweezer-type electrodes (model 520, 7 mm diameter, BTX, San Diego) briefly soaked in PBS were placed to softly hold the heads of the pups, and five square pulses of 50-ms duration with 950-ms intervals were applied by using a pulse generator, CUY21 (Nepagene, Chiba, Japan) or ECM830 (BTX). For rat newborn pups, 100-V pulses were applied, and for mouse newborn pups, 80-V pulses were applied. Usually DNA was transfected only into right eyes.
Replication incompetent retroviruses were produced by transfecting pLIA (17) into Phoenix-E packaging cells, concentrated, and titrated on NIH 3T3 cells. Concentrated virus (1 × 107 colony-forming units/ml) was injected into the subretinal space of newborn rats as described for DNA injection.
In Vitro Electroporation and Retinal Explant Culture. Dissected retinae were transferred to a micro electroporation chamber (Nepagene, model CUY532, 3 mm × 10 mm × 5 mm) filled with a DNA solution (1 μg/μl in Hanks' balanced salt solution), and five square pulses (30 V) of 50-ms duration with 950-ms intervals were applied by using pulse generator CUY21. Electroporated retinae were cultured at 37°C on Nucleopore polycarbonate filters (Whatman, 0.2 μm pore size) with Neurobasal Medium (Invitrogen) containing 1× B-27 serum-free supplement (Invitrogen).
Preparation of Retinal Sections. Electroporated retinae were harvested 2–50 days after electroporation and dissected under a fluorescent microscope (Leica, MZFL III) to select the GFP- or DsRed-positive retinae. Dissected retinae were fixed with 4% paraformaldehyde in PBS for 20 min at room temperature, cryoprotected in PBS containing 30% sucrose for several hours at 4°C, and embedded in OCT compound (Sakura, Torrance, CA) on dry ice. Cryosections (20 or 30 μm) were cut on a cryostat.
Immunostaining of Dissociated Retinal Cells. Retinae were dissociated into single cells essentially as described (18), except that papain (Worthington, final 50 ng/μl) was used instead of trypsin, and stained with the following antibodies: anti-rhodopsin (Rho4D2 obtained from R. Molday, University of British Columbia, Vancouver, ref. 19), anti-Crx (obtained from S. Chen, Washington University School of Medicine, St. Louis, ref. 20), anti-Nrl (obtained from A. Swaroop, University of Michigan, Ann Arbor, ref. 21), anti-GAPDH (Ambion 4300), anti-Chx10 (developed by our laboratory, unpublished work), anti-protein kinase Cα (Oncogene Science OP74), anti-glutamine synthetase (Chemicon MAB302), anti-HPC-1 (Santa Cruz Biotechnology sc-12736), anti-Calbindin (Sigma C9848), anti-Gt2α (Santa Cruz Biotechnology sc-390), and anti-Thy-1 (Santa Cruz Biotechnology sc-19614).
Results
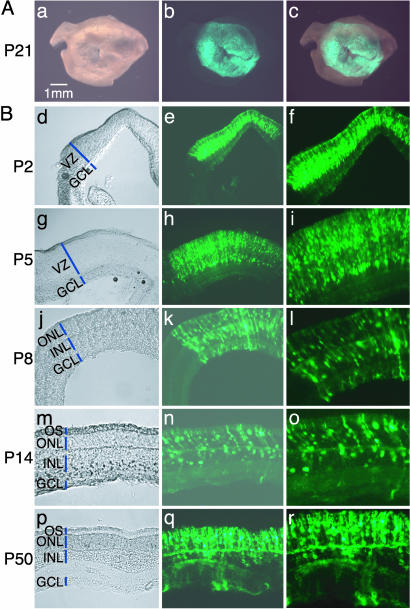
In Vivo Electroporation. DNA constructs were injected into the subretinal space of newborn mouse or rat pups (for detailed protocol, see Fig. 8, which is published as supporting information on the PNAS web site). Tweezer-type electrodes were then placed on the heads of pups, and five electric pulses of 50-ms duration (100 V for rats, 80 V for mice) were applied to the eyes in the direction shown in Fig. 8A by using a square wave electroporator. The DNA was transduced into the scleral side of the retina, where undiffentiated mitotic and newly postmitotic cells exist. Almost all operated pups survived and were apparently healthy after electroporation. When a GFP expression vector driven by the CAG (chicken β-actin promoter with CMV enhancer, ref. 10) promoter, a strong ubiquitous promoter, was electroporated into the retinae on postnatal day 0 (P0), an average of ≈80% rat retinae and ≈50% mouse retinae expressed GFP. In a good transfection, GFP expression was observed in a wide area of the retina (Fig. 1A).
Fig. 1.
In vivo electroporated rat retinae harvested at various developmental stages. (A) Whole-mount preparation of rat retina in vivo electroporated with pCAG-GFP at P0 and harvested at P21. Pictures were taken from the scleral side. (B) Rat retinae in vivo electroporated with pCAG-GFP at P0 were harvested at P2 (d–f), P5 (g–i), P8 (j–l), P14 (m–o), or P50 (p–r), and cryosections were prepared.
A GFP expression vector transfected at P0 allowed clear visualization of the morphologies of retinal cells throughout development (Fig. 1B). At P2, GFP was detected in retinal progenitor cells in the ventricular zone (VZ) (Fig. 1B d–f). At P5, GFP was observed in two different cell populations, one forming the future outer nuclear layer (ONL) and the other forming the inner nuclear layer (INL), seen as beginning to segregate in the former VZ area (Fig. 1B g–i). At P8, GFP labeled immature rod photoreceptors (PRs) in the ONL, and immature bipolar cells, differentiating Müller glial cells, and/or progenitor cells, whose cell bodies were located in the INL (Fig. 1B j–l). At P14, the majority of the GFP-positive cells were differentiated into rod PR located in the ONL, and the rest became bipolar cells and Müller glial cells (Fig. 1B m–o). Rod outer segments (OSs) were also clearly labeled by GFP. The GFP expression was observed at the latest time point examined (P50, Fig. 1B p–Ir). However, it appeared that the GFP expression level was gradually decreasing by 3–4 weeks after electroporation.
The distribution of GFP-positive cells in the P14 differentiated rat retinae was examined after DNA introduction at P0 (Fig. 2). Judging based on cell morphology and location, ≈80% of the GFP-positive cells were rod PRs, ≈15% were bipolar cells, ≈3% were Müller glial cells, and <1% were amacrine cells (Fig. 2 A). These values are largely comparable to those of retinal cells infected with a replication-incompetent MLV retrovirus (LIA) carrying an alkaline phosphatase reporter (Fig. 2 A purple bars). This result was further confirmed by staining the dissociated retinal cells with several retinal cell type-specific antibodies. Approximately 75% of the GFP-positive cells expressed rhodopsin (a marker for rods), ≈20% expressed Chx10 (a marker for bipolars), ≈4% expressed glutamine synthetase (a marker for Müller glia), and <1% expressed HPC1 (a marker for amacrines) (Fig. 2B). Very few GFP-positive (<1%) cells were positive for Gt2α (a marker for cones), but we could not detect GFP-positive cells expressing calbindin (a marker for horizontals) or Thy-1 (a marker for ganglion cells).
Fig. 2.
Cell type composition of retinal cells labeled by in vivo electroporation. (A) Cell type composition determined based on morphologies and locations in the retina. Rat retinae electroporated in vivo with pCAG-GFP, or infected in vivo with the replication-incompetent LIA retrovirus at P0, were harvested at P14 and sectioned. The LIA-infected retinae expressing alkaline phosphatase (AP) were stained histochemically for AP activity. Green bars represent the retinal cells electroporated with the pCAG-GFP plasmid. Purple bars represent the retinal cells infected with the LIA retrovirus. (B) Cell type composition determined by immunostaining. Rat retinae electroporated in vivo with pCAG-GFP at P0 were harvested and dissociated into single cells at P14. Dissociated cells were stained with anti-rhodopsin (rod PR), anti-Chx10 (bipolar), anti-protein kinase Cα (rod bipolar), anti-glutamine synthetase (Müller glia), anti-HPC-1 (amacrine), anti-calbindin (horizontal), anti-Gt2α (cone PR), or anti Thy-1 (GC), and the numbers of positive cells were scored. Both GFP-positive (green bars) and GFP-negative (yellow bars) cells were analyzed.
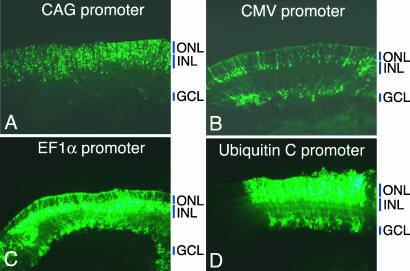
Several ubiquitous promoters, including CMV (22), human EF1α (11), and human ubiquitin C (23) promoters, were also tested in the developing rat retina. EF1α and ubiquitin C promoters exhibited higher GFP expression than CMV or CAG. When sectioned at P10, CMV and EF1α promoters appeared to label the cells whose cell bodies were in the INL with processes extending to the ONL and ganglion cell (GC) layer (GCL), more than those in the ONL, suggesting a relative lack of activity, perhaps caused by silencing of these two promoters in PRs (Fig. 3 B and C). The GFP expression pattern driven by the ubiquitin C promoter was similar to that by the CAG promoter (Fig. 3D), and the cell type composition labeled by the ubiquitin promoter, determined by immunostaining of dissociated cells, was comparable to that by the CAG promoter (data not shown). These results indicate that some “ubiquitous” promoters are not suitable for the studies of mammalian retinal development when PRs need to be labeled.
Fig. 3.
Comparison of ubiquitous promoters in the developing rat retina. Rat retinae were electroporated in vivo at P0 with the GFP expression vectors driven by CAG promoter (A), CMV promoter (B), human EF1α promoter (C), or human ubiquitin C promoter (D) and sectioned at P10.
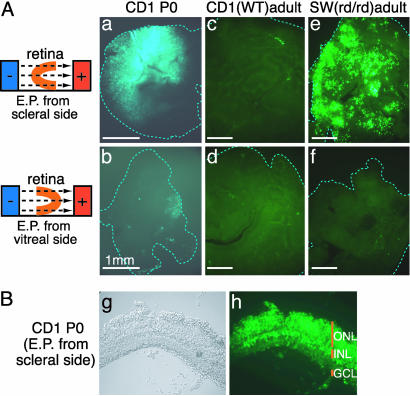
In Vitro Electroporation. To label GCs, which line the surface of the retina facing the vitreous body, a GFP expression vector (pCAG-GFP) was injected into the vitreous of P0 rat eyes and electric pulses were applied in the direction opposite to that used for Fig. 1. However, few GFP-positive cells were detected (data not shown), suggesting that unlike progenitor cells in the VZ, GCs are not highly transfectable. To further test this possibility, DNA was electroporated into CD1 mouse retinae in vitro from the scleral side or from the vitreal side, using a micro chamber (shown in Fig. 8C). A significant number of retinal cells (5–20% of total cells) became GFP-positive (Fig. 4Aa) when electroporated from the scleral side, with the distribution in sections (Fig. 4B) similar to that of in vivo electroporated retinae (Fig. 1). In contrast, only a few cells became GFP-positive when the vector was transfected from the vitreal side (Fig. 4Ab). A similar result was observed when the ubiquitin C promoter was used to express GFP (data not shown).
Fig. 4.
In vitro electroporated mouse retinal explants. (A) Mouse retinae of P0 CD1 (a and b), adult CD1 (c and d), or adult Swiss–Webster having a retinal degeneration mutation (e and f) were in vitro electroporated with pCAG-GFP from the scleral side (a, c, and e) or from the vitreal side (b, d, and f) and cultured for 5 days. Similar results were observed 16 h after electroporation. (B) A section of CD1 mouse retina in vitro electroporated with pCAG-GFP at P0 and cultured for 10 days.
Using the in vitro electroporation technique, we examined whether DNA could be electroporated into adult mouse retina. In the normal retina of adult CD1 mice, few GFP-positive cells were observed even if the vector was transfected from the scleral side (Fig. 4Ac). DNA was also not efficiently transfected from the vitreal side (Fig. 4Ad). Interestingly, many GFP-positive cells were detected when adult Swiss–Webster mouse retinae, having a retinal degeneration mutation, were transfected from the scleral side (Fig. 4Ae). Immunostaining with anti-glutamine synthetase antibody showed that most of the GFP-positive cells were Müller glial cells (data not shown). These results suggest that it is not easy to transfect DNA into mature rod PRs, as well as GCs, by electroporation.
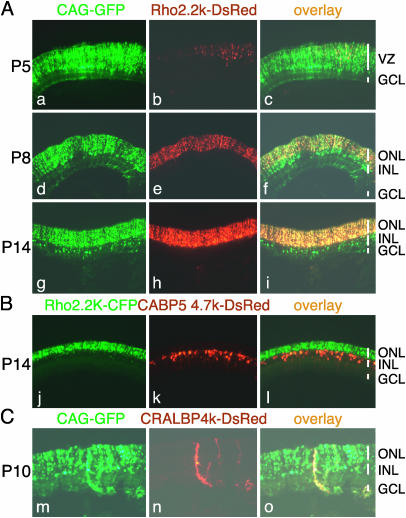
Promoter Analysis. One application of in vivo electroporation is the mapping of transcriptional regulatory elements that can be influenced by viral elements and also frequently need to be larger than the allowance of viral vectors. To evaluate this application, we chose one characterized and two uncharacterized regulatory regions. The 2.2-kDa fragment (from –2174 to +70) of bovine rhodopsin promoter has been shown to direct PR-specific expression in transgenic mice (13). The bovine rhodopsin 2.2-kDa promoter, used to express DsRed, was cotransfected into P0 rat retinae with pCAG-GFP. As shown in Fig. 5A, at P5, the expression of DsRed was detected only in a small population of cells located at the upper part of the VZ (Fig. 5A a–c). At P8, the DsRed expression was detected exclusively in the ONL, and most of the GFP-positive cells in the ONL coexpressed DsRed (Fig. 5A d–f). At P14, the expression of DsRed in the ONL became much stronger (Fig. 5A g–i), consistent with rhodopsin expression profiles (3, 13).
Fig. 5.
Cell type-specific labeling using rhodopsin, CABP5, and CRALBP promoters. (A) Rat retinae were coelectroporated in vivo with pCAG-GFP (3μg/μl) and bovine rhodopsin promoter 2.2K-DsRed (pRho-2.2K-DsRed, 3 μg/μl) at P0, harvested at P5 (a–c), P8 (d–f), or P14 (g–i), and sectioned. (B) Rat retinae were coelectroporated in vivo with pRho-2.2K-CFP (3 μg/μl) and mouse CABP5 promoter 4.7K-DsRed (pCABP5-4.7K-DsRed, 3 μg/μl) at P0, harvested at P14, and sectioned. (C) Rat retinae were coelectroporated in vivo with pCAG-GFP (3 μg/μl) and mouse CRALBP promoter 4.0K-DsRed (pCRALBP-4.0K-DsRed, 3 μg/μl) at P0, harvested at P10, and sectioned. Some DsRed-positive Müller glial cells were truncated in the process of cutting sections.
To determine whether other retinal cell types could be labeled by this technique, we tested an uncharacterized sequence from the promoter region of mouse CABP5 (from –4534 to +148) and a partially characterized promoter sequence of CRALBP (from –3932 to + 71, ref. 24) obtained by genomic PCR. CABP5 is expressed by rod bipolar cells and a subset of cone bipolar cells (25, 26), whereas CRALBP is expressed by Müller glial cells. The CABP5 promoter led to expression of DsRed only in the cells in the INL, consistent with labeling of bipolar cells (Fig. 5B). On the other hand, CRALBP promoter led to expression of DsRed only in Müller glial cells (Fig. 5C).
Gain-of-Function Analysis. Using a bicistronic expression vector containing the CAG promoter and an IRES-GFP cassette (pCAGIG, Fig. 6A), we tested whether the electroporation technique is useful for functional analysis of candidate genes. We focused on a homeobox transcription factor Rax (15) [also called Rx (27)] whose functions had been characterized in the developing rat retina with a replication-incompetent retrovirus vector (28). Rax is expressed by proliferating retinal progenitor cells and differentiating Müller glial cells, and its forced expression in the P0 rat retina with an MLV vector leads to the generation of cells resembling Müller glia (28).
Fig. 6.
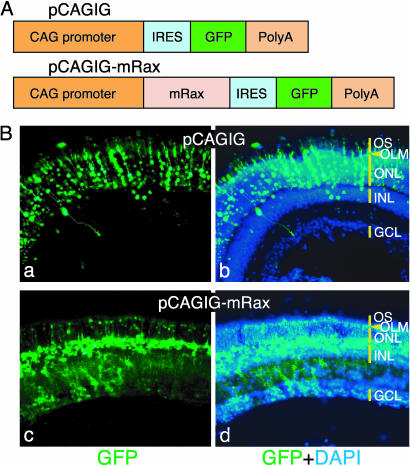
Ectopic expression of Rax using the IRES-GFP vector. (A) Structures of pCAGIG and pCAGIG-mRax vectors. (B) Rat retinae in vivo electroporated with pCAGIG (a and b) or pCAGIG-mRax (c and d) at P0 were harvested at P21 and sectioned. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI).
When P0 rat retinae electroporated with the pCAGIG-mRax expression vector were analyzed at P21, almost all of the GFP-positive cells had cell bodies in the INL, with processes extending to the outer limiting membrane and/or to the GCL (Fig. 6B c and d). This morphology is characteristic of differentiating Müller glial cells or retinal progenitor cells, confirming the previous results obtained with a retrovirus vector (28). In contrast, in the rat retina transfected with the pCAGIG empty vector, most GFP-positive cells were rod PRs in the ONL, and a few cells were bipolar and Müller glial cells (Fig. 6B).
Loss-of-Function Analysis. Recently, several groups have developed DNA-based RNAi vectors that stably produce double-stranded small interfering RNAs in mammalian cells (16, 29–34). Using such RNAi vectors, we tested whether the in vivo electroporation technique would be useful for loss-of-function analyses in the retina. In this experiment, we chose two retinal transcription factors, Crx (35–37) and Nrl (38), as their loss-of-function phenotypes were known through studies of conventional knockout (KO) mice (39, 40). Both of these mutant mice have a normal complement of retinal cell types other than PRs. The Crx KO mice initially have approximately normal levels of PR but lack OSs, whereas the Nrl KO mice lack rods and have more than the normal number of cones, with abnormal OSs. Significant reduction of many rod PR-specific genes, including rhodopsin, was reported for both mutant mice.
Several RNAi vectors were designed based on the mouse Crx and Nrl nucleotide sequences. We wanted to have as a control an RNAi vector that would target a retinal RNA, rather than an ineffective double-stranded RNA, in the event that a successful targeting vector would trigger a cellular reaction that would nonspecifically alter PR development. To this end, a control RNAi vector that suppressed the expression of mouse GAPDH was designed. We found that vectors producing the double-stranded RNAs corresponding to the coding regions of mCrx (408–428), mNrl (306–326), and mGAPDH (393–413) efficiently and selectively suppressed the expression of mCrx:GFP, mNrl:GFP, and mGAPDH:GFP fusion proteins, respectively, in 293T cells (Fig. 9, which is published as supporting information on the PNAS web site).
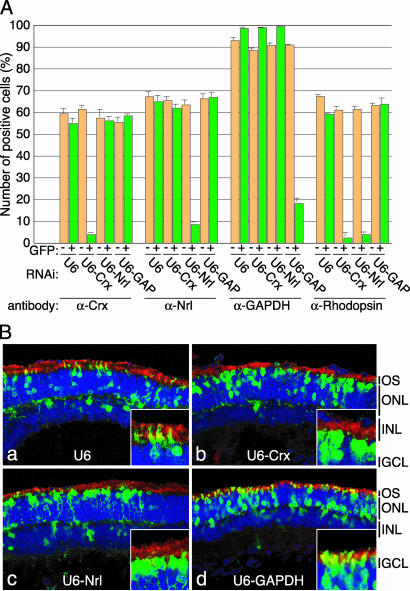
To determine whether these RNAi vectors could work in the retina, they were coelectroporated in vivo with pCAG-GFP into CD1 mouse retinae at P0. When electroporated retinae were dissociated into single cells at P10, and stained with antibodies, the numbers of Crx-, Nrl-, and GAPDH-positive cells were significantly and specifically reduced in the GFP-positive fractions of the retinae transfected with Crx-, Nrl-, and GAPDH-RNAi vectors (U6-Crx, U6-Nrl, and U6-GAPDH), respectively (Fig. 7A). In the retinae transfected with U6-Crx and U6-Nrl, the numbers of rhodopsin-positive cells were also significantly reduced. An empty RNAi vector (U6) had no apparent effects.
Fig. 7.
Knockdown of Crx and Nrl expressions in the mouse retina using RNAi vectors. (A) Specific down-regulation of the expression of Crx, Nrl, GAPDH, and rhodopsin in the RNAi vector transfected mouse retinae. CD1 mouse retinae were coelectroporated in vivo with pCAG-GFP (2 μg/μl) and an RNAi vector (U6, U6-Crx, U6-Nrl or U6-GAPDH; 4 μg/μl) at P0, harvested at P10, and dissociated into single cells. The dissociated cells were stained with anti-Crx, anti-Nrl, anti-GAPDH, or anti-rhodopsin antibody, and the numbers of positive cells were scored. Both GFP-positive (green bars) and GFP-negative (yellow bars) cells were analyzed. (B) Morphology of the retinal cells transfected with RNAi vector. CD1 mouse retinae were coelectroporated in vivo with pCAG-GFP and an RNAi vector at P0, harvested at P20, and sectioned. Sections were stained with anti-rhodopsin antibody (red) and 4′,6-diamidino-2-phenylindole (blue), and images were taken with a confocal microscope (Zeiss LSM 510). (Insets) Higher-magnification views of OSs. (Magnifications: ×800.)
When the sections of electroporated retinae were analyzed at P20, the ratios of GFP-positive cells in the ONL to those in the INL were almost comparable among the retinae transfected with U6-empty, U6-Crx, U6-Nrl, and U6-GAPDH vectors (Fig. 7B). However, in the retinae transfected with U6-Crx and U6-Nrl, most GFP-positive cells in the ONL did not have clear OS structures (Fig. 7B b and c), consistent with the reported phenotypes of knockout mice. Moreover, in the retinae transfected with U6-Nrl, the nuclei of most GFP-positive cells in the ONL were localized right below the outer limiting membrane, as are cone PR. A similar tendency was observed in the Crx-RNAi vector-transfected retinae, although it was not as significant. In the retinae transfected with U6-empty or U6-GAPDH vectors, the GFP-positive cells in the ONL had clear OSs (Fig. 7B a and d).
Discussion
We demonstrate that DNA can be readily introduced into retinal cells of newborn mouse and rat by electroporation in vivo and in vitro. In vivo electroporation has been applied to a variety of tissues of various animal species including mouse, chicken, and frog (9). For the mammalian central nervous system, this technique has been used to introduce genes into the fetal mouse brain, targeting the neural progenitor cells in the VZ (41–43). It has also been used for the retina in that Dezawa et al. (44) recently reported the introduction of DNA into retinal GCs of the adult rat from the vitreous. However, at least in our experiments, we could see only a few GFP-positive cells when the GFP expression vector was injected into the vitreous of newborn rat eyes, followed by electroporation. Moreover, we could detect few GFP-positive cells when the vector was electroporated into the vitreal side of newborn and adult mouse retinae in vitro. It is likely that electroporation from the vitreal side is less efficient, requiring further improvement of the transfection conditions. The presence of a basal laminae at the inner limiting membrane might impede vector penetration. Nonetheless, this could also be a very useful technique for the introduction of genes into GC, where it is difficult to transduce using other methods.
The electroporation technique described here has several advantages over conventional methods to deliver genes into the rodent retina. First, this method is rapid and safe. Second, the DNA transduction efficiency can be remarkably high when the injection of DNA is performed accurately. Third, it is possible to introduce various types of DNA constructs with a size limitation that is significantly larger than that of viral vectors. These constructs include cell type-specific promoters used to express reporter genes as well as recently developed DNA-based RNAi vectors. Fourth, multiple DNA constructs can be introduced into single retinal cells. Recently, it has been reported that a combination of multiple transcription factors, rather than a single transcription factor, is important for retinal cell type specification (45, 46). For example, coexpression of Math1 or Math3 with Chx10 efficiently induces rod bipolar cell genesis in retinal explants, whereas misexpression of either Math1/Math3 or Chx10 alone cannot (45). Thus, the fourth point would be especially helpful to analyze the functions of retinal transcription factors.
Unlike retrovirus vectors that integrate into the host genome and stably express foreign genes for a long time period, gene expression from DNA constructs introduced by in vivo electroporation should not be so stable. Although we did not determine exactly how long expression of introduced genes is maintained in the retina, it is unlikely that the gene expression persists for more than several months. For this reason, in vivo electroporation may not be suitable for therapeutic uses, such as gene therapy of inherited retinal diseases. Nonetheless, we did find that the GFP expression is visible for at least 50 days, enough for the study of retinal development, including maturation of differentiated retinal cells (e.g., elongation of rod OS).
Recently, we have identified a large number of candidate genes involved in retinal development and disease, by microarray analysis and serial analysis of gene expression (SAGE) (1, 3). The in vivo and in vitro electroporation techniques, together with the conventional virus-mediated gene transfer system, would greatly contribute to the elucidation of the functions of these candidate genes.
Supplementary Material
Acknowledgments
We thank Drs. D. Zack, Y. Shi, J. Miyazaki, S. Sugano, T. Kitamura, and C. Lois for kindly providing plasmids; S. Chen, A. Swaroop, and R. Molday for antibodies; and T. Young for help in RNAi experiments. Confocal microscopic analyses were done at the Harvard Center for Neurodegeneration and Repair. T.M. was supported by a Japan Society for the Promotion of Science Postdoctoral fellowship for research abroad. This work was supported by National Institutes of Health Grant EY08064 and the Howard Hughes Medical Institute.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 30, 2002.
Abbreviations: MLV, Moloney murine leukemia virus; CABP5, calcium binding protein 5; CRALBP, cellular retinaldehyde binding protein; GC, ganglion cell; GCL, GC layer; INL, inner nuclear layer; ONL, outer nuclear layer; OS, outer segment; VZ, ventricular zone; PR, photoreceptor; RNAi, RNA interference; CMV, cytomegalovirus; Pn, postnatal day n; EF, elongation factor; IRES, internal ribosome entry site.
See accompanying Biography on page 14.
References
- 1.Livesey, F. J., Furukawa, T., Steffen, M. A., Church, G. M. & Cepko, C. L. (2000) Curr. Biol. 10, 301–310. [DOI] [PubMed] [Google Scholar]
- 2.Farjo, R., Yu, J., Othman, M. I., Yoshida, S., Sheth, S., Glaser, T., Baehr, W. & Swaroop, A. (2002) Vision Res. 42, 463–470. [DOI] [PubMed] [Google Scholar]
- 3.Blackshaw, S., Fraioli, R. E., Furukawa, T. & Cepko, C. L. (2001) Cell 107, 579–589. [DOI] [PubMed] [Google Scholar]
- 4.Price, J., Turner, D. & Cepko, C. (1987) Proc. Natl. Acad. Sci. USA 84, 156–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner, D. L. & Cepko, C. L. (1987) Nature 328, 131–136. [DOI] [PubMed] [Google Scholar]
- 6.Turner, D. L., Snyder, E. Y. & Cepko, C. L. (1990) Neuron 4, 833–845. [DOI] [PubMed] [Google Scholar]
- 7.Miyoshi, H., Takahashi, M., Gage, F. H. & Verma, I. M. (1997) Proc. Natl. Acad. Sci. USA 94, 10319–10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flannery, J. G., Zolotukhin, S., Vaquero, M. I., LaVail, M. M., Muzyczka, N. & Hauswirth, W. W. (1997) Proc. Natl. Acad. Sci. USA 94, 6916–6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swartz, M., Eberhart, J., Mastick, G. S. & Krull, C. E. (2001) Dev. Biol. 233, 13–21. [DOI] [PubMed] [Google Scholar]
- 10.Niwa, H., Yamamura, K. & Miyazaki, J. (1991) Gene 108, 193–199. [DOI] [PubMed] [Google Scholar]
- 11.Kim, D. W., Uetsuki, T., Kaziro, Y., Yamaguchi, N. & Sugano, S. (1990) Gene 91, 217–273. [DOI] [PubMed] [Google Scholar]
- 12.Lois, C., Hong, E. J., Pease, S., Brown, E. J. & Baltimore, D. (2002) Science 295, 868–872. [DOI] [PubMed] [Google Scholar]
- 13.Zack, D. J., Bennett, J., Wang, Y., Davenport, C., Klaunberg, B., Gearhart, J. & Nathans, J. (1991) Neuron 6, 187–199. [DOI] [PubMed] [Google Scholar]
- 14.Nosaka, T., Kawashima, T., Misawa, K., Ikuta, K., Mui, A. L. & Kitamura, T. (1999) EMBO J. 18, 4754–4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furukawa, T., Kozak, C. A. & Cepko, C. L. (1997) Proc. Natl. Acad. Sci. USA 94, 3088–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sui, G., Soohoo, C., Affar, E. B., Gay, F., Shi, Y., Forrester, W. C. & Shi, Y. A. (2002) Proc. Natl. Acad. Sci. USA 99, 5515–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bao, Z. Z. & Cepko, C. L. (1997) J. Neurosci. 17, 1425–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrow, E. M., Belliveau, M. J. & Cepko, C. L. (1998) J. Neurosci. 18, 3738–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molday, R. S. (1989) Prog. Ret. Res. 8, 173–209. [Google Scholar]
- 20.La Spada, A. R., Fu, Y. H., Sopher, B. L., Libby, R. T., Wang, X., Li, L. Y., Einum, D. D., Huang, J., Possin, D. E., Smith, A. C., et. al. (2001) Neuron 31, 913–927. [DOI] [PubMed] [Google Scholar]
- 21.Swain, P. K., Hicks, D., Mears, A. J., Apel, I. J., Smith, J. E., John, S. K., Hendrickson, A., Milam, A. H. & Swaroop, A. (2001) J. Biol. Chem. 276, 36824–36830. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt, E. V., Christoph, G., Zeller, R. & Leder, P. (1990) Mol. Cell. Biol. 10, 4406–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schorpp, M., Jager, R., Schellander, K., Schenkel, J., Wagner, E. F., Weiher, H. & Angel, P. (1996) Nucleic Acids Res. 24, 1787–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennedy, B. N., Huang, J., Saari, J. C. & Crabb, J. W. (1998) Mol. Vis. 4, 14–21. [PubMed] [Google Scholar]
- 25.Haeseleer, F, Sokal, I, Verlinde, C. L., Erdjument-Bromage, H., Tempst, P., Pronin, A. N., Benovic, J. L., Fariss, R. N. & Palczewski, K. (2000) J. Biol. Chem. 275, 1247–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haverkamp, S., Ghosh, K. K., Hirano, A. A. & Wassle, H. (2003) J. Comp. Neurol. 455, 463–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathers, P. H., Grinberg, A., Mahon, K. A. & Jamrich, M. (1997) Nature 387, 603–607. [DOI] [PubMed] [Google Scholar]
- 28.Furukawa, T., Mukherjee, S., Bao, Z. Z., Morrow, E. M. & Cepko, C. L. (2000) Neuron 26, 383–394. [DOI] [PubMed] [Google Scholar]
- 29.Yu, J. Y., DeRuiter, S. L. & Turner, D. L. (2002) Proc. Natl. Acad. Sci. USA 99, 6047–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyagishi, M. & Taira, K. (2002) Nat. Biotechnol. 20, 497–500. [DOI] [PubMed] [Google Scholar]
- 31.Lee, N. S., Dohjima, T., Bauer, G., Li, H., Li, M. J., Ehsani, A., Salvaterra, P. & Rossi, J. (2002) Nat. Biotechnol. 20, 500–5005. [DOI] [PubMed] [Google Scholar]
- 32.Paul, C. P., Good, P. D., Winer, I. & Engelke, D. R. (2002) Nat. Biotechnol. 20, 505–508. [DOI] [PubMed] [Google Scholar]
- 33.Brummelkamp, T. R., Bernards, R. & Agami, R. (2002) Science 296, 550–553. [DOI] [PubMed] [Google Scholar]
- 34.Paddison, P. J., Caudy, A. A., Bernstein, E., Hannon, G. J. & Conklin, D. S. (2002) Genes Dev. 16, 498–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furukawa, T., Morrow, E. M. & Cepko, C. L. (1997) Cell 91, 531–541. [DOI] [PubMed] [Google Scholar]
- 36.Freund, C. L., Gregory-Evans, C. Y., Furukawa, T., Papaioannou, M., Looser, J., Ploder, L., Bellingham, J., Ng, D., Herbrick, J. A., Duncan, A., et al. (1997) Cell 91, 543–553. [DOI] [PubMed] [Google Scholar]
- 37.Chen, S., Wang, Q. L., Nie, Z., Sun, H., Lennon, G., Copeland, N. G., Gilbert, D. J., Jenkins, N. A. & Zack, D. J. (1997) Neuron 19, 1017–1030. [DOI] [PubMed] [Google Scholar]
- 38.Swaroop, A., Xu, J. Z., Pawar, H., Jackson, A., Skolnick, C. & Agarwal, N. (1992) Proc. Natl. Acad. Sci. USA 89, 266–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Furukawa, T., Morrow, E. M., Li, T., Davis, F. C. & Cepko, C. L. (1999) Nat. Genet. 23, 466–470. [DOI] [PubMed] [Google Scholar]
- 40.Mears, A. J., Kondo, M., Swain, P. K., Takada, Y., Bush, R. A., Saunders, T. L., Sieving, P. A. & Swaroop, A. (2001) Nat. Genet. 29, 447–452. [DOI] [PubMed] [Google Scholar]
- 41.Saito, T. & Nakatsuji, N. (2001) Dev. Biol. 240, 237–246. [DOI] [PubMed] [Google Scholar]
- 42.Tabata, H. & Nakajima, K. (2001) Neuroscience 103, 865–872. [DOI] [PubMed] [Google Scholar]
- 43.Ohtsuka, T., Sakamoto, M., Guillemot, F. & Kageyama, R. (2001) J. Biol. Chem. 276, 30467–30474. [DOI] [PubMed] [Google Scholar]
- 44.Dezawa, M., Takano, M., Negishi, H., Mo, X., Oshitari, T. & Sawada, H. (2002) Micron 33, 1–6. [DOI] [PubMed] [Google Scholar]
- 45.Hatakeyama, J., Tomita, K., Inoue, T. & Kageyama, R. (2001) Development (Cambridge, U.K.) 128, 1313–1322. [DOI] [PubMed] [Google Scholar]
- 46.Inoue, T., Hojo, M., Bessho, Y., Tano, Y., Lee, J. E. & Kageyama, R. (2002) Development (Cambridge, U.K.) 129, 831–824. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.